Abstract
Objective
The aim of this study was to assess the HLA haplotype frequencies and genetic profiles of the Kuwaiti population.
Materials and Methods
Whole venous blood was obtained from 595 healthy, unrelated Kuwaiti volunteers. The study population was genotyped for HLA class I (HLA-A, HLA-B, and HLA-C) and class II (HLA-DRB1 and HLA-DQB1) loci using sequence-specific oligonucleotide (SSO) probe-based hybridization and high-resolution HLA genotyping. Haplotype frequencies were estimated using an implementation of the expectation maximization algorithm that resolves both phase and allelic ambiguity. The Kuwaiti population was compared with other populations from the US National Marrow Donor Program (NMDP), by running a principal component analysis (PCA) on the relevant haplotype frequencies.
Results
The most common HLA class I alleles in Kuwait were HLA-A*02:01g, HLA-C*06:02g, and HLA-B*50:01g with frequencies of 16, 14, and 12%, respectively. The most common HLA class II alleles in Kuwait were HLA-DQB1*02:01g and HLA-DRB1*07:01 with frequencies of 29.7 and 16.5%, respectively. The most common Kuwaiti haplotype observed was HLA-A*02:01g∼HLA-C*06:02g∼HLA-B*50:01g∼HLA-DRB1*07:01∼HLA-DQB1*02:01g at a frequency of 2.3%. The PCA demonstrated close genetic proximity of the Kuwaiti population with Middle Eastern, Southeast Asian, and North African populations in the NMDP.
Conclusion
Identifying the haplotype diversity in the Kuwaiti population will contribute to the selection of an HLA-match for HSCT, disease associations, pharmacogenomics, and knowledge of population HLA diversity.
Keywords: Human leukocyte antigen, Kuwait, HLA haplotype frequency, Genetic profile
Significance of the Study
We describe here the HLA haplotype diversity in a Kuwaiti Arab population. It is the first attempt to describe the HLA genetic profile of Kuwaiti natives and investigate the Kuwaiti genetic profile by means of high-resolution analysis at the 5 -locus level of A∼C∼B∼DRB1∼DQB1.
Introduction
Hematopoietic stem-cell transplantation (HSCT) is a curative treatment for many life-threatening diseases, including blood cancers. The extent of human leukocyte antigen (HLA) matching between the donor and the recipient largely influences the outcome of related and unrelated donor HSCT. In addition, the outcome of HSCT is significantly improved for HLA fully matched donor-patient pairs compared with mismatched cases, even if only a single HLA allele is different [1]. In general, an HLA-matched identical sibling is the ideal donor; however, only an average of 30% of patients can benefit from the related donor option [2]. When a matched sibling is not available, the second alternative is an unrelated donor. Due to the extreme diversity of HLA alleles and the resulting number of potential genotypic combinations, the chances of finding a matched donor are low when searching international registries as patients with certain ethnic and racial backgrounds are underrepresented.
The HLA region on human chromosome 6p21 is the most polymorphic region in the human genome [3, 4, 5]. In different ethnic groups, HLA polymorphisms and variations in the HLA allele and haplotype distributions are usually reflective of the origin, migration, and degree of admixture in these populations. In addition, linkage disequilibrium (LD) among different alleles and the predominant ancestral haplotypes vary widely across different human populations [6]. Knowledge of population HLA diversity can contribute to the selection of an HLA-match for HSCT, disease associations, and pharmacogenomics.
The Arabian Peninsula, situated at the nexus of Africa, Europe, and Asia, was an early human migration route out of Africa and part of the intercontinental trade route [7, 8, 9, 10]. The State of Kuwait is located at the northwestern tip of the Arabian Gulf and is bordered by Iraq and Saudi Arabia. Kuwait was established by settlers from the Najd region of eastern Saudi Arabia in the 17th and 18th centuries [11]. The current population is about 3.5 million and is made up of 1.5 million Kuwaiti Arab nationals as well as expatriates, mainly Arabs and South Asians (Indians, Pakistanis, Filipinos, etc.). Kuwaiti Arab-national males represent 15.7% of the population and females represent 16.3% (a male-to-female ratio of 1:1). Thalassemia major and other hemoglobinopathies are prevalent in the Kuwaiti population, with 80% of the cases resulting from consanguineous marriages. This represents an important cause of morbidity and mortality in Kuwait [12, 13, 14].
To our knowledge, HLA diversity in the ethnic Kuwaiti population has not been previously studied. Most previous reports were related to the role of HLA, disease associations, and other populations [15, 16, 17]. This is a considerable disadvantage for unrelated HSCT locally and also for predicting the size of a national unrelated stem cell registry. The aim of this study was to assess the HLA haplotype frequencies and evaluate the genetic profiles of the Kuwaiti population.
Materials and Methods
Study Population
Whole venous blood was obtained from 595 healthy, unrelated Kuwaiti volunteers. The study population was evenly distributed geographically to obtain a representative sample of the 6 governorates of Kuwait. All individuals who participated in this study were Kuwaiti Arab citizens. The QIAamp DNA blood mini kit (Qiagen, Hilden, Germany) was used to extract DNA. Volunteers were selected independently of any patient need.
HLA Typing
Two-thirds of the study population (n = 395) were genotyped for HLA class I (HLA-A, HLA-B, and HLA-C) and class II (HLA-DRB1 and HLA-DQB1) loci by a sequence-specific oligonucleotide (SSO) probe-based hybridization technique, using the One Lambda LABtype sequence-specific oligonucleotide kit, which provided intermediate resolution. High-resolution HLA genotyping, up to the third-nomenclature field, was obtained for 200 individuals with DNA sequence-based typing (SBT), by using the Illumina MiSeq and targeting exon 2 and 3 for class I and exon 2 for class II.
Statistical Analysis
To minimize the loss of information from the use of allele codes, SSO primary data were directly interpreted into HLA assignments based on the IPD-IMGT/HLA database (v3.15.0) and then formatted as genotype-list strings [18]. Alleles are described with the first 2 fields of HLA allele nomenclature, representing protein level assignment. Alleles with nucleotide sequences that translated to amino acids identical at the antigen recognition site (ARS) were combined. They were indicated by the lowest-numbered allele followed by “g,” since these sets of alleles are often not distinguished by current typing systems, and genomic regions are not defined outside the ARS for many of the alleles [3, 19].
Haplotype frequencies were estimated using an implementation of the expectation maximization (EM) algorithm that resolves both phase and allelic ambiguity [20]. It computes the most probable set of haplotypes that explain the unphased genotype input data via a maximum-likelihood estimation. Starting from arbitrary initial haplotype frequencies, it calculates genotype frequencies under the assumption of the Hardy-Weinberg equilibrium (expectation step). After normalizing, these genotype frequencies are used to estimate haplotype frequencies (maximization step). The expectation and maximization steps are repeated until a stop criterion with a predefined value is fulfilled [19].
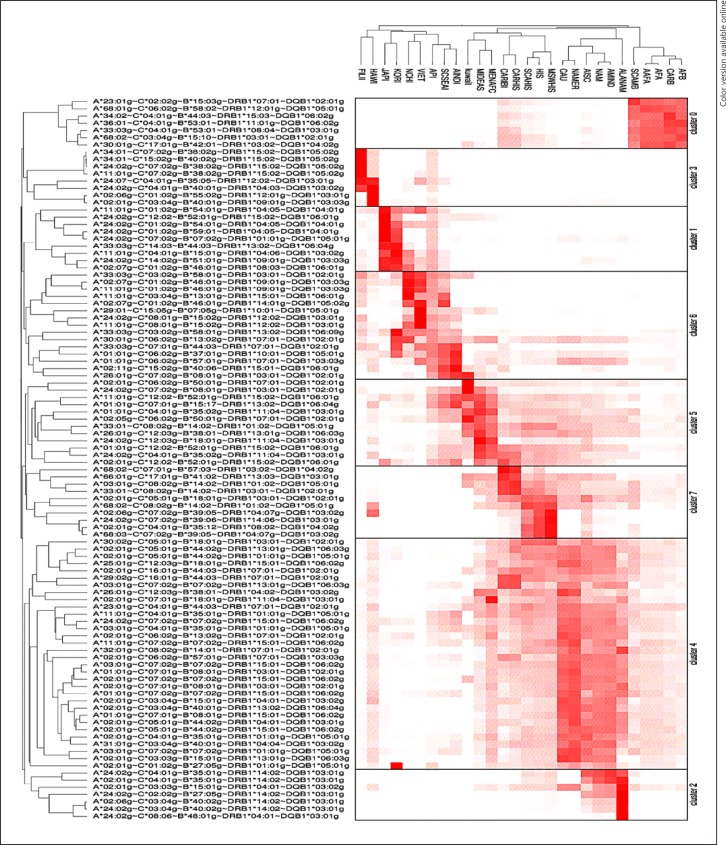
The Kuwaiti population was compared with other populations from the US National Marrow Donor Program (NMDP) using principal component analysis (PCA) of the relevant haplotype frequencies [21]. The PCA was performed using MATLAB (MATLAB and Statistics Toolbox release 2015b, MathWorks, Inc. Natick, MA, USA). Comparisons were also performed using clustering analysis by means of CLUTO software [22]. This was done by clustering the top 100 haplotypes (rows) in the Kuwaiti population together with NMDP populations (columns) [21, 22] (Fig. 1), separating the total set of generated haplotypes into 9 prespecified clusters based on haplotype similarity across columns. Rows were clustered according to frequency similarities across populations, with bright-red entries corresponding to higher haplotype frequencies and the brightness of the entries decreasing as their value decreased.
Fig. 1.
Clusters of the top 100 haplotypes in the Kuwaiti population with some of the NMDP populations. Darker color indicates higher haplotype frequency.
Results
HLA Allele Frequencies: Class I and Class II
We found the most common HLA class I alleles in Kuwait wereHLA-A*02:01g, HLA-C*06:02g, and HLA-B*50:01g with frequencies of 16, 14, and 12%, respectively. The most common class II alleles in Kuwait were HLA-DQB1*02:01g and HLA-DRB1*07:01 with frequencies of 29.7 and 16.5%, respectively. The top 10 most common HLA class I and class II alleles are depicted in Tables 1 and 2.
Table 1.
Top 10 HLA class I allele frequencies in the Kuwaiti population
| HLA-A | Frequency, % | HLA-B | Frequency, % | HLA-C | Frequency, % | |
| 1 | A*02:01g | 15.89 | B*50:01g | 12.02 | C*06:02g | 13.96 |
| 2 | A*24:02g | 9.37 | B*51:01g | 10.49 | C*04:01g | 13.20 |
| 3 | A*01:01g | 7.73 | B*08:01g | 7.23 | C*07:02g | 12.89 |
| 4 | A*11:01g | 6.48 | B*52:01g | 4.20 | C*15:02g | 9.20 |
| 5 | A*68:01g | 5.79 | B*41:01 | 4.03 | C*07:01g | 7.29 |
| 6 | A*02:05g | 4.78 | B*35:01g | 3.74 | C*17:01g | 5.38 |
| 7 | A*26:01g | 4.44 | B*07:02g | 3.70 | C*12:03g | 5.13 |
| 8 | A*03:01g | 4.28 | B*18:01g | 3.61 | C*12:02 | 4.45 |
| 9 | A*31:01g | 4.21 | B*40:06 | 3.61 | C*14:02g | 3.78 |
| 10 | A*33:03g | 3.82 | B*58:01g | 3.19 | C*08:02g | 3.19 |
Table 2.
Top 10 HLA class II allele frequencies in the Kuwaiti population
| HLA-DRB1 | Frequency, % | HLA-DQB1 | Frequency, % | |
|---|---|---|---|---|
| 1 | DRB1*07:01 | 16.4 | DQB1*02:01g | 29.69 |
| 2 | DRB1*03:01 | 13.24 | DQB1*03:01g | 12.58 |
| 3 | DRB1*13:01g | 5.29 | DQB1*05:02g | 8.62 |
| 4 | DRB1*13:02 | 4.72 | DQB1*05:01g | 8.57 |
| 5 | DRB1*16:02 | 4.71 | DQB1*03:02g | 8.33 |
| 6 | DRB1*15:01 | 4.62 | DQB1*06:02g | 7.12 |
| 7 | DRB1*11:01g | 4.04 | DQB1*06:03g | 5.23 |
| 8 | DRB1*11:04 | 3.85 | DQB1*06:01g | 4.76 |
| 9 | DRB1*15:02 | 2.99 | DQB1*06:04g | 3.76 |
| 10 | DRB1*10:01 | 2.94 | DQB1*04:02g | 1.98 |
HLA Haplotype Frequencies
The haplotype A*02:01g∼C*06:02g∼B*50:01g∼DRB1*07:01∼DQB1*02:01g was the most common in our study population, with a frequency of 2.3%. This haplotype is the 159th and 65th most common haplotype in the US European Caucasian (i.e., individuals of European ancestry) (EURCAU) and Middle Eastern/North African Coast/Mediterranean (MENAFC) populations, respectively [21]. The second most common haplotype was A*26:01g∼C*07:02g∼B*08:01g∼DRB1*03:01∼DQB1*02:01g, at a frequency of 1.6%. This is the 1923rd and 66th most common haplotype in the US EURCAU and MENAFC populations, respectively. The third most common Kuwaiti haplotype was A*02:05g∼C*06:02g∼B*50:01g∼DRB1*07:01∼DQB1*02:01g, at a frequency of 1.5%. This haplotype is the 34th and 10th most common haplotype in the US EURCAU and MENAFC populations, respectively. The top 10 most common haplotypes are shown in Table 3.
Table 3.
The 10 most common haplotype frequencies in the Kuwaiti population
| Haplotype | Frequency, % | |||||
|---|---|---|---|---|---|---|
| 1 | A*02:01g | C*06:02g | B*50:01g | DRB1*07:01 | DQB1*02:01g | 2.26 |
| 2 | A*26:01g | C*07:02g | B*08:01g | DRB1*03:01 | DQB1*02:01g | 1.58 |
| 3 | A*02:05g | C*06:02g | B*50:01g | DRB1*07:01 | DQB1*02:01g | 1.5 |
| 4 | A*33:03g | C*03:02g | B*58:01g | DRB1*03:01 | DQB1*02:01g | 1.15 |
| 5 | A*24:02g | C*07:02g | B*08:01g | DRB1*03:01 | DQB1*02:01g | 1.1 |
| 6 | A*33:01 | C*08:02g | B*14:02 | DRB1*01:02 | DQB1*05:01g | 1.09 |
| 7 | A*31:01g | C*15:02g | B*51:01g | DRB1*13:01g | DQB1*06:03g | 0.98 |
| 8 | A*11:01g | C*15:02g | B*40:06 | DRB1*16:02 | DQB1*05:02g | 0.92 |
| 9 | A*11:01g | C*12:02 | B*52:01g | DRB1*15:02 | DQB1*06:01g | 0.92 |
| 10 | A*23:01g | C*06:02g | B*50:01g | DRB1*07:01 | DQB1*02:01g | 0.88 |
Regular LD and asymmetric LD (ALD) are described. The definition of LD is the nonrandom association of alleles at 2 loci. The LD can assess the degree to which pairs of alleles are likely to be observed on the same haplotype and has important implications in analyzing immunogenetic data for population and disease-association studies [23]. The ALD measures are symmetric and equivalent to the correlation coefficient r when both loci are biallelic. When the numbers of alleles at the 2 loci differ, the ALD measures capture this asymmetry and provide additional detail about the LD structure [24].
Population Genetics Analysis
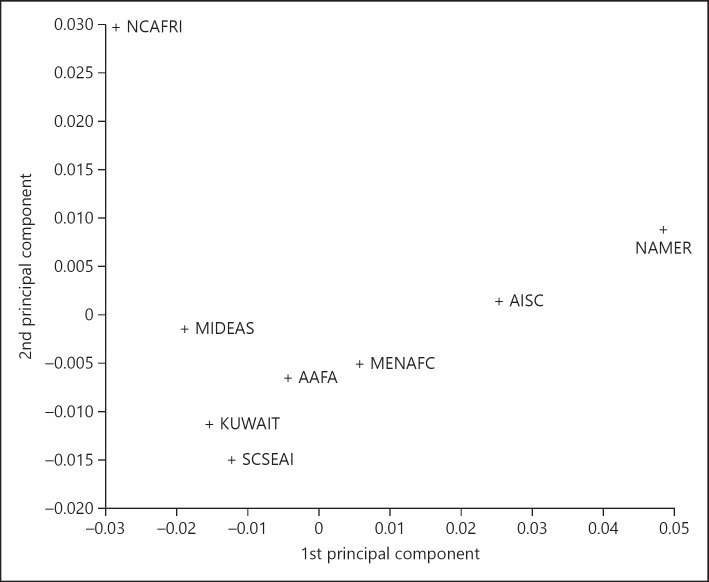
The result of the PCA showed that the first 7 principal components (PCs) account for all the variance in the Kuwaiti haplotype frequencies with the first 3 PCs accounting for >95% of the variances (69.4, 20.9, and 5.8% variance for the first 3 PCs) (Fig. 2). The first and second PCs demonstrate the close genetic proximity of the Kuwaiti population with the NMDP Middle Eastern (MIDEAS), Southeast Asian (SCSEAI), and combined MENAFC populations.
Fig. 2.
First and second principal components of Kuwaiti and NMDP population clusters. NCAFRI, North African Coast; MIDEAS, Middle East; MENAFC, Middle Eastern/North African Coast/Mediterranean; SCSEAI, Southeast Asian; AAFA, African-American; NAMER, North American; AISC, American Indian-South or Central.
The clustering of the top 100 haplotypes (rows) in the Kuwaiti population together with NMDP populations [21, 22] is shown in Figure 1. The total set of generated haplotypes is separated into 8 clusters based on haplotype similarity across columns. Rows are clustered by similarity in frequency across populations. The clustering analysis showed trends similar to the PCA, specifically the clustering of the Kuwaiti population with the NMDP MIDEAS, MENAFC, and SCSEAI populations (cluster 5). Cluster 4 shows some overlap between the Kuwaiti and US populations of European descent, albeit at lower frequencies (indicated by the lighter color in the heat map).
Discussion
This study describes HLA haplotype diversity in a Kuwaiti Arab population. To our knowledge, this is the first attempt to describe the HLA genetic profile of Kuwaiti natives and investigate Kuwaiti genetic profiles using high-resolution (i.e., the first 2 nomenclature fields) HLA haplotype frequencies at 5 loci (A∼C∼B∼DRB1∼DQB1). We are aware that there is limited haplotype frequency information available from surrounding Arab populations at the same level of resolution studied here. However, keeping this resolution discrepancy in mind, we observed that the most common Kuwaiti haplotypes were closely related to neighboring populations in Saudi Arabia [25, 26], Tunisia [27], and Jordan [28]. In particular, the third most common Kuwaiti haplotype, A*02:05g∼C*06:02g∼B*50:01g∼DRB1*07:01∼DQB1*02:01g, is common in NMDP Middle Eastern populations and also one of the most common in eastern Mediterranean populations [25, 29, 30, 31]. Hajeer et al. [26] demonstrated, at the 4-locus level, some overlap between the most common Saudi and Kuwaiti haplotypes, i.e., A*24:02:01:01∼B*08:01:01∼C*07:02:01:01∼DRB1*03:01:01:01 and A*23:01:01∼B*50:01:01∼C*06:02:01:01∼DRB1*07:01:01:01. Additionally, we found some overlap with the HLA haplotype frequencies observed in Arab populations in allelefrequencies.net, albeit at a much lower resolution.
Our PCA and clustering analyses showed that the Kuwaiti population has genetic proximity with the NMDP MIDEAS, MENAFC, and SCSEAI populations due to some shared haplotypes: A*11:01g∼C*12:02∼B*52:01g∼DRB1*15:02∼DQB1*06:01g, A*01:01g∼C*07:01g∼B*15:17∼DRB1*13:02∼DQB1*06:04g, A*24:02g∼C*07:02g∼B*08:01g∼DRB1*03:01∼DQB1*02:01g, A*11:01g∼C*12:02∼B*52:01g∼DRB1*15:02∼DQB1*06:01g, A*01:01g∼C*07:01g∼B*15:17∼DRB1*13:02∼DQB1*06:04g as well as A*02:05g∼C*06:02g∼B*50:01g∼DRB1*07:01∼DQB1*02:01g (common throughout the eastern Mediterranean region). While the haplotype overlap with the NMDP Middle Eastern population was expected, the genetic proximity with the SCSEAI population can possibly be explained by a recent genetic flow from the 37% South Asian immigrant population in Kuwait [10, 32]. The proximity to the AAFA (African-American) population is better explained as shared alleles. The Kuwaiti population shares some alleles with the AAFA population (A*02:01g, A*33:03g, A*68:01g, B*08:01g, B*07:02g, C*07:02g, C*06:02g, DRB1*03:01, DRB1*07:01, DRB1*15:01, DQB1*02:01g, and 05:01g). Moreover, one of the top ranked AAFA and Kuwaiti haplotypes is A*30:01g∼C*17:01g∼B*42:01∼DRB1*03:02∼DQB1*04:02g.
The results of this study are important for supporting research investigating HLA linkage in diseases with etiologies, susceptibilities, or frequencies that are related to the major histocompatibility complex, and also to facilitate the search for HLA-matched unrelated donors for patients in need of HSCT. Currently, patients of Arab or Kuwaiti descent who require HSCT face challenges in the identification of matched unrelated donors because of the lower representation of donors of Middle Eastern heritage in international bone marrow registries and cord blood banks. Donors of predominantly European descent are unable to fully meet the needs of the Kuwaiti patient population. This study provides information about a relatively underrepresented population and can potentially provide much-needed insight into their genomics.
Conclusion
Identifying the haplotype diversity in the Kuwaiti population will contribute to the selection of HLA-matches for HSCT, disease associations, and pharmacogenomics as well as more knowledge of HLA diversity in the population.
Statement of Ethics
Only those who signed the informed consent form were recruited. The Ethics Committee of the Health Sciences Center approved the design of the study.
Funding Source
This research was supported by a grant from the Kuwait Foundation for Advancement of Science (KFAS) (grant No: 2012-130-204), Kuwait.
Acknowledgements
We acknowledge Michael Halagan and Abeer Madboulyfor their help in reviewing the manuscript and providing insights into the implementation of the EM algorithm.
References
- 1.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006 Apr;354((17)):1813–26. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- 2.Rosenmayr A, Pointner-Prager M, Mitterschiffthaler A, Bozic L, Pelzmann B, Tüchler H, et al. What are a patient's current chances of finding a matched unrelated donor? Twenty years' central search experience in a small country. Bone Marrow Transplant. 2012 Feb;47((2)):172–80. doi: 10.1038/bmt.2011.67. [DOI] [PubMed] [Google Scholar]
- 3.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010 Apr;75((4)):291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, et al. The DNA sequence and analysis of human chromosome 6. Nature. 2003 Oct;425((6960)):805–11. doi: 10.1038/nature02055. [DOI] [PubMed] [Google Scholar]
- 5.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015 Jan;43((Database issue)):D423–31. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, Sanchez-Mazas A, et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. 2008 Jul;69((7)):443–64. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armitage SJ, Jasim SA, Marks AE, Parker AG, Usik VI, Uerpmann HP. The southern route “out of Africa”: evidence for an early expansion of modern humans into Arabia. Science. 2011 Jan;331((6016)):453–6. doi: 10.1126/science.1199113. [DOI] [PubMed] [Google Scholar]
- 8.Cabrera VM, Abu-Amero KK, Larruga JM, González AM, The Arabian peninsula: Gate for human migrations out of africa or cul-de-Sac? A Mitochondrial DNA Phylogeographic Perspective . The Evolution of Human Populations in Arabia: Paleoenvironments, Prehistory and Genetics. In: Petraglia MD, Rose JI, editors. Dordrecht: Springer Netherlands; 2010. pp. pp. 79–87. [Google Scholar]
- 9.Ghirotto S, Penso-Dolfin L, Barbujani G. Genomic evidence for an African expansion of anatomically modern humans by a Southern route. Hum Biol. 2011 Aug;83((4)):477–89. doi: 10.3378/027.083.0403. [DOI] [PubMed] [Google Scholar]
- 10.Thareja G, John SE, Hebbar P, Behbehani K, Thanaraj TA, Alsmadi O. Sequence and analysis of a whole genome from Kuwaiti population subgroup of Persian ancestry. BMC Genomics. 2015 Feb;16((1)):92. doi: 10.1186/s12864-015-1233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abu-Hakima AM. London: Probsthain; 1988. History of eastern Arabia, 1750-1800: the rise and development of Bahrain, Kuwait and Wahhabi Saudi Arabia. [Google Scholar]
- 12.Adekile AD, Azab AF, Al-Sharida SI, Al-Nafisi BA, Akbulut N, Marouf RA, et al. Clinical and molecular characteristics of non-transfusion-dependent thalassemia in Kuwait. Hemoglobin. 2015;39((5)):320–6. doi: 10.3109/03630269.2015.1053489. [DOI] [PubMed] [Google Scholar]
- 13.Adekile AD, Gu LH, Baysal E, Haider MZ, al-Fuzae L, Aboobacker KC, et al. Molecular characterization of alpha-thalassemia determinants, beta-thalassemia alleles, and beta S haplotypes among Kuwaiti Arabs. Acta Haematol. 1994;92((4)):176–81. doi: 10.1159/000204216. [DOI] [PubMed] [Google Scholar]
- 14.al-Fuzae L, Aboolbacker KC, al-Saleh Q. beta-Thalassaemia major in Kuwait. J Trop Pediatr. 1998 Oct;44((5)):311–2. doi: 10.1093/tropej/44.5.311. [DOI] [PubMed] [Google Scholar]
- 15.ElAoud S, Kamoun A, Mahfoudh N, Charfi A, Snoussi M, Hachicha H, et al. Beyond human leukocyte antigen class I antigens: hereditary hemochromatosis gene mutations in recurrent aphthous oral ulcers and Behcet disease in the South of Tunisia. Med Princ Pract. 2017;26((5)):427–32. doi: 10.1159/000481782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pekiner FN, Aytugar E, Demirel GY, Borahan MO. HLA-A, B (Class I) and HLA-DR, DQ (Class II) antigens in Turkish patients with recurrent aphthous ulceration and Behçet's disease. Med Princ Pract. 2013;22((5)):464–8. doi: 10.1159/000348366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiftikcioglu BI, Uludag IF, Zorlu Y, Pirim İ, Sener U, Tokucoglu F, et al. Human leucocyte antigen B50 is associated with conversion to generalized myasthenia gravis in patients with pure ocular onset. Med Princ Pract. 2017;26((1)):71–7. doi: 10.1159/000452954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milius RP, Mack SJ, Hollenbach JA, Pollack J, Heuer ML, Gragert L, et al. Genotype List String: a grammar for describing HLA and KIR genotyping results in a text string. Tissue Antigens. 2013 Aug;82((2)):106–12. doi: 10.1111/tan.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schäfer C, Schmidt AH, Sauter J. Hapl-o-Mat: open-source software for HLA haplotype frequency estimation from ambiguous and heterogeneous data. BMC Bioinformatics. 2017 May;18((1)):284. doi: 10.1186/s12859-017-1692-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kollman C, Maiers M, Gragert L, Müller C, Setterholm M, Oudshoorn M, et al. Estimation of HLA-A, -B, -DRB1 haplotype frequencies using mixed resolution data from a National Registry with selective retyping of volunteers. Hum Immunol. 2007 Dec;68((12)):950–8. doi: 10.1016/j.humimm.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Gragert L, Madbouly A, Freeman J, Maiers M. Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum Immunol. 2013 Oct;74((10)):1313–20. doi: 10.1016/j.humimm.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Karypis G. CLUTO-a clustering toolkit. Minnesota Univ Minneapolis Dept of Computer Science. 2002 [Google Scholar]
- 23.Mack SJ, Gourraud PA, Single RM, Thomson G, Hollenbach JA. Analytical methods for immunogenetic population data. Methods Mol Biol. 2012;882:215–44. doi: 10.1007/978-1-61779-842-9_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Single RM, Strayer N, Thomson G, Paunic V, Albrecht M, Maiers M. Asymmetric linkage disequilibrium: tools for assessing multiallelic LD. Hum Immunol. 2016 Mar;77((3)):288–94. doi: 10.1016/j.humimm.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Al-Awwami M, Aljurf M, Al-Humidan H, El-Solh H, Almeshari K, Al-Seraihy A, et al. 85-P: HLA Haplotype frequencies in Saudi Arabia for design of a Saudi Stem-Cell registry. Hum Immunol. 2012;73:106. [Google Scholar]
- 26.Hajeer AH, Al Balwi MA, Aytül Uyar F, Alhaidan Y, Alabdulrahman A, Al Abdulkareem I, et al. HLA-A, -B, -C, -DRB1 and -DQB1 allele and haplotype frequencies in Saudis using next generation sequencing technique. Tissue Antigens. 2013 Oct;82((4)):252–8. doi: 10.1111/tan.12200. [DOI] [PubMed] [Google Scholar]
- 27.Hajjej A, Almawi WY, Hattab L, El-Gaaied A, Hmida S. HLA class I and class II alleles and haplotypes confirm the Berber origin of the present day Tunisian population. PLoS One. 2015 Aug;10((8)):e0136909. doi: 10.1371/journal.pone.0136909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elbjeirami WM, Abdel-Rahman F, Hussein AA. Probability of finding an HLA-matched donor in immediate and extended families: the Jordanian experience. Biol Blood Marrow Transplant. 2013 Feb;19((2)):221–6. doi: 10.1016/j.bbmt.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Bishara A, Israel S, Halagan M, Madbouly A, Fernandez Vina M, Maiers M, et al. HLA Allele and haplotype frequencies for christian and muslim arab donors in Hadassah Registry: P197. Tissue Antigens. 2015;85:400–1. [Google Scholar]
- 30.Halagan M, Stein J, Manor S, Madbouly A, Gragert L, Shriki N, et al. HLA diversity in the Ezer mizion Registry: P199. Tissue Antigens. 2015;85:401–2. [Google Scholar]
- 31.Klitz W, Gragert L, Maiers M, Fernandez-Viña M, Ben-Naeh Y, Benedek G, et al. Genetic differentiation of Jewish populations. Tissue Antigens. 2010 Dec;76((6)):442–58. doi: 10.1111/j.1399-0039.2010.01549.x. [DOI] [PubMed] [Google Scholar]
- 32.Abu-Hakima AM. Intl Book Centre. 1st English Edition edition. 1983. June 1. The modern history of Kuwait, 1750-1965; p. p 226. [Google Scholar]