Abstract
Background:
Coverage of viral load testing remains low with only half of patients in need having adequate access. Alternative technologies to high throughput centralized machines can be used to support viral load scale-up; however, clinical performance data are lacking. We conducted a meta-analysis comparing the Cepheid Xpert HIV-1 viral load plasma assay to traditional laboratory-based technologies.
Methods:
Cepheid Xpert HIV-1 and comparator laboratory technology (including the Abbott m2000, bioMerieux NucliSENS, and Roche COBAS TaqMan assays) plasma viral load results were provided from 13 of the 19 eligible studies, which accounted for a total of 3,784 paired data points. We used random effects models to determine the accuracy and misclassification at various treatment failure thresholds (detectable, 200, 400, 500, 600, 800 and 1000 copies/ml).
Results:
Thirty percent of viral load test results were undetectable, while 45% were between detectable and 10,000 copies/ml and the remaining 25% were above 10,000 copies/ml. The median Xpert viral load was 119 copies/ml and the median comparator viral load was 157 copies/ml, while the log10 bias was 0.04 (0.02 – 0.07). The sensitivity and specificity to detect treatment failure were above 95% at all treatment failure thresholds, except for detectable, at which the sensitivity was 93.33% (95% CI: 88.2 – 96.3%) and specificity was 80.56% (95%CI: 64.6 – 90.4%).
Conclusions:
The Cepheid Xpert HIV-1 viral load plasma assay results were highly comparable to laboratory-based technologies with limited bias and high sensitivity and specificity to detect treatment failure. Alternative specimen types and technologies that enable decentralized testing services can be considered to expand access to viral load.
INTRODUCTION
Viral load access is critical to both patient and public health care. Currently, 59% of patients on ART have access to viral load testing [1]; however, three countries in particular (Kenya, South Africa, and Uganda) account for approximately 50% of viral load tests performed in 2017 suggesting that viral load coverage outside of these countries is significantly lower. Challenges with cold chain and specimen transport, particularly in ensuring blood samples reach the laboratory within six to 24 hours per manufacturer guidelines have hindered viral load access and scale-up.
Alternative technologies, such as near point-of-care and point-of-care assays, may support increased access to viral load testing. Several technologies are in development or on the market [2]. POC technologies have previously been found to significantly reduce test turnaround times and support faster treatment decisions. When CD4 was still used to determine treatment eligibility, a systematic review and meta-analysis found that POC CD4 testing improved linkage to care and timeliness of ART initiation [3]. Furthermore, two recent studies from Malawi and Mozambique have shown significantly reduced test turnaround times and improved treatment initiation rates when POC early infant diagnosis technologies were used compared to laboratory-based technologies [4,5]. Decentralizing testing for both diagnosis and monitoring closer to the patient may increase access to care, improve both personal and public health, and support more sustainable, inclusive health systems [6].
Monitoring patients on treatment is critical to understand treatment effectiveness and identify patients in need of increased adherence support or regimen switch. While rapid or immediate test result return using point-of-care and near point-of-care technologies may not be necessary for all patients in need of viral load testing, several populations may most benefit. Pregnant and breastfeeding women to prevent vertical transmission, infants and children who often have higher levels of drug resistance [7,8], patients suspected of failing treatment, or patients with advanced HIV disease may all be in need of more rapid testing and clinical action to address unsuppressed viral loads. As laboratory systems continue to be scaled up and improved, ensuring swift test turnaround times to facilitate high quality clinical services continues to prove challenging. New technologies that can be placed closer to the patient may prove beneficial, especially to support patients in more critical need of rapid test results and clinical decision-making.
Several diagnostic accuracy studies have been conducted measuring the performance of the Cepheid Xpert HIV-1 viral load plasma assay [9-27] and a systematic review was generated to provide initial analytical data [28]; however, clinical utility data of the Cepheid Xpert HIV-1 viral load plasma assay are lacking. The Cepheid Xpert HIV-1 viral load assay was approved by the European Union in 2017 and included on the WHO list of prequalified in vitro diagnostic products in August 2017 [29]. This assay can provide HIV-1 viral load results within two hours, does not require the same specimen transportation logistics of laboratory-based technologies, and has already been procured and implemented significantly for tuberculosis testing across high burden regions [2,29,30,31]. We, therefore, consolidated primary data from diagnostic accuracy studies [12,14-19,21,23-27] to determine how the Cepheid Xpert HIV-1 viral load assay performed when used within the WHO treatment failure algorithm [32,33].
METHODS
Search Strategy and Study Selection
The search strategy was previously published [28]. An updated search was conducted to identify studies published since the previous systematic review. Briefly, PubMed, EMBASE, Google Scholar, and Medline databases were used to identify peer-reviewed original research with appropriate data for this meta-analysis on July 30, 2018. Conference abstracts from Conference on Retroviruses and Opportunistic Infections (CROI), International Conference on AIDS and STIs in Africa (ICASA), International AIDS Society (IAS), and AIDS Conferences as well as extensive bibliography and grey literature were screened for possible inclusion. Studies were included if they included technical evaluation data comparing the Cepheid Xpert HIV-1 viral load plasma assay to a laboratory-based viral load plasma assay and were performed using HIV-positive clinical samples. Studies were excluded if they used spike blood samples or panels, performed a qualitative analysis, or the comparator was a sample type other than plasma. We contacted the corresponding authors of all studies that met the inclusion criteria to explain the analysis plan and request original data. Each corresponding author was contacted at least twice.
Statistical Analyses
Data from studies willing to share primary data were pooled to compare the performance of Cepheid Xpert HIV-1 viral load plasma assay with comparator laboratory technology. Study-specific data were generated for each study providing primary data including the median values, distribution, R2, and bias (95% CI). As WHO guidelines suggest the use of a threshold to support treatment monitoring, several treatment failure thresholds for viral load were assessed including detectable (defined as any result above the limit of detection indicates treatment failure), 200, 400, 500, 600, 800, and 1000 copies/mL. Each treatment failure threshold for Cepheid Xpert was compared to the same treatment failure threshold value for the comparator technology. Analyses were conducted by pooling all comparators together as well as disaggregating by comparator (three comparators were included in submitted data: Abbott RealTime HIV-1, bioMerieux NucliSENS EasyQ HIV-1, and Roche COBAS TaqMan HIV-1 v2.0). The limit of detection varied slightly by technology: Abbott RealTime HIV-1 = 40 copies/ml, bioMerieux NucliSENS EasyQ HIV-1 = 25 copies/ml, Cepheid Xpert HIV-1 viral load = 40 copies/ml, and Roche COBAS TaqMan HIV-1 v2.0 = 20 copies/ml and notified the definition of the detectable threshold analysis. Using these treatment failure thresholds, the sensitivity, specificity, upward and downward misclassification rates, and positive and negative predictive values were calculated. Definitions were previously described [34]. Bivariate random effects models were used to estimate summary sensitivity, specificity and other accuracy measures to simultaneously determine the pooled estimates, accounting for the covariance of sensitivity and specificity as well as study specific heterogeneity [35,36]. Analyses were completed using the lme4 R package in R version 3.4.3 (The R Foundation).
Protocol
The prepared protocol was reviewed by the World Health Organization.
RESULTS
A systematic review was previously conducted including 12 articles and 13 studies, in which the quality of studies and study characteristics were described [28]. Since the previous search, an additional six studies were published which met the inclusion criteria [10,21,22,25-27] (Table 1). After contacting each corresponding author, a total of 13 studies provided 14 primary datasets out of the total of 19 eligible Cepheid Xpert plasma viral load studies resulting in a total of 3,790 paired plasma viral load results [12,14-19,21,23-27]. Approximately 55% of the paired data points were from studies conducted in Africa [12,14,15,21,26,27], of which 50% were from each of the Southern African Development Community region [12,14,15,26,27] and the East African Community region [21,25]. Just over 30% of paired data points were from studies conducted in Europe or USA [16,17], while 11% were from studies conducted in India [18,23,24]. The study-specific characteristics such as sample size, viral load medians, and patient viral load distributions are summarized in Table 2.
Table 1.
Study characteristics of additional studies found beyond those incuded in [28].
| Reference | Year | Sample size | Location | Patient population | Age | % female | |
|---|---|---|---|---|---|---|---|
| Beard | 10 | 2016 | 292 | Botswana | HIV+, naïve and ART | NR | NR |
| Mosha | 21 | 2016 | 753 | Tanzania | HIV+, naïve and ART | >70% btw 15-48 | 69.6% |
| Mwau | 22 | 2017 | 430 | Kenya | HIV+, naïve and ART | >18 years | 73.0% |
| Umviligihozo | 25 | 2016 | 331 | Rwanda | HIV+, naïve and ART | NR | NR |
| ZEHRP | 26 | NR | 37 | Zambia | HIV+ | NR | NR |
| Zinyowera | 27 | 2015 | 376 | Zimbabwe | HIV+, on ART | median: 40 years | 58.90% |
NR: not reported
grey shading: studies included in the meta-analysis
Table 2.
Analytical (r2, bias, median viral loads) and patient viral load distributions for each study included in the meta-analysis.
| Comparator | Reference | Sample size | R2 | bias (95% CI) | Median comparator |
Median Xpert |
Difference in medians |
Proportion undetectable |
Proportion 1 - 10,000 cp/ml |
Proportion > 10,000 cp/ml |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ceffa | Abbott m2000rt RealTime HIV-1 | 12 | 300 | 0.8527 | −0.01 (−0.09,0.07) | 3.35 | 3.3 | 0.05 | 20.7% | 50.0% | 21.7% |
| Garrett | Roche COBAS Ampliprep/TaqMan v2 | 14 | 42 | 0.9411 | 0.10 (−0.04,0.27) | 3.88 | 3.94 | −0.06 | 28.6% | 23.8% | 47.6% |
| Gous A | Abbott m2000rt RealTime HIV-1 | 15 | 155 | 0.7017 | 0.69 (0.55,0.83) | 1.59 | 0.00 | 1.59 | 57.7% | 67.1% | 7.4% |
| Gous B | Roche COBAS Ampliprep/TaqMan v2 | 15 | 155 | 0.6868 | −0.09 (−0.22,0.04) | 1.59 | 1.70 | −0.11 | 20.0% | 64.5% | 10.3% |
| Gueudin | Abbott m2000rt RealTime HIV-1 | 16 | 295 | 0.8156 | −0.21 (−0.30,−0.12) | 1.88 | 2.00 | −0.12 | 10.2% | 56.6% | 22.0% |
| Jordan | Abbott m2000rt RealTime HIV-1 | 17 | 724 | 0.9209 | 0.02 (−0.02,0.06) | 1.82 | 1.97 | −0.15 | 31.8% | 58.0% | 20.6% |
| Kulkarni | Abbott m2000rt RealTime HIV-1 | 18 | 219 | 0.9189 | 0.03 (−0.05,0.11) | 3.93 | 3.76 | 0.17 | 20.1% | 36.5% | 40.2% |
| Mor | Abbott m2000rt RealTime HIV-1 | 19 | 188 | 0.5797 | 0.68 (0.49,0.86) | 2.12 | 1.59 | 0.53 | 39.8% | 65.9% | 15.3% |
| Mosha | Roche COBAS Ampliprep/TaqMan v2 | 21 | 753 | 0.8846 | −0.07 (−0.12,−0.02) | 1.59 | 1.45 | 0.14 | 34.8% | 33.6% | 28.5% |
| Nash | Roche COBAS Ampliprep/TaqMan v2 | 23 | 115 | 0.8801 | −0.52 (−0.65,−0.38) | 1.59 | 1.28 | 0.31 | 0.0% | 29.6% | 29.6% |
| Swathirajan | Abbott m2000rt RealTime HIV-1 | 24 | 100 | 0.5364 | 0.26 (0.14,0.35) | 3.88 | 3.62 | 0.26 | 0.0% | 77.8% | 21.2% |
| Umviligihozo | Roche COBAS Ampliprep/TaqMan v2 | 25 | 331 | 0.9070 | 0.06 (0.00,0.12) | 3.81 | 3.79 | 0.02 | 13.8% | 40.2% | 43.3% |
| ZEHRP | Roche COBAS Ampliprep/TaqMan v2 | 26 | 37 | 0.7886 | −0.03 (−0.22,0.16) | 1.59 | 1.28 | 0.31 | 27.0% | 58.3% | 5.4% |
| Zinyowera | bioMerieux NucliSens HIV-1 QT | 27 | 376 | 0.9027 | 0.16 (0.10,0.22) | 1.59 | 0.00 | 1.59 | 51.9% | 35.9% | 17.6% |
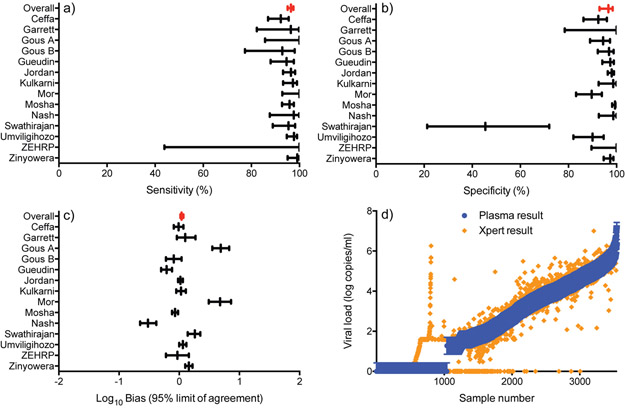
Figure 1 shows the distribution of the viral load values from all studies. Thirty percent of viral load test results were undetectable, while 45% were between detectable and 10,000 copies/ml and the remaining 25% were above 10,000 copies/ml. The median comparator viral load was 157 copies/ml and median Xpert viral load was 119 copies/ml, while the overall log10 bias was 0.04 log copies/ml (95% CI: 0.02 – 0.07) (Table 3 and Figure 2). The R2 was 0.941 (p<0.001). When disaggregated by the comparator used, the median and bias results were similarly consistent and within 0.2 log copies/ml.
Figure 1.
Patient plasma viral load (copies/ml) distribution using the comparator assay from all studies included in the meta-analysis.
Table 3.
Meta-analysis of clinical metrics (median viral loads, mean bias, sensitivity, specificity, and misclassification) overall and for each comparator technology, across various treatment failure thresholds.
| Comparator used | |||||
|---|---|---|---|---|---|
| All together | Abbott RealTime HIV-1 | bioMerieux NucliSENS EasyQ HIV-1 |
Roche COBAS TaqMan HIV-1 |
||
| n | 3,790 | 1,981 | 376 | 1,433 | |
| Cepheid Xpert | |||||
| median viral load (copies/ml) | 119 | 188 | 39 | 63 | |
| Comparator | |||||
| median viral load (copies/ml) | 157 | 219 | 1 | 133 | |
| difference in medians (copies/ml) | −38 | −31 | 38 | −70 | |
| mean bias (log copies/ml) | 0.04 (0.02-0.07) | 0.11 (0.07-0.14) | 0.16 (0.10-0.22) | −0.08 (−0.11- −0.04) | |
| Threshold comparisons | |||||
| Sensitivity (UCL-LCL) | 1000:1000 | 96.47 (95.10-97.47) | 95.86 (93.59-97.35) | 99.10 (95.07-99.95) | 96.56 (94.54-97.85) |
| 800:800 | 96.92 (95.80-97.75) | 96.96 (95.27-98.06) | 98.26 (93.88-99.52) | 96.66 (94.58-97.96) | |
| 600:600 | 96.86 (95.74-97.69) | 97.24 (95.82-98.19) | 97.54 (93.02-99.16) | 95.71 (95.70-95.73) | |
| 500:500 | 96.74 (95.60-97.59) | 96.82 (95.41-97.80) | 97.58 (93.13-99.17) | 96.44 (94.05-97.89) | |
| 400:400 | 96.04 (94.80-96.98) | 96.24 (94.73-97.33) | 95.28 (90.08-97.82) | 95.61 (92.51-97.47) | |
| 200:200 | 95.36 (93.37-96.77) | 95.12 (92.92-96.66) | 91.16 (85.46-94.76) | 96.20 (90.71-98.50) | |
| detectable | 93.33 (88.24-96.31) | 95.92 (91.41-98.12) | 96.13 (92.23-98.11) | 88.42 (75.29-95.03) | |
| Specificity (UCL-LCL) | 1000:1000 | 96.59 (92.90-98.39) | 94.18 (86.10-97.69) | 97.36 (94.65-98.71) | 98.22 (94.68-99.42) |
| 800:800 | 96.75 (92.58-98.61) | 94.23 (82.60-98.25) | 96.93 (94.07-98.44) | 98.22 (95.24-99.34) | |
| 600:600 | 95.88 (91.86-97.96) | 92.98 (84.68-96.94) | 97.64 (94.94-98.91) | 98.03 (98.03-98.04) | |
| 500:500 | 95.35 (90.22-97.85) | 92.06 (81.55-96.82) | 97.62 (94.90-98.90) | 97.61 (92.83-99.23) | |
| 400:400 | 96.00 (92.66-97.86) | 94.91 (89.73-97.55) | 94.78 (91.27-96.92) | 97.95 (93.60-99.36) | |
| 200:200 | 97.69 (94.56-99.04) | 96.24 (89.71-98.68) | 96.51 (93.26-98.22) | 98.63 (94.23-99.69) | |
| detectable | 80.56 (64.63-90.39) | 69.31 (40.83-88.09) | 86.15 (80.60-90.31) | 83.02 (66.46-92.34) | |
| Total Misclassification (UCL-LCL) | 1000:1000 | 3.53 (2.52-4.93) | 4.69 (3.13-6.96) | 2.13 (1.08-4.14) | 2.67 (1.73-4.10) |
| 800:800 | 3.13 (2.22-4.41) | 3.97 (2.53-6.16) | 2.66 (1.45-4.83) | 2.53 (1.61-3.94) | |
| 600:600 | 3.58 (2.67-4.79) | 4.30 (2.95-6.23) | 2.39 (1.26-4.49) | 3.15 (2.08-4.74) | |
| 500:500 | 3.84 (2.90-5.06) | 4.66 (3.26-6.63) | 2.39 (1.26-4.49) | 3.21 (2.21-4.65) | |
| 400:400 | 3.91 (2.98-5.12) | 4.15 (2.74-6.26) | 5.05 (3.26-7.76) | 3.29 (2.47-4.37) | |
| 200:200 | 3.78 (2.63-5.41) | 4.55 (2.86-7.17) | 5.59 (3.68-8.39) | 2.85 (1.78-4.54) | |
| detectable | 11.19 (6.59-18.36) | 11.76 (5.49-23.42) | 9.04 (6.54-12.37) | 12.74 (5.69-26.11) | |
| Upward Misclassification (UCL-LCL) | 1000:1000 | 3.41 (1.61-7.10) | 5.82 (2.31-13.90) | 2.64 (1.29-5.35) | 1.78 (0.58-5.32) |
| 800:800 | 3.25 (1.39-7.42) | 5.77 (1.75-17.40) | 3.07 (1.56-5.93) | 1.78 (0.66-4.76) | |
| 600:600 | 4.12 (2.04-8.14) | 7.02 (3.06-15.32) | 2.36 (1.09-5.06) | 1.97 (1.96-1.97) | |
| 500:500 | 4.65 (2.15-9.78) | 7.94 (3.18-18.45) | 2.38 (1.10-5.10) | 2.39 (0.77-7.17) | |
| 400:400 | 4.00 (2.14-7.34) | 5.09 (2.45-10.27) | 5.22 (3.08-8.73) | 2.05 (0.64-6.40) | |
| 200:200 | 2.31 (0.96-5.44) | 3.76 (1.32-10.29) | 3.49 (1.78-6.74) | 1.37 (0.31-5.77) | |
| detectable | 19.44 (9.61-35.37) | 30.69 (11.91-59.17) | 13.85 (9.69-19.40) | 16.98 (7.66-33.54) | |
| Downward Misclassification (UCL-LCL) | 1000:1000 | 3.53 (2.53-4.90) | 4.14 (2.65-6.41) | 0.90 (0.05-4.93) | 3.44 (2.15-5.46) |
| 800:800 | 3.08 (2.25-4.20) | 3.04 (1.94-4.73) | 1.74 (0.48-6.12) | 3.34 (2.04-5.42) | |
| 600:600 | 3.14 (2.31-4.26) | 2.76 (1.81-4.18) | 2.46 (0.84-6.98) | 4.29 (4.27-4.30) | |
| 500:500 | 3.26 (2.41-4.40) | 3.18 (2.20-4.59) | 2.42 (0.83-6.87) | 3.56 (2.11-5.95) | |
| 400:400 | 3.96 (3.02-5.20) | 3.76 (2.67-5.27) | 4.72 (2.18-9.92) | 4.39 (2.53-7.49) | |
| 200:200 | 4.64 (3.23-6.63) | 4.88 (3.34-7.08) | 8.84 (5.24-14.54) | 3.80 (1.50-9.29) | |
| detectable | 6.67 (3.69-11.76) | 4.08 (1.88-8.59) | 3.87 (1.89-7.77) | 11.58 (4.97-24.71) | |
| PPV (UCL-LCL) | 1000:1000 | 95.37 (92.07-97.33) | 93.29 (87.10-96.63) | 94.02 (88.16-97.07) | 97.17 (93.19-98.86) |
| 800:800 | 96.04 (92.83-97.85) | 94.43 (87.89-97.54) | 93.39 (87.49-96.61) | 97.50 (93.80-99.02) | |
| 600:600 | 95.40 (92.24-97.31) | 93.45 (87.35-96.72) | 95.20 (89.92-97.78) | 97.19 (93.17-98.87) | |
| 500:500 | 95.25 (91.93-97.24) | 93.25 (86.85-96.65) | 95.28 (90.08-97.82) | 96.78 (91.68-98.79) | |
| 400:400 | 95.97 (92.84-97.77) | 95.30 (88.40-98.18) | 90.30 (84.11-94.24) | 97.35 (95.00-98.62) | |
| 200:200 | 98.00 (95.03-99.21) | 97.88 (89.24-99.61) | 94.37 (89.28-97.12) | 98.34 (95.10-99.45) | |
| detectable | 96.19 (89.68-98.65) | 94.24 (78.74-98.64) | 86.57 (81.16-90.60) | 97.12 (90.04-99.21) | |
| NPV (UCL-LCL) | 1000:1000 | 97.66 (94.83-98.96) | 97.96 (88.62-99.66) | 99.61 (97.85-99.98) | 97.53 (96.12-98.44) |
| 800:800 | 97.85 (96.51-98.69) | 97.89 (95.01-99.12) | 99.22 (97.19-99.78) | 97.46 (96.02-98.39) | |
| 600:600 | 97.51 (96.22-98.37) | 97.82 (91.92-99.44) | 98.80 (96.55-99.59) | 96.63 (94.85-97.81) | |
| 500:500 | 97.25 (95.53-98.32) | 97.28 (86.95-99.48) | 98.80 (96.52-99.59) | 96.82 (95.33-97.84) | |
| 400:400 | 96.56 (94.53-97.86) | 96.60 (84.57-99.33) | 97.52 (94.70-98.86) | 96.21 (94.41-97.45) | |
| 200:200 | 95.32 (94.02-96.35) | 94.09 (87.27-97.37) | 94.44 (90.73-96.72) | 96.28 (94.26-97.60) | |
| detectable | 69.76 (41.88-88.08) | 78.08 (46.47-93.60) | 96.00 (91.97-98.05) | 55.28 (14.32-90.14) | |
UCL: upper confidence level
LCL: lower confidence level
Figure 2.
Clinical performance of the Xpert HIV-1 viral load assay compared to the comparator.
The sensitivity and specificity to detect treatment failure were above 95% at all treatment failure thresholds, except for detectable. The sensitivity and specificity at 1,000 copies/ml were 96.47% (95% CI: 95.1 – 97.5%) and 96.59% (95% CI: 92.9 – 98.4%), respectively (Table 3 and Figure 2). At a treatment failure threshold of detectable, the sensitivity was 93.33% (95% CI: 88.2 – 96.3%) and specificity was 80.56% (95%CI: 64.6 – 90.4%). The positive and negative predictive values were also high at all treatment failure thresholds, except the negative predictive value at a detectable treatment failure threshold was 69.76% (95% CI: 41.9 – 88.1%). Upward and downward misclassification rates were below 5% at all treatment failure thresholds, except for detectable. At a treatment failure threshold of detectable, the upward misclassification rate was 19.44% (95% CI: 9.6 – 35.4%) and downward misclassification rate was 6.67% (95% CI: 3.7 – 11.8%). When comparing the analytical performance between specific comparator technologies (Abbott m2000, bioMerieux NucliSENS, and Roche TaqMan), the differences were negligible (data not shown).
Quantitative polymerase chain reaction inherently has a level of variability in test results, generally +/− 0.3 log copies/ml [37]. We, therefore, sought to understand if the performance observed with each technology was within the inherent assay variability limits. Nearly 65% of Cepheid Xpert HIV-1 viral load results were within +/−0.3 log10 copies/ml of the comparator viral load (Figure 2d).
Finally, the proportion of invalid rates was also recorded. The comparator technologies had an invalid rate of 0.5%, while the Xpert viral load assay had an invalid rate of 1.66%. However, it is possible that not all investigators provided invalid rates for both technologies.
DISCUSSION
The performance of the Cepheid Xpert HIV-1 viral load plasma assay was comparable to laboratory-based assays. The clinical accuracy when measured considering programmatic use (the ability to correctly diagnose patients as failing treatment or suppressed) was notable in that very few patients (<5%) would have been misclassified. Furthermore, the proportion of invalid tests within these studies was low (<2%). This meta-analysis, therefore, provides clear insights into the clinical diagnostic accuracy of this technology to support viral load scale-up.
There was some heterogeneity across studies observed. The mean bias, for example, was beyond +/− 0.3 log copies/ml in three studies [15,19,23]. Furthermore, the difference in comparator and Xpert medians was significant, greater than 1 log copy/ml, in two studies [15,27]. It is unknown why these differences were observed; however, these studies had significantly higher proportions of patients who were undetectable included in the primary studies. This may have skewed the bias and medians as any variability within the smaller subset of detectable results could have been accentuated.
We also reviewed the performance of the Cepheid Xpert HIV-1 viral load assay to correctly classify patients as above or below lower treatment failure thresholds. One published study has suggested that patients with low level viremia on efavirenz-based treatment may have or be at risk for drug resistance [38]. The Cepheid Xpert HIV-1 viral load assay had good clinical performance (>95% sensitive and specific) at all lower treatment failure thresholds, except detectable. This would indicate that this technology could be used for countries considering a lower treatment failure threshold than the current standard of 1,000 copies/ml. For those using or considering a treatment failure threshold of detectable, it should be noted that nearly 20% of patients who are actually suppressed will be misclassified as failing treatment. While this could be resolved with follow-up testing, upward misclassification may result in unnecessarily switching patients to more expensive second line treatment. Furthermore, the role of low level viremia in the context of optimized treatment, such as dolutegravir, is unknown and may be different from efavirenz.
While the accuracy of the Xpert HIV-1 viral load assay is clear, the programmatic feasibility and operational acceptability remain to be studied. Though it can be used in decentralized settings near the point-of-care, centrifugation of whole blood into plasma is required. Thus far, a whole blood HIV-1 viral load assay has not yet been developed that can allow for direct sample application and greater decentralization. Few health care facilities have centrifuges and/or the necessary skills to process samples. Additionally, this technology requires consistent electricity and temperature-controlled rooms, further limiting decentralization; however, new technologies are being developed, by this supplier and others, to tackle these challenges [2].
Though the patient impact of point-of-care assays has been well documented for CD4 and early infant diagnosis [3-5], the impact of point-of-care or near point-of-care technologies for viral load testing is unclear. For example, the clinical utility, reductions in morbidity and/or mortality, and improvements in and faster switching to second line treatment will help to determine whether global recommendations should be made. In the meantime, consideration for point-of-care and near point-of-care viral load technologies may be focused on the most critical and vulnerable patient populations. Several countries are prioritizing point-of-care and near point-of-care technologies for use in maternity settings to support reduction of mother to child transmission as well as HIV care settings for patients suspected of failing treatment, infants and children who are typically at greater risk for drug resistance, and patients with advanced HIV disease. These patient populations may be in need of more rapid clinical decision-making that can be supported with point-of-care and near point-of-care technology; however, further research is needed.
Furthermore, there is limited analysis on the affordability and cost-effectiveness of point-of-care and near point-of-care technologies for viral load testing. Several studies have found viral load monitoring in general to be cost-effective [39], particularly when coupled with differentiated care programs. The few studies on the cost-effectiveness of point-of-care viral load were done before any such technology was on the market or implemented [40-42]. The models, including assumptive characteristics and pricing, indicated that point-of-care viral load testing was cost-effective due to reductions in unnecessary switches to second-line antiretroviral therapy. Shortening the delays associated with laboratory-based testing resulted in small benefits. Updated cost-effectiveness and affordability studies would be of great benefit, including current and future test pricing, as well as incorporating upcoming patient impact data.
Our study has several limitations. First, possible bias exists as not all primary data were shared and included in this meta-analysis. Unfortunately, this will always be a significant limitation to such meta-analysis. However, nearly 70% of studies were included in this meta-analysis and limited heterogeneity was observed. The total sample size was large with nearly 4,000 paired data points and confidence intervals tight, indicating that additional data may not change the results significantly. Unfortunately, the comparators used were variable across studies. Seven studies used the Abbott RealTime HIV-1 assay, one used the bioMerieux NucliSENS EasyQ HIV-1 assay, and six used the Roche COBAS TaqMan HIV-1 assay. While differences between comparator assays should be subtle, this is not ideal within a meta-analysis. Fortunately, however, the results were relatively consistent across comparators and when analyzed together. Finally, a similar study reviewing the clinical accuracy of laboratory-based assays has not been done to provide a clear comparison for new, alternative technologies such as point-of-care assays.
While systematic reviews and meta-analyses using the results already included in primary manuscripts is the most conventional method of analysis, they can often lack in providing clear clinical data. Furthermore, it is frequent for studies to measure and include different parameters within manuscripts, making consolidation of data and comparisons difficult. This was especially observed in the original systematic review where Table 3 highlighted the diversity of measures of agreement between included studies – no one measurement was included within all [28]. Meta-analyses using primary data can allow for richer data analysis and consider not just analytical but more importantly clinical review. For example, though viral load is a continuous variable requiring analysis using Bland-Altman, linear regression, etc. analyses, it is often used in many high burden programmatic settings as a binary value to determine whether patients may be failing treatment or are suppressed. Being able to analyze the data in the way the technology will be applied is critical to support country access and uptake. Ideally, systematic reviews and meta-analyses would review and include both analytical (bias, correlation coefficient, R2, etc) and clinical data (sensitivity, specificity, misclassification).
The Cepheid Xpert HIV-1 viral load assay performed well compared to traditional laboratory-based comparators. Further diagnostic accuracy studies of the Cepheid Xpert HIV-1 viral load assay are unlikely to yield additional data or information; however, operational feasibility, acceptability, and affordability remain unknown. The GeneXpert technology has already been widely used for tuberculosis testing [30,31] with over 10,000 Xpert devices procured in Africa and Asia by the end of 2017. A range of technologies that can test for multiple disease or assays at the point-of-care and within laboratories currently exist, both on the market and in development [43]. The ability to use a single technology to diagnose and monitor multiple diseases, called multiplexing, multi-disease testing or diagnostics integration, is a novel and new innovation that should be considered by national programs as it may lead to significant programmatic, clinical, and financial efficiencies [44]. Furthermore, alternative, more stable sample types, such as dried blood spot specimens and plasma separating products can offer additional approaches to enable increased access to viral load testing. Selection of appropriate technologies to support viral testing needs requires consideration of not only diagnostic accuracy, but also patient impact, costs, supply chain, ease of use and implementation context. As with all diagnostic technologies, post-market surveillance and quality assurance are critical to ensure high quality test results.
ACKNOWLEDGEMENTS
This work was supported by the Gates Foundation and NIH R01AI122991.
Disclaimer: The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention.
Footnotes
CONFLICTS OF INTEREST STATEMENTS
The authors declare no conflicting interests.
REFERENCES
- 1.Clinton Health Access Initiative. 2018 HIV Market Report. Accessed January 25, 2019: https://clintonhealthaccess.org/content/uploads/2018/09/2018-HIV-Market-Report_FINAL.pdf
- 2.Unitaid. HIV/AIDS Diagnostics Landscape. 2015. Geneva. [Google Scholar]
- 3.Vojnov L, Markby J, Boeke C, Harris L, Ford N, Peter T. POC CD4 improves linkage to care and timeliness of ART initiation in a public health approach: a systematic review and meta-analysis. PLoS ONE. 2016; 11(5): e0155256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mwenda R, Fong Y, Magombo T, Saka E, Midiani D, Mwase C, et al. Significant patient impact observed upon implementation of point-of-care early infant diagnosis technologies in an observations study in Malawi. Clin Infect Dis. 2018;67(5):701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jani IV, Meggi B, Loquiha O, Tobaiwa O, Mudenyanga C, Zitha A, et al. Effect of point-of-care early infant diagnosis on antiretroviral therapy initiation and retention of patients. AIDS. 2018;32(11):1453–1463. [DOI] [PubMed] [Google Scholar]
- 6.Jani IV and Peter TF. How point-of-care testing could drive innovation in global health. NEJM. 2013;368;24. [DOI] [PubMed] [Google Scholar]
- 7.Nuttall J, Pillay V. Antiretroviral resistance patterns in children with HIV infection. Curr Infect Dis Rep. 2019;21(2):7. [DOI] [PubMed] [Google Scholar]
- 8.Huibers MHW, Moons P, Cornelissen M, Zorgdrager F, Maseko N, Gushu MB, et al. High prevalence of virological failure and HIV drug mutations in a first-line cohort of Malawian children. J Antimicrob Chemother. 2018;73(12):3471–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avidor B, Matus N, Girshengorn S, Achsanov S, Gielman S, Zeldis I, et al. Comparison between Roche and Xpert in HIV-1 RNA quantitation: a high concordance between the two techniques except for a CRF02_AG subtype variant with high viral load titters detected by Roche but undetected by Xpert. J Clin Virol. 2017; 93:15–19. [DOI] [PubMed] [Google Scholar]
- 10.Beard R, Mathoma A, Hurlston M, Nyepetsi N, Basotli J, Meswele O, et al. Expanding viral load through partnerships: performance evaluation of Cepheid GeneXpert HIV-1 viral load assay in Botswana. ASLM 2016. [Google Scholar]
- 11.Bruzzone B, Caligiuri P, Nigro N, Arcuri C, Delucis S, Di Biagio A, et al. Xpert HIV-1 viral load assay and versant HIV-1 RNA 1.5 assay: a performance comparison. J Acquired Immune Defic Syndr. 2017;74:e86–e88. [DOI] [PubMed] [Google Scholar]
- 12.Ceffa S, Luhanga R, Andreotti M, Brambilla D, Erba F, Jere H, et al. Comparison of the Cepheid GeneXpert and Abbott M2000 HIV-1 real time molecular assays for monitoring HIV-1 viral load and detecting HIV-1 infection. J Virol Methods. 2016;229:35–39. [DOI] [PubMed] [Google Scholar]
- 13.Fidler S, Lewis H, Meyerowitz J, Kuldanek K, Thornhill J, Muir D, et al. A pilot evaluation of whole blood finger-prick sampling for point-of-care HIV viral load measurement: the UNICORN study. Sci Rep. 2017;7(1):13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrett NJ, Drain PK, Werner L, Samsunder N, Abdool Karim SS. Diagnostic accuracy of the point-of-care xpert HIV-1 viral load assay in a South African HIV clinic. J Acquir Immune Defic Syndr. 2016;72:e45–e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gous N, Scott L, Berrie L, Stevens W. Options to expand HIV viral load testing in South Africa: evaluation of the GeneXpert HIV-1 viral load assay. PLoS One. 2016;11:e0168244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gueudin M, Baron A, Alessandri-Gradt E, Lemee V, Mourez T, Etienne M, et al. Performance evaluation of the new HIV-1 quantification assay, Xpert HIV-1 viral load, on a wide panel of HIV-1 variants. J Acquir Immune Defic Syndr. 2016;72:521–526. [DOI] [PubMed] [Google Scholar]
- 17.Jordan JA, Plantier JC, Templeton K, Wu AHB. Multi-site clinical evaluation of the Xpert HIV-1 viral load assay. J Clin Virol. 2016;80:27–32. [DOI] [PubMed] [Google Scholar]
- 18.Kulkarni S, Jadhav S, Khopkar P, Sane S, Londhe R, Chimanpure V, et al. 2017. GeneXpert HIV-1 quant assay, a new tool for scale up of viral load monitoring in the success of ART programme in India. BMC Infect Dis. 2017;17:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mor O, Gozlan Y, Wax M, Mileguir F, Rakovsky A, Noy B, et al. Evaluation of the realtime HIV-1, Xpert HIV-1, and Aptima HIV-1 Quant Dx assays in comparison to the NucliSens EasyQ HIV-1 v20 assay for quantification of HIV-1 viral load. J Clin Microbiol. 2015;53:3458–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moyo S, Mohammed T, Wirth KE, Prague M, Bennett K, Pretorius Holme M, et al. Point-of-care Cepheid Xpert HIV-1 viral load test in rural African communities is feasible and reliable. J Clin Microbiol. 2016;54:3050–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosha F Evaluation of HIV Point-of-Care testing assays for the monitoring of HIV patients (Cepheid near POC device evaluation report for viral load testing). 2016. [Google Scholar]
- 22.Mwau M Performance and usability evaluation of Cepheid Xpert for HIV-1 viral load assay. 2017. [Google Scholar]
- 23.Nash M, Ramapuram J, Kaiya R, Huddart S, Pai M, Baliga S. Use of the GeneXpert tuberculosis system for HIV viral load testing in India. Lancet Glob Health. 2017;5:e754–e755. [DOI] [PubMed] [Google Scholar]
- 24.Swathirajan CR, Vignesh R, Boobalan J, Solomon SS, Saravanan S, Balakirshnan P. Performance of point-of-care Xpert HIV-1 plasma viral load assay at a tertiary HIV care centre in Southern India. J Med Microbiol. 2017;66(10):1379–1382. [DOI] [PubMed] [Google Scholar]
- 25.Umviligihozo G, Uwera G. Use of the Cepheid GeneXpert instrument in comparison with the Roche COBAS Ampliprep/COBAS TaqMan HIV-1 for HIV-1 diagnosis and HIV-1 viral load monitoring. [Google Scholar]
- 26.ZEHRP-NDOLA GeneXpert HIV-1 qualitative and HIV-1 viral load techniques validation report.
- 27.Zinyowera S, Ndlovu Z, Mbofana E, Kao K, Orozco D, Fajardo E, et al. Laboratory evaluation of the GeneXpert HIV-1 viral load assay in Zimbabwe. 23rd Conference on Retroviruses & Opportunistic Infectious (CROI) Boston, MA 2016; abstract no 512. [Google Scholar]
- 28.Nash M, Huddart S, Badar S, Baliga S, Saravu K, Pai M. Performance of the Xpert HIV-1 viral load assay: a systematic review and meta-analysis. J Clin Microbiol. 2018;56:e01673–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WHO Prequalification of In Vitro Diagnostics Public report for Xpert HIV-1 Viral Load with GeneXpert Dx, version 3.0. August 2017. Accessed January 21, 2019: https://www.who.int/diagnostics_laboratory/evaluations/pq-list/hiv-vrl/170830_amended_final_pqpr_0192_0193_0194_0195_070_00_v3.pdf?ua=1.
- 30.Albert H, Nathavitharana RR, Isaacs C, Pai M, Denkinger CM, Boehme CC. Development, roll-out and impact of Xpert MTB/RIF for tuberculosis: what lessons have we learnt and how can we do better? Eur Respir J. 2016;48(2):516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cazabon D, Pande T, Kik S, Van Gemert W, Sohn H, Denkinger C, et al. Market penetration of Xpert MTB/RIF in high tuberculosis burden countries: A trend analysis from 2014-2016. Gates Open Res. 2018;2:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection: Recommendations for a Public Health Approach. 2013. Geneva. [PubMed] [Google Scholar]
- 33.World Health Organization. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection: Recommendations for a Public Health Approach. 2016. Geneva. [PubMed] [Google Scholar]
- 34.Vojnov L, Carmona S, Zeh C, Markby J, Boeras D, Prescott MR, et al. The performance of using dried blood spot specimens for HIV-1 viral load testing: a systematic review and meta-analysis. PLoS Medicine. 2019. Manuscript in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arends L, Hamza T, Van Houwelingen J, Heijenbrok-Kal MH, Hunink MG, Stijnen T. Bivariate random effects meta-analysis of ROC curves. Medical Decision Making. 2008;28(5):621–638. [DOI] [PubMed] [Google Scholar]
- 36.Dahabreh IJ, Trikalinos TA, Lau J, Schmid C. An Empirical Assessment of Bivariate Methods for Meta-Analysis of Test Accuracy. Methods Research Report. Agency for Healthcare Research and Quality. 2012;12(13):EHC136–EF. [PubMed] [Google Scholar]
- 37.Cobb BR, Vaks JE, Do T, Vilchez RA. Evolution in the sensitivity of quantitative HIV-1 viral load tests. J Clin Virol. 2011;52 Suppl 1:S77–82. [DOI] [PubMed] [Google Scholar]
- 38.Hermans LE, Moorhouse M, Carmona S, Grabbee DE, Hofstra LM, Richman DD, et al. Effect of HIV-1 low-level viraemia during antiretroviral therapy on treatment outcomes in WHO-guided South African treatment programmes: a multicentre cohort study. Lancet Infect Dis 2018;18:188–97. [DOI] [PubMed] [Google Scholar]
- 39.Barnabas RV, Revill P, Tan N, Phillips A. Cost-effectiveness of routine viral load monitoring in low- and middle-income countries: a systematic review. JIAS 2017;20(S7):e25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estill J, Egger M, Blaser N, Vizcaya LS, Garone D, Wood R, et al. Cost-effectiveness of point-of-care viral load monitoring of antiretroviral therapy in resource-limited settings: mathematical modelling study. AIDS 2013;27(8):1483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pham QD, Wilson DP, Nguyen TV, Do NT, Truong LX, Nguyen LT, et al. Projecting the epidemiological effect, cost-effectiveness and transmission of HIV drug resistance in Vietnam associated with viral load monitoring strategies. J Antimicrob Chemother 2016;71(5):1367–79. [DOI] [PubMed] [Google Scholar]
- 42.Phillips A, Shroufi A, Vojnov L, Cohn J, Roberts T, Ellman T, et al. Sustainable HIV treatment in Africa through viral load-informed differentiated care. Nature 2015;528(7580):S68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Unitaid. Multi-disease diagnostic landscape for integrated management of HIV, HCV, TB and other coinfections. 2018. Geneva. [Google Scholar]
- 44.World Health Organization. Considerations for adoption and use of multidisease testing devices in integrated laboratory networks. 2017. Geneva. [Google Scholar]