Abstract
Purpose
To date, many kinds of classical swine fever (CSF) vaccines have been developed to protect against this disease. However, the efficacy of these vaccines to protect the pig against field CSF strains needs to be considered, based on circulating strains of classical swine fever virus (CSFV).
Materials and Methods
Recombinant E2-CSFV protein produced by baculovirus/insect cell system was analyzed by western blots and immunoperoxidase monolayer assay. The effect of CSFV-E2 subunit vaccines was evaluated in experimental pigs with three genotypes of CSFV challenge. Anti-E2 specific and neutralizing antibodies in experimental pigs were analyzed by blocking enzyme-linked immunosorbent assay and neutralization peroxidize-linked assay.
Results
The data showed that CSFV VN91-E2 subunit vaccine provided clinical protection in pigs against three different genotypes of CSFV without noticeable clinical signs, symptoms, and mortality. In addition, no CSFV was isolated from the spleen of the vaccinated pigs. However, the unvaccinated pigs exhibited high clinical scores and the successful virus isolation from spleen. These results showed that the E2-specific and neutralizing antibodies induced by VN91-E2 antigen appeared at day 24 after first boost and a significant increase was observed at day 28 (p<0.01). This response reached a peak at day 35 and continued until day 63 when compared to controls. Importantly, VN91-E2 induced E2-specific and neutralizing antibodies protected experimental pigs against high virulence of CSFVs circulating in Vietnam, including genotype 1.1, 2.1, and 2.2.
Conclusion
These findings also suggested that CSFV VN91-E2 subunit vaccine could be a promising vaccine candidate for the control and prevention of CSFV in Vietnam.
Keywords: Classical swine fever, Genotype, Vaccine, Vietnam
Introduction
Classical swine fever (CSF) is a highly contagious and one of the most dangerous and emerging swine disease worldwide with causing substantial economic losses to the swine industry in many countries and one of the Office International des Epizooties (OIE) notifiable diseases [1]. Classical swine fever virus (CSFV) is a single-stranded, positive-sense, enveloped RNA virus that belongs to the Pestivirus genus of the Flaviviridae family [2]. The genome of CSFV has an approximately 12.3 kb containing a single large open reading frame coding for a polyprotein of 3,898 amino acids flanked by 50 and 30 non-translated regions [2]. The translated polyprotein is cleaved by both viral and host proteases to yield 12 mature proteins, including four structural proteins (C, E0 or Erns, E1, and E2) and eight nonstructural viral proteins (Npro, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) [3,4]. The genotypes analysis of CSFV has classified into three significant genotypes with eleven sub-genotypes. Genotype 1 contains primarily outbreak virus strains that have been isolated from many regions of the world and includes all live-attenuated vaccine strains and include four sub-genotypes 1.1, 1.2, 1.3, and 1.4. Genotype 2 includes three sub-genotype 2.1, 2.2, and 2.3, and contains most of the currently circulating virus strains, whose prevalence has increased and caused epidemic infection since 1980. Genotype 3 (3.1, 3.2, 3.3, and 3.4) contains most of the strains distributed in separated geographic regions [3,4,5]. The clinical signs of CSFV includes high fever, huddling, weakness, drowsiness, anorexia, ataxia, hemorrhage, purple discoloration or cyanosis of the skin, conjunctivitis, and constipation followed by diarrhea [1,3,5]. The morbidity and mortality of pigs infected with CSFV have been reported up to 100% [1].
To date, CSFV vaccines have been developed, and three types of commercially vaccine are frequently used: (1) modified live virus (MLV) vaccines which are manufactured and widely used in local CSF endemic countries [6,7], (2) subunit vaccines based on CSF viral envelope protein E2 [8,9,10,11,12], and (3) a chimeric live recombinant viral vector vaccine [13,14]. Use of the MLV CSFV vaccine such as live attenuated vaccines is still practiced in some regions of the world such as Vietnam, in which the virus is enzootic, to prevent and control the spread of the CSFV. However, attenuated vaccines are not safe enough and do not allow differentiation of infected from vaccinated animals (DIVA) [11,14]. A chimeric CSFV vaccine has been constructed using a modified genome of live bovine viral diarrhea virus to express the CSFV E2 gene and was approved by the European Union in 2015. However, this chimeric CSFV vaccine is recommended only for emergency vaccination [10]. Subunit recombinant marker vaccines have been considered safer and cheaper alternatives against CSFV that are designed from two envelope glycoproteins Erns and E2, and have been targeted for vaccine development to meet the DIVA [10,15]. Many kinds of subunit vaccines based on E2 have been generated using different expression systems, and the most commercial subunit vaccines were based on baculovirus-expressed E2 protein in insect cell line [5,9,15]. Previous studies have established that the CSFV glycoprotein E2 is the most immunodominant protein in protecting against CSF disease with multiple identified epitopes which induce CSFV neutralizing antibodies [11,15,16]. The disadvantage of subunit CSFV E2 recombinant vaccines which induce clinical protection 14 days after vaccination, and with total prevention of horizontal transmission, only provide incomplete prevention of vertical transmission. In second time vaccination with subunit CSFV E2 recombinant vaccines, the neutralizing antibodies can be detected after 7 days of second dose, suggesting that the efficacy of the E2 subunit vaccines is lower than that of MLV [7,10,16,17].
MLV CSFV vaccine is used as a national program to control and prevent the spread of CSFV in Vietnam. Recently, some outbreaks of CSFV frequently appeared in the whole country of Vietnam with different genotypes. In this study, we identified the genotype of CSFV in recent outbreaks and describe a new potential CSFV subunit vaccine based on a recombinant CSFV VN91-E2 protein in baculovirus expressed in an insect cell. We also provide the first report in Vietnam that pigs immunized with two-dose the subunit VN91-E2 recombinant vaccine are protected clinically from CSFV challenge with three different genotypes that are isolated in field of Vietnam.
Materials and Methods
Virus and cells
CSFV-VN91 virus strain was isolated from died pig in Hung Yen province in 1991 (GenBank # LC374604) [18] and Regional Animal Health Office No. 3 (RAHO3)/NA strain was isolated from died pig in Nghe An province in 2013, and CSFV-NAM DINH strain was isolated from the outbreak in NAM DINH in 2018. CSFV isolated strains were passaged 4 times in PK15a cell line (ATCC, Manassas, VA, USA) in DMEM (Thermo Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Thermo Scientific) and 1% Penicillin-streptomycin solution (Thermo Scientific). For recombinant E2 production in insect cells, Spodoptera frugiperda (Sf21AE; ATCC) and TN5 insect cells were grown in SF900 III SFM medium (Sigma-Aldrich, Louis, MO, USA) supplemented with 10% FBS.
Whole-genome sequence of field classical swine fever strains isolated in Vietnam
The whole-genome sequence of CSFV in Vietnam was previously described [18]. Briefly, the RAHO3/NA and NAM DINH strains were propagated in the culture of PK15a cells for 4 to 5 days at 37℃, and total RNAs were extracted from the cell supernatant by using High Pure Viral Nucleic Acid Kit (Roche Diagnostics, Rotkreuz, Switzerland) according to the manufacturer's instruction. The RNAs were subjected to a reverse transcription reaction with the use of random oligonucleotide primers (Applied Biosystems, Waltham, MA, USA) and were amplified by polymerase chain reaction (PCR) with various sets of primers designed based on the genome sequences of previously known CSFV strains to prepare 25 overlapping cDNA fragments spanning the entire lengths of the genome for sequencing analysis. Determination of the 5′- and 3′-termini of the genome was performed by PCR in combination with the ligation of a 5′-phosphorylated oligonucleotide tag, as previously reported [19,20], with slight modification in the tag sequence. The chromatograms received after sequencing were analyzed using the Lasergene software (DNASTAR Inc., Madison, WI, USA). BLAST was performed with the sequences within the non-redundant nucleotide database (http://www.ncbi.nlm.nih.gov/Blast) to confirm the presence of the gene-specific to CSFV and then compared with nucleotide sequences of other CSFV isolates in Vietnam that are available in the GenBank database. The multiple sequence alignment was performed using the Lasergene software (DNASTAR Inc., Madison, WI, USA). Phylogenetic analyses of nucleotides sequences of whole-genome sequences of CSFV was constructed using the neighbor-joining method with a bootstrap value of 1,000 in the MEGA7 program (https://www.megasoftware.net/) [21].
Production of recombinant E2 protein using field VN91 strain as “template”
PCR-amplified full length of CSFV E2 genes from CSFV-VN91 virus (GenBank # LC374604, Genotype 1.1) was cloned into a pFastBac1 vector (Invitrogen, Carlsbad, CA, USA) Baculovirus Expression System plasmid vector using the following restriction enzyme-anchored primers: EcoRI-anchored forward primer 5′-CGCGGATCCATGCGGCTAGCCTGCAAGGAAGATTACA-3′ and BamHI-anchored reverse primer 5′-CCGGAATTC-TCATCAATCTGAGTGGCGGTCAGTCACGTC-3′. The freshly PCR products were transformed into DH10Bac Escherichia coli (Invitrogen) host strain that contains a baculovirus shuttle vector (bacmid) and a helper plasmid. Upon screening of colonies, positive E. coli transformants with recombinant E2 bacmid was upscaled by overnight culture in liquid media and the bacmid was prepared. Briefly, bacterial cells were suspended in 0.25 mL of solution I (50 mM glucose, 25mM Tris-HCl [pH 8.0], 10 mM ethylenediaminetetraacetic acid [EDTA]) and lysed by adding 0.25 mL of solution II (0.2 N NaOH, 1% sodium dodecyl sulfate). The lysed cell fluid was neutralized by adding 0.35 mL of solution III (5 M KOH, 11.5 mL glacial acetic acid, 28.5 mL DW), then centrifuged ×14,000 g for 10 minutes. To precipitate bacmid DNA, the cleared lysate was mixed with an equal volume of 2-propanol, then centrifuged ×14,000 g at 40℃ for 20 minutes. Bacmid DNA was dried briefly and resolved in 40 µL of TE. To generate recombinant baculovirus stock for E2 expression, Sf21AE insect cells were transfected using Cellfectin II Reagent (Thermo Scientific) and passaged 3 times to amplify the E2 bearing recombinant baculovirus. At passage 3, the supernatant of Sf21AE cell was collected and clarified by centrifugation at 500× g for 5 minutes to obtain the baculovirus stock that was used to infect Sf21AE Cells for VN91-E2 expression.
Western blot analysis
The supernatants (100 µL) were concentrated with a centrifugal filter (Microcon YM-30; Merck, Darmstadt, Germany) by centrifuge ×14,000 g at 25℃ for 30 minutes, then solubilized in 2× sample buffer and treated at100℃ for 5 minutes before use. The E2 CSFV recombinant protein (10 µL) was reduced with 2.5% β-mercaptoethanol. Proteins were either stained with Coomassie Brilliant Blue Stain (Thermo Scientific) or transferred to a polyvinylidene difluoride (PVDF) membrane (Invitrogen) for western blotting using Mini Trans-Blot cell system (BioRad, Hercules, CA, USA). The transferred PVDF membrane was blocked with 5% nonfat milk (Sigma-Aldrich) dissolved in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST). The membrane was incubated with mAb anti-CSFV antibody (The National Institute of Animal Health, Tsukuba, Japan) for 1 hour at 25℃, washed 3 times with PBST. Then, the membrane was incubated with POD-anti-mIgG (H+L) (GE Healthcare, Chalfont St. Giles, UK) for 1 hour at 25℃ and washed 3 times with PBST. Detection of immunoreactive bands was performed using Western Lightning Plus-ECL substrate (Thermo Scientific) as per the supplier's instructions.
Immunoperoxidase monolayer assay
TN5 cells were seeded in 96-well cell culture plates, inoculated with recombinant baculovirus containing E2 gene for 24 houts at room temperature. Subsequently, the culture medium was removed, cells were washed with PBS and fixed in 4% paraformaldehyde for 10 minutes and then washed twice with PBS. A solution of 1% H2O2 in methanol was added to the cells for 5 minutes, and cells were subsequently washed twice with PBS. The anti-CSFV antibody in phosphate-buffered saline supplemented with 1% Tween 80 (PBS-T) and negative SPF pig serum was added to the infected TN5 cells and cells was incubated for 1 hour at 37℃. Subsequently, serum dilutions were removed, and cells were washed with PBS-T. Cells were then incubated with peroxidase-labeled goat anti-swine immunoglobulin G antibodies (Sigma-Aldrich) at 37℃ for 1 hour. Finally, the plates were washed with PBS-T, and 50 µL of a substrate solution of 5% 3-amino-9-ethyl-carbazole in 0.05 M Na-acetate buffer (pH 5) supplemented with 0.05% H2O2 (Sigma-Aldrich) was added to each well and incubated for 20 minutes at room temperature. The reaction was stopped by replacing the substrate solution with Na-acetate buffer, and the staining was analyzed via light microscopy.
Viral plaque assay
Virus titers were expressed in plaque-forming units (pfu/mL) as previously described [22]. Briefly, Sf21AE insect cells were grown SF900 III SFM medium (Sigma-Aldrich) supplemented with 10% FBS to obtain a culture at ≥1×106 cells/mL. Cells were added to each 6-well culture plate (Thermo Scientific, approximately 2.5×106 cells/well) and incubated at room temperature for 60 minutes to allow cells to attach. For each virus sample, appropriate dilutions were prepared using SF900 III SFM media. After 60 minutes, the media was removed, without disturbing the cell layer. Plates were inoculated with 1 mL of the diluted virus sample of each selected dilution in three appropriately labeled well. Positive and negative control wells were inoculated at the same time with 1 mL of the appropriate control. Each well was immediately rocked 3–5 times to ensure uniform distribution of the virus and then incubated at room temperature for 60 minutes with manual rocking every 15 minutes. A 50% mixture of SF900 III SFM medium/Agarose (Sigma-Aldrich) overlay solution was prepared at 40℃ and then added to each well, avoiding disturbance of the cell layer. The wells were allowed to rest upright for 45 minutes to allow the agarose overlayer to solidify. The wells were then inserted inside a plastic container with a dampened absorbent towel and incubated at 27℃ for 6 days after which wells were examined under low magnification with a microscope to count the number of plaques. To elucidate subunit VN91-E2 recombinant protein activity and induce specific CSFV antibody, pigs were stimulated with 107 pfu/mL with an equal volume of adjuvant. The final concentration was based on pilot experiments that demonstrated a significantly higher effect of these concentrations of subunit VN91-E2 recombinant proteins on the activation of the immune response in comparison to lower or higher levels.
Pigs, VN91/E2-subunit vaccine, and vaccination
Conventional Large White crossbred weaned specific-pathogen-free piglets (3 weeks of age) were purchased from the Pig Research Centre of National Institute of Animal Science, Vietnam. The pigs were fed a standard commercial diet, and the study was conducted in compliance with the institutional rules for the care and use of laboratory animals and using a protocol approved by the relevant committee at National Institute of Veterinary Research in Vietnam. The CSFV E2 subunit vaccine (VN91-E2) was inactivated by 10 nM binary ethylenimine (BEI) in 48 hours as described previously [23] and was prepared by ultrasonic of VN91-E2 with a Freund's Adjuvant Complete (Sigma-Aldrich) as described by the manufacturer as shown in Table 1. The pigs were randomly allotted into 10 groups (n=5 for each group) with four control groups, three VN91-E2 vaccinated groups, and three commercial E2 vaccinated groups (Table 1). The control group, pigs were injected intramuscularly with 2 mL of PBS. Second doses were applied 2 weeks post 1st dose (2 mL/dose), the VN91-E2 recombinant protein was prepared with a Freund's Adjuvant Incomplete (Sigma-Aldrich). Five weeks after the second vaccination, pigs were challenged with 5×105 TCID50 CSFV isolates: VN91, RAHO3/NA, and NAM DINH strain (2 mL intramuscularly). The CSFV VN91, RAHO3/NA, and NAM DINH strains were evaluated as a high virulent strain in our previous study (data not shown).
Table 1. Summarized the experiment in this study.
| Group name | No. of pigs | Dose 1 (2 mL) | Dose 2 (2 mL) | Classical swine fever virus challenge at D49 | ||
|---|---|---|---|---|---|---|
| VN91 | RAHO3/NA | NAM DINH | ||||
| None | 5 | D0 | D28 | |||
| Control G1 | 5 | D0 | D28 | + | ||
| Control G2 | 5 | D0 | D28 | + | ||
| Control G3 | 5 | D0 | D28 | + | ||
| VxE2-VN91 | 5 | D0 | D28 | + | ||
| VxE2-RAHO3/NA | 5 | D0 | D28 | + | ||
| VxE2-NAMDINH | 5 | D0 | D28 | + | ||
| VxInt-VN91 | 5 | D0 | D28 | + | ||
| VxInt-RAHO3/NA | 5 | D0 | D28 | + | ||
| VxInt-NAMDINH | 5 | D0 | D28 | + | ||
The VN91-E2 subunit recombinant protein was prepared with a Freund's Adjuvant Complete in dose 1 and with Freund's Adjuvant Incomplete in dose 2 of vaccination.
Clinical examination
Following the challenge, clinical signs were recorded daily according to the protocol by Mittelholzer et al. [24]. Cumulative clinical scores (CS) were calculated over time for individual pigs and the respective groups. In addition, the body temperature was measured on a daily basis from 1 day before vaccination until 14 days post-challenge. A body temperature of >40.0℃ for at least two consecutive days was recorded as fever.
Sample collection and analysis
The serum and EDTA blood samples were collected from the piglets on study day 0 (dose 1), day 7 and day 14 (dose 2), and every 3 days after dose 2 until the end of this study. For white blood cells (WBCs) analysis, slides were fixed in pure methanol and stained by Giemsa solution (azure B/azure II, eosin and methylene blue) according to the manufacturer's protocol (Sigma-Aldrich). WBCs were counted under the light microscope at 40× magnification in a random sequence. The samples were analyzed on the day of acquisition. Detection of viral RNA was performed on spleen samples using the CSFV specific reverse transcription PCR (RT-PCR) assays described in the previous section.
Antibody analyses
Serum samples were examined for neutralizing antibodies to CSFV through a direct virus neutralization peroxidize-linked assay, according to OIE with some modifications. In brief, serum samples collected at day 35 vaccination, day 49 (D0 day post-challenge [DPC]) and 14DPC were first diluted five-fold and inactivated for 30 minutes at 56℃, and then serially diluted two-fold. The diluted serum samples (in duplicate) were incubated with 200 TCID50 of CSFV CSFV VN91, RAHO3/NA, or NAM DINH in DMEM with 10% FBS for 1–2 hours at 37℃. After incubation, PK-15 cells were added, and the plates were incubated 3 days in 96-well plate. The cell-monolayers were fixed and stained as previously described [25], and viral titers were calculated [26]. Anti-E2 specific antibodies were determined by using PrioCHECK CSFV Ab Kit (Thermo Scientific) as described by the manufacturer.
Statistical analysis
Statistical analysis was performed by using IBM SPSS software for Windows ver. 25.0 (IBM Corp., Armonk, NY, USA). A p-value <0.05 was considered to be statistically significant. Differences between the groups were tested by Duncan's multiple comparison methods.
Results
The analysis of the whole genome sequences of classical swine fever viruses isolated in Vietnam
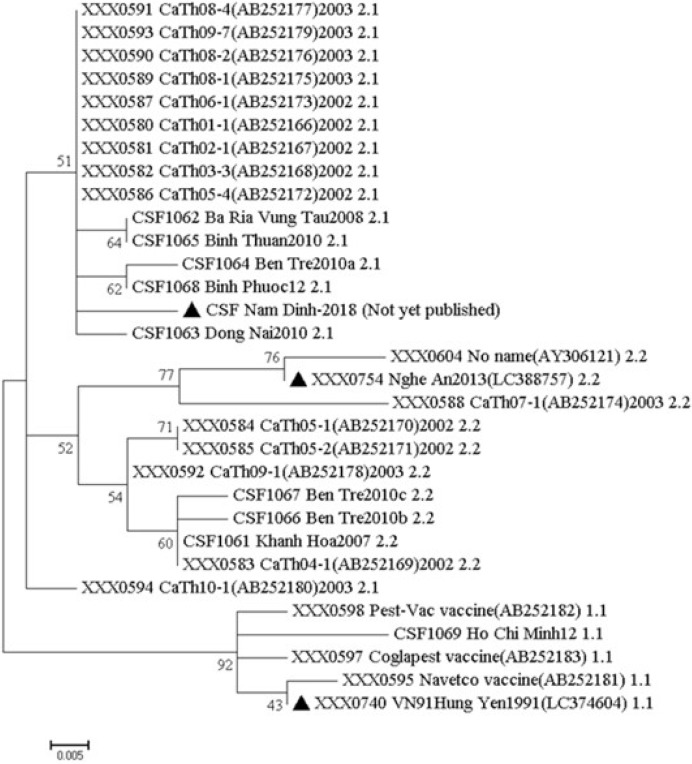
The whole-genome sequences were analyzed by BLAST, the sequences obtained in the study showed identity with sequences of other Vietnam isolates of CSFV and references genomes. The maximum identity was to sub-genotype 1.1 isolates with VN91, sub-genotype 2.1 isolates with NAM DINH and sub-genotype 2.2 isolates with RAHO3/NA. First, the identity of whole-genome sequences of three CSFV strains with other CSFV in Vietnam sequences ranged from 91% to 98%. Second, the identification of whole-genome sequences between CSFV isolates VN91, NAM DINH, and RAHO3/NA ranged from 94%–95%. The phylogenetic analysis was carried out using whole-genome sequences as shown in Fig. 1. As shown in Fig. 1, the VN91 strain indicated 72.0% to 99.0% genetic similarities with other viruses belonging to genotype 1.1, showing a strikingly high similarity (99.0%) with an attenuated vaccine strain, HCLV (GenBank accession no., AF091507) [18]. The RAHO3/NA and NAM DINH CSFV strain indicated 94%–100% and 92%–100% genetic similarities with other viruses belonging to genotype 2.2 and 2.1, respectively. Moreover, the genetic similarity between RAHO3/NA, NAM DINH CSFV strain with an attenuated vaccine strain indicated 92% and 94.1%, respectively. These results indicate three circulating genotypes in Vietnam, including 1.1, 2.1, and 2.2. The vaccine currently, being used in Vietnam is derived from the CSFV strain that should be isolated in Vietnam and studies have to be carried out to ascertain whether this strain of CSFV provides complete protection against those currently circulating CSF viruses in Vietnam. The whole-genome sequences of VN91 and RAHO3/NA CSFV strain have published in GenBank as an accession number LC374604 and LC388757, respectively. For NAM DINH strain sequence is not yet published.
Fig. 1. Phylogenetic tree based on the gene sequences of three classical swine fever virus that are isolated from recently outbreak in Vietnam. The tree was constructed using the neighbor-joining method with a bootstrap value of 1,000 in the MEGA7 program.
Field VN91 strain served as “template” for production of recombinant E2 proteins
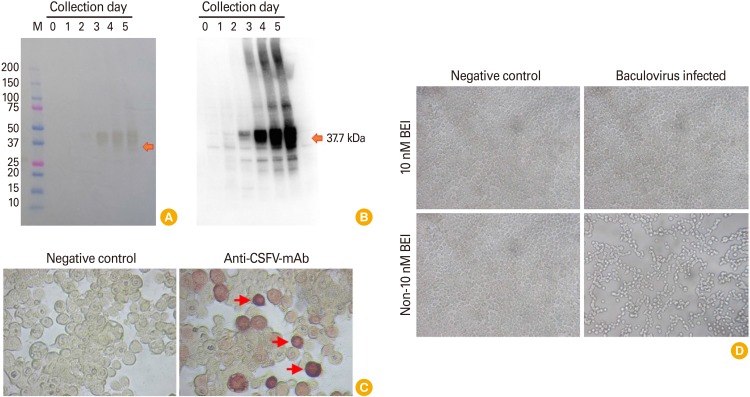
Field VN91 strain served as “template” to produce recombinant E2 protein using the baculovirus expression system. The biological properties of E2 protein were further evaluated by western blot assay and immunoperoxidase monolayer assay (IPMA) in TN5 insect cell, respectively. As shown in Fig. 2A and B, recombinant VN91-E2 protein in the supernatant appeared at day 2 after recombinant baculovirus infection into SF21AE cells and a higher affinity to mAb CSFV on day 5 culture than other days and recombinant VN91-E2 protein appeared to be approximately 40 kDa.
Fig. 2. Production and characterization of recombinant CSFV VN91-E2 protein. (A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis. From left to right: lane 1: protein marker; lane 2–7: Sf21AE cell culture supernatant was collected at day 0, 1, 2, 3, 4, and 5 after infected with baculovirus contained VN91-E2 gene. (B) Western blot of VN91-E2 protein. From left to right: lane 1–6: Sf21AE cell culture supernatant was collected at day 0, 1, 2, 3, 4, and 5 after infected with baculovirus contained VN91-E2 gene. E2-specific mAb CSFV was used for the western blot. (C) Identification of baculovirus contained E2 CSFV at 3 days in TN5 cell cultured by Immunoperoxidase Monolayer Assay (×200). Specific cytoplasmic was shown in the TN5 cells infected with baculovirus contained E2 CSFV (arrows). (D) Sf21AE infected baculovirus contained VN91-E2 at 5 days was inactivated by 10 nM BEI in 48 hours. CSFV, classical swine fever virus; BEI, binary ethylenimine.
Further analysis by IPMA in TN5 cells was also performed as seen in Fig. 2C in which a strong positive result was noted in the well incubated with anti-CSFV antibodies whereas no signal was observed in negative control using SPF serum (Fig. 2C), reflecting the biological characterizes of recombinant E2 immunogen. Further examination on immunological sites is required to ensure that this protein is a candidate for subunit vaccine production against three circulating genotypes in Vietnam, including 1.1, 2.1, and 2.2.
Baculovirus contains CSFV VN91-E2 gene expression in insect cell were inactivated by 10 nM BEI in 48 hours (Fig. 2D). The analysis by microscopy resulted in the detection of pfu of CSFV VN91-E2 gene in an insect cell and that the insect cell infected with baculovirus and inactivated by 10 nM BEI in 48 hours were similar to the untreated negative controls (Fig. 2D). Antigenicity was also preserved, according to the results obtained by western blot (data not shown). These results indicate the adequacy of BEI to maintain the recombinant E2 protein. The absence of any residual active baculovirus was further confirmed by viral experiments. Sf21AE cells were incubated with inactivated baculovirus contain CSFV VN91-E2 diluted 1:10 in the culture medium. After 3–4 days, the supernatant was collected and used to infect fresh Sf21AE cell monolayers. At the end of the third passage, the culture medium was titrated, but no baculovirus plaques were detected, indicating total inactivation of the virus.
VN91-E2-immunized pigs were clinical, pathological, and hematological protected from classical swine fever disease
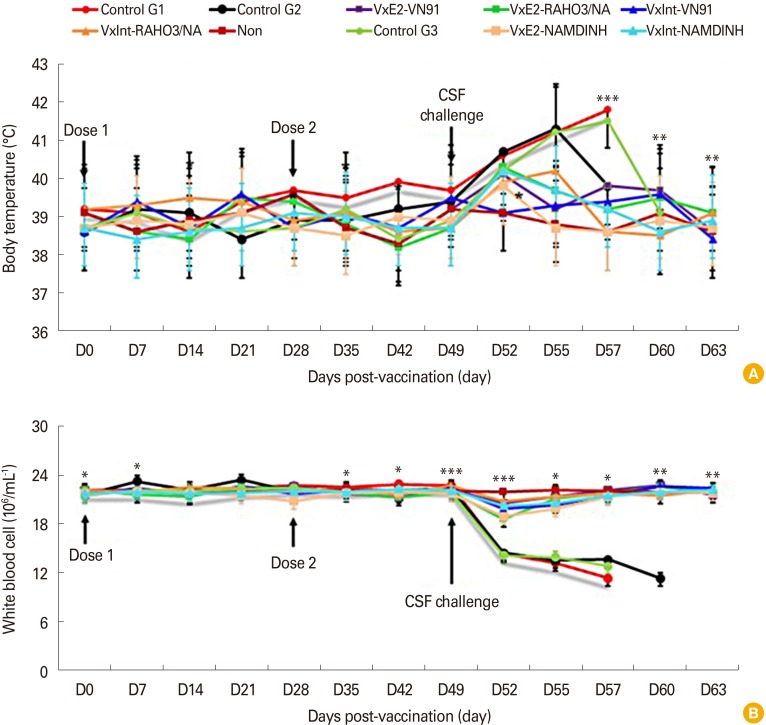
To test the efficacy of CSFV VN91-E2 subunit vaccine, three groups of pigs were immunized with VN91-E2 subunit vaccine with 107 pfu/mL dose of inactivated recombinant baculovirus in the presence of Freund adjuvants and three groups of the pig were immunized with commercial CSFV vaccine, and four groups of the pig were not immunized with an antigen. Pigs were challenged with 1×105 TCID50 of CSFV VN91, RAHO3/NA, and NAM DINH strain at 5 weeks after second dose post-vaccination (D49) (Table 1). The clinical signs and body temperature were monitored during the first 7 days of first dose vaccination and 3 days of second dose vaccination. As shown in Fig. 3A, all VN91-E2 and commercial vaccinated pigs remained healthy and did not develop fever throughout the challenge with three strains of CSFV (Fig. 3A), and only slightly increased body temperature (maximum 39.7℃) after 3-day virus challenge. No body temperature abnormalities were detected before the end of the trial.
Fig. 3. Pigs immunized with VN91-E2 subunit vaccine were protected clinically from CSFV challenge. Pigs were immunized with VN91-E2 on day 0 for the first dose group and a second dose on day 14. Five weeks after the second vaccination (D49), pigs were challenged with 5×105 TCID50 CSFV strain VN91, RAHO3/NA, and NAM DINH. (A) Body temperature was monitored daily after CSFV infection. Vaccinated pigs did not have body temperatures higher than 40℃. (B) Pigs vaccinated with VN91-E2 were protected from CSFV-induced white blood cells. Each symbol represents the mean±standard error. CSF, classical swine fever; CSFV, classical swine fever virus. The significant difference in the body temperatures, number of white blood cell between vaccinated and control groups were indicated with *p<0.01, **p<0.001, and ***p<0.0001.
In contrast, the control pigs developed fever with temperature values, above 40℃ between 3 and 10 days after challenge, as well as clinical signs (diarrhea, hemorrhages in skin, loss of body weight, unable to walk, anorexia, depression, paresis, and paralysis posterior). In addition, the control group G1 challenge with CSFV-VN91 had to be euthanized prior from 7 to 10 days after virus infection (Table 2). Coversely, the control group G2 and G3 challenge with CSFV RAHO3/NA and NAM DINH were killed when moribund from 7 to 12 days after challenge (Table 3). Organ samples from all pigs in three control groups were positive by virus isolation and RT-PCR from spleen samples indicating the presence of CSFV (Table 2). WBC counts in three control groups of pigs challenged with three strain of CSFV were dramatically reduced after CSFV inoculation and then deceased until before being euthanized (Fig. 3B). In contrast, there was a slightly decreased in the numbers of WBC in pigs vaccinated with VN91-E2 and commercial vaccine at D52; however, the numbers of these cells in both of vaccinated pigs increased significantly at D55 and then after (Fig. 3B). Taken together, our data suggest that VN91-E2 subunit vaccine immunized pigs are clinically and hematological protected against three genotypes of CSFV that isolated in Vietnam including genotype 1.1, 2.1, and 2.2 at least 2 weeks post-vaccination.
Table 2. CSFV-neutralizing antibody titers in pig sera before and after CSFV challenge.
| Group | Pig # | D0 | D28 | D35 | DPC0 | DPC7 | DPC14 | Daya) |
|---|---|---|---|---|---|---|---|---|
| VxE2+VN91 | 21 | 0 | 32 | 128 | 256 | 512 | 14 | |
| 22 | 0 | 32 | 128 | 256 | 512 | 14 | ||
| 23 | 0 | 16 | 64 | 256 | 256 | 14 | ||
| 24 | 0 | 64 | 128 | 512 | >512 | 14 | ||
| 25 | 0 | 32 | 64 | 128 | 512 | 14 | ||
| VxE2+RAHO3/NA | 26 | 0 | 64 | 128 | 512 | 512 | 14 | |
| 27 | 0 | 64 | 256 | 512 | >512 | 14 | ||
| 28 | 0 | 32 | 64 | 256 | 256 | 14 | ||
| 29 | 0 | 16 | 64 | 256 | 512 | 14 | ||
| 30 | 0 | 32 | 128 | 256 | 512 | 14 | ||
| VxE2+Nam Ðịnh | 31 | 0 | 64 | 128 | 128 | 256 | 14 | |
| 32 | 0 | 64 | 128 | 512 | >512 | 14 | ||
| 33 | 0 | 32 | 64 | 128 | 512 | 14 | ||
| 34 | 0 | 32 | 64 | 256 | 512 | 14 | ||
| 35 | 0 | 64 | 128 | 256 | 512 | 14 | ||
| Non | 41 | 0 | 0 | 0 | 0 | 0 | 14 | |
| 42 | 0 | 0 | 0 | 0 | 0 | 14 | ||
| 43 | 0 | 0 | 0 | 0 | 0 | 14 | ||
| 44 | 0 | 0 | 0 | 0 | 0 | 14 | ||
| 45 | 0 | 0 | 0 | 0 | 0 | 14 | ||
| Control G1+VN91 | 46 | 0 | 0 | 0 | 0 | 0 | 8 | |
| 47 | 0 | 0 | 0 | 0 | 0 | 7 | ||
| 48 | 0 | 0 | 0 | 0 | 8 | 10 | ||
| 49 | 0 | 0 | 0 | 0 | 0 | 8 | ||
| 50 | 0 | 0 | 0 | 0 | 8 | 9 | ||
| Control G1+RAHO3/NA | 51 | 0 | 0 | 0 | 0 | 8 | 8 | |
| 52 | 0 | 0 | 0 | 0 | 0 | 7 | ||
| 53 | 0 | 0 | 0 | 0 | 8 | 9 | ||
| 54 | 0 | 0 | 0 | 0 | 0 | 8 | ||
| 55 | 0 | 0 | 0 | 0 | <8 | 11 | ||
| Control G1+Nam Ðịnh | 56 | 0 | 0 | 0 | 0 | 0 | 8 | |
| 57 | 0 | 0 | 0 | 0 | 0 | 8 | ||
| 58 | 0 | 0 | 0 | 0 | <8 | 12 | ||
| 59 | 0 | 0 | 0 | 0 | 0 | 7 | ||
| 60 | 0 | 0 | 0 | 0 | 0 | 7 |
CSFV, classical swine fever virus; DPC, day post-challenge.
a)The last day of serum in vaccinated and unvaccinated pig.
Table 3. Results obtained from control group G1, G2, and G3 (non-vaccinated and challenged at 7 weeks of age) at euthanization.
| Group | Pig # | Euthanization | Virus isolation from spleen | RNA viral from spleen | Note | |||
|---|---|---|---|---|---|---|---|---|
| Day | Body temperature (℃) | Clinical scores | White blood cell (106/mL-1) | |||||
| Control G1 | 46 | 8 | 41.2 | 16 | 14.3 | Positive | Positive | No vaccine, challenge at day 49 with VN91 |
| 47 | 7 | 40.9 | 16 | 14.1 | Positive | Positive | ||
| 48 | 10 | 41.9 | 15 | 13.5 | Positive | Positive | ||
| 49 | 8 | 41.2 | 16 | 13.6 | Positive | Positive | ||
| 50 | 9 | 41.8 | 17 | 11.2 | Positive | Positive | ||
| Control G2 | 51 | 8 | 40.7 | 15 | 11.1 | Positive | Positive | No vaccine, challenge at day 49 with RAHO3/NA |
| 52 | 7 | 40.8 | 9 | 13.1 | Positive | Positive | ||
| 53 | 9 | 41.4 | 9.8 | 13.7 | Positive | Positive | ||
| 54 | 8 | 40.6 | 11 | 14.1 | Positive | Positive | ||
| 55 | 11 | 40.5 | 16 | 11.5 | Positive | Positive | ||
| Control G3 | 56 | 8 | 40.6 | 15 | 13.5 | Positive | Positive | No vaccine, challenge at day 49 with NAM DINH |
| 57 | 8 | 40.8 | 10 | 12.6 | Positive | Positive | ||
| 58 | 12 | 41.1 | 11 | 13.5 | Positive | Positive | ||
| 59 | 7 | 40.2 | 13 | 13.5 | Positive | Positive | ||
| 60 | 7 | 40.7 | 16 | 12.8 | Positive | Positive | ||
The VN91-E2 subunit and commercial vaccinated group did not exhibit adverse reactions post-vaccination and virus challenge. Slight depression and reduced appetite were observed in two piglets on day 3 virus challenge. These mild symptoms were observed up to day 5 virus challenge. The maximum mean CS of 1.1 was reached at day 7 virus challenge (Fig. 4A). None of the piglets from three groups of VN91-E2 subunit and commercial vaccine displayed signs of disease during the 63 days of the experiment (Fig. 4A, B). The three mock-vaccinated groups were severely affected post-challenge, and all piglets of control group G1 infected with CSFV/VN91 were euthanized before the termination of the experiment and the maximum mean CS of 17 was reached at day 10 virus challenge (Fig. 4A, B, Table 2). The control group G2 and G3 infected with CSFV/RAHO3/NA and CSFV/NAM DINH respectively were euthanized and killed when moribund from 7 to 12 days after challenge (Fig. 4A, B, Table 2). The maximum mean CS of 15 and 16 was reached at day 11, and 12 viruses challenge in the control group G2 and G3, respectively (Fig. 4A, B, Table 2).
Fig. 4. Clinical scores (A) and survived (B) recorded the classical swine fever virus challenge. Each symbol represents the mean±standard error. The significant difference in the body temperatures, number of white blood cell between vaccinated and control groups were indicated with *p<0.01, **p<0.001, and ***p<0.0001.
Pigs vaccinated with VN91-E2 developed high levels of E2-specific and neutralizing antibodies before and after classical swine fever virus challenge
Sera collected from the vaccinated and non-vaccinated pigs were analyzed by blocking enzyme-linked immunosorbent assay (Fig. 5). As shown in Fig. 5, all vaccinated pigs developed E2-specific antibody after immunization and antibody levels appeared early at day 24, and a significant increase was observed at day 28 (p<0.05). This response reached the pick at day 35 and continued until day 63 when compared to control (p<0.001). As expected, pigs immunized with commercial vaccine-induced antibodies at day 24 and continued until day 63 whereas no signal was noted in control group (Fig. 5). Moreover, the level of specific E2-antibody of all vaccinated pigs was significantly increased after challenge with three genotype CSFV virus (Fig. 5), and E2-specific antibody was not detected in control pigs before or after the challenge with three genotypes of CSFV (Fig. 5). Importantly, our results indicated that VN91-E2 subunit vaccinated pig groups did not show CS, change of WBC or no live circulating of CSFV and our data may be demonstrated that the levels of E2 antibodies in VN91-E2 pigs vaccinated were sufficient for clinical protection against different circulating genotypes of CSFVs isolated in Vietnam including genotype 1.1, 2.1, and 2.2 at least 2 weeks post-vaccination.
Fig. 5. Results obtained by blocking enzyme-linked immunosorbent assay and depicted as average OD% with a cut-off value of 40 (red dashed line). Results ≥40 are positive for CSFV antibodies and results <40 are negative for CSFV antibodies. Each symbol represents the mean±standard error. OD, optical density; CSF, classical swine fever; CSFV, classical swine fever virus. The significant difference in the OD% between vaccinated and control groups were indicated with *p<0.01, **p<0.001, and ***p<0.0001.
Since an essential factor in preventing and control CSFV infection is the production of neutralizing antibodies that block CSFV infection to target cells, we measured the level of CSFV-neutralizing antibodies from vaccinated or unvaccinated animals in each group (Table 2). All vaccinated groups had produced CSFV-neutralizing antibodies on the day of before challenge (D35) and day of challenge (D49) that persisted throughout the day of post-challenge (D63/DPC14) with three genotypes of CSFV challenge (CSFV VN91, RAHO3/NA, and NAM DINH strains). Pigs in the control groups had no detectable neutralizing antibody at day 35 vaccination, day of challenge (D49) and 7&14DPC (Table 2). Thus, our results indicated that the high correlation between anti-CSFV neutralizing antibody titers and levels of E2-specific antibodies in pigs after VN91-E2 vaccination and three genotypes of CSFV challenge.
Virus isolation and viral RNA detection by reverse transcription polymerase chain reaction
In the control groups, CSF virus was isolated from the spleen of five piglets from each of control group G1, G2, and control group G3 on the day of euthanized after challenge with CSFV VN91, RAHO3/NA, and NAM DINH strain (Table 3). In contrast, no CSFV was isolated from the spleen of the pig vaccinated VN91-E2 subunit or commercial vaccine when they were added to PK15 cells for virus isolation (data not shown). Based on the RT-PCR, viral CSF RNA could not be detected in all of the pig vaccinated with VN91-E2 subunit and commercial vaccines (data not shown). Viral CSFV RNA was detected in the spleen samples of all piglets from the three control groups G1, G2, and G3 on day euthanized after challenge with CSFV VN91, RAHO3/NA, and NAM DINH strain (Table 3). No viral CSFV RNA was detected in the remaining piglets from group Non at the end of the experiment (data not shown).
Discussion
CSF is one of the major viral diseases affecting livestock in Vietnam. The outbreak of the disease is notifiable to the OIE, and it has a devastating effect on the swine industry in the world. The high mortality and reduced pig production of the CSF disease that causes the loss of economic to the farmers involved in pig farming [5]. CSFV outbreaks can be effectively controlled and managed only by the timely detection of the disease in the field [27]. Molecular epidemiological studies on CSF are performed by assessing the genetic dissimilarities of the different CSFV isolates. Amplification of the CSFV nucleic acid was performed by RT-PCR, nucleotide sequencing and genetic typing that are commonly used to the analysis of the CSF isolates. The different regions of the CSFV genome such as E2, 5′ UTR, and NS5B are widely used for the study of phylogenetic trees and subsequent typing [28,29]. Our results indicated that CSFV-VN91, RAHO3/NA, and NAM DINH isolate was found to have 72%–99%, 94%–100%, and 92%–100% identity at nucleotide level among the isolates of genotypes 1.1, 2.1, and 2.2 in Vietnam, respectively, and 99%, 92%, and 94.1% nucleotide identity with an attenuated vaccine strain, HCLV (GenBank accession no., AF091507) used in the country for control of the disease. The test isolates CSFV-RAOH/NA and NAM DINH were segregated to genotypes 2.1 and 2.2 and did not seem to be related to the vaccine strains (genotype 1.1) used in the country which indicate the lower matches with the live vaccine virus strain being used in the country.
The CSFV commercially subunit vaccines are available and only in a few countries used it to prevent, and control of CSFV that include Porcilis Pesti vaccine made from Intervet, Boxmeer and BAYOVAC CSF E2 produced from Bayer, Leverkusen, Germany [12,30]. The BAYOVAC-CSFV-E2 antigen was used genotype 1.2 of CSFV Brescia strain [31]. With a single dose of vaccination and when challenged with homologous CSFV strain, it can dramatically reduce mortality of pigs up to 13 months but did not protect pigs from developing a fever [32,33]. On the other hand, the E2 antigen in Porcilis Pesti was cloned from genotype 2.3 of CSFV Alfort/Tubingen strain [34] and a single dose of vaccination with Porcilis Pesti shown that the pigs were failed to protect and prevent horizontal transmission of CSFV genotype 1.1 strain Alfort/187, to unvaccinated sentinel pigs [30]. The attenuated CSFV vaccine HCLV strain is well-known for its efficacy, but it's not safe enough and does not allow DIVA, so the CSFV E2 gene of genotype 1.1 was cloned and expressed with the insect cell/baculovirus system in this study. Our results have demonstrated that CSF VN91-E2, composed of a recombinant E2 protein that cloned from genotype 1.1 isolated in Vietnam and expressed by insect cells, and a complete/incomplete adjuvant, is safe to administer and can effectively stimulate protective immunity against CSFV with a two-dose vaccination. Moreover, our present data showing that two-dose vaccination of VN91-E2 antigen can protect pigs from different genotype (1.1, 2.1, and 2.2) CSFV challenge which is circulating in Vietnam. Pigs vaccinated with VN91-E2 did not exhibit any fever, clinical or mortality after CSFV challenge with three genotypes until day 14 of a challenge. CSFV was not detected in the spleen of vaccinated pigs 14 days post-challenge by viral isolation or RT-PCR. Our report demonstrates that subunit vaccine VN91-E2 is safe and effective at stimulating immunity against heterologous and geographically with different genotypes are circulating in Vietnam. It has been reported recently, that E2 homodimers have higher affinity to E2-specific mAb CSF than do the monomers, suggesting that glycosylation of E2 protein is important for the induction of protective immune response and neutralizing antibodies [10]. Moreover, other research demonstrated that sows vaccinated with E2 proteins from CSFV genotype 1.2 were better protected against clinical CSFV than sows vaccinated with E2 proteins from CSFV genotype 2.3 when these pigs were challenged with CSFV genotype 2.1 [35]. Our results demonstrated that the specific antibodies and CSFV-neutralizing antibodies responses induced by VN91-E2 antigen appeared at day 24 after first boot and a significant increase was observed at day 28 and reached the pick at day 35 and continued until day 49 when compared to control. Importantly, VN91-E2 induced specific antibodies and neutralizing antibodies protected experimental pigs against high virulence of CSFVs circulating in Vietnam, including genotype 1.1, 2.1, and 2.2. We suggest that the high levels of VN91-E2-specific antibodies and CSFV-neutralizing antibodies prior to challenge in pigs may accelerate the inactivation and removal of circulating CSFV post-challenge. This may lead to less or fewer, clinical signs and live CSFV available for further immunological stimulation. In future work, we will carefully examine the early onset of protection, duration of immunity, kinds of pig, viral shedding, and typical criteria for evaluating complete protection in vaccinated pigs. Evaluation of those aspects will be significantly critical in a CSFV outbreak where viral replication needs to be fully controlled, prevent, and eliminated quickly.
In conclusion, the CSFV VN91-E2 protein was successfully generated by insect cell/baculovirus expression system and formulated with a Freund's Adjuvant Complete or Incomplete Adjuvant as a CSF subunit vaccine (VN91-E2) for pigs. Pigs immunized with two-dose vaccination of VN91-E2 can be clinical, symptom and mortality protected from of CSFV challenge with three different genotypes (1.1, 2.1, and 2.2) which are circulating in Vietnam. VN91-E2 subunit vaccine will candidate for pig national vaccination program because which have been targeted for vaccine development and designed to meet the DIVA. VN91-E2 subunit vaccine can be manufactured in CSF-free in Vietnam and is suitable for CSF prevention and control in the endemic area and emergency outbreaks of CSF in Vietnam.
Footnotes
No potential conflict of interest relevant to this article was reported.
This work was supported by the Ministry of Agriculture and Rural Development of Vietnam through the National Program of Biotechnology Application on Agriculture and Aquaculture (726/2017/HÐ-KHCN-CNSH) to Ha Thi Thanh Tran. The author also thanks Andrew G. Yersin, Ph.D. of Kemin Industries, Inc. Scott Avenue Des Moines, IA 50317, USA, for critical review and English improved of this manuscript.
References
- 1.Moennig V, Floegel-Niesmann G, Greiser-Wilke I. Clinical signs and epidemiology of classical swine fever: a review of new knowledge. Vet J. 2003;165:11–20. doi: 10.1016/s1090-0233(02)00112-0. [DOI] [PubMed] [Google Scholar]
- 2.Paton DJ, McGoldrick A, Greiser-Wilke I, et al. Genetic typing of classical swine fever virus. Vet Microbiol. 2000;73:137–157. doi: 10.1016/s0378-1135(00)00141-3. [DOI] [PubMed] [Google Scholar]
- 3.Gomez-Villamandos JC, Carrasco L, Bautista MJ, et al. African swine fever and classical swine fever: a review of the pathogenesis. Dtsch Tierarztl Wochenschr. 2003;110:165–169. [PubMed] [Google Scholar]
- 4.Paton DJ, Greiser-Wilke I. Classical swine fever: an update. Res Vet Sci. 2003;75:169–178. doi: 10.1016/s0034-5288(03)00076-6. [DOI] [PubMed] [Google Scholar]
- 5.Blome S, Staubach C, Henke J, Carlson J, Beer M. Classical swine fever-an updated review. Viruses. 2017;9 doi: 10.3390/v9040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blome S, Meindl-Böhmer A, Loeffen W, Thuer B, Moennig V. Assessment of classical swine fever diagnostics and vaccine performance. Rev Sci Tech. 2006;25:1025–1038. [PubMed] [Google Scholar]
- 7.Van Oirschot JT. Vaccinology of classical swine fever: from lab to field. Vet Microbiol. 2003;96:367–384. doi: 10.1016/j.vetmic.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Ganges L, Barrera M, Nunez JI, et al. A DNA vaccine expressing the E2 protein of classical swine fever virus elicits T cell responses that can prime for rapid antibody production and confer total protection upon viral challenge. Vaccine. 2005;23:3741–3752. doi: 10.1016/j.vaccine.2005.01.153. [DOI] [PubMed] [Google Scholar]
- 9.Laughlin RC, Madera R, Peres Y, et al. Plant-made E2 glycoprotein single-dose vaccine protects pigs against classical swine fever. Plant Biotechnol J. 2019;17:410–420. doi: 10.1111/pbi.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madera R, Gong W, Wang L, et al. Pigs immunized with a novel E2 subunit vaccine are protected from subgenotype heterologous classical swine fever virus challenge. BMC Vet Res. 2016;12:197. doi: 10.1186/s12917-016-0823-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munoz-Gonzalez S, Sordo Y, Perez-Simo M, et al. Corrigendum to “Efficacy of E2 glycoprotein fused to porcine CD154 as a novel chimeric subunit vaccine to prevent classical swine fever virus vertical transmission in pregnant sows”. Vet Microbiol. 2018;213:143–149. doi: 10.1016/j.vetmic.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Ahrens U, Kaden V, Drexler C, Visser N. Efficacy of the classical swine fever (CSF) marker vaccine Porcilis Pesti in pregnant sows. Vet Microbiol. 2000;77:83–97. doi: 10.1016/s0378-1135(00)00265-0. [DOI] [PubMed] [Google Scholar]
- 13.Blome S, Wernike K, Reimann I, Konig P, Mob C, Beer M. A decade of research into classical swine fever marker vaccine CP7_E2alf (Suvaxyn(R) CSF Marker): a review of vaccine properties. Vet Res. 2017;48:51. doi: 10.1186/s13567-017-0457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eble PL, Geurts Y, Quak S, et al. Efficacy of chimeric Pestivirus vaccine candidates against classical swine fever: protection and DIVA characteristics. Vet Microbiol. 2013;162:437–446. doi: 10.1016/j.vetmic.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez O, Barrera M, Farnos O, et al. Effectiveness of the E2-classical swine fever virus recombinant vaccine produced and formulated within whey from genetically transformed goats. Clin Vaccine Immunol. 2014;21:1628–1634. doi: 10.1128/CVI.00416-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Aarle P. Suitability of an E2 subunit vaccine of classical swine fever in combination with the E(rns)-marker-test for eradication through vaccination. Dev Biol (Basel) 2003;114:193–200. [PubMed] [Google Scholar]
- 17.Suarez M, Sordo Y, Prieto Y, et al. A single dose of the novel chimeric subunit vaccine E2-CD154 confers early full protection against classical swine fever virus. Vaccine. 2017;35:4437–4443. doi: 10.1016/j.vaccine.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 18.Tran HT, Dang HV, Nguyen DT, Miyazawa K, Kokuho T. Complete genome sequencing of a classical swine fever virus strain endemic in Vietnam. Genome Announc. 2018;6 doi: 10.1128/genomeA.00307-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Troutt AB, McHeyzer-Williams MG, Pulendran B, Nossal GJ. Ligation-anchored PCR: a simple amplification technique with single-sided specificity. Proc Natl Acad Sci U S A. 1992;89:9823–9825. doi: 10.1073/pnas.89.20.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becher P, Orlich M, Thiel HJ. Complete genomic sequence of border disease virus, a pestivirus from sheep. J Virol. 1998;72:5165–5173. doi: 10.1128/jvi.72.6.5165-5173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roldao A, Oliveira R, Carrondo MJ, Alves PM. Error assessment in recombinant baculovirus titration: evaluation of different methods. J Virol Methods. 2009;159:69–80. doi: 10.1016/j.jviromet.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Rueda P, Fominaya J, Langeveld JP, Bruschke C, Vela C, Casal JI. Effect of different baculovirus inactivation procedures on the integrity and immunogenicity of porcine parvovirus-like particles. Vaccine. 2000;19:726–734. doi: 10.1016/s0264-410x(00)00259-0. [DOI] [PubMed] [Google Scholar]
- 24.Mittelholzer1 C, Moser2 C, Tratschin JD, Hofmann MA. Analysis of classical swine fever virus replication kinetics allows differentiation of highly virulent from avirulent strains. Vet Microbiol. 2000;74:293–308. doi: 10.1016/s0378-1135(00)00195-4. [DOI] [PubMed] [Google Scholar]
- 25.Meyer D, Loeffen W, Postel A, Fritsche S, Becher P. Reduced specificity of E(rns) antibody ELISAs for samples from piglets with maternally derived antibodies induced by vaccination of sows with classical swine fever marker vaccine CP7_E2alf. Transbound Emerg Dis. 2018;65:e505–e508. doi: 10.1111/tbed.12795. [DOI] [PubMed] [Google Scholar]
- 26.Biacchesi S, Skiadopoulos MH, Yang L, Murphy BR, Collins PL, Buchholz UJ. Rapid human metapneumovirus microneutralization assay based on green fluorescent protein expression. J Virol Methods. 2005;128:192–197. doi: 10.1016/j.jviromet.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Penrith ML, Vosloo W, Mather C. Classical swine fever (hog cholera): review of aspects relevant to control. Transbound Emerg Dis. 2011;58:187–196. doi: 10.1111/j.1865-1682.2011.01205.x. [DOI] [PubMed] [Google Scholar]
- 28.Rios L, Coronado L, Naranjo-Feliciano D, et al. Deciphering the emergence, genetic diversity and evolution of classical swine fever virus. Sci Rep. 2017;7:17887. doi: 10.1038/s41598-017-18196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang DL, Gong WJ, Li RC, et al. Phylogenetic analysis using E2 gene of classical swine fever virus reveals a new subgenotype in China. Infect Genet Evol. 2013;17:231–238. doi: 10.1016/j.meegid.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Ziegler U, Kaden V. Vaccination of weaner pigs against classical swine fever with the subunit vaccine “Porcilis Pesti”: influence of different immunization plans on excretion and transmission of challenge virus. Berl Munch Tierarztl Wochenschr. 2002;115:267–273. [PubMed] [Google Scholar]
- 31.Hulst MM, Westra DF, Wensvoort G, Moormann RJ. Glycoprotein E1 of hog cholera virus expressed in insect cells protects swine from hog cholera. J Virol. 1993;67:5435–5442. doi: 10.1128/jvi.67.9.5435-5442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouma A, de Smit AJ, de Kluijver EP, Terpstra C, Moormann RJ. Efficacy and stability of a subunit vaccine based on glycoprotein E2 of classical swine fever virus. Vet Microbiol. 1999;66:101–114. doi: 10.1016/s0378-1135(99)00003-6. [DOI] [PubMed] [Google Scholar]
- 33.De Smit AJ, Bouma A, de Kluijver EP, Terpstra C, Moormann RJ. Duration of the protection of an E2 subunit marker vaccine against classical swine fever after a single vaccination. Vet Microbiol. 2001;78:307–317. doi: 10.1016/s0378-1135(00)00306-0. [DOI] [PubMed] [Google Scholar]
- 34.Dong XN, Chen YH. Marker vaccine strategies and candidate CSFV marker vaccines. Vaccine. 2007;25:205–230. doi: 10.1016/j.vaccine.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 35.Depner KR, Bouma A, Koenen F, et al. Classical swine fever (CSF) marker vaccine: trial II: challenge study in pregnant sows. Vet Microbiol. 2001;83:107–120. doi: 10.1016/s0378-1135(01)00410-2. [DOI] [PubMed] [Google Scholar]