Abstract
Cilia serve as cellular antennae that translate sensory information into physiological responses. In the sperm flagellum, a single chemoattractant molecule can trigger a Ca2+ rise that controls motility. The mechanisms underlying such ultra‐sensitivity are ill‐defined. Here, we determine by mass spectrometry the copy number of nineteen chemosensory signaling proteins in sperm flagella from the sea urchin Arbacia punctulata. Proteins are up to 1,000‐fold more abundant than the free cellular messengers cAMP, cGMP, H+, and Ca2+. Opto‐chemical techniques show that high protein concentrations kinetically compartmentalize the flagellum: Within milliseconds, cGMP is relayed from the receptor guanylate cyclase to a cGMP‐gated channel that serves as a perfect chemo‐electrical transducer. cGMP is rapidly hydrolyzed, possibly via “substrate channeling” from the channel to the phosphodiesterase PDE5. The channel/PDE5 tandem encodes cGMP turnover rates rather than concentrations. The rate‐detection mechanism allows continuous stimulus sampling over a wide dynamic range. The textbook notion of signal amplification—few enzyme molecules process many messenger molecules—does not hold for sperm flagella. Instead, high protein concentrations ascertain messenger detection. Similar mechanisms may occur in other small compartments like primary cilia or dendritic spines.
Keywords: cilium, electron tomography, fertilization, quantitative mass spectrometry, signaling
Subject Categories: Neuroscience, Signal Transduction
Reversed enzyme‐messenger ratios in sperm cilia ensure high‐sensitivity, low noise messenger detection.
Introduction
Sensory systems command over exquisite sensitivity and precision. Photoreceptors in the eye can detect single photons (Baylor et al, 1979; Yau & Hardie, 2009; Gross et al, 2015); chemosensory neurons in the antennae of insects and vomero‐nasal organs of mammals can detect a few or even single pheromone molecules (Kaissling, 1986; Leinders‐Zufall et al, 2000); algae and sperm from marine invertebrates can register single chemoattractant molecules (Boland et al, 1982; Starr et al, 1995; Kaupp et al, 2003). These sensory feats happen in specialized cellular compartments, called cilia or flagella: long, slender filaments that emanate from the cell body (Marshall & Basto, 2017). Cilia and flagella serve as antennae that translate chemical and physical stimuli into electrical signals that eventually evoke a cellular or behavioral response. In recent years, a concept of physiological significance has emerged that cilia and flagella are specialized cell organelles, whose ionic milieu and inventory of signaling proteins are unique (Corbit et al, 2005; DeCaen et al, 2013; Delling et al, 2013; Kaupp & Strünker, 2017; Balbach et al, 2018). Factors that may contribute to the exquisite sensory sensitivity of cilia or flagella are the inventory, concentrations, and topographical arrangement of signaling proteins, and the compartmentalization of cellular reactions. How each factor contributes to ultra‐sensitivity and plays out in the minuscule reaction volume is largely unknown.
Here, we provide the first absolute quantification of the inventory of a flagellar chemosensory signaling pathway from sperm of the sea urchin Arbacia punctulata; this pathway controls chemotactic steering (Kaupp & Strünker, 2017). It is an optimal model system that meets all requirements for reliable absolute quantification. Sperm constitute a homogenous cell population, and flagella can be prepared purely. Moreover, most signaling proteins are sperm‐specific; therefore, potential contamination by other cell types does not compromise quantification. Importantly, most signaling proteins have been identified—from the chemoattractant receptor to the Ca2+ channel that controls chemotactic steering. Absolute mass spectrometric quantification using the QconCAT (quantification concatemers) technology (Scott et al, 2016) reveals astoundingly high copy numbers of all 11 signaling proteins in the flagellum. Compared with a typical cell soma, the enzyme‐to‐substrate ratio is reversed by several orders of magnitude. Using kinetic opto‐chemical techniques, we probed the physiological consequences for the dynamics of cellular messengers. Quantification of signaling proteins and cellular responses provides new insights into the molecular underpinnings of chemosensory ultra‐sensitivity.
Results
The cGMP‐signaling pathway
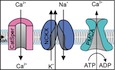
Chemosensation in the flagellum of sea urchin sperm involves changes in the membrane potential (V m) and the intracellular concentration of cGMP, cAMP, Na+, Ca2+, and H+ (Fig 1A). A signaling pathway encodes these cellular events (Fig 1B) (for reviews, see; Kaupp & Alvarez, 2016; Wachten et al, 2017). The pathway is triggered by resact, a chemoattractant peptide that binds to a chemoreceptor guanylate cyclase (GC) and stimulates cGMP synthesis (Garbers, 1976; Singh et al, 1988; Kaupp et al, 2003; Pichlo et al, 2014). The rise of cGMP opens K+‐selective cyclic nucleotide‐gated (CNGK) ion channels (Strünker et al, 2006; Galindo et al, 2007; Bönigk et al, 2009). The ensuing hyperpolarization activates a voltage‐gated Na+/H+ exchanger (sNHE) (Lee, 1985; Windler et al, 2018) and a hyperpolarization‐activated, cyclic nucleotide‐gated (HCN) pacemaker channel (Gauss et al, 1998; Galindo et al, 2005). Na+/H+ exchange increases pHi, which primes CatSper Ca2+ channels to open during the recovery from hyperpolarization probably driven by HCN channels that carry an inward current (Seifert et al, 2015; Espinal‐Enriquez et al, 2017). Changes in free Ca2+ concentration ([Ca2+]i) control the swimming path.
Figure 1. Signaling pathway controlling chemotactic navigation.
- Schematic representation of signaling events. Left: time course of changes in cAMP (dotted line) and cGMP (solid line); right: time course of changes in V m (green), pHi (red); [Na+]i (orange), and [Ca2+]i (pink). Vertical dotted lines indicate time of stimulation.
- GC, receptor guanylate cyclase; CNGK, cyclic nucleotide‐gated K+‐selective channel; sNHE (SLC9C1), sperm‐specific Na+/H+ exchanger; HCN1, HCN2, hyperpolarization‐activated, and cyclic nucleotide‐gated channels; CatSper, cation channel of sperm; NCKX, Na+/Ca2+‐K+ exchanger; PMCA, plasma membrane Ca2+‐ATPase; PDE5, phosphodiesterase type 5; PDE10, phosphodiesterase type 10; sAC, soluble adenylate cyclase. For clarity, sAC is placed in the cytosol, although it is associated with the membrane; the same may apply to PDE10.
The chemoattractant also stimulates cAMP synthesis (Hansbrough & Garbers, 1981; Harumi et al, 1992; Kaupp et al, 2003), presumably by a soluble adenylate cyclase (sAC) (Vacquier et al, 2014). cAMP modulates the activity of sNHE and HCN channels, which both carry a cyclic nucleotide‐binding domain (CNBD) (Gauss et al, 1998; Wang et al, 2003; Windler et al, 2018). For signal termination, a cGMP‐specific phosphodiesterase (PDE5) (Su & Vacquier, 2006) breaks down cGMP and, finally, a Na+/Ca2+‐K+ exchanger (NCKX) (Su & Vacquier, 2002) and/or a plasma membrane Ca2+‐ATPase (PMCA) (Gunaratne et al, 2006) restore resting Ca2+ levels.
Mass spectrometric (MS) analysis of purified flagella from A. punctulata sperm identified all previously known signaling components, but also additional proteins that might be involved in the signaling pathway (Appendix Table S1): (i) the Ca2+‐activated Cl− channel TMEM16 of olfactory cilia, which has been predicted to exist in sperm from pharmacological and modeling studies (Guerrero et al, 2013; Espinal‐Enriquez et al, 2017); (ii) a dual‐specific PDE10 that, unlike PDE5, can also hydrolyze cAMP, and, together with sAC, may control cAMP metabolism; (iii) K+/Cl− co‐transporters KCC1 (SLC12A4) and/or KCC3 (SLC12A5); (iv) a Na+/HCO3 − co‐transporter (SLC4A11); (v) a sperm‐specific Na+/K+‐ATPase α subunit; and (vi) two members (TMC 5 and 7) of a mechano‐sensitive channel family (Zhao & Müller, 2015; Pan et al, 2018). Sperm respond to mechanical stimulation with a Ca2+ response (Kambara et al, 2011). The TMC channels may endow sperm with mechano‐sensitivity. Using antibodies directed against their mammalian homologues, several transmembrane adenylate cyclases (tmAC 1, 2, 5, and 9) were detected in sea urchin sperm (Beltrán et al, 2007; Vacquier et al, 2014). Although we identified 45 different peptides for sAC, we could not detect peptides for tmACs.
For absolute quantification, we selected 11 signaling proteins shown in Fig 1B along with their subunits, a total of 19 polypeptides (Appendix Table S2). The newly discovered proteins molecules, except for PDE10 and Na+/K+‐ATPase, were not considered for absolute quantification because their function in chemotactic signaling remains to be elucidated.
Absolute quantification by QconCAT technology
The abundance of proteins was quantified by selected reaction monitoring (SRM) using a synthetic, isotope‐labeled protein as a standard (Rivers et al, 2007; Brownridge et al, 2011; Holman et al, 2012). For the synthesis of the standard protein, we selected three individual tryptic peptides from most of the 19 signaling proteins; a few proteins were covered by two or four peptides (Appendix Table S2). The presence of the peptides after trypsin digestion of the standard protein was verified by both MALDI‐TOF and LC‐ESI‐MS/MS (SRM). These control experiments failed to retrieve three peptides (Appendix Table S2), which, therefore, were not used for quantification. Consequently, quantification of NCKX and CatSper3 rested on a single peptide, which might underestimate the protein abundance. For absolute quantification, we used the receptor GC, the most abundant protein of the flagellar membrane, as a reference protein. Previous work determined 200,000–400,000 GC molecules/flagellum by quantitative SRM (using a PSAQ‐GC as a standard) (Pichlo et al, 2014), Coomassie Blue densitometry (Pichlo et al, 2014), and high‐resolution electron tomography (Farci, 2017). We consider the GC density determined by electron tomography as the most precise estimate. Therefore, protein stoichiometry was calculated assuming 400,000 GC molecules (Table 1); all protein ratios from SRM analyses are given in Dataset Table EV1. Moreover, as another internal benchmark of quantification, we included creatine kinase (CK) that, according to gel densitometry, is almost as abundant as the receptor GC (Pichlo et al, 2014). By SRM, we observed a GC:CK ratio close to unity, underscoring the high accuracy and suitability of using a synthetic labeled protein for MS determination of protein stoichiometries.
Table 1.
Copy number, concentration, and membrane density of signaling proteins
| Proteins | Molecules/Flagelluma | Concentrationb (μM) | Densityc (molecules/μm2) |
|---|---|---|---|
| GC | 400,000 | 400 | 11,976 |
| CNGK | 21,653 ± 4,038 | 21.7 ± 4 | 648 |
| sNHE | 53,979 ± 20,659 | 54 ± 20.7 | 1,616 |
| sAC | 6,718 ± 2,630 | 6.7 ± 2.6 | 201 |
| HCN1 | 10,258 ± 901 | 10.3 ± 0.9 | 307 |
| HCN2 | 18,337 ± 887 | 18.3 ± 0.9 | 549 |
| CatSper1 | 1,853 ± 147 | 1.9 ± 0.1 | 56d |
| CatSper2 | 2,125 ± 138 | 2.1 ± 0.1 | |
| CatSper3e | 587 ± 296 | 0.6 ± 0.3 | |
| CatSper4 | 1,208 ± 113 | 1.2 ± 0.1 | |
| CatSper β | 1,690 ± 547 | 1.7 ± 0.5 | |
| CatSper γ | 1,169 ± 1,395 | 1.2 ± 1.4 | |
| CatSper δ | 2,071 ± 392 | 2.1 ± 0.4 | |
| PDE5 | 48,567 ± 6,525 | 48.6 ± 6.5 | |
| PDE10 | 4,211 ± 752 | 4.2 ± 0.8 | |
| NCKXe | 120,795 ± 41,520 | 120.8 ± 41.5 | 3,617 |
| Ca2+‐ATPase | 1,991 ± 749 | 2 ± 0.7 | 60 |
| Na+/K+‐ATPase | 48,280 ± 16,788 | 48.3 ± 16.8 | 1,446 |
| Creatine Kinase | 364,465 ± 29,428 | 364.5 ± 29.4 | 10,912 |
Mean ± SD from six animals.
Calculated from column 2, assuming a matrix volume V ma of 1.7 femtoliter.
Calculated from column 2, assuming a flagellar surface area of 33.4 μm2 (r = 0.133 μm; l = 40 μm); only membrane‐bound proteins are considered.
Mean calculated from all CatSper subunits, except CatSper3, assuming a 1:1 stoichiometry of subunits.
Data from a single peptide only.
Determination of flagellar volume by cryo‐electron tomography
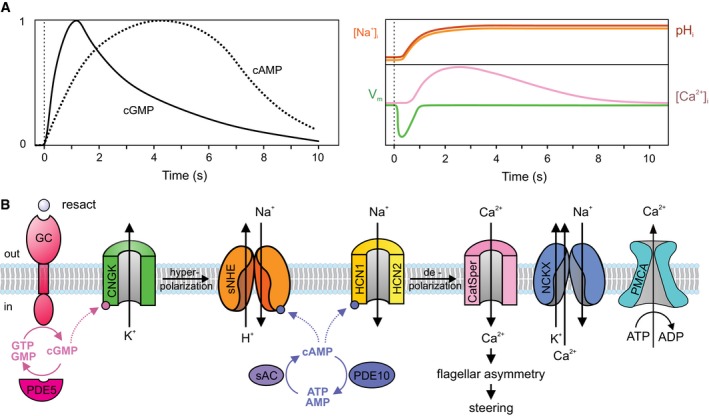
For comparison, we calculated protein concentrations from copy numbers in the flagellum. The total volume inside motile cilia and flagella (V t) contains three distinct compartments: the matrix volume (V ma) that is accessible to diffusing or transported ciliary proteins, the volume occupied by the [9 + 2] axonemal scaffold (V a), and the lumen (V l) of the nine doublet microtubules and the two singlet microtubules of the central pair complex. The microtubules are only accessible to small molecules like ions, nucleotides, and ligands (e.g., Taxol) (Fig 2). Thus, V t = V ma + V a + V l, and the volume accessible to small molecules is V asm = V ma = + V l (Fig 2F and F′). We measured these volumes using cryo‐electron tomography reconstructions of rapidly frozen flagella for two evolutionary distant species, i.e., from the unicellular green algae Chlamydomonas reinhardtii and from sperm of the sea urchin Strongylocentrotus purpuratus (Carbajal‐Gonzalez et al, 2013; Lin & Nicastro, 2018; preprint: Fu et al, 2019; Lin et al, 2019) (Fig 2A–D).
Figure 2. Determination of flagellar volumes.
-
A–DLongitudinal (A and C, proximal view on left) and cross‐sectional (B and D, proximal‐to‐distal view) tomographic slices of representative intact flagella from wild‐type Chlamydomonas reinhardtii (C.r.) flagella (A and B) and sea urchin Strongylocentrotus purpuratus (S.p.) sperm (C and D). Note that in Chlamydomonas, the flagellar membrane (m) is surrounded by a glycocalyx (g). Other labels: central pair complex (c) and doublet microtubule (d). Scale bar, 100 nm.
-
EDiameter of intact flagella from Chlamydomonas and sea urchin sperm measured on cross‐sectional views of cryo‐tomographic reconstructions. Numbers indicate average diameters ± SD (n = 84 for sea urchin (S.p.) and n = 18 for Chlamydomonas (C.r.)).
-
F′, F″Schematic diagrams of Chlamydomonas flagella in cross‐sectional view. The yellow area (F′) indicates the matrix volume V ma that is accessible to proteins, and the cyan area (F″) represents the volume that is accessible to small molecules V asm (i.e., the combination of the matrix volume and the microtubule lumen).
For calculating the V t, we measured the mean diameter (d) of intact flagella for Chlamydomonas as 247 ± 13 nm and for sea urchin sperm as 266 ± 13 nm (Fig 2E). Thus, the mean V t per 1‐μm flagellar length is for Chlamydomonas 0.0479 μm3 and for sea urchin sperm 0.0557 μm3; i.e., for an average length of sea urchin flagella of 40 μm, the total volume V t is 2.2 fl (see Materials and Methods section for details). Overall, the sub‐volumes per 1‐μm flagellar length were very similar between the two species (Table 2). We used the matrix volume (for 40 μm long S. purpuratus flagella) V ma = 1.7 fl to calculate protein concentrations in Table 1. Surprisingly, volumes accessible to proteins (V ma) and small molecules (V asm) are 77 and 86% of the total volume.
Table 2.
Diameter and volumesa of flagella from the green alga Chlamydomonas reinhardtii and of sperm from the sea urchin Strongylocentrotus purpuratus
| d b | V ma | V a | V l | V asm | V t | |
|---|---|---|---|---|---|---|
| C. reinhardtii | 247 ± 13 | 3.66/76.3 | 0.70/14.6 | 0.43/9.1 | 4.09/85.4 | 4.79/100 |
| S. purpuratus | 266 ± 13 | 4.31/77.4 | 0.81/14.5 | 0.45/8.1 | 4.76/85.5 | 5.57/100 |
Sub‐volumes in ×10−2 μm3/percentage of total volume V t.
Flagellar diameter d in nm.
Signaling proteins are orders of magnitude more abundant than free messengers
To compare protein abundance, Table 1 lists copy numbers, protein concentrations, and the densities of integral membrane proteins. We find that signaling proteins in the flagellum (Fig 1B) are up to 1,000‐fold more abundant than the respective cellular messengers like H+, Ca2+, cAMP, and cGMP (Table 1). For example, the flagellum harbors 21,700 CNGK channels that are key targets of cGMP. Each CNGK channel is activated by a single molecule of cGMP (Bönigk et al, 2009), and, at low and moderate chemoattractant stimulation of sperm, only ten to a few hundred cGMP molecules are synthesized by the receptor GC (Bönigk et al, 2009; Pichlo et al, 2014). The large number of CNGK channels suggests that this stage of signal transduction, i.e., capturing of cGMP, is highly efficient. The downstream targets of the CNGK channel, sNHE exchangers and HCN channels (Fig 1B) that are activated by hyperpolarization, are present at 54,000 and 28,000 molecules per flagellum, respectively. Remarkably, the sNHE is not involved in pH homeostasis at rest; only upon chemoattractant stimulation, it tightly couples membrane voltage and cAMP to generate rapid alkalization (latency < 300 ms) that is necessary for CatSper channel activation (Seifert et al, 2015; Windler et al, 2018). In mammalian sperm, sAC exists as a full‐length form (sACfl; 180 kDa) and a truncated form (sACt; 55 kDa). We chose four peptides, two from sACt and two from sACfl to estimate the relative abundance of each form. All four peptides occurred at similar frequencies, suggesting that only full‐length sAC exists in sea urchin sperm. Approximately 1,700 CatSper channels generate a rise of [Ca2+]i, and a remarkably high number of 120,000 NCKX exchanger proteins subsequently clear the flagellum from Ca2+ and restore the resting state. Because the time derivative of the Ca2+ response modulates the flagellar beat (Alvarez et al, 2012), tightly balanced Ca2+ influx and efflux are key to successful navigation. Finally, we determined 48,500 PDE5 molecules and 4,200 PDE10 molecules, which hydrolyze cGMP and cAMP, respectively, and terminate chemotactic signaling. In summary, the concentrations of signaling proteins range from 2 to 400 μM, whereas their respective substrates cAMP, cGMP, protons, and Ca2+ are present at much lower free concentrations.
The prodigiously high copy numbers of signaling proteins raise several paradigmatic questions about signaling in ciliary compartments. How does the high density affect the speed and efficacy of signaling at each stage along the transduction pathway? Does the high density confine signaling events to the flagellum? Do signaling proteins serve as buffers for messengers? In the following, using opto‐chemical techniques, we address these questions experimentally and assess the results by theory and numerical simulation. Thereby, we gain mechanistic insight into the physiological network of chemotactic signaling.
Tracking of cGMP by the CNGK channel and PDE5
The CNGK‐mediated hyperpolarization is the first key signaling event; when hyperpolarization is abolished, downstream events are forestalled (Harumi et al, 1992; Pichlo et al, 2014; Seifert et al, 2015). For maximum efficacy, the CNGK channel must capture any cGMP molecule before it is wasted by hydrolysis or diffusion to the sperm head. Using rapid‐mixing techniques and flash photolysis of caged cGMP (Hamzeh et al, 2019), we addressed two questions: How fast and efficiently does the CNGK channel entrap cGMP molecules, and how fast is the channel cleared from cGMP by PDE5 hydrolysis?
As read‐out for cGMP dynamics, we recorded the CNGK‐mediated V m changes using fluorescent potentiometric indicators FluoVolt and Di‐8‐ANEPPS, which detect changes in V m by photo‐induced electron transfer (PeT) and electrochromism, respectively. For the physiological cGMP‐induced hyperpolarization, this read‐out is likely contaminated by opening of HCN channels that carry a depolarizing inward Na+ current (Gauss et al, 1998). Therefore, we recorded V m in the presence of 100 mM external [K+]o (100KASW), which clamps the reversal potential V rev for CNGK to approximately −37 mV, i.e., more positive than the resting voltage V rest of approximately −50 mV (Strünker et al, 2006; Seifert et al, 2015). Consequently, at high [K+]o, CNGK opening should depolarize rather than hyperpolarize sperm. At V m > −37 mV, the open probability of HCN channels is low, and HCN channels are unlikely to contribute much to V m (Gauss et al, 1998). Furthermore, high [K+]o abolishes changes in pHi, [Ca2+]i, and cAMP evoked by the chemoattractant or cGMP (Pichlo et al, 2014; Seifert et al, 2015); therefore, putative pH‐, Ca2+‐, or cAMP‐regulated conductances, like HCN channels, are unlikely to interfere with changes in V m evoked by CNGK activity.
Sperm were loaded with DEACM‐caged cGMP. This neutral hydrophobic compound readily equilibrates across the cell membrane; therefore, photorelease of cGMP inside cells can be calibrated precisely. Changes in V m were followed with the voltage‐sensitive PeT dye FluoVolt (Miller et al, 2012; Hamzeh et al, 2019). Sperm were rapidly mixed in a stopped‐flow apparatus with 100KASW and then stimulated with a brief flash of light that photo‐released cGMP. A 2‐ms flash (releasing 52 nM of cGMP) gave rise to an almost instantaneous depolarization that relaxed to baseline levels in < 1 s (Fig 3A). A fit of the rise and relaxation of the V m signal using equation [Link] (in Materials and Methods—data analysis and simulations) yielded time constants τrise = 16.6 ± 7.3 ms and τrelax = 384 ± 107 ms (n = 3 experiments) (Fig 3A). The mean τrise is compatible with estimates of the rate of cGMP binding to CNGK (τbinding = 1–2 ms) or the RC time constant of the membrane (5–20 ms), which represent lower bounds of τrise (Materials and Methods section).
Figure 3. Dynamics of CNGK activation and cGMP hydrolysis.
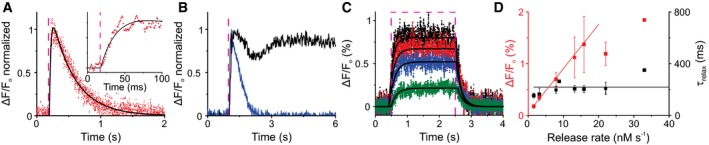
- Time course of the change in voltage V m evoked by a 2‐ms light flash in sperm that was loaded with caged cGMP (15 μM) and recorded in 100KASW. Inset: extended timescale showing the rise of V m. Fit of data (black) using two exponentials with time constants τrise = 17.4 ms and τrelax = 354 ms. Light flash in all panels is depicted as dashed magenta line.
- Time course of the changes in V m evoked by 100‐ms flash in 100KASW using caged cGMP (blue) and caged 8‐Br‐cGMP (black). The flashes released 260 nM cGMP or 2,600 nM 8‐Br‐cGMP.
- Time course of changes in V m in 100KASW evoked by 2‐s flashes at different light energy that corresponds to 440, 220, 75, and 8 nM/s of cGMP. Data were fitted using a heuristic model described by equation [Link] (Materials and Methods section) (black lines).
- Amplitude (red) and decay time τrelax (black) for different light energies and release rates for experiments as shown in panel C. τrelax was determined using a single exponential fit. The linear fit of the V m amplitude was restricted to low light levels; at higher levels, the V m response saturates (see panel C). Points indicate mean ± SD from five experiments.
Because CNGK channels are not voltage dependent and do not desensitize in the presence of cGMP (Bönigk et al, 2009), the relaxation time τrelax might reflect the closure of CNGK due to the removal of cGMP. We tested this presumption using a DEACM‐caged form of 8‐Br‐cGMP (Hagen et al, 2003), a hydrolysis‐resistant analogue that also activates CNG channels (Zimmerman et al, 1985; Kaupp & Seifert, 2002). Release of 8‐Br‐cGMP by a light flash evoked a rapid depolarization that, by contrast to cGMP, was persistent and did not relax—except for a small transient dip (Fig 3B). The 8‐Br‐cGMP experiment shows that cGMP hydrolysis is ultimately required for closure of CNGK channels and rapid recovery of V m.
What limits the rate of cGMP hydrolysis: the rate of cGMP dissociation from CNGK or the catalytic power of PDE5? We determined the V m/light energy relation using 2‐s light pulses over a 50‐fold range of light energies. Invariably, during the light pulse, V m rose to a stable plateau that was graded with increasing light energy (Fig 3C and D), suggesting that PDE5 is exactly counterbalancing steady cGMP release. The steady‐state amplitude of ∆V m increased linearly for low light energies, consistent with the idea that PDE5 is constitutively active (Rybalkin et al, 2003) (Fig 3C and D). At the end of a light pulse, V m returned to V rest with a characteristic time τrelax that was constant for a 50‐fold range of cGMP‐photolysis rates and that was similar to τrelax of signals evoked by short light flashes (220 ms vs. 384 ms) (Fig 3C). The independence of τrelax from cGMP levels suggests that dissociation of cGMP from CNGK determines the rate of V m recovery rather than PDE5's catalytic power. We scrutinized this notion by mathematical calculation and numerical simulations.
Estimates of cGMP turnover kinetics
We calculated the V m recovery kinetics for three different reaction schemes (Fig 4A) (see Materials and Methods section and Appendix). Scheme 1 involves the CNGK channel alone; scheme 2 involves the PDE alone; and scheme 3 involves CNGK and PDE together, recapitulating the in vivo situation. Schemes assume that cGMP must first dissociate from CNGK before PDE5 can degrade it. Therefore, the rate of cGMP dissociation provides an upper bound for τrelax. The calculated dissociation rate is k off = 0.68/s or τoff = 1.48 s, assuming a k on = 2.6 × 107/M/s and a constant of half‐maximal activation K1/2 = 2.6 × 10−8 M (Bönigk et al, 2009; Peuker et al, 2012). Accounting for the effect of ionic strength on second‐order rate constants (Peuker et al, 2012) and assuming that the ionic strength of seawater and the cytosol is similar, k on rates might be twofold larger (k on = 5 × 107/M/s; τoff = 0.75 s) (Fig 4B). Intriguingly, the shortest estimate of τoff is about twofold to fourfold longer than experimental τrelax for V m recovery.
Figure 4. Discrete‐event simulations of cGMP dynamics.
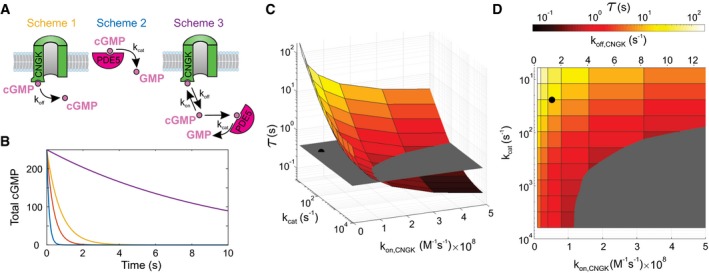
- Schemes illustrate three different situations defined by chemical reactions (2) to (5) in the Materials and Methods section. Colors of schemes refer to the simulations shown in panel (B).
- Characteristic decay of V m in experiments shown in Fig 3A and C (orange; τrelax = 0.384 s) is compared with cGMP hydrolysis by PDE5 in the absence of CNGK channels (blue; characteristic time τ = 0.131 s), dissociation of cGMP from CNGK at the rate k off (yellow; τ = 0.748 s), and cGMP hydrolysis in the presence of CNGK channels and PDE (violet; τ = 9.73 s). Simulations were done using those parameters listed in Appendix Table S3. cGMP is given in molecules.
- Simulations of the rate of cGMP hydrolysis upon varying the catalytic rate k cat of active PDE and the k on rate of cGMP binding to the CNBD of the CNGK channel. The gray plane indicates the experimental relaxation time (τrelax). The black dot represents k cat (24/s) and k on (5.2 × 107/M/s) compatible with previous studies (Appendix Table S3). The pairs of parameters compatible with the experimental τrelax are positioned at the intersection of the colored surface and the gray plane.
- Top view of panel C showing k off values compatible with k on rates used for simulations (assuming K D,CNGK = 25.7 nM; see Appendix Table S3).
Second, we estimated a lower bound for τrelax if the turnover number k cat of PDE5 were rate‐limiting (Fig 4A, PDE alone). For constitutively active PDE5 (k cat = 24/s and NPDE5 = 48,600), the time constant of cGMP hydrolysis was estimated to be 131 ms (Materials and Methods section) (Fig 4B). Thus, the hydrolytic power of PDE5 is not rate‐limiting.
Finally, we simulated cGMP hydrolysis under in vivo conditions, i.e., in the presence of both PDE5 and CNGK. In this scenario, cGMP hydrolysis was considerably slowed down (τ = 9.6 s) (Fig 4B), primarily due to rebinding of cGMP to the high‐affinity site of CNGK. We searched the parameter space for values of k cat (PDE) and k on/k off (CNGK) that are compatible with experimental τrelax (Fig 4C). To achieve τrelax, high values for both k on (CNGK) and k cat (PDE5) are required (Fig 4D, gray area). Such high k on and k cat values have not been observed for CNBD domains and for sea urchin or mammalian PDE5, respectively (Kaupp et al, 2003; Rybalkin et al, 2003). In conclusion, given the high CNGK concentration and the high k on value, hydrolysis of cGMP free in solution, for a wide range of parameters, fails to explain the extraordinary fast cGMP breakdown. To solve this conundrum, we propose substrate “channeling” between CNGK and PDE as a mechanism that might accelerate cGMP dissociation from the channel and, ultimately, hydrolysis (see Discussion).
Simulations provided additional insight regarding the potential role of the allosteric GAF‐A domain that is conserved in A. punctulata PDE5. Mammalian PDE5 displays basal activity that is about threefold enhanced by binding of cGMP to the GAF‐A domain (Rybalkin et al, 2003). The cGMP‐stimulated activity scheme yielded the same kinetics as the basal activity‐only scenario: For physiological stimulation levels, the enhanced activity of a few PDE5 molecules is negligibly small compared with the basic activity of ten thousand of active PDE5 molecules. Although GAF‐A domains display a relatively high cGMP affinity (K D ~200 nM; Francis et al, 2011), they do not compete with CNGK or the catalytic PDE domain for cGMP, because its k on is almost 1,000‐ and 100‐fold lower than k on for cGMP binding to the CNGK channel and to the catalytic PDE5 domain, respectively (Appendix Table S3). Thus, putting the kinetic argument aside, even at equilibrium, the CNGK channel captures ≥ 90% of cGMP molecules.
Control of V m recovery by HCN channels
In many cells, HCN channels counteract hyperpolarization and, thereby, serve several different functions. In the retina, HCN channels extend the frequency response and operational light range of rod and cone photoreceptors (Fain et al, 1978; Barrow & Wu, 2009). In neurons and heart cells, HCN channels sustain oscillatory activity and modulate resting V m (Pape, 1996). In sperm, the precise function of HCN channels has been elusive. Two different HCN channel subunits (HCN1 and HCN2) exist in sea urchin sperm (Gauss et al, 1998; Galindo et al, 2005). Do these subunits form homomeric or heteromeric channels? What is their physiological function? We detected in the flagellum 10,000 and 18,000 copies of HCN1 and HCN2 channel subunits, respectively. Do HCN channels counteract the chemoattractant‐induced hyperpolarization? We compared the rate of V m recovery after weak and strong cGMP stimulation, i.e., when HCN channel activity is low and high, and for both depolarizing (100KASW) and hyperpolarizing (ASW) responses. For strong hyperpolarization, V m completely recovered within 110 ms, while a large fraction of CNGK channels was still open (Fig 5A, compare blue and red traces). Thus, HCN channels clearly outperform CNGK channels. For weak stimuli, i.e., for small depolarization or hyperpolarization, the V m response relaxed with similar time constants of about 300 ms (Fig 4A, compare pink and navy traces). The 1.5‐fold larger amplitude of the hyperpolarizing compared with the depolarizing signal is due to the larger electrochemical driving force. We systematically determined the recovery time over a wide range of stimulus strength (Appendix Fig S1A). The recovery time was similar (τrelax = 280 ms) (Fig 5B inset) when ρ 30 cGMP molecules (equivalent to ≤ 3 chemoattractant molecules) were produced, and hastened gradually with increasing hyperpolarization, i.e., HCN channel activation (Fig 5B). In conclusion, HCN channels accelerate recovery—except for weak stimuli.
Figure 5. Recovery from hyperpolarization.
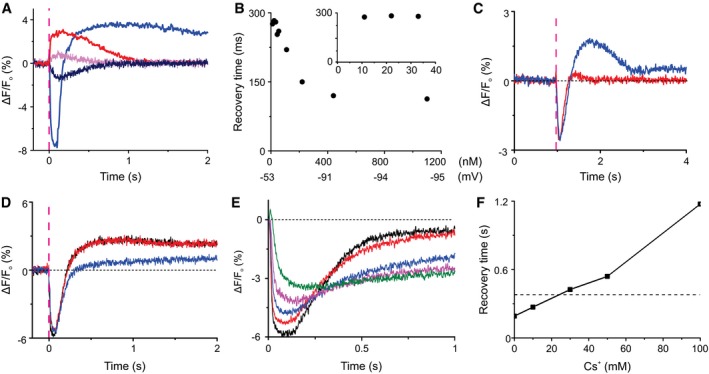
- Time course of cGMP‐evoked changes in V m in 100KASW (red, 500 nM cGMP; pink, 9 nM cGMP) and normal ASW (blue, 500 nM cGMP; navy, 9 nM cGMP). The time constants of recovery after cGMP release (9 nM) were 289 ms (depolarization) and 303 ms (hyperpolarization). Signals in panels (A–C) were detected using the potentiometric probe FluoVolt; dashed magenta lines indicate the timing of the photolysis flash.
- Representative measurement of the dependence of recovery‐time constant on the amount of released cGMP. Inset: Decay time was constant for low stimulation levels equivalent to ρ 30 cGMP molecules or ≤ 3 resact molecules.
- Voltage response evoked by photolysis of DEACM‐caged cGMP (blue) or 8‐Br‐cGMP (red). The light energy was adjusted such that V m response amplitudes became similar; for 8‐Br‐cGMP, about five times more light energy was required. Light pulse 10 ms.
- V m responses evoked by cGMP in the presence (blue) and absence (red) of the HCN channel blocker ZD7288 (50 μM).
- cGMP‐evoked V m response in ASW (black), in the presence of IBMX (1 mM) (red), and in Ca2+‐free ASW ([Ca2+] ≤ 500 nM) in the presence of IBMX (1 mM) (blue). Signals in panels (D–F) were detected using the potentiometric probe Di‐8‐ANEPPS in the ratio mode.
- Normalized responses from panel (E).
We scrutinized this conclusion by inhibiting either cGMP hydrolysis or HCN activity. The relaxation times of hyperpolarizing responses evoked by photolysis of caged 8‐Br‐cGMP or caged cGMP were similar (Fig 5C), except for a depolarizing overshoot. Similarly, inhibition of PDE activity by high concentrations of IBMX (1 mM) did not retard recovery (Fig 5D). The depolarizing voltage overshoot was abolished either in the absence of Ca2+ (Fig 5D) or when 8‐Br‐cGMP was used as a stimulus (Fig 5C), suggesting that recovery is first driven by a Na+ inward current carried by HCN channels, followed by Ca2+ currents carried by CatSper channels. Finally, inhibition of the HCN channels by the drug ZD7288 (Postea & Biel, 2011) strongly slowed down recovery (Fig 5E and F). In the presence of the drug, V m signals also became smaller (Fig 5E), because the drug also shifted V rest to more negative values, which compressed the operational range of hyperpolarization (Appendix Fig S1B). Of note, in the presence of ZD7288, the recovery time is similar to that at 100KASW, i.e., in the absence of HCN channel activity. Collectively, these results show that except for low stimulation levels, HCN channel activation controls the time of recovery from hyperpolarization.
Components of Ca2+ signaling
Three different signaling proteins control Ca2+ homeostasis: the Ca2+ channel CatSper, a NCKX exchanger, and a Ca2+‐ATPase (Fig 1B). Their quantitative stoichiometry provides several insights. First, CatSper in mammalian sperm comprises four homologous pore‐forming subunits (CatSper 1‐4) (Navarro et al, 2008) and at least five auxiliary subunits (CatSper β, γ, δ, ɛ, and ζ) (Liu et al, 2007; Wang et al, 2009; Chung et al, 2011, 2017). CatSper1‐4, β, γ, and δ were also detected in Arbacia sperm (Seifert et al, 2015). Except for CatSper3, each of the subunits was equally abundant, suggesting that each subunit contributes one copy to a large CatSper complex. The CatSper 3 quantification relied on one peptide only; thus, the estimate is less reliable than for the other CatSper subunits.
Second, the NCKX exchanger is one of the most abundant signaling molecule, only second to the receptor GC (Table 1). By contrast, the PMCA pump is 50‐fold less abundant, showing that Na+/Ca2+/K+ exchange is the major if not only mechanism of Ca2+ clearance from the flagellum. In fact, PMCA is more abundant in the head than the flagellum (Gunaratne et al, 2006). The NCKX exchanger is also about 50‐fold more abundant than the CatSper channel. This high ratio is required because of the vastly different transport numbers of ion channels and ion exchangers: Ca2+ channels can carry > 1,000/ions/s (Hille, 2004), whereas NCKX exchangers can transport about 100–300 Ca2+/ions/s (Jalloul et al, 2016).
NCKX transporters use inward Na+‐ and outward K+ gradients for Ca2+ extrusion (Cervetto et al, 1989; Schnetkamp, 2013). The exchange is electrogenic (4Na+:1Ca2+:1K+) and therefore enhanced at negative membrane potentials (Cervetto et al, 1989). The involvement of Na+‐ and K+ gradients and the electrogenic mechanism suggests rapid ion‐exchange kinetics and very low Ca2+ levels at rest. NCKX is primarily expressed in sensory neurons, in particular in rod and cone photoreceptors (Schnetkamp et al, 2014). The resting [Ca2+]i in rods is 100 nM and, during hyperpolarization, can reach levels as low as 5–10 nM (Woodruff et al, 2002). We assume that [Ca2+]i in sperm can also adopt such low levels.
Third, its high density argues that NCKX can restore basal Ca2+ levels in the flagellum before Ca2+ diffuses to the head compartment. We tested this presumption by recording cGMP‐evoked Ca2+ changes, using the fluorescent Ca2+ indicator GFP‐certified FluoForte, in the flagellum and the head separately. A flash of light, producing 28 cGMP molecules (equivalent to approximately 3 resact molecules), evoked a short Ca2+ pulse in the flagellum, but no significant change in the head (Fig 6A). For stronger stimulation, a small delayed Ca2+ signal in the head became apparent (Fig 6B). In conclusion, for low stimulation levels, the Ca2+ response is restricted to the flagellum; for higher stimulus strengths, the signal partially propagates from the flagellum to the head.
Figure 6. Compartmentalization of Ca2+ and pHi signals. Changes in relative fluorescence ∆F/F recorded from the head and the flagellum are shown in blue and black, respectively.
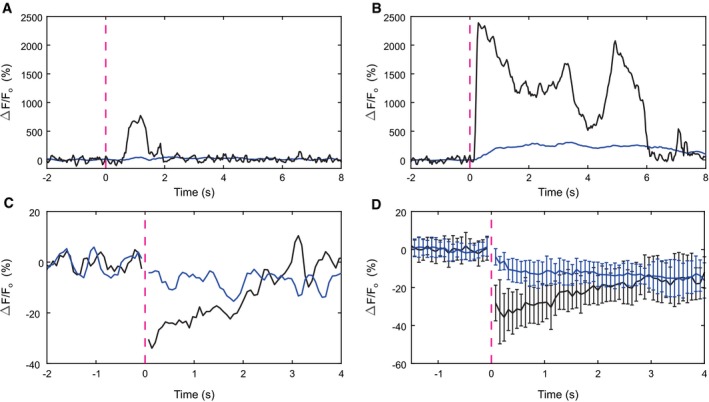
-
A, BCa2+ signals elicited by cGMP release and recorded with FluoForte. The UV flash released 28 (A) and 215 B) cGMP molecules. Light flashes are indicated by dashed magenta lines.
-
CpHi signals, elicited by the release of approximately 1,400 cGMP molecules, were recorded with pHrodo Red. An increase in pHi is indicated by a decrease in ∆F/F. Exemplary recording from a single cell.
-
DAverage of pHi signals recorded from n = 25 cells. Error bars represent SD See Appendix Fig S2 for a longer time course.
Changes in pHi are compartmentalized
A voltage‐activated Na+/H+ exchanger (SLC9C1 or sNHE) is the only molecule known in sperm to regulate pHi. Its exquisitely high density (Table 1) explains why changes in pHi occur on a millisecond time scale (Seifert et al, 2015; Windler et al, 2018). We examined whether cGMP‐evoked changes in pHi, similar to Ca2+ signals, are restricted to the flagellum. Changes in pHi were measured, using the fluorescent pH indicator pHrodo Red, in the flagellum and the head separately (Fig 6C and D). The pHi rose rapidly in the flagellum and declined again within 3 s (Fig 6C). By contrast, the alkalization in the head was much smaller and slower. After about 4 s, the pHi in flagellum and head was equal and slightly elevated. This result is consistent with the idea that Na+/H+ exchange in the flagellum elevates pHi (Windler et al, 2018) and that this change slowly propagates to the head.
Discussion
In a typical cell soma, signaling proteins are less or about equally abundant than their respective substrates; processing of many substrate molecules by a single active enzyme lies at the heart of cellular signal amplification. For example, in the ERK and AKT pathways concentrations of signaling proteins range between 10 and 1,000 nM (Adlung et al, 2017). By contrast, the enzyme‐to‐substrate ratio in the sperm flagellum is reversed: Signaling proteins are up to 1,000‐fold more abundant (micromolar range) than their free substrates (nanomolar range). Simply put, a large number of enzymes, transporters, and channels compete for few substrate molecules, and a single enzymatic turnover by a minute fraction of signaling proteins suffices to initiate or upend a cell response. Clearly, this minuscule reaction vessel is designed for a specific purpose: low‐noise, perfect molecule detection rather than signal amplification.
The high‐affinity CNGK channel acts as a close‐to‐perfect cGMP detector that avidly adsorbs cGMP molecules and translates binding events into an incremental voltage response. Binding happens faster than spreading of cGMP along the flagellum by unrestricted diffusion (about 1 s) (Pichlo et al, 2014). Furthermore, PDE5 is unlikely to kinetically compete for cGMP binding. Thus, the single‐molecule sensitivity of sperm ultimately rests not only on the exquisitely high density and affinity of the receptor GC (Pichlo et al, 2014), but also on the CNGK channel as well.
The target proteins of cAMP, Ca2+, and H+ are also orders of magnitude more abundant than the free messengers themselves. The high density serves two different functions. Rapid sequestering of cellular messengers attenuates their diffusional exchange between flagellum and head. Kinetic compartmentalization enhances antennal sensitivity and shields acrosomal exocytosis in the head, which is also controlled by cAMP, Ca2+, and pHi (Buffone, 2016), from signaling events in the flagellum. In addition, the time derivative of the Ca2+ response d[Ca2+]i/dt rather than absolute [Ca2+]i determines the curvature of the swimming path (Alvarez et al, 2012). Therefore, kinetic control of the Ca2+ influx/efflux balance is key to chemotactic navigation. Transport rates of solute carriers are orders of magnitude slower than those of ion channels. The 60‐fold higher NCKX density compared with CatSper ascertains that the Ca2+ rise can be rapidly counterbalanced. The same argument holds for Na+/H+ exchange by sNHE that, within milliseconds, can alkalize the flagellum—a key event that primes Ca2+ entry (Seifert et al, 2015; Windler et al, 2018).
Slow dissociation of cGMP from its high‐affinity site at CNGK is at odds with fast 380‐ms cGMP turnover. This discrepancy is reminiscent of a similar conundrum about cAMP recycling in the protein kinase A (PKA) system. The regulatory subunit R binds cAMP with extremely high affinity (K D = 2–10 nM); consequently, cAMP recycling is expected to be slow. The decade‐long riddle (Ramirez‐Sarmiento, 2017) was recently unraveled by proposing a “substrate‐channeling” mechanism: cAMP, PDE8, and RIα form a ternary complex; cAMP is transferred between active sites of RIα and PDE8 by restricted diffusion without release into the cytosol (Krishnamurthy et al, 2014, 2015; Ramirez‐Sarmiento, 2017; Tulsian et al, 2017). The allosteric active‐site coupling tweaks cAMP dissociation from RIα and, thereby, accelerates hydrolysis. Notably, PDE8 is only active in the RIα‐cAMP‐PDE8 complex (Tulsian et al, 2017), a mechanism that prevents breakdown of free cAMP. Two lines of evidence suggest that CNGK and PDE5 form a similar “channeling” complex (Fig 7A): cGMP turnover is much faster than estimated from reaction kinetics in free solution, and the CNGK/PDE5 ratio is 1:1 (assuming PDE5 forms dimers via the GAF‐B domain; Francis and Corbin (2010)). Future work needs to establish this mechanism for PDE5. The “channeling” mechanism renders cGMP binding to CNGK essentially irreversible; before cGMP has a chance to dissociate, it is hydrolyzed. Within the framework of chemosensation physics (Berg & Purcell, 1977; Endres & Wingreen, 2008), the inner surface of the flagellar membrane constitutes a perfect absorber rather than a monitoring surface that allows dissociation and rebinding of a ligand. The sensing precision of a perfect absorber is 2.5‐fold enhanced compared with a monitoring surface (Endres & Wingreen, 2008).
Figure 7. Channeling model of cGMP turnover.
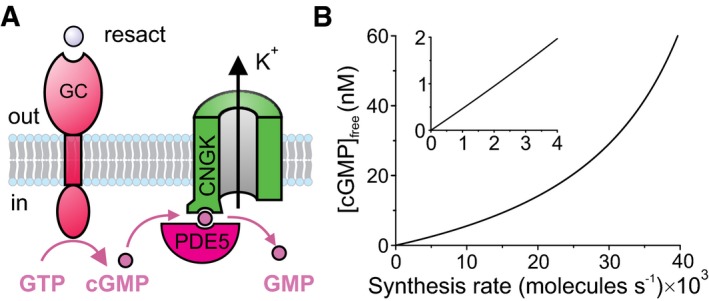
- cGMP molecules synthesized by the GC receptor rapidly bind to the CNGK channel. CNGK, PDE5, and cGMP form a ternary complex, and cGMP bound to CNGK is subsequently transferred to PDE5 for hydrolysis without being released to the cytosol.
- Relationship between cGMP synthesis rate and free cGMP concentration. We assume that a single GC molecule can synthesize 10 cGMP molecules during its lifetime of 150 ms (Pichlo et al, 2014). Inset: expanded scale.
A far‐reaching consequence of rapid cGMP binding to CNGK and “channeling” to PDE5 is that the mean resting‐free cGMP concentration is almost zero and that the cGMP action happens on the membrane surface rather than the cytosol. Several back‐of‐the‐envelope estimates illustrate this conclusion. The free cGMP concentration is determined by the rates of synthesis and hydrolysis. To maintain a free cGMP concentration of 1 nM (equivalent to one cGMP molecule/flagellum) in the face of a basal hydrolysis rate of 2.6/s requires the synthesis of 2,103 cGMP molecules/s by 210 active GC molecules/s (calculated from a turnover rate of 72 cGMP/s/GC and a lifetime of 150 ms of active GC; Pichlo et al (2014)). Such basal activity would be equivalent to 30 pM of the chemoattractant; at this concentration, 210 chemoattractant molecules/s hit the flagellum; Berg, 1993; Pichlo et al, 2014). When PDE activity is blocked, the resting cGMP level should rise to high micromolar concentrations. In fact, elevated cGMP levels were not observed in such experiments (Kaupp et al, 2003), arguing that non‐stimulated GC activity at rest is negligible. Moreover, such high background activity would seriously compromise if not abolish single‐molecule detection of sperm. For comparison, single‐photon sensitivity in rod photoreceptors is limited by spontaneous thermal activation of roughly one rhodopsin molecule/min (Luo et al, 2011). Finally, at chemical equilibrium with 1 nM free cGMP, approximately 800 cGMP molecules are bound to CNGK channels [calculated from K D = 26 nM (Bönigk et al, 2009) and cCNGK = 22 μM; Table 1)]. Because binding of a single cGMP molecule can open CNGK (Bönigk et al, 2009), 800 CNGK channels would be permanently open at rest. By comparison, the opening of maximally 300 CNGK channels by the chemoattractant saturates the V m response (Strünker et al, 2006). In conclusion, free cGMP concentration is virtually zero at rest.
We propose that the GC/CNGK/PDE5 triad is designed to encode rates of cGMP production rather than free cGMP concentrations. Over a 40,000‐fold range of cGMP synthesis rates, the free cGMP concentration increases almost linearly (Fig 7B). Of note, even for strong stimulation, a rate‐encoding mechanism never saturates the CNGK channel with cGMP and allows cells to operate over a wide dynamic range. Another important benefit of rate‐detection is the speed with which a cell response can be initiated: No binding equilibrium has to be reached, and no diffusional times have to be awaited. A cell can rapidly and continuously sample a stimulus landscape and respond in real time. The mechanism is optimal to probe a chemical gradient on periodic paths with 1–2 Hz frequency.
Finally, our study also illustrates fundamental limits to track cellular messenger concentrations in small compartments using genetically encoded or chemical sensors. The sensitivity of available cAMP/cGMP sensors (ranging between 70 nM; Mukherjee et al (2016) and 3 μM; Halls and Canals (2018)) would be not sufficient to detect changes in free cyclic nucleotide concentrations in sperm. Enhancing a sensor's ligand affinity creates another dilemma because sensors become inherently slow and are expected to seriously interfere with the in situ kinetics and steady‐state concentrations of messengers. These challenges are exacerbated in primary cilia (volume about 0.2 fl), where one cAMP molecule is equivalent to 10‐nM concentration, and a single molecule of adenylate cyclase can produce 100 cAMP molecules/s (equivalent to 1 μM/s). Using intrinsic sensors as read‐out for cAMP/cGMP signaling combined with quantitative photonic control of cAMP/cGMP, as shown here, may overcome this fundamental problem.
The mechanisms of kinetic compartmentalization of cilia seem to depend on the nature of the messenger. The porous cilia base allows small proteins to freely enter or exit the cilium (Kee et al, 2012; Breslow et al, 2013). Accordingly, Ca2+ ions can freely diffuse from the primary cilium into the cell soma. Nevertheless, the ciliary [Ca2+] is threefold to fivefold higher than that in the soma (DeCaen et al, 2013; Delling et al, 2013). The high concentration is maintained by Ca2+‐permeant channels in the cilia membrane despite steady Ca2+ diffusion into the cytoplasm at the ciliary base (Delling et al, 2013). Thus, the kinetic balance of Ca2+ entry and exit determines ciliary Ca2+ levels. Considering that the soma volume is up to 50,000‐fold larger than the ciliary volume, the steady Ca2+ flux into the soma does not change the cytosolic [Ca2+]i appreciably.
In a model of the cAMP‐regulated sonic hedgehog (Hh) pathway, cAMP leaves the cilium and activates PKA at the ciliary base or in the soma (Mukhopadhyay & Rohatgi, 2014). However, a recent study using proximity‐labeling MS identifies several cAMP‐signaling proteins in cilia of IMCD‐3 cells (Mick et al, 2015), including AC5, AC6, and RIα and RIIβ subunits of PKA, suggesting that activation of the transcription factors GLI2 and GLI3 by PKA phosphorylation occurs inside the cilium during transient visits of the respective targets (Mick et al, 2015). The absolute and relative abundance of ciliary ACs and PKA is largely unknown (for a comprehensive discussion see Mick et al, 2015). The ciliary free cAMP concentration reportedly is high (4 μM), i.e., fivefold higher than in the soma of embryonic fibroblasts (Moore et al, 2016). How such high concentrations can be reconciled with a PKA sensitivity of < 10 nM cAMP remains enigmatic. We envisage that similar scenarios of high‐abundance signaling proteins, kinetic compartmentalization, substrate‐channeling, and reaction‐rate sensing may endow primary cilia with exquisite sensitivity, speed, and a wide dynamic range. Rigorous quantification of signaling repertoires combined with rapid photonic techniques, as outlined here, may provide unprecedented insights not only for ciliary signaling, but also for other small subcellular compartments like neuronal dendrites and spines.
Materials and Methods
Preparation of flagella
The collection of A. punctulata sperm and the preparation of flagella were as described in Seifert et al (2015) with one modification: Instead of shearing with a 24‐G needle, the sperm suspension was sheared 20 times by centrifugation for 30 s at 75× g and 4°C through a 40‐μm mesh of a cell strainer (BD Biosciences, USA). Flagella were washed in artificial seawater (ASW), containing (in mM): 423 NaCl, 9.27 CaCl2, 9 KCl, 22.94 MgCl2, 25.5 MgSO2, 0.1 EDTA, and 10 HEPES, adjusted to pH 7.8 with NaOH, and stored as pellet.
Analysis of solubilization efficacy by Coomassie staining and Western blotting
The precise determination of copy numbers and protein stoichiometries relies on the efficacy of protein solubilization by detergents. Therefore, we tested for different solubilization conditions the fraction of proteins in the supernatant and the pellet by SDS–PAGE and Western blotting of selected membrane‐associated proteins (Appendix Fig S3). Flagella were resuspended in solubilization buffer [10 mM Tris–HCl, pH 7.6; 140 mM NaCl, 1 mM EDTA, 1% dodecyl‐maltopyranoside (DDM), protease inhibitor cocktail 1:500 (P8340, Sigma‐Aldrich, USA)]. The protein concentration determined by BCA assay was 1.5 μg/μl. The solubilizate was split into three samples for parallel incubation. Tubes 1 and 2 were incubated on ice for 30 min, and tube 3 was incubated on ice for 120 min. Flagella in tube 2 were sonicated three times for 30 s (Sonifier 450, Branson, Danbury, CT). After incubation, supernatant and pellet were separated by centrifugation (10,000× g at 4°C).
To compare the different solubilization conditions, proteins in the supernatant and pellet were separated on a 7.5% SDS gel for Coomassie staining or on an 8% SDS precast gel (SurePAGE, Genscript, NJ, USA) for Western blotting. Each lane was loaded with the equivalent of 8 μg of flagellar protein. Pre‐stained Protein Marker VI (AppliChem, Darmstadt, Germany) was used as molecular weight marker.
Proteins were transferred onto an Immobilon FL PVDF membrane (Merck Millipore, Darmstadt, Germany) and consecutively probed with various antibodies. After each probing, the Western blot was analyzed using the Odyssey Imaging System (LI‐COR, Bad Homburg, Germany). All figure panels were taken from the same Western blot. Figures were prepared using CorelDraw X6 and Photo‐Paint X6 software (both from Corel Corporation). Primary antibodies were monoclonal: rat GCN 3D10 (anti‐GC, 1:200) (Pichlo et al, 2014), polyclonal rabbit ETK rb1 (anti‐ApsAC, 1:200), monoclonal rat ApNHE 14E1 (anti‐NHE, 1:200), monoclonal rat AP47G4 (anti‐ApCNGK (repeat 4), 1:20) (Bönigk et al, 2009), and monoclonal mouse α‐tubulin B‐5‐1‐2; (1:5,000, Sigma, T5168). Antibodies have been produced in rat (monoclonal) or rabbit by the Helmholtz Zentrum München (German Research Center for Environmental Health, Monoclonal Antibody Core Facility, Germany) and LifeTein (Somerset, NJ, USA), respectively. Secondary antibodies were as follows: anti‐rat or anti‐rabbit IRDye680 antibody (1:25,000, red channel) and anti‐rat or anti‐mouse IRDye800 antibody (1:25,000, green channel) from LI‐COR.
The major integral membrane protein, the receptor guanylate cyclase (GC), and the equally abundant membrane‐associated creatine kinase (CK) were almost completely transferred to the supernatant. For the GC, we estimated that < 10% remained in the pellet (Appendix Fig S3A). A similar result was obtained for the soluble adenylate cyclase (sAC), the CNGK channel, and the Na+/H+ exchanger (sNHE) (Appendix Fig S3B). The proteins in the pellet were largely axonemal proteins, e.g., tubulin (55 kDa) and an intermediate chain of the motor protein dynein (75 kDa). Furthermore, copy number ratios in the DDM‐extracted supernatant were compared with ratios in flagella solubilized in DDM plus SDS‐sample buffer. Except for PDE10, these ratios were similar within experimental error (Appendix Table S4). In conclusion, for quantitative MS, aliquots of flagella from six animals were resuspended and incubated for 30 min on ice in solubilization buffer. After centrifugation at 10,000× g and 4°C, the supernatant was transferred to a new tube.
Selection of candidate peptides for protein quantification
Candidate peptides were identified from previous data using MudPIT and one‐dimensional sodium dodecyl sulfate–polyacrylamide gel‐electrophoresis followed by liquid chromatography (GeLC‐MS) (Seifert et al, 2015). For MudPIT, flagella were washed twice with 0.1 M (NH4)2CO3 and sedimented by ultracentrifugation (100,000× g, 30 min, 4°C). Membrane pellets were resuspended, sonicated, and processed by tryptic in‐solution digestion (sequencing grade‐modified trypsin, Promega) in a methanol and NH4HCO3 buffer (Fischer & Poetsch, 2006). After removal of membranes by ultracentrifugation, samples were desalted using Spec PT C18 AR tips (Varian). For GeLC‐MS, 45 μg flagella protein was loaded on a 10% SDS–PAGE gel. The gel was stained with colloidal Coomassie G‐250, then destained with acetic acid and the separation gel divided into 12 pieces. Each piece was cut into gel cubes and digested (Pichlo et al, 2014) with 6.25 ng/μl trypsin in a NH4HCO3 buffer (20 mM at pH 8.6). Spectra obtained from GeLC‐MS and MudPIT were searched against the latest target sequences (to be published) using SEQUEST algorithm, embedded in Proteome Discoverer™ (Rev. 1.4.1.14, Thermo Fisher Scientific, USA). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez‐Riverol et al, 2019) partner repository with the dataset identifier PXD015332. Additionally, an in silico prediction using Skyline software (Version 3.7) (MacLean et al, 2010) was performed to complete the list of candidate peptides for quantification. Only peptides without methionine, cysteine, and without arginine or lysine motifs like KK, KR, RK, or RR were validated in subsequent SRM measurements of A. punctulata sperm flagella. The 53 most suitable peptides (Table 2) were selected for quantification of the 19 polypeptides involved in chemotactic signaling. The corresponding 13C, 15N Arg/Lys‐labeled QconCAT standard protein (Mw 65.42 kDa; concentration: 1.22 mg/ml; 15.2 nmol/mg) was designed and custom‐synthesized by PolyQuant GmbH (Germany). Importantly, the isotope‐labeled protein was used to confirm the identity of the previously selected 53 peptides by manual pairwise comparison of retention times during LC and by comparison of fragment ions of the respective sample and standard peptides. For three peptides, no correct light/heavy pairs were observed; these peptides were excluded from quantitative analysis (Table 2). Ultimately, 50 peptides were used for protein quantification.
Quantification of chemotaxis signaling proteins by selected reaction monitoring (SRM) mass spectrometry
For quantification, an aliquot of 20 μg sperm flagella protein was spiked with either 0.15, 1.5, or 15 pmol isotope‐labeled standard protein to account for different concentrations of sperm target proteins. The standard protein/sample mixtures were subjected to 12.5% (v/v) SDS–PAGE. The SDS–PAGE was stopped after the sample had migrated about 1 cm into the separation gel, and the gel was stained with Coomassie Blue. The about 1‐cm‐long gel piece was cut into 1–2 mm‐sized pieces. In‐gel digestion was performed as described (Pichlo et al, 2014) with 6.25 ng/μl trypsin in a NH4HCO3 buffer (40 mM at pH 8.6).
The LC‐MS/MS system consisted of a TSQ Vantage Triple Quad MS (Thermo Fisher Scientific, USA) interfaced with a nanoAcquity UHPLC system (Waters, USA). LC used an Acquity UPLC M‐Class Symmetry C18 trap column (5 μm particle size, 180 μm × 20 mm, 2D, V/M) and an Acquity UPLC M‐Class HSS T3 analytical column (1.8 μm particle size, 75 μm × 150 mm) (Waters, USA). LC conditions (66‐min multistep gradient from 1 to 99% acetonitrile/0.1% formic acid at a flow rate of 0.4 μl/min) were as described (Pichlo et al, 2014). SRM parameters for the TSQ Vantage and data analysis with Skyline 3.7 were as described (Pichlo et al, 2014) with the following changes: The two most intense and specific fragment ions (transitions) were used for quantification, and a measuring time window of ± 2.5 min from each peak apex and a 3‐s cycle time were set. For optimal ion transmission, the following S‐lens RF values were fixed: 53 at 182.082 m/z, 109 at 508.208 m/z, and 220 at 997.398 m/z. The collision energy was first predicted by Skyline 3.7 and then individually optimized for each precursor ion. Heavy‐isotope incorporation was determined by in‐solution digestion of the standard protein, followed by SRM analysis as described above. Heavy‐isotope incorporation was 97% (arginine) and 98% (lysine).
Calculation of copy numbers and statistical analysis
All SRM data were manually inspected to ensure correct peak identification. Furthermore, from three different amounts of 0.15, 1.5, or 15 pmol standard protein per 20 μg flagella protein, individual L/H ratios were selected to exclude extreme values. The peak area of each light (sperm sample) and heavy standard peptide was calculated using Skyline 3.7 and exported to an Excel sheet for further data processing. The total area of each heavy peptide was corrected for the percentage of heavy‐isotope incorporation. For example:
| Total area heavy | Heavy‐isotope incorporation | Total area heavy corrected | ||
|---|---|---|---|---|
| 5,558,109 | / | 0.971 | = | 5,724,108 |
The total area of each light peptide was corrected for the percentage of light‐isotope incorporation in the isotope‐labeled standard protein:
| Total area light | Total area heavy corrected | Light‐isotope incorporation | Total area light corrected | |||
|---|---|---|---|---|---|---|
| 556,239 | – | (5,724,108 | * | 0.029) | = | 390,240 |
The ratios between the corrected peak areas of each light and heavy peptide were calculated using Excel. Based on these ratios, the numbers of molecules of the light peptides were calculated using Avogadro's number. For the different peptides assigned to the same protein, the median and the standard deviation were calculated. For each replicate, the ratios between the different proteins, e.g., guanylate cyclase‐to‐creatine kinase ratio, and the mean ± SD of all replicates belonging to the same animal were calculated. Representative results are presented in Dataset Table EV2, and the underlying RAW files and Skyline analyses have been made publicly available at ProteomeXchange with identifier PXD015502. Furthermore, the ratios between signaling proteins and the mean ± SD of the replicates belonging to the same animal are provided in Dataset Table EV1.
Processing of electron microscopy images
For the measurement of flagellar diameters and axonemal protein volumes, the following previously published cryo‐electron tomography and subtomogram averaging data were used: the three‐dimensional (3D) reconstructions of intact wild‐type Chlamydomonas reinhardtii and sperm flagella from the sea urchin Strongylocentrotus purpuratus (Lin & Nicastro, 2018) for flagellar diameter, the 3D structures of the averaged DMT‐associated (Lin et al, 2019) and central pair complex repeats (preprint: Fu et al, 2019) of wild‐type Chlamydomonas, as well as the averaged DMT‐associated repeats (Lin & Nicastro, 2018), and central pair complex repeats of sea urchin sperm flagella (Carbajal‐Gonzalez et al, 2013).
The IMOD software (Kremer et al, 1996) and ImageJ (Schneider et al, 2012) were used for visualization of the tomographic slices and measurement of the diameters (measured at the inside of the flagellar membrane), respectively. Only tomograms with straight and non‐ or mildly compressed flagella were used for diameter measurements. In total, 6 tomograms of Chlamydomonas flagella and 19 tomograms of sea urchin sperm flagella were selected, and their diameters were measured at three different positions: proximal end, midpoint, and distal end.
The UCSF chimera package (Pettersen et al, 2004) was used for estimating the volumes occupied by axonemal proteins using the subtomogram averages of the 96‐nm DMT‐associated repeat (V DMT) and the [3 × 32 nm] central pair complex repeat (V CPC). The isosurface‐rendering threshold for volume measurements was determined using the average density of 1.43 g/cm3 (Quillin & Matthews, 2000) for proteins and normalizing the threshold so that the measured molecular mass of microtubule protofilaments and dynein heads matched the calculated molecular masses for a 96‐nm long protofilament (~1,200 kDa) and dynein head domain (~350 kDa). For instance, the surface threshold for the averaged 96‐nm DMT‐associated axonemal repeat of Chlamydomonas was set to 125.1, resulting in estimated molecular weights for the IDA a dynein head and protofilament B7 of 343 and 1,210 kDa, respectively.
Volume calculations
The measured average diameters (d = 2r) of the Chlamydomonas and sea urchin sperm flagella were used to calculate the total volumes V t for a flagellar length (l) as V t = π × r 2 × l. The volumes of the axonemal proteins were calculated for 96‐nm length as V a = (9 × V DMT) + V CPC and then scaled to 1‐μm flagellar length. For measuring the luminal volumes V l inside the DMTs and central pair microtubules, we “filled” the microtubule lumen in the subtomogram averages of the 96‐nm DMT‐associated repeat and the [3 × 32 nm] central pair complex repeat by setting the pixel values inside the microtubules to zero (i.e., black like protein) using a mask or the IMOD command imodmop. Using the same chimera thresholds as for estimating the volumes V DMT and V CPC before, we estimated the volumes of the “filled” averages V* DMT and V* CPC and calculated V l = (9 × V* DMT) + V* CPC – V a for 96‐nm length, which was then scaled to 1‐μm flagellar length.
Stopped‐flow techniques
Resact‐ and cGMP‐induced changes in membrane voltage were measured with a voltage‐sensitive dye (FluoVolt, Thermo Fisher) in a stopped‐flow device (SFM‐400; Bio‐Logic) as described previously (Hamzeh et al, 2019). Dry sperm was suspended 1:6 (v/v) in loading buffer containing ASW and 5 μM fluorescent dye. After incubation for at least 20 min at 18 °C, the sample was diluted 1:20 with ASW in the presence of 15 μM caged DEACM‐cGMP and 0.05% Pluronic F127 (Sigma‐Aldrich). In the stopped‐flow device, sperm were rapidly mixed 1:1 with either ASW or ASW containing 191 mM K+. Mixing with 191 mM K+ ASW yielded a final K+ concentration of 100 mM (referred to as 100KASW). Fluorescence was excited by a Spectra X Light Engine (Lumencor). Emission was recorded by photomultiplier modules (H9656‐20; Hamamatsu Photonics). Data acquisition was performed with a data acquisition board (PCI‐6221; National Instruments) and Bio‐Kine software v. 4.49 (Bio‐Logic). The excitation light was passed through a BrightLine 513/17 nm filter (Semrock). The excitation light was operated at 1–10 kHz. Emission was recorded through a 542/20 nm filter by a photomultiplier module and filtered through a lock‐in amplifier. Signals represent the average of at least two recordings and are depicted as the percent change in fluorescence (ΔF) relative to the mean of the first 5–10 data points before the onset of the signal (F o). The control (ASW) ΔF/F o signal was subtracted from the resact‐ or cGMP‐induced signals. The data obtained were analyzed and plotted using OriginPro 9.0 (OriginLab Corporation).
Caged compounds and flash photolysis
Caged cGMP was photolyzed using a 365‐nm LED (M365LP1, Thorlabs). The LED was coupled to a liquid light guide and delivered to the cuvette (FC‐15 Bio‐Logic). The fluorescence background resulting from the 365‐nm LED was removed using lock‐in detection. The waveform, timing, and triggering of the photolysis light were controlled through the interface of a self‐made LabVIEW program using a DAQ card (NIUSB‐625; National Instruments). The photolytic release of cGMP was calibrated via the increase in fluorescence upon photolysis (Hamzeh et al, 2019).
Data analysis and simulations
Simulations of cGMP dynamics in the flagellum were based on the following reaction schemes:
|
|
(1) | |
|
|
(2) | |
|
|
(3) | |
|
|
(4) | |
|
|
(5) |
These reactions correspond to the release of cGMP from its caged derivative (reaction 1), binding (unbinding) of cGMP to (from) the CNGK channel (reaction 2), binding (unbinding) of cGMP to (from) the allosteric GAF‐A domain of the inactive PDE (PDEi) to produce active PDE (PDEa; reaction 3), cGMP binding to the catalytic domain of PDE of either PDEi or PDEa (PDEi,a) (reaction 4), and cGMP hydrolysis by PDE (reaction 5). Reaction rates were taken from previous studies (see Appendix Table S3). The catalytic rate for hydrolysis (k cat) of cGMP by PDEi is assumed to be about one‐third of that of PDEa, as reported for mammalian PDE5 (Rybalkin et al, 2003). The apparent k on for cGMP binding to the catalytic domain of PDE (reaction 4) was estimated assuming that the reaction is irreversible (k off ~0/s), and using the Michaelis–Menten relation: K M = (k off + k cat)/k on, wherein K M is the Michaelis–Menten constant. The maximal catalytic rate (k cat) was estimated from cGMP measurements of sperm stimulated with saturating resact concentrations (Appendix) (Kaupp et al, 2003).
Discrete‐event simulations were performed using the direct Gillespie method (Gillespie, 1977). Stochastic reaction rates were derived from the deterministic rates listed in Appendix Table S3. The on rates for the second‐order reactions 2, 3, and 4 were normalized by the flagellar volume as described (Gillespie, 1977). The flagellum was approximated by a cylinder 40 μm in length and a radius of 125 nm. Turnover rates were obtained by fitting of a mono‐exponential decay function to the average of 100 simulations.
Fitting of depolarizing V m signals
Depolarization signals resulting from 2‐ms flashes in the presence of 100KASW were fitted with the equation:
| (1) |
Estimate of time constant of cGMP binding
The time constant of cGMP binding to CNGK was calculated using:
| (2) |
Fitting of light‐evoked V m responses
A macroscopic heuristic model was used for fitting voltage data. The change in free cGMP concentration ([cGMP]) was derived for the mass‐balance equation:
| (3) |
where in k rel represents the rate of cGMP release by light and [PDE], [PDE·cGMP], and [CNGK·cGMP] are the free concentrations of PDE, PDE bound to cGMP, and CNGK bound to cGMP, respectively. For simplicity, we assume that [CNGK·cGMP] and V m are proportional to [cGMP] and that binding of cGMP to PDE is irreversible. Additionally, for low light levels, [PDE] and [CNGK] can be approximated by a constant.
Equation [Link] was solved for a square pulse stimulation (Fig 3C). All fits were performed using nonlinear least‐square methods. For a square release waveform, the release rate can be approximated by
| (4) |
where in k is a constant and t on and t off correspond to the time points where the UV light was switched on and off, respectively. Solving equation [Link] yields the following:
| (5) |
with A and k eff being constants.
Time constant of cGMP binding to CNGK
The time constant of cGMP binding to the CNGK channel was estimated to be between 1 and 2 ms (equation [Link]) using a lower and upper bounds of k on. A k on = 2.6 × 107/M/s was determined for a CNBD of similar affinity (Peuker et al, 2012). The k on is moderately dependent on ionic strength; it is maximally twofold larger at the high ionic strength of ASW compared with a Ringer solution (Peuker et al, 2012). CNGK concentration is 21.6 μM. This exercise shows that cGMP binding is at least an order of magnitude faster than τrise of cGMP‐induced ∆V m.
Estimate of membrane time constant
The τRC of sperm was estimated to be 5–20 ms assuming an input resistance of mouse sperm of about 10–20 GΩ (Navarro et al, 2007; Zeng et al, 2013). As membrane capacitance, we used Cm = 1 pF. In mouse, Cm was measured to be 2.5 pF (Navarro et al, 2007). Thus, the Cm is estimated to be 1.25 pF in sea urchin sperm, i.e., half of the value measured in mouse sperm, whose flagellum is about twofold longer (100 μm) than that of A. punctulata sperm. The time constant of CNGK activation is not known, but is of the order of a few milliseconds for ion channels (Hille, 2004). Thus, τrise of cGMP‐induced ∆V m agrees well with these estimates.
Estimate of the rate of cGMP by PDE5
The rate k cat for PDE5 was estimated based on previous studies where the cGMP kinetics was evaluated by quench‐flow techniques on intact sperm. In brief, sperm were mixed with saturating resact concentrations (250 nM) and the suspension was allowed to age. After different time intervals, the suspension was mixed with a denaturing agent (perchloric acid) stopping all biological reactions, and the cGMP concentration in the cell was determined using a radioimmunoassay (Kaupp et al, 2003). The total cGMP synthesized could be estimated from experiments performed in the presence of the PDE5 inhibitor IBMX to be about 110 pmol cGMP per 108 cells, i.e., about 6.6 × 105 cGMP molecules per sperm cell (or 660 μM), assuming a flagellar volume of 1.7 fl. At such high cGMP concentrations, all binding domains for cGMP are fully saturated. From the time difference between synthesis and total cGMP(t) in the cell, a hydrolysis rate of about 40 pmol cGMP per 108 cells in 0.2 s can be derived. This rate is equivalent to a k cat of 24/s. This rate is larger than that observed for mammalian PDE5 (approximately 4/s) (Francis & Corbin, 2010). Of note, this estimate for k cat could underestimate the catalytic rate of the sea urchin PDE5, if enzyme inhibition by IBMX was incomplete.
Single‐cell Ca2+ and pHi measurements
Sperm cells were suspended 1:100 (v/v) in ASW supplemented with 0.5% Pluronic F127 (Sigma‐Aldrich) and either 30 μM GFP‐certified FluoForte‐AM (Enzo Life Sciences; Ca2+ imaging) or 20 μM pHrodo Red (Molecular Probes; pH imaging). Samples were incubated for 45 min at room temperature; 10 min prior to the measurements, DEACM‐caged cGMP (15 μM) was included in the incubation. Fluorescence was recorded from single cells with an inverted microscope (Olympus IX71) equipped with a 20× objective lens (UPlanSApo 20×, 0.75 NA; Olympus) and two dichroic mirrors to image sperm that were loaded with GFP‐certified FluoForte (520LP; FF520‐Di02; Semrock) or pHrodo Red (560 DCXR; Chroma). For sharp imaging of the rapidly moving sperm flagellum, 2‐ms pulses of excitation light were used (Spectra X Light Engine; Lumencor). For Ca2+ imaging, Teal light was used in combination with the excitation band‐pass filter (513/17; FF01‐513/17; Semrock) and a long‐pass filter (BLP01‐532R; Semrock). Due to the large difference in emission intensity between head and flagellum, imaging with pHrodo Red resulted either in a poor dynamical range for imaging of the flagellum or in a saturating head signal. To overcome this problem, we used two alternating excitation wavelengths with different pHrodo Red excitation efficacies: Green excitation (Spectra X Light Engine; Lumencor) in combination with a band‐pass excitation filter (ET545/30; Chroma) was used to image the sperm flagellum, and Teal excitation (Spectra X Light Engine; Lumencor) in combination with an excitation filter (FF01‐513/17 nm; Semrock) for imaging of the head. Emission light from pHrodo Red was filtered using a band‐pass filter (BA575‐625; Olympus). Emitted light was collected using a back‐illuminated electron‐multiplying charge‐coupled device camera (DU‐897D; Andor Technology). Photolysis of DEACM‐caged cGMP was accomplished with either 365 nm LED (10 ms flashes; M365LP1, Thorlabs) or the 390‐nm LED from the Spectra X Light Engine (20‐ms flashes; Lumencor). Quantification of fluorescence signals was done using custom‐made software written in MATLAB (MathWorks) (Alvarez et al, 2012). Signal smoothing was done using a moving average of three points, corresponding to a time window of 208 ms (pH imaging) or 107 ms (Ca2+ imaging).
Author contributions
UBK, CT, HH, LA, RS, and TS contributed to the design of the project; CT and AP performed and analyzed mass spectrometry experiments; HH, UBK, TS, and RS performed stopped‐flow experiments; WB performed sequence analysis; HGK and AM prepared and characterized flagella preparations; LA designed and programmed numerical simulation; LA, FL, and RP performed Ca2+ and pH measurements on single cells; AR synthesized all caged compounds; DN and LG determined the flagellar volumes from cryo‐tomograms; UBK, HH, LA, RS, DN, and AP wrote the manuscript. All authors edited the manuscript. The authors declare no competing financial interests.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Dataset Table EV1
Dataset Table EV2
Review Process File
Acknowledgements
We thank Heike Krause for preparing the manuscript. Financial support by the Deutsche Forschungsgemeinschaft (DFG) via the priority program SPP 1726 “Microswimmers” and the Cluster of Excellence 1023 “ImmunoSensation” is gratefully acknowledged. We thank D. Stoddard for management of the UTSW cryo‐electron microscope facility, which is funded in part by a Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Award (RP170644). This study was supported by HHS|National Institutes of Health (NIH) grant R01 GM083122 and by CPRIT grant RR140082 to D. Nicastro.
The EMBO Journal (2020) 39: e102723
[The copyright line of this article was changed on 23 January 2020 after original online publication.]
Contributor Information
Christian Trötschel, Email: christian_troetschel@yahoo.de.
Ansgar Poetsch, Email: ansgar.poetsch@rub.de.
U Benjamin Kaupp, Email: u.b.kaupp@caesar.de.
Data availability
Shotgun proteomics data have been uploaded to PRIDE with identifier PXD015332, http://www.ebi.ac.uk/pride/archive/projects/PXD015332. Targeted proteomics data can be accessed at PRIDE with identifier PXD015502 and PANORAMA with URL https://panoramaweb.org/arbacia_flagella.url.
References
- Adlung L, Kar S, Wagner MC, She B, Chakraborty S, Bao J, Lattermann S, Boerries M, Busch H, Wuchter P et al (2017) Protein abundance of AKT and ERK pathway components governs cell type‐specific regulation of proliferation. Mol Syst Biol 13: 904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez L, Dai L, Friedrich BM, Kashikar ND, Gregor I, Pascal R, Kaupp UB (2012) The rate of change in Ca2+ concentration controls sperm chemotaxis. J Cell Biol 196: 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbach M, Beckert V, Hansen JN, Wachten D (2018) Shedding light on the role of cAMP in mammalian sperm physiology. Mol Cell Endocrinol 468: 111–120 [DOI] [PubMed] [Google Scholar]
- Barrow AJ, Wu SM (2009) Low‐conductance HCN1 ion channels augment the frequency response of rod and cone photoreceptors. J Neurosci 29: 5841–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylor DA, Lamb TD, Yau K‐W (1979) Responses of retinal rods to single photons. J Physiol 288: 613–634 [PMC free article] [PubMed] [Google Scholar]
- Beltrán C, Vacquier VD, Moy G, Chen Y, Buck J, Levin LR, Darszon A (2007) Particulate and soluble adenylyl cyclases participate in the sperm acrosome reaction. Biochem Biophys Res Commun 358: 1128–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC, Purcell EM (1977) Physics of chemoreception. Biophys J 20: 193–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC (1993) Random walks in biology. Princeton, NJ: Princeton University Press; [Google Scholar]
- Boland W, Terlinden R, Jaenicke L, Muller DG (1982) Binding‐mechanism and sensitivity in gamete chemotaxis of the phaeophyte Cutleria multifida . Eur J Biochem 126: 173–179 [DOI] [PubMed] [Google Scholar]
- Bönigk W, Loogen A, Seifert R, Kashikar N, Klemm C, Krause E, Hagen V, Kremmer E, Strünker T, Kaupp UB (2009) An atypical CNG channel activated by a single cGMP molecule controls sperm chemotaxis. Sci Signal 2: ra68 [DOI] [PubMed] [Google Scholar]
- Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV (2013) An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol 203: 129–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownridge P, Holman SW, Gaskell SJ, Grant CM, Harman VM, Hubbard SJ, Lanthaler K, Lawless C, O'Cualain R, Sims P et al (2011) Global absolute quantification of a proteome: challenges in the deployment of a QconCAT strategy. Proteomics 11: 2957–2970 [DOI] [PubMed] [Google Scholar]
- Buffone MG (2016) Sperm acrosome biogenesis and function during fertilization. New York, NY: Springer; [Google Scholar]
- Carbajal‐Gonzalez BI, Heuser T, Fu X, Lin J, Smith BW, Mitchell DR, Nicastro D (2013) Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton 70: 101–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervetto L, Lagnado L, Perry RJ, Robinson DW, McNaughton PA (1989) Extrusion of calcium from rod outer segments is driven by both sodium and potassium gradients. Nature 337: 740–743 [DOI] [PubMed] [Google Scholar]
- Chung JJ, Navarro B, Krapivinsky G, Krapivinsky L, Clapham DE (2011) A novel gene required for male fertility and functional CATSPER channel formation in spermatozoa. Nat Commun 2: 153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JJ, Miki K, Kim D, Shim SH, Shi HF, Hwang JY, Cai X, Iseri Y, Zhuang X, Clapham DE (2017) CatSperzeta regulates the structural continuity of sperm Ca2+ signaling domains and is required for normal fertility. Elife 6: e23082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF (2005) Vertebrate Smoothened functions at the primary cilium. Nature 437: 1018–1021 [DOI] [PubMed] [Google Scholar]
- DeCaen PG, Delling M, Vien TN, Clapham DE (2013) Direct recording and molecular identification of the calcium channel of primary cilia. Nature 504: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE (2013) Primary cilia are specialized calcium signalling organelles. Nature 504: 311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres RG, Wingreen NS (2008) Accuracy of direct gradient sensing by single cells. Proc Natl Acad Sci USA 105: 15749–15754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinal‐Enriquez J, Priego‐Espinosa DA, Darszon A, Beltran C, Martinez‐Mekler G (2017) Network model predicts that CatSper is the main Ca2+ channel in the regulation of sea urchin sperm motility. Sci Rep 7: 4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain GL, Quandt FN, Bastian BL, Gerschenfeld HM (1978) Contribution of a caesium‐sensitive conductance increase to the rod photoresponse. Nature 272: 467–469 [DOI] [PubMed] [Google Scholar]
- Farci D (2017) Spatial organization of a chemoreceptor guanylate cyclase in the flagellum of Arbacia punctulata sperm. University of Cologne. PhD Thesis
- Fischer F, Poetsch A (2006) Protein cleavage strategies for an improved analysis of the membrane proteome. Proteome Sci 4: 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SH, Corbin JD (2010) Handbook of cell signaling. Amsterdam, The Netherlands: Elsevier; [Google Scholar]
- Francis SH, Blount MA, Corbin JD (2011) Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions. Physiol Rev 91: 651–690 [DOI] [PubMed] [Google Scholar]
- Fu G, Zhao L, Dymek E, Hou Y, Song K, Phan N, Shang Z, Smith EF, Witman GB, Nicastro D (2019) Structural organization of the C1a‐e‐c supercomplex within the ciliary central apparatus. bioRxiv 10.1101/773416 [PREPRINT] [DOI] [PMC free article] [PubMed]
- Galindo BE, Neill AT, Vacquier VD (2005) A new hyperpolarization‐activated, cyclic nucleotide‐gated channel from sea urchin sperm flagella. Biochem Biophys Res Commun 334: 96–101 [DOI] [PubMed] [Google Scholar]
- Galindo BE, de la Vega‐Beltrán JL, Labarca P, Vacquier VD, Darszon A (2007) Sp‐tetraKCNG: a novel cyclic nucleotide gated K+ channel. Biochem Biophys Res Commun 354: 668–675 [DOI] [PubMed] [Google Scholar]
- Garbers DL (1976) Sea urchin sperm guanylate cyclase. J Biol Chem 251: 4071–4077 [PubMed] [Google Scholar]
- Gauss R, Seifert R, Kaupp UB (1998) Molecular identification of a hyperpolarization‐activated channel in sea urchin sperm. Nature 393: 583–587 [DOI] [PubMed] [Google Scholar]
- Gillespie DT (1977) Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81: 2340–2361 [Google Scholar]
- Gross OP, Pugh EN Jr, Burns ME (2015) cGMP in mouse rods: the spatiotemporal dynamics underlying single photon responses. Front Mol Neurosci 8: 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero A, Espinal J, Wood CD, Rendon JM, Carneiro J, Martinez‐Mekler G, Darszon A (2013) Niflumic acid disrupts marine spermatozoan chemotaxis without impairing the spatiotemporal detection of chemoattractant gradients. J Cell Sci 126: 1477–1487 [DOI] [PubMed] [Google Scholar]
- Gunaratne HJ, Neill AT, Vacquier VD (2006) Plasma membrane calcium ATPase is concentrated in the head of sea urchin spermatozoa. J Cell Physiol 207: 413–419 [DOI] [PubMed] [Google Scholar]
- Hagen V, Frings S, Wiesner B, Helm S, Kaupp UB, Bendig J (2003) [7‐(Dialkylamino)coumarin‐4‐yl]methyl‐caged compounds as ultrafast and effective long‐wavelength phototriggers of 8‐bromo‐substituted cyclic nucleotides. Chem Biol Chem 4: 434–442 [DOI] [PubMed] [Google Scholar]
- Halls ML, Canals M (2018) Genetically encoded FRET biosensors to illuminate compartmentalised GPCR signalling. Trends Pharmacol Sci 39: 148–157 [DOI] [PubMed] [Google Scholar]
- Hamzeh H, Alvarez L, Strünker T, Kierzek M, Brenker C, Deal PE, Miller EW, Seifert R, Kaupp UB (2019) Kinetic and photonic techniques to study chemotactic signaling in sea urchin sperm. Methods Cell Biol 151: 487–517 [DOI] [PubMed] [Google Scholar]
- Hansbrough JR, Garbers DL (1981) Sodium‐dependent activation of sea urchin spermatozoa by speract and monensin. J Biol Chem 256: 2235–2241 [PubMed] [Google Scholar]
- Harumi T, Hoshino K, Suzuki N (1992) Effects of sperm‐activating peptide I on Hemicentrotus pulcherrimus spermatozoa in high potassium sea water. Dev Growth Differ 34: 163–172 [DOI] [PubMed] [Google Scholar]
- Hille B (2004) Ionic channels of excitable membranes. Sunderland, MA: Sinauer Associates; [Google Scholar]
- Holman SW, Sims PF, Eyers CE (2012) The use of selected reaction monitoring in quantitative proteomics. Bioanalysis 4: 1763–1786 [DOI] [PubMed] [Google Scholar]
- Jalloul AH, Szerencsei RT, Schnetkamp PP (2016) Cation dependencies and turnover rates of the human K+‐dependent Na+‐Ca2+ exchangers NCKX1, NCKX2, NCKX3 and NCKX4. Cell Calcium 59: 1–11 [DOI] [PubMed] [Google Scholar]
- Kaissling KE (1986) Chemo‐electrical transduction in insect olfactory receptors. Annu Rev Neurosci 9: 121–145 [DOI] [PubMed] [Google Scholar]
- Kambara Y, Shiba K, Yoshida M, Sato C, Kitajima K, Shingyoji C (2011) Mechanism regulating Ca2+‐dependent mechanosensory behaviour in sea urchin spermatozoa. Cell Struct Funct 36: 69–82 [DOI] [PubMed] [Google Scholar]
- Kaupp UB, Seifert R (2002) Cyclic nucleotide‐gated ion channels. Physiol Rev 82: 769–824 [DOI] [PubMed] [Google Scholar]
- Kaupp UB, Solzin J, Hildebrand E, Brown JE, Helbig A, Hagen V, Beyermann M, Pampaloni F, Weyand I (2003) The signal flow and motor response controling chemotaxis of sea urchin sperm. Nat Cell Biol 5: 109–117 [DOI] [PubMed] [Google Scholar]
- Kaupp UB, Alvarez L (2016) Sperm as microswimmers ‐ navigation and sensing at the physical limit. Eur Phys J Special Topics 225: 2119–2139 [Google Scholar]
- Kaupp UB, Strünker T (2017) Signaling in sperm: more different than similar. Trends Cell Biol 27: 101–109 [DOI] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ (2012) A size‐exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol 14: 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three‐dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, Moorthy BS, Xin Xiang L, Xin Shan L, Bharatham K, Tulsian NK, Mihalek I, Anand GS (2014) Active site coupling in PDE:PKA complexes promotes resetting of mammalian cAMP signaling. Biophys J 107: 1426–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, Tulsian NK, Chandramohan A, Anand GS (2015) Parallel allostery by cAMP and PDE coordinates activation and termination phases in cAMP signaling. Biophys J 109: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC (1985) The voltage‐sensitive Na+/H+ exchange in sea urchin spermatozoa flagellar membrane vesicles studied with an entrapped pH probe. J Biol Chem 260: 10794–10799 [PubMed] [Google Scholar]
- Leinders‐Zufall T, Lane AP, Puche AC, Ma W, Novotny MV, Shipley MT, Zufall F (2000) Ultrasensitive pheromone detection by mammalian vomeronasal neurons. Nature 405: 792–796 [DOI] [PubMed] [Google Scholar]
- Lin J, Nicastro D (2018) Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science 360: eaar1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Le TV, Augspurger K, Tritschler D, Bower R, Fu G, Perrone C, O'Toole ET, Mills KV, Dymek E et al (2019) FAP57/WDR65 targets assembly of a subset of inner arm dyneins and connects to regulatory hubs in cilia. Mol Biol Cell 30: 2659–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xia J, Cho KH, Clapham DE, Ren D (2007) CatSperbeta, a novel transmembrane protein in the CatSper channel complex. J Biol Chem 282: 18945–18952 [DOI] [PubMed] [Google Scholar]
- Luo DG, Yue WW, Ala‐Laurila P, Yau KW (2011) Activation of visual pigments by light and heat. Science 332: 1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W, Basto R (2017) Cilia. Cold Spring Harbor, NY: Cold Spring Harbor Perspectives in Biology; [Google Scholar]
- Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, Nachury MV (2015) Proteomics of primary cilia by proximity labeling. Dev Cell 35: 497–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EW, Lin JY, Frady EP, Steinbach PA, Kristan WB Jr, Tsien RY (2012) Optically monitoring voltage in neurons by photo‐induced electron transfer through molecular wires. Proc Natl Acad Sci USA 109: 2114–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BS, Stepanchick AN, Tewson PH, Hartle CM, Zhang J, Quinn AM, Hughes TE, Mirshahi T (2016) Cilia have high cAMP levels that are inhibited by Sonic Hedgehog‐regulated calcium dynamics. Proc Natl Acad Sci USA 113: 13069–13074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Jansen V, Jikeli JF, Hamzeh H, Alvarez L, Dombrowski M, Balbach M, Strünker T, Seifert R, Kaupp UB et al (2016) A novel biosensor to study cAMP dynamics in cilia and flagella. Elife 5: e14052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Rohatgi R (2014) G‐protein‐coupled receptors, Hedgehog signaling and primary cilia. Semin Cell Dev Biol 33: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro B, Kirichok Y, Clapham DE (2007) KSper, a pH‐sensitive K+ current that controls sperm membrane potential. Proc Natl Acad Sci USA 104: 7688–7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro B, Kirichok Y, Chung JJ, Clapham DE (2008) Ion channels that control fertility in mammalian spermatozoa. Int J Dev Biol 52: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Akyuz N, Liu XP, Asai Y, Nist‐Lund C, Kurima K, Derfler BH, Gyorgy B, Limapichat W, Walujkar S et al (2018) TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron 99: 736–753 e736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape H‐C (1996) Queer current and pacemaker: the hyperpolarization‐activated cation current in neurons. Annu Rev Physiol 58: 299–327 [DOI] [PubMed] [Google Scholar]
- Perez‐Riverol Y, Csordas A, Bai J, Bernal‐Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M et al (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Peuker S, Cukkemane A, Held M, Noe F, Kaupp UB, Seifert R (2012) Kinetics of ligand‐receptor interaction reveals an induced‐fit mode of binding in a cyclic nucleotide‐activated protein. Biophys J 104: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlo M, Bungert‐Plümke S, Weyand I, Seifert R, Bönigk W, Strünker T, Kashikar ND, Goodwin N, Müller A, Pelzer P et al (2014) High density and ligand affinity confer ultrasensitive signal detection by a guanylyl cyclase chemoreceptor. J Cell Biol 206: 541–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postea O, Biel M (2011) Exploring HCN channels as novel drug targets. Nat Rev Drug Discov 10: 903–914 [DOI] [PubMed] [Google Scholar]
- Quillin ML, Matthews BW (2000) Accurate calculation of the density of proteins. Acta Crystallogr D Biol Crystallogr 56: 791–794 [DOI] [PubMed] [Google Scholar]
- Ramirez‐Sarmiento CA (2017) “Riddle Me This”: substrate channeling solves the paradigms of cAMP‐dependent activation of PKA. Biophys J 112: 2451–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers J, Simpson DM, Robertson DH, Gaskell SJ, Beynon RJ (2007) Absolute multiplexed quantitative analysis of protein expression during muscle development using QconCAT. Mol Cell Proteomics 6: 1416–1427 [DOI] [PubMed] [Google Scholar]
- Rybalkin SD, Rybalkina IG, Shimizu‐Albergine M, Tang XB, Beavo JA (2003) PDE5 is converted to an activated state upon cGMP binding to the GAF A domain. EMBO J 22: 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetkamp PP (2013) The SLC24 gene family of Na+/Ca2+‐K+ exchangers: from sight and smell to memory consolidation and skin pigmentation. Mol Aspects Med 34: 455–464 [DOI] [PubMed] [Google Scholar]
- Schnetkamp PP, Jalloul AH, Liu G, Szerencsei RT (2014) The SLC24 family of K+‐dependent Na+‐Ca2+‐exchangers: structure‐function relationships. Curr Top Membr 73: 263–287 [DOI] [PubMed] [Google Scholar]
- Scott KB, Turko IV, Phinney KW (2016) QconCAT: internal standard for protein quantification. Methods Enzymol 566: 289–303 [DOI] [PubMed] [Google Scholar]
- Seifert R, Flick M, Bönigk W, Alvarez L, Trötschel C, Poetsch A, Müller A, Goodwin N, Pelzer P, Kashikar ND et al (2015) The CatSper channel controls chemosensation in sea urchin sperm. EMBO J 34: 379–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Lowe DG, Thorpe DS, Rodriguez H, Kuang W‐J, Dangott LJ, Chinkers M, Goeddel DV, Garbers DL (1988) Membrane guanylate cyclase is a cell‐surface receptor with homology to protein kinases. Nature 334: 708–712 [DOI] [PubMed] [Google Scholar]
- Starr RC, Marner FJ, Jaenicke L (1995) Chemoattraction of male gametes by a pheromone produced by female gametes of Chlamydomonas. Proc Natl Acad Sci USA 92: 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strünker T, Weyand I, Bönigk W, Van Q, Loogen A, Brown JE, Kashikar N, Hagen V, Krause E, Kaupp UB (2006) A K+ ‐selective cGMP‐gated ion channel controls chemosensation of sperm. Nat Cell Biol 8: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Su Y‐H, Vacquier VD (2002) A flagellar K+‐dependent Na+/Ca2+ exchanger keeps Ca2+ low in sea urchin spermatozoa. Proc Natl Acad Sci USA 99: 6743–6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Vacquier VD (2006) Cyclic GMP‐specific phosphodiesterase‐5 regulates motility of sea urchin spermatozoa. Mol Biol Cell 17: 114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulsian NK, Krishnamurthy S, Anand GS (2017) Channeling of cAMP in PDE‐PKA complexes promotes signal adaptation. Biophys J 112: 2552–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier VD, Loza‐Huerta A, Garcia‐Rincon J, Darszon A, Beltran C (2014) Soluble adenylyl cyclase of sea urchin spermatozoa. Biochim Biophys Acta 1842: 2621–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachten D, Jikeli JF, Kaupp UB (2017) Sperm sensory signaling. Cold Spring Harb Perspect Biol 9: a028225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, King SM, Quill TA, Doolittle LK, Garbers DL (2003) A new sperm‐specific Na+/H+ exchanger required for sperm motility and fertility. Nat Cell Biol 5: 1117–1122 [DOI] [PubMed] [Google Scholar]
- Wang H, Liu J, Cho KH, Ren D (2009) A novel, single, transmembrane protein CATSPERG is associated with CATSPER1 channel protein. Biol Reprod 81: 539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windler F, Bönigk W, Körschen HG, Grahn E, Strünker T, Seifert R, Kaupp UB (2018) The solute carrier SLC9C1 is a Na+/H+‐exchanger gated by an S4‐type voltage‐sensor and cyclic‐nucleotide binding. Nat Comm 9: 2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff ML, Sampath AP, Matthews HR, Krasnoperova NV, Lem J, Fain GL (2002) Measurement of cytoplasmic calcium concentration in the rods of wild‐type and transducin knock‐out mice. J Physiol 542: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau KW, Hardie RC (2009) Phototransduction motifs and variations. Cell 139: 246–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng XH, Navarro B, Xia XM, Clapham DE, Lingle CJ (2013) Simultaneous knockout of Slo3 and CatSper1 abolishes all alkalization‐ and voltage‐activated current in mouse spermatozoa. J Gen Physiol 142: 305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Müller U (2015) The elusive mechanotransduction machinery of hair cells. Curr Opin Neurobiol 34: 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AL, Yamanaka G, Eckstein F, Baylor DA, Stryer L (1985) Interaction of hydrolysis‐resistant analogs of cyclic GMP with the phosphodiesterase and light‐sensitive channel of retinal rod outer segments. Proc Natl Acad Sci USA 82: 8813–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Dataset Table EV1
Dataset Table EV2
Review Process File
Data Availability Statement
Shotgun proteomics data have been uploaded to PRIDE with identifier PXD015332, http://www.ebi.ac.uk/pride/archive/projects/PXD015332. Targeted proteomics data can be accessed at PRIDE with identifier PXD015502 and PANORAMA with URL https://panoramaweb.org/arbacia_flagella.url.


