Abstract
The Hsp70 family of heat shock proteins plays a critical function in maintaining cellular homeostasis within various subcellular compartments. The human mitochondrial Hsp70 (HSPA9) has been associated with cellular death, senescence, cancer and neurodegenerative diseases, which is the rational for the name mortalin. It is well documented that mortalin, such as other Hsp70s, is prone to self-aggregation, which is related to mitochondria biogenesis failure. Here, we investigated the assembly, structure and function of thermic aggregates/oligomers of recombinant human mortalin and Hsp70–1A (HSPA1A). Summarily, both Hsp70 thermic aggregates have characteristics of supramolecular assemblies. They display characteristic organized structures and partial ATPase activity, despite their nanometric size. Indeed, we observed that the interaction of these aggregates/oligomers with liposomes is similar to monomeric Hsp70s and, finally, they were non-toxic over neuroblastoma cells. These findings reveled that high molecular mass oligomers of mortalin and Hsp70–1A preserved some of the fundamental functions of these proteins.
Keywords: Hsp70, mortalin, protein interaction, aggregation, oligomerization
Introduction
The Hsp70 family is one of the most conserved group of proteins between prokaryotes and eukaryotes [1–6]. These proteins are ubiquitous and can be found in almost all cellular compartments, exhibiting a plethora of functions that ensure the preservation of proteostasis and protein quality control [3,6,7]. They can act at different levels, including nascent protein folding, transport, signaling, assembly/disassembly of protein complexes, degradation, among others [4–6,8–13].
Hsp70s are modular and flexible proteins with canonical structure formed by two domains: a N-terminal domain containing an adenosine nucleotide hydrolysis activity (NBD – nucleotide binding domain), and a C-terminal domain characterized by binding client proteins (PBD – protein binding domain), both connected by a conserved short hydrophobic inter-domain linker[3,5,14–17]. These domains work as a bidirectional heterotrophic allosteric system[3]. Moreover, Hsp70 functional cycle is assisted by a myriad of co-chaperones such as J-proteins and NEFs [3,5,13,17,18].
There are several humans Hsp70 isoforms [13,19], among of them the mitochondrial Hsp70 (mtHsp70) is also named mortalin (HSPA9) due to its role in cell death in mammals [20,21]. Studies in the literature shown that mtHsp70 is critical for mitochondria homeostasis [22–24]. It acts in the folding and importing of proteins from the cytosol into the mitochondrial matrix, being the only member of the translocate motor of the complex PAM (pre-sequence translocase-associated motor) with ATPase activity [25,26]. Mortalin is also found in almost all cellular compartments, working in folding, transport of proteins; protection against oxidative stress, among others [20,27–29]. It interacts and arrests p53 in the cytosol blocking the p53 suppressor effects over oncogenes [24,30]. Besides, it has been reported that mortalin is an important biological target, once it is involved in several types of cancers and age diseases such as Parkinson’s and Alzheimer’s [30,31]. It has been shown that its expression is elevated in many human tumors in cell and in vitro studies [27,30–32]. Moreover, overexpression of mortalin was sufficient to increase breast cancer, suggesting that the increase in its expression plays an important role to the formation and maintenance of tumors [20,27].
It has been well-shown that Hsp70s are prone to undergo self-oligomerization or self-aggregation not only in vitro but also in vivo [33–38]. Here, we have unveiled the formation of aggregates or supramolecular assemblies obtained from human recombinant mortalin (HSPA9) and Hsp70–1A (HSPA1A) monomers after a thermal treatment. Our findings indicate that recombinant human mortalin and Hsp70–1A aggregation processes depend on the temperature and concentration. We also show that mortalin and Hsp70–1A aggregates were structured, have ATPase activity, interact with negatively charged liposomes, and exert no toxicity over mammalian cells, indicating functionality. The relevance of such Hsp70 supramolecular assemblies is discussed.
MATERIAL AND METHODS
Protein expression and purification
The recombinant human mortalin was produced by co-expressing it with hHep1 in the Escherichia coli BL21(DE3) strain as previously described [39,40]. Recombinant human cytoplasmic Hsp70–1A was produced as reported in [41]. All proteins were prepared in TKP buffer (25 mmol L−1 Tris–HCl (pH 7.5), 50 mmol L−1 NaCl, 5 mmol L−1 sodium phosphate, 5 mmol L−1 KCl, 2 mmol L−1 MgCl2 and 2 mmol L−1 β-mercaptoethanol). Protein concentrations were determined applying the Eldelhock method as previously described [39,41] and protein purity was attested by SDS-PAGE.
Mortalin and Hsp70–1A aggregation kinetics
The thermal aggregation propensity of recombinant human mortalin and Hsp70–1A were analyzed at different temperatures and protein concentration (from 2 up to 10 μmol L−1). The assays were performed using a 1 cm quartz cuvette cell, for 80 min and monitored by light scattering at 340 nm using a UV-Vis Spectrophotometer UV-2600 containing a Cell Positioner CPS-240A (Shimadzu). In addition, assuming a first-order kinetic, it was possible to adjust the aggregation curves at the various temperatures applied for exponential curves and, thereby, calculate an experimental aggregation constant (kobs) and limit absorbance (ALim) [42].
Protein aggregates preparation and isolation
In order to produce and isolate the mortalin and Hsp70–1A thermal aggregates, recombinant protein aliquots were subjected to 43 °C for 2 hours. Then, the aggregated proteins were subjected to preparative size exclusion chromatography in TKP buffer at room temperature and the eluated at the column void were collected. After, the protein was concentrated and used in the experiments.
Hydrodynamic characterization
The monomeric and aggregated samples, after being subjected at different temperatures for 80 min, were analyzed by analytical size exclusion chromatography (aSEC), as previously described [39].
Dynamic light scattering (DLS) experiments were performed in a Zetasizer Nano ZS (Malvern). The samples containing monomeric mortalin or Hsp70–1A at 4 μmol L−1 were prepared in TKP buffer and placed in the thermostatic sample chamber. Both were submitted to a heat gradient, from 15 up to 70 °C, with reading gaps of 5 °C and 2 min for stabilizing time between readings. The data were then threated evaluating the Stokes radius (Rs) from the scattering particles in the course of the temperature increasing. These tests were performed in triplicate.
Transmission electron microscopy analysis using negatively stained samples
For samples visualization by negative stain transmission electron microscopy (TEM), mortalin and Hsp70–1A samples (~0.5 mg mL−1), in monomeric or in aggregates forms, were prepared in TKP buffer. The samples were deposited (~ 3 μL) onto the holey carbon-coated grids (Ultrathin Carbon film 400 mesh, Agar Scientific), at neutral charge, for 30 s followed by two washing steps with 10 mmol L−1 HEPES buffer (pH 7.5). After, the grids were stained with 3 μL of 2% uranyl acetate for 30 s, then submitted to blotting and air-drying for 5 min. Images were recorded at −3.5 μm defocus using 120 kx magnification at JEOL 2100 LaB6 microscope operating at 200 kV with TVIPS 16 Mpi camera (IQSC/USP).
The acquired images were processed using ImageJ [43], resulting the Gaussian blur dataset (cut-off < 10) after the images filtering. The particles were measured, and the size statistical analysis were performed using Origin 2015 software [44].
Spectroscopy studies
Circular dichroism (CD) measurements were performed with a J-815 spectropolarimeter (Jasco Inc.) coupled to the Peltier system PFD 425S for temperature control. Recombinant proteins (recombinant human mortalin/Hsp70–1A and their thermic aggregates) were tested in TKP buffer at final concentrations between 5 and 10 μmol L−1 in a 0.2 mm circular path-length cuvette. The presence of adenosine nucleotides (200 μmol L−1) were also tested. The thermal-induced unfolding for mortalin, Hsp70–1A or their thermic aggregates (5–10 μmol L−1) was evaluated following the CD signal at 222 nm in the temperature range of 15 up to 90 °C using a 1.0 nm path-length cuvette. All data were normalized to the mean residue ellipticity ([Θ]). The temperature at the midpoint of the unfolding transition (Tm) were estimated by sigmoidal fitting of the unfolding transition.
Intrinsic fluorescence emission measurements were performed in an F-4500 fluorescence spectrophotometer (Hitachi), using a 10.0 mm × 2.0 mm path length quartz cuvette with mortalin, Hsp70–1A and their thermic aggregates solved in TKP buffer (5–10 μmol L−1), at 25 °C. The fluorescence emission spectra were measured from 315 to 420 nm, after excitation at 295 nm, for proteins and buffers. After buffer spectra subtraction, the maximum fluorescence emission wavelength (λmax) and spectral center of mass (<λ>) were calculated [39,41].
The Thioflavin-T (Th-T) assay was performed in the same equipment, with mortalin, Hsp70–1A and their thermic aggregates solved in TKP buffer at 5 μmol L−1, with addition of 25 μmol L−1 of Th-T. SEPT6G was used as positive control, since it is known to form amyloid fibrils [45], in the same conditions of Hsp70 proteins. Measurements were performed exposing the samples at temperatures of 20 °C and 40 °C for 120 min. The fluorescence emission spectra were measured from 468 to 600 nm, after excitation at 450 nm [46–48].
ANS (8-Anilino-1naphthalene-sulphonic acid) fluorescence assay was performed on a Varioskan™ Lux Microplate Reader (Thermofisher), using a microlon microplate of 96 well (Greiner). Mortalin, Hsp70–1A and their aggregates at 5 μmol L−1 in the presence of 30 μmol L−1 of ANS, solved in TKP buffer, were tested in a time dependent manner. Experiments were conducted at 20 °C, measuring fluorescence emission from 400 to 600 nm after excitation at 350 nm [46,47].
Differential scanning calorimetry
Differential scanning calorimetry (DSC) measurements of thermal-induced unfolding were performed in a Nano DSC (TA Instruments). We performed measurements of recombinant human mortalin/Hsp70–1A and their thermic aggregates at 1 mg mL−1. The presence of nucleotides (200 μmol L−1) and magnesium chloride (1 mmol L−1) were also tested. The proteins were prepared in TKP buffer after extensive dialysis without the presence of β-mercaptoethanol. The scan rate tested was of 1.0 °C min−1 at the 15 up to 90 °C temperature range. The experimental thermograms were collected with DSC run software [49] (TA Instruments) and analyzed using the NanoAnalyze software [50] (TA Instruments) in order to estimate the Tm as the peak of the unfolding transition. The baselines were calculated from the pre- and post-transition temperature regions.
Liposome preparation and Isothermal titration calorimetry
Liposomes were prepared using the extrusion method [51]. Palmitoyl-oleoyl phosphoserine (POPS) and Cardiolipin (CL) (Avanti Polar Lipids) were prepared in CHCl3 at 10 mg Ml−1 and dried in N2. After, dry lipid film was re-suspending in 50 mmol L−1 of Tris-HCl (pH 7.4) and vortexed 6 times in 30 min. Finally, the resuspended material was extruded using a 100 nm membrane filter.
In other to assess the interaction of mortalin and Hsp70–1A thermal aggregates with POPS and CL liposomes it was used isothermal titration calorimetry (ITC), as previously described thermal aggregated proteins samples were dialyzed in 50 mmol L−1 of Tris-HCl (pH 7.4) over night. Summarily, the titrations were performed at 25 °C in an iTC200 microcalorimeter (GE Healthcare Life Sciences), in which seventeen 2-μL aliquots of POPS or CL at 3 mmol L−1 were injected into 203.8 μL of 10–15 μmol L−1 protein thermal aggregates. All solutions were prepared in 50 mmol L−1 Tris-HCl (pH 7.4) buffer. The experimental isotherms curves were analyzed to obtained the association constant (KA) (the dissociation constant (KD) is 1/KA), apparent enthalpy change (ΔHapp) and stoichiometric coefficient (n), as previously described [39,52,53]. By the relation ΔGapp = −RTlnKA, were R is the gas constant and T the absolute temperature, we calculated the apparent Gibbs energy change (ΔGapp). The apparent entropy change (ΔSapp) was determined by the equation:.
ATPase activity
In order to access the ATPase activity of proteins the PiColorLock™ kit (Innova Biosciences) was used. Summarily, it is a spectroscopic method that allows the quantification of the inorganic phosphate (Pi) released from ATP hydrolysis by the enzyme. Mortalin, Hsp70–1A and their thermal aggregates (~2 μmol L−1, respectively) were prepared in TKP buffer (without phosphate ions) and incubated with ATP (0 to 2 mmol L−1) for 40 min at 37 °C. The negative control was prepared without the recombinant protein. After, the samples containing the Pi hydrolyzed from ATP were incubated with malachite dye-solutions. Due to generation of phosphomolybdate complexes, it was possible to measure the quantity of Pi released by the reaction by absorbance at 620 nm. The amount of Pi released per min (i.e., V0 in μmol L−1 min−1) was plotted against the ATP concentration, and a Michaelis-Menten fitting routine was used (Origin software [44]) to obtain the kinetic parameters KM (Michaelis constant) and Vmax (maximum velocity). The kcat was calculated by the ratio of Vmax to the protein concentration.
Cell viability by flow cytometry
N2A neuroblastoma cells were incubated or not with mortalin, Hsp70 or their thermal aggregates at 10 μmol L−1 at 37 °C for 48 h in DMEM medium containing 1% fetal bovine serum (1% FBS). Cells were then harvested and stained with APC-conjugated annexin V and propidium iodide (PI). Flow cytometry was performed using a FACSCanto II flow cytometer with FACSDiVa [54] software (BD Biosciences). The data were analyzed using FlowJo software [55]v.10.1 (Tree Star).
RESULTS AND DISCUSSION
Mortalin and Hsp70–1A form aggregates depending on concentration and temperature
Previous fluorescence studies showed that mortalin displayed two transition phases with the raise of temperature: a blue and then a red shift [39]. The blue shift is likely associated with an alteration in the microenvironments around a single tryptophan present in NBD of the human mortalin that became less exposed to solvent. In contrast, Hsp70–1A only displayed red-shift transitions in response to the increase in temperature indicating a higher exposure of two tryptophan residues to the solvent [39].
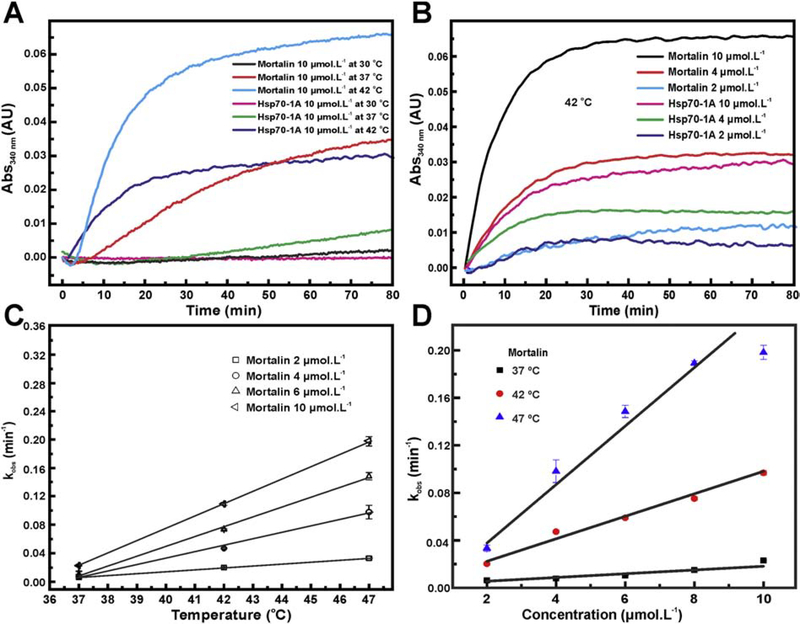
To further investigate the aggregation process of mortalin and Hsp70–1A, we followed the thermal aggregation of these two proteins (10 μmol L−1) at 30, 37 and 42 °C by light scattering at 340 nm. No aggregation was detected for mortalin at 30 °C whereas the aggregation process was rapidly activated after incubation at 37 or 42 °C (Figure 1a). The thermal aggregation was also increased with increments in the mortalin concentration (Figure 1b). In contrast, Hsp70–1A only showed some relevant level of aggregation at 42 °C (Figure 1a), which was also concentration dependent (Figure 1b).
Figure 1: Mortalin and Hsp70–1A aggregation profile is dependent on protein concentration and temperature.
a) Mortalin showed a pronounced aggregation propensity with increasing temperature. The same was observed for Hsp70–1A. However, mortalin showed aggregation even at temperatures at 37 °C. b) The aggregation process is dependent on the protein concentrations. The process is again more evident for mortalin. c) and d) Assuming an exponential fit for mortalin aggregation curves in the concentrations (2, 4, 6 and 10 μmol L−1) and temperatures (37, 42 and 47 °C) tested, it is possible to calculate the kobs (see text for details). Both values follow a linear dependence on the temperature and concentration.
The aggregation profile of Hsp70–1A also showed that this protein is more stable at high temperature as compared with mortalin. While mortalin aggregate substantially at 37 °C, the level of Hsp70–1A aggregation was very reduced at such temperature (Figure 1a). At 42 °C, mortalin aggregation yielded twice scattering signal when compared to Hsp70–1A (Figure 1b). These data indicated that the thermal aggregation processes for mortalin and Hsp70–1A are indeed different [39], even that they are similar proteins.
Next we calculated the kobs-values to all the mortalin aggregation curves as a function of temperature (Figure 1c) or protein concentration (Figure 1d) [42]. The experimental values for kobs, which represent the rates of mortalin aggregation, followed a linear pattern (Figure 1c and 1d) indicating that mortalin thermal aggregation is mainly dependent on both temperature and protein concentration (Figure 1d). Similar results were observed for Hsp70–1A (data not shown).
Human mortalin and Hsp70–1A thermal aggregates have structural elements and behave as a polydisperse system
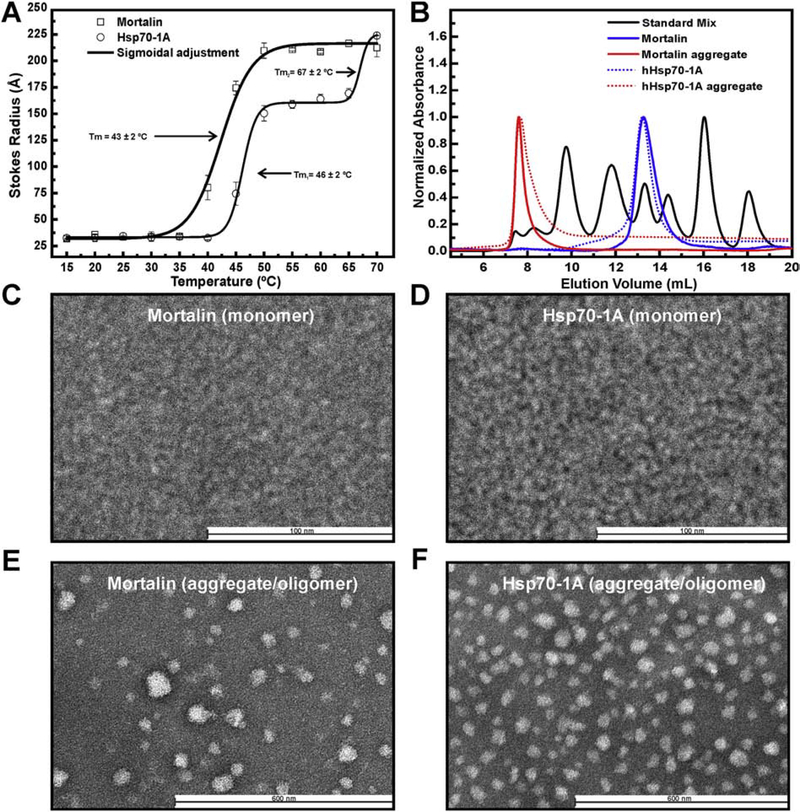
Despite the tendency of Hsp70s to form oligomers/aggregates both in vitro and in vivo [33–36,38], there is not information on the structure, function and assembly mechanisms for these oligomers/aggregates. In order to understand how temperature influences the formation of mortalin and Hsp70–1A oligomers/aggregates, DLS assays were performed. The mechanism of mortalin aggregation is slightly different from the one of Hsp70–1A (Figure 2a). Indeed, mortalin initiates the aggregation process at around 35 °C with a single Tm 43 ± 2 °C. In addition, we observed the appearance of stable species with a Stokes radius (Rs) around 220 Å between 50 and 70 °C. In other hands, the Hsp70–1A thermal aggregation process presented at least two transitions, the first centered at 46 ± 2 °C that leaded to the formation of relatively stable particles (Rs of about 160 Å between 50 and 60 °C) and a second transition centered at 67 ± 2 °C with the formation of particles with Rs of approximately 220 Å at 70 °C. These data are in accordance with our previously published results obtained by fluorescence analysis [39]. Although the process of aggregation appears to be different between mortalin and Hsp70–1A, the final thermal aggregates observed at 70 °C seem to be of similar weight-average size.
Figure 2: Mortalin and Hsp70–1A aggregates behave as a polydisperse systems.
a) Mortalin and Hsp70–1A were submitted to DLS analysis. Thermal induced aggregation for mortalin led to the formation of stable larger aggregates in a wide temperature range and seemed to be different as compared to Hsp70–1A. b) Mortalin, Hsp70–1A and their thermal aggregates were submitted to an aSEC experiments. The results indicate that mortalin and Hsp70–1A thermal aggregates are stable and eluates in the column void. (c-f) Negative stain TEM images. Results obtained for monomeric mortalin (c), Hsp70–1A (d), mortalin aggregates (e) and Hsp70–1A aggregates (f), respectively. Mortalin and Hsp70–1A thermal aggregates are composed by large and heterogeneous particles.
The size of mortalin and Hsp70–1A before and after thermal aggregation were further evaluated by aSEC (Figure 2b). At 25 °C, mortalin and Hsp70–1A behave as monomeric species (blue curves) as previously shown [39,41,56]. After thermal aggregation at 42 °C, aSEC analysis indicated that the molecular mass of mortalin and Hsp70–1A aggregates exceeds 600 kDa (red curves), corresponding to the exclusion limit of our aSEC column. These results also implied that, once the thermal aggregates were formed, they remained stable 1n solution. Furthermore, analytical ultracentrifugation data revealed that mortalin aggregates form a polydisperse system, with at least three different high molecular mass species (2 MDa, 13 MDa and 26 MDa; data not shown). Interestingly, Steel at al. [38] observed the in vivo formation of polymeric structures of Hsp70–1A and Hsc70 which suggest that the formation of these thermal aggregates with high molecular mass would be an intrinsic process for Hsp70s.
To further characterize mortalin and Hsp70–1A aggregates, we used negatively-stained TEM. Mortalin and Hsp70–1A in their monomeric state showed the presence of particles with homogenous dimensions (diameter about 10 nm in average) with high affinity to the carbon foil (Figures 2c and 2d). This dimension is compatible with the size of Hsp70 monomers observed by small angle X-ray scattering analysis in similar buffer conditions [39,52]. Images obtained for mortalin or Hsp70–1A thermal aggregates clearly revealed the presence of very large particles (diameter of ~50 nm in average) presenting high polydispersity (higher for mortalin aggregates when compared to Hsp70–1A aggregates) and with no morphological pattern (Figures 2e and 2f). These data are in agreement with the results obtained by DLS (Figure 2a), aSEC (Figure 2b) and analytical ultracentrifugation (data not shown).
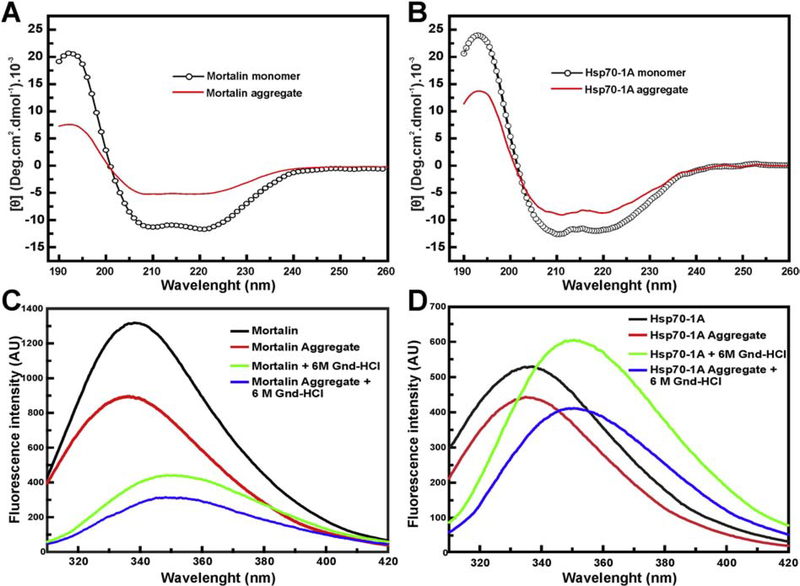
To study the secondary and tertiary structure of mortalin and Hsp70–1A aggregates, we used CD and the intrinsic fluorescence of tryptophan. The CD analysis reveals that mortalin (Figure 3a) and Hsp70–1A (Figure 3b) aggregates showed secondary structure with about half of the [Θ] found in the respective monomeric protein. In addition, (Figure 3c) showed that the aggregation of mortalin results in a blue shift event indicating that the single tryptophan of mortalin become less exposed to the solvent, as previously reported [39]. The monomeric mortalin presented a λmax and a <λ> of about 338 nm and 346.1 nm, respectively (Table 1). These values were significantly modified after thermal aggregation to 336 nm and 345.6 nm for λmax and <λ>, respectively. These results suggest that the NBD of mortalin may be involved in the aggregation process. For Hsp70–1A, a red shift in both parameters was observed due to thermal aggregation (Table 1). The presence of guanidinium hydrochloride induced chemical unfolding for both mortalin and Hsp70–1A (monomers and thermal aggregates) indicating that these two proteins do not behave like amyloid-like structures (Figure 3 and Table 1). Besides, we also investigated if the Hsp70 aggregates interact with Th-T, used as a probe for β-amyloid structures. The results presented in Figure S1 indicated that both Hsp70 aggregates interacted little with such probe independently of the temperature and time, mainly in comparison with SEPT6G that was used as a positive control. Altogether, these spectroscopic data indicated that the thermal aggregate preparations for mortalin and Hsp70–1A have secondary and tertiary structural contents. Based on the structural information obtained for mortalin and Hsp70–1A aggregates, we hypothesize that they may constitute supramolecular assemblies rather than amorphous aggregates.
Figure 3: Hsp70 thermic aggregates retain secondary and local tertiary structures.
CD experiments of Mortalin, Hsp70–1A and their aggregates. All results were normalized to the [Θ]. a) and b) CD data indicate that the mortalin and Hsp70–1A aggregates, respectively, produced at 43 °C, retain secondary structure content (see text for more details). Intrinsic Fluorescence emission of tryptophan measurements were also performed. c) and d) Intrinsic fluorescence emission results suggest that mortalin and Hsp70–1A aggregates possess local tertiary structure and their chemical denaturation led them to unfolding (see text for more details).
Table 1:
Mortalin, Hsp70–1A and their thermal aggregates spectroscopic parameters.
| Sample | CD [Θ] (Deg.cm2.dmol−1).10−3 | Emission Fluorescence€ | ||
|---|---|---|---|---|
| [Θ]208 nm | [Θ]222 nm | λmax (nm) | <λ> (nm) | |
| Mortalin | −11.0 ± 0.5 | −11.5 ± 0.5 | 338 ± 1 | 346.2 ± 0.1 |
| Mortalin aggregate | −5.2 ± 0.2 | −5.0 ± 0.2 | 336 ± 1 | 345.6 ± 0.1 |
| Mortalin + Gnd-HCl | - | - | 349 ± 1 | 351.5 ± 0.1 |
| Mortalin aggregate + Gnd-HCl | - | - | 348 ± 1 | 351.3 ± 0.1 |
| Hsp70–1A | −12.5 ± 0.6 | −11.5 ± 0.6 | 335 ± 1 | 345.2 ± 0.1 |
| Hsp70–1A aggregate | −8.0 ± 0.4 | −8.2 ± 0.4 | 337 ± 1 | 345.7 ± 0.1 |
| Hsp70–1A + Gnd-HCl | - | - | 350 ± 1 | 351.4 ± 0.1 |
| Hsp70–1A aggregate + Gnd-HCl | - | - | 351 ± 1 | 351.4 ± 0.1 |
Values calculated from averaging of 6 independent measures.
Mortalin and Hsp70–1A thermal aggregates interact with adenosine nucleotides
In order to further investigate the structural organization of both human Hsp70 and their thermal aggregates, we used thermal induced unfolding strategies since Hsp70s are formed by domains of different stabilities [39,41,57]. In addition, the Tm of the first transition is influenced by the presence of adenosine nucleotides [39,41], offering an opportunity to monitor the propensity of the thermal aggregates to interact with those binders.
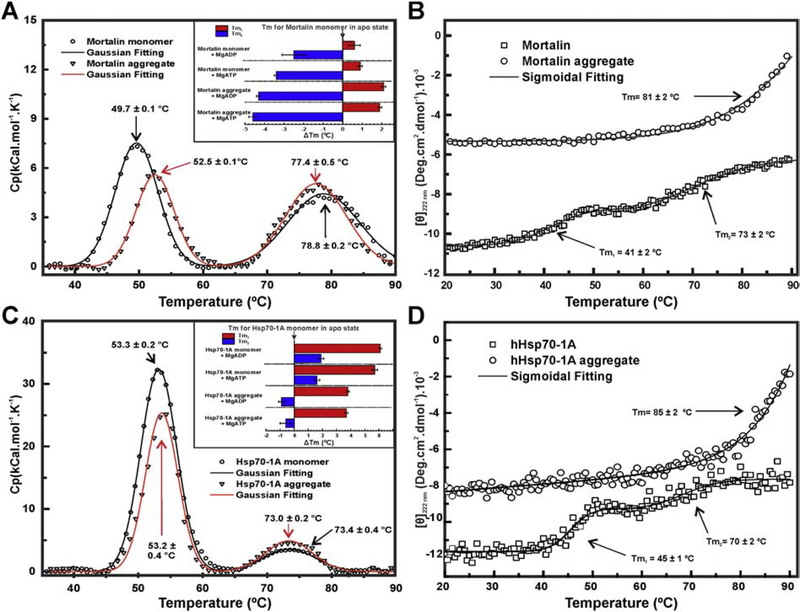
We first analyzed mortalin and Hsp70–1A in their monomeric and apo state (without adenosine nucleotides). Mortalin thermal unfolding assays performed by DSC indicated the existence of two transitions: one at 49.7 °C (Tm1) and another at 78.8 °C (Tm2) (Figure 4a). These data were partially in agreement with thermal unfolding performed by CD222nm (Figure 4b), which showed two transitions: Tm1 = 41 °C and Tm2 = 73 °C, as also previously reported [39]. Hsp70–1A also unfolded following two well defined transitions as observed by DSC (Tm1 at 53.3 °C and a Tm2 at 73.4 °C; Figure 3c) and CD222nm (Tm1 at 45 °C and a Tm2 at 70 °C; Figure 3d). Despite these values were similar to the ones previously reported [41], a third unfolding transition for Hsp70–1A was not observed here.
Figure 4: Mortalin/Hsp70–1A and their aggregates thermal unfolding.
DSC measurements of monomeric proteins and their aggregates. a) and c) Data indicated that mortalin and Hsp70–1A monomers, respectively, had two transitions: at 50 °C and at 71 °C. Mortalin aggregate also showed both thermal-transitions observed to monomeric form. Insets: graphs of the variation of the Tms, expressed as ΔTm, in relation to the monomeric Hsp70 in the apo state. b) and d) Thermal unfolding accompanied by CD222 nm. The Tm were estimated by sigmoidal fitting of the unfolding transition. In addition, the secondary structure loss process was not complete. Both mortalin and Hsp70–1A aggregates thermal-induced unfolding, accompanied by CD222 nm, showed a single transition at temperatures >80 °C and almost total loss of structure (see text for details).
Tm discrepancies observed between DSC and CD222nm are likely the product of differences in the probes used in each technical approach. DSC probes changes in the heat capacity involved in hydration of hydrophobic amino acids and therefore can be considered as a probe for global unfolding, while CD222nm probes mainly α-helix elements and can be considered a local probe. In other words, the unfolding events related to 43 °C, the temperature used for aggregate preparations, must involve the local unfolding of the NBD forming a partially stable structure. These data allowed us to hypothesize that the protein aggregates formation at 43 °C involves some folding intermediate of the NBD, since the first thermal induced transition of both mortalin and Hsp70–1A is influenced by adenosine nucleotide presence [39,41]. The presence of adenosine nucleotides should cause some conformational change which increased the aggregation propensity of both mortalin and Hsp70–1A. The second transition involves part of the PBD, as previously described [41,57].
As previously reported for monomeric Hsp70–1A [41] and monomeric mortalin [39], the presence of MgATP or MgADP caused substantial changes in the Tm recorded by DSC (Table 2). For monomeric Hsp70–1A, the recorded increment in the Tm1 and Tm2 were, respectively, of about 6 °C and 2 °C (Fig. 4c, inset). For monomeric mortalin, the increment in the Tm1 was about 1 °C while Tm2 showed a reduction of 3–4 °C (Fig. 4a, inset). These results indicated that both monomeric proteins were capable of binding to adenosine nucleotides leading the NBD to a more stable state. Besides, a small change in the unfolding apparent enthalpy (ΔHapp) was also observed for the first transition (ΔHapp1; Table 2).
Table 2:
Thermal stabilities of mortalin, Hsp70–1A and their thermal aggregates.
| Thermodynamic property | Protein preparation | Protein state | |||
|---|---|---|---|---|---|
| apo | MgATP | MgADP | |||
| Tm1 (°C) | Mortalin | monomer | 49.7 ± 0.1 | 50.6 ± 0.1 | 50.3 ± 0.3 |
| aggregate | 52.5 ± 0.1 | 51.6 ± 0.1 | 51.8 ± 0.1 | ||
| Hsp70–1A | monomer | 53.3 ± 0.2 | 59.0 ± 0.2 | 59.4 ± 0.1 | |
| aggregate | 53.2 ± 0.4 | 57.0 ± 0.1 | 57.1 ± 0.1 | ||
| Tm2 (°C) | Mortalin | monomer | 78.8 ± 0.2 | 75.4 ± 0.1 | 76.3 ± 0.6 |
| aggregate | 77.4 ± 0.5 | 74.2 ± 0.2 | 74.5 ± 0.1 | ||
| Hsp70–1A | monomer | 73.4 ± 0.4 | 75.0 ± 0.2 | 75.3 ± 0.2 | |
| aggregate | 73.0 ± 0.2 | 72.8 ± 0.4 | 72.5 ± 0.2 | ||
| ΔHapp1 (kcal.mol−1) | Mortalin | monomer | 270 ± 30 | 380 ± 30 | 310 ± 20 |
| aggregate | 160 ± 10 | 220 ± 20 | 120 ± 20 | ||
| Hsp70–1A | monomer | 940 ± 50 | 1010 ± 60 | 1100 ± 50 | |
| aggregate | 640 ± 30 | 550 ± 30 | 610 ± 20 | ||
| ΔHapp2 (kcal.mol−1) | Mortalin | monomer | 260 ± 20 | 70 ± 10 | 70 ± 10 |
| aggregate | 250 ± 20 | 80 ± 10 | 50 ± 10 | ||
| Hsp70–1A | monomer | 150 ± 20 | 120 ± 20 | 90 ± 20 | |
| aggregate | 190 ± 20 | 160 ± 20 | 120 ± 20 | ||
Mortalin and Hsp70–1A aggregate preparations also presented two transitions when thermally unfolded by DSC (Figure 4a and 4c). For mortalin aggregates, the Tm1 showed an increment of about 2–3 °C, while Tm2 was reduced by about 4 °C compared to the monomeric apo form, depending on the presence of adenosine nucleotides (Figure 4a, inset). In contrast, Hsp70–1A monomers and aggregates presented similar Tm in apo state. The presence of adenosine nucleotides induced an increase of about 4 °C in the Tm1 and a decrease of about 1 °C in the Tm2 comparing to the monomeric apo form (Figure 4c, inset). These results also display some differences in the behavior of Hsp70 aggregates/oligomers, as evidenced by others experiments like interaction with ANS (Figure S2), which indicated that both monomeric and aggregated proteins present differences in exposition of hydrophobic clefts. A substantial reduction in the ΔHapp1 of mortalin and Hsp70–1A aggregates in comparison to the monomeric forms was observed, indicating changes in the structural organization of the NBD (Table 2).
Since DSC data for both thermal aggregates presented the first thermal transition influenced by adenosine nucleotides (likely observed for the monomers [39,41,57]), we can conclude that NBD structure should be, totally or partially, preserved in the protein aggregates. Similar conclusions can be done for the PBD that partially unfolds in the second transition [41,57]. However, the first transition was not observed when the thermal unfolding of both Hsp70 aggregates were monitored by CD222nm (Figure 3b and 3d), which suggests that a stable form is reached by the NBD due to the thermic treatment.
The thermal-induced unfolding followed by CD222nm (Figure 4b and 4d) also showed that both monomeric proteins did not undergo a complete unfolding, even at 90 °C [39]. Interestingly, the thermal transition for both protein aggregate preparations started around 80 °C to reach a complete unfolding at 90 °C (CD222nm ≈ 0). Therefore, the Hsp70 aggregates, once formed, had a lower thermal stability than monomeric protein at high temperatures, since temperatures around 80 °C induced further loss of secondary structure and consequently the complete unfolding of the aggregates (Figure 4b and 4d).
Altogether, these thermal induced unfolding experiments indicated that mortalin and Hsp70–1A aggregates retain some structures with similar stability as observed for the monomeric forms. Besides, the insets of the (Figure 4a and 4c) show that the presence of adenosine nucleotides caused changes in the Tm of both Hsp70 aggregates preparations as they did for the monomeric Hsp70s.
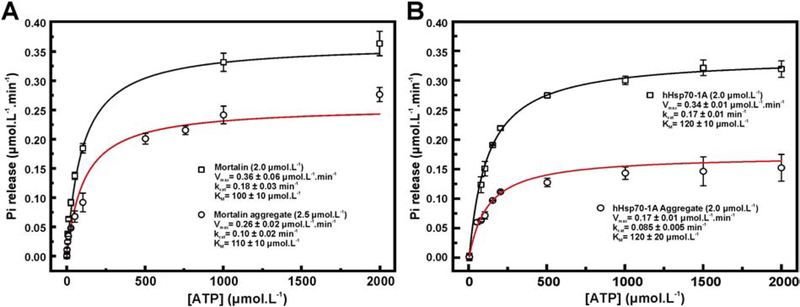
Hsp70 aggregate preparations present partial ATPase activity
We investigated the functionality of mortalin, Hsp70–1A and their thermal aggregates by evaluating their ATPase activity (Figure 5). Data were analyzed by using Michaelis-Menten nonlinear fitting and the parameters are summarized in (Table 2). Kinetic parameters obtained for monomeric mortalin and Hsp70–1A were similar to those previously reported as well as for other Hsp70 orthologues [39].
Figure 5: Mortalin and Hsp70–1A aggregates present partial ATPase activity.
a) Michaelis-Menten fitting to the ATPase activity for mortalin and its thermal aggregate and b) Hsp70–1A and its thermal aggregate. The data indicate that the protein aggregates retain about 50% of the kcat observed for both monomeric states (see text for details).
Previous work indicated that Hsp70 aggregates including Escherichia coli DnaK [36] and human Hsp70–1A in a hydrostatic aggregate [37] have ATPase activity. The thermal aggregates for mortalin and Hsp70–1A also presented ATPase activity (Figure 5a) with turnover numbers (kcat) of about 50 to 60% of that shown for the monomeric protein (Table 3). Therefore, Hsp70 aggregates presenting ATPase activity seems to be a general property of Hsp70. The Michaelis constant (KM) measured for the Hsp70 aggregates (Table 3) were similar to those recorded for the monomeric proteins indicating that the first part of the ezyme reaction (formation of the enzyme-substrate complex) was not influenced by the thermal aggregation. Therefore, Hsp70 monomers and thermal aggregates interact with ATP similarly, as shown by the DSC experiments. However, thermal aggregates most likely have some limitations to reach the transition state necessary for ATP hydrolysis.
Table 3:
Kinetics properties of mortalin and Hsp70–1A thermic aggregates
| Protein | Sample | KM (μmol L−1) | kcat (min−1) | kcat ratio& |
|---|---|---|---|---|
| Mortalin | Monomer ¥ | 100 ± 20 | 0.18 ± 0.03 | 0.6 ± 0.1 |
| Aggregate€ | 110 ± 10 | 0.10 ± 0.02 | ||
| Hsp70–1A | Monomer ¥ | 120 ± 10 | 0.17 ± 0.01 | 0.50 ± 0.03 |
| Aggregate ¥ | 120 ± 20 | 0.085 ± 0.005 |
Assays performed with 2.0 μmol L−1.
Assays performed with 2.5 μmol L−1.
ratio of aggregate kcat by kcat for the monomeric state.
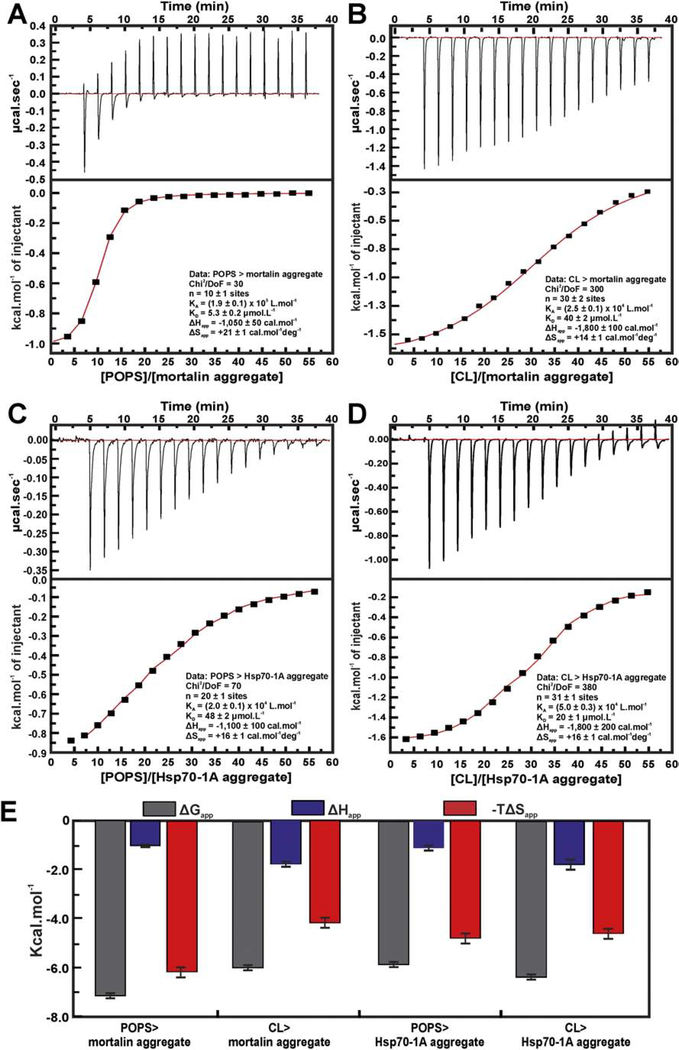
Mortalin and Hsp701A thermal aggregates interact with anionic liposomes
Previous studies indicated that Hsp70 have the ability to interact with negatively charged membranes [51,58]. We have shown that monomeric mortalin and Hsp70–1A strongly interact with liposomes made with POPS and CL lipids ([59]-Biochemistry, submitted). By means of ITC experiments, we obtained the thermodynamic signature for such interactions and estimated the apparent dissociation constant (KDapp) of about 20–40 μmol L−1, which indicates specific interaction.
Here, we investigated if liposomes made of POPS and CL interact with both Hsp70 aggregates by ITC. Our results, presented in (Figure 6), indicated that the mortalin and Hsp70–1A thermal aggregates interacted with the tested liposomes with ΔG in the same order of magnitude of those found for the monomeric proteins ([59]-Biochemistry, submitted). The interaction of Hsp70 aggregates with negatively charged lipids were driven by both entropy and enthalpy and showed similar affinities (Figure 6e and Table 4). However, the thermodynamic signature of mortalin or Hsp70–1A aggregates showed some differences in the −TΔSapp magnitude which indicate some subtle differences in the aggregates organization when interacting with liposomes.
Figure 6: Mortalin and Hsp70–1A aggregates interact with anionic liposomes.
ITC experiments. a) Mortalin aggregates interaction with POPS liposomes showed KDapp of 5.3 ± 0.3 μmol L−1; b) CL liposomes interaction with mortalin aggregate was driven by entropy and enthalpy and KDapp of 40 ± 1 μmol.L−1. c) Hsp70–1A aggregate interaction with POPS liposomes presented KDapp of 50 ± 2 μmol L−1. d) Hsp70–1A interaction with CL liposomes presented KDapp of 20 ± 1 μmol L−1. e) The thermodynamic signatures for mortalin and Hsp70–1A aggregates interaction with POPS and CL indicating that all interactions were driven by enthalpy and entropy.
Table 4 –
Thermodynamic signature of the interaction between mortalin and Hsp70–1A aggregates and POPS or CL liposomes.
| Thermodynamic parameters | Mortalin aggregates | Hsp70–1A aggregates | ||
|---|---|---|---|---|
| POPS | CL | POPS | CL | |
| KDapp (μmolL−1) | 5.3 ± 0.2 | 40 ± 2 | 48 ± 2 | 20 ± 1 |
| ΔGapp (cal mol−1) | −7,200 ± 100 | −6,000 ± 100 | −5,900 ± 100 | −6,400 ± 100 |
| ΔHapp (cal mol−1) | −1,050 ± 50 | −1,800 ± 100 | −1,100 ± 100 | −1,800 ± 200 |
| ΔSapp (cal mol−1 deg−1) | +21 ± 1 | +14 ± 1 | +16 ± 1 | +16 ± 1 |
| -TΔSapp (cal mol−1 deg−1) | −6,200 ± 200 | −4,200 ± 200 | −4,800 ± 200 | −4,600 ± 200 |
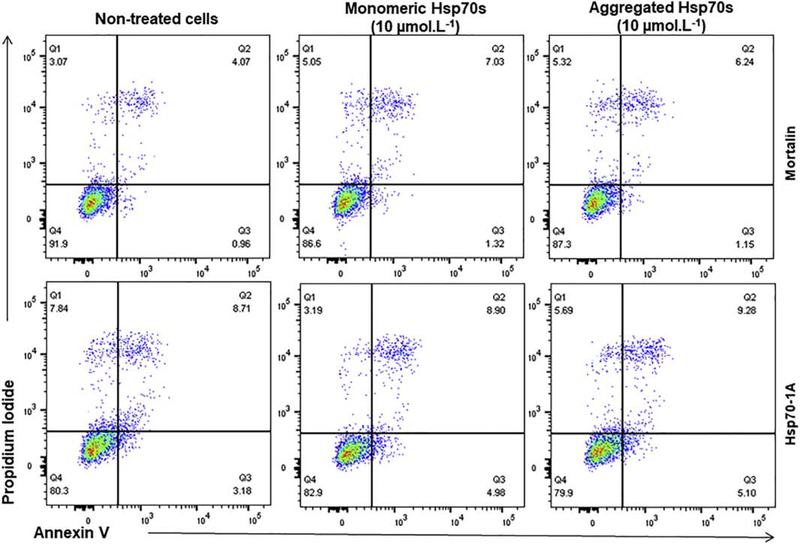
Hsp70 thermal aggregates cell toxicity
It is now well established that several members of the Hsp70 family can be found in the extracellular environment where they can exert a myriad of functions [60]. However, some level of toxicity has been reported [61]. To evaluate if Hsp70 thermal aggregates induced cell toxicity, we exposed N2A neuroblastoma cells to recombinant human mortalin and Hsp70–1A in monomeric or thermal aggregate states for 48 h and analyzed cell death by flow cytometry using annexin V and propidium iodide (PI) staining. A similar number of annexin V positive (early apoptotic) or annexin V/PI double positive (late apoptotic/dead) cells was detected in all conditions tested indicating that neither the monomeric mortalin and Hsp70–1A proteins nor their thermal aggregates induced toxicity in N2A neuroblastoma cells when added in the extracellular medium (Figure 7).
Figure 7: Mortalin or Hsp70–1A aggregates had no toxicity over N2A neuroblastoma cells.
The results indicated that mortalin, Hsp70–1A and their aggregates did not induce toxicity in N2A neuroblastoma cells.
CONCLUDING REMARKS
Here, we describe the production and purification of thermal aggregates from recombinant human mortalin and Hsp70–1A: two important Hsp70 found mainly in mitochondria and cytoplasm of human cells, respectively. We then demonstrate that these thermal aggregate preparations are stable aggregates of supramolecular size that retain partial structural elements, are able to interact with adenosine nucleotides and have partial ATPase activity. In addition, we show that these Hsp70 aggregates, like their monomeric counterparts, are capable of interacting with negatively charged liposomes. Finally, we provide evidence that these thermal aggregates are not toxic for cells when added in the extracellular environment. Altogether, our results indicate that Hsp70 aggregates behave as supramolecular assemblies and may have some physiological functions. Although mortalin and Hsp70–1A display about 50% identity in their amino acid sequence (76% similarity) and are prepared using the same methodology, it is notable that their aggregates have some differences in their physical-chemical properties. This observation implies that Hsp70 aggregates may have different cellular functions.
Our observations are very consistent with prior observations. Mortalin and Hsp70–1A, as other Hsp70s, are prone to self-oligomerize or self-aggregate [33–37]. Several reports indicated that mortalin self-oligomerization, self-aggregation or even misfolding is involved in mitochondria biogenesis failure, cancer, senescence and neurodegenerative diseases[12]. Angelidis et al. demonstrates that both Hsp70 and Hsc70 isoforms oligomerize or aggregate in a temperature-dependent manner, and that the presence of adenosine nucleotides and J-proteins influence that process [35]. Moreover, the aggregated Hsp70 and Hsc70 are incapable of exerting chaperone activity on client proteins or interacting with the J-proteins [35]. Thompson et al. reports that E. coli DnaK forms multiple oligomers in vitro, that also retains ATPase activity and holder activity on luciferase, indicating that such oligomers could maintain certain cellular function. However, the oligomerized DnaK loses the ability to interact with DnaJ (J-protein) and has no foldase activity [36]. Similarly, a human Hsp70–1A oligomer formed by hydrostatic pressure retains ATPase activity [37]. Thus, it was proposed that ATP, GrpE, J-proteins and client proteins recover DnaK from oligomers and reestablish its foldase activity. Furthermore, Steel et al. reported the in vivo acquisition of thermal oligomers from Hsp70.1 and Hsc70, which behaves as polymeric structures and are located into the cytoplasm and nuclei. It is shown that the polymeric structures are quickly formed even at small heating doses, and that the formation of such structures are related to the increase of cell resistance to thermal killing [38].
Summarizing, the Hsp70 temperature-dependent oligomerization/aggregation leading to formation of supramolecular assemblies is a general property to this group of protein. We can hypothesize that it would be one way to protect client proteins from aggregation during thermal stress, or even to Hsp70 protect themselves from irreversible aggregation [36]. In this direction, the identification and understanding of the elements that may mobilize or cause the dissociation of Hsp70 aggregates into monomeric forms is an important perspective. It might help to understand the importance and possible applications of the Hsp70 supramolecular assemblies.
Supplementary Material
Acknowledgements
This work was supported by: i) Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Pesquisa e Desenvolvimento (CNPq) grants: #2007/05001-4, #2011/23110-0, #2012/50161-8, #2014/07206-6, #2017/07335-9, #2017/26131-5 and #471415/2013-8. ii) National Institutes of Health (NIH) grants: R01 GM098455-04 and R01 GM114473-01. P.R. Dores-Silva also thanks FAPESP for financial support (grants: #2014/16646-0 and #2016/22447-1). Additionally, we would like to thank the Grupo de Química Inorgânica e Analítica for providing the use of the UV-Vis Spectrophotometer UV-2600 (Shimadzu) and Cell Positioner CPS-240A (Shimadzu) and the NEQUIMED group for providing the use of the Nano DSC (TA Instruments).
Footnotes
Competing interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference list
- [1].Bukau B, Horwich AL, The Hsp70 and Hsp60 chaperone machines, Cell 92 (1998) 351–366. [DOI] [PubMed] [Google Scholar]
- [2].Daugaard M, Rohde M, Jaattela M, The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions, FEBS Lett. 581 (2007) 3702–3710. 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- [3].Silva KP, Borges JC, The molecular chaperone Hsp70 family members function by a bidirectional heterotrophic allosteric mechanism, Protein Pept Lett 18 (2011) 132–142. [DOI] [PubMed] [Google Scholar]
- [4].Moran Luengo T, Mayer MP, Rudiger SGD, The Hsp70-Hsp90 Chaperone Cascade in Protein Folding, Trends Cell Biol 29 (2019) 164–177. 10.1016/j.tcb.2018.10.004. [DOI] [PubMed] [Google Scholar]
- [5].Mayer MP, Gierasch LM, Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones, J Biol Chem 294 (2019) 2085–2097. 10.1074/jbc.REV118.002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].De Maio A, Heat shock proteins: facts, thoughts, and dreams, Shock 1-1 (1999) 112 10.1097/00024382-199901000-00001. [DOI] [PubMed] [Google Scholar]
- [7].Tiroli-Cepeda AO, Ramos CHI, An Overview of the Role of Molecular Chaperones in Protein Homeostasis, Protein Pept. Lett. 18 (2011) 101–109. [DOI] [PubMed] [Google Scholar]
- [8].Mayer MP, Brehmer D, Gassler CS, Bukau B, Hsp70 chaperone machines, Adv Protein Chem 59 (2001) 1–44. [DOI] [PubMed] [Google Scholar]
- [9].Hartl FU, Hayer-Hartl M, Protein folding - Molecular chaperones in the cytosol: from nascent chain to folded protein, Science 295 (2002) 1852–1858. DOI 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- [10].Young JC, Hoogenraad NJ, Hartl FU, Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70, Cell 112 (2003) 41–50. [DOI] [PubMed] [Google Scholar]
- [11].Mayer MP, Bukau B, Hsp70 chaperones: Cellular functions and molecular mechanism, Cell.Mol.Life Sci. 62 (2005) 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Londono C, Osorio C, Gama V, Alzate O, Mortalin, apoptosis, and neurodegeneration, Biomolecules 2 (2012) 143–164. 10.3390/biom2010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kampinga HH, Craig EA, The HSP70 chaperone machinery: J proteins as drivers of functional specificity, Nat. Rev. Mol. Cell Biol. 11 (2010) 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Young JC, Mechanisms of the Hsp70 chaperone system, Biochem. Cell Biol. 88 (2010) 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hartl FU, Bracher A, Hayer-Hartl M, Molecular chaperones in protein folding and proteostasis, Nature 475 (2011) 324–332. 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- [16].Borges JC, Seraphim TV, Dores-Silva PR, Barbosa LRS , A review of multi-domain and flexible molecular chaperones studies by small-angle X-ray scattering, Biophys. Rev. 8 (2016) 107–120. 10.1007/s12551-016-0194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mayer MP, Hsp70 chaperone dynamics and molecular mechanism, Trends Biochem. Sci. 38 (2013) 507–514. 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- [18].Cyr DM, Ramos CH, Specification of Hsp70 Function by Type I and Type II Hsp40, in: Blatch GL, Edkins AL (Eds.) The Networking of Chaperones by Co-chaperones, Springer International Publishing2015, pp. 91–102. [Google Scholar]
- [19].Radons J, The human HSP70 family of chaperones: where do we stand?, Cell Stress Chaperones 21 (2016) 379–404. 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kaul SC, Deocaris CC, Wadhwa R, Three faces of mortalin: a housekeeper, guardian and killer, Exp Gerontol 42 (2007) 263–274. 10.1016/j.exger.2006.10.020. [DOI] [PubMed] [Google Scholar]
- [21].Wadhwa R, Kaul SC, Ikawa Y, Sugimoto Y, Identification of a novel member of mouse hsp70 family. Its association with cellular mortal phenotype, J.Biol.Chem. 268 (1993) 6615–6621. [PubMed] [Google Scholar]
- [22].Sichting M, Mokranjac D, Azem A, Neupert W, Hell K, Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1, EMBO J 24 (2005) 1046–1056. 10.1038/sj.emboj.7600580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Deocaris CC, Kaul SC, Wadhwa R, Mortalin’s Machinery, in: Kaul SC, Wadhwa R (Eds.) Mortalin biology: life, stress and death, Dordrecht: Springer, c2012., New York, 2012, pp. 21–30. [Google Scholar]
- [24].Wadhwa R, Yaguchi T, Hasan K, Mitsui Y, Reddel RR, Kaul SC, Hsp70 family member, mot-2/mthsp70/GRP75, binds to the cytoplasmic sequestration domain of the p53 protein, Exp.Cell Res. 274 (2002) 246–253. [DOI] [PubMed] [Google Scholar]
- [25].Bohnert M, Pfanner N, van der Laan M, A dynamic machinery for import of mitochondrial precursor proteins, FEBS Lett. 581 (2007) 2802–2810. [DOI] [PubMed] [Google Scholar]
- [26].Fan ACY, Young JC, Function of Cytosolic Chaperones in Tom70-Mediated Mitochondrial Import, Protein Pept. Lett. 18 (2011) 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wadhwa R, Takano S, Kaur K, Deocaris CC, Pereira-Smith OM, Reddel RR, Kaul SC, Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis, Inter.J.Cancer 118 (2006) 2973–2980. [DOI] [PubMed] [Google Scholar]
- [28].Kaul SC, Reddel RR, Sugihara T, Mitsui Y, Wadhwa R, Inactivation of p53 and life span extension of human diploid fibroblasts by mot-2, FEBS Lett. 474 (2000) 159–164. [DOI] [PubMed] [Google Scholar]
- [29].Ran QT, Wadhwa R, Kawai R, Kaul SC, Sifers RN, Bick RJ, Smith JR, Pereira-Smith OM, Extramitochondrial localization of mortalin/mthsp70/PBP74/GRP75, Biochem. Biophys. Res. Comm. 275 (2000) 174–179. [DOI] [PubMed] [Google Scholar]
- [30].Lu WJ, Lee NP, Kaul SC, Lan F, Poon RTP, Wadhwa R, Luk JM, Mortalin-p53 interaction in cancer cells is stress dependent and constitutes a selective target for cancer therapy, Cell Death and Differentiation 18 (2011) 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jin JH, Hulette C, Wang Y, Zhang T, Pan C, Wadhwa R, Zhang J, Proteomic identification of a stress protein, mortalin/mthsp70/GRP75 - Relevance to Parkinson disease, Mol.Cell.Proteomics 5 (2006) 1193–1204. [DOI] [PubMed] [Google Scholar]
- [32].Na Y, Kaul SC, Ryu J, Lee JS, Ahn HM, Kaul Z, Kalra RS, Li L, Widodo N, Yun CO, Wadhwa R, Stress chaperone mortalin contributes to epithelial-mesenchymal transition and cancer metastasis, Cancer Res 76 (2016) 2754–2765. 10.1158/0008-5472.CAN-15-2704. [DOI] [PubMed] [Google Scholar]
- [33].Benaroudj N, Batelier G, Triniolles F, Ladjimi MM, Self-Association of the Molecular Chaperone Hsc70, Biochemistry 34 (1995) 15282–15290. Doi 10.1021/Bi00046a037. [DOI] [PubMed] [Google Scholar]
- [34].King C, Eisenberg E, Greene L, Polymerization of 70-kDa heat shock protein by yeast DnaJ in ATP, J Biol Chem 270 (1995) 22535–22540. [DOI] [PubMed] [Google Scholar]
- [35].Angelidis CE, Lazaridis I, Pagoulatos GN, Aggregation of hsp70 and hsc70 in vivo is distinct and temperature-dependent and their chaperone function is directly related to non-aggregated forms, The FEBS journal 259 (1999) 505–512. DOI 10.1046/j.1432-1327.1999.00078.x. [DOI] [PubMed] [Google Scholar]
- [36].Thompson AD, Bernard SM, Skiniotis G, Gestwicki JE, Visualization and functional analysis of the oligomeric states of Escherichia coli heat shock protein 70 (Hsp70/DnaK), Cell Stress Chaperones 17 (2012) 313–327. 10.1007/s12192-011-0307-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Araujo TL, Borges JC, Ramos CH, Meyer-Fernandes JR, Oliveira Junior RS, Pascutti PG, Foguel D, Palhano FL, Conformational changes in human Hsp70 induced by high hydrostatic pressure produce oligomers with ATPase activity but without chaperone activity, Biochemistry 53 (2014) 2884–2889. 10.1021/bi500004q. [DOI] [PubMed] [Google Scholar]
- [38].Steel R, Cross RS, Ellis SL, Anderson RL, Hsp70 architecture: the formation of novel polymeric structures of Hsp70.1 and Hsc70 after proteotoxic stress, PLoS One 7 (2012) e52351 10.1371/journal.pone.0052351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dores-Silva PR, Barbosa LR, Ramos CH, Borges JC, Human mitochondrial Hsp70 (mortalin): shedding light on ATPase activity, interaction with adenosine nucleotides, solution structure and domain organization, PLoS One 10 (2015) e0117170 10.1371/journal.pone.0117170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dores-Silva PR, Minari K, Ramos CHI, Barbosa LRS, Borges JC, Structural and stability studies of the human mtHsp70-escort protein 1: An essential mortalin co-chaperone, Int. J. Biol. Macromol. 56 (2013) 140–148. [DOI] [PubMed] [Google Scholar]
- [41].Borges JC, Ramos CHI, Spectroscopic and thermodynamic measurements of nucleotide-induced changes in the human 70-kDa heat shock cognate protein, Arch. Biochem. Biophys. 452 (2006) 46–54. [DOI] [PubMed] [Google Scholar]
- [42].Kurganov BI, Kinetics of protein aggregation. Quantitative estimation of the chaperone-like activity in test-systems based on suppression of protein aggregation, Biochemistry (Mosc) 67 (2002) 409–422. [DOI] [PubMed] [Google Scholar]
- [43].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nature methods 9 (2012) 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Corporation O, Origin, Northampton, MA, USA., 2015. [Google Scholar]
- [45].Kumagai PS, Martins CS, Sales EM, Rosa HVD, Mendonça DC, Damalio JCP, Spinozzi F, Itri R, Araujo APU, Correct partner makes the difference: Septin G-interface plays a critical role in amyloid formation, Int J Biol Macromol 133 (2019) 428–435. 10.1016/j.ijbiomac.2019.04.105. [DOI] [PubMed] [Google Scholar]
- [46].Alam P, Siddiqi MK, Malik S, Chaturvedi SK, Uddin M, Khan RH, Elucidating the inhibitory potential of Vitamin A against fibrillation and amyloid associated cytotoxicity, Int J Biol Macromol 129 (2019) 333–338. 10.1016/j.ijbiomac.2019.01.134. [DOI] [PubMed] [Google Scholar]
- [47].Furkan M, Siddiqi MK, Zakariya SM, Khan FI, Hassan MI, Khan RH, An In Vitro elucidation of the antiaggregatory potential of Diosminover thermally induced unfolding of hen egg white lysozyme; A preventive quest for lysozyme amyloidosis, Int J Biol Macromol 129 (2019) 1015–1023. 10.1016/j.ijbiomac.2019.02.107. [DOI] [PubMed] [Google Scholar]
- [48].Majid N, Siddiqi MK, Khan AN, Shabnam S, Malik S, Alam A, Uversky VN, Khan RH, Biophysical Elucidation of Amyloid Fibrillation Inhibition and Prevention of Secondary Nucleation by Cholic Acid: An Unexplored Function of Cholic Acid, ACS Chem Neurosci 10 (2019) 4704–4715. 10.1021/acschemneuro.9b00482. [DOI] [PubMed] [Google Scholar]
- [49].Instruments T, DSC run software TA Instruments. [Google Scholar]
- [50].Instruments T, NanoAnalyze software, 2015. [Google Scholar]
- [51].Lopez V, Cauvi DM, Arispe N, De Maio A, Bacterial Hsp70 (DnaK) and mammalian Hsp70 interact differently with lipid membranes, Cell Stress Chaperones 21 (2016) 609–616. 10.1007/s12192-016-0685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dores-Silva PR, Nishimura LS, Kiraly VT, Borges JC, Structural and functional studies of the Leishmania braziliensis mitochondrial Hsp70: Similarities and dissimilarities to human orthologues, Arch Biochem Biophys 613 (2017) 43–52. 10.1016/j.abb.2016.11.004. [DOI] [PubMed] [Google Scholar]
- [53].Batista FA, Almeida GS, Seraphim TV, Silva KP, Murta SM, Barbosa LR, Borges JC, Identification of two p23 co-chaperone isoforms in Leishmania braziliensis exhibiting similar structures and Hsp90 interaction properties despite divergent stabilities, The FEBS journal 282 (2015) 388–406. 10.1111/febs.13141. [DOI] [PubMed] [Google Scholar]
- [54].Verwer B, FACSDiVa Option, bdbiosciences.com, White paper, 2002.
- [55].Treestar, FlowJo software, Ashland, OR [Google Scholar]
- [56].Borges JC, Ramos CHI , Characterization of nucleotide-induced changes on the quaternary structure of human 70 kDa heat shock protein Hsp70.1 by analytical ultracentrifugation, BMB Rep. 42 (2009) 166–171. [DOI] [PubMed] [Google Scholar]
- [57].Montgomery DL, Morimoto RI, Gierasch LM, Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling, J. Mol. Biol. 286 (1999) 915–932. 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- [58].Armijo G, Okerblom J, Cauvi DM, Lopez V, Schlamadinger DE, Kim J, Arispe N, De Maio A, Interaction of heat shock protein 70 with membranes depends on the lipid environment, Cell Stress Chaperones 19 (2014) 877–886. 10.1007/s12192-014-0511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dores-Silva PR, Cauvi DM, Kiraly VTR, Borges JC, De Maio A, Human mtHsp70 (mortalin, HSPA9) interacts with lipid bilayers resembling the inner mitochondrial membrane., Biochemistry Submitted (2019). [DOI] [PubMed] [Google Scholar]
- [60].De Maio A, Extracellular Hsp70: Export and Function, Curr. Protein Pept. Sci. 15 (2014) 225–231. 10.2174/1389203715666140331113057. [DOI] [PubMed] [Google Scholar]
- [61].Arispe N, Doh M, Simakova O, Kurganov B, De Maio A, Hsc70 and Hsp70 interact with phosphatidylserine on the surface of PC12 cells resulting in a decrease of viability, FASEB J 18 (2004) 1636–1645. 10.1096/fj.04-2088com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.