Abstract
Despite the lack of confirmed reports of an exogenous Simian betaretrovirus (SRV) isolated from baboons (Papio sp.), reports of simian endogenous gammaretrovirus (SERV) in baboons with complete genomes suggest that such viruses may be potentially infectious. In addition, serologic tests have repeatedly demonstrated antibody reactivity to SRV in baboons from multiple colonies. These findings complicate the management and use of such animals for research. To provide further insight into this situation, we performed in vitro and in vivo studies to determine if baboons are or can be infected with SRV. In our initial experiment, we were not able to isolate SRV from 6 seropositive or sero-indeterminate baboons by coculturing their peripheral blood mononuclear cells (PBMC) with macaque PBMC or permissive cell lines. In a subsequent experiment, we found that baboon PBMC infected in vitro with high dose SRV were permissive to virus replication. To test in vivo infectibility, groups of naive baboons were infused intravenously with either (i) the same SRV tissue culture virus stocks used for the in vitro studies, (ii) SRV antibody positive and PCR positive macaque blood, (iii) SRV antibody positive or indeterminate, but PCR negative baboon blood, or (iv) SRV antibody and PCR negative baboon blood. Sustained SRV infection, as defined by reproducible PCR detection and/or antibody seroconversion, was confirmed in 2 of 3 baboons receiving tissue culture virus but not in any recipients of transfused blood from seropositive macaques or baboons. In conclusion, the data indicate that even though baboon cells can be infected experimentally with high doses of tissue culture grown SRV, baboons that are repeatedly SRV antibody positive and PCR negative are unlikely to be infected with exogenous SRV and thus are unlikely to transmit a virus that would threaten the SPF status of captive baboon colonies.
Abbreviations: SRV, Simian betaretrovirus; BAEV, Baboon endogenous retrovirus; SERV, Simian endogenous retrovirus; PBMC, peripheral blood mononuclear cells
Both exogenous simian Betaretroviruses (SRVs) and simian endogenous retroviruses (SERV) are members of the Retroviridae family. When infectious, these enveloped RNA viruses exhibit a type D retrovirus morphology: an icosahedral capsid composed of an envelope-associated outer shell and an inner ribonucleoprotein core. These viruses were previously known as Simian Retrovirus, type D. As is typical for type D retroviruses, their genome is organized into 4 main coding genes: gag (group specific antigen), Prt (viral protease), Pol (polymerase and endonuclease/integrase enzymes), and Env (external envelope spike and transmembrane glycoproteins). The virus replicates by sequential steps of reverse transcription, integration, transcription, translation, assembly and viral budding from the cell membrane.19,26
While endogenous SRVs have not been associated with active infection, exogenous SRVs have been. In Asian macaques, naturally acquired SRVs (along with the later described simian immunodeficiency viruses introduced from African species) are etiologic agents for simian acquired immunodeficiency syndrome (SAIDS). SRV-3 (also known as Mason–Pfizer Virus) was the first reported SRV prototype. It was isolated from rhesus macaque mammary carcinoma tissue in 1970.6 Since then, at least 6 related serotypes have been isolated from macaques and sufficiently sequenced to confirm their close genetic relation.11,30 SRV serotypes 1, 3, and 5 tend to predominate in rhesus macaques (M. mulatta) of Indian and Asian origin; and SRV-2 is most common in SE Asian island-origin cynomologus macaques (M. fascicularis) and pigtailed macaques (M. nemestrina).20 SRV-4, SRV-8 and other variants have been isolated from cynomologus macaques of mainland SE Asian origin.8,24,31 Although the serotypes share at least some serologic crossreactivity and tissue culture characteristics, each can be distinguished by neutralization as well as serotype specific PCR genetic amplification methods.17,27
Overt SRV infection of macaques is manifested as a wide range of clinical sequelae including anemia, granulocytopenia, lymphopenia, thrombocytopenia, diarrhea, weight loss, splenomegaly, and lymphadenopathy. Likewise, these animals display immunologic perturbations such as suppression of T and B lymphocyte function leading to the downregulation of MHC Class II antigen expression, reduced mitogen-induced proliferation, decreased immunoglobulin production, and other functional defects. Retroperitoneal fibromatosis herpesvirus and Epstein-Barr virus-related lymphocryptovirus coinfections have been reported to result in the SRV-associated tumors such as cutaneous fibrosarcoma-retroperitoneal fibromatosis and B cell lymphomas.11,13 SRV can be readily transmitted horizontally by direct and indirect contact in macaque colonies.13,30 Previous studies have reported prevalence rates ranging from 0% to 50%.19,26 The prevalence of SRV is highly variable, and greatly influenced by the geographic origin of the monkeys, testing program, management, and husbandry practices of a colony. Because of its ability to compromise both NHP colony health and experimental research study data, SRV was one of the original agents targeted for exclusion from SPF (SPF) macaque colonies in the mid-1980s. Elimination of SRV has improved animal health, removed a significant confounding research variable, and reduced exposure risk to animal care personnel.14,30
However, while eradicating SRV from macaque colonies remains the ultimate goal, the natural history of SRV poses a number of challenges and research opportunities to better understand virus-host interactions. Like other persistent infections, after an initial period of viremia lasting weeks to months, SRV can latently infect host cells and become either undetectable, or very rare in the peripheral blood.29 The interval between initial infection and overt disease is characterized by a long asymptomatic stage during which virus can be shed intermittently and transmitted to other animals.16 We observed periodic immune responses to bursts of viral replication leading to increases in antibody titers. Thus antibody can serve as a marker to indicate that the infectious agent is present, even when no virus can be detected. The breadth and intensity of the antibody response to SRV can vary widely over time and differs among animals. In the majority of infected animals, a strong antibody response correlates with persistent, latent infection with very low or undetectable viral DNA in peripheral blood cells. Thus, an antibody positive / virus negative profile in a macaque is considered as evidence of exogenous SRV infection when confirmed by other diagnostic, clinical, or historical findings.30 In a small portion of infected animals, (less than 10%), the virus can replicate to extremely high levels while the host mounts a weak and sometimes undetectable antibody response.15,16 Therefore, as suggested in published guidelines, testing for SRV in colonies of NHPs must include both antibody and direct virus detection.20,30 Specifically, molecular methods such as polymerase chain reaction (PCR) or culture techniques should be used for virus detection, and host antibody responses to the virus should be detected using a combination of a screening test (for example ELISA or multiplex) and a secondary, more stringent confirmatory test (for example immunoblot, or immunofluorescence). Testing at multiple time points is recommended. These diagnostic algorithms have been developed recognizing that closely related endogenous retrovirus sequences can potentially confound detection of exogenous SRVs due to regions of high homology in the genomes, particularly in the core and transmembrane envelope coding regions. Even with these methods, the accurate diagnosis of simian betaretrovirus (SRV) infection is an ongoing diagnostic challenge- even in macaques for which the reagents and assays were designed.29
In contrast to our rather extensive knowledge about SRV in Asian macaques, there is less published information and more uncertainty about SRV in non-Asian macaque species. Other retroviruses including baboon, langur and squirrel monkey endogenous viruses and exogenous SRV-6, 7 have been reported.21-23 Although there are no confirmed reports of an exogenous simian betaretrovirus (SRV) isolated from baboons (Papio sp.), there are reports of endogenous gammaretrovirus (SERV) in baboons with complete genomes. These endogenous viruses have the potential to be infectious.7,25 To date, no SRV isolates have been reported from African primate species, including baboons.
Using reagents developed and validated for SRV 1 to 5 in macaques, the authors (and other NHP testing laboratories) have repeatedly observed apparent antibody reactivity in serum or plasma from baboons from various colonies with no detectable virus or other signs of SRV disease. These findings raise many questions about how to interpret and apply such data toward the management and utility of these valuable animals for research studies. Using the same algorithm developed to diagnose SRV in macaques for baboons could lead to false positive reports of SRV infection and unnecessary exclusion from colony groups and research studies. In efforts to identify any potential infections in colonies that are either SRV negative or have very low prevalence, current SRV antibody diagnostic methods are designed to be very sensitive. However, increasing sensitivity and decreasing prevalence lowers the statistical positive predictive value and may result in increased numbers of false positive results.30 There is a possibility that the host baboon could be making an immune response to endogenous virus, or to a new or baboon specific SRV serotype detectable by cross-reaction with current SRV serology but not molecular reagents.
In an attempt to understand better the meaning and significance of our laboratory findings, both the California and Washington National Primate Research Center laboratories have performed in vitro and in vivo studies to determine if antibody reactivity in baboon species is indicative of infection. The data from these experiments suggest that even though baboon cells can be infected experimentally with SRV, they are not very susceptible to infection in vivo. Thus, baboons that are SRV antibody positive and PCR negative are unlikely to be infected with transmissible, exogenous SRV and do not need to be removed from captive SPF colonies without additional evidence.
Materials and Methods
Animals and procedures.
Male (n = 5) and female (n = 7) juvenile baboons, approximately 2.5 y of age, were used for this study. Groups 1 to 3 included both males and females. Control Group 4 was comprised of 2 females. All animals were maintained in fully AAALAC-accredited facilities in accordance with the Animal Welfare Act, Regulations, and the Guide for the Care and Use of Laboratory Animals.1,2,9 All procedures involving animals used in this study were approved by each institution's IACUC and performed in animal biosafety level 2 (ABSL2) containment facilities in accordance with the Centers for Disease Control and Prevention Biosafety in Microbiologic Laboratories (BMBL) guidelines.4 Animals were fed standard monkey chow twice daily, as well as receiving daily food supplements (fruit and forage) and environmental enrichment (including group housing) among study groups. Trained animal care and veterinary staff monitored animal health daily. Increased monitoring was provided during the 48 h period after blood transfusion. Baboons were sedated at the indicated time points with ketamine (10 mg/kg) and acepromazine (0.05 mg/kg) for blood transfusions, virus administration and/or sample collections. Blood samples were collected via venipuncture of either the saphenous, cephalic, or femoral veins. Transfusion of citrated pooled blood from either olive baboons (Groups 3 and 4) or rhesus macaques (Group 2), 20 mL total, was administered through an intravenous catheter over a time period of 15 to 20 min. Diphenhydramine (5 mg/kg) was administered prior to transfusion to help prevent transfusion reaction to cross-species or unmatched blood types. At the designated time post transfusion, the animals were euthanized according to the recommendations of the American Veterinary Medical Association (2013 panel on euthanasia).3
Antibody.
Microbead Fluorescent ImmunoAssay (MFIA) panels and Western blot (WB) assays were used to detect and semiquantitate antibody. SRV specific antigen targets developed and validated in the California and Washington National Primate Center10 laboratories or available commercially (Charles River Laboratories, Wilmington, MA)5 were used to screen for SRV antibody reactivity using MFIA liquid microbead arrays (Luminex Corp, Austin, TX). These assays use simian betaretrovirus viral lysate and recombinant (rgp20 transmembrane, rgp70 outer membrane, and rgp90 env precursor) antigens. In our laboratories, we have validated that the viral lysate, the rgp 20 and the rgp90 react with SRV 1 to 5 serotypes and the rgp70 is serotype specific. Microbead sets distinguished by specific fluorescent signatures are coated with the target antigens. The microbeads bind antibody that is subsequently detected by binding phycoerythrin conjugated goat antihuman IgG.10 WB strips were prepared using SRV1 and SRV2 viral lysate electrophoresed through a 4% to 15% gradient gel and transblotted onto nitrocellulose membranes. Bound antibody was detected by subsequent incubation with Alkaline Phosphatase conjugated goat antihuman IgG and BCIP/NBT (5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium) and reactivity patterns were compared with molecular weight standards and known positive and negative control sera.
PCR.
Real time PCR was performed using protocols validated on both the BioRad CFX96 (Hercules, CA) and the ABI Thermo Fisher Quant Studio (Foster City, CA). The ABI assay uses primers and probes validated to detect SRV1-5 serotypes and incorporates the housekeeping gene Oncostatin M. The same primers are used in a sybrgreen + melt curve assay on BioRad CFX96. Both assays have limits of detection of one to one hundred copies for SRV1-5. These probes, primers, and cycling conditions have been validated and published and are used regularly for diagnostic work.27
Coculture.
Fresh whole blood was processed using ficoll hypaque (Sigma–Aldrich, St Louis, MO) gradient centrifugation. The resulting peripheral blood mononuclear cells (PBMCs) were washed, and counted and resuspended at 2 × 10+4 cells per ml in RPMI 1640 and cocultured with either Raji cells at 5 × 10+5 in RPMI1640 + 10% Fetal Calf Serum + Pen/strep or macaque PBMCs were supplemented with 20% fetal bovine serum. One set of cultures was stimulated by the addition of 0.5 µg/mL Staphylococcus Enterotoxin A (Toxin Technology, Sarasota, FL); while a second set was not stimulated. After 3 d, the stimulated culture volume was doubled with the addition of either fresh Raji, SupT, or donor PBMC at 5 × 10+5 cells in RPMI1640 with 10% fetal bovine serum.13 Thus, coculture with PBMC, Raji, and SupT cells were attempted on each donor PBMC. The cocultures were observed twice weekly for cytopathic effect (CPE) and supernatant was collected for PCR for up to 8 wk, or until a reproducible positive signal at multiple time points was detected. Fresh media and / or cells were added as needed to maintain healthy conditions for propagation.
Study Outline.
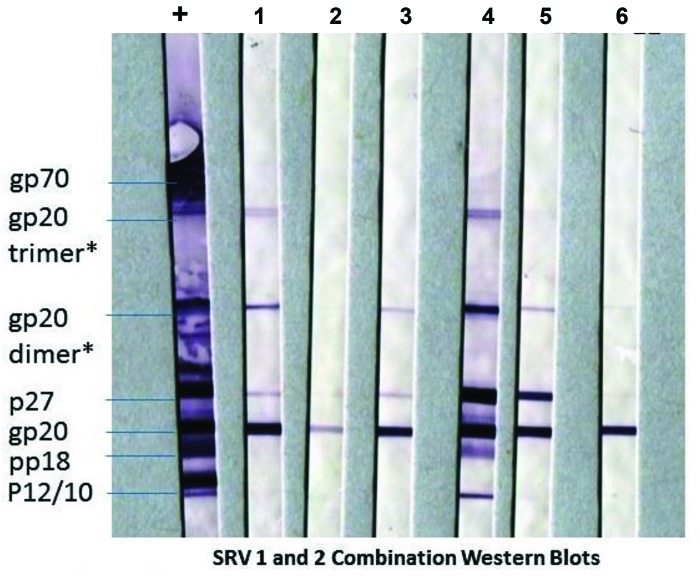
The first phase of this study was an attempt to culture virus from repeatedly seropositive or indeterminate but PCR negative baboons. WB patterns are shown in Figure 1. PBMC from these 6 baboons were cocultured with known uninfected macaque PBMCs, Raji, or SupT cells known to be susceptible to SRV infection. Cultures were observed for CPE and sampled for PCR twice weekly for 8 wk.
Figure 1.
SRV seroreactivity patterns of baboons with unclear infection status. Western blots demonstrate the SRV antibody reactivity of the antibody reactive (positive or indeterminate), but PCR negative baboon donors from which PBMC were initially cultured in an attempt to isolate a virus. The target antigen on the blots is a mixture of SRV1 and SRV2 viral lysate. Each strip was reacted with either serum of an SRV positive macaque control (+) or of individual baboons that were used for the PBMC culture experiment (1-6). All the baboon sera reacted against the transmembrane gp20 and core p27. Although there is not clear reactivity to env gp70 this pattern is sufficient to interpret as indeterminate or positive in most routine SRV testing. *Hypothesized due to reactivity of these bands to mAb specific for gp20.
In the next in vitro phase, PBMCs from 2 SRV antibody and PCR negative baboon blood donors were isolated, stimulated, and inoculated with SRV1 or SRV 2 tissue culture virus in RPMI 1640 media supplemented with 5% fetal calf serum.13 These inocula were pooled from tissue culture supernatants with titers of 100 to 1,000 50% tissue culture infectious doses (TCID50) per mL, that had been used to infect rhesus macaques by intravenous inoculation in past experiments. Fresh media and cells were added as needed; and after 2 wk, the remaining cells were washed twice with PBS and subcultured with fresh, uninfected baboon PBMCs in fresh media and new flasks for an additional week to ensure that any virus detected was not residual input inoculum. The cultures were observed for CPE and tested by SRV PCR twice weekly.
In the final in vivo phase of the study, the infection was attempted via intravenous inoculation as outlined in Figure 2. Three naïve baboons in Group 1 each received 10 mL of tissue culture virus supernatant pooled from the same SRV1 and SRV2 virus stocks used in the in vitro phase. Three naïve baboons in Group 2 were each transfused with 20 mL of ACD anticoagulated blood from one or 2 (pooled) SRV antibody and PCR positive rhesus macaque donors. Group 3 was comprised of 4 naïve baboon recipients that were each transfused with 20 mL of citrated blood from one of 3 antibody-positive or indeterminate but PCR negative baboon donors that were cultured in phase one. (Two recipients received blood from the same donor.) As negative controls, 2 naïve baboons in Group 4 were transfused with 20 mL each of SRV antibody and PCR negative baboon blood from the same single donor. Animals were monitored continuously for 8 h post transfusion/inoculation, multiple times a day for the next 40 h, twice daily for 1 wk, and then daily for the remainder of the study to ensure no adverse reactions occurred that would require immediate clinical intervention after transfusion/inoculation. Blood samples for SRV antibody and PCR testing were collected at days 0, 4, 7, 10, 14, 21, 28, 36, 43, 49, 56 and at the conclusion of the experiment prior to necropsy (greater than 71 d). Coculture with Raji and / or SupT cells was performed on necropsy samples from Group 1 and 2 baboons.
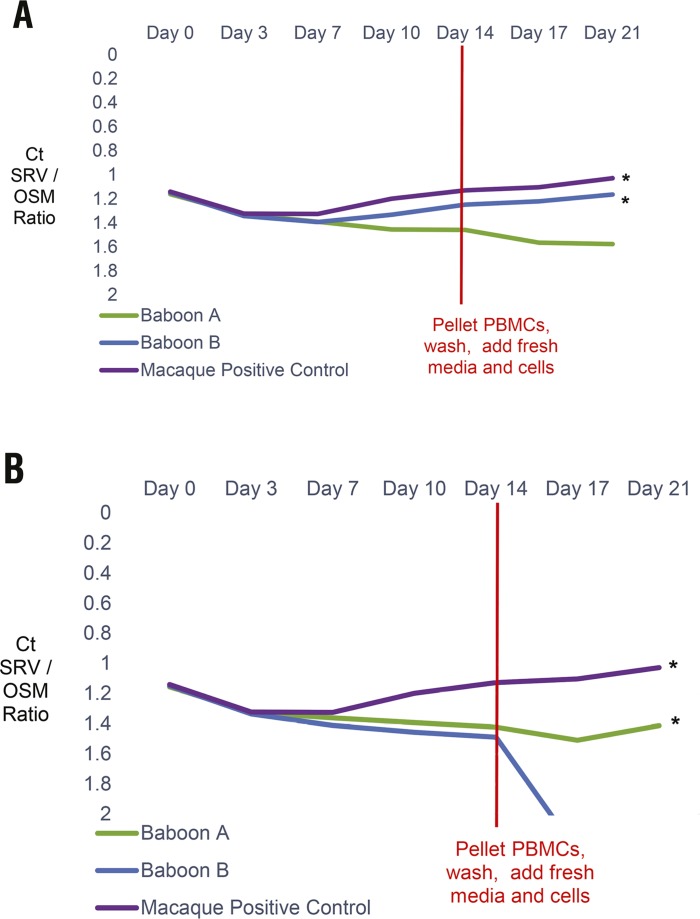
Figure 2.
In vitro infection experiment of baboon PBMC. (A) SRV PCR remained positive for both baboon PBMC cultures inoculated with SRV1 and (B) for one of 2 baboon PBMC cultures inoculated with SRV2, even after the cells were washed and subcultured into fresh cells and media. Rhesus macaque (MMU) PBMC were also inoculated as positive controls. SRV DNA qPCR results are shown as the ratio of cycling threshold (Ct) values for SRV DNA normalized against the Oncostatin M (OSM) internal housekeeping gene Ct values in the same sample and plotted over time. SRV Ct values greater than 55 are interpreted as negative for SRV DNA. A decline in Ct ratio of SRV/OSM, as indicated by the * on the graph, indicates increasing amounts of SRV DNA.
Results
Inability to isolate SRV from baboon PBMC.
In the initial experiments, PBMC from 6 baboons were cultured, observed and sampled for 8 wk. Although some mild cytopathologic effects were noted in a few cultures (Figure S1), no positive SRV-specific PCR signals were detected in any of the culture samples. The animals selected for these culture studies had historically been tested at multiple time points and found to be PCR negative, while simultaneously displaying the western blot antibody indeterminate or positive band patterns shown in Figure 1.
SRV 1 and SRV 2 can infect baboon PBMC in vitro.
Cocultures of PBMCs from 2 SRV antibody and PCR negative baboon donors, inoculated with SRV1 or SRV2 tissue culture supernatant, were observed and tested for SRV by PCR for 2 wk. Even after washing twice and subculturing with fresh uninfected PBMCs in fresh media and new flasks at day 14 to remove any residual input virus, DNA PCR remained positive for SRV1 for both baboon PBMC new subcultures and positive for SRV2 for one of the 2 baboon PBMC new subcultures monitored for an additional week. The SRV viral load continued to increase over time when the SRV cycling threshold (Ct) values were normalized against the housekeeping gene Oncostatin M in the same samples. Rhesus macaque PBMCs were also infected in parallel as a positive control (Figure 2).
In vivo SRV inoculation experiments in baboons.
The results of the experimental in vivo infection attempts are summarized in Figure 3.
Figure 3.
Overview of experimental design and outcome of in vivo study. Four groups of baboons were inoculated with various potential sources of SRV on day 0, with subsequent sample collection. Necropsy (Nx) was performed after day 71. Culture on necropsy samples was performed on samples from Groups 1 and 2. No cultures (NT- not tested) were done on Groups 3 and 4.
SRV viral DNA was detected by PCR at multiple time points in the Group 1 baboons 1A and 1B that received pooled SRV1 and SRV2 tissue culture supernatant. These same 2 baboons sero-converted at day 36. Baboon 1 C was PCR positive only on day 14 and did not seroconvert. The necropsy cocultures with Raji and SupT cells observed for CPE and sampled for PCR for up to 8 wk were negative for 1 B and 1 C and positive for 1 A with SupT cells.
Antibody was first detected immediately after inoculation in the Group 2 baboons that received SRV antibody and PCR positive blood from rhesus macaque donors. However, consistent with a gradual decline of passively administered antibodies, the antibody signal waned to baseline against all antigens by day 36 in baboons 2 A and 2 C; and against all antigens except viral lysate for 2 B. Reactivity to viral lysate in the absence of specific reactivity to any of the recombinant proteins is not interpreted as being SRV specific in the testing algorithms used in our laboratories for macaques. Baboons 2 A and 2 B were PCR positive only at day 10; and no PCR signal was detected in 2C. Baboons 2 A and 2 B were recipients of blood from donor 1 and pooled blood from donors 1 and 2, respectively. Animal 2 C received blood from donor 2. SRV permissive Raji and Sup T cell cocultures with samples taken at necropsy were negative for all 3 baboons.
No SRV positive PCR signals or antibody were detected at any point from day 0 through necropsy (71 d or longer) in any of the Group 3 baboons that received either SRV antibody positive or suspect, but PCR negative, blood. Likewise, no detectable SRV or antibody was found in the Group 4 control baboons that received SRV antibody and PCR negative blood.
Discussion
The goal of the studies reported here was to gain better insights into the virus-host relationship of exogenous SRV in baboons to assist in interpreting unexpected findings during routine virus testing of SPF baboon colonies. In the initial experiments, we demonstrated that baboons that test positive for SRV-binding antibodies did not actually harbor infectious SRV. In subsequent experiments, we were able to experimentally infect baboon cells with SRV in vitro; but, unlike macaques, were not able to productively and sustainably infect seronegative baboons with SRV in vivo by inoculating infected blood. These results are important observations of the possible responses of baboons to attempted SRV infection. However, no further statistical analysis or conclusions related to male/female or any other demographic variables can be made with such small study group sizes. The variable response among the research animals in each group is similar to what has been seen in practice when screening baboons for SRV; and a reason for this study. This important difference between baboons and macaques has direct colony management implications. In macaques, the detection of SRV antibodies is a key indicator of an infection which can give rise to significant pathology and immune perturbation, confounding any experimental results gleaned from studies using these animals. Although SRV reactive antibodies were found in baboons, they did not correlate with infection and there was no evidence of detectable virus or disease.
SRV is an exogenous β-retrovirus pathogen that can be transmitted horizontally in Asian macaques in natural and captive settings; however, the range of species infected by these viruses is not well studied. Given the wide distribution of the β-γ retrovirus envelope receptor, the transmissibility of SRV in bodily fluids and the permissiveness of many species cells in vitro, it is possible that SRV could be transmitted to other species in wild and captive settings, but such widespread distribution has not been reported.
As in humans, there are no reports of naturally SRV infected baboons. In this study, our in vitro attempts to culture SRV from baboon blood with either indeterminate or positive antibody reactivity did not yield any virus. In vivo, we found no culture-able virus, no viral DNA and no ability to transmit an SRV infection from 2 different “seropositive” baboons by IV transfusion of whole blood to 4 negative baboons. We directly assayed for virus using PCR and cell culture, and we assayed for antibody using Luminex bead-based assays and immunoblots over multiple time points. Despite the very similar antibody band patterns and intensity (as shown in Figure 1), our findings indicate that these seropositive baboons are not infected with SRV; they have an unexplained specific pattern of reactivity that is shown by the robust reactivity of their serum antibodies to purified viral proteins on immunoassay in the absence of any evidence of virus infection.
Baboons and other African primate species are not natural hosts of SRV and exposure to macaques in today's strict breeding or research settings is unlikely. However, antibodies that react to purified SRV proteins, specifically the transmembrane glycoprotein, gp20, and the major capsid protein, p27, are present in many baboons, including 2 donor baboons used in the current study. A review of SRV antibody testing results from samples submitted to one of our laboratories has shown that approximately, 13% of baboons from all sources possess antibodies that would be considered SRV positive if detected in macaques. Thus, even some baboons raised in SPF colonies for 2 or 3 generations could appear to be SRV seropositive although they are not known to have had any contact with macaques. An alignment of the env regions of SRV1, SRV2, Baboon endogenous retrovirus (BAEV), and pigtail Simian endogenous retrovirus (SERV) reveals close homology at the C-terminal, leading us to speculate that crossreactivity between antibodies to these targets is possible. A review of the scientific literature documents that BAEV and other related viruses can be infectious in vitro.7,25
It is known that primate chromosomes contain thousands of copies of endogenous retroviral (ERV) genes that comprise more than 8% of the genome and that some of the ERVs in macaques and baboons are homologous to SRV.18,25 Work in rhesus has shown that immune responses to ERVs are present and can be measured.28 Therefore it is possible that baboons are raising antibodies to ERVs that are related to SRV and those antibodies could cross-react in our assays . Additional work is needed to determine the significance of the ERV ENV genes, proteins and antibodies.
Our study does reveal that baboons can be infected with tissue culture virus inoculum but not blood from infected macaques under the experimental conditions of this study. Infection is supported by the detection of positive signals beginning at day 14 for PCR and day 36 for antibody in 2 of the baboons receiving tissue culture virus persisting across multiple time points and even at necropsy in one baboon. In the baboons that received blood from SRV infected macaques, the antibody detected immediately after infusion had waned to baseline and the single positive PCR signal at day 10 in the baboons could most likely represent passive transfer, although one cannot exclude a transient or abortive infection that was not sustained. Future studies should consider the possible role of virus in body fluids such as urine and saliva as well as the host T lymphocyte immune response in transmission attempts. Although infection may be possible; however, it does not appear to be very efficient. It is known that SRV can be grown in a wide variety of mammalian cell lines, including nonhuman and human primates, dogs, cats, mice, bats and more, therefore caution is advised when SRV is present. It is likely that many other species could support SRV infection given the opportunity.
In the process of raising SPF NHP it is necessary to repeatedly test for the presence of select infectious agents and to use the test results to apply careful barrier management in breeding and rearing operations.12,20,30 Of primary importance for elimination from the NHP colonies are the persistent pathogens in the Herpesvirus and Retrovirus families as well as the bacteria in the M. tuberculosis complex. The detection of these infectious agents has multiple challenges, mostly related to the persistent, latent nature of these infections and extremely low levels of the infecting agent. In this study we focused on the host response to SRV and the potential for cross-species infections from macaques to baboons to give a better understanding of the risks associated with the presence of SRV-reactive antibodies in some SPF baboons. Our data indicate that baboons that are serologically reactive to SRV do not harbor detectable SRV. We believe this information will contribute to ensuring that baboons in SPF colonies are not carrying specific infectious agents with potential to interfere with study data or threaten animal or human health.
Supplemental Materials
Summary of cocultures with permissible Raji and Sup T cell lines and SRV antibody reactive, PCR negative donor PBMCs.
Acknowledgments
We thank the members of the laboratory and animal care staff, including Amanda Carpenter, Rebeca Huebner, Robin Watanabe, Nicole Reuter, and Alisha Preno, at our institutions for their project support. This study was partially supported by the Office of Research Infrastructure Programs/OD (P51 OD011107-54, P51 OD010425, P40 OD010431, P40 OD10988, 5U42OD010990).
References
- 1.Animal Welfare Act as Amended. 2013. 7 USC §2131–2159.
- 2.Animal Welfare Regulations. 2013. 9 CFR § 3.129.
- 3.American Veterinary Medical Association. [Internet]. 2013. AVMA guidelines on euthanasia, 2013 ed. [Cited 01 March 2019]. Available at: https://www.avma.org/KB/Policies/Documents/euthanasia.pdf
- 4.Centers for Disease Control. [Internet]. 2018. Centers for disease control and prevention biosafety in microbiological laboratories (BMBL) guidelines. [Cited 01 March 2019]. Available at: https://www.cdc.gov/labs/BMBL.html
- 5.Charles River Laboratories. [Internet]. 2016. Serologic methods manual: Multiplexed Fluorometric ImmunoAssay (MFIA). [Cited 01 March 2019]. Available at: https://www.criver.com/sites/default/files/resources/SerologicMethodsManualMultiplexedFluorometricImmunoAssay%C2%AEMFIA%C2%AE.pdf
- 6.Chopra HC, Mason MM. 1970. A new virus in a spontaneous mammary tumor of a rhesus monkey. Cancer Res 30:2081–2086. [PubMed] [Google Scholar]
- 7.Grant RF, Windsor SK, Malinak CJ, Bartz CR, Sabo A, Benveniste RE, Tsai CC. 1995. Characterization of infectious type D retrovirus from baboons. Virology 207:292–296. 10.1006/viro.1995.1080. [DOI] [PubMed] [Google Scholar]
- 8.Hara M, Sata T, Kikuchi T, Nakajima N, Uda A, Fujimoto K, Baba T, Mukai R. 2005. Isolation and characterization of a new simian retrovirus type D subtype from monkeys at the Tsukuba Primate Center, Japan. Microbes Infect 7:126–131. 10.1016/j.micinf.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Institute for Laboratory Animal Research. 2011. Guide for the care and use of laboratory animals, 8th ed Washington (DC): National Academies Press. [Google Scholar]
- 10.Kuller L, Watanabe R, Anderson D, Grant R. 2005. Development of a whole-virus multiplex flow cytometric assay for antibody screening of a specific pathogen-free primate colony. Diagn Microbiol Infect Dis 53:185–193. 10.1016/j.diagmicrobio.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Lerche NW. 2010. Simian retroviruses: infection and disease—implications for immunotoxicology research in primates. J Immunotoxicol 7:93–101. 10.3109/15476911003657406. [DOI] [PubMed] [Google Scholar]
- 12.Lerche NW, Cotterman RF, Dobson MD, Yee JL, Rosenthal AN, Heneine WM. 1997. Screening for simian type-D retrovirus infection in macaques, using nested polymerase chain reaction. Lab Anim Sci 47:263–268. [PubMed] [Google Scholar]
- 13.Lerche NW, Marx PA, Osborn KG, Maul DH, Lowenstine LJ, Bleviss ML, Moody P, Henrickson RV, Gardner MB. 1987. Natural history of endemic type D retrovirus infection and acquired immune deficiency syndrome in group-housed rhesus monkeys. J Natl Cancer Inst 79:847–854. [PubMed] [Google Scholar]
- 14.Lerche NW, Osborn KG. 2003. Simian retrovirus infections: potential confounding variables in primate toxicology studies. Toxicol Pathol 31 Suppl:103–110. 10.1080/01926230390174977. [DOI] [PubMed] [Google Scholar]
- 15.Lerche NW, Yee JL, Jennings MB. 1994. Establishing specific retrovirus-free breeding colonies of macaques: an approach to primary screening and surveillance. Lab Anim Sci 44:217–221. [PubMed] [Google Scholar]
- 16.Letvin NL, Daniel MD, Sehgal PK, Chalifoux LV, King NW, Hunt RD, Aldrich WR, Holley K, Schmidt DK, Desrosiers RC. 1984. Experimental infection of rhesus monkeys with type D retrovirus. J Virol 52:683–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marx PA, Bryant ML, Osborn KG, Maul DH, Lerche NW, Lowenstine LJ, Kluge JD, Zaiss CP, Henrickson RV, Shiigi SM. 1985. Isolation of a new serotype of simian acquired immune deficiency syndrome type D retrovirus from Celebes black macaques (Macaca nigra) with immune deficiency and retroperitoneal fibromatosis. J Virol 56:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer J, Meese E. 2005. Human endogenous retroviruses in the primate lineage and their influence on host genome. Cytogenet Genome Res 110:448–456. 10.1159/000084977. [DOI] [PubMed] [Google Scholar]
- 19.Montiel NA. 2010. An updated review of simian betaretrovirus (SRV) in macaque hosts. J Med Primatol 39:303–314. 10.1111/j.1600-0684.2010.00412.x. [DOI] [PubMed] [Google Scholar]
- 20.Morton WR, Agy MB, Capuano SV, Grant RF. 2008. Specific pathogen-free macaques: definition, history, and current production. ILAR J 49:137–144. 10.1093/ilar.49.2.137. [DOI] [PubMed] [Google Scholar]
- 21.Nandi JS, Bhavalkar-Potdar V, Tikute S, Raut CG. 2000. A novel type D simian retrovirus naturally infecting the Indian Hanuman langur (Semnopithecus entellus). Virology 277:6–13. 10.1006/viro.2000.0567. [DOI] [PubMed] [Google Scholar]
- 22.Nandi JS, Tikute SA, Chhangani AK, Potdar VA, Tiwari-Mishra M, Ashtekar RA, Kumari J, Walimbe A, Mohnot SM. 2003. Natural infection by simian retrovirus-6 (SRV-6) in Hanuman langurs (Semnopithecus entellus) from 2 different geographical regions of India. Virology 311:192–201. 10.1016/S0042-6822(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 23.Nandi JS, Van Dooren S, Chhangani AK, Mohnot SM. 2006. New simian beta retroviruses from rhesus monkeys (Macaca mulatta) and langurs (Semnopithecus entellus) from Rajasthan, India. Virus Genes 33:107–116. 10.1007/s11262-005-0032-x. [DOI] [PubMed] [Google Scholar]
- 24.Takano J, Leon A, Kato M, Abe Y, Fujimoto K. 2013. Isolation and DNA characterization of a simian retrovirus 5 from a Japanese monkey (Macaca fuscata). J Gen Virol 94:955–959. 10.1099/vir.0.047621-0. [DOI] [PubMed] [Google Scholar]
- 25.van der Kuyl AC, Mang R, Dekker JT, Goudsmit J. 1997. Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J Virol 71:3666–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voevodin AF, Marx PA. 2009. Simian virology. Ames (IA): Wiley–Blackwell.
- 27.White JA, Todd PA, Rosenthal AN, Yee JL, Grant R, Lerche NW. 2009. Development of a generic real-time PCR assay for simultaneous detection of proviral DNA of simian betaretrovirus serotypes 1, 2, 3, 4 and 5 and secondary uniplex assays for specific serotype identification. J Virol Methods 162:148–154. 10.1016/j.jviromet.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu HL, Leon EJ, Wallace LT, Nimiyongskul FA, Buechler MB, Newman LP, Castrovinci PA, Johnson PR, Gifford RJ, Jones RB, Sacha JB. 2016. Identification and spontaneous immune targeting of an endogenous retrovirus K envelope protein in the Indian rhesus macaque mode of human disease. Retrovirology 13:1–9.10.1186/s12977-01600238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yee JL, Grant R, Van Rompay KK, Kuller L, Carpenter A, Watanabe R, Huebner R, Agricola B, Smedley J, Roberts JA. 2017. Emerging diagnostic challenges and characteristics of simian betaretrovirus infections in captive macaque colonies. J Med Primatol 46:149–153. 10.1111/jmp.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yee JL, Vanderford TH, Didier ES, Gray S, Lewis A, Roberts J, Taylor K, Bohm RP. 2016. Specific pathogen free macaque colonies: a review of principles and recent advances for viral testing and colony management. J Med Primatol 45:55–78. 10.1111/jmp.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zao CL, Armstrong K, Tomanek L, Cooke A, Berger R, Estep JS, Marx PA, Trask JS, Smith DG, Yee JL, Lerche NW. 2010. The complete genome and genetic characteristics of SRV-4 isolated from cynomolgus monkeys (Macaca fascicularis). Virology 405:390–396. 10.1016/j.virol.2010.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of cocultures with permissible Raji and Sup T cell lines and SRV antibody reactive, PCR negative donor PBMCs.