Highlights
-
•
The hippocampus, the sagittal stratum, the cingulum, the parahippocampal gyrus, the superior temporal white matter and the fusiform white matter are among the top ranked regions with memory impairment and microstructural changes in AD.
-
•
The hippocampus, the sagittal stratum left, the parahippocampal gyrus right and the cingulum (hippocampus) are significantly associated with clinical/cognitive traits as well as amyloid beta accumulation in AD.
-
•
Hippocampus right and sagittal stratum left are the two top regions shared in our ranking highly correlated with memory scores as well as AD-pathology endophenotypes.
-
•
Though fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (L1) and radial diffusivity (RD) features were highly correlated with clinical/cognitive/pathology endophenotypes, planarity of the tensor (PLA) and mode of the tensor (MOD) features are not informative to predict white matter changes in AD.
-
•
An immune response, interferon signaling and cytokine signaling enriched gene subnetwork derived from the matched blood gene expression data is most correlated with DTI features across all the brain regions.
-
•
CELF1, a GWAS AD risk gene, is significantly associated with white matter microstructural changes in the hippocampus.
Keywords: Alzheimer's disease, Diffusion tensor imaging, White matter, Brain regions, CELF1, Immune response, Gene expression, Multiscale embedded gene coexpression network analysis
Abstract
Introduction
Microstructural abnormalities in white matter (WM) are often reported in Alzheimer's disease (AD). However, it is unclear which brain regions have the strongest WM changes in presymptomatic AD and what biological processes underlie WM abnormality during disease progression.
Methods
We developed a systems biology framework to integrate matched diffusion tensor imaging (DTI), genetic and transcriptomic data to investigate regional vulnerability to AD and identify genetic risk factors and gene subnetworks underlying WM abnormality in AD.
Results
We quantified regional WM abnormality and identified most vulnerable brain regions. A SNP rs2203712 in CELF1 was most significantly associated with several DTI-derived features in the hippocampus, the top ranked brain region. An immune response gene subnetwork in the blood was most correlated with DTI features across all the brain regions.
Discussion
Incorporation of image analysis with gene network analysis enhances our understanding of disease progression and facilitates identification of novel therapeutic strategies for AD.
1. Introduction
Alzheimer's disease (AD) is the most common type of dementia, affecting 5.7 million people in the U.S. alone, with the number of new diagnoses increasing dramatically each year (Weiner et al., 2015; Alzheimer's, 2016, Weiner et al., 2017). AD patients display hippocampal atrophy, memory impairment, and other cognitive and axonal loss. The ultimate outcome of AD is neuronal loss and catastrophic memory deficits. While many studies have focused on amyloid and neurofibrillary pathology and cortical atrophy in the cortex and subcortical gray matter, less attention has been paid to the significant white matter (WM) abnormalities, such as demyelination, microglial activation, loss of oligodendrocytes and reactive astrocytosis, that have been characterized recently (Acosta-Cabronero and Nestor, 2014; Dean et al., 2017).
Magnetic resonance imaging (MRI) technology provides a path to more thoroughly explore associations between white matter abnormalities and AD related phenotypes. With increasingly high-resolution characterizations of brain structure, MRI now plays an important role in the diagnosis of AD (Knight et al., 2016; Rathore et al., 2017). Diffusion tensor imaging (DTI) detects abnormal changes in neuronal fibers at the microstructural level in mild cognitive impairment (MCI) and AD, and provides data on the integrity of brain circuitry (Rathore et al., 2017; Shen et al., 2017). DTI detects the 3-dimensional (3D) diffusion of water molecules along the main fiber directions (Alexander et al., 2007; Basser and Pierpaoli, 2011), giving rise to specific features such as mean diffusivity (MD) and fractional anisotropy (FA) in white matter regions that have been demonstrated to be strongly correlated WM damage and cognitive impairment (Naggara et al., 2006; Lee et al., 2017). However, despite the roles well-established AD biomarkers such as amyloid-β deposition and inflammation are known to play in disrupting WM integrity during the early stages of AD, only a small number of studies to date have focused on specific DTI features to identify associations between CSF-biomarkers and changes in WM integrity in AD (Bendlin et al., 2012; Melah et al., 2016). Given this lack of attention, several key questions remain to be explored regarding variations in WM associated with AD-related traits, including the identification of brain regions in which WM changes most strongly correlated with presymptomatic AD and the identification of molecular and biological processes that underlie WM abnormalities that develop as a result of disease progression.
To help address these critical questions, we systematically interrogated DTI, genetic, gene expression and clinical data generated on participants in the Alzheimer's Disease Neuroimaging Initiative (ADNI). Given amyloid beta depositions and neurofibrillary tangle pathology are associated with a range of pathophysiologic changes in the brain, including axonal degeneration, disruption of cytoskeletal equilibrium, and synaptic dysfunction (Mandelkow et al., 2003; Cardenas et al., 2012), we sought to characterize covariation structures between DTI-derived features and AD-related traits such as CSF tau/p-tau and Abeta levels, episodic memory scores, and metabolic activity and amyloidosis (PET) during disease progression. Specifically, we employed a systems biology framework to organize, process, and integrate these data and then characterize the associations among the different dimensions (imaging, molecular, and clinical) of data. Our framework provided for a completely data driven approach to first rank-order all brain regions with respect to their ability, through DTI-derived features, to discriminate between AD cases and controls. Then, from the top ranked brain regions, we systematically modeled white matter microstructural abnormalities in AD to identify 1) regional differences in microstructural changes; 2) correlations between AD-pathology related endophenotypes and brain region specific DTI-features; 3) key biological processes underlying WM pathology by integrating DTI and gene expression data; and 4) AD susceptibility loci associated with microstructural changes in AD. From this integrative analysis, we identified several DTI-derived features that were not only strongly associated with AD-related traits, but that were associated with known genetic risk loci in known AD-risk genes such as CELF1. In addition we identified an immune-associated gene network in blood that was strongly associated with these DTI features across all brain regions, providing a complementary view of disease progression and therapeutic strategies for AD.
2. Methods
2.1. Study participants
We obtained the data used in this study from the ADNI database (adni.loni.usc.edu). ADNI was launched in 2003 by a public-private partnership led by Michael W. Weiner. Participants were recruited from more than 50 sites across the United States and Canada. ADNI participants consist of older individuals, aged 55–90, who are cognitively normal (CN), or who have significant memory concerns (SMC), mild cognitive impairment (MCI) or clinically diagnosed AD (http://www.adni-info.org/). The ADNI dataset includes structural MRI and PET scans, longitudinal CSF markers, and performance on neuropsychological and clinical assessments. In addition, the ADNI data include APOE and genome-wide genotyping generated on study participants. We analyzed diffusion tensor imaging scans from 269 individuals, including 57 CN older individuals, 33 individuals with SMC, 76 individuals diagnosed with EMCI, 27 individuals with LMCI, and 76 individuals diagnosed with AD. We note that not all individuals included in the analysis of the DTI data had matched clinical and pathological data (Supplementary Table S1). Clinical and neuroimaging procedures and the other information about the ADNI cohort can be found at http://www.adni-info.org/.
2.1.1. Study participants for each analysis
Since not all participants in ADNI had clinical and cognitive information, we used different sample sizes for each analysis (Supplementary Table S2). The largest set was comprised of 269 individuals. However, since only 255 or fewer individuals had their phenotype information such as memory scores, cerebrospinal fluid (CSF) amyloid beta and tau levels and CDR score (more information can be found in Supplementary Table S1), we opted to use all available samples (Table 2 and Supplementary Table S1) for the correlation analyses involving each DTI feature and the neuropathological traits. For the co-expression analysis we used 735 individuals (CN=258, EMCI=212, LMCI=225, AD=40) from gene expression profiling data in ADNI. For correlation analysis of co-expression network and DTI features we used 105 individuals (CN=34, MCI=56 and AD=15) who had matching blood expression and DTI data. For the genetic association analysis we used 225 individuals (CN=46, SMC=29, EMCI=62, LMCI=25, AD=63) from ADNIGO/2 who had both DTI scans and genotyping data.
Table 2.
The number of participants considered for the imaging-clinical/cognition correlation analyses across the five disease categories.
| Phenotypes | CN | SMC | EMCI | LMCI | AD |
|---|---|---|---|---|---|
| Clinical | |||||
| ADAS13 (N=254) | 57 | 31 | 72 | 25 | 69 |
| MMSE (N=255) | 57 | 31 | 73 | 25 | 69 |
| MOCA (N=248) | 57 | 31 | 71 | 25 | 64 |
| RAVLT_immediate (N=251) | 56 | 31 | 73 | 25 | 66 |
| RAVLT_learning (N=251) | 56 | 31 | 73 | 25 | 66 |
| RAVLT_forgetting (N=250) | 56 | 31 | 73 | 25 | 65 |
| FAQ (N=251) | 56 | 31 | 69 | 25 | 70 |
| EcogPtTotal (N=249) | 57 | 31 | 72 | 25 | 64 |
| EcogSPTotal (N=246) | 56 | 30 | 65 | 25 | 70 |
| CDRSB (251) | 56 | 30 | 70 | 25 | 70 |
| AD-pathology related | |||||
| CSF_ABETA (N=50) | 12 | 12 | 6 | 4 | 16 |
| CSF_TAU (N=62) | 14 | 17 | 10 | 5 | 16 |
| CSF_PTAU (N=62) | 14 | 17 | 10 | 5 | 16 |
| [18F]Florbetapir PET (AV45) (N=134) | 36 | 25 | 30 | 12 | 31 |
| FDG_PET (N=44) | 13 | 4 | 10 | 3 | 14 |
2.2. Acquisition of the diffusion weighted MRI data
Diffusion weighted MRI (DWI) data were downloaded from the ADNI website. Typical DWI acquisition parameters for these data were as follows: Field Strength=3.0 T; Flip Angle=90.0 degree; Gradient Directions=41.0; Manufacturer=GE MEDICAL SYSTEMS; Matrix X=256.0 pixels; Matrix Y=256.0 pixels; Matrix Z=3588.0; Pixel Size X=1.37 mm; Pixel Size Y=1.37 mm; Pulse Sequence=EP/SE; Slice Thickness=2.70 mm; TE=57.205 ms; TR=9050.0 ms.
2.3. Diffusion tensor imaging processing
The diffusion weighted MRI (DWI) data were first corrected for Eddy current distortion through affine registration to a reference volume (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/eddy). Next, using the Brain Extraction Tool (BET, https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BET), we removed the skull from each DWI image. We computed the DWI image features based on two different reconstruction models: 1) a single tensor model (DTI) and 2) neurite orientation dispersion and density imaging (NODDI) model. Altogether, 9 DWI features were computed, 8 of them are based on DTI model, and the remaining one is based on NODDI model. Details of these 9 features based on regarding these two models are described below, (Table 1).
Table 1.
Demographic and clinical characteristics of the ADNI participants
| CN | SMC | EMCI | LMCI | AD | |
|---|---|---|---|---|---|
| N | 57 | 33 | 76 | 27 | 76 |
| Age (SD) | 75.91 (6.765) | 75.24 (5.432) | 75.58 (8.211) | 76 (6.884) | 76.43 (7.643) |
| Gender (M/F) | 25/32 | 12/21 | 44/32 | 18/9 | 49/27 |
| APOE (ε4−/ε4+) | 37/19 | 21/12 | 45/29 | 10/16 | 22/53 |
2.3.1. Single tensor model
In the single tensor model, an ellipsoid (tensor) for each 3D image voxel is computed to quantify the magnitude and anisotropy of water diffusion at the corresponding spatial location. Eigenvalues (λ1, λ2, λ3) are then computed from each 3D tensor and used to generate eight DTI features: 1) Fractional anisotropy (FA), 2) Mean diffusivity (MD), 3) Axial diffusivity (L1), 4) Radial diffusivity (RD), 5) Linearity of the tensor (LIN), 6) Sphericity of the tensor (SPH), 7) Planarity of the tensor (PLA) and 8) Mode of the tensor (MOD) using FSL (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki). These features are computed by the following formulae:
-
1)Fractional anisotropy (FA), which indicates the degree of anisotropy of a diffusion process:
-
2)Mean diffusivity (MD), which represents the average rate of diffusivity in all directions:
-
3)Axial diffusivity (L1), which indicates diffusivity parallel to the majority of axonal fibers:
-
4)Radial diffusivity (RD), which captures the average diffusivity perpendicular to axial diffusivity:
-
5)Linearity of the tensor (LIN), which evaluates the uniformity of the diffusion direction along the main fiber direction (largest eigenvalue):
-
6)Sphericity of the tensor (SPH), which measures a less uniform isotropic diffusion process that implies more isotropic diffusion:
-
7)Planarity of the tensor (PLA), which indicates the existence of dispersed fibers along just two dimensions:
-
8)Mode of the tensor (MOD), which differentiates three types of anisotropy, including diffusion along a geometric plane, diffusion along a single direction, or isotropic diffusion:
2.3.2. Neurite orientation dispersion and density imaging model (NODDI)
Although the single tensor model has been widely used to extract DTI image features, the single model based measures all rely on the assumption that there is only one axon orientation in each voxel. However, this may not be the case for all brain regions. For example, it has been reported that axonal bundles spread, bend, or cross one another in white matter between the cerebral cortex and nuclei (Zhang et al., 2011). With the NODDI model, the dispersion in axon orientation can be explicitly represented. Therefore, in addition to the 8 DTI-based features, the orientation dispersion index (ODI) (Zhang et al., 2012) was chosen as the NODDI-based features in our analysis for its ability to quantify angular variation of neurite orientation and the extent of orientation dispersion about the mean orientation. Higher ODI values indicate larger axonal dispersion.
2.3.3. Brain region parcellation
After all nine DWI features were computed for each subject, they were registered to a common template image so that the voxels in each image could be transformed into a common coordinate system. In our study, the image registration involves an affine registration step (FLIRT, https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FLIRT), followed by a non-linear registration step (FNIRT, https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FNIRT). We chose the FA image of John Hopkins University Eve atlas (https://www.mristudio.org/) as the common template image and registered the FA feature image of each subject to this template. Using the transformation parameters that had been thus obtained, the other 8 feature images were also transformed into the same coordinate system. In addition, we used the JHU-MNI SS Type-I Mori Atlas (https://www.slicer.org/wiki/Slicer3:Mori-Atlas_labels_JHU-MNI_SS_Type-I) to parcellate the whole brain of each subject into a total of 176 brain regions (Supplementary Fig. S2), which were subsequently evaluated for relevance to AD severity.
2.4. Endophenotypes
All AD-related endophenotypes, including amyloid beta levels, Fluorodeoxyglucose (18F) (FDG)-PET and Florbetapir (AV-45) PET were obtained from the ADNIGO/2 database (http://adni.loni.usc.edu). CSF measurements and quality control data were downloaded from the LONI website as “UPENN CSF Biomarkers Elecsys”. The complete descriptions of the collection and process protocols are provided in the ADNI procedural manual at www.adni-info.org. Alzheimer's Disease Assessment Scale (ADAS-Cog), Mini–Mental State Examination (MMSE), Rey Auditory Verbal Learning Test (RAVLT), Functional Activities Questionnaire (FAQ), and Montreal Cognitive Assessment (MoCA) and Clinical Dementia Rating Sum of Boxes (CDRSB), clinical and cognitive performance scores were downloaded as representations of cognitive test scores (Crane et al., 2012; Johnson et al., 2012). MMSE and MOCA, which measure memory, recall, and attention, were used as cognitive screening tests (Trzepacz et al., 2015). For all score types, those scores determined closest in time to the diffusion MRI scan were chosen for our analyses. Since not all participants in ADNIGO/2 have cognitive and clinical recorded scores, Table 2a summarizes how many participants were included for each correlation-based analysis.
Table 2a.
Known pathways enriched in the gene modules significantly correlated with DTI-features in specific brain regions. The column Corrected p-value shows corrected p values for the Spearman's rank correlation between the indicated DTI feature and gene expression module.
| Region | DTI Feature | Coexpression Module | Enriched Pathways | Corrected p-value |
|---|---|---|---|---|
| Superior occipital gyrus right | FA | M30 | immune response, response to cytokine, cytokine-mediated signaling pathway, innate immune response, regulation of immune system process | 1.34 × 10−2 |
| Supramarginal gyrus left | PLA | M30 | immune response, response to cytokine, cytokine-mediated signaling pathway, innate immune response, regulation of immune system process | 1.62 × 10−2 |
| Supramarginal gyrus right | PLA | M30 | immune response, response to cytokine, cytokine-mediated signaling pathway, innate immune response, regulation of immune system process | 2.16 × 10−2 |
| Supramarginal gyrus left | LIN | M30 | immune response, response to cytokine, cytokine-mediated signaling pathway, innate immune response, regulation of immune system process | 2.70 × 10−2 |
| Postcentral gyrus right | PLA | M30 | immune response, response to cytokine, cytokine-mediated signaling pathway, innate immune response, regulation of immune system process | 2.97 × 10−2 |
| Supramarginal gyrus right | MD | M30 | immune response, response to cytokine, cytokine-mediated signaling pathway, innate immune response, regulation of immune system process | 3.24 × 10−2 |
| Supramarginal gyrus right | RD | M30 | immune response, response to cytokine, cytokine-mediated signaling pathway, innate immune response, regulation of immune system process | 3.38 × 10−2 |
| Corticospinal tract left | ODI | M18 | Sensory perception, nervous system process and G-protein coupled receptor signaling pathway | 3.92 × 10−2 |
| Inferior fronto occipital fasciculus right | ODI | M145 | Cell-cell adhesion, IRE1-mediated unfolded protein response, RNA interference and posttranscriptional regulation of gene expression | 4.32 × 10−2 |
| Supramarginal gyrus right | L1 | M30 | immune response, response to cytokine, cytokine-mediated signaling pathway, innate immune response, regulation of immune system process | 4.32 × 10−2 |
| Fusiform WM left | MD | M26 | immune response, response to cytokine, cytokine-mediated signaling pathway, innate immune response, regulation of immune system process | 4.46 × 10−2 |
| Medulla right | MOD | M23 | cell surface receptor signaling pathway, neurotrophin TRK receptor signaling pathway, B cell activation, cell differentiation, regulation of response to stimulus and cellular developmental process | 8.51 × 10−3 |
2.5. A systems biology framework for the integrative analysis
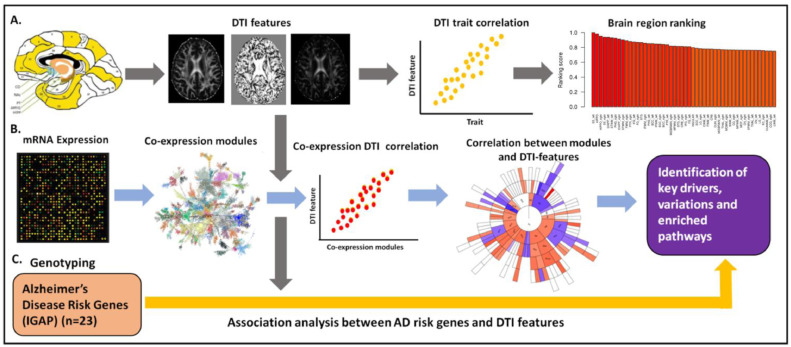
We developed a systems biology framework to integrate brain region specific DTI data with matched genetic and gene expression data to identify genetic risk factors and gene subnetworks underlying white matter abnormalities in AD (Fig. 1).
Fig. 1.
A systems biology framework to integrate DTI, genetic, gene expression, and clinical data in AD. (A) DTI features were extracted and correlated with the clinical and cognitive traits, to systematically rank order 176 brain regions by relevance to AD severity. (B) Multiscale gene coexpression network analysis was then performed on the gene expression data to identify co-expressed gene modules that were enriched for genes that are correlated with DTI features, providing insights into molecular processes associated with WM abnormalities. (C) Gene- and SNP-based association analyses were performed across 23 AD GWAS genes and nine DTI-derived features to assess causal links between AD, AD-associated traits, gene expression traits, and DTI features. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.5.1. Correlation analysis of brain region specific DTI-features and neuropathological traits
As described above, along with the DTI scans, a number of clinical/cognitive scores and AD pathology biomarkers (Aβ, glucose metabolism and CSF-tau) were measured as surrogates of AD status for the ADNI samples. We stratified the traits into two sets as clinical/cognitive scores (ADAS13, MMSE, MOCA, RAVLT, FAQ, Ecog scores, CDR) and AD-pathology (CSF-ABETA, CSF-TAU, FDG-PET and AV45-PET) that reflect different aspects of AD and investigate the relationship between DWI measures with them separately. We elucidated the correlation structure between the imaging feature endophenotypes and clinical/cognitive and pathology biomarker variables to investigate whether any brain region-specific imaging endophenotypes were predictive of clinical outcomes.
Since DTI features are quantified at the unit of voxel, and given there are hundreds of thousands of voxels in each brain region, we first reduced the dimensionality of the data by computing principal components (PCs) from the voxel-level data for each region-specific DTI feature in each brain region. Principal component analysis (PCA) is a well-established method in neuroimage analysis to reduce data dimensionality. As a data-reduction algorithm, PCA generates a set of new variables or principal components (PCs) that are orthogonal linear combinations of a given dataset to maximally explain the variance of the dataset (Ghodadra et al., 2016; Jolliffe and Cadima, 2016). In this study, we performed principal component analysis with the voxel-level data for each DTI feature in each brain region, and took the first right-singular vector (i.e. the first principal component, PC1) of the standardized voxel-level data as the representative-voxel, called eigen-voxel in this paper. For each brain region, the voxel-wise DTI measurements were treated as a two-dimensional matrix with samples in the rows and voxels in the columns. We consider the voxels as individual variables. PCA was performed on the two-dimensional data using the prcomp function in R program language. We then used a respective PC1 as the representative of each region-specific DTI feature. As a result of this dimensionality reduction, we computed 9 eigen-voxels for each brain region, corresponding to the 9 DTI features. We then computed Spearman's correlation coefficient rijk between each clinical/cognitive (i) and each eigen-voxel (j) in each brain region (k). P value significance (Pijk) of the correlation coefficient was computed via the asymptotic t approximation. Significant correlations were defined as those with Bonferroni adjusted P value less than 0.05 (adjusted by the i*j*k number of correlations computed).
In addition to characterizing the correlation between DTI features and AD-related cognitive traits, we sought to prioritize the different brain regions with respect to their relevance to AD by comparing the magnitude of the correlations between the imaging features and cognitive/clinical measures. For this purpose, the various eigen-voxel and clinical/cognitive trait correlations for a brain region (k) were assembled into a composite importance score defined as , where n denotes the number of correlation values (i.e. number of traits times 9 features). The importance score essentially computes the mean of the absolute value of the correlation coefficients across traits and DTI features. We have previously used this type of composite score to ranking the importance of key driver genes identified in gene networks across multiple brain regions (Zhang et al., 2013). However, instead of ranking based on p values as was done in this previous work, we sought to use a quantitative sorting measure from the magnitude of the association in order to incorporate more information into this sorting measure. Finally, the composite importance scores (Sk) were normalized by dividing by the maximum score, and the brain regions were subsequently rank ordered by this normalized importance score. The brain region with the highest composite importance score was ranked as the top one, while the brain region with the lowest composite importance score was ranked as the bottom one. In this analysis, we used all available clinical/cognitive traits to give an unbiased ranking of the brain regions as only few participants have AD-pathology variables. However, we tested whether each top ranked region was also highly correlated with AD-pathology endophenotypes to predict white matter changes in AD.
ROI analysis is a simple and effective means to investigate white matter changes in small, well defined regions on good quality data. In the ROI analysis, the average signal of all the voxels in a region is correlated with clinical outcomes. However, the previous studies have showed that this technique is prone to error and not suitable for the investigation of structures with complex boundaries or poorly defined changes in white matter microstructure (Hermoye et al., 2006; Froeling et al., 2016). To compare the performance of ROI and PCA, we performed a large scale correlation analysis between each voxel with PC1 and ROI from each DTI feature in each region. In all the 176 regions, 67.4% of the voxels are better correlated with PC1s than ROIs. The distributions of correlation coefficients from ROIs and PC1s are shown in the Supplementary Fig. S3.
2.5.2. Correlation analysis of gene co-expression network and DTI features
To further characterize the correlation structure between DTI features, clinical/cognitive traits, and biological processes, we constructed coexpression networks using the quality-controlled and normalized gene expression profiling data generated on the blood samples collected on ADNI participants (N=744). These data were downloaded from the ADNI LONI website (http://adni.loni.usc.edu) (Saykin et al., 2015). These gene expression data were generated using the Affymetrix Human Genome U219 platform (Affymetrix, Santa Clara, CA). Probe sets defined on this platform that did not map to any gene or that mapped to multiple genes were excluded from further analyses. For genes represented by multiple probe sets, the probe set giving rise to the most expression variation across all samples was selected as the representative for the gene. The purpose of choosing the probe sets with most variation is to keep the probe sets with signals (e.g. differentially regulated) among individuals and conditions (low variation means no difference between different conditions). So, with highest variation we can retain genes/isoforms have difference between case and control and exclude those genes have little/no difference (Mancarci et al., 2017). In total, 17,849 genes were included for the gene expression analysis. We excluded 9 samples that were not associated with gender and age information, given gender and age information are needed to appropriately adjust the gene expression data for our analyses. This resulted in 735 samples being used for the co-expression network analysis.
A gene co-expression network for the ADNI blood expression data was constructed using Multiscale Embedded Gene co-Expression Network Analysis (MEGENA) (Song and Zhang, 2015). This process involves first constructing a planar filtered network in MEGENA from significantly correlated gene pairs. The output of this step is then input into a multiscale clustering analysis to identify co-expression modules using different scales of compactness of modular structures controlled by a resolution parameter. The modules identified from this step are then compared to random Planar Filtered Network (PFN) modules generated by shuffling the link weights of the parent cluster to calculate statistical significance. Finally, to elucidate the structure of the significant coexpression modules, a multiscale hub analysis is run to identify highly connected hub nodes (genes) for each significant module. MEGENA modules identified via this process containing less than 50 genes were excluded from further analysis. Principal component analysis was performed on the modules to obtain the principal component vectors (eigen-genes) used in the gene expression-DTI feature correlation analysis. In addition, gene ontology enrichment analysis (Wang et al., 2012) was performed on the significant gene modules to identify representative biological processes associated with the modules. Pearson and Spearman correlation analyses were carried out on the module eigen-genes and the DTI features across the 176 brain regions. To control for multiple testing, we employed a Bonferroni correction procedure, and adjusted p values < 0.05 were considered statistically significant.
2.6. Genotyping data and quality control
GWAS and APOE genotyping data generated on the ADNI participants were downloaded from LONI. ADNIGO/2 samples were genotyped according to manufacturer's protocol (Illumina, Inc., San Diego, CA) using the Human OmniExpress BeadChip. SNP quality control procedures of GWAS data such as SNP call rate < 95%, Hardy–Weinberg equilibrium test p < 1 × 10−6, and frequency filtering (MAF ≥ 5%) were performed using PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/), version 1.07 (Purcell et al., 2007; Saykin et al., 2010; Hohman et al., 2014; Ramanan et al., 2014). For sample quality procedures, due to limitations of population stratification in this cohort, only non-Hispanic Caucasian participants were selected for this analysis by genetic clustering with CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) and TSI (Tuscans in Italy) populations using HapMap 3 genotype data and multidimensional scaling (MDS) analysis after performing standard quality control (QC) procedures for genetic markers and participants (Ramanan et al., 2014). Markov Chain Haplotyping software based on the 1000 Genomes Project as a reference panel was used to impute un-genotyped SNPs (Ramanan et al., 2014).
2.7. Association analysis
For association analysis we used 225 individuals (CN=46, SMC=29, EMCI=62, LMCI=25, AD=63) from ADNIGO/2 who have both DTI scans and genotyping data. Set-based association tests were performed in an additive genetic model using PLINK (Purcell et al., 2007) for 23 International Genomics of Alzheimer's Project (IGAP) genes (Lambert et al., 2013). For each of the 23 gene regions, SNPs falling into untranslated regions, 3′ UTR, 5′ UTR, coding regions, intronic regions, and regulatory regions (±100 kb of upstream and downstream regions) were considered. A total of 12,438 variants passed filters and QC from the 23 gene regions. A mean test statistic for each SNP for each set (gene) was computed to determine if other SNPs were in linkage disequilibrium (LD; i.e., r2 > 0.5). For each SNP within each set a quantitative trait analysis (QT) was then performed, with age and sex included as covariates. Then, the top independent SNPs in each set were selected if their p-values were less than 0.05 in the QT analysis. From these subsets of SNPs, the statistic for each set was calculated as the mean of these single SNP statistics. Empirical p values for each set were calculated from 20,000 permutations. In ADNIGO/2, a conservative significance threshold (p < 0.00217) was used based on Bonferroni correction for 23 IGAP genes.
2.8. Construction of CELF1 centered co-expression gene network in AD
Previously, CUGBP Elav-Like Family Member 1 (CELF1) is identified as GWAS significant gene in AD (Hinney et al., 2014). CELF1 co-localizes with other CELF proteins and regulates pre-mRNA alternative splicing, mRNA editing and translation (Dasgupta and Ladd, 2012). Since we identified that CELF1 was significantly associated with the establishment of WM integrity in the hippocampus, for further functional analysis we constructed CELF1 centered co-expression gene network and identified functional pathways enriched in the CELF1-centered subnetwork. We assembled 6 gene expression datasets from 6 different brain regions spanning 3 human postmortem brain cohorts, including the Mount Sinai Brain Bank (MSBB) AD RNA-seq (4 cortex regions) (Wang et al., 2018a), MSBB AD microarray (hippocampus region) (Haroutunian et al., 2009; Wang et al., 2016), and the Religious Order Study and Memory Aging Project (ROSMAP) RNA-seq (DLPFC) (David et al., 2012a; David et al., 2012b). The number of subjects for each human postmortem brain cohort can be found in Supplementary Table S3. The MSBB AD cohort contained brain specimens obtained from the Mount Sinai/JJ Peters VA Medical Center Brain Bank, which holds over 1,700 samples. This cohort was assembled after applying stringent inclusion/exclusion criteria and represents the full spectrum of disease severity ranging from cognitively normal to severe dementia. RNA-sequencing gene expression profiles were generated across 4 cortex brain regions from approximately 360 brains (Wang et al., 2018b). In addition, microarray gene expression data generated on 19 different cortical regions, including hippocampus, isolated from 55 brains from MSBB were also used (Wang et al., 2016). The ROSMAP dataset included two prospective cohort studies of aging, i.e. the Religious Order Study (ROS) and the Memory and Aging Project (MAP) (De Jager et al., 2018a). While all subjects were without known dementia at study entry, ~60% of the subjects presented a pathologic diagnosis of AD at autopsy. Postmortem RNA-sequencing data were generated from dorsolateral prefrontal cortex (DLPFC) of over 600 brains (De Jager et al., 2018b). The MSBB AD microarray data was obtained from gene expression omnibus (GEO) (accession GSE84422). For the MSBB AD RNA-seq and ROSMAP RNA-seq cohorts, the preprocessed expression data matrices were downloaded from the AMP-AD knowledge portal at Synapse (Wang et al., 2018a). For each dataset, a Pearson's correlation matrix was derived by calculating correlation coefficients between CELF1 and each of the remaining individual genes. For simplicity, we focused only on the set of genes that were present in all 6 datasets. Finally, the genes showing significant correlation with CELF1 (FDR-corrected pvalues ≤ 0.05) in all datasets with consistent correlation direction were used to define the consensus CELF1-centered correlation network.
3. Results
3.1. Regional DTI features are significantly correlated with AD clinical outcomes
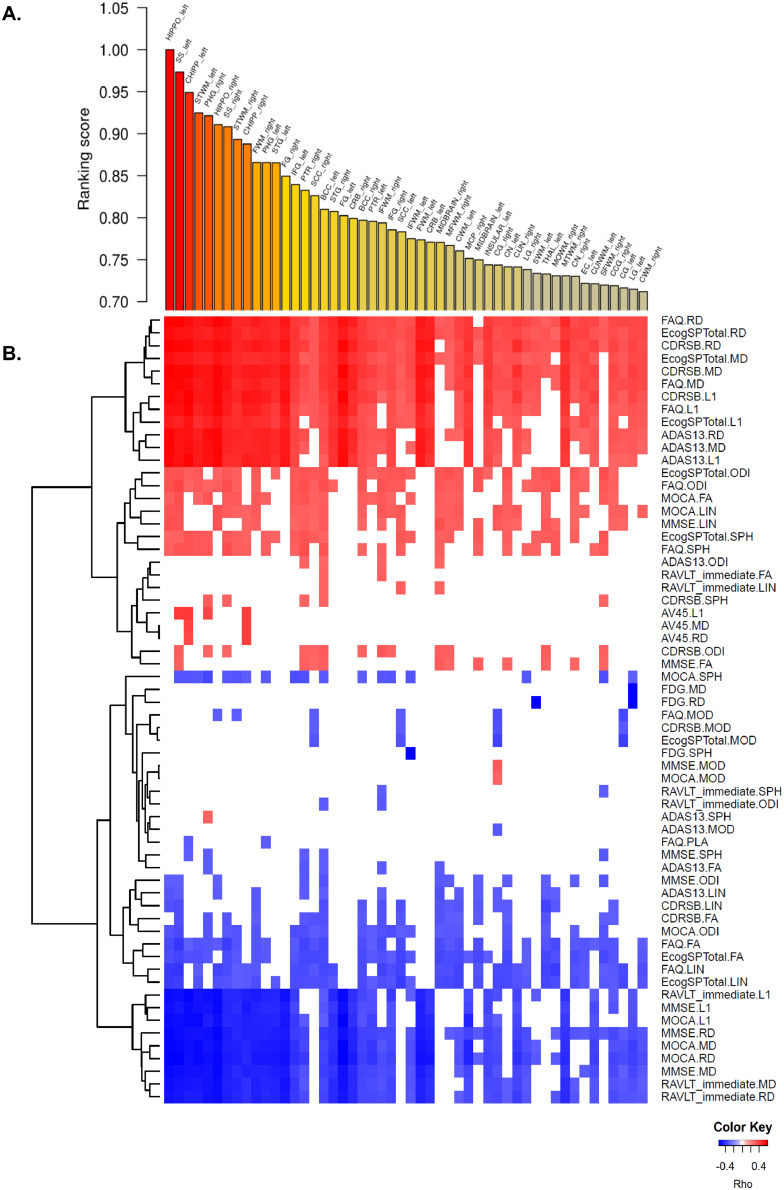
We stratified the traits into two sets as clinical/cognitive scores (ADAS13, MMSE, MOCA, RAVLT, FAQ, Ecog scores, CDR) and AD-pathology scores (CSF-ABETA, CSF-TAU, CSF-PTAU FDG-PET and AV45-PET) that reflect different aspects of AD and investigated the association of DWI measures with cognitive and pathological scores separately. To characterize association of DTI-derived features, which define the structure in the human brain, with AD-related clinical/cognitive traits and pathology endophenotypes, we computed correlations between the first principal component of each of the 9 DTI features in each of the 176 regions and cognitive traits, and then rank-ordered these brain regions according to the extent of association of each region to the cognitive features. The top 10 ranked regions with cognitive scores in the descending order were the hippocampus left (HIPPO-left), the sagittal stratum left (SS-left), the hippocampal cingulum left (CHIPP-left), the superior temporal white matter left (STWM-left), the parahippocampal gyrus right (PHG-right), the HIPPO-right, the SS-right, the STWM-right, the CHIPP-right and the fusiform white matter right (FWM-right) (Fig. 2A; Supplementary Table S4). The PHG, one top region identified, was recently identified as the most affected region with respect to transcriptomic changes among 19 brain regions in an independent AD cohort from the Mount Sinai Brain Bank (Wang et al., 2016). Since AD-pathology endopenotypes were available for only few individuals, they were not used for ranking regions. Instead, we tested how AD-pathology traits were correlated with the DTI-derived measurements of the top ranked regions (Supplementary Table S5).
Fig. 2.
A) Rank-ordered brain regions according to the extent of association to the clinical/cognitive features. B) Heatmap of the correlations between clinical/cognitive traits and DTI-derived features. Cognitive and pathological traits with DTI-derived features are listed on the right axis, while the top 50 brain regions are listed across the top axis. The intensity of the color in each cell indicates the magnitude of the Spearman's rank correlation coefficient between the corresponding row and column variables, for those correlations with adjusted pvalues < 0.05. Red indicates positive correlation; blue indicates negative correlation. Fluorodeoxyglucose (18F) (FDG)-PET; Florbetapir (AV-45) PET, Alzheimer's Disease Assessment Scale (ADAS-Cog), Mini–Mental State Examination (MMSE), Rey Auditory Verbal Learning Test (RAVLT), Functional Activities Questionnaire (FAQ), and Montreal Cognitive Assessment (MoCA) and Clinical Dementia Rating Sum of Boxes (CDRSB), clinical and cognitive performance scores, self (PT)- and informant (SP)- everyday cognition (ECog) memory scores, Fluorodeoxyglucose (18F) (FDG)-PET and Florbetapir (AV-45) PET. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For the top 50 brain regions, we performed a more expansive correlation analysis between the first principal components of the 9 DTI-derived features in each region and various clinical/cognitive traits. Higher values of some clinical features such as CDR, ADAS13, Ecog memory scores were associated with poor outcomes while higher scores of the others clinical features such as MOCA, MMSE and RAVLT were associated with improved outcomes (Fig. 2B). Our results show that MD, RD and L1 features were all predictive of poor outcomes, and they were highly correlated with the clinical dimentia rating (CDR) score as well as composite memory scores, such as RAVLT, MMSE, MOCA, and ADAS 13, across all top 10 brain regions. In contrast, the FA and LIN DTI-derived features were predictive of improved outcomes, although the correlations with AD endophenotypes in these instances were not consistent across the top 10 brain regions. FA score is highly associated with MOCA, FAQ and ECogSP scores in the hippocampus left, the saggital stratum and STWM regions. Informant Ecog memory score, are highly correlated with DTI-derived features except PLA and MOD across 10 brain regions (Fig. 2B). MOD and PLA are not informative of clinical/cognitive outcomes in AD.
Our correlation analysis between the first principal components of the 9 DTI-derived features in top ten ranked regions and various AD pathology traits revealed that the hippocampus right and left, the sagittal stratum left, the parahippocampal gyrus right and the cingulum right and left were significantly associated with amyloid beta pathology while MD, RD and L1 features in the HIPPO right and left, the SS left, the PHG right, the CHIPP right and left were highly correlated with amyloid beta accumulation (AV45 PET) (corrected p-value < 0.05) (Fig. 2B; Supplementary Table S5). Given L1 (axial diffusivity) is associated with axonal injury (Acosta-Cabronero et al., 2010) and RD (radial diffusivity) is highly associated with demyleination (Bosch et al., 2012; Mayo et al., 2018) and both DTI features increase in AD patients, these results support amyloid pathology as highly associated with white matter microstructural damage in the hippocampus, the cingulum (hippocampus), the sagittal stratum left as well as the parahippocampal gyrus right.
3.2. Co-expression-diffusion network analysis establishes a link between DTI features and a blood transcriptional network associated with AD
To connect the imaging and clinical feature data with molecular data that can help elucidate the biological processes associated with variations in the DTI features, we constructed gene coexpression networks from molecular profiling data generated on the blood of ADNIGO/2 participants. Applying MEGENA (Song and Zhang, 2015) to these data captured interactions among 17,849 genes and identified 150 gene co-expression modules (Supplementary Table S6) comprised of highly interconnected (coregulated) genes. A total of 105 ADNIGO/2 participants had DTI scans that were matched with blood gene expression data (Supplementary Table S7). We correlated the eigengenes computed for each module with the 9 DTI features for each brain region (Supplementary Table S8), identifying all significant correlations at a Bonferroni adjusted significance threshold of p < 3.7 × 10−5 (Table 2). We also carried out gene ontology enrichment analysis to annotate the MEGENA modules with representative biological process categories (Supplementary Table S9). Among the significant expression module-DTI feature correlations identified, a module enriched for immune response, cytokine signaling was found to be most correlated with demyelination and axonal injury in left fusiform white matter and right superior occipital gyrus, right supramarginal gyrus and postcentral gyrus. In addition, modules associated with cell surface receptor signaling, neurotrophin TRK receptor signaling, B cell activation, cell differentiation, regulation of response to stimulus, actin filament bundle assembly, glomerulus development and cellular developmental process, were significantly correlated with DTI data in the right medulla. Modules enriched for cell-cell adhesion, IRE1-mediated unfolded protein response, RNA interference and post-transcriptional regulation of gene expression were correlated with the right inferior fronto occipital fasciculus, while the modules associated with sensory perception, nervous system process and G-protein coupled receptor signaling were strongly correlated with DTI features in the left corticospinal tract.
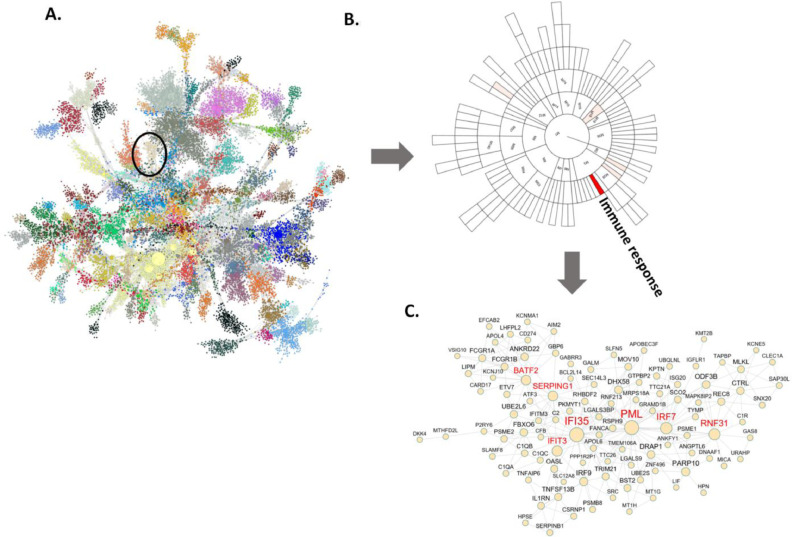
To further characterize the degree of association between the gene coexpression modules and DTI features, we rank-ordered the modules based on the degree of association between each module and the DTI features in all the regions (Supplementary Table S10). Fig. 3 depicts the correlation summary for all the modules. The module M30, which is associated with immune response pathways, is the module most associated with multiple brain regions and DTI features (Fig. 3B). Fig. 3C shows the network structure of M30 and highlights 7 highly connected (hub) genes including SERPING1, BATF2, PML, IRF7, IFI35, IFIT3 and RNF31.
Fig. 3.
Gene co-expression network analysis of the gene expression data from the blood samples of the AD and normal control subjects identifies an immune enriched gene module associated with the DTI features. (A) The global gene co-expression network is comprised of highly co-expressed modules which are highlighted in different colors. Co-expression network is constructed based on the significant correlations among gene expression profiles and then used to identify clusters or modules of highly interconnected genes. Associations between the DTI features and the module eigenvectors (the principal components of modules) are calculated to rank order the gene modules. The most significant module (M30) is highlighted in a black circle. (B) A sunburst plot showing module hierarchy and the associations between the modules and the DTI features across all the brain regions. Each block represents a module and the color intensity of a block indicates the correlation strength between the corresponding module eigenvector and the DTI features (FA, PLA, LIN, MD and RD) with the solid red color standing for the strongest correlation (M30) and the white color for no significant correlation. The M30 was ranked the top based on the summary of all the significant correlations with 9 DTI features. (C) The network plot of the module (M30) that is most significantly associated with the DTI features. The nodes with red labels are the hubs in the network. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Genetic links between DTI features and AD risk loci
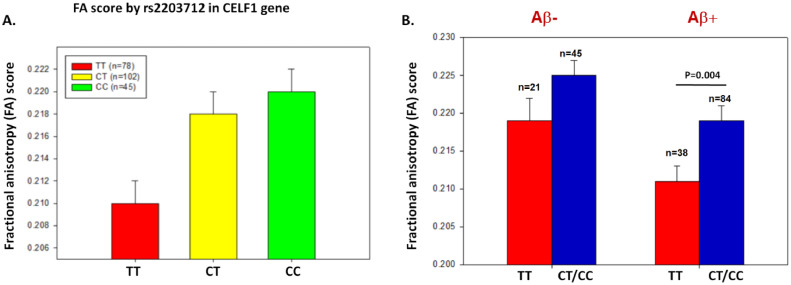
While the gene coexpression network analysis provides a way to link coherent molecular interactions in blood with the brain imaging data, pleiotropic relationships between imaging data and genetic risk loci for AD reflect potentially more direct causal links. To identify such links we analyzed the DTI scans from the participants’ most recent visits and their association with the genotyping data in the ADNIGO/2 dataset (N=225) (Supplementary Table S11). Given that the hippocampus left was the first ranked region with clinical/cognitive outcomes and was most strongly related to AD-associated traits, we performed gene-based association analysis between twenty-three AD risk loci and the fractional anisotropy (FA) feature in the hippocampus. We identified CELF1 (CUGBP Elav-Like Family Member 1) as the most significantly associated loci with the DTI-derived FA feature in the hippocampus (Bonferroni corrected p = 8 × 10−4; Table 3). An intronic SNP (rs2203712) in the CELF1 gene was identified as the most significantly associated with FA, LIN, and the ODI DTI features in the hippocampus (p = 6.34 × 10−4, 1.4 × 10−3, 7 × 10−4, respectively; Table 4). At this loci, the participants with no copies of the minor allele (n = 78; TT genotype) had a smaller FA value in the hippocampus compared to those with one copy of the minor allele (n = 102; CT genotype) or two copies of minor allele (n = 45; CC genotype) (Fig. 4).
Table 3.
Set-based association for 23 IGAP loci and the FA feature using all the common variants (MAF ≥ 0.05) in the neighborhood of the identified risk gene corresponding to each AD risk locus. Empirical p-values were calculated using 20,000 permutations in PLINK.
| Gene | Number of SNPs in gene | Number of significant SNPs (p < 0.05, r2 < 0.5) | Empirical gene-based p-value | List of significant SNP |
|---|---|---|---|---|
| CELF1 | 346 | 4 | 8 × 10−4 | rs7118178|rs7925299| rs2203712|rs71475924 |
| CLU | 539 | 3 | 0.0117 | rs11135998|rs59953408| rs532754 |
| SORL1 | 540 | 5 | 0.01405 | rs664424|rs11600686| rs4372460|rs12276905| rs10790451 |
| PICALM | 687 | 13 | 0.1529 | rs471470|rs67598967| rs6592271|rs11234568| rs598561|rs11607590| rs56302636|rs11234555| rs3862786|rs10792829| rs11234549|rs12277986| rs11825598 |
| FERMT2 | 486 | 1 | 0.2276 | rs62003479 |
| HLA-DRB5 | 373 | 2 | 0.2783 | rs9271176|rs680151 |
| SLC24A4 | 1134 | 10 | 0.2957 | rs79544202|rs12434183| rs9972287|rs34723497| rs1108161|rs55840245| rs4904955|rs12436499| rs66753927|rs55866218 |
| HLA-DRB1 | 399 | 2 | 0.3195 | rs9271176|rs680151 |
| EPHA1 | 450 | 3 | 0.3443 | rs73154208|rs55893042| rs7810606 |
| CASS4 | 494 | 3 | 0.3445 | rs6024817|rs6024937| rs11906849 |
| RIN3 | 1093 | 5 | 0.3584 | rs12434183|rs55977629|rs55840245|rs4904955| rs1242105 |
| CD33 | 320 | 13 | 0.3916 | rs10425414|rs273692| rs2134068|rs7253654| rs34564964|rs2411329| rs3887787|rs35982135| rs79254320|rs1710353| rs10409348|rs73055137| rs4802772 |
| INPP5D | 577 | 6 | 0.4419 | rs72984219|rs7581670| rs3792106|rs59199559| rs62192899|rs62192926 |
| BIN1 | 908 | 3 | 0.4477 | rs6716819|rs6431230| rs13031473 |
| PTK2B | 779 | 13 | 0.4514 | rs17375582|rs2565045| rs2322607|rs11780471| rs11994882|rs2565048| rs2241657|rs7000615| rs1367088|rs187228254| rs7013346|rs2271920| rs2741341 |
| ZCWPW1 | 234 | 1 | 0.5396 | rs118119933 |
| CR1 | 617 | 3 | 0.6456 | rs61821132|rs1570564| rs71515116 |
| NME8 | 750 | 3 | 0.6471 | rs1668347|rs2198050| rs1668357 |
| ABCA7 | 672 | 7 | 0.7435 | rs2240615|rs35500465| rs4147940|rs349310| rs8106918|rs4147912| rs7260506 |
| MEF2C | 373 | 3 | 0.7944 | rs7728694|rs214134| rs75034114 |
| CD2AP | 884 | 1 | 0.8546 | rs16876369 |
| DSG2 | 491 | 1 | 0.8602 | rs36032521 |
| MS4A6A | 403 | 0 | 1 | NA |
Table 4.
Association of rs2203712 in CELF1 with neuroimaging phenotypes
| rs2203712 | p-value after adjusting for diagnosis | p-value before adjusting for diagnosis |
|---|---|---|
| FA | 6.34 × 10−4 | 5.12 × 10−4 |
| LIN | 1.4 × 10−3 | 1.17 × 10−3 |
| ODI | 7 × 10−4 | 5.97 × 10−4 |
Fig. 4.
rs2203712 in the CELF1 gene region is associated with higher fractional anisotropy (FA) in amyloid-positive participants, as classified by CSF Aβ levels. (A) The presence of at least one copy of the minor allele (T) of rs2203712 was significantly associated with increased FA in hippocampus (p = 0.001). (B) The effect of the common C allele on fractional anisotropy was present in both amyloid-negative (left column) and amyloid-positive (right column) participants. rs2203712 is significantly associated with FA in amyloid-positive participants (p = 0.004). Each participant was classified by their amyloid status (positive versus negative) at the last visit (determined by standard cutoffs on CSF amyloid levels).
We also examined the effect of the minor allele (rs2203712-C) on FA in both amyloid-negative and amyloid-positive participants classified as positive or negative by CSF Aβ1-42 levels. Hippocampal FA values were significantly higher in Aβ positive participants carrying at least one minor allele (p = 0.004) (Fig. 4).
3.4. Constructing CELF1 centered co-expression gene network in AD
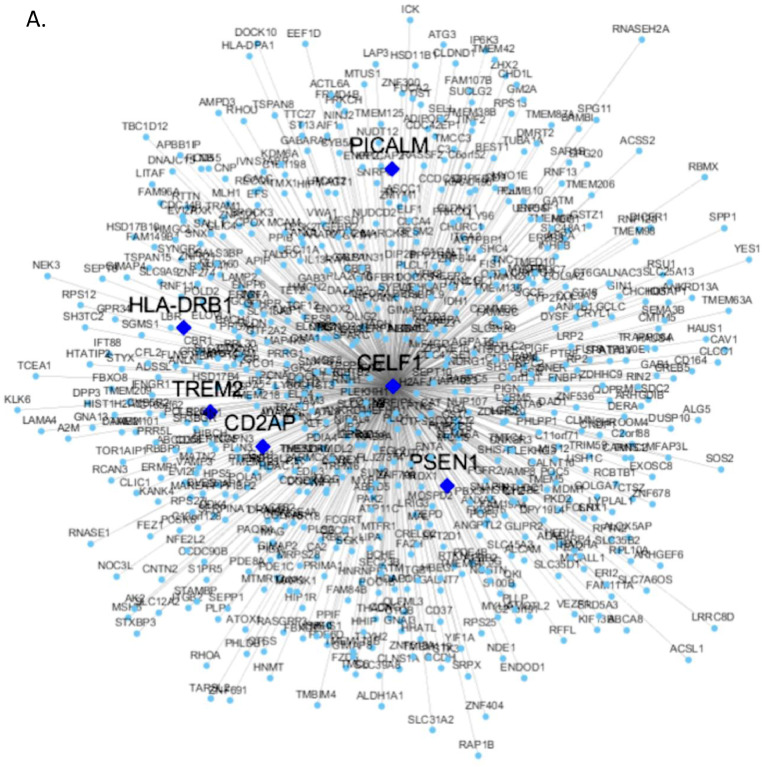
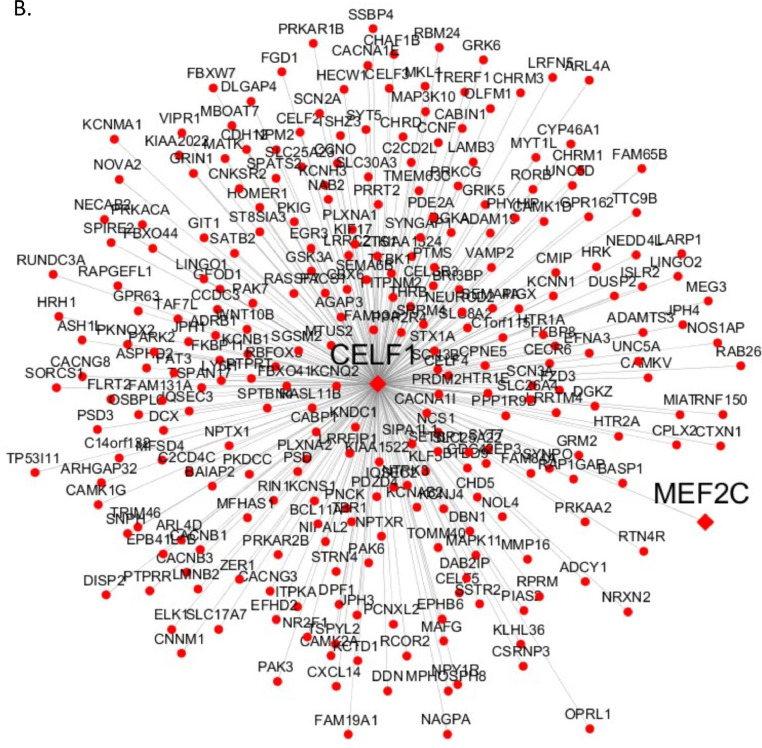
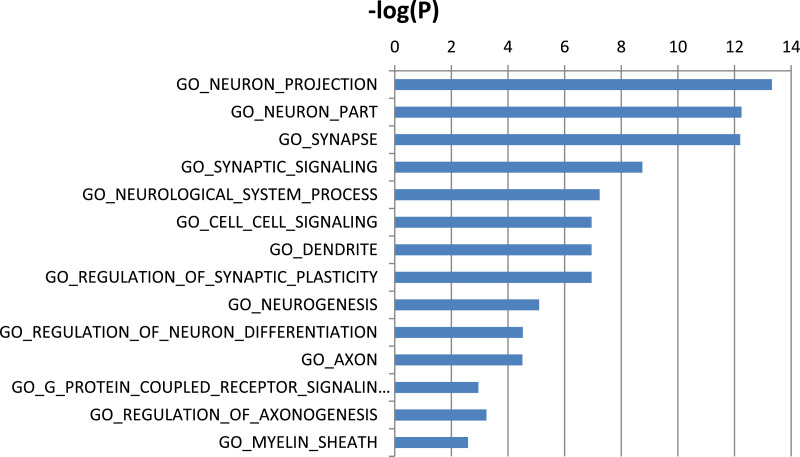
To understand the functional context of CELF1 operation in AD, we constructed a CELF1 centered network using 6 gene expression datasets from 6 different brain regions in three human postmortem brain cohorts, including Mount Sinai Brain Bank (MSBB) AD RNA-seq (4 cortex regions) (Wang et al., 2018b), MSBB AD microarray (hippocampus region) (Haroutunian et al., 2009; Wang et al., 2016), and ROSMAP RNA-seq (DLPFC) (David et al., 2012a; David et al., 2012b). The genes significantly correlated (positively or negatively) with CELF1 in all 6 datasets formed the consensus network (Fig. 5 and Supplementary Table S12). This subnetwork was enriched for a number of neuronal system related pathways such as synapse, neuron differentiation, synaptic signaling, synaptic transformation, myelin sheath, axonogenesis and neurogenesis (Supplementary Table S13) (Fig. 6). Moreover, the network harbors several well-known AD risk genes including MEF2C, PSEN1, CD2AP, PICALM, HLA-DRB1 and TREM2.
Fig. 5.
CELF1 centered consensus gene network in AD. (A) Genes negatively correlated with CELF1 represented by blue circle nodes and (B) genes positively correlated with CELF1 represented by red circle nodes. In both network plots, the diamond nodes are AD risk genes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
MSigDB GO and canonical pathways enriched in the CELF1 centered consensus gene network (union of the Fig. 5A and B). Blue bars represent –log value of adjusted p-values.
4. Discussion
Most previous studies involving imaging data in an AD context have focused on specific tensor-based metrics such as FA (a measurement of overall directionality of the diffusion of water molecules) and MD (a measurement of the average rate of diffusivity) features to maximize the power of diffusion MRI scans to identify disease related microstructural abnormalities in the WM of AD related regions (Kantarci et al., 2011; Maggipinto et al., 2017; Nir et al., 2017). In this study, we employed a data driven approach to rank order 176 brain regions with respect to their correlation with region specific DTI features and disease severity. The hippocampus, the cingulum, the parahippocampal gyrus right, the sagittal stratum, the superior temporal white matter and the fusiform white matter (FWM) right were the most correlated with clinical/cognitive traits. Most of these regions have been previously shown to be associated with AD related phenotypes, with some well demonstrated to play a central role in AD pathogenesis and progression (Kuczynski et al., 2010; Nir et al., 2013). In addition, we identified that the parahippocampal gyrus is highly associated with white matter microstructural changes in AD. The hippocampus plays an important role in the formation of episodic and spatial memory and is associated with many neurodegenerative diseases (Axmacher et al., 2008). A pattern of increased MD has previously been found in the hippocampus of AD patients compared to normal individuals (Lin et al., 2016). The cingulum on the other hand is enriched for WM fibers and is known to play an important role in in construction of memory networks by carrying axons that project pyramidal neurons in CA1 and CA3 sub-regions of the hippocampus (Kantarci et al., 2011; O'Dwyer et al., 2011; Nowrangi et al., 2013; Sali et al., 2013; Nowrangi and Rosenberg, 2015). The WM microstructural changes and myelin specific pathology in the parahippocampal region have been identified in MCI and AD patients (Salat et al., 2010; Solodkin et al., 2013). In another recent study carried out by our group, the parahippocampal gyrus was identified as the most affected region in the transcriptomic analysis of 19 brain regions in an independent AD cohort from the Mount Sinai Brain Bank (Wang et al., 2016). Fractional anisotropy reduces specifically in cingulum in hippocampus and sagittal tracts in AD patients, suggesting that disruption of WM tracts occur near the medial temporal lobe (Qiu et al., 2010). A three year follow-up study showed changes in WM in the cingulum fibers and hippocampus (Fu et al., 2014), the top two regions by our study. We further discovered here that the L1, MD and RD DTI features were all highly correlated with amyloid beta pathology as well as memory composite scores in regions most affected by AD. Fusiform gyrus plays a crucial role in facial recognition (Hargrave et al., 2002), and lower FA scores in fusiform gyrus were significantly associated with lower MoCA scores in the MCI group relative to normal controls (Cooley et al., 2015). We also revealed a positive correlation between MoCA scores and myelination in the fusiform white matter right. Moreover, we found that the MMSE, MOCA, RAVLT, ADAS13, and CDRSB scores were all highly correlated with the L1, MD and RD DTI features, while informant everyday cognition (EcogSP) memory scores were highly correlated with all DTI-derived features in the parahippocampal gyrus right, the superior temporal white matter, and the fusiform white matter right.
These results demonstrate that our findings are in agreement with previous findings of impaired WM integrity in the hippocampus, the parahippocampal gyrus and the fusiform regions of the brains with MCI and AD. Most of the brain regions in our findings are more generic to cognitive impairment in general, and may not be specific for AD. However, our correlation analysis with AD-pathology biomarker variables revealed that L1, MD and RD features in the HIPPO-left, the HIPPO-right, the SS-left, the PHG-right, the CHIPP-left and the CHIPP-right are highly associated with amyloid burden but not any of CSF tau measurements. Previous studies showed that there was a sharp increase in λ1, which represents L1 measurement in AD (Acosta-Cabronero et al., 2012; Acosta-Cabronero and Nestor, 2014). In AD patients, it has been observed that regions with significant increase of λ1 that were concordant to those of RD and MD had substantial anisotropy changes (Acosta-Cabronero et al., 2010). Since the degeneration leads to increase in λ1 or axial diffusivity, higher level of L1 is associated with worse memory scores and higher CDR score. The parahippocampal gyrus, the hippocampal cingulum and the sagittal stratum are the regions most impacted by axial degeneration and myelination during disease progression and thus they have the decreased white matter integrity (Liu et al., 2011; Sun et al., 2014). Further research is needed to explore the connectivity of the other top-ranked regions to confirm the disruption of WM integrity.
The integration of gene coexpression network analysis with the DTI data led to identification of an immune-response gene subnetwork that was significantly correlated with demyelination and axonal injury, especially in the fusiform WM, one of the top regions we identified. Many studies have showed that immune response including the activation of microglia, release of inflammatory cytokines/chemokines and enhanced oxidative stress, is associated with AD (Britschgi and Wyss-Coray, 2007; Frank-Cannon et al., 2009). Microglia exhibit region specific patterns of gene expression as well as phenotypic diversity in different brain regions associated with neurogenesis, neuroinflammation and aging (Olah et al., 2011; Grabert et al., 2016). Increased microglial activity was found mostly in WM regions relative to other brain regions in AD animal models (Raj et al., 2017). The immune enriched subnetwork was also correlated with WM changes. Further studies related to imaging of glial cell activation and WM changes in specific brain regions could potentially reveal a mechanistic understanding of the role of microglia response in WM microstructural changes. After examining some of the structural features of the immune enriched subnetwork, we identified SERPING1, a gene known to regulate inflammatory processes (a C1 inhibitor) during inflammation (Glezer et al., 2007), as one of the most highly connected nodes (hub nodes) in this subnetwork. SERPING1 is known to play a crucial role in cortical development (Gorelik et al., 2017) and age-related macular degeneration (Ennis et al., 2008). We also identified another important gene, IRF7 which is interferon regulatory transcription factor, modulates immune responses against Herpes simplex virus type 1 by effecting IFN pathway in Alzheimer's Disease (Costa et al., 2017). Our other hub gene PML (Promyelocytic Leukemia) plays important role in plasticity, circadian rhythms (Manners et al., 2018) and polymorphism in this gene was significantly associated with neuroimaging biomarkers in hippocampus and insular cortex (Moon et al., 2015).
In addition to the characterization of gene expression and AD phenotypes associated with DTI-derived features, we explored the genetics of DTI-derived features in the hippocampus with respect to the known AD susceptibility loci and identified CELF1, which is contained in a risk locus recently identified in a large-scale AD GWAS (Lambert et al., 2013), as a potential genetic regulator. CELF1 co-localizes with other CELF proteins and regulates pre-mRNA alternative splicing, mRNA editing and translation (Dasgupta and Ladd, 2012). We observed that CELF1 was significantly associated with the establishment of WM integrity in hippocampus. CELF1 may play important role in myelination process and axonal injury. Of particular note is that the most significantly AD-associated SNP in this locus, rs2203712, is highly associated with higher FA levels, a score that reflects fiber density and myelination in WM. In IGAP, the SNP rs10838725 in CELF1 is associated with the expression of MTCH2 and FNBP4 expression levels (Karch et al., 2016), and more recently it was found to be associated with delayed AD onset and decreased expression of SPI1 in monocytes and macrophages. The SNP rs2203712 we identified, is contained in the same LD (Linkage Disequilibrium) block (97%) as the GWAS SNP rs10838725, and from the Brain eQTL Almanac (BRAINEAC) brain tissue microarray-based gene expression database (http://www.braineac.org/) and as with rs10838725, rs2203712 is significantly assciated with FNBP4 expression levels in the hippocampus (p-value = 8.9 × 10−3).
To better understand the functional pathways underlying CELF1’s regulation of AD pathogenesis, we identified a CELF1 centered consensus gene co-expression network based on three AD gene expression datasets including the Mount Sinai Brain Bank (MSBB) AD RNA-seq data (4 brain regions) (Wang et al., 2018a), the MSBB AD microarray data in the hippocampus region (Wang et al., 2016), and ROSMAP RNA-seq data from the dorsolateral prefrontal cortex (DLPFC) (De Jager et al., 2018a). The consensus network was associated with neuronal functions such as synaptic signaling, transformation, myelin sheath, axonogenesis and neurogenesis. In addition, this network contained known AD risk genes such as PSEN1 and TREM2. Given that the hippocampus is comprised mainly of grey matter and very sensitive to early changes in MCI and AD, our findings with respect to the hippocampus may represent secondary, indirect association effects. CELF1 and its associated gene network not only provide novel insights into disease pathophysiology, but they represent potential therapeutic targets. Interestingly, CELF1 was also contained in one of the top coexpression network modules associated with DTI-derived features from the inferior fronto occipital fasciculus right region.
The results from the DTI data and those from our previous transcriptomic data from 19 brain regions (Wang et al., 2016) pinpointed the PHG as one of the most modified brain regions in AD. Therefore, PHG may serve as a potential important indicator for AD progression. More focused imaging and multi-Omics studies of the PHG may unlock some key mechanisms with respect to AD pathogenesis. Our study not only quantifies the vulnerability of 176 human brain regions to AD but also reveals their potential upstream genetic regulators as well as downstream manifestation in blood. However, there are important limitations of our study. First, while ADNI still remains the best and largest cohort with matched multi-omics and DTI data, the study contains only a moderate number of subjects and a very limited number of subjects have longitudinal DTI data. Therefore this cohort is less suitable for causal inference. To overcome this limitation, we used quantitative traits (QT) as phenotypes to substantially increase the detection power of variant association analysis over a traditional case-control setup (Shen et al., 2014). Further, while our study involved a great many traits and many associations, we applied very rigorous processes to contain false discovery rates. As such, the most prominent findings reported here are statistically significant after rigorous corrections for multiple testing. Another limitation with this study is the lack of inference of trajectories or causal sequences as ADNI currently has a very limited number of subjects with longitudinal DTI data. Moreover, this study is the lack of replication in the gene-based analyses. Meta-analysis results would increase our detection power. Further studies are needed to explore functional evidence of the CELF1 SNP, which was found to be associated with DTI features and gene expression in AD. Notwithstanding these limitations, this study provides novel insights into the biological processes and key regulators related to white matter microstructural changes and the findings could lead to novel therapeutic strategies for AD as well as other neurodegenerative diseases.
Funding
This work was supported in parts by grants from the National Institutes of Health/National Institute on Aging (U01AG046170, RF1AG054014, RF1AG057440, R01AG057907 and F32AG064904).
CRediT authorship contribution statement
Emrin Horgusluoglu-Moloch: Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing - original draft, Writing - review & editing. Gaoyu Xiao: Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing - original draft, Writing - review & editing. Minghui Wang: Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing - original draft, Writing - review & editing. Qian Wang: Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing - original draft, Writing - review & editing. Xianxiao Zhou: Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing - original draft, Writing - review & editing. Kwangsik Nho: Writing - review & editing. Andrew J. Saykin: Writing - review & editing. Eric Schadt: Writing - review & editing. Bin Zhang: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors of this manuscript have no conflict of interest to declare.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.nicl.2020.102203.
Appendix. Supplementary materials
References
- Acosta-Cabronero J., Alley S., Williams G.B., Pengas G., Nestor P.J. Diffusion tensor metrics as biomarkers in Alzheimer's disease. PLoS One. 2012;7(11):e49072. doi: 10.1371/journal.pone.0049072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Cabronero J., Nestor P.J. "Diffusion tensor imaging in Alzheimer's disease: insights into the limbic-diencephalic network and methodological considerations. Front. Aging Neurosci. 2014;6:266. doi: 10.3389/fnagi.2014.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Cabronero J., Williams G.B., Pengas G., Nestor P.J. "Absolute diffusivities define the landscape of white matter degeneration in Alzheimer's disease. Brain. 2010;133(Pt 2):529–539. doi: 10.1093/brain/awp257. [DOI] [PubMed] [Google Scholar]
- Alexander A.L., Lee J.E., Lazar M., Field A.S. "Diffusion tensor imaging of the brain. Neurotherapeutics. 2007;4(3):316–329. doi: 10.1016/j.nurt.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer's A. 2016 Alzheimer's disease facts and figures. Alzheimers Dement. 2016;12(4):459–509. doi: 10.1016/j.jalz.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Axmacher N., Elger C.E., Fell J. Memory formation by refinement of neural representations: the inhibition hypothesis. Behav. Brain Res. 2008;189(1):1–8. doi: 10.1016/j.bbr.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Basser P.J., Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. 1996. J. Magn. Reson. 2011;213(2):560–570. doi: 10.1016/j.jmr.2011.09.022. [DOI] [PubMed] [Google Scholar]
- Bendlin B.B., Carlsson C.M., Johnson S.C., Zetterberg H., Blennow K., Willette A.A., Okonkwo O.C., Sodhi A., Ries M.L., Birdsill A.C., Alexander A.L., Rowley H.A., Puglielli L., Asthana S., Sager M.A. CSF T-Tau/Abeta42 predicts white matter microstructure in healthy adults at risk for Alzheimer's disease. PLoS One. 2012;7(6):e37720. doi: 10.1371/journal.pone.0037720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B., Arenaza-Urquijo E.M., Rami L., Sala-Llonch R., Junque C., Sole-Padulles C., Pena-Gomez C., Bargallo N., Molinuevo J.L., Bartres-Faz D. Multiple DTI index analysis in normal aging, amnestic MCI and AD. relationship with neuropsychological performance. Neurobiol. Aging. 2012;33(1):61–74. doi: 10.1016/j.neurobiolaging.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Britschgi M., Wyss-Coray T. Systemic and acquired immune responses in Alzheimer's disease. Int. Rev. Neurobiol. 2007;82:205–233. doi: 10.1016/S0074-7742(07)82011-3. [DOI] [PubMed] [Google Scholar]
- Cardenas A.M., Ardiles A.O., Barraza N., Baez-Matus X., Caviedes P. Role of tau protein in neuronal damage in Alzheimer's disease and Down syndrome. Arch. Med. Res. 2012;43(8):645–654. doi: 10.1016/j.arcmed.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Cooley S.A., Cabeen R.P., Laidlaw D.H., Conturo T.E., Lane E.M., Heaps J.M., Bolzenius J.D., Baker L.M., Salminen L.E., Scott S.E., Paul R.H. Posterior brain white matter abnormalities in older adults with probable mild cognitive impairment. J. Clin. Exp. Neuropsychol. 2015;37(1):61–69. doi: 10.1080/13803395.2014.985636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A.S., Agostini S., Guerini F.R., Mancuso R., Zanzottera M., Ripamonti E., Racca V., Nemni R., Clerici M. Modulation of immune responses to herpes simplex virus Type 1 by IFNL3 and IRF7 polymorphisms: a study in Alzheimer's disease. J. Alzheimers Dis. 2017;60(3):1055–1063. doi: 10.3233/JAD-170520. [DOI] [PubMed] [Google Scholar]
- Crane P.K., Carle A., Gibbons L.E., Insel P., Mackin R.S., Gross A., Jones R.N., Mukherjee S., Curtis S.M., Harvey D., Weiner M., Mungas D., Alzheimer's Disease Neuroimaging I. Development and assessment of a composite score for memory in the Alzheimer's Disease Neuroimaging Initiative (ADNI) Brain Imaging Behav. 2012;6(4):502–516. doi: 10.1007/s11682-012-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T., Ladd A.N. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip Rev. RNA. 2012;3(1):104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A.B., Julie A.S., Aron S.B., Lisa L.B., Patricia A.B., Robert S.W. Overview and findings from the rush memory and aging project. Curr. Alzheimer Res. 2012;9(6):646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A.B., Julie A.S., Zoe A., Robert S.W. Overview and findings from the religious orders study. Curr. Alzheimer Res. 2012;9(6):628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager P.L., Ma Y., McCabe C., Xu J., Vardarajan B.N., Felsky D., Klein H.-U., White C.C., Peters M.A., Lodgson B., Nejad P., Tang A., Mangravite L.M., Yu L., Gaiteri C., Mostafavi S., Schneider J.A., Bennett D.A. A multi-omic atlas of the human frontal cortex for aging and Alzheimer's disease research. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager P.L., Ma Y., McCabe C., Xu J., Vardarajan B.N., Felsky D., Klein H.U., White C.C., Peters M.A., Lodgson B., Nejad P., Tang A., Mangravite L.M., Yu L., Gaiteri C., Mostafavi S., Schneider J.A., Bennett D.A. A multi-omic atlas of the human frontal cortex for aging and Alzheimer's disease research. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D.C., 3rd, Hurley S.A., Kecskemeti S.R., O'Grady J.P., Canda C., Davenport-Sis N.J., Carlsson C.M., Zetterberg H., Blennow K., Asthana S., Sager M.A., Johnson S.C., Alexander A.L., Bendlin B.B. Association of amyloid pathology with myelin alteration in preclinical Alzheimer disease. JAMA Neurol. 2017;74(1):41–49. doi: 10.1001/jamaneurol.2016.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis S., Jomary C., Mullins R., Cree A., Chen X., Macleod A., Jones S., Collins A., Stone E., Lotery A. Association between the SERPING1 gene and age-related macular degeneration: a two-stage case-control study. Lancet. 2008;372(9652):1828–1834. doi: 10.1016/S0140-6736(08)61348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Cannon T.C., Alto L.T., McAlpine F.E., Tansey M.G. Does neuroinflammation fan the flame in neurodegenerative diseases? Mol. Neurodegener. 2009;4:47. doi: 10.1186/1750-1326-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froeling M. P.P., Leemans A. DTI Analysis methods: region of interest analysis. Diffus. Tens. Imaging. 2016 [Google Scholar]
- Fu J.L., Liu Y., Li Y.M., Chang C., Li W.B. Use of diffusion tensor imaging for evaluating changes in the microstructural integrity of white matter over 3 years in patients with amnesic-type mild cognitive impairment converting to Alzheimer's disease. J. Neuroimaging. 2014;24(4):343–348. doi: 10.1111/jon.12061. [DOI] [PubMed] [Google Scholar]
- Ghodadra A., Alhilali L., Fakhran S. Principal Component analysis of diffusion tensor images to determine white matter injury patterns underlying postconcussive headache. AJNR Am. J. Neuroradiol. 2016;37(2):274–278. doi: 10.3174/ajnr.A4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glezer I., Chernomoretz A., David S., Plante M.M., Rivest S. Genes involved in the balance between neuronal survival and death during inflammation. PLoS One. 2007;2(3):e310. doi: 10.1371/journal.pone.0000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik A., Sapir T., Woodruff T.M., Reiner O. Serping1/C1 Inhibitor affects cortical development in a cell autonomous and non-cell autonomous manner. Front. Cell Neurosci. 2017;11:169. doi: 10.3389/fncel.2017.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabert K., Michoel T., Karavolos M.H., Clohisey S., Baillie J.K., Stevens M.P., Freeman T.C., Summers K.M., McColl B.W. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 2016;19(3):504–516. doi: 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrave R., Maddock R.J., Stone V. Impaired recognition of facial expressions of emotion in Alzheimer's disease. J. Neuropsychiatry Clin. Neurosci. 2002;14(1):64–71. doi: 10.1176/jnp.14.1.64. [DOI] [PubMed] [Google Scholar]
- Haroutunian V., Katsel P., Schmeidler J. Transcriptional vulnerability of brain regions in Alzheimer's disease and dementia. Neurobiol. Aging. 2009;30(4):561–573. doi: 10.1016/j.neurobiolaging.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermoye L., Saint-Martin C., Cosnard G., Lee S.K., Kim J., Nassogne M.C., Menten R., Clapuyt P., Donohue P.K., Hua K., Wakana S., Jiang H., van Zijl P.C., Mori S. Pediatric diffusion tensor imaging: normal database and observation of the white matter maturation in early childhood. Neuroimage. 2006;29(2):493–504. doi: 10.1016/j.neuroimage.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Hinney A., Albayrak O., Antel J., Volckmar A.L., Sims R., Chapman J., Harold D., Gerrish A., Heid I.M., Winkler T.W., Scherag A., Wiltfang J., Williams J., Hebebrand J., Consortium G., Consortium I., Consortium G. Genetic variation at the CELF1 (CUGBP, elav-like family member 1 gene) locus is genome-wide associated with Alzheimer's disease and obesity. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2014;165B(4):283–293. doi: 10.1002/ajmg.b.32234. [DOI] [PubMed] [Google Scholar]
- Hohman T.J., Koran M.E., Thornton-Wells T.A., Alzheimer's Neuroimaging I. Genetic variation modifies risk for neurodegeneration based on biomarker status. Front. Aging Neurosci. 2014;6:183. doi: 10.3389/fnagi.2014.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.K., Gross A.L., Pa J., McLaren D.G., Park L.Q., Manly J.J., Alzheimer's Disease Neuroimaging I. Longitudinal change in neuropsychological performance using latent growth models: a study of mild cognitive impairment. Brain Imaging Behav. 2012;6(4):540–550. doi: 10.1007/s11682-012-9161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe I.T., Cadima J. Principal component analysis: a review and recent developments. Philos. Trans. A Math. Phys. Eng. Sci. 2016;374(2065) doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarci K., Senjem M.L., Avula R., Zhang B., Samikoglu A.R., Weigand S.D., Przybelski S.A., Edmonson H.A., Vemuri P., Knopman D.S., Boeve B.F., Ivnik R.J., Smith G.E., Petersen R.C., Jack C.R., Jr. Diffusion tensor imaging and cognitive function in older adults with no dementia. Neurology. 2011;77(1):26–34. doi: 10.1212/WNL.0b013e31822313dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch C.M., Ezerskiy L.A., Bertelsen S., Alzheimer's Disease Genetics C., Goate A.M. Alzheimer's disease risk polymorphisms regulate gene expression in the ZCWPW1 and the CELF1 Loci. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0148717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M.J., McCann B., Kauppinen R.A., Coulthard E.J. Magnetic Resonance Imaging to Detect early molecular and cellular changes in Alzheimer's disease. Front. Aging Neurosci. 2016;8:139. doi: 10.3389/fnagi.2016.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski B., Targan E., Madison C., Weiner M., Zhang Y., Reed B., Chui H.C., Jagust W. White matter integrity and cortical metabolic associations in aging and dementia. Alzheimers Dement. 2010;6(1):54–62. doi: 10.1016/j.jalz.2009.04.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., Russo G., Thorton-Wells T.A., Jones N., Smith A.V., Chouraki V., Thomas C., Ikram M.A., Zelenika D., Vardarajan B.N., Kamatani Y., Lin C.F., Gerrish A., Schmidt H., Kunkle B., Dunstan M.L., Ruiz A., Bihoreau M.T., Choi S.H., Reitz C., Pasquier F., Cruchaga C., Craig D., Amin N., Berr C., Lopez O.L., De Jager P.L., Deramecourt V., Johnston J.A., Evans D., Lovestone S., Letenneur L., Moron F.J., Rubinsztein D.C., Eiriksdottir G., Sleegers K., Goate A.M., Fievet N., Huentelman M.W., Gill M., Brown K., Kamboh M.I., Keller L., Barberger-Gateau P., McGuiness B., Larson E.B., Green R., Myers A.J., Dufouil C., Todd S., Wallon D., Love S., Rogaeva E., Gallacher J., St George-Hyslop P., Clarimon J., Lleo A., Bayer A., Tsuang D.W., Yu L., Tsolaki M., Bossu P., Spalletta G., Proitsi P., Collinge J., Sorbi S., Sanchez-Garcia F., Fox N.C., Hardy J., Deniz Naranjo M.C., Bosco P., Clarke R., Brayne C., Galimberti D., Mancuso M., Matthews F., European Alzheimer's Disease I., Genetic, D. Environmental Risk in Alzheimer's, C. Alzheimer's Disease Genetic, H. Cohorts for, E. Aging Research in Genomic. Moebus S., Mecocci P., Del Zompo M., Maier W., Hampel H., Pilotto A., Bullido M., Panza F., Caffarra P., Nacmias B., Gilbert J.R., Mayhaus M., Lannefelt L., Hakonarson H., Pichler S., Carrasquillo M.M., Ingelsson M., Beekly D., Alvarez V., Zou F., Valladares O., Younkin S.G., Coto E., Hamilton-Nelson K.L., Gu W., Razquin C., Pastor P., Mateo I., Owen M.J., Faber K.M., Jonsson P.V., Combarros O., O'Donovan M.C., Cantwell L.B., Soininen H., Blacker D., Mead S., Mosley T.H., Jr., Bennett D.A., Harris T.B., Fratiglioni L., Holmes C., de Bruijn R.F., Passmore P., Montine T.J., Bettens K., Rotter J.I., Brice A., Morgan K., Foroud T.M., Kukull W.A., Hannequin D., Powell J.F., Nalls M.A., Ritchie K., Lunetta K.L., Kauwe J.S., Boerwinkle E., Riemenschneider M., Boada M., Hiltuenen M., Martin E.R., Schmidt R., Rujescu D., Wang L.S., Dartigues J.F., Mayeux R., Tzourio C., Hofman A., Nothen M.M., Graff C., Psaty B.M., Jones L., Haines J.L., Holmans P.A., Lathrop M., Pericak-Vance M.A., Launer L.J., Farrer L.A., van Duijn C.M., Van Broeckhoven C., Moskvina V., Seshadri S., Williams J., Schellenberg G.D., Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P., Ryoo H., Park J., Jeong Y., Alzheimer's Disease Neuroimaging I. Morphological and microstructural changes of the hippocampus in early MCI: a study utilizing the Alzheimer's disease neuroimaging initiative database. J. Clin. Neurol. 2017;13(2):144–154. doi: 10.3988/jcn.2017.13.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.H., Hsu W.C., Ng S.H., Cheng J.S., Khegai O., Huang C.C., Chen Y.L., Chen Y.C., Wang J.J. Increased water diffusion in the parcellated cortical regions from the patients with amnestic mild cognitive impairment and Alzheimer's disease. Front. Aging Neurosci. 2016;8:325. doi: 10.3389/fnagi.2016.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Spulber G., Lehtimaki K.K., Kononen M., Hallikainen I., Grohn H., Kivipelto M., Hallikainen M., Vanninen R., Soininen H. Diffusion tensor imaging and tract-based spatial statistics in Alzheimer's disease and mild cognitive impairment. Neurobiol. Aging. 2011;32(9):1558–1571. doi: 10.1016/j.neurobiolaging.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Maggipinto T., Bellotti R., Amoroso N., Diacono D., Donvito G., Lella E., Monaco A., Antonella Scelsi M., Tangaro S. DTI measurements for Alzheimer's classification. Phys. Med. Biol. 2017;62(6):2361–2375. doi: 10.1088/1361-6560/aa5dbe. [DOI] [PubMed] [Google Scholar]
- Mancarci B.O., Toker L., Tripathy S.J., Li B., Rocco B., Sibille E., Pavlidis P. Cross-laboratory analysis of brain cell type transcriptomes with applications to interpretation of bulk tissue data. eNeuro. 2017;4(6) doi: 10.1523/ENEURO.0212-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E.M., Stamer K., Vogel R., Thies E., Mandelkow E. Clogging of axons by tau, inhibition of axonal traffic and starvation of synapses. Neurobiol. Aging. 2003;24(8):1079–1085. doi: 10.1016/j.neurobiolaging.2003.04.007. [DOI] [PubMed] [Google Scholar]
- Manners H.N., Roy S., Kalita J.K. Intrinsic-overlapping co-expression module detection with application to Alzheimer's Disease. Comput. Biol. Chem. 2018;77:373–389. doi: 10.1016/j.compbiolchem.2018.10.014. [DOI] [PubMed] [Google Scholar]
- Mayo C.D., Garcia-Barrera M.A., Mazerolle E.L., Ritchie L.J., Fisk J.D., Gawryluk J.R., Alzheimer's Disease Neuroimaging I. Relationship between DTI metrics and cognitive function in Alzheimer's disease. Front. Aging Neurosci. 2018;10:436. doi: 10.3389/fnagi.2018.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melah K.E., Lu S.Y., Hoscheidt S.M., Alexander A.L., Adluru N., Destiche D.J., Carlsson C.M., Zetterberg H., Blennow K., Okonkwo O.C., Gleason C.E., Dowling N.M., Bratzke L.C., Rowley H.A., Sager M.A., Asthana S., Johnson S.C., Bendlin B.B. Cerebrospinal fluid markers of Alzheimer's disease pathology and microglial activation are associated with altered white matter microstructure in asymptomatic adults at risk for Alzheimer's disease. J. Alzheimers Dis. 2016;50(3):873–886. doi: 10.3233/JAD-150897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.W., Dinov I.D., Kim J., Zamanyan A., Hobel S., Thompson P.M., Toga A.W. Structural Neuroimaging Genetics Interactions in Alzheimer's Disease. J. Alzheimers Dis. 2015;48(4):1051–1063. doi: 10.3233/JAD-150335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naggara O., Oppenheim C., Rieu D., Raoux N., Rodrigo S., Dalla Barba G., Meder J.F. Diffusion tensor imaging in early Alzheimer's disease. Psychiatry Res. 2006;146(3):243–249. doi: 10.1016/j.pscychresns.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Nir T.M., Jahanshad N., Villalon-Reina J.E., Isaev D., Zavaliangos-Petropulu A., Zhan L., Leow A.D., Jack C.R., Jr., Weiner M.W., Thompson P.M., Alzheimer's Diseaase Neuroimaginng I. Fractional anisotropy derived from the diffusion tensor distribution function boosts power to detect Alzheimer's disease deficits. Magn. Reson. Med. 2017;78(6):2322–2333. doi: 10.1002/mrm.26623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir T.M., Jahanshad N., Villalon-Reina J.E., Toga A.W., Jack C.R., Weiner M.W., Thompson P.M., Alzheimer's Disease Neuroimaging I. Effectiveness of regional DTI measures in distinguishing Alzheimer's disease, MCI, and normal aging. Neuroimage Clin. 2013;3:180–195. doi: 10.1016/j.nicl.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrangi M.A., Lyketsos C.G., Leoutsakos J.M., Oishi K., Albert M., Mori S., Mielke M.M. Longitudinal, region-specific course of diffusion tensor imaging measures in mild cognitive impairment and Alzheimer's disease. Alzheimers Dement. 2013;9(5):519–528. doi: 10.1016/j.jalz.2012.05.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrangi M.A., Rosenberg P.B. The fornix in mild cognitive impairment and Alzheimer's disease. Front. Aging Neurosci. 2015;7:1. doi: 10.3389/fnagi.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dwyer L., Lamberton F., Bokde A.L., Ewers M., Faluyi Y.O., Tanner C., Mazoyer B., O'Neill D., Bartley M., Collins D.R., Coughlan T., Prvulovic D., Hampel H. Multiple indices of diffusion identifies white matter damage in mild cognitive impairment and Alzheimer's disease. PLoS One. 2011;6(6):e21745. doi: 10.1371/journal.pone.0021745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah M., Biber K., Vinet J., Boddeke H.W. Microglia phenotype diversity. CNS Neurol. Disord. Drug Targets. 2011;10(1):108–118. doi: 10.2174/187152711794488575. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A., Oishi K., Miller M.I., Lyketsos C.G., Mori S., Albert M. Surface-based analysis on shape and fractional anisotropy of white matter tracts in Alzheimer's disease. PLoS One. 2010;5(3):e9811. doi: 10.1371/journal.pone.0009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj D., Yin Z., Breur M., Doorduin J., Holtman I.R., Olah M., Mantingh-Otter I.J., Van Dam D., De Deyn P.P., den Dunnen W., Eggen B.J.L., Amor S., Boddeke E. Increased white matter inflammation in aging- and Alzheimer's disease brain. Front. Mol. Neurosci. 2017;10:206. doi: 10.3389/fnmol.2017.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V.K., Risacher S.L., Nho K., Kim S., Swaminathan S., Shen L., Foroud T.M., Hakonarson H., Huentelman M.J., Aisen P.S., Petersen R.C., Green R.C., Jack C.R., Koeppe R.A., Jagust W.J., Weiner M.W., Saykin A.J., Alzheimer's Disease Neuroimaging I. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol. Psychiatry. 2014;19(3):351–357. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore S., Habes M., Iftikhar M.A., Shacklett A., Davatzikos C. A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer's disease and its prodromal stages. Neuroimage. 2017;155:530–548. doi: 10.1016/j.neuroimage.2017.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat D.H., Tuch D.S., van der Kouwe A.J., Greve D.N., Pappu V., Lee S.Y., Hevelone N.D., Zaleta A.K., Growdon J.H., Corkin S., Fischl B., Rosas H.D. White matter pathology isolates the hippocampal formation in Alzheimer's disease. Neurobiol. Aging. 2010;31(2):244–256. doi: 10.1016/j.neurobiolaging.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali D., Verganelakis D.A., Gotsis E., Toulas P., Papatriantafillou J., Karageorgiou C., Thomaides T., Kapsalaki E.Z., Hadjigeorgiou G., Papadimitriou A. Diffusion tensor imaging (DTI) in the detection of white matter lesions in patients with mild cognitive impairment (MCI) Acta Neurol. Belg. 2013;113(4):441–451. doi: 10.1007/s13760-013-0197-3. [DOI] [PubMed] [Google Scholar]
- Saykin A.J., Shen L., Foroud T.M., Potkin S.G., Swaminathan S., Kim S., Risacher S.L., Nho K., Huentelman M.J., Craig D.W., Thompson P.M., Stein J.L., Moore J.H., Farrer L.A., Green R.C., Bertram L., Jack C.R., Jr., Weiner M.W., Alzheimer's Disease Neuroimaging I. Alzheimer's disease neuroimaging initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin A.J., Shen L., Yao X., Kim S., Nho K., Risacher S.L., Ramanan V.K., Foroud T.M., Faber K.M., Sarwar N., Munsie L.M., Hu X., Soares H.D., Potkin S.G., Thompson P.M., Kauwe J.S., Kaddurah-Daouk R., Green R.C., Toga A.W., Weiner M.W., Alzheimer's Disease Neuroimaging I. Genetic studies of quantitative MCI and AD phenotypes in ADNI: progress, opportunities, and plans. Alzheimers Dement. 2015;11(7):792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Thompson P.M., Potkin S.G., Bertram L., Farrer L.A., Foroud T.M., Green R.C., Hu X., Huentelman M.J., Kim S., Kauwe J.S., Li Q., Liu E., Macciardi F., Moore J.H., Munsie L., Nho K., Ramanan V.K., Risacher S.L., Stone D.J., Swaminathan S., Toga A.W., Weiner M.W., Saykin A.J., Alzheimer's Disease Neuroimaging I. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging Behav. 2014;8(2):183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Finn E.S., Scheinost D., Rosenberg M.D., Chun M.M., Papademetris X., Constable R.T. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat. Protoc. 2017;12(3):506–518. doi: 10.1038/nprot.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solodkin A., Chen E.E., Van Hoesen G.W., Heimer L., Shereen A., Kruggel F., Mastrianni J. In vivo parahippocampal white matter pathology as a biomarker of disease progression to Alzheimer's disease. J. Comp. Neurol. 2013;521(18):4300–4317. doi: 10.1002/cne.23418. [DOI] [PubMed] [Google Scholar]
- Song W.M., Zhang B. Multiscale embedded gene co-expression network analysis. PLoS Comput. Biol. 2015;11(11) doi: 10.1371/journal.pcbi.1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Salat D., Upchurch K., Deason R., Kowall N., Budson A., Alzheimer's Disease Neuroimaging I. Destruction of white matter integrity in patients with mild cognitive impairment and Alzheimer disease. J. Investig Med. 2014;62(7):927–933. doi: 10.1097/JIM.0000000000000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzepacz P.T., Hochstetler H., Wang S., Walker B., Saykin A.J., Alzheimer's Disease Neuroimaging I. Relationship between the montreal cognitive assessment and mini-mental state examination for assessment of mild cognitive impairment in older adults. BMC Geriatr. 2015;15:107. doi: 10.1186/s12877-015-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhou X., Zhu J., Gu Y., Zhao W., Zou J., Guo Z. GO-function: deriving biologically relevant functions from statistically significant functions. Brief Bioinform. 2012;13(2):216–227. doi: 10.1093/bib/bbr041. [DOI] [PubMed] [Google Scholar]
- Wang M., Beckmann N.D., Roussos P., Wang E., Zhou X., Wang Q., Ming C., Neff R., Ma W., Fullard J.F., Hauberg M.E., Bendl J., Peters M.A., Logsdon B., Wang P., Mahajan M., Mangravite L.M., Dammer E.B., Duong D.M., Lah J.J., Seyfried N.T., Levey A.I., Buxbaum J.D., Ehrlich M., Gandy S., Katsel P., Haroutunian V., Schadt E., Zhang B. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer's disease. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Beckmann N.D., Roussos P., Wang E., Zhou X., Wang Q., Ming C., Neff R., Ma W., Fullard J.F., Hauberg M.E., Bendl J., Peters M.A., Logsdon B., Wang P., Mahajan M., Mangravite L.M., Dammer E.B., Duong D.M., Lah J.J., Seyfried N.T., Levey A.I., Buxbaum J.D., Ehrlich M., Gandy S., Katsel P., Haroutunian V., Schadt E., Zhang B. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer's disease. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Roussos P., McKenzie A., Zhou X., Kajiwara Y., Brennand K., DeLuca G.C., Crary J.F., Casaccia P., Buxbaum J., Ehrlich M., Gandy S., Goate A., Katsel P., Schadt E., Haroutunian V., Zhang B. Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer's disease. Genome Med. 2016;8:104. doi: 10.1186/s13073-016-0355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Roussos P., McKenzie A., Zhou X., Kajiwara Y., Brennand K.J., De Luca G.C., Crary J.F., Casaccia P., Buxbaum J.D., Ehrlich M., Gandy S., Goate A., Katsel P., Schadt E., Haroutunian V., Zhang B. Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer's disease. Genome Med. 2016;8(1):104. doi: 10.1186/s13073-016-0355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner M.W., Veitch D.P., Aisen P.S., Beckett L.A., Cairns N.J., Cedarbaum J., Donohue M.C., Green R.C., Harvey D., Jack C.R., Jr., Jagust W., Morris J.C., Petersen R.C., Saykin A.J., Shaw L., Thompson P.M., Toga A.W., Trojanowski J.Q., Alzheimer's Disease Neuroimaging I. Impact of the Alzheimer's disease neuroimaging initiative, 2004 to 2014. Alzheimers Dement. 2015;11(7):865–884. doi: 10.1016/j.jalz.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner M.W., Veitch D.P., Aisen P.S., Beckett L.A., Cairns N.J., Green R.C., Harvey D., Jack C.R., Jr., Jagust W., Morris J.C., Petersen R.C., Saykin A.J., Shaw L.M., Toga A.W., Trojanowski J.Q., Alzheimer's Disease Neuroimaging I. Recent publications from the Alzheimer's disease neuroimaging initiative: reviewing progress toward improved AD clinical trials. Alzheimers Dement. 2017;13(4):e1–e85. doi: 10.1016/j.jalz.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., Fluder E., Clurman B., Melquist S., Narayanan M., Suver C., Shah H., Mahajan M., Gillis T., Mysore J., MacDonald M.E., Lamb J.R., Bennett D.A., Molony C., Stone D.J., Gudnason V., Myers A.J., Schadt E.E., Neumann H., Zhu J., Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153(3):707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Hubbard P.L., Parker G.J., Alexander D.C. Axon diameter mapping in the presence of orientation dispersion with diffusion MRI. Neuroimage. 2011;56(3):1301–1315. doi: 10.1016/j.neuroimage.2011.01.084. [DOI] [PubMed] [Google Scholar]
- Zhang H., Schneider T., Wheeler-Kingshott C.A., Alexander D.C. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61(4):1000–1016. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.