Abstract
Objectives
Current machine learning models aiming to predict sepsis from electronic health records (EHR) do not account 20 for the heterogeneity of the condition despite its emerging importance in prognosis and treatment. This work demonstrates the added value of stratifying the types of organ dysfunction observed in patients who develop sepsis in the intensive care unit (ICU) in improving the ability to recognize patients at risk of sepsis from their EHR data.
Materials and Methods
Using an ICU dataset of 13 728 records, we identify clinically significant sepsis subpopulations with distinct organ dysfunction patterns. We perform classification experiments with random forest, gradient boost trees, and support vector machines, using the identified subpopulations to distinguish patients who develop sepsis in the ICU from those who do not.
Results
The classification results show that features selected using sepsis subpopulations as background knowledge yield a superior performance in distinguishing septic from non-septic patients regardless of the classification model used. The improved performance is especially pronounced in specificity, which is a current bottleneck in sepsis prediction machine learning models.
Conclusion
Our findings can steer machine learning efforts toward more personalized models for complex conditions including sepsis.
Keywords: sepsis, sepsis subtypes, sepsis prediction, machine learning, artificial intelligence in medicine
INTRODUCTION
Sepsis, defined by a life-threatening response to infection and potentially leading to multiple organ failure, is 1 of the most significant causes of worldwide morbidity and mortality.1 Sepsis is implicated in 6 million deaths annually with costs totaling $24 billion in the USA alone.2
Early identification of sepsis is a crucial factor in improving outcomes.3–5 Yet, traditional score-based screening tools lack the specificity needed to identify and elevate the care of potentially septic patients.6–10 In response, machine learning (ML) algorithms have been developed to recognize sepsis onset from vital signs data. A select number of ML models have shown improved predictions by taking advantage of computational power and large-scale data mining11–13 or attempting to optimize the feature set required for prediction.14,15 Nevertheless, current ML models have shown mixed results reflecting the heterogeneity of sepsis,16–18 populations,19 and methodologies.20
The objective of this paper is to highlight the importance of classifying the clinical heterogeneity of sepsis in enhancing our ability to anticipate onset, with focus on sepsis developed in the ICU. By analyzing routine clinical data of patients who develop sepsis in the ICU, we show that: 1) the clinical presentation of sepsis is underpinned by distinct combinations of dysfunction patterns that are mostly independent of etiology, 2) these patterns exhibit associations with distinct variations in vital signs and laboratory tests obtained within 24 hours of ICU admission, 3) using the relevant vitals and tests for each pattern as features in classification experiments produces highly sensitive and specific predictions regardless of the classification algorithm used, reflecting the relevance of the features to the clinical outcome. The results advocate that future sepsis prediction ML models can be guided toward better discriminative power by reformulating the sepsis prediction task to target the recognition of the different clinical manifestations of sepsis as opposed to the classic definitions currently in use. Such task will prioritize the features used for prediction using preprocessing steps that map patients’ routinely collected clinical data to previously derived subpopulations. Although this work does not aim to devise a sepsis prediction algorithm, it advocates a methodological shift in ML sepsis prediction supported by recent findings of reproducible clinical phenotypes of sepsis.21
MATERIALS AND METHODS
Data and preprocessing
We used the data of ICU stays between 2001 and 2012 obtained from the anonymized Medical Information Mart for Intensive Care III (MIMICIII) database.22 We extracted ICU stays of adults scoring a Sequential Organ Failure (SOFA) severity score ≥ 2 with neither a primary sepsis diagnosis or suspected sepsis recorded in the ICU admission notes; we further processed the data to exclude the following: stays shorter than 24 hours (1756 records), records with incomplete administrative information (581 records), and records with more than 15% missing vital signs as they can lead to inadequate imputation (3962 records). We used pattern matching to identify paragraphs within the admission notes containing mentions of sepsis. Records with no mention of sepsis were automatically included in our cohort (7823 records), while the extracted paragraphs of records with sepsis mentions (27 041 paragraphs) were manually validated over a 1-year period to exclude records with ambiguous mentions or suspected or confirmed sepsis. The final dataset (SQL scripts for recreating the dataset using the MIMICIII database are available at https://github.com/KHP\-Informatics/sepsis) contained 13 728 ICU stays, with 31% (4256) of the records having the primary outcome of sepsis (ICD-9 codes 995.91, 995.92, 785.52) (1976) in the discharge records or satisfying the Third International Definition of Sepsis and Septic Shock (Sepsis-3) of a life-threatening organ dysfunction (identified as a SOFA score ≥ 2) caused by a dysregulated host response to infection and confirmed by positive cultures (2280 records).8
We extracted admission details, comorbidity indices, etiology details, and the precalculated respiratory, cardiovascular, renal, hepatic, central nervous system, and coagulation subcomponents of the SOFA score (Supplementary Appendix A). We calculated 63 vitals and laboratory tests aggregated over the first 24 hours of admission (Supplementary Appendix B). As in similar studies,23 we extended the window forward by 24 hours for the infrequently sampled laboratory measurements to improve data completion. We imputed missing data using k-nearest neighbor (k = 7).
Experimental design
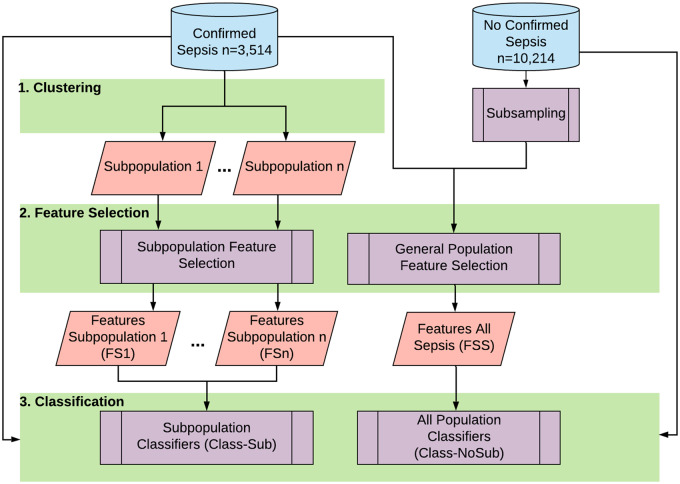
Figure 1 shows our approach. The idea is to cluster the ICU records with confirmed sepsis using the SOFA subcomponents to uncover subpopulations with distinct organ dysfunction patterns and then to perform feature selection on the individual clusters to identify subsets of the 63 vitals with the highest variance in each subpopulation. We compare classification performance with sepsis diagnosis as outcome using 2 feature sets selected from: a) the entire septic population, and b) individual subpopulations.
Figure 1.
The overall flow of subtype-based sepsis identification.
Sepsis subpopulations
We used self-organization maps (SOM)24 and clustering to obtain a 2-dimensional visualization of the confirmed sepsis records based on organ dysfunction patterns as done in.16 A SOM is a powerful ML model that maps highly dimensional data into a 2-dimensional grid of neurons, each corresponding to records with extremely similar features. We trained a 17x17 SOM to aggregate the 4256 records into 289 neurons—each representing 5–35 ICU stays extremely similar in organ dysfunction types. SOM parameters were chosen heuristically by minimizing the number of empty nodes and balancing the number of records mapping to each node.25
Consistent with existing biomedical literature citing SOM superiority,26–30 we did not compare SOM clustering with other techniques. As our aim is to use the SOM-based similarity to discover any underlying clusters, we used a distance-based clustering, mainly hierarchical, to minimize the within-cluster variance in SOM-generated distances. We determined 4 as the optimal number of clusters by examining hierarchical clustering dendograms over 1000 iterations. We used the R SOM package Kohonen31 and NBClust32 for clustering. We report summary statistics as median/interquartile range or count/percentage, as appropriate. We compared the central tendencies of the features using the Kruskal-Wallis test, using a cutoff value of P = .01.
Feature selection
Using the 63 predictors and 4256 records of diagnosed sepsis, we performed feature selection to identify the most relevant features for each subpopulation. We used Random Forests (RF)33 with conditional permutation variable importance, to account for spurious correlations among predictors.34,35 For each RF, the target cluster assignment designated positive outcomes (ie, records with class label 1 for RF1). Additionally, we performed feature selection using all 13 728 records with sepsis diagnosis as outcome.
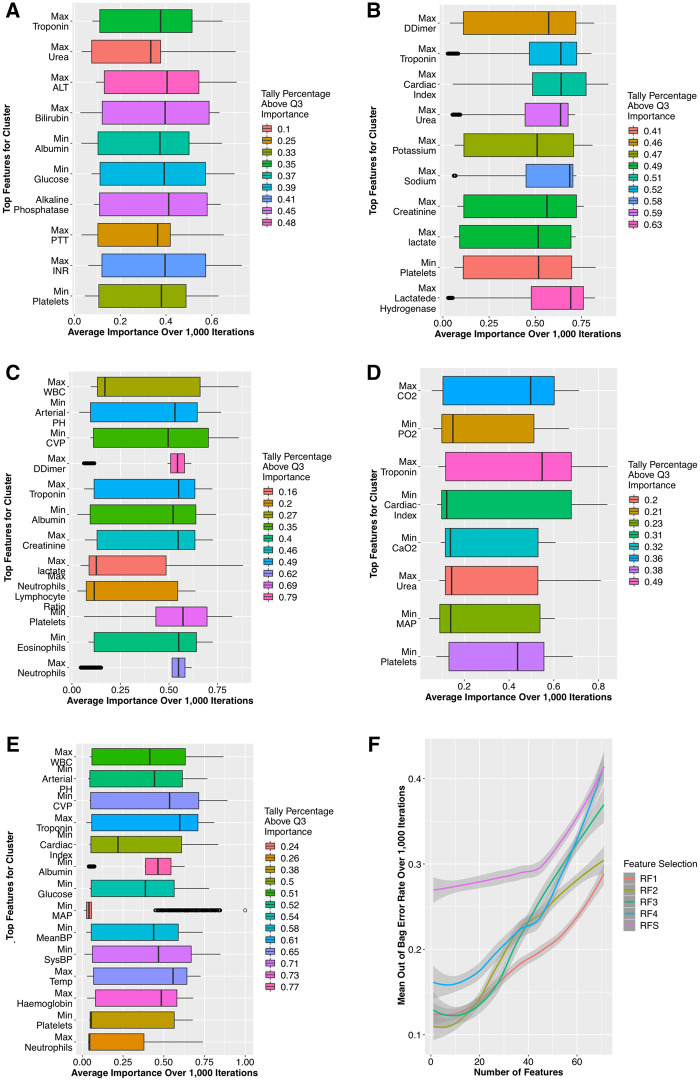
We trained each RF over 1000 bootstrapped iterations to avoid overfitting and computed the importance of the 63 predictors after each. To achieve robustness against statistical fluctuations, the best features for every RF were the top quartile of a rank-invariant tally for the number of iterations a feature’s importance was above the mean importance. The resulting feature sets are FS1–FS4 corresponding to each subpopulation and FSS for the entire septic population (Figure 2a–e).
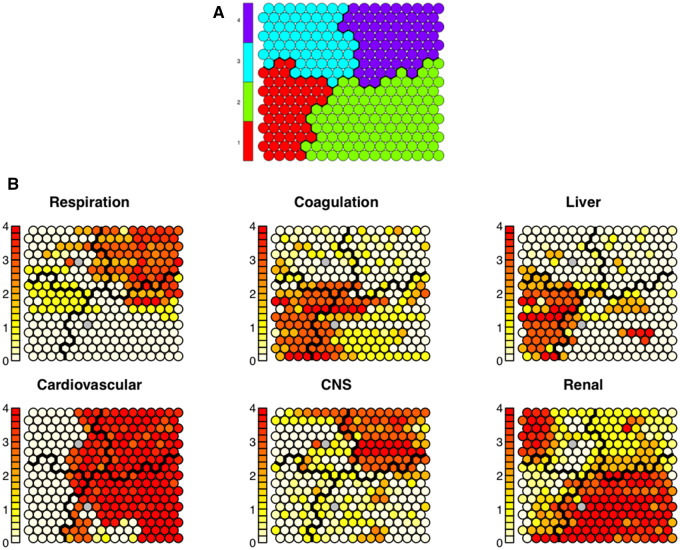
Figure 2.
(A) The resulting Kohonen Self-Organizing Map (SOM) depicting overall clusters—color-coded and separated by black lines. Each SOM node contains 5–35 intensive care unit (ICU) stays extremely similar to each other. (D) Shows those same clusters but with depictions of individual sequential organ failure (SOFA) subscores of the nodes within each cluster. Within each node, the given value is represented by the darkness of the color in the node. Each node is shaded from white to red, where darker colors represent higher average values (ie, higher SOFA subscores) among the ICU stays in the given node. The distribution of the SOFA subcomponents shows clear distinctions among the 4 clusters. The patterns visible in (B) suggest that the 4 clusters represent: 1) liver disease, 2) cardiogenic dysfunction with elevated creatinine, 3) minimal organ dysfunction, and 4) cardiogenic dysfunction with hypoxemia and altered mental status.
Sepsis identification
Using the 13 728 ICU records (9472 patients without sepsis and 4256 with sepsis), we performed 2 binary classification tasks with sepsis diagnosis as outcome. The first task (Class-NoSub) uses the feature set selected using the entire septic population (FSS), while the second (Class-Sub) uses the combined feature sets discovered from the septic subpopulations (FS1–FS4).
Each of the classification tasks was implemented using 3 different algorithms via the R package MLR36: Random forest, eXtreme gradient boosting (XGBoost) and support vector machines (SVM) using a Gaussian kernel to accommodate nonlinearity in the feature space. We optimized each classifier’s parameters through a bootstrapped grid search over the respective classifier’s hyperparameter space. All classifiers were trained over 1000 iterations of a 10-fold cross-validation.
RESULTS
The clustering procedure uncovered 4 subpopulations with distinct organ dysfunction patterns in septic patients in the MIMICIII database (Figure 2c). Further characteristics of the 4 clusters are given in Table 1. The clusters have been found to represent (1) liver disease, (2) cardiogenic and renal dysfunction, (3) minimal organ dysfunction, and (4) cardiogenic dysfunction with hypoxemia and altered mental status. The subpopulations identified are mostly independent of the origins of sepsis, with etiology being widely distributed across clusters. All predictors showed significant difference across clusters (all P values < .01).
Table 1.
Cluster Descriptive Statistics of the clusters formed using 31% of the records with an ICD-9 diagnosis of sepsis
| Liver Disease | Cardio & Renal Dysfunction | Minimal Organ Dysfunction | Cardio, Resp. & CNS Dysfunction | All Septic Population | |
|---|---|---|---|---|---|
| Count | 983 | 1441 | 953 | 879 | 4256 |
| Age (IQR) | 62 (51,72) | 69 (59,79.1) | 69 (57,81) | 76 (68,86) | 68 (58,81) |
| Female | 43% (412) | 39% (533) | 39% (371) | 46% (404) | 40% (1720) |
| Total SOFA (IQR) | 11.2 (8.5,12.3) | 11.8 (11.1,16.9) | 6.7 (3.4,8.2) | 12.1 (9.5,15.7) | 11.1 (4,15) |
| Pulmonary SOFA | 0.3 (0,0.6) | 0.8 (0,1.2) | 1.1 (0,2.4) | 3.2 (2,4) | 1.2 (0,3) |
| Coagulation SOFA | 3.1 (1.8,4) | 1.5 (0,2.1) | 1 (0,2.1) | 0.5 (0,1) | 1.53 (0,2.9) |
| Hepatic SOFA | 3.3 (2.5,4) | 0.7 (0,1.1) | 0.7 (0,1.1) | 0.4 (0,1) | 0.9 (0,1.2) |
| Cardio SOFA | 0.8 (0,2) | 3.5 (3,4) | 1.8 (0,4) | 3.8 (3,4) | 2.9 (0,4) |
| CNS SOFA | 0.3 (0,1) | 0.6 (0,1) | 0.9 (0,2) | 3 (0,4) | 0.7 (0,2) |
| Renal SOFA | 1.7 (0,2) | 3.1 (1,4) | 1.4 (0,2.1) | 1.3 (0,2) | 2.6 (1,3.1) |
| Comorbidity Elixhauser Index (IQR) | 12.4 (5,18.9) | 16 (8.7,21.8) | 7.3 (5,16) | 14 (6,19) | 14.1 (6.6,21) |
| 30-day Mortality | 28% (220) | 55% (724) | 25% (238) | 37% (325) | 35% (1507) |
| Length of stay (IQR) | 4.3 (1.1,4.8) | 5.6 (2,6.1) | 3.8 (1.8,4.2) | 8.5(3.2,11.9) | 5.4 (1.8,6.2) |
| Etiology (%) | |||||
| Pneumonia | 30% (295) | 31% (447) | 35% (334) | 43% (379) | 38% (1617) |
| Urinary Tract | 21% (206) | 32% (461) | 27% (257) | 13% (115) | 23% (979) |
| Abdominal | 9% (88) | 8% (115) | 7% (67) | 11% (98) | 8% (341) |
| Biliary | 12% (118) | 2% (29) | 2% (19) | 2% (19) | 3% (128) |
| Soft Tissue | 12% (118) | 15% (216) | 11% (104) | 9% (80) | 11% (468) |
| Other | 16% (158) | 12% (173) | 18% (172) | 21% (186) | 17% (723) |
Abbreviations: CNS, central nervous system; IQR, interquartile range; SOFA, sequential organ failure.
Comparing the central tendencies using the Kruskal–Wallis test and found that the clusters significantly differ in length of stay (P value < .01), Elixhauser-Quan comorbidity score (P value < .01) and in-hospital 30-day mortality (P value < .01). Etiology values showed no significant differences (P value ≥ .01)
The identified feature sets FS1–FS4 exhibit direct relevance to the types of organ dysfunction prevalent in the corresponding cluster (detailed in Figure 3a–e). In contrast, the feature set FSS includes a subset of the combined FS1–FS4 but largely consists of general signs of deterioration (eg, systolic blood pressure, white blood cell count) which have been found to be good indicators of acuity but nondiscriminative between septic and nonseptic patients.37 The feature selection performance (Figure 3f) indicates the promising contribution of the respective high-resolution feature sets in reducing the error rates of their respective classifiers compared to the low-resolution features.
Figure 3.
Feature selection results: (A) FS1 comprises high-profile hepatic and coagulation indicators (including bilirubin, Alanine transaminase [ALT], and the International Normalised Ratio [INR]); (B) FS2 features mainly comprise cardiovascular (troponin, lactate, d-dimer) and renal (creatinine, potassium, and urea) indicators; (C) FS3 features are relevant to all SOFA subcomponents, reflecting the heterogeneity of the minimal organ dysfunction; (D) FS4 features are of respiratory (CO2, CaO2, and PO2), and cardiovascular relevance (including mean arterial pressure [MAP]). (E) In contrast, FSS includes a portion FS1–FS4, but largely consists of general signs of deterioration (eg, systolic blood pressure, temperature, white blood cell count [WBC]), which have been found to be good indicators of mortality but nondiscriminative between septic and nonseptic patients.31 (F) shows the out-bag-error rates over 1000 iterations of bootstrapped feature selection.
The classification results comparing Class-Sub and Class-NoSub using RFs, SVMs, and XGBoost are presented in Table 2. As the table shows, while XGBoost outperformed other algorithms in all cases, all classifiers showed notably improved performance in the Class-Sub classification task (using the combined feature set FS1–FS4). The improved performance is especially pronounced in the specificity of predictions in which classification without subpopulation features consistently underperforms by not distinguishing septic patients from those with inflammations and comorbidities—a general bottleneck in ML sepsis prediction.11
Table 2.
Performance metrics for the 2 classification tasks Class-NoSub (using features selected without distinguishing subpopulations) and Class-Sub (using features selected from the individual subpopulations), using Random Forests (RF), Support Vector Machines (SVM) and Extreme Gradient Boosting (XGBoost)
| Classifier | Model | Sensitivity (95% CI) | Specificity (95% CI) | AUC | PLR (95% CI) | NLR (95% CI) |
|---|---|---|---|---|---|---|
| RF | ClassNoSub | 0.85 (0.81–0.88) | 0.79 (0.74–0.78) | 0.8 | 4 (4.9–4.2) | 0.18 (0.16–0.19) |
| Class-Sub | 0.94 (0.92–0.95) | 0.93 (0.91–0.95) | 0.94 | 13.4 (20.34–22.29) | 0.06 (0.03–0.08) | |
| XGBoost | Class-NoSub | 0.87 (0.85–0.91) | 0.81 (0.80–0.4) | 0.84 | 4.6 (4.1–4.9) | 0.16 (0.1–0.21) |
| Clas-Sub | 0.96 (0.94–0.97) | 0.95 (0.93–0.97) | 0.96 | 19.12 (18.6–46.18) | 0.04 (0.02–0.05) | |
| SVM | Class-NoSub | 0.86 (0.88–0.91) | 0.83 (0.84–0.86) | 0.84 | 5.1(4.5–6.2) | 0.17 (0.13-0.2) |
| Class-Sub | 0.95 (0.93–0.96) | 0.93 (0.92–0.95) | 0.94 | 13.6 (11.51–16.65) | 0.07 (0.02–0.12) |
Abbreviations: AUC, area under the curve; CI, confidence interval; PLR, positive likelihood ratio; NLR, negative likelihood ratio.
DISCUSSION
The principle goal of this work is to evaluate the importance of accounting for the heterogeneity of sepsis in improving early diagnosis. Current ML prediction models operate by linking vital signs to sepsis without acknowledging the heterogeneity of the condition. The subpopulations we derived here exhibit distinct patterns of organ dysfunction and outcome distributions and are mostly independent of severity or etiology of sepsis. Instead, there exist significant correlations in each sepsis subpopulation between specific combinations of vital sign values and sepsis diagnoses. Because the results show the superiority of subpopulation-specific features in discriminating septic from nonseptic patients, further research in early sepsis diagnosis can improve prediction quality by focusing on the recognition of the patterns of vital sign changes of specific relevance to sepsis subpopulations as opposed to the generic features currently used in ML prediction models.
It is important to place our findings in the context of the goal of this work: The clusters identified are representative of ICU admissions in a single hospital and are only used to support our central hypothesis—not to suggest the discovery of new sepsis subtypes. Similarly, although the experiments show an improved predictive power, they do not constitute a new sepsis prediction algorithm; doing the latter requires examining the temporal evolution of predictors, which we chose to exclude from our evaluations as it adds a level of complexity not needed to evaluate our premise. Nevertheless, by choosing the most abnormal value for each variable, we single out the maximal physiological derangement connecting the given feature to the outcome. In addition, our findings are not intended to discover new rules for manual scoring systems, but to direct future development of EHR-integrated decision support tools. Besides early diagnosis, the direction provided by the subpopulations can be further developed to issue justifications and explanations to clinicians including embedded visualizations.
The particular nature of MIMICIII renders the results specific to sepsis developed in the ICU. While early diagnosis of sepsis in ICU settings is an important problem,38 our findings have not been verified in non-ICU wards where more than 50% of sepsis cases are identified. However, by obtaining aggregate values of patient vitals over the first 24 hours of ICU admission, we aimed to obtain a clinical picture similar to the worst value obtained over the last hours in hospital wards. Nonetheless, our ongoing work targets the generalization of the model to non-ICU settings.
Finally, suspicion of sepsis, as defined by the co-occurrence of culture and antibiotic, is systematically underrepresented in a large portion of MIMICIII records.15 Therefore, although ICD codes do not constitute a perfect representation of the true incidence of sepsis in EHR records,39 we used them in conjunction with culture results to identify septic records.
CONCLUSIONS
Current sepsis prediction tools are yet to take into account the known heterogeneity of the condition. Our study found that accounting for clinically meaningful subpopulations within a large ICU sepsis cohort highly improved predictive power. The findings of our work can guide future sepsis prediction toward more accurate and explainable models. However, more multi-center studies are warranted on the etiological and vital variations within septic patients, and for evaluation.
FUNDING
This work was supported by the following sources:
1. ZI and RD are supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at the South London and Maudsley NHS Foundation Trust, King’s College London and University College London Hospitals.
2. RD is supported by the Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation, and Wellcome Trust.
3. HW is funded by MRC grants (MR/S004149/1 and MC_PC_18029).
AUTHOR CONTRIBUTIONS
AA and ZI conceived the research idea. ZI and HW designed the machine learning pipeline. LS performed auxiliary analyses to isolate the machine learning models to be used in this work. ZI and AA performed data acquisition. AH performed manual verification of the septic cohort from the MIMICIII clinical admission notes. All authors made substantial contributions to the analysis and interpretation of the results. ZI drafted the manuscript with all authors contributing to the critical revision for important intellectual content and addressing the reviewers’ comments. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of Interest statement
None declared.
Supplementary Material
REFERENCES
- 1. Fleischmann C, Scherag A, Adhikari N, et al. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med 2016; 193 (3): 259–72. [DOI] [PubMed] [Google Scholar]
- 2. Gaieski D, Edwards M, Carr B.. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit Care Med 2013; 41 (5): 1167–74. [DOI] [PubMed] [Google Scholar]
- 3. Dellinger M, Rhodes L, Annane D, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock. Intensive Care Med 2013; 39 (2): 165–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferrer R, Martin-Loeches I, Phillips G, et al. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program. Crit Care Med 2014; 42 (8): 1749–55. [DOI] [PubMed] [Google Scholar]
- 5. Seymour C, Gesten F, Prescott H, et al. Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med 2017; 376 (23): 2235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maitra S, Som A, Bhattacharjee S.. Accuracy of quick sequential organ failure assessment (qSOFA) score and systemic inflammatory response syndrome (SIRSs) criteria for predicting mortality in hospitalized patients with suspected infection: a meta-analysis of observational studies. Clin Microbiol Infect 2018; 24 (11): 1123–9. [DOI] [PubMed] [Google Scholar]
- 7.Editorial. Crying wolf: the growing fatigue around sepsis alerts. Lancet Respir Med 2018; 6 (3): 161. [DOI] [PubMed] [Google Scholar]
- 8. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). J Am Med Assoc 2016; 315 (8): 801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Askim A, Moser F, Gustad L, et al. Poor performance of quick-sofa (qSOFA) score in predicting severe sepsis and mortality-a prospective study of patients admitted with infection to the emergency department. Scand J Trauma Resusc Emerg Med 2017; 25 (1): 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Capan M, Hoover S, Miller K, et al. Data-driven approach to early warning score-based alert management. BMJ Open Qual 2018; 7 (3): e000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Islam MM, Nasrin T, Walther BA, et al. Prediction of sepsis patients using machine learning approach: a meta-analysis. Comput Methods Programs Biomed 2019; 170: 1–9. [DOI] [PubMed] [Google Scholar]
- 12. Olenick EM, Zimbro KS, D'lima GM, et al. Predicting sepsis risk using the “sniffer” algorithm in the electronic medical record. J Nurs Care Qual 2017; 32 (1): 25–31. [DOI] [PubMed] [Google Scholar]
- 13. Kam HJ, Kim HY.. Learning representations for the early detection of sepsis with deep neural networks. Comput Biol Med 2017; 89 (1): 248–55. [DOI] [PubMed] [Google Scholar]
- 14. Mao Q, Jay M, Hoffman JL, et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open 2018; 8 (1): e017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desautels T, Calvert J, Hoffman J, et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inform 2016; 4 (3): e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knox DB, Lanspa MJ, Kuttler KJ, et al. Phenotypic clusters within sepsis-associated multiple organ dysfunction syndrome. Intensive Care Med 2015; 41 (5): 814–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vincent JL. The clinical challenge of sepsis identification and monitoring. PLoS Med 2016; 13 (5): e1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Churpek MM, Snyder A, Sokol S, et al. Investigating the impact of different suspicion of infection criteria on the accuracy of quick sepsis-related organ failure assessment, systemic inflammatory response syndrome, and early warning scores. Crit Care Med 2017; 45 (11): 1805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McCoy A, Das R.. Reducing patient mortality, length of stay and readmissions through machine learning-based sepsis prediction in the emergency department, intensive care unit and hospital floor units. BMJ Open Qual 2017; 6 (2): e000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Downing NL, Rolnick J, Poole SF, et al. Electronic health record-based clinical decision support alert for severe sepsis: a randomised evaluation. BMJ Qual Saf 2019; 28 (9): 762–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seymour C, Kennedy J, Wang S, et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. J Am Med Assoc 2019; 321 (20): 2003–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson A, Pollard T, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson A, Pollard T, Mark R, et al. Reproducibility in critical care: a mortality prediction case study. In: proceedings of the 2nd Machine Learning for Healthcare Conference;18–19 August, 2017; Boston, Massachusetts, USA. [Google Scholar]
- 24. Kohonen K. The self-organizing map. Proc IEEE 1990; 78 (9): 1464–80. [Google Scholar]
- 25. Tian J, Azarian M, Pecht M.. Anomaly detection using self-organizing maps-based k-nearest neighbor algorithm. In: the Second European Conference of the Prognostics and Health Management Society; July 8–10, 2014; Nantes, France; 2014: 407–15. [Google Scholar]
- 26. Sebelius F, Eriksson L, Holmberg H, et al. Classification of motor commands using a modified self-organising feature map. Med Eng Phys 2005; 27 (5): 403–13. [DOI] [PubMed] [Google Scholar]
- 27. Nikkilä J, T Ör Önen P, Kaski S, et al. Analysis and visualization of gene expression data using self-organizing maps. Neural Netw 2002; 15 (8-9): 953–66. [DOI] [PubMed] [Google Scholar]
- 28. Boudier A, Curjuric I, Basagaña X, et al. Ten-year follow-up of cluster-based asthma phenotypes in adults: a pooled analysis of three cohorts. Am J Respir Crit Care Med 2013; 188 (5): 550–60. [DOI] [PubMed] [Google Scholar]
- 29. Vanfleteren LE, Spruit MA, Groenen M, et al. Clusters of comorbidities based on validated objective measurements and systemic inflammation in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2013; 187 (7): 728–35. [DOI] [PubMed] [Google Scholar]
- 30. Lötsch J, Ultsch A.. A machine- learned knowledge discovery method for associating complex phenotypes with complex genotypes application to pain. J Biomed Inform 2013; 46 (5): 921–8. [DOI] [PubMed] [Google Scholar]
- 31. Wehrens R, Kruisselbrink J.. Flexible self-organizing maps in Kohonen 3.0. J Stat Softw 2018; 87 (7): 1–18. [Google Scholar]
- 32. Charrad M, Ghazzali N, Boiteau V, et al. NbClust: an R package for determining the relevant number of clusters in a data set. J Stat Softw 2014; 61 (6): 1–36. [Google Scholar]
- 33. Breiman L. Random forests. Mach Learn 2001; 45 (1): 5–32. [Google Scholar]
- 34. Strobl C, Boulesteix AL, Kneib T, et al. Conditional variable importance for random forests. BMC Bioinformatics 2008; 9 (1): 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strobl C, Boulesteix AL, Zeileis A, et al. Bias in random forest variable importance measures: illustrations, sources, and a solution. BMC Bioinformatics 2007; 8 (1): 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bischl B, Lang M, Kotthoff L.. MLR: machine learning in R. J Mach Learn Res 2016; 17 (170): 1–5. [Google Scholar]
- 37. Rothman M, Levy M, Dellinger P, et al. Sepsis as 2 problems: identifying sepsis at admission and predicting onset in the hospital using an electronic medical record-based acuity score. J Crit Care 2017; 38: 237–44. [DOI] [PubMed] [Google Scholar]
- 38. Shashikumar S, Stanley M, Sadiq I, et al. Early sepsis detection in critical care patients using multiscale blood pressure and heart rate dynamics. J Electrocardiol 2017; 50 (6): 739–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iwashyna T, Odden A, Rohde J, et al. Identifying patients with severe sepsis using administrative claims: patient-level validation of the Angus implementation of the international consensus conference definition of severe sepsis. Med Care 2014; 52 (6): e39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.