Abstract
Precise diagnosis and subtyping of kidney tumors are imperative to optimize and personalize treatment decision for patients. Patients with the most common benign renal tumor, renal oncocytomas (RO), may be over-treated with surgical resection due to limited pre-operative diagnostic methods that can accurately identify the benign condition with certainty. In this study, desorption electrospray ionization (DESI) mass spectrometry imaging was applied to study the metabolic and lipid profiles of various types of renal tissues, including normal kidney, RO, and renal cell carcinomas (RCC). A total of 73,992 mass spectra from 71 patient samples were obtained and used to build predictive models using the least absolute shrinkage and selection operator (lasso). Overall accuracies of 99.47% per-pixel and 100% per-patient for prediction of the three tissue types was achieved. In particular, RO and chromophobe renal cell carcinoma (chRCC), which present the most significant morphologic overlap and are sometimes indistinguishable using histology alone, were also investigated and the predictive models built yielded 100% accuracy in discriminating these tumor types. Discrimination of three subtypes of RCC was also achieved based on DESI-MS imaging data. Importantly, several small metabolites and lipids species were identified as characteristic of individual tissue types and chemically characterized using tandem MS and high mass accuracy measurements. Collectively, our study shows that the metabolic data acquired by DESI-MS imaging in conjunction with statistical modeling allows discrimination of renal tumors, and thus has the potential to be used in the clinical setting to improve treatment of kidney tumor patients.
Introduction
The incidence of renal tumors has been steadily increasing in the past few decades worldwide, especially in Europe, Australia, and USA.(1,2) Renal cell carcinoma (RCC) accounts for more than 80% of all kidney neoplasms, within which the subtypes clear cell RCCs (ccRCCs), papillary RCCs (ppRCCs), and chromophobe RCCs (chRCCs) constitute 80–90%, 10–15%, and 4–5% of cases respectively.(3) Renal oncocytoma (RO), on the other hand, is a common benign neoplasm of kidney which most often occurs in men who are over age of 50, and accounts for 5–9% of all renal tumors. Due to its asymptomatic nature, diagnosis of RO often occurs incidentally during regular medical exams. Radiological imaging using ultrasound (US)(4), magnetic resonance imaging (MRI)(5), computed tomography (CT)(6) and positron emission tomography (PET)(7) are useful for early screening of renal tumors. While these diagnostic imaging tests are powerful to visualize and identify suspicious renal tumors, they are limited in their ability to accurately differentiate between benign and malignant lesions. With the increased use of imaging, more renal tumors are identified incidentally which lead to increased incidence of surgery. Core needle biopsies have thus been increasingly applied to improve the diagnosing accuracy in renal masses.(8) Yet, an obstacle preventing wider use of preoperative core needle biopsies of renal lesions is diagnostic uncertainty in the differentiation of RO from malignant chRCC using cytological/histological evaluation due to significant histologic and immunophenotypic overlap between these tumor types. (9,10) Hybrid tumors with features of both RO and chRCC also occur, although rare. Consequently, patients with RO are subjected to surgical excision of the renal tumor, although these asymptomatic benign tumors do not necessarily require treatment. Therefore, differentiation of ROs from RCCs, especially the more closely-related subtypes such as chRCCs, is crucial for assure appropriate management of patients with renal neoplasms.
Mass spectrometry (MS) imaging has been increasingly applied as a tool to image and diagnose human cancer tissues. In particular, ambient ionization MS techniques are appealing for clinical use in cancer diagnosis due to their minimal sample preparation requirements and simplified experimental workflows.(11) Desorption electrospray ionization mass spectrometry (DESI-MS), for example, was first introduced in 2004 and has been widely explored by different research groups for tissue imaging and cancer diagnosis for various tissue types including brain, gastric, lung, prostate, ovarian, thyroid, and breast tumors.(12) Other ambient ionization MS techniques have also been investigated for cancer diagnosis (13,14) and intraoperative tools for surgical guidance (15–18). Molecular differentiation of normal kidney and kidney cancer by ambient ionization MS has been previously explored. Dill et al., for example, used DESI-MS imaging and multivariate statistical analysis to visualize and classify the differences between RCC and adjacent normal tissue on the basis of glycerophospholipid (GP) profiles.(19) In the study, changes in the lipid profiles detected allowed discrimination of RCC (combining ppRCC and ccRCC) from normal tissues with 97.8% accuracy using statistical analysis. Most recently, Alfaro et al. used touch spray mass spectrometry (TS-MS) for rapid diagnosis between RCC and healthy kidney tissue.(20) Statistical analysis using linear discriminant analysis (LDA) on the PCA-compressed data provided sensitivity and specificity of 98 and 95 %, respectively, using 35 specimens from 21 patents. More recently, Tamura et al. used DESI-MS imaging to identify lipid markers that can predict disease progression in patients with ccRCC, revealing that fatty acids were often more abundant in cancerous tissues.(21) While these previous studies have shown the capability of ambient ionization MS to diagnose RCC from normal tissues, differentiation between normal kidney, benign tumor, and subtypes of malignant kidney tumors is important in clinical practice and has not been yet fully explored.
Here, we explore the use of DESI-MS imaging to discriminate RO from normal kidney and RCC tissue samples with the expectation of improving diagnostic accuracy. The lasso method was applied to the large dataset obtained to provide robust statistical models for multi-class disease diagnosis. Importantly, differentiation of RO from chRCC, as well as from other cancer subtypes, was achieved based on the biological molecular information obtained from DESI-MS imaging. Further, several metabolites and lipid markers characteristic of normal, benign and cancerous kidney tissues were tentatively identified using high mass accuracy measurements and tandem MS data, in comparison to information from online database engine searching. Collectively, our study provides evidence that DESI-MS imaging may be a valuable tool to accurately diagnose renal neoplasms to improve clinical management of patients.
Materials and Methods
Tissue Samples
A total of 81 banked frozen human tissue samples including uninvolved 20 kidney with cortex and/or medullar, 15 renal oncocytoma and 46 RCC samples (11 ccRCCs, 12 chRCCs, 16 ppRCCs, and 2 unclassified) were obtained from Cooperative Human Tissue Network under approved IRB protocol. For more information, please see Supporting Information.
DESI-MS imaging
A 2D Omni stage (Prosolia Inc., Indianapolis, IN) was used with a homebuilt DESI sprayer for tissue imaging with a spatial resolution of 150 μm. DESI-MS imaging was performed in the negative ion mode from m/z 100–1500, using a hybrid LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific, San Jose, CA. Histologically compatible solvent system dimethylformamide:acetonitrile (v/v 1:1) was used for analysis at 1.2 μL/min (22).
Tissue staining and evaluation
The same tissue sections analyzed by DESI-MSI were stained using standard H&E staining protocol. Pathologic evaluation was performed using light microscopy by Dr. Wendong Yu. After the evaluation of 81 samples, 10 of them were excluded due to the abnormal appearance of the histological/cytological patterns of the tissue sections. In total, 71 samples, including 16 kidneys with cortex and/or medulla (5 out of 16 normal kidney tissues contain both cortex and medulla, and the rest only have cortex), 14 RO and 41 RCC samples (11 ccRCCs, 12 chRCCs, 16 ppRCCs, and 2 unidentified) were used for data evaluation and statistical analysis.
Molecular identification
Structures of molecular species were identified using high mass accuracy measurements and collision induced dissociation tandem (CID)/Higher-energy collisional dissociation (HCD) mass spectrometry analysis. The Human Metabolome Database (HMDB), CFM-ID and Lipid Maps databases were used to confirm the tentatively assigned molecules. For more information, please see Supporting Information.
2D imaging data extraction and Statistical analyses
Data were extracted and then uploaded into the open source imaging software packages BioMAP (Novartis) for visualization.(23) For statistical analyses, we applied the lasso method (multiclass-logistic regression with L1 penalty) and principle component analysis (PCA).(24) For more details on data pre-processing and statistical methods, please see Supporting Information.
Results
Molecular characterization of normal kidney
First, we used DESI-MS imaging to investigate the spatial distribution of lipids and other small metabolites in normal kidney tissues. Kidney tissues present two major structures: the outer renal cortex where ultrafiltration and erythropoietin production occur, and the innermost renal medulla which is divided into a number of sections and contains the nephrons that are important for maintaining salt and water balance of the blood. The total lipid content of normal human kidney is ~3% of its wet weight; cortical lipids are composed of 58% phospholipids and 42% neutral lipids, while the medulla contains less phospholipids (37.7%) and more neutral lipids (67.3%).(25)
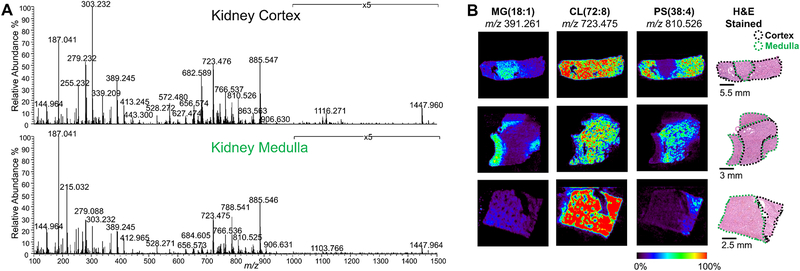
As shown in Fig. 1A, rich molecular information was obtained from normal kidney tissue in both medullar and cortex regions, including many ions characterized as free fatty acids (FFA) and monoradylglycerolipids (MG) in the m/z 100 to 400 range, FA dimers and ceramides (Cer) in m/z 400 to 700 range, and glycerophospholipids (PL) in the m/z 700 to 1000 range. Fig. 1B shows representative DESI-MS ion images from tissue sections of three patients containing both renal cortex and medulla regions. Differences in the relative abundances and spatial distribution of several ions were clearly observed when comparing cortex and medulla regions of the kidney tissues. For example, PS(20:4_18:0) at m/z 810.525 (Supplementary Fig. S1) was more abundant in the cortex regions from all the three patients, while MG(18:1) at m/z 391.261 was more abundant in the medulla (Fig. 1b). The doubly charged CL(18:2_18:2_18:2_18:2) ion at m/z 723.475 (identified by MS/MS, Supplementary Fig. S2) showed relatively homogenous distribution in both cortical and medullar regions with high relative abundances. The DESI-MS images were compared with the hematoxylin and eosin (H&E) stained slides of the same tissue section analyzed, and clear overlap between the distributions of ions in DESI-MS images and the specific histologic regions of the tissues annotated through pathological evaluation was observed.
Figure 1.
DESI-MS analysis of normal kidney tissues. A) Representative DESI mass spectra of renal cortex and medulla. B) DESI-MS ion images of m/z 391.261, m/z 723.475 and m/z 810.526 from three normal kidney samples which contain both cortical and medullar regions. Corresponding optical images of the annotated H&E-stained tissue section are also shown.
Changes in the inter- and intra-patient mass spectra profiles were investigated (Supplementary Fig. S3a,b and S4a,b). When qualitatively comparing the mass spectra profiles of renal cortex and medulla present within the same tissue piece from the same patient, and between two patient tissues (Supplementary Fig. S4), characteristics trends in molecular patterns were observed. For instance, for patient 2, higher relative abundances of Cer(d42:2) at m/z 682.589 (mass error of −2.9 ppm) and PS(20:4_18:0) at m/z 810.525 were clearly seen in cortex regions, while higher relative abundances of MG(18:1) m/z 391.261 (mass error of −2.6ppm) and SHexCer(t42:1) at m/z 906.631 (mass error of −3.3ppm), were found in medulla regions for patient 2. To collectively evaluate differences in molecular profiles between cortex and medullar kidney tissue regions, unsupervised principle component analysis (PCA) was carried out by averaging the mass spectra from the cortex and medulla regions of sixteen normal kidney tissues (n=5 medulla, and n=16 cortex), yielding clear separation between the mass spectra from the tissue regions (Supplementary Fig. S5a,b). When analyzing the loadings plot, (Supplementary Fig. S6a–f), the two ions with the highest and lowest coefficients in loading 2 are m/z 887.56 (in loadings 2, the value is 0.11), assigned as PI (20:3_18:0) (Supplementary Fig. S7), and m/z 216.04 (in loadings 2, the value is −0.13), assigned as a chorine adduct of hexose (Supplementary Fig. S8 and Table S1). Interestingly, m/z 810.52 and m/z 391.26 followed a similar trend in abundance change to what observed qualitatively (m/z 810.52 is high in cortex and m/z 391.26 is high in medulla), however, with less contribution for the PCA separation of the two regions (Supplementary Fig. S6c and S6d). The lipid PS(38:3) at m/z 812.53, which has one less double bond and is less abundant than PS(38:4) m/z 810.52, showed a greater difference in peak intensity between cortex and medulla regions (Supplementary Fig. S6f), with 13 fold- change in abundance detected between cortex and medulla. These results demonstrate that the metabolomic/lipid profiles obtained from normal kidney samples by DESI-MSI allows differentiation between the cortex and medulla. Importantly, note that the distinct molecular information linked to the sub-regions of the kidney tissues would have been lost using traditional metabolomics approaches such as HPLC-MS of the entire normal kidney tissues, thus potentially leading to confusion when building statistical classifiers to discriminate between tissue types. With MS imaging, precise data extraction of histologically validated ROIs is performed, providing reliable data to build robust statistical models.
Molecular characterization of benign and malignant tumor
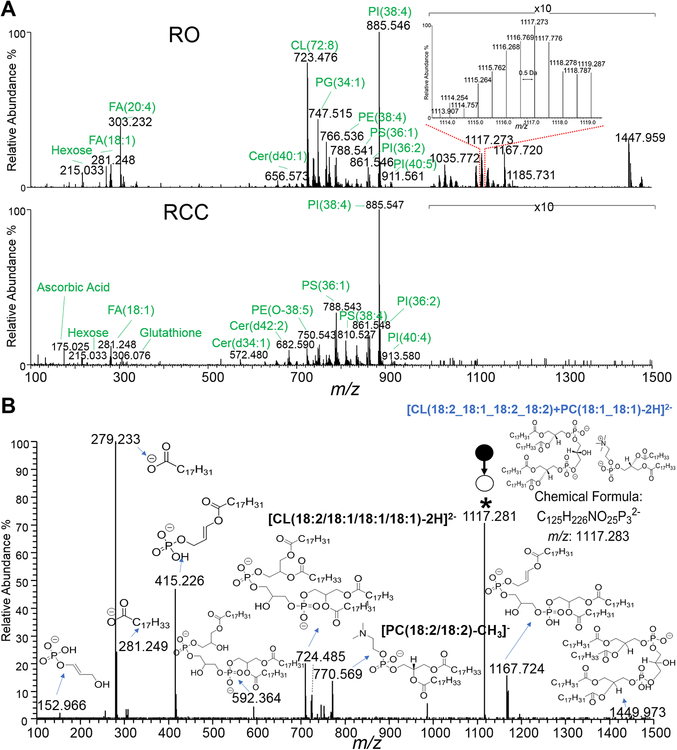
RO is one of the most common types of benign kidney tumors. It is usually not life-threatening, but commonly overtreated as cancerous due to the lack of pre-operative methods that can provide accurate identification. Therefore, there is significant need to better discriminate benign RO tumors from malignant renal tumors. To evaluate if the molecular information obtained by DESI-MS allows discrimination between benign RO and malignant kidney tumors, we used DESI-MS imaging to analyze RO and RCC tissue sections. Representative DESI mass spectra from two kidney tumor samples, one benign RO and one malignant RCC, are shown in Fig. 2A. As observed in the mass spectra, the profiles of RO and RCC were distinct in both the low m/z 100–400 region where metabolites and FA are detected, and the complex lipid region from m/z 600–1500, while fewer and less abundant peaks were detected from m/z 400–600. Contrasting molecular patterns between the two tumor tissue types can be even more clearly seen in the zoomed-in mass spectra (amplified by 10 times for both mass spectra) from m/z 1000–1500. In the mass range from m/z 100–400, several metabolites such as ascorbic acid (m/z 175.025, Supplementary Fig. S9), hexose (m/z 215.033), and free FA such as FA(18:1) (m/z 281.248, mass error at −2.1 ppm), and FA (20:4) (m/z 303.232, mass error at −3.1 ppm) were found at different relative abundances in RO compared to RCC. In the mass range of m/z 600–1000, several lipid species, such as PI (20:4_18:0) (m/z 885.547, Supplementary Fig. S10), PG(18:1_18:1)/PG(16:0_20:2) (m/z 773.532, Supplementary Fig. S11), and PE (18:0_20:4) (m/z 766.536, Supplementary Fig. S12) were detected with similar relative abundances in both tissues. On the other hand, higher intensity of cardiolipin (CL) ions, such as m/z 723.476 and m/z 737.490 in the m/z range of 700–750, were clearly observed in RO tissue. Note that we have previously reported that CLs, including various oxidized and lyso-CL species, are biomarkers of oncocytic thyroid neoplasms.(26) Here, similar molecular trends were observed in RO compared to RCC. Higher relative abundances of singly charged CL species in the mass range of m/z 1000–1500, such as m/z 1447.960 and m/z 1469.940, as well as doubly charged lipid dimers (CL+DG or CL+PC) which are either CL coupled with diacylglycerol (DG) or phosphatidylcholines (PC) (as shown in the inserted Fig. 2A), such as m/z 1035.772 (identified as CL(72:5)+DG(36:2), Supplementary Fig. S13) and m/z 1117.273, were seen in the RO mass spectra. Fig. 2B provides a detailed explanation of the fragmentation pattern of CL(18:2_18:1_18:2_18:2)+PC(18:1_18:1) at m/z 1117.273.
Figure 2.
Representative DESI mass spectra of RO and RCC tissues, and tandem mass spectrum of m/z 1117.3. A) Comparison between RO (on the top) and RCC (at the bottom) mass spectrum in the m/z 100 to m/z 1500 range. B) The m/z 1117.3 ion was assigned as CL(18:2_18:1_18:2_18:2)+PC(18:1_18:1) based on the fragmentation pattern. (* indicates the parent ion). The parent ion fragments of both CL(18:2_18:1_18:2_18:2) and PC(18:1_18:1) with loss of CH3 could be observed.
Discrimination of Normal, RO and RCC in Kidney using DESI-MS imaging
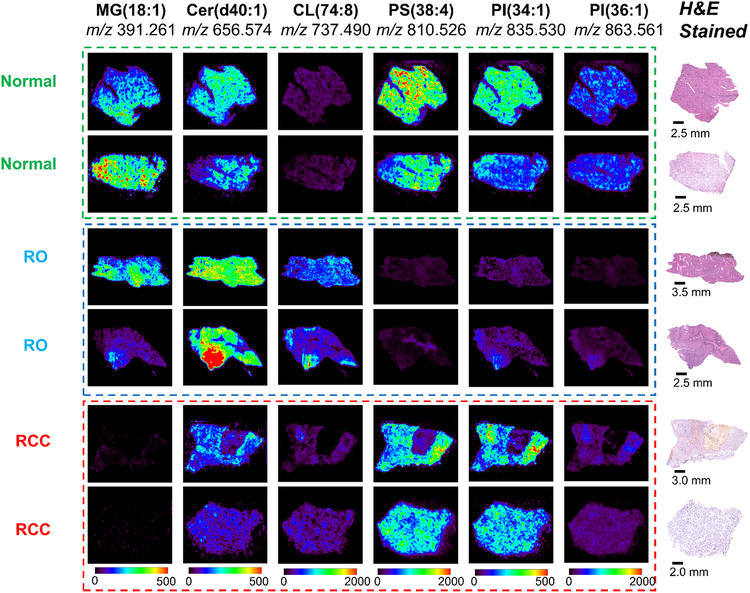
We next evaluated the different in mass spectral profiles between normal kidney tissues, benign RO tissues, and malignant RCC tissues. Fig. 3 presents selected DESI-MS ion images from two normal kidney tissues, two RO tissues, and two RCC tissue samples. Considering the distinct molecular patterns observed in normal renal cortex and medulla, the mass spectra shown for the normal kidney samples are from renal cortex tissues only, to provide a more fair comparison considering ROs originate from the renal cortex region.(27) As can be seen in the DESI-MS images of m/z 391.261, m/z 656.574, m/z 737.490, m/z 810.526, m/z 835.530, and m/z 863.561, different abundances of these ions were observed when comparing the different tissue types, while consistent patterns were observed between the two samples of the same type. In normal renal tissues, all the six ions presented except for m/z 737.490 (identified as CL(20:2_18:2_18:2_18:2), Supplementary Fig. S14) have relatively high abundances. In contrast, the two RO samples showed lower abundances of m/z 810.527 (PS(20:4_18:0)), m/z 835.531(PI(34:1)) and m/z 863.561 (PI(36:1)), but higher abundance of m/z 737.490 when compared to RCC and normal kidney tissues. For RCCs, m/z 656.574 (Cer(d40:1)), m/z 810.527, and m/z 835.531 showed similar abundance with normal samples, but lower abundance of m/z 391.229 and m/z 863.561 when compared to both RO and normal tissues.
Figure 3.
Representative DESI-MS ion images of normal kidney, RO, and RCC tissues. To better visualize the differences among the three tissue types for the six ions selected, the scale bars used are in unit of absolute intensity, and were amplified by a factor of 4 (0–500 absolute intensity) for m/z 391.260, m/z 656.574 and m/z 835.530 of all the samples.
Interestingly, four samples from RO disease contained both normal and tumor regions within the same tissue piece, as shown in the selected DESI-MS images in Fig. 4. Histopathological examination of the H&E stained tissue sections was carefully performed to mark the regions of normal and tumor in each tissue. Different distributions of the six selected ions can be observed in the normal and RO tissue regions, which clearly demonstrates the capability of DESI-MS imaging in separating normal and tumor regions within the same tissue section based on the distribution of molecular species. Three ions, including MG(18:1) at m/z 391.260, PS(34:1) at m/z 760.513, and PS(38:4) at m/z 810.524, showed higher abundance in the normal kidney tissue regions, which was in agreement with the area outlined by the pathologist. On the other hand, CL(20:2_18:2_18:2_18:2) at m/z 737.490 and PI (20:4_18:0) at m/z 885.550 presented an opposite pattern, with higher relative abundance in RO region, which was also in agreement with pathological evaluation. Shown in the middle of the Fig. 4, CL(18:2_18:2_18:2_18:2) at m/z 723.475 was distributed relatively homogeneously in normal and tumor regions, similar to what was observed in normal cortex and medulla tissues.
Figure 4.
Selected DESI-MS ion images from four RO tissues containing both normal and tumor regions. DESI-MS ion images are shown for m/z 391.260, m/z 760.514, m/z 810.524, m/z 723.475, m/z 737.490, and m/z 885.550. Pathologic annotations on the H&E stained optical images of the same tissue section analyzed by DESI-MS are also provided, allowing clear visualization of the marginal regions between normal and tumor margins.
Lasso Prediction of Normal, RO and RCC
We next used statistical analysis to explore the possibility of predicting normal, RO, and RCC diagnosis in kidney tissues, considering all the 73,992 pixels or mass spectra acquired from the three different tissue types. Lasso was employed to construct predictive models that are highly interpretable by performing both variable selection and regularization. Various data-preprocessing strategies were tested for statistical analysis (please see Supplementary Information for more details). The optimal results reported here were obtained using TIC normalization with log transformation in full mass range (m/z 100–1500). A total of 183 ions, which are important in characterizing the three classes, were selected by lasso and their corresponding weights for each class are shown in Table S2. From all the selected m/z values, 53 m/z values with positive lasso weights and 11 m/z values with negative lasso weights were chosen by the classifier as important features of ROs. For normal kidney prediction, 16 m/z values with positive lasso weights and 33 m/z values with negative lasso weights were selected, while 34 m/z values with positive lasso weights and 57 m/z values with negative lasso weights were selected to characterize RCCs. Fig. 5A shows a graphical representation of the model obtained including the weights given by the lasso for each of the selected mass spectral features (normal in green, RO in blue, and RCC in red), and the prediction results for the three groups. Remarkably, as shown in Fig. 5B, an overall accuracy of 99.47% was achieved per-pixel, and 100% accuracy was achieved per-patient for 71 samples analyzed, including 16 normal, 14 ROs, and 41 RCCs. Note that although the identification of the selected molecular features is not required for tissue diagnosis using DESI-MS/Lasso, evaluation of tandem MS data and high mass accuracy measurements in conjunction with online databases searches was meticulously performed to characterize ion species and provide insights of the metabolic dysfunctionality in kidney tumors. For example, based on characteristic fragmentation patterns of PE, PA and PS, the ions m/z 464.314, m/z 697.482, and m/z 842.589 were assigned as [PE(P-18:0)-H]−, [PA(18:1_18:2)-H]− and [PS(18:1_22:1)-H]−, respectively (Supplementary Fig. S15a–c).
Figure 5.
The Lasso method yields a model with parsimonious sets of features for discriminating between normal kidney, RO, and RCC tissues. A) The mathematical weights for each statistically informative feature selected by the lasso model are shown in red for RCC, blue for RO, and green for normal class, depending on the importance of the abundance of that peak in characterizing a certain class. Note that features that do not contribute to characterizing any of the class receive a weight of zero and are disregarded. B) Lasso per-pixel and per-patient prediction results are provided for normal kidney, RO, and RCC.
Discrimination of RO and chRCC using DESI-MS imaging
Differentiating ROs from chRCCs through histopathology is challenging as these tumor subtypes share several characteristics as both originate from the intercalated cells of the renal collecting duct, and have similar gross appearance in the cut-surface. Owing to the prominent component of cells with granular eosinophilic cytoplasm, chRCC sometimes resembles RO with overlapping histologic features. Mutations in mitochondrial genes have been reported in both RO and chRCC.(28,29) Because chRCC has metastatic potential, the distinction of these two renal tumor subtypes is particularly important.
To evaluate the ability to discriminate chRCC and RO using DESI-MS imaging data, we first performed unsupervised PCA was performed on the extracted data from the two classes, achieving complete separation between RO and chRCC samples (Fig. 6A). The ions m/z 1169.73 (assigned as lyso-CL(54:5)) and m/z 1128.27 (assigned as CL+PC(110:14)), which have the top two values in loadings 1 (Fig. 6B), showed significantly higher intensities in RO tissues compared with chRCC ones, while m/z 418.19 (unidentified) and m/z 201.11(assigned as [C10H18O4]−), which have the top 2 values in loadings 2, presented higher intensities in chRCC tissues. Next, lasso models were built to evaluate prediction performance of RO versus chRCC using DESI-MS data. Cross validation was performed on the training set of data comprising of 29093 pixels or mass spectra obtained from 12 chRCC and 14 RO tissues, yielding an overall per pixel accuracy of 99.64% (95% Confidence Interval, 95.72%−99.98%), RO recall of 99.66%, RCC recall of 99.60%, receiver operating characteristic area under curve (ROC AUC) of 1, and precision recall area under curve (PR AUC) of 1 (Table 1). Notably, 100% accuracy was achieved per patient. A total of 88 ions were selected by the lasso for the chRCC versus RO model (Table S3), 60 of which were given a negative weight for characterizing chRCC, and 28 were given a positive weight for characterizing chRCC. Among those, m/z 421.23, which was tentatively assigned as 2-methylacetophenone (mass error is −4.7 ppm, identified by matching Metaspace database) has the highest lasso weight for chRCC prediction. Several other species, such as glutathione at m/z 306.08 (Supplementary Fig. S16) and PS(16:0_18:2) at m/z 758.50, were also given positive weights for characterizing chRCC. On the other hand, all 14 CL related species (Table S4), such as ox-CL(72:9) at m/z 730.47 and CL+DG (108:9) at m/z 1034.75, and all eight PE related lipid species, such as LPE(18:0) at m/z 480.31 and PE(40:5) at m/z 792.56, were given positive weights to characterize RO tissues.
Figure 6.
PCA results of DESI-MS data acquired from RO and chRCC tissues. A) PCA separation of the two types using PC1 and PC2. B) Box and whisker plots are shown for the top 2 values of loadings 1 (m/z 1169.72 and m/z 1128.27) and the top 2 values of loadings 2 (m/z 418.19 and m/z 201.11), respectively.
Table 1.
Lasso per pixel and per patient prediction results for ROs and chRCCs (top), and lasso per-pixel and per-patient prediction results for RCC subtypes (bottom).
| Model | per pixel | per patient | |||||||
|---|---|---|---|---|---|---|---|---|---|
| RO | chRCC | recall | RO | chRCC | recall | ||||
| RO vs chRCC | RO | 16187 | 55 | 99.66% | RO | 14 | 0 | 100% | |
| chRCC | 38 | 9449 | 99.60% | chRCC | 0 | 12 | 100% | ||
| Overall Accuracy: 99.64% | Overall Accuracy: 100% | ||||||||
| ccRCC | chRCC | ppRCC | recall | ccRCC | chRCC | ppRCC | recall | |||
|---|---|---|---|---|---|---|---|---|---|---|
| RCC subtypes | ccRCC | 6130 | 76 | 816 | 87.30% | ccRCC | 10 | 0 | 1 | 91.91% |
| chRCC | 363 | 7324 | 1800 | 77.20% | chRCC | 0 | 9 | 3 | 75.00% | |
| ppRCC | 1505 | 156 | 18261 | 91.66% | ppRCC | 1 | 0 | 15 | 93.75% | |
| Overall Accuracy: 87.05% | Overall Accuracy: 87.18% | |||||||||
Lasso Prediction of RCC Subtypes
Precise subtyping of renal tumors is critical to assure appropriate treatment options for patients. Due to the recent success of new treatment options for advanced and metastatic disease, renal cancer subtypes management has been imbued with new interest.(30) Many studies focused on diagnosing RCC subtypes have been reported using staining methods such as histological evaluation, immunohistochemistry (IHC), and gene sequencing.(31) Recent developments in IHC of percutaneous biopsy analysis, has gained interest, and this minimally invasive method has accuracy of 70%−90%.(32)
We further explored the capability of DESI-MS and lasso to predict RCC subtypes, including ccRCC, chRCC, and ppRCC, which are the three most common histological subtypes and account for 90% of all renal cancers. As shown in Table 1, overall accuracies of 87.05 % per-pixel and 87.18% per-patient were achieved for the analysis of 11 ccRCC, 12 chRCC, and 16 ppRCC samples. Five out of 39 renal cancer tissues were classified incorrectly, which is comparable with the accuracies of IHC methods. A total of 35 m/z values were selected as important features for discriminating between the three subtypes (Table S5). Among the features selected, five CL related species (m/z 711.98, m/z 712.48 (Fig. S17), m/z 713.49, m/z 713.99, m/z 714.51) have positive lasso weights in chRCC samples.
Discussion
In this study, we used DESI-MS imaging to investigate the molecular profiles of normal and tumor kidney tissues, and build classification models that are predicate of disease state. First, we demonstrated the capability of DESI-MS imaging in discriminating distinct histologic regions of normal kidney, the cortex and medulla, based on the metabolic and lipid information acquired (please see Supporting Information and Fig. S18 for additional discussion). Following on the successful results in discriminating histologic regions of kidney tissues, we then studied two tumor types: RO and RCC. RO are predominantly benign neoplasms with dramatic accumulation of defective and mutated mitochondria, even though the biological processes leading to mitochondria accumulation and dysregulation are not fully known in any oncocytic tumor.(33) By comparing the mass spectra from RO and RCC, significantly higher abundances of CL species were detected in RO, including CL related lipid dimers in the higher m/z range of the mass spectra. Further, comparison between selected DESI images obtained for the three tissue types was performed, revealing consistent and distinct molecular patterns in each tissue type. Interestingly, although normal kidney tissue generally present high relative abundance of CLs in the DESI mass spectra, specific CL species such as CL(20:2_18:2_18:2_18:2) at m/z 737.490 were observed in high abundances in RO tissues, likely associated to the defective mitochondria processes known to occur in oncocytic tumors, as we previously discussed.(26)
Statistical analysis using lasso was performed to predict normal kidney, RO and RCC tissues using cross-validation. Excellent overall accuracy per-pixel (99.47%) and per-patient (100%) was obtained, for all the 73,992 pixels evaluated. Interestingly, within the 41 RCC samples, 12 tissues were of chRCC subtype, which are histologically similar to RO considering the abundant, eosinophilic cytoplasm and densely packed mitochondria present in both tissues. Nevertheless, the lasso models built allowed discrimination of the benign RO and malignant RCC class with minimal confusion. The majority of the ions selected by the lasso as statistically significant were tentatively assigned as biologically relevant metabolites and lipids. Within the features selected by lasso, glutathione (at m/z 306.08) was given the lowest lasso weight (−0.17) for predicting normal kidney, 0 lasso weight for predicting RO, and the third highest lasso weight (0.076) for predicting RCC. Interestingly, Li et al. previously reported that RCC tumors display significant accumulation of reduced GSH, with about 150 fold-of-change of tumor/normal.(34) Moreover, when evaluating all the m/z selected by lasso (Table S1), the top 10 positive lasso weights in the selected features for RO were characterized as five PE related species (LPE(18:0), PE(40:5), PE(40:5), PE(40:4) and PE(40:4)), two CL related species (CL+PC(108:9), and CL+PC(108:8)), two FAs (FFA(20:0) and FFA(22:1)), and one ceramide lipid (Cer (d38:1)). These results indicate that PE lipids may be implicated in the dysregulated metabolism of RO. Also, CLs were the most frequently selected lipid class by lasso, with positive/zero lasso weight for RO and negative/zero weight for both normal and RCC. In total, 134 different molecuels were identified including 16 FAs, 5 DGs, 30 CLs, 4 Cers, 6 MGs, 13 PEs, 7 PGs, 7 PIs, 11 PSs, 6 PCs, 4 SHexCers, and 34 other small metabolites.
Previous studies have reported that both RO and chRCC both have high levels of expression of mitochondrial and oxidative phosphorylation genes.(35) We performed a direct comparison of the two tumor types which have with similar cytological features. From a total of 26 samples, PCA analysis revealed clear separation between RO and chRCC tissues (Fig. 6). Interestingly, when grouping RO and chRCC together to compare the two tumor types to the other renal tumor subtypes (ccRCC and ppRCC), clear separation could also be obtained with PCA (Supplementary Fig. S19a,b and Table S6). Careful evaluation of the PCA results (Supplementary Fig. S19a,b) revealed that although the chRCC datapoints were more dispersed in the PCA plot than the other three tumor types, they were much closer to the RO samples than to the ccRCCs and ppRCCs. These results suggest that RO and chRCC have distinct molecular patterns between each other, however, these two renal tumor subtypes also carry similarities when compared to other tumor tissues.
Finally, we investigated the performance of DESI-MS imaging in discriminating the three RCC subtypes. Overall ~87% accuracy was achieved for the 39 samples (total of 36,431 pixels), which is promising. Note that ppRCC tissues, which is thought to arise from the epithelium of the proximal tubule, were classified with the highest prediction accuracy compared with the other two subtypes.(32,36) Interestingly, glutathione (m/z 306.08), which was selected as a significant metabolite for differentiating normal and benign tissues from malignant tissues, was not selected in the statistical model for RCC subtypes.
In summary, in this study we evaluated the potential usefulness of DESI-MS imaging combined with lasso statistical analysis to distinguish normal and various subtypes of kidney tumors. From a diagnostic perspective, two major tumor types, RO and RCC, were investigated and clear differences in the mass spectra profiles between the two groups were observed, allowing lasso classification with excellent accuracy. Remarkably, two renal tumor subtypes that are indistinguishable by histopathologic analysis, RO and chRCC, were studied and the high prediction accuracies were also obtained. Lastly, DESI-MS allowed discrimination of RCC subtypes, including ccRCC, chRCC and ppRCC, with an overall accuracy of 87.18% for the 39 samples. While the results obtained provide evidence of the diagnostic potential of DESI-MS imaging, further studies are planned to validate the results described here with a larger sample set. Studies using core biopsy tissues will also be pursued to show potential clinical value in guiding treatment decisions for patients. Altogether, the results presented in our study strongly suggest that DESI-MS can be potentially used as a clinical approach to diagnose and subtype renal tumors and improve management for cancer patients.
Supplementary Material
Statement of Significance:
Metabolic data acquired by mass spectrometry imaging in conjunction with statistical modeling allows discrimination of renal tumors, and has the potential to be used in the clinic to improve treatment of patients.
Acknowledgments
This work was supported by the NIH/NCI (grant 4R00CA190783-02) and the Alfred P. Sloan Foundation (FG-2018-10608). Tissue samples were provided by the Cooperative Human Tissue Network which is funded by the NCI.
Footnotes
Disclosure of Potential Conflicts of Interest
Livia S. Eberlin and Jialing Zhang have ownership in MS Pen Technologies, Inc. in which Livia S. Eberlin serves as an unpaid member of the board of directors and Jialing Zhang serves as an unpaid consultant.
References
- 1.Ljungberg B, Campbell SC, Choi HY, Jacqmin D, Lee JE, Weikert S, et al. The epidemiology of renal cell carcinoma. Eur Urol 2011;60:615–21 [DOI] [PubMed] [Google Scholar]
- 2.Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J, et al. Epidemiology of Renal Cell Carcinoma. European Urology 2019;75:74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ridge CA, Pua BB, Madoff DC. Epidemiology and Staging of Renal Cell Carcinoma. Semin Intervent Rad 2014;31:3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McArthur C, Baxter GM. Current and potential renal applications of contrast-enhanced ultrasound. Clin Radiol 2012;67:909–22 [DOI] [PubMed] [Google Scholar]
- 5.Morrell GR, Zhang JL, Lee VS. Magnetic Resonance Imaging of the Fibrotic Kidney. J Am Soc Nephrol 2017;28:2565–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishigami K, Jones AR, Dahmoush L, Leite LV, Pakalniskis MG, Barloon TJ. Imaging spectrum of renal oncocytomas: a pictorial review with pathologic correlation. Insights Imaging 2015;6:53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith CJ, Wang MX, Feely M, Otto B, Grajo JR. Oncocytoma: A Differential Consideration for an Incidentally Detected FDG-Avid Renal Mass on PET/CT. J Radiol Case Rep 2017;11:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yusenko MV. Molecular pathology of renal oncocytoma: a review. Int J Urol 2010;17:602–12 [DOI] [PubMed] [Google Scholar]
- 9.Wobker SE, Williamson SR. Modern Pathologic Diagnosis of Renal Oncocytoma. J Kidney Cancer Vhl 2017;4:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng KL, Rajandram R, Morais C, Yap NY, Samaratunga H, Gobe GC, et al. Differentiation of oncocytoma from chromophobe renal cell carcinoma (RCC): can novel molecular biomarkers help solve an old problem? Journal of Clinical Pathology 2014;67:97–104 [DOI] [PubMed] [Google Scholar]
- 11.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004;306:471–3 [DOI] [PubMed] [Google Scholar]
- 12.Ifa DR, Eberlin LS. Ambient Ionization Mass Spectrometry for Cancer Diagnosis and Surgical Margin Evaluation. Clinical Chemistry 2016;62:111–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandal MK, Saha S, Yoshimura K, Shida Y, Takeda S, Nonami H, et al. Biomolecular analysis and cancer diagnostics by negative mode probe electrospray ionization. Analyst 2013;138:1682–8 [DOI] [PubMed] [Google Scholar]
- 14.Pirro V, Llor RS, Jarmusch AK, Alfaro CM, Cohen-Gadol AA, Hattab EM, et al. Analysis of human gliomas by swab touch spray-mass spectrometry: applications to intraoperative assessment of surgical margins and presence of oncometabolites. Analyst 2017;142:4058–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balog J, Sasi-Szabo L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, et al. Intraoperative Tissue Identification Using Rapid Evaporative Ionization Mass Spectrometry. Sci Transl Med 2013;5 [DOI] [PubMed] [Google Scholar]
- 16.Woolman M, Kuzan-Fischer CM, Ferry I, Kiyota T, Luu B, Wu MG, et al. Picosecond Infrared Laser Desorption Mass Spectrometry Identifies Medulloblastoma Subgroups on Intrasurgical Timescales. Cancer Res 2019;79:2426–34 [DOI] [PubMed] [Google Scholar]
- 17.Fatou B, Saudemont P, Leblanc E, Vinatier D, Mesdag V, Wisztorski M, et al. In vivo Real-Time Mass Spectrometry for Guided Surgery Application. Scientific Reports 2016;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang JL, Rector J, Lin JQ, Young JH, Sans M, Katta N, et al. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci Transl Med 2017;9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dill AL, Eberlin LS, Zheng C, Costa AB, Ifa DR, Cheng LA, et al. Multivariate statistical differentiation of renal cell carcinomas based on lipidomic analysis by ambient ionization imaging mass spectrometry. Anal Bioanal Chem 2010;398:2969–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfaro CM, Jarmusch AK, Pirro V, Kerian KS, Masterson TA, Cheng L, et al. Ambient ionization mass spectrometric analysis of human surgical specimens to distinguish renal cell carcinoma from healthy renal tissue. Anal Bioanal Chem 2016;408:5407–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Horikawa M, Sato S, Miyake H, Setou M. Discovery of lipid biomarkers correlated with disease progression in clear cell renal cell carcinoma using desorption electrospray ionization imaging mass spectrometry. Oncotarget 2019;10:1688–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cheng L, Cooks RG. Non-Destructive, Histologically Compatible Tissue Imaging by Desorption Electrospray Ionization Mass Spectrometry. ChemBioChem 2011;12:2129–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robichaud G, Garrard KP, Barry JA, Muddiman DC. MSiReader: An Open-Source Interface to View and Analyze High Resolving Power MS Imaging Files on Matlab Platform. Journal of the American Society for Mass Spectrometry 2013;24:718–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong J, Soufan O, Li C, Caraus I, Li SZ, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 2018;46:W486–W94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Druilhet RE, Overturf ML, Kirkendall WM. Cortical and medullary lipids of normal and nephrosclerotic human kidney. Int J Biochem 1978;9:729–34 [DOI] [PubMed] [Google Scholar]
- 26.Zhang JL, Yu WD, Ryu SW, Lin J, Buentello G, Tibshirani R, et al. Cardiolipins Are Biomarkers of Mitochondria-Rich Thyroid Oncocytic Tumors. Cancer Res 2016;76:6588–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren A, Cai F, Shang YN, Ma ES, Huang ZG, Wang W, et al. Differentiation of Renal Oncocytoma and Renal Clear Cell Carcinoma Using Relative CT Enhancement Ratio. Chinese Medical Journal 2015;128:175–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi S, Tolkunov D, Aviv H, Hakimi AA, Yao M, Hsieh JJ, et al. The Genomic Landscape of Renal Oncocytoma Identifies a Metabolic Barrier to Tumorigenesis. Cell Reports 2015;13:1895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis CF, Ricketts CJ, Wang M, Yang LX, Cherniack AD, Shen H, et al. The Somatic Genomic Landscape of Chromophobe Renal Cell Carcinoma. Cancer Cell 2014;26:319–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutcher JP. Recent developments in the treatment of renal cell carcinoma. Ther Adv Urol 2013;5:338–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F, Zhang Y, Senbabaoglu Y, Ciriello G, Yang L, Reznik E, et al. Multilevel Genomics-Based Taxonomy of Renal Cell Carcinoma. Cell Rep 2016;14:2476–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muglia VF, Prando A. Renal cell carcinoma: histological classification and correlation with imaging findings. Radiol Bras 2015;48:166–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasparre G, Romeo G, Rugolo M, Porcelli AM. Learning from oncocytic tumors: Why choose inefficient mitochondria? Bba-Bioenergetics 2011;1807:633–42 [DOI] [PubMed] [Google Scholar]
- 34.Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature 2014;513:251–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klomp JA, Petillo D, Niemi NM, Dykema KJ, Chen J, Yang XJ, et al. Birt-Hogg-Dube renal tumors are genetically distinct from other renal neoplasias and are associated with up-regulation of mitochondrial gene expression. BMC Med Genomics 2010;3:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cairns P Renal cell carcinoma. Cancer Biomark 2010;9:461–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.