Abstract
Purpose:
Identifying imaging phenotypes and understanding their relationship with prognostic markers and patient outcomes can allow for a non-invasive assessment of cancer. The purpose of this study was to identify and validate intrinsic imaging phenotypes of breast cancer heterogeneity in pre-operative breast Dynamic Contrast Enhanced Magnetic Resonance Imaging (DCE-MRI) scans and evaluate their prognostic performance in predicting 10-year recurrence.
Experimental Design:
Pre-treatment DCE-MRI scans of 95 women with primary invasive breast cancer with at least 10-years of follow-up from a clinical trial at our institution (2002-2006) were retrospectively analyzed. For each woman, a signal enhancement ratio (SER) map was generated for the entire segmented primary lesion volume from which 60 radiomic features of texture and morphology were extracted. Intrinsic phenotypes of tumor heterogeneity were identified via unsupervised hierarchical clustering of the extracted features. An independent sample of 163 women diagnosed with primary invasive breast cancer (2002-2006), publicly available via The Cancer Imaging Archive, was used to validate phenotype reproducibility.
Results:
Three significant phenotypes of low, medium, and high heterogeneity were identified in the discovery cohort and reproduced in the validation cohort (p<0.01). Kaplan-Meier curves showed statistically significant differences (p < 0.05) in recurrence-free survival (RFS) across phenotypes. Radiomic phenotypes demonstrated added prognostic value (c=0.73) predicting RFS.
Conclusions:
Intrinsic imaging phenotypes of breast cancer tumor heterogeneity at primary diagnosis can predict 10-year recurrence. The independent and additional prognostic value of imaging heterogeneity phenotypes suggests that radiomic phenotypes can provide a non-invasive characterization of tumor heterogeneity to augment personalized prognosis and treatment.
Keywords: Radiomics, Breast cancer prognosis, Imaging biomarkers
Introduction:
Breast cancer inter-tumor and intra-tumor heterogeneity can be seen in gene expression, histopathology, and macroscopic structure(1). In particular, intra-tumor heterogeneity can manifest both spatially and temporally(2–7). Spatial heterogeneity is thought to originate from variable, micro-environment-specific stresses and branched evolution from a common ancestor cell population into divergent subclonal populations(2). Temporal heterogeneity can arise from the dynamic progression and growth of cancer cells as well as in response to systemic therapy(2). Recent studies suggest that increased intra-tumor heterogeneity is associated with adverse clinical outcomes, posing a challenge for accurate patient prognosis and prediction(8). Specifically, more aggressive tumor sub-regions may drive disease progression, expanding to inhabit recurrent tumors(9–12). Additionally, treatments such as chemotherapy, radiotherapy, or targeted agents may apply dynamic stresses to select subpopulations of the tumor causing resistance to treatment and subsequent recurrent growth(11,13–16).
Currently, critical disease treatment decisions are made on the basis of markers acquired from tissue samples, typically obtained via core biopsy or surgical excision. Histopathologic assessment of this sample determines common prognostic markers including tumor size, shape, grade, nodal status, and metastasis; expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) status are determined via immunohistochemistry. Commercial prognostic tests such as MammaPrint® (Agendia BV, Amsterdam, the Netherlands) and Oncotype DX ® (Genomic Health, Inc., Redwood City, CA, USA) have been developed to measure mRNA and assess gene expression profiles respectively, but they are clinically limited by use in only specific breast cancer molecular subtypes(17). In addition, they are expensive, their assessment can vary depending upon the tissue sample, and they are not always implemented in routine diagnosis(18,19). The prognostic and predictive markers derived from the limited diagnostic tissue samples may under-sample spatially heterogeneous breast tumors as well as overlook temporal shifts due to breast cancer progression or exposure to therapy. Therefore, there is a clinical need to develop prognostic and predictive markers of intra-tumor heterogeneity that may augment established biomarkers for personalized disease diagnosis, staging, management, and to assess treatment response to neoadjuvant therapy.
Medical imaging is currently used for breast cancer diagnosis, staging, and treatment response assessment, providing a means for longitudinal, non-invasive, whole-tumor evaluation of disease burden(20–22). Dynamic contrast enhanced magnetic resonance imaging (DCE-MRI), in particular, is highly sensitive for primary lesion detection and staging, with the ability to assess tumor vascularization with contrast enhancement(21,23). The field of “Radiomics” leverages high throughput data extracted from medical images and has shown promise in quantifying the imaging presentation of underlying tumor biology(24–28). Identifying intrinsic radiomic phenotypes of breast cancer and understanding their relationship with patient outcomes and other histopathologic factors could complement current prognostic and predictive biomarkers. The purpose of this study was to identify and validate such intrinsic DCE-MRI radiomic phenotypes of breast cancer tumor heterogeneity, and evaluate their independent prognostic performance in predicting 10-year recurrence, and their performance in augmenting established, histopathologic prognostic factors.
Methods:
Discovery cohort: Study population and imaging protocol
Breast DCE-MRI scans were retrospectively analyzed from a previously completed, multimodality imaging trial conducted at our institution (2002-2006; National Institutes of Health; P01CA85484) designed to evaluate an array of different breast imaging modalities in cancer staging, diagnosis, and screening. The study was Health Insurance Portability and Accountability Act-compliant, approved by the institutional review board at our institution, and in accordance with U.S. Common Rule. The trial originally recruited 901 women, including women with newly diagnosed breast cancer presenting for staging, women with a mammographically detected suspicious finding or suspicious palpable mass directed to biopsy, and women eligible for high-risk screening. Informed written consent was obtained prior to trial participation. From these, 317 women were diagnosed with primary breast cancer, of which 231 were diagnosed with invasive breast cancer. From this subset of 231 women, 177 women had DCE-MRI images available for analysis. An additional 72 women were excluded for not receiving a consistent imaging protocol of fat-suppressed, T1-weighted DCE-MRI with at least two post contrast scans available for analysis. Lastly, 3 women were excluded on the basis of image quality, determined by biopsy artifacts and fiduciary markers, or the presence of diffuse disease in order to prevent inaccurate ROI segmentation, and 7 women were excluded due software algorithm output resulting in incomplete values during radiomic feature extraction. Therefore, a total of 95 women diagnosed with primary invasive breast cancer and imaged with a consistent T1-weighted DCE-MRI protocol with a first and second post-contrast acquisition, prior to any treatment, were included in our analysis. For this retrospective analysis, the requirement of informed consent was waived under institutional review board approval.
Following intravenous administration of gadolinium contrast, DCE-MRI images were acquired sagitally via a T1-weighted 3D protocol. Images were acquired with a 45-degree flip angle over a 16-18 cm field of view, with 2-2.5 mm slice thickness. Women subsequently underwent surgery for tumor removal. Histopathologic analysis of surgical specimens evaluated hormone receptor (HR) status, consisting of ER and PR status, HER2 status, clinical stage, size (cm) as determined from pathology sample, and surgical margins. Stage, Modified Bloom Richardson grade (MBRG), lymph invasion status, nuclear grade, and presence of ductal carcinoma in situ (DICS) were also documented. Post-surgery therapy included a variable combination of chemotherapy, hormone therapy, and radiation. Recurrence-free survival (RFS) was monitored for all women over a 10-year follow-up period. Survival was determined as the date of breast cancer diagnoses to death or more recent follow-up. Patients without an event were censored at the date of last follow-up. In the discovery cohort, 11 women (12%) had recurrence events, and 84 women (88%) were event-free until their last available follow-up (Table 1). Clinical stage was statistically significantly associated with recurrence events (p = 0.02) (Table 1).
Table 1.
Summary of patient characteristics from the discovery cohort
| Primary Invasive Cancers (n=95) | |||
|---|---|---|---|
| Non-recurrent cases at the time of last follow-up 84 (88% of total) |
Recurrent cases 11 (12%) |
Significance tested using Chi-square analysis | |
| Malignant Pathology | p=0.48 | ||
| Invasive Ductal Carcinoma (IDC) | 65 (77% of NR) | 8 (73% of R) | |
| Invasive Lobular Carcinoma (ILC) | 8 (10%) | 1 (7%) | |
| IDC/ILC | 9 (11%) | 2 (14%) | |
| Receptor Status | p= 0.37 | ||
| Hormone Receptor Positive | 61(73%) | 9 (82%) | |
| HER2 Positive | 20 (24%) | 2 (18%) | |
| Triple Negative | 11 (13%) | 1 (9%) | |
| Clinical Stage | p=0.02 | ||
| Early Stage (1) | 35 (41%) | 1 (9%) | |
| Advanced Stage (2-3) | 45 (54%) | 10 (91%) | |
| DCIS | p=1 | ||
| Present | 67 (80%) | 9 (82%) | |
| Margins | p=0.07 | ||
| Positive | 40 (48%) | 6 (55%) | |
| Negative | 40 (48%) | 5 (45%) |
Validation cohort: Study population and imaging protocol
An independent validation cohort was acquired from a subset of the ISPY-1/ACRIN 6657 trial (2002-2006)(29). Women diagnosed with T3 breast tumors measuring 3cm or larger were enrolled in this trial, and underwent anthracycline-based neoadjuvant chemotherapy. DCE-MRI scans were acquired for women in this study as previously described(30). The pre-treatment and pre-operative DCE-MRI images of 222 women were publicly available via The Cancer Imaging Archive(31). From this, 15 women were excluded for having incomplete DCE acquisition or variability in imaging protocol. A further 43 women were excluded for having missing histopathologic data, RFS outcome, or pre-treatment DCE-MRI scans, and 1 woman was excluded due software algorithm output resulting in incomplete values during radiomic feature extraction. In all, 163 women were included in the validation cohort for this study; validation analysis utilized the scans that were both pre-treatment and pre-operative. Clinical information including HR status and HER2 status were available for each woman in the validation cohort. RFS status, defined as the time between first chemotherapy treatment and disease recurrence, was also available. A total of 44 women in the validation cohort (27%) had recurrent tumors (Supplemental figure 1). A comparison between the two cohorts via chi-square analysis indicated a statistically significant difference between number of recurrent cases (p = 0.02), number of HR positive cases (p = 0.02), and clinical stage of tumors (p < 0.001) (Supplemental figure 1).
Radiomic feature extraction
For each woman in the discovery cohort, the primary lesion was selected by a radiologist from the pre-treatment, pre-operative DCE-MRI scan, and manually segmented from the most representative slice, as determined by the largest tumor volume. This manual segmentation served as the initialization for 3-D tumor volume segmentation, which was performed using a previously validated, automated method (32) and visually verified by an expert after segmentation. Images were pre-processed using N3 bias-field normalization(33) and histogram normalization in order to correct for low frequency bias field signal or outliers that may induce artifacts within the image. Using the first (I1) and second (I2) post-contrast images, a signal enhancement ratio (SER) map was generated(23) for the entire tumor volume, defined as the voxel-wise ratio between the first and second post contrast images:
The first and second post-contrast images were acquired in succession at an average approximation of 90 seconds after contrast injection and first-post contrast scan, respectively. A multi-parametric, radiomic feature vector was extracted from the SER map for each woman (Supplemental figure 2), including a) previously validated morphologic features of tumor perimeter, area, ellipticity, and convexity, shown to be associated with disease progression(22,34), and b) radiomic features capturing structural(35), run-length(36–38), co-occurrence matrix(39), gray-level histogram, and gray-level size zone matrix textures (40), which were extracted and summarized over the primary lesion. Briefly, structural features capture intensity variations between central voxels and neighboring voxels. Run-length features measure the coarseness of an image in specific linear directions. Co-occurrence features analyze the spatial distribution of voxel intensity values by capturing frequency information of gray-level intensity values within a neighborhood of voxels in a specific linear orientation. Gray level histogram features are first order statistical features assessing the distribution of gray-level voxel intensities within an image. Gray level size zone features capture the connectedness of varying intensity levels within an image. Additionally, the mean and standard deviation of SER values of the tumor were calculated. Consequently, a total of 60 radiomic features were extracted (Appendix). All features were extracted using the publically available software, Cancer Imaging Phenomics Toolkit (CaPTk) (ver. 1.7.1, Univ. of Pennsylvania) (https://cbica.github.io/CaPTk/) (41).
Discovery of intrinsic imaging phenotypes
Prior to phenotype identification, the multi-parametric radiomic features extracted from the pre-operative DCE-MRI scans were z-score normalized(42). Furthermore, z-scored features with extreme skewness or extremely low variations in distributions across women, defined as interquartile range (IQR) < 1 or kurtosis > 15, were excluded from further analysis to prevent a biased analysis. This resulted in a total of 22 features concatenated to form the final feature vector. Given the definition of each radiomic feature in the final feature vector, features were standardized such that a greater feature value indicates greater image heterogeneity. A tumor heterogeneity index was then generated for each woman, defined as the statistical average of z-score normalized, standardized features in the final feature vector. Thus, a higher heterogeneity index corresponds to higher intra-tumor heterogeneity whereas a lower heterogeneity index corresponds to increased intra-tumor homogeneity.
To identify intrinsic imaging phenotypes, unsupervised hierarchical clustering was performed on the extracted, multi-parametric feature vectors for women in the discovery cohort(43). The k clusters obtained from the unsupervised hierarchical clustering algorithm are interpreted as intrinsic imaging phenotypes in the population. Briefly, an agglomerative approach was used to create a hierarchical clustering of women, using Euclidean distance for the distance between feature vectors and Ward’s minimum variance method as the clustering criterion(44). The optimal number of distinct phenotypes, k, was determined by assessing the stability and significance of each phenotype for each value of k that was considered. The optimal number of stable phenotypes was determined using consensus clustering(45), where the dataset was subsampled and cluster arrangements were determined using varying numbers of k. For each number of k phenotypes, the proportion that two women occupied the same phenotype cluster out of the number of times they appeared in the same subsample was determined and stored in a symmetric consensus matrix, from which a cumulative distribution function (CDF) was determined. Cluster stability, as determined by the area under the CDF curve, was evaluated for each increase in k phenotype, with a change in stability less than 10% deemed insignificant. Statistical significance of the identified, stable phenotypes was evaluated using the SigClust(46) method. Here, the significance of the cluster index, defined as the sum of within-cluster sums of squares about the cluster-mean divided by the total sum of squares about the overall mean, was tested against a null distribution, simulated using 10,000 samples from a Gaussian distribution fit to the data. The test was performed at each phenotype split to determine statistical significance (p<0.05).
Independent validation of intrinsic imaging phenotypes
Tumor segmentation for cases from the validation cohort was performed per the ISPY-1/ACRIN 6657 protocol(30). The 22 features identified from the discovery cohort were extracted from segmented tumors in the validation cohort using the same feature pre-processing steps outlined above, to form the final feature vectors for hierarchical unsupervised clustering analysis. These features were normalized using the mean and standard deviation values of each respective feature’s distribution in the discovery cohort, to standardize feature ranges. To validate identified phenotype reproducibility, women in the validation cohort were assigned to the discovery cohort-identified phenotypes by minimizing the Euclidian distance between each validation cohort feature vector and the discovery cohort-identified phenotype centroid. The significance and reproducibility of phenotype assignment in the validation cohort was assessed using Consensus Clustering and the SigClust methods (Figure 1).
Figure 1.
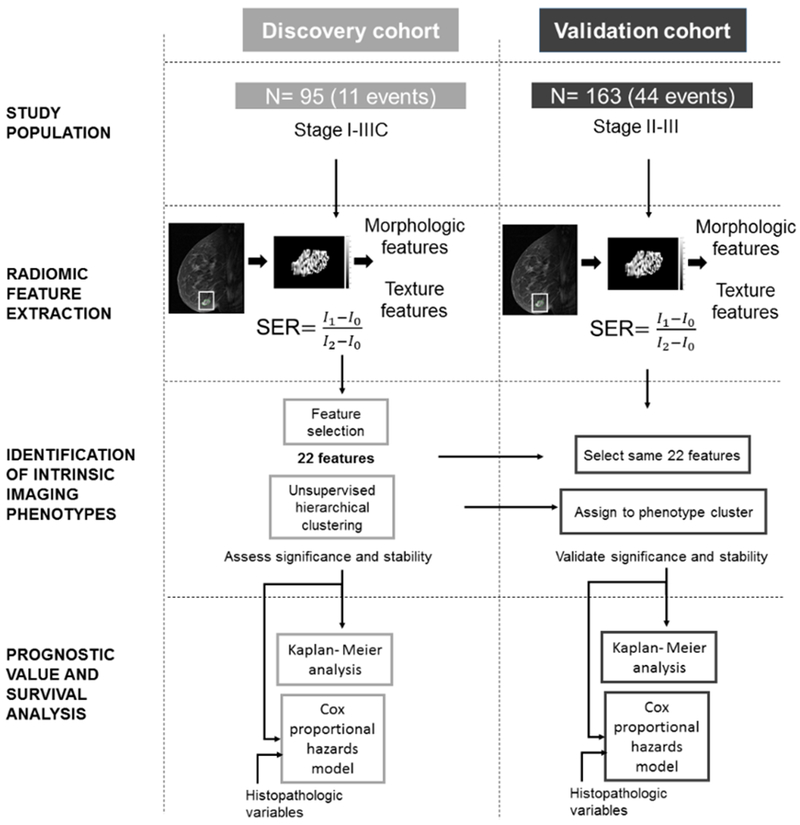
Study design. Radiomic features were extracted from SER maps generated from pre-operative DCE-MRI scans from women in the discovery cohort (n = 95). Feature selection resulted in a 22-feature feature vector. Unsupervised hierarchical clustering was used to identify intrinsic imaging phenotypes, which were assessed for statistical significance and stability. The same 22 features were extracted from the pre-operative DCE-MRI scans of 163 women in an independent validation cohort. Women in the validation cohort were assigned to a phenotype identified in the discovery cohort by minimizing the distance between their 14-feature feature vector and the corresponding phenotype centroids. The independent and additional prognostic values of heterogeneity phenotypes were assessed via Kaplan-Meier RFS analysis and Cox proportional hazards models when compared to a baseline model of established histopathologic biomarkers.
Prognostic value of imaging phenotypes
We assessed the distribution of histopathologic, prognostic covariate values for women assigned to each heterogeneity phenotype using chi-square tests for categorical biomarker values and Kruskal-Wallis tests for continuous biomarker values. The distributions of post-surgery therapy received by each woman (i.e., chemotherapy, hormone therapy, and radiation therapy) and recurrence-free survival were assessed across phenotypes to identify any associations between therapy and RFS or heterogeneity phenotypes.
RFS probabilities across heterogeneity phenotypes within the discovery and validation cohorts were evaluated using Kaplan-Meier curves, with log-likelihood statistical tests used to assess their significance and determine their independent prognostic value. To determine the additional value categorizing tumor heterogeneity phenotypes, a baseline Cox proportional hazards model was built using the established histological prognostic factors of HR and HER2 status. Performance of this model in predicting RFS was tested both with and without phenotype cluster assignment, coded as a categorical variable.
Results
Discovery of intrinsic imaging phenotypes
Three statistically significant phenotypes were identified in the discovery cohort via unsupervised hierarchical clustering and found to be statistically significant via the SigClust methods (p < 0.01) (Figure 2, Supplemental figure 3). Ordering the heterogeneity indices of the corresponding centroids for the identified phenotypes in ascending order allowed for the identified phenotypes to be interpreted as phenotypes of low, medium, and high intra-tumor heterogeneity. The number of recurrences were statistically significantly different across the heterogeneity phenotypes via chi-square analysis (p = 0.01).
Figure 2.
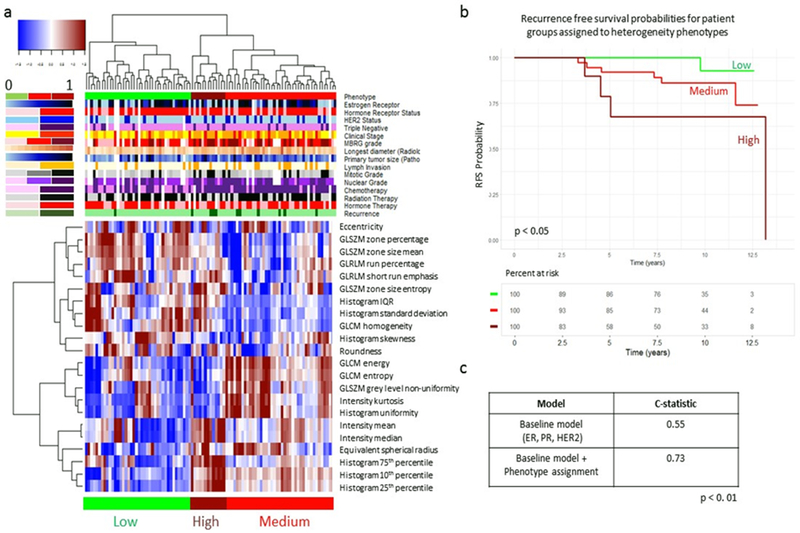
Identification of intrinsic imaging phenotypes of tumor heterogeneity. Unsupervised hierarchical clustering of SER features identifies three intrinsic phenotypes in the discovery cohort (A). RFS curves for women stratified by imaging heterogeneity phenotype show that heterogeneity phenotype is statistically significant (p < 0.05) when predicting RFS (B). Adding phenotype information to Cox regression model shows an improvement in C-statistic when predicting recurrence events (C).
Kaplan-Meier RFS curves for women stratified by heterogeneity phenotype assignment were found to be statistically significantly different, as determined using the log-rank test (p < 0.05). A baseline Cox-proportional hazards model consisting of HR status and HER2 status resulted in a c-statistic of 0.55 when predicting 10-year RFS. Adding heterogeneity phenotype assignment to the baseline model resulted in a c-statistic of 0.73 A log-likelihood test showed statistically significant improvement in the augmented model performance (p = 0.007) (Figure 2).
Analysis of clinical covariate significance across heterogeneity phenotype status showed that differences in tumor MBRG, estrogen receptor percentage, and tumor mitotic stage were statistically significant across heterogeneity phenotypes (p = 0.03, p=0.001, and p=0.02, respectively). Of poorly differentiated (MBRG 8-9) tumors, 80% were assigned to the medium or high heterogeneity phenotypes, and 20% were assigned to the low heterogeneity phenotype (Figure 3a). Tumors assigned to the low and medium heterogeneity phenotypes had median estrogen receptor percentages of 75% and 70% respectively, while tumors assigned to the high heterogeneity phenotype had median estrogen receptor percentages of 40% (Figure 3b). Of tumors with high mitotic stages, 81% were assigned to the medium or high heterogeneity phenotypes, and 19% were assigned to the low heterogeneity phenotype (Figure 3c).
Figure 3.
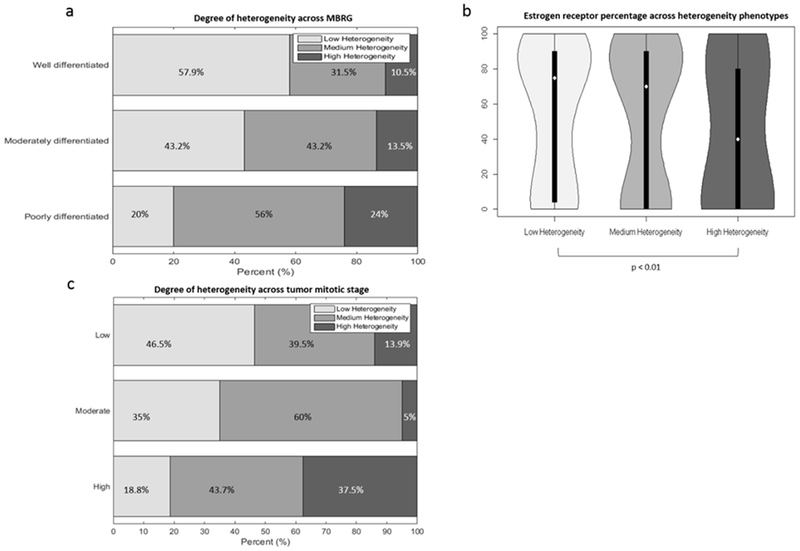
Associations between histopathologic prognostic markers and heterogeneity phenotypes. Associations between histopathologic prognostic markers and heterogeneity phenotypes identified in the discovery cohort. Degree of phenotypic heterogeneity in well, moderately, and poorly differentiated tumors (A). Percent of estrogen receptor distribution for women in low, medium, and high heterogeneity phenotypes (B). Degree of phenotypic heterogeneity in tumors with low, moderate and high mitotic stage (C).
Validation of intrinsic imaging phenotypes
Women in the discovery cohort assigned to the low, medium, and high heterogeneity phenotypes had average heterogeneity indices of −0.09, −0.03, and 0.16, respectively (Figure 4a). Women in the validation cohort assigned to the low, medium, and high heterogeneity phenotypes had average heterogeneity indices of −0.05, −0.24, and 0.21, respectively (Figure 4b).
Figure 4.
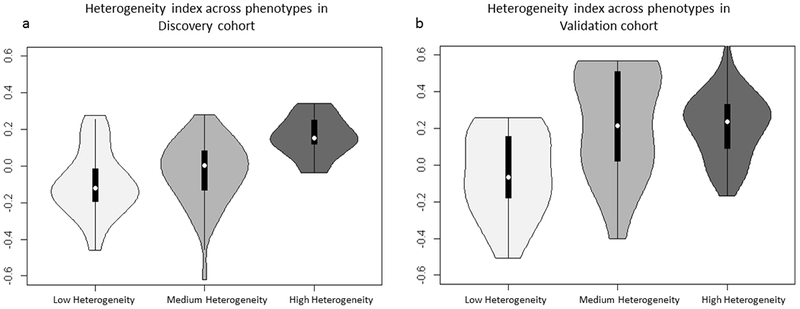
Heterogeneity index* distributions of women in low, medium, and high heterogeneity phenotypes in the discovery (A) and validation (B) cohorts.
* defined as the statistical average of z-score normalized, heterogeneity standardized features in the final feature vector for each tumor in the discovery and validation cohorts.
The heterogeneity phenotypes were found to be reproducible and statistically significant in the validation set using the SigClust methods (p = 0.01). Kaplan-Meier RFS curves for women stratified by phenotype clustering assignment were statistically significantly different (p = 0.01 (Supplemental figure 4). A baseline Cox-proportional hazards model consisting of HR status and HER2 status resulted in a c-statistic of 0.61 when predicting 10-year RFS. Adding heterogeneity phenotype assignment to the baseline model resulted in a c-statistic of 0.67 A log-likelihood test showed statistically significant improvement in the augmented model performance (p = 0.01).
Analysis of clinical covariate significance across heterogeneity phenotype status showed that differences in progesterone receptor status were statistically significant across heterogeneity phenotypes (p = 0.03).
Discussion
Our results indicate that distinct imaging phenotypes exist within invasive breast tumors which correspond to different degrees of intra-tumor heterogeneity, suggesting that radiomic features can non-invasively characterize such heterogeneity patterns. The identification and validation of distinct image heterogeneity phenotypes show that the phenotype clusters identified are both interpretable and meaningful. Most notably, the validated heterogeneity phenotypes show independent prognostic value when predicting 10-year RFS, indicating that intrinsic imaging phenotypes can potentially identify intra-tumor heterogeneity features driving aggressive tumor behavior. Women assigned to the high heterogeneity phenotype demonstrated decreased probabilities of RFS over the 10-year follow-up period, corroborating the hypothesis that heterogeneous tumors are associated with aggressive tumor behavior and treatment resistance.
Of particular note, the medium heterogeneity phenotype encompasses a wide range of tumors, as evidenced by the differences in Kaplan-Meier survival analysis between women assigned to the medium heterogeneity phenotype in the discovery and validation cohorts. While both cohorts represent populations of women diagnosed with invasive breast cancer, all women in the validation cohort were diagnosed with advanced stage disease, and therefore eligible for neoadjuvant therapy. As the discovery cohort consists of more diverse disease stages, women in the validation cohort assigned to the low heterogeneity phenotype may have higher degrees of tumor heterogeneity as compared to women in the discovery cohort also assigned to the low heterogeneity phenotypes and may therefore be more similar to tumors assigned in the discovery cohort as having a medium heterogeneity phenotype. This is supported by the similarity of the average heterogeneity indices for women in the validation cohort assigned to the low heterogeneity phenotype versus women in the discovery cohort assigned to the medium heterogeneity phenotype (−0.05 and −0.03, respectively). Consequently, survival probabilities for women in the validation cohort assigned to the low heterogeneity phenotype may be more similar to women in the discovery cohort originally assigned to the medium heterogeneity phenotype, as compared to women in the low heterogeneity phenotype. A statistical comparison between cohorts suggests women in the validation cohort had a statistically significantly higher proportion of HR negative tumors, thereby suggesting more aggressive tumor behavior and outcome (Supplemental figure 1).
Imaging phenotypes of intra-tumor heterogeneity also provide additional prognostic value when augmenting established histopathologic prognostic biomarkers. The independent and additional prognostic value of phenotype assignment suggests that imaging phenotypes can provide unique information about underlying tumor behavior, and therefore, complement clinically utilized prognostic markers for personalized prognosis and decision making.
Higher degrees of imaging phenotype heterogeneity were shown to be associated with poorly defined tumors as per the MBRG and tumors with higher mitotic grades. The increased mitotic grade of tumors in the higher heterogeneity phenotypes may contribute to the genetic diversity and subclonal evolution thought to increase intratumor heterogeneity (2). These results suggest that tumor characteristics such as nuclear pleomorphism and increased mitotic rates, which are characteristic of aggressive tumor behavior, may be captured by the imaging phenotypes of tumor heterogeneity. Additionally, tumors with lower percentages of estrogen receptors displayed statistically significantly higher imaging phenotype heterogeneity, correlating with established hypotheses that estrogen positive tumors are associated with more positive prognoses(47,48). The statistically significant associations of histopathologic prognostic covariate distributions across heterogeneity phenotypes indicate that image heterogeneity may correlate with underlying tumor biology.
We have identified intrinsic imaging phenotypes of intra-tumor heterogeneity in primary invasive breast cancer that can independently predict 10-year recurrence and have validated these findings in an independent cohort. While previous studies have utilized hierarchical clustering analysis to identify imaging phenotypes or have investigated relationships between radiomic features and histopathologic and genomic tumor characteristics, most of these studies have used surrogate measures of recurrence or were limited by a lack of independent validation(28,49).
Limitations to our study should be noted. For this exploratory analysis, we chose a fixed set of radiomic features. The independent validation cohort utilized for this study consisted of only advanced stage tumor diagnoses with a limited availability of histopathologic prognostic biomarkers, as opposed to the discovery cohort which consisted of both early and advanced stage tumors and a wide array of histopathologic prognostic biomarker information available for each woman. As both cohorts included women diagnosed with invasive breast cancer and follow-up information, we determined that validating our heterogeneity phenotypes with a more niche cohort can still demonstrate the added value and generalizability of tumor imaging heterogeneity phenotypes. We also aim to expand our analysis to a larger cohort. Lastly, we will seek to further explore the added prognostic value of imaging phenotypes to that of emerging prognostic molecular profiling assays.
In conclusion, our results demonstrate that intrinsic imaging phenotypes of tumor heterogeneity exist, with independent and additional prognostic value in predicting RFS. Additionally, these heterogeneity phenotypes show associations with established histopathologic prognostic biomarkers, suggesting that image heterogeneity phenotypes non-invasively capture underlying tumor biology.
Supplementary Material
Translational Relevance:
Breast cancer is a heterogeneous disease, with known inter-tumor and intra-tumor heterogeneity. Established histopathologic prognostic biomarkers generally acquired from tumor biopsy may be limited by sampling variation. Radiomics is an emerging field with the potential to leverage the whole tumor via non-invasive sampling afforded by medical imaging to extract high throughput, quantitative features for personalized tumor characterization. In this exploratory study, the identified and independently validated intrinsic radiomic phenotypes of tumor heterogeneity provide independent and additional prognostic value when predicting 10-year breast cancer recurrence. Radiomic phenotypes have the potential to provide a non-invasive characterization of tumor heterogeneity to augment personalized clinical decision making for breast cancer.
Acknowledgements
ACKNOWLEDGEMENT OF RESEARCH SUPPORT: This study was supported by the National Cancer Institute at the National Institutes of Health:
5R01CA197000-03: R. Chitalia, E. McDonald, L. Pantalone, E. Cohen, M. Feldman, D. Kontos
R01CA223816-01A1: R. Chitalia, E. McDonald, L. Pantalone, E. Cohen, M. Feldman, D. Kontos
U24-CA189523: A. Gastounioti, D. Kontos
Footnotes
The authors declare no potential conflicts of interest
References
- 1.Polyak K Heterogeneity in breast cancer. The Journal of clinical investigation 2011;121(10):3786–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nature Reviews Clinical Oncology 2017. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. science 2013;339(6127):1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murugaesu N, Wilson GA, Birkbak NJ, Watkins TB, McGranahan N, Kumar S, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer discovery 2015;5(8):821–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nature medicine 2015;21(7):751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiley C, de Bruin EC, McGranahan N, Swanton C. Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome biology 2014;15(8):453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer cell 2015;27(1):15–26. [DOI] [PubMed] [Google Scholar]
- 8.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer 2010;1805(1):105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris LG, Riaz N, Desrichard A, Şenbabaoğlu Y, Hakimi AA, Makarov V, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 2016;7(9):10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012;481(7382):506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014;343(6167):189–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turke AB, Zejnullahu K, Wu Y-L, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer cell 2010;17(1):77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486(7404):532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyo-eun CB, Ruddy DA, Radhakrishna VK, Caushi JX, Zhao R, Hims MM, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nature medicine 2015;21(5):440. [DOI] [PubMed] [Google Scholar]
- 15.Kwak EL, Ahronian LG, Siravegna G, Mussolin B, Borger DR, Godfrey JT, et al. Molecular heterogeneity and receptor coamplification drive resistance to targeted therapy in MET-amplified esophagogastric cancer. Cancer discovery 2015;5(12):1271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer discovery 2016;6(2):147–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Győrffy B, Hatzis C, Sanft T, Hofstatter E, Aktas B, Pusztai L. Multigene prognostic tests in breast cancer: past, present, future. Breast cancer research 2015;17(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozick Z, Hashmi A, Dove J, Hunsinger M, Arora T, Wild J, et al. Disparities in compliance with the Oncotype DX breast cancer test in the United States: A National Cancer Data Base assessment. The American Journal of Surgery 2018;215(4):686–92. [DOI] [PubMed] [Google Scholar]
- 19.Kwa M, Makris A, Esteva FJ. Clinical utility of gene-expression signatures in early stage breast cancer. Nature Reviews Clinical Oncology 2017;14(10):595. [DOI] [PubMed] [Google Scholar]
- 20.Gavenonis SC, Roth SO. Role of magnetic resonance imaging in evaluating the extent of disease. Magnetic Resonance Imaging Clinics 2010;18(2):199–206. [DOI] [PubMed] [Google Scholar]
- 21.Weinstein S, Rosen M. Breast MR imaging: current indications and advanced imaging techniques. Radiologic Clinics 2010;48(5):1013–42. [DOI] [PubMed] [Google Scholar]
- 22.Tse GM, Chaiwun B, Wong KT, Yeung DK, Pang AL, Tang AP, et al. Magnetic resonance imaging of breast lesions--a pathologic correlation. Breast cancer research and treatment 2007;103(1):1–10 doi 10.1007/s10549-006-9352-3. [DOI] [PubMed] [Google Scholar]
- 23.Hylton NM. Vascularity assessment of breast lesions with gadolinium-enhanced MR imaging. Magnetic resonance imaging clinics of North America 2001;9(2):321–32, vi. [PubMed] [Google Scholar]
- 24.Ashraf A, Gaonkar B, Mies C, DeMichele A, Rosen M, Davatzikos C, et al. Breast DCE-MRI kinetic heterogeneity tumor markers: preliminary associations with neoadjuvant chemotherapy response. Translational oncology 2015;8(3):154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clinical Cancer Research 2015;21(2):249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahrooghy M, Ashraf AB, Daye D, Mies C, Feldman M, Rosen M, et al. Heterogeneity wavelet kinetics from DCE-MRI for classifying gene expression based breast cancer recurrence risk. 2013. Springer; p 295–302. [DOI] [PubMed] [Google Scholar]
- 27.Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ breast cancer 2017;3(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Zhu Y, Burnside ES, Drukker K, Hoadley KA, Fan C, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology 2016;281(2):382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newitt D HN, on behlaf of the I-SPY 1 Network and ACRIN 6657 Trial Team. Multi-center breast DCE-MRI data and segmentations from patients in the I-SPY 1/ACRIN 6657 trials. The Cancer Imaging Archive. The Cancer Imaging Archive 2016. [Google Scholar]
- 30.Hylton NM, Gatsonis CA, Rosen MA, Lehman CD, Newitt DC, Partridge SC, et al. Neoadjuvant Chemotherapy for Breast Cancer: Functional Tumor Volume by MR Imaging Predicts Recurrence-free Survival-Results from the ACRIN 6657/CALGB 150007 I-SPY 1 TRIAL. Radiology 2016;279(1):44–55 doi 10.1148/radiol.2015150013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. Journal of digital imaging 2013;26(6):1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashraf AB, Gavenonis S, Daye D, Mies C, Feldman M, Rosen M, Kontos D. A multichannel Markov random field approach for automated segmentation of breast cancer tumor in DCE-MRI data using kinetic observation model. InInternational Conference on Medical Image Computing and Computer-Assisted Intervention 2011. (pp. 546–553). [DOI] [PubMed] [Google Scholar]
- 33.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE transactions on medical imaging 1998;17(1):87–97. [DOI] [PubMed] [Google Scholar]
- 34.Szabo BK, Aspelin P, Kristoffersen Wiberg M, Tot T, Bone B. Invasive breast cancer: correlation of dynamic MR features with prognostic factors. European radiology 2003;13(11):2425–35 doi 10.1007/s00330-003-2000-y. [DOI] [PubMed] [Google Scholar]
- 35.Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Transactions on pattern analysis and machine intelligence 2002;24(7):971–87. [Google Scholar]
- 36.Tang X Texture information in run-length matrices. IEEE transactions on image processing 1998;7(11):1602–9. [DOI] [PubMed] [Google Scholar]
- 37.Galloway MM. Texture analysis using grey level run lengths. NASA STI/Recon Technical Report N 1974;75. [Google Scholar]
- 38.Chu A, Sehgal CM, Greenleaf JF. Use of gray value distribution of run lengths for texture analysis. Pattern Recognition Letters 1990;11(6):415–9. [Google Scholar]
- 39.Haralick RM, Shanmugam K. Textural features for image classification. IEEE Transactions on systems, man, and cybernetics 1973(6):610–21. [Google Scholar]
- 40.Thibault G, Angulo J, and Meyer F. Advanced statistical matrices for texture characterization: application to cell classification. IEEE transactions on bio-medical engineering, 2014, 61(3):630–7. [DOI] [PubMed] [Google Scholar]
- 41.Davatzikos C, Rathore S, Bakas S, Pati S, Bergman M, Kalarot R, Sridharan P, Gastounioti A, Jahani N, Cohen E, Akbari H, Tunc B, Doshi J, Parker D, Hsieh M, Sotiras A, Li H, Ou Y, Doot RK, Bilello M, Fan Y, Shinohara RT, Yushkevich P, Verma R, Kontos D, Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome, J Med Imaging, 5(1):011018, 2018. DOI: 10.1117/1.JMI.5.1.011018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman J, Hastie T, Tibshirani R. The elements of statistical learning. Springer series in statistics; New York; 2001. [Google Scholar]
- 43.Perou CM, Sørlie T, Eisen MB, Van De Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature 2000;406(6797):747. [DOI] [PubMed] [Google Scholar]
- 44.Ward JH Jr. Hierarchical grouping to optimize an objective function. Journal of the American statistical association 1963;58(301):236–44. [Google Scholar]
- 45.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Machine learning 2003;52(1-2):91–118. [Google Scholar]
- 46.Liu Y, Hayes DN, Nobel A, Marron J. Statistical significance of clustering for high-dimension, low–sample size data. Journal of the American Statistical Association 2008;103(483):1281–93. [Google Scholar]
- 47.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast cancer research 2007;9(1):R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher B, Redmond C, Fisher ER, Caplan R. Relative worth of estrogen or progesterone receptor and pathologic characteristics of differentiation as indicators of prognosis in node negative breast cancer patients: findings from National Surgical Adjuvant Breast and Bowel Project Protocol B-06. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 1988;6(7):1076–87 doi 10.1200/JCO.1988.6.7.1076. [DOI] [PubMed] [Google Scholar]
- 49.Ashraf AB, Daye D, Gavenonis S, Mies C, Feldman M, Rosen M, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: preliminary associations with gene expression profiles. Radiology 2014;272(2):374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


