Abstract
Purpose
The emergence of clarithromycin resistance is a challenge in treating Mycobacterium abscessus infections. Known mechanisms that contribute to intrinsic clarithromycin resistance focus on rrl gene-related mutations, but resistant clinical isolates often exhibit an inconsistent rrl genotype.
Patients and Methods
In this study, 194 clinical Mycobacterium abscessus isolates were collected from patients with lung infections and the whole genome of each isolate was sequenced. A comprehensive examination of the molecular mechanisms underlying intrinsic clarithromycin resistance was performed, combining MIC determination, comparative genome sequence analysis and qRT-PCR.
Results
Of the 194 isolates, 13 (6.7%) were clarithromycin resistant; only seven of these harbored a rrl 2270/2271 mutation. The remaining six resistant isolates did not exhibit a specific resistance-associated mutation in the clarithromycin target-site genes, rrl, rplC, rplD and rplV, or in the rrl modification gene erm(41). qRT-PCR analysis showed that the increased expression of the efflux pump genes, MAB_2355c, MAB_1409c and MAB_1846, as well as their positive regulatory gene whiB7, consistently correlated with increased clarithromycin resistance. The presence of efflux pump inhibitors significantly decreased the MIC of clarithromycin for nonsusceptible isolates, especially the intrinsic resistant isolates that exhibited no rrl 2270/2271 mutation.
Conclusion
These findings indicate that efflux pumps play a prominent role in the intrinsic resistance of M. abscessus to clarithromycin, complementing other known resistance mechanisms.
Keywords: Mycobacterium abscessus, clarithromycin resistance, efflux pumps
Introduction
Infections caused by nontuberculous mycobacteria (NTM) have increased dramatically worldwide in recent years.1–4 Mycobacterium abscessus is one of the most common NTM detected. M. abscessus infections are difficult to manage because M. abscessus is intrinsically resistant to a variety of antimicrobials currently available in clinical practice; infections that are refractory to antibiotic therapy frequently result in morbidity and mortality.5–7 Recently, human-to-human M. abscessus transmission was reported, making the problem even more disconcerting.8
Clarithromycin (CLA), which is effective in treating M. abscessus lung disease, is recommended as the core agent for treatment of infections although effective therapeutic options are evolving.9,10 The emergence of CLA resistance, however, is challenging. Currently, a 2270/2271 point mutation (Escherichia coli 2058/2059) in the 23S rRNA (rrl) gene and a full-length erm(41) gene with thymine located at position 28 [erm(41)full-T28 sequevar] are the best described mechanisms conferring CLA resistance. The erm(41)full-T28 sequevar confers induced CLA resistance and a rrl 2270/2271 point mutation confers intrinsic CLA resistance in the M. abscessus isolates assessed.11 The rrl mutation, however, fails to account fully for the intrinsic resistance to CLA exhibited by M. abscessus.12–15 In addition to a rrl 2270/2271 mutation, the expression of a variety of efflux pump genes correlates with CLA resistance in mycobacterium.16–24 Almost all the homologous, efflux pump genes are found in M. abscessus; whether these efflux pump genes are involved in the resistance of clinical M. abscessus isolates to CLA remains to be determined.
In this study, the MIC of CLA was determined for 194 clinical M. abscessus isolates collected from patients with lung diseases. A comprehensive exploration of the molecular mechanisms of CLA resistance was performed by combining comparative genome sequence analysis and qRT-PCR; the efflux pump genes were a specific focus. In addition to mutations in the CLA target-site genes, efflux pump genes MAB_2355c, MAB_1409c and MAB_1846 were found to play an important role in CLA resistance. To our knowledge, this is the first comprehensive mechanistic investigation of intrinsic CLA resistance in a large number of clinical, M. abscessus isolates. This work extends our understanding of the factors that affect the resistance of M. abscessus to CLA and suggests novel approaches to treating CLA-resistant infections.
Materials and Methods
Bacterial Isolation and Identification
Between January 2014 and December 2017, 194 M. abscessus isolates were collected at Shanghai Pulmonary Hospital from sputum and bronchoalveolar lavage fluid samples of patients with M. abscessus lung infections. All isolates were preserved in the Clinical Microbiology Laboratory. Species were preliminarily screened for NTM by MGIT960 medium culture and the p-nitrobenzoic acid test, followed by molecular identification of M. abscessus by sequencing the rpoB and erm(41) genes.25 Each isolate was then stored at −80°C until use.
Clarithromycin Susceptibility Test
CLA susceptibility was determined by the broth microdilution method according to Clinical and Laboratory Standards Institute (CLSI) guidelines; the breakpoints were interpreted according to (CLSI)-M24-A2, as follows: susceptible, MIC ≤ 2 mg/L; intermediate, MIC = 4 mg/L; and resistant, MIC ≥ 8 mg/L. The intrinsic resistance of M. abscessus to CLA was assessed after 3 days exposure. Mycobacterium peregrinum (ATCC 700686; American Type Culture Collection, Manassas, VA, USA) and Staphylococcus aureus (ATCC 29213; American Type Culture Collection, Manassas, VA, USA) served as control reference strains.
Whole-Genome Sequencing and Comparison of Resistance Genes
One hundred ninety-four isolates were sequenced. DNA extraction, library construction and sequencing were conducted by methods previously reported by us.26 The full genome sequence of each isolate has been published and is available at DDBJ/ENA/GenBank (96 sequences found under bioproject PRJNA488058, 35 under bioproject PRJNA448987 and 63 under bioproject PRJNA398137).26,27 Sequence homologs were identified by BLAST and aligned with the homologous sequences of the M. abscessus reference strain ATCC19977 (NC_010397.1). Ribosome structural genes, rrl, rplC, rplD and rplV, and rrl modification gene erm(41) were also included in this analysis.
RNA Extraction and Quantitative Reverse Transcription PCR (qRT-PCR)
Seven homolog efflux pump genes and the regulatory gene whiB7 (MAB_3508c) correlate with macrolides resistance in mycobacteria, were selected and analyzed. Relative expression of the genes was assessed by comparing the quantity of mRNA expressed by the organism cultured in the presence and absence of CLA using the same technical approach reported previously.28 A culture incubated in the presence of half its MIC of CLA was shaken at 37°C for 3 h; the RNA was then extracted according to the protocols described by Medjahed et al29 cDNA was synthesized using the HiScript III RT SuperMix with gDNA wiper (Vazyme Biotech Co., Ltd). qRT-PCR was performed using ChamQ Universal SYBR Master Mix (Vazyme Biotech Co., Ltd) on a QS6 Real-Time PCR System (Applied Biosystems, Carlsbad, CA). SigA was chosen as the endogenous reference gene. All PCR primer pairs used for amplification are shown in Supplementary Table 1. Calculation of fold change was described previously in detail.30 Reactions were repeated in triplicate; genes with expression levels ≥4 were considered overexpressed
Efflux Pump Inhibition Assay
The MIC of CLA used in combination with efflux pump inhibitors, phenylalanine-arginine β-naphthylamide (PaβN), carbonyl cyanide 3-chlorophenylhydrazone (CCCP) or verapamil (VP), was determined as previously described.31,32 A 2-fold decrease in MIC in the presence of an inhibitor was considered significant. The efflux pump inhibition test was performed in triplicate.
Results
Clarithromycin Susceptibility and Resistance Profiles
One hundred ninety-four clinical, M. abscessus isolates were collected; 148 isolates belonged to subsp. abscessus and 46 isolates belonged to subsp. massiliense. The MICs of CLA ranged from 0.06 to 256 mg/L for these isolates; MIC50 and MIC90 at 3 days were 0.5 and 2 mg/L, respectively.
A rrl 2270/2271 mutation confers intrinsic resistance to CLA (MIC ≥8 mg/L accessed after 3 days). Accordingly, a comprehensive analysis of the distribution of this resistant genotype and resistance profile among the 194 isolates was performed. A total of 13 (6.7%, 8 subsp. abscessus and 5 subsp. massiliense) resistant isolates were found (Table 1). Among these, only 7 (53.8%) of the isolates harbored a rrl 2270/2271 mutation while 6 isolates with no rrl 2270/2271 mutation also exhibited intrinsic resistance.
Table 1.
MIC of Clarithromycin for 194 Clinical M. abscessus Isolates
| M. abscessus | No. of Isolates | Number of Isolates Exhibiting the MIC (mg/L) of CLA Indicated | No. of Resistant Isolates (%) | Resistant Isolates Harboring a rrl | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 2270/2271 Mutationa | |||
| subsp. abscessus | 148 | 16 | 17 | 10 | 30 | 34 | 27 | 6 | 2 | 1 | 1 | 2 | 2 | 0 | 8 (5.4) | 4 |
| subsp. massiliense | 46 | 23 | 10 | 3 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 5 (10.9) | 3 |
Note: aE. coli numbering 2058 and 2059.
Gene Comparison of M. abscessus Isolates
CLA target-site genes rrl, rplC, rplD and rplV, encoding the ribosome structure, were further analyzed among all 194 isolates. Besides the 2270/2271 mutation, 23 additional mutations were observed in rrl although none specifically presented in CLA-susceptible or -nonsusceptible isolates (Supplementary Table 2). Moreover, no meaningful mutation in ribosome protein genes rplC, rplD and rplV was found in the resistant isolates. The rrl modification gene erm(41) was also analyzed; no specific sequence changes related to CLA resistance were found.
Transcriptional Analysis of Efflux Pumps
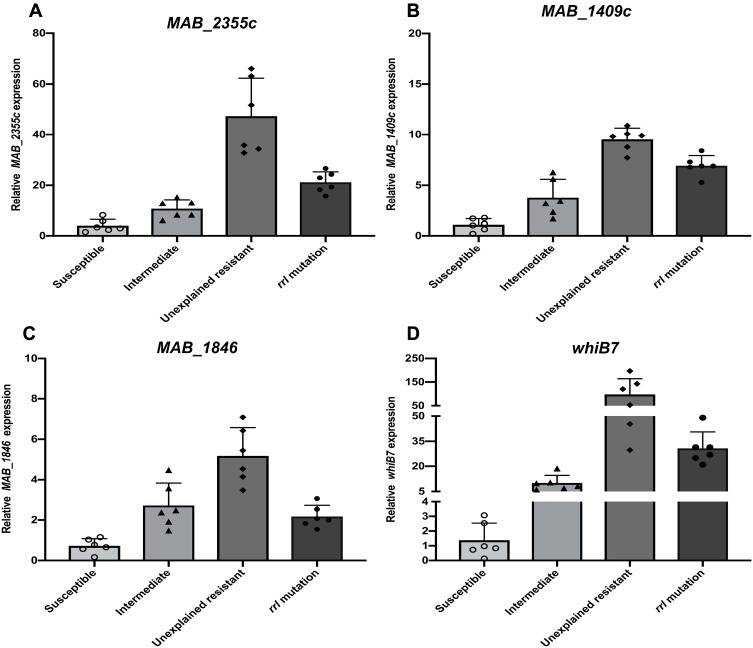
The putative efflux pump genes in M. abscessus were identified by literature review and analysis of homology to genes found in other Mycobacterium strains (Table 2). Six CLA-resistant isolates (MIC ≥8 mg/L) harboring a rrl mutation and 6 resistant isolates with no rrl mutation, along with 6 CLA sensitive (MIC ≤2 mg/L) and 6 CLA intermediate (MIC ≤4 mg/L) isolates were selected at random. The expression of efflux pump genes was quantified by qRT-PCR. Among the 7 putative efflux pump genes analyzed, the transcriptional levels of MAB_2355c (Figure 1A), MAB_1409c (Figure 1B) and MAB_1846 (Figure 1C), as well as their positive regulatory gene whiB7 (Figure 1D), showed consistent increases that correlated inversely with decreases in clarithromycin susceptibility (Figure 1). No significant changes in expression of the four remaining putative efflux pump genes were observed after CLA exposure (data not shown). The efflux pump genes in the unexplained (rrl mutation-negative) intrinsic resistant isolates showed the highest level of expression. Among the efflux pump genes, for example, MAB_2355c exhibited >40-fold and >20-fold increases in transcription in the rrl mutation-negative and rrl mutation-positive isolates, respectively.
Table 2.
Blast Search of the M. abscessus Genome with Reference Gene Sequences Corresponding to Efflux Pumps Conferring or Putatively Involved in CLA Resistance
| Targeted Gene | Sequence in Reference Strain (NC_010397.1)a | Gene Description | Reference Gene | Reference(s) | |
|---|---|---|---|---|---|
| Homolog | Strain | ||||
| MAB_2355c | 2410335-1993 | Putative ABC transporter ATP-binding protein | Rv1473 MSMEG_3140 MAV_3306 |
M. tuberculosis M. smegmatis M. avium |
15,16 |
| MAB_1409c | 1412431-3702 | Putative drug antiporter protein precursor | Rv1258c (tap) MAV_1406 |
M. tuberculosis M. avium |
17,18 |
| MAB_1846 | 1844817-6403 | Putative ABC transporter ATP-binding protein | Rv2477c | M. tuberculosis | 18,19 |
| MAB_3142c | 3180588-2105 | Putative transmembrane efflux protein | Rv2846c(efpA) | M. tuberculosis | 20,21 |
| MAB_2807 | 2854903-6447 | Major facilitator superfamily MFS_1 | Rv1410c (p55) | M. tuberculosis | 22,23 |
| MAB_0970c | 976402-8414 | Probable drug resistance transporter | Rv1877 | M. tuberculosis | 20,21 |
| MAB_1560 | 1587054-8727 | Probable ABC transporter (macrolide-transport) ATP-binding protein | MAV_1695 | M. avium | 16 |
Note: aComplete nucleotide sequence in the M. abscessus genome NCBI database.
Figure 1.
Efflux pump gene expression by clinical, M. abscessus isolates following exposure to CLA stress. Isolates that were CLA-susceptible, -intermediate and resistant [without (unexplained) or with a rrl 2270/2271 mutation] are shown. MAB_2355c (A), MAB_1409c (B), MAB_1846 (C) and whiB7 (D), message levels were quantified by qRT-PCR. Open circles, triangles, diamonds, and solid circles indicate the distribution of mRNA expression levels in isolates comprising the sensitive, intermediate, unexplained drug resistance, and rrl 2270/2271 mutation groups, respectively. Experiments were repeated >3 times for each isolate; error bars represent the standard error of the mean for each data set.
Efflux Pump Inhibition Assay
Previously, three highly expressed genes (MAB_2355c, MAB_1409c and MAB_1846) were found to encode efflux pumps capable of extruding CLA in mycobacteria (15–19). An efflux pump inhibition test was conducted to evaluate the role of these efflux pumps in intrinsic CLA resistance exhibited by M. abscessus. The addition of efflux pump inhibitors PAβN, CCCP and VP significantly decreased the MIC of CLA 47.4%, 52.6% and 63.2%, respectively, for the nonsusceptible isolates while there was no significant effect on the CLA sensitive isolates (Table 3). This decrease was more pronounced in resistant isolates that did not harbor a rrl 2270/2271 mutation. Although the level of efflux pump gene expression was significantly elevated, the inhibitory effect of efflux pump inhibitors was nearly absent in resistant isolates characterized by a rrl 2270/2271 mutation.
Table 3.
Efflux Pump Inhibition Decreases the MIC of Clarithromycin for Non-Susceptible Isolatesa
| Treatment | Number (%) of Isolates Exhibiting a Significant Decrease in CLA Resistanceb | |||
|---|---|---|---|---|
| Intermediate (n = 6) | Resistant with rrl Mutation (n =7) | Resistant Without rrl Mutation (n = 6) | Total (n=19) | |
| CLA + PaβN | 4 (66.7) | 0 (0) | 5 (83.3) | 9 (47.4) |
| CLA + CCCP | 5 (83.3) | 0 (0) | 5 (83.3) | 10 (52.6) |
| CLA + VP | 5 (83.3) | 1 (14.2) | 6 (100) | 12 (63.2) |
Notes: aIsolates exhibiting MIC ≥4 mg/L after 3 days incubation were considered non-susceptible. bAfter the addition of the inhibitor, a decrease in MIC of ≥2-fold was considered significant.
Abbreviations: PaβN, phenylalanine-arginine β-naphthylamide; CCCP, Carbonyl cyanide 3-chlorophenylhydrazone; VP, verapamil.
Discussion
CLA remains the core antibiotic used to treat M. abscessus infections. Treatment options and outcomes differ significantly among patients infected with CLA-resistant versus -sensitive M. abscessus genotypes.33 In a certain proportion of clinical M. abscessus isolates, resistance cannot be explained by the genotype,12–15 alternate mechanisms exist. In this study, 194 isolates were obtained from M. abscessus lung infection cases in mainland China. In addition to CLA-target site mutations, efflux pumps played an important role in the intrinsic resistance of M. abscessus to CLA.
Almost all known mechanisms of intrinsic resistance to CLA involve changes in the CLA binding site due to mutations in rrl (i.e., 2270/2271) or adjacent sites.11–15 The results reported provide additional support to indicate that the rrl genotype correctly predicts the CLA-resistant phenotype of most (181/194, 96.9%) clinical isolates. However, 6 (3.1%) intrinsic resistant isolates in this study exhibited an inconsistent, rrl mutation-negative, genotype. Recently, Lipworth and colleagues identified eight potential new resistance-conferring single nucleotide polymorphisms (SNPs) present only in the rrl and erm(41) genes of antibiotic-resistant, but not sensitive, isolates.14 None of the predicted SNPs or other resistance-conferring SNPs were found in rrl in the current study. Reportedly, mutations in ribosomal protein genes rplC, rplD and rplV are also associated with structural resistance to macrolides.34 Mutations in these genes, however, were absent among all the CLA-resistant isolates collected in this study.
Efflux was an anticipated factor in the drug resistance exhibited by clinical M. abscessus isolates collected here. Multi-drug efflux pumps directly mediate the resistance of a number of mycobacteria species to CLA.16–24 Reportedly, efflux inhibitors reduce the MIC of CLA for M. abscessus.31,35 However, CLA-related efflux pumps have not been systematically demonstrated and validated in clinical, M. abscessus isolates specifically. In the present study, three putative efflux pumps (an ATP binding cassette efflux pump encoded by MAB_2355c, a tap like efflux pump encoded by MAB_1409c, and an ATP binding cassette efflux pump encoded by MAB_1846) contributed to CLA resistance exhibited by M. abscessus. The ATP binding cassette efflux pump encoded by MAB_2355c was the most notable. MAB_2355c is a homolog of genes found in: M. avium (MAV_3306), M. tuberculosis (Rv1473) and M. smegmatis (MSMEG_3140). Schmalstieg and colleagues first reported that exposure to azithromycin over a 3-day period led to a 56-fold increase in expression of MAV_3306, which encodes the ATP binding cassette efflux pump in M. avium.17 They envisioned that this increase was the first step in a general pathway to drug resistance, which eventually leads to high-level, chromosome mutation-related resistance. Macrolide resistance mediated by the ATP binding cassette efflux pump encoded by Rv1473 was further characterized in 2019.16 Rv1258c (MAB_1409c homolog) is associated with multiple drug efflux in M. tuberculosis.18 We also found this efflux pump was closely related to amikacin resistance in M. abscessus.36 Recently, Vianna and colleagues reported the association between MAB_1409c overexpression and CLA resistance in M. abscessus, a finding consistent with that reported herein.37 In contrast to their results, however, we found no correlation between MAB_3142c and CLA resistance; presumably, sample bias contributes to this difference. Purportedly, Rv2477c (MAB_1846 homolog) was involved in mycobacterial protein translation, and resistance to tetracyclines and macrolides in M. tuberculosis.20 This finding is consistent with our results showing MAB_1846 overexpression is associated with macrolides (CLA) resistance. Notably, MAB_2355c, MAB_1409c and MAB_1846 encode factors that belong to the intrinsic macrolides resistance network which is positively regulated by whiB7. WhiB7 is a transcription factor that promotes intrinsic CLA resistance through self-activation and the activation of downstream genes involved in CLA efflux and the protection of ribosomes against antibiotic stress.16 Taken together, these findings are consistent with the results reported herein.
It is generally believed that the induction of efflux pumps is the first event leading to increased antibiotic resistance expressed by Mycobacterium, which usually exhibits low or moderate resistance prior to drug exposure.17 Similarly, most resistant isolates that lacked the rrl 2270/2271 mutation exhibited low or moderate resistance in the study reported here (ie, MIC ≤16mg/L, Supplementary Table 3), but exhibit relative higher expression of efflux pumps than do resistant isolates with rrl 2270/2271 mutation. Moreover, efflux pump inhibitors exerted a greater effect on resistant isolates that lack a rrl 2270/2271 mutation. As such, we speculate that a change in the target-site (e.g., a rrl 2270/2271 mutation) is the ultimate event that culminates in a high level of CLA resistance. Efflux pump overexpression, on the other hand, is a reversible, compensatory drug resistance mechanism making efflux pumps a potentially better target for clinical intervention. Whether efflux pump expression affects macrolide resistance induced by functioning erm(41) is currently unknown. Inasmuch as increases in both erm(41) and efflux pump expression reduce CLA sensitivity; however, we speculate that efflux pump expression can further enhance macrolide resistance dependent upon the degree of resistance induced by functional erm(41).
Conclusion
The present study identified three efflux pumps associated with the intrinsic resistance of M. abscessus to clarithromycin. Identification of these pumps adds to our current understanding of the factors that affect drug resistance, and suggests novel approaches to treating CLA-resistant M. abscessus infections.
Acknowledgments
We sincerely thank Dr. Stephen H. Gregory (Providence, Rhodes Island, USA) who helped write and edit this manuscript.
Funding Statement
This work was funded by grants provided by the: National Natural Science Foundation of China (Nos. 81672063, 81971973 and 81800003), Natural Science Foundation of Shanghai Municipal Science and Technology Commission (Nos. 18ZR1431600 and 19ZR1442800), Medical Guide Program of Shanghai Science and Technology Committee (Nos.18411970600 and 19411969600), New Frontier Technology Joint Project of Municipal Hospital, Shanghai Shenkang Hospital Development Center (No. SHDC12017113), Shanghai Health and Family Planning Commission Excellent Talents Training Program (No. 2018YQ55), General Project of Shanghai Health and Family Planning Commission (201940229), and Project of Top Clinical Medicine Centers and Key Disciplines Construction in Shanghai (No. 2017ZZ02012).
Ethics Approval and Informed Consent
Ethical approval was not required because the isolates were collected for routine diagnostic testing.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cowman S, van Ingen J, Griffith DE, Loebinger MR. Non-tuberculous mycobacterial pulmonary disease. Eur Respir J. 2019;54(1):1900250. doi: 10.1183/13993003.00250-2019 [DOI] [PubMed] [Google Scholar]
- 2.Hoefsloot W, van Ingen J, Andrejak C, et al. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J. 2013;42(6):1604–1613. doi: 10.1183/09031936.00149212 [DOI] [PubMed] [Google Scholar]
- 3.Lin C, Russell C, Soll B, et al. Increasing prevalence of nontuberculous mycobacteria in respiratory specimens from US-affiliated Pacific Island Jurisdictions. Emerg Infect Dis. 2018;24(3):485–491. doi: 10.3201/eid2403.171301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H, Myung W, Koh WJ, Moon SM, Jhun BW. Epidemiology of nontuberculous mycobacterial infection, South Korea, 2007-2016. Emerg Infect Dis. 2019;25(3):569–572. doi: 10.3201/eid2503.181597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffith DE. Mycobacterium abscessus and antibiotic resistance: same as it ever was. Clinical Infectious Diseases. 2019;69(10):1687–1689. doi: 10.1093/cid/ciz071 [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Zhao L, Mao Y, et al. Clinical efficacy and adverse effects of antibiotics used to treat Mycobacterium abscessus pulmonary disease. Front Microbiol. 2019;10:1977. doi: 10.3389/fmicb.2019.01977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother. 2012;67(4):810–818. doi: 10.1093/jac/dkr578 [DOI] [PubMed] [Google Scholar]
- 8.Bryant JM, Grogono DM, Rodriguez-Rincon D, et al. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science. 2016;354(6313):751–757. doi: 10.1126/science.aaf8156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haworth CS, Banks J, Capstick T, et al. British Thoracic Society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). Thorax. 2017;72(Suppl2):ii1–ii64. [DOI] [PubMed] [Google Scholar]
- 10.Griffith DE, Aksamit T, Brown-Elliott BA, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367–416. doi: 10.1164/rccm.200604-571ST [DOI] [PubMed] [Google Scholar]
- 11.Wallace RJ Jr, Meier A, Brown BA, et al. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother. 1996;40(7):1676–1681. doi: 10.1128/AAC.40.7.1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida S, Tsuyuguchi K, Kobayashi T, et al. Discrepancies between the genotypes and phenotypes of clarithromycin-resistant Mycobacterium abscessus complex. Int J Tuberc Lung Dis. 2018;22(4):413. doi: 10.5588/ijtld.17.0673 [DOI] [PubMed] [Google Scholar]
- 13.Mougari F, Bouziane F, Crockett F, et al. Selection of resistance to clarithromycin in Mycobacterium abscessus subspecies. Antimicrob Agents Chemother. 2017;61(1). doi: 10.1128/AAC.00943-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipworth S, Hough N, Leach L, et al. Whole-Genome sequencing for predicting clarithromycin resistance in Mycobacterium abscessus. Antimicrob Agents Chemother. 2019;63(1). doi: 10.1128/AAC.00400-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H, Liu D, Lu J, et al. Genetic correlation of antibiotic susceptibility and resistance genotyping for the Mycobacterium abscessus Group. Antimicrob Agents Chemother. 2019;63(1). doi: 10.1128/AAC.00779-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan W, Li X, Ge Y, et al. Mycobacterium tuberculosis Rv1473 is a novel macrolides ABC efflux pump regulated by WhiB7. Future Microbiol. 2019;14(1):47–59. doi: 10.2217/fmb-2018-0207 [DOI] [PubMed] [Google Scholar]
- 17.Schmalstieg AM, Srivastava S, Belkaya S, et al. The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrob Agents Chemother. 2012;56(9):4806–4815. doi: 10.1128/AAC.05546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma S, Kumar M, Sharma S, Nargotra A, Koul S, Khan IA. Piperine as an inhibitor of Rv1258c, a putative multidrug efflux pump of Mycobacterium tuberculosis. J Antimicrob Chemother. 2010;65(8):1694–1701. doi: 10.1093/jac/dkq186 [DOI] [PubMed] [Google Scholar]
- 19.Miranda-CasoLuengo AA, Staunton PM, Dinan AM, Lohan AJ, Loftus BJ. Functional characterization of the Mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence. BMC Genomics. 2016;17. doi: 10.1186/s12864-016-2868-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniel J, Abraham L, Martin A, Pablo X, Reyes S. Rv2477c is an antibiotic-sensitive manganese-dependent ABC-F ATPase in Mycobacterium tuberculosis. Biochem Bioph Res Co. 2018;495(1):35–40. doi: 10.1016/j.bbrc.2017.10.168 [DOI] [PubMed] [Google Scholar]
- 21.De Rossi E, Arrigo P, Bellinzoni M, et al. The multidrug transporters belonging to major facilitator superfamily (MFS) in Mycobacterium tuberculosis. Mol Med. 2002;8(11):714–724. doi: 10.1007/BF03402035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li XZ, Zhang L, Nikaido H. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob Agents Chemother. 2004;48(7):2415–2423. doi: 10.1128/AAC.48.7.2415-2423.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramon-Garcia S, Martin C, Thompson CJ, Ainsa JA. Role of the Mycobacterium tuberculosis P55 efflux pump in intrinsic drug resistance, oxidative stress responses, and growth. Antimicrob Agents Chemother. 2009;53(9):3675–3682. doi: 10.1128/AAC.00550-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hohl M, Remm S, Eskandarian HA, et al. Increased drug permeability of a stiffened mycobacterial outer membrane in cells lacking MFS transporter Rv1410 and lipoprotein LprG. Mol Microbiol. 2019;111(5):1263–1282. doi: 10.1111/mmi.2019.111.issue-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macheras E, Roux AL, Bastian S, et al. Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (Sensu Lato) strains. J Clin Microbiol. 2011;49(2):491–499. doi: 10.1128/JCM.01274-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Yang S, Chu H, et al. Relationship between Antibiotic susceptibility and genotype in Mycobacterium abscessus clinical isolates. Front Microbiol. 2017;8:1739. doi: 10.3389/fmicb.2017.01739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Ye M, Guo Q, et al. Determination of MIC distribution and mechanisms of decreased susceptibility to bedaquiline among clinical isolates of Mycobacterium abscessus. Antimicrob Agents Chemother. 2018;62(7). doi: 10.1128/AAC.00175-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machado D, Couto I, Perdigao J, et al. Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS One. 2012;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medjahed H, Singh AK. Genetic manipulation of Mycobacterium abscessus. Curr Protoc Microbiol. 2010;10:Unit 10D 12. [DOI] [PubMed] [Google Scholar]
- 30.Viveiros M, Dupont M, Rodrigues L, et al. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS One. 2007;2(4):e365. doi: 10.1371/journal.pone.0000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramis IB, Vianna JS, Silva L, et al. In silico and in vitro evaluation of tetrahydropyridine compounds as efflux inhibitors in Mycobacterium abscessus. Tuberculosis. 2019;118:101853. doi: 10.1016/j.tube.2019.07.004 [DOI] [PubMed] [Google Scholar]
- 32.Ye MP, Xu LY, Zou YZ, et al. Molecular analysis of linezolid-resistant clinical isolates of Mycobacterium abscessus. Antimicrob Agents Chemother. 2019;63(2). doi: 10.1128/AAC.00779-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Q, Chu HQ, Ye MP, et al. The Clarithromycin susceptibility genotype affects the treatment outcome of patients with Mycobacterium abscessus lung disease. Antimicrob Agents Chemother. 2018;62(5):e02360–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doucet-Populaire F, Buriankova K, Weiser J, Pernodet JL. Natural and acquired macrolide resistance in mycobacteria. Curr Drug Targets Infect Disord. 2002;2(4):355–370. [DOI] [PubMed] [Google Scholar]
- 35.Vianna JS, Ramis IB, Bierhals D, et al. Tetrahydropyridine derivative as efflux inhibitor in Mycobacterium abscessus. J Glob Antimicrob Resist. 2019;17:296–299. doi: 10.1016/j.jgar.2018.12.020 [DOI] [PubMed] [Google Scholar]
- 36.Wu M, Li B, Guo Q, et al. Detection and molecular characterisation of amikacin-resistant Mycobacterium abscessus isolated from patients with pulmonary disease. J Glob Antimicrob Resist. 2019;19:188–191. doi: 10.1016/j.jgar.2019.05.016 [DOI] [PubMed] [Google Scholar]
- 37.Vianna JS, Machado D, Ramis IB, et al. The contribution of efflux pumps in Mycobacterium abscessus complex resistance to clarithromycin. Antibiotics. 2019;8(3):153. doi: 10.3390/antibiotics8030153 [DOI] [PMC free article] [PubMed] [Google Scholar]