Abstract
Objective
Consumers increasingly turn to the internet in search of health-related information; and they want their questions answered with short and precise passages, rather than needing to analyze lists of relevant documents returned by search engines and reading each document to find an answer. We aim to answer consumer health questions with information from reliable sources.
Materials and Methods
We combine knowledge-based, traditional machine and deep learning approaches to understand consumers’ questions and select the best answers from consumer-oriented sources. We evaluate the end-to-end system and its components on simple questions generated in a pilot development of MedlinePlus Alexa skill, as well as the short and long real-life questions submitted to the National Library of Medicine by consumers.
Results
Our system achieves 78.7% mean average precision and 87.9% mean reciprocal rank on simple Alexa questions, and 44.5% mean average precision and 51.6% mean reciprocal rank on real-life questions submitted by National Library of Medicine consumers.
Discussion
The ensemble of deep learning, domain knowledge, and traditional approaches recognizes question type and focus well in the simple questions, but it leaves room for improvement on the real-life consumers’ questions. Information retrieval approaches alone are sufficient for finding answers to simple Alexa questions. Answering real-life questions, however, benefits from a combination of information retrieval and inference approaches.
Conclusion
A pilot practical implementation of research needed to help consumers find reliable answers to their health-related questions demonstrates that for most questions the reliable answers exist and can be found automatically with acceptable accuracy.
Keywords: consumer health questions, question answering, natural language processing, deep learning, artificial intelligence
INTRODUCTION
Finding quality health information is not easy even for professionals with extensive background knowledge and excellent information-seeking skills. Consumers may lack both and need help finding good quality answers to their health- and lifestyle-related questions. Consumers often consult online sources first, searching the internet for health information and finding answers of varying degrees of quality that they are often unable to judge.1 To provide access to high quality health-related information online and support customer services, the National Library of Medicine (NLM) started the Consumer Health Information and Question Answering (CHiQA) project. The project’s goal is to develop question-answering approaches able of providing answers from reliable sources to both the short questions frequently typed into the search box of NLM’s consumer-oriented resource MedlinePlus2 and the longer descriptions of information needs sent to customer services, shown in Box 1. To test the viability of question-answering approaches in an end-to-end task, we developed a prototype Consumer Health Information and Question Answering system called CHiQA.
Box 1.
Examples of a short question submitted to MedlinePlus search box (A) and a long request sent to NLM customer services (B)
| A. what medicine i will take if i have tension type headache |
| B. I have been suffering from digestive problems for 30 years. It has recently flared up bad and I am looking for ideas to try to heal. I was diagnosed with IBS at 13 in 1989. I was curious about the servings in the C-IBS Formulation. How's much of each in mgs? How many times a day? I want to take a more holistic approach as I'm going to be 42 and i am tired of suffering and the side effects of my anti nausea meds that left me with an eye tic. If any information can be given I would be greatly appreciative. |
While much preliminary work was done on understanding and answering consumer health questions automatically3–5 and several online biomedical question answering systems do exist,6–8 to the best of our knowledge, CHiQA is the first online specialized question-answering system for providing reliable answers from patient-oriented resources to health questions asked by consumers.
OBJECTIVE
This article presents an overview of our approaches to: 1) understanding consumers’ questions, 2) building data sets for training and testing the approaches to question analysis and finding reliable answers, 3) building a functional online consumer health question answering system, as well as 4) the system architecture, and 5) several evaluations of the overall system performance.
MATERIALS AND METHODS
We first present the CHiQA online system (https://CHiQA.nlm.nih.gov) and its architecture, and then the details of the specific processes and the evaluation.
CHiQA architecture
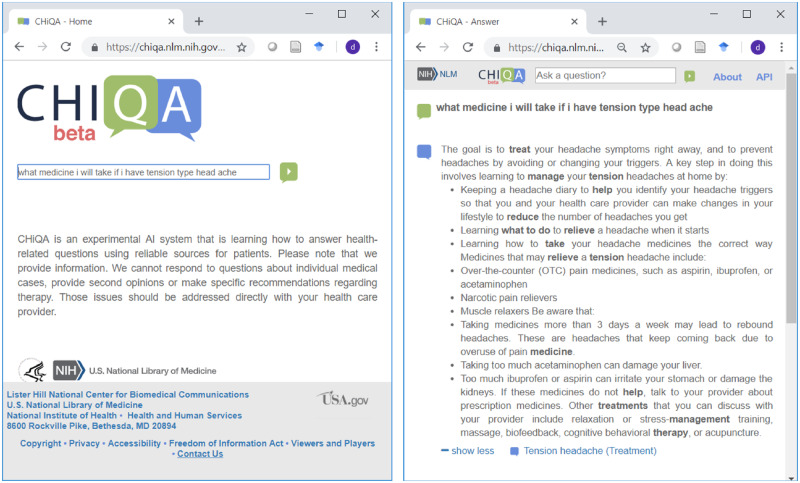
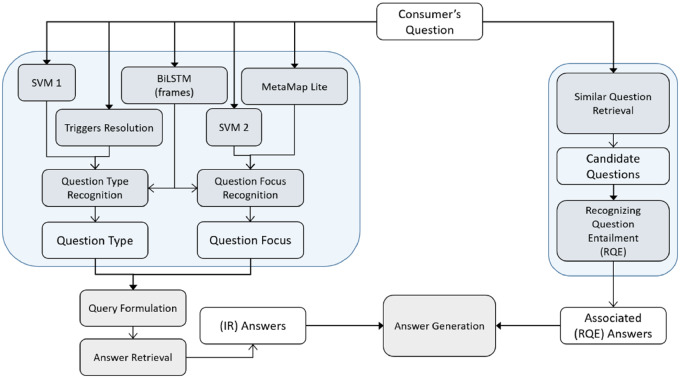
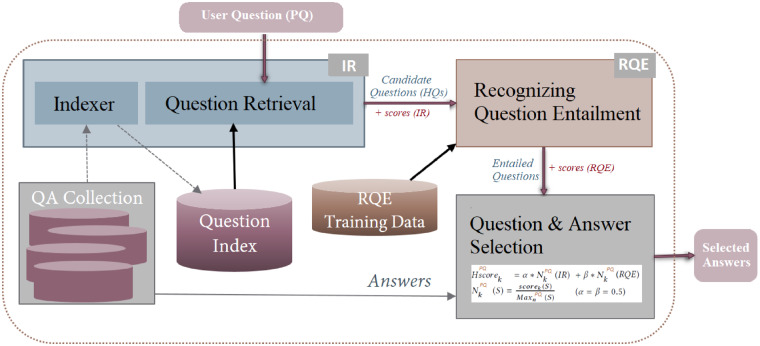
The beta version of the system released in August 2018 is shown in Figure 1. The system consists of a responsive web interface and a back end schematically shown in Figure 2. The back end consists of a preprocessing module, which at present is limited to spelling correction,9 a question understanding module, two complementary answer retrieval modules, and an answer generation module. Figure 2 also shows the specific implementation of the two answer retrieval modules: the traditional information retrieval (IR)-based module, and the module based on recognizing question entailment (RQE)10 that leverages the National Institutes of Health (NIH) FAQs for patients. These modules are described in more detail later.
Figure 1.
CHIQA landing page and answer to the short question shown in Box 1.
Figure 2.
Schematic representation of CHIQA architecture.
Some of the modules, such as question classification, are still under development and are not included in the publicly available system. All modules are constantly improving based on the system evaluation, users’ feedback, and the latest research findings in natural language processing.
Data sets
We created and distributed several data sets that enable question understanding and answer retrieval. Since knowing the focus and type of the question is sufficient to find up to 65% of the answers in consumer-oriented sources,11 we prepared several collections of questions annotated with focus and type,12 other entities,13 and spelling corrections9,14 that might improve question understanding. In addition, we created two types of question–answer pair data sets: 1) naturally occurring questions paired with manually found reference answers15,16 and 2) automatically generated pairs of NIH FAQs for patients, combined with patient education information converted to question–answer pairs using rules for representing section headers as questions and sections as answers.10 The descriptions of the data sets and their locations are shown in Table 1.
Table 1.
Data sets created to develop CHiQA. All data sets are publicly available
| Collection | Description | Question Category | Location |
|---|---|---|---|
| GARD question type | 1476 consumer health questions submitted to the Genetic and Rare Disease Information Center (GARD) manually labeled with question types17,18 | Short and long well-formed real-life consumer questions | https://ceb.nlm.nih.gov/ridem/infobot_docs/GARD_qdecomp.master.03.qtd.xml |
| GARD question decomposition | GARD manually labeled with question decomposition annotations.17–19 | Short and long well-formed real-life consumer questions | https://ceb.nlm.nih.gov/ridem/infobot_docs/GARD_qdecomp.v1.zip |
| CHiQA named entity | 1548 consumer health questions submitted to NLM, deidentified and annotated with named entities from 15 broad categories, including medical problems, drug/supplements, anatomy, and procedures.12 | Short and long mostly ungrammatical consumer questions | https://ceb.nlm.nih.gov/ridem/infobot_docs/CHQA-NER-Corpus_1.0.zip |
| CHiQA corpus 1 | 2614 consumer health questions annotated with named entities, question topic, question triggers, and question frames13 | Short and long, mostly ungrammatical consumer questions | https://bionlp.nlm.nih.gov/CHIQAcollections/CHQA-Corpus-1.0.zip. |
| Spelling corrections 1 | 471 consumer health questions with 24 837 tokens, 1008 annotation tags and 774/964 instances of non-word/real-word corrections14 | Long, mostly ungrammatical consumer questions | https://ceb.nlm.nih.gov/ridem/infobot_docs/CHQA_SpellCorrection_Dataset.zip |
| Spelling corrections 2 | 224 questions, 16 707 tokens, 1946 annotation tags and 974/1178 instances of non-word/real-word corrections9 | Long, mostly ungrammatical consumer questions | https://umlslex.nlm.nih.gov/cSpell |
| Recognizing Question Entailment | Training: A collection of 8588 clinical question–question pairs. Test: A collection of 302 medical pairs of NLM-questions and NIH-FAQs22 |
|
https://github.com/abachaa/RQE_Data_AMIA2016 |
| MedQuAD(Question Answering) | 47 457 medical question-answer pairs created from 12 NIH websites10 | Automatically derived well-formed short questions | https://github.com/abachaa/MedQuAD |
| LiveQA-Med |
|
Short and long, mostly ungrammatical consumer questions | https://github.com/abachaa/LiveQA_MedicalTask_TREC2017 |
| MEDIQA-QA | 2 QA data sets: LiveQA-Med and Alexa (QA pairs developed as Alexa skill by MedlinePlus staff), with 2000 judged and reranked answers23 |
|
https://github.com/abachaa/MEDIQA2019/tree/master/MEDIQA_Task3_QA |
| Drug questions | 674 question–answer pairs annotated with question types, question foci, answer sources, and text snippets containing the answers16 | Short and long, mostly ungrammatical consumer questions | https://github.com/abachaa/Medication_QA_MedInfo2019 |
| MeQSum (Question Summarization) | 1000 consumer health questions and their summaries24 | Long consumer health questions | https://github.com/abachaa/MeQSum |
Question understanding
The question-understanding unit applies a variety of methods to extract the question focus and the question type. It combines knowledge-based, traditional machine learning, and deep learning approaches. The candidate results are then ranked with an ensemble method, and the best results are selected as the question focus and the question type.
To recognize the question type, we combine three methods:
A multi-class Support Vector Machine (SVM1 in Figure 2) trained on a data set of 1,400 curated consumer health questions from the Genetic and Rare Diseases Information Center (GARD) using the features described in Roberts et al.17 This method returns only one candidate type for a given question.
A rule-based method relying on a set of regular expressions. For example, a question matching the regular expression “.* sign of .*” is identified as a potential question about Symptoms. This method can return multiple candidate types for a given question.
A question frame extraction method based on the deep learning module described below, which provides only one question type for a given question.
To recognize the question focus, we combine 3 methods:
A multi-class Support Vector Machine (SVM2 in Figure 2) trained on the GARD data set using the features described in Roberts et al.18 This method returns only one candidate focus for a given question.
Medical entity extraction using MetaMap Lite.20 We consider each medical entity as a candidate for the question focus.
A question frame extraction method based on the deep learning module described below, which provides only one question focus for a given question.
In our deep learning approach, we translate the problem of extracting the question focus and the question type to a frame extraction task using joint named entity recognition. The mention of the question focus is annotated with the Beginning, Inside, Outside, End, and Single (BIOES) token tagging format. The question type is added with a special token (#QType) at the end of each question. Figure 3 shows an example representation of a consumer question.
Figure 3.
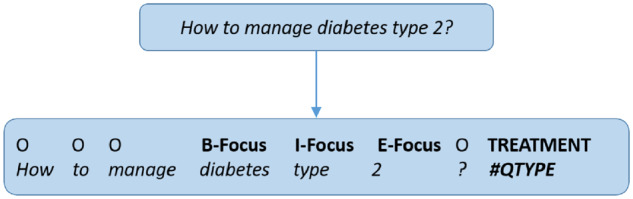
Question translated to BIOES format for BiLSTM training.
Word embeddings
We tested several embedding spaces to recognize the question focus and question type, including GloVe embeddings,21 binary vector encoding of part-of-speech tags for each token, and several embeddings we built from the Unified Medical Language System (UMLS)25 semantic types.
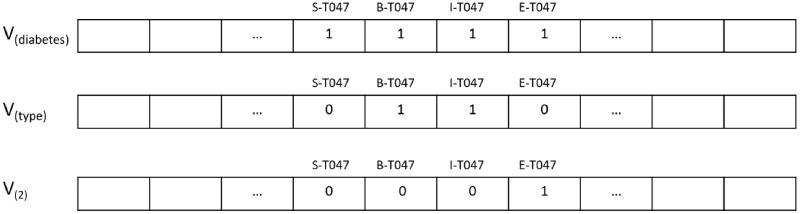
The UMLS variants embedding shown in Figure 4 relies on vector representations with each dimension representing a BIOES tag for a given semantic type.
Figure 4.
Binary vector representation for the term diabetes type 2 that has the semantic type T047 (Disease or Syndrome). The values in the vectors are computed offline from terms associated with the semantic type in the UMLS Metathesaurus. The terms that generated the embedding for the word sequence diabetes type 2 include, for instance, Diabetes, Alloxan Diabetes, diabetes type 2, and type 1 diabetes.
We experimented with different values for the vectors’ weights, including
Term frequency–inverse document frequency scores computed by considering each term as a document
Raw word frequency values
Binary values indicating whether or not a token is present (see Figure 3)
We also tested a distinction between the headwords (the head of a term [phrase] is the word that provides semantic information and determines the syntactic category of that phrase) and the content words of a term, building different vector spaces for each of them.
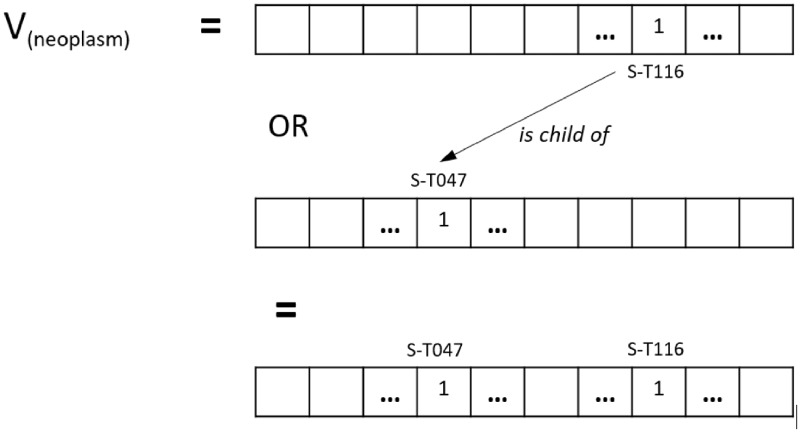
We took into consideration the fact that the semantic types in the UMLS have different levels of granularity. For instance, the semantic type associated with some disease names is not Disease or Syndrome but one of its child types in the UMLS semantic network, such as Neoplastic Process or Mental or Behavioral Dysfunction. These distinctions can create substantial data sparsity and irregularities that can have a substantial negative impact on training neural models.
Simple addition of the parent types in the BIOES vector, however, will lead to a loss of fine-grained information from the more specific semantic types. To address this dichotomy, we tested an additional variant of the embedding vectors by propagating the values from the child types to the parent types. The final value of a given dimension (eg, B-T047) is obtained by summing all the contributions from its direct terms and inherited from its child terms, multiplied by the reduction factor, as shown in Figure 5 for the term neoplasm. For binary vectors, contributions are aggregated with an OR operator. For numeric vectors, the contribution of child types is weighted by a 0.5 coefficient.
Figure 5.
Propagation of binary embeddings based on the UMLS semantic network.
To extract question frames, we trained the network architecture on the 800 short questions submitted by anonymous users to the MedlinePlus website. These questions are a part of the CHiQA corpus 1 collection described in Table 1.
Network architecture
We use Bi-directional Long Short-Term Memory (Bi-LSTM) networks26 for the frame extraction task. We use the state-of-the-art recurrent neural network (RNN) architecture proposed by Ma and Hovy.27 The system includes a first Bi-LSTM network to build character-level embeddings, and a second Bi-LSTM taking as input both the character-level embeddings built during training with the first Bi-LSTM layer and our word embeddings described earlier. The token labels are generated by a final conditional random fields (CRF) layer. We tested all the embedding variants described earlier as well as many combinations (concatenations) of them. After more than 200 runs with these different options, we selected the best performing combination with the following elements:
Binary embeddings of part-of-speech tags
GloVe embeddings of 100 dimensions from the 8 billion model
Propagated UMLS embeddings-based content words frequency
Propagated binary UMLS embeddings for headwords
Answer retrieval
IR-based answer retrieval
We indexed the updated MedQuAD collection that consists of NIH consumer-oriented Web pages (see Table 1) using Apache Solr.28 The documents in the MedQuAD collection are split into sections and subsections based on the formatting of the Web pages. The MedQuAD documents are indexed by subsection, keeping meta-information about the topic and the section header in a way that enables faceted searches.
For retrieval, the question focus and the type are given large weights in a search query. Other terms found in the question are added to the query with smaller weights. We preserve the Solr BM25 similarity-based ranking for the answers and use this information to generate the final ranking that combines the IR and entailment-based answers.
Entailment-based answer retrieval
The RQE module operates on the assumption that a question A entails a question B if every answer to B is also a complete or partial answer to A.20 To recognize entailment between question pairs, we apply a feature-based classifier using logistic regression, which provided the best performance in previous experiments with RQE data and outperformed deep learning models when trained on the clinical-RQE data set.10
We use a list of 9 features, selected after several experiments on textual entailment data sets. We compute 5 similarity measures between the questions and use their values as features (Word Overlap, Dice coefficient, and Cosine, Levenshtein, and Jaccard similarities). The feature list also includes the maximum and average values obtained with these measures, the question length ratio, and the number of common nouns and verbs between the questions.
Classifying the full question–answer (QA) collection for each test question is not feasible for real-time applications. To reduce the search space for the RQE system, we use the Terrier search engine29 to find similar questions among the MedQuAD collection of 47 000 QA pairs.10 For a wider coverage we added the synonyms of the question focus and the triggers of the question type to each indexed question.
For each user question, we retrieve the top 100 questions from Terrier then apply the logistic regression model to classify them as entailed (or not) by the user/test question. The final ranking of the candidate questions is obtained by a weighted combination of the search engine score and the RQE classification score. Figure 6 presents the overall architecture of the RQE module.
Figure 6.
Architecture of the entailment-based question answering system (RQE module).
Answer generation
The answers of the IR and RQE systems are combined using conventional team–draft interleaving30 and the top 5 answers are shown to the users. Answers found by the RQE system alone are displayed separately as related questions. For example, if the users ask about treatments for common cold, the related questions will be “What is Common cold?” and “What causes Common cold?”
Evaluation and results
The components of the system were thoroughly evaluated in our earlier work.9,10,17,18,20 In this paper, we evaluate the overall system performance using the MEDIQA-QA collection, which consists of the LiveQA-Med 2017 and Alexa MedlinePlus collections described in Table 1. The LiveQA-Med questions are randomly selected from the consumer health questions received by the NLM customer services from all over the world and cover different question types such as Treatment, Diagnosis, Indications, Ingredient, and Side Effects. The Alexa data set consists of 104 short simple questions about the most popular health topics in MedlinePlus searches. These question–answer pairs were developed as a skill for Amazon Alexa31 by MedlinePlus staff who prepared template questions about the most frequently searched disorders. The templates address the most frequent question types such as Treatment and Diagnosis and pair them with 1–2 sentences long answers. The Alexa collection allows us to measure the system’s performance on the most basic short questions. Reusing the LiveQA-Med 2017 collection of real-life questions allows us to gauge the system against the state-of-the-art established in the community-wide evaluation.
We evaluated the answers returned by the IR method, the RQE method, and the hybrid QA method (IR+RQE).
We used the same judgment scores as the LiveQA Track:
Correct and Complete Answer (4)
Correct but Incomplete (3)
Incorrect but Related (2)
Incorrect (1)
We computed 3 measures:
Average Score: this is the main score used to rank LiveQA participating systems. The average score evaluates the first retrieved answer for each test question (transfers 1–4 level grades to 0–3 scores).
Mean Average Precision (MAP): the mean of the average precision scores for each question, and
Mean Reciprocal Rank (MRR): the average of the reciprocal ranks of results for each question.
We consider answers rated as “Correct and Complete Answer” or “Correct but Incomplete” as correct answers.
Table 2 presents the average scores, MAP, and MRR results.
Table 2.
End-to-end evaluation of CHiQA and its IR and RQE question answering units. The best Average Score in the official TREC evaluation was 0.637
| Metric | LiveQA-Med Data set |
Alexa Data set |
||||
|---|---|---|---|---|---|---|
| IR | RQE | IR+RQE | IR | RQE | IR+RQE | |
| Average Score (First Answer) | 1.183 | 0.827 | 1.308 | 2.375 | 2.365 | 2.336 |
| MAP@10 | 0.405 | 0.311 | 0.445 | 0.787 | 0.752 | 0.766 |
| MRR@10 | 0.438 | 0.333 | 0.516 | 0.879 | 0.862 | 0.866 |
The Average Score of 1.308 on LiveQA-Med collection indicates that the automatic answers returned by the system are fair on average (the system gets 0 for poor answers; 1 for fair; 2 for good; and 3 for excellent.) For the simple short questions, on average, the answers returned by all architectures are good-to-excellent.
In addition to evaluating the overall performance on question answering, we evaluated the question understanding module’s ability to recognize the question type and the question focus on the same two components of MEDIQA-QA data set, using Recall, Precision, and F1 score. We observed a significant drop in performance for both tasks between the simple questions and the real-life consumer health questions; going from F1 scores of 98.6% and 96.3%, respectively, in question type recognition and partial span identification of the question focus on the Alexa simple questions to only 48.1% and 56.1% on the LiveQA-Med data set, as shown in Table 3.
Table 3.
Evaluation of the question understanding module performance on question focus and type recognition
| Question | LiveQA-Med Data set |
Alexa Data set |
||||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | |
| Focus (exact) | 28.9% | 41.5% | 34.1% | 95.1% | 69.5% | 80.3% |
| Focus (partial) | 56.4% | 55.9% | 56.1% | 96.3% | 96.4% | 96.3% |
| Type | 55.5% | 42.5% | 48.1% | 98.6% | 98.6% | 98.6% |
DISCUSSION
We present the first publicly available specialized question answering system that leverages the existing reliable sources to provide health related information to consumers. The system achieves 1.308 average score, which indicates the answers found by the system are fair and, at the moment, compares favorably with the experimental systems that participated in LiveQA.15 A prospective study is needed to compare the system to the existing commercial search engines that are converging toward question answering in the form of featured snippets.32 We will also actively seek users’ feedback.
We observed sizable differences in both the overall system performance and the recognition of question focus and type in the simple questions of Alexa data set and the real-life questions submitted to NLM. Several factors can explain the drop in performance:
The real-life consumer health questions often contain several subquestions with different types and foci, while the questions used to train the system had only 1 focus and 1 type per question and potentially different syntactic and lexical features.
The external supervision using MetaMap Lite to recognize medical entities and trigger words for the question types is less likely to succeed on the long questions due to the presence of several peripheral medical entities and type triggers related to a description of the patient’s history or to other background information.
Despite the complexity of the task and the low recognition of focus and type in real-life questions, we note that CHiQA still achieved the best performance known to date on finding relevant answers for the consumer health questions.
Two factors can explain these results:
CHiQA’s answer retrieval approach worked well as a failover strategy when the question type and question focus were misidentified (ie, the system relies on a hybrid answer retrieval strategy that combines free text search with the structured search over the focus and type information).
In 35% of the cases where the question type was not identified correctly, the system returned “INFORMATION” as the question type, which often led to document sections summarizing various aspects of the central topic (eg, known treatments and causes for a disease), and therefore containing the right answer.
In the process of preparing answers for the evaluations, we realized that they might not be always available from reliable sources, not available at all, or hard to understand for the consumers due to highly specialized language.33 For example, some rare diseases might only be discussed in the professional literature, which will require translating or simplifying the answer to a question about this disease. To some questions, the answer is indicated by the absence of specific printed resources. For example, if the patient asks if a drug interacts with specific food and the food is not listed in the drug label or side-effect databases, the inferred answer is “no, to the best of our knowledge,” but our system is not yet ready to provide such an answer.
Our work has several limitations that we are actively addressing in the ongoing research. The system handles questions about diseases and medications that constitute over 80% of the questions submitted to NLM. There is, however, a long tail of other question types13 we have not addressed yet. More research needs to be done to determine if the current system can handle these questions satisfactorily or if new modules need to be developed for each question type. Another limitation that needs to be resolved in the future is the need for a question classification module. The current prototype system assumes everything is a question and notifies the users if the question could not be answered. Although a stand-alone module might not be needed if our question understanding is improved, it might make the system more robust. The same step can address the current limitation of treating all requests as single questions with a single focus and type. Decomposing the complex requests into simpler questions might improve the performance from fair to good that we observed for Alexa questions. Question summarization, an alternative approach to question decomposition and simplification, could also lead to improved answer retrieval.22
In addition to better question understanding, we plan to summarize and simplify the answers and provide illustrations from patient-oriented sources.
CONCLUSION
Our work is an initial practical application of research needed to help consumers find reliable answers to their health-related questions. We demonstrate that for most questions the reliable answers exist and can be found automatically with acceptable accuracy. Several promising research directions are arising: question summarization, answer simplification, deeper question understanding, and better answer generation.
FUNDING
This work was supported by the intramural research program at the US National Library of Medicine, National Institutes of Health.
AUTHOR CONTRIBUTIONS
DDF conceptualized the study, manually reviewed data, analyzed data, oversaw study design and implementation, and contributed to writing and editing of manuscript. ABA and YM built the systems and the data collections, performed data analysis, deep learning experiments and evaluations, and contributed to writing and editing of the manuscript.
Conflict of interest statement
None declared.
REFERENCES
- 1.National Network of Libraries of Medicine. The Consumer Health Reference Interview and Ethical Issues. https://nnlm.gov/initiatives/topics/ethics Accessed August 14, 2019.
- 2.MedlinePlus. https://medlineplus.gov/ Accessed August 14, 2019.
- 3. Roberts K, Demner-Fushman D.. Interactive use of online health resources: a comparison of consumer and professional questions. J Am Med Inform Assoc 2016; 23 (4): 802–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu F, Antieau LD, Yu H.. Toward automated consumer question answering: Automatically separating consumer questions from professional questions in the healthcare domain. J Biomed Inform 2011; 44 (6): 1032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crangle CE, Bradley C, Carlin PF, et al. Exploring patient information needs in type 2 diabetes: A cross sectional study of questions. PLoS ONE 2018; 13 (11): e0203429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.askHERMES. http://www.askhermes.org/index2.html Accessed August 14, 2019.
- 7.EAGLi. http://eagl.unige.ch/EAGLi/ Accessed August 14, 2019.
- 8.LODQA. Question-Answering over Linked Open Data. http://lodqa.org/ Accessed August 14, 2019.
- 9. Lu CJ, Aronson AR, Shooshan SE, Demner-Fushman D.. Spell checker for consumer language (CSpell). J Am Med Inform Assoc 2019; 26 (3): 211–8. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ben Abacha A, Demner-Fushman D. A question-entailment approach to question answering. arXiv: 1901.08079 [cs.CL], 2019. [DOI] [PMC free article] [PubMed]
- 11. Deardorff A, Masterton K, Roberts K, Kilicoglu H, Demner-Fushman D.. A protocol-driven approach to automatically finding authoritative answers to consumer health questions in online resources. J Assoc Inf Sci Technol 2017; 68 (7): 1724–36. [Google Scholar]
- 12. Kilicoglu H, Ben Abacha A, Mrabet Y, et al. Annotating named entities in consumer health questions In: proceedings of the LREC; May 23–28, 2016, Portorož; Slovenia. [Google Scholar]
- 13. Kilicoglu H, Ben Abacha A, Mrabet Y, et al. Semantic annotation of consumer health questions. BMC Bioinform 2018; 19 (1): 34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kilicoglu H, Fiszman M, Roberts K, Demner-Fushman D.. An ensemble method for spelling correction in consumer health questions. AMIA Annu Symp Proc 2015; 2015: 727–36. [PMC free article] [PubMed] [Google Scholar]
- 15. Ben Abacha A, Agichtein E, Pinter Y, Demner-Fushman D. Overview of the medical QA task @ TREC 2017 LiveQA track. In: TREC, Gaithersburg, MD; 2017.
- 16. Ben Abacha A, Mrabet Y, Sharp M, Goodwin T, Shooshan SE, Demner-Fushman D. Bridging the gap between consumers’ medication questions and trusted answers. In: proceedings of the 17th World Congress of Medical and Health Informatics MEDINFO, 2019. [DOI] [PubMed]
- 17. Roberts K, Kilicoglu H, Fiszman M, Demner-Fushman D.. Automatically classifying question types for consumer health questions. AMIA Annu Symp Proc 2014; 2014: 1018–27. [PMC free article] [PubMed] [Google Scholar]
- 18. Roberts K, Kilicoglu H, Fiszman M, Demner-Fushman D. Decomposing consumer health questions. In proceedings of the 2014 Workshop on Biomedical Natural Language Processing (BioNLP), June 26 –27, 2014; Baltimore, MD.
- 19. Mrabet Y, Kilicoglu H, Roberts K, Demner-Fushman D.. Combining open-domain and biomedical knowledge for topic recognition in consumer health questions. AMIA Annu Symp Proc 2017; 2017: 914–23. [PMC free article] [PubMed] [Google Scholar]
- 20. Demner-Fushman D, Rogers WJ, Aronson SR.. MetaMap Lite: an evaluation of a new Java implementation of MetaMap. J Am Med Inform Assoc 2017: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pennington J, Socher R, Manning C. Glove: global vectors for word representation. In: proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), October 25–29, 2014; Doha, Qatar.
- 22. Ben Abacha A, Demner-Fushman D.. Recognizing question entailment for medical question answering. AMIA Annu Symp Proc 2017; 2017: 310–8. [PMC free article] [PubMed] [Google Scholar]
- 23. Ben Abacha A, Shivade C, Demner-Fushman D. Overview of the MEDIQA 2019 shared task on textual inference, question entailment and question answering. In: proceedings of the BioNLP 2019 Workshop; August 1, 2019; Association for Computational Linguistics, Florence, Italy.
- 24. Ben Abacha A, Demner-Fushman D. On the summarization of consumer health questions. In: 57th Annual Meeting of the Association for Computational Linguistics (ACL), Florence, Italy, 2019.
- 25. Lindberg DA, Humphreys BL, McCray AT.. The Unified Medical Language System. Methods Inf Med 1993; 32 (4): 281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Named Entity Recognition (LSTM + CRF). https://github.com/guillaumegenthial/sequence_tagging Accessed August 14, 2019.
- 27. Ma X, Hovy E. End-to-end sequence labeling via bi-directional LSTM-CNNS-CRF. arXiv preprint arXiv: 1603.01354, 2016.
- 28.Apache Solr. http://lucene.apache.org/solr/ Accessed August 14, 2019.
- 29.Terrier http://terrier.org/. Accessed August 14, 2019.
- 30. Chuklin A, Schuth A, Zhou K, Rijke MD.. A comparative analysis of interleaving methods for aggregated search. ACM Trans Inf Syst 2015; 33 (2): 1. [Google Scholar]
- 31.Amazon Alexa. https://developer.amazon.com/alexa Accessed August 14, 2019.
- 32.Google’s featured snippets. https://support.google.com/websearch/answer/9351707 Accessed August 14, 2019.
- 33. Ben Abacha A, Demner-Fushman D. On the role of question summarization and information source restriction in consumer health question answering. In: proceedings of the AMIA Informatics Summit, March 25–28, 2019; San Francisco, CA. [PMC free article] [PubMed]