Abstract
Objective
We developed medExtractR, a natural language processing system to extract medication information from clinical notes. Using a targeted approach, medExtractR focuses on individual drugs to facilitate creation of medication-specific research datasets from electronic health records.
Materials and Methods
Written using the R programming language, medExtractR combines lexicon dictionaries and regular expressions to identify relevant medication entities (eg, drug name, strength, frequency). MedExtractR was developed on notes from Vanderbilt University Medical Center, using medications prescribed with varying complexity. We evaluated medExtractR and compared it with 3 existing systems: MedEx, MedXN, and CLAMP (Clinical Language Annotation, Modeling, and Processing). We also demonstrated how medExtractR can be easily tuned for better performance on an outside dataset using the MIMIC-III (Medical Information Mart for Intensive Care III) database.
Results
On 50 test notes per development drug and 110 test notes for an additional drug, medExtractR achieved high overall performance (F-measures >0.95), exceeding performance of the 3 existing systems across all drugs. MedExtractR achieved the highest F-measure for each individual entity, except drug name and dose amount for allopurinol. With tuning and customization, medExtractR achieved F-measures >0.90 in the MIMIC-III dataset.
Discussion
The medExtractR system successfully extracted entities for medications of interest. High performance in entity-level extraction provides a strong foundation for developing robust research datasets for pharmacological research. When working with new datasets, medExtractR should be tuned on a small sample of notes before being broadly applied.
Conclusions
The medExtractR system achieved high performance extracting specific medications from clinical text, leading to higher-quality research datasets for drug-related studies than some existing general-purpose medication extraction tools.
Keywords: natural language processing, medication extraction, real world data, medication population study
INTRODUCTION
Electronic health records (EHRs) are a rich source of data for clinical research when information can be extracted accurately and efficiently. Medication information from EHRs can be used in many studies from aiding in defining phenotypes to determining drug exposure.1 In particular, detailed medication information is required to perform pharmacokinetic studies that are useful to determine patient characteristics affecting drug exposure including genotypes via pharmacogenomic studies.2
Information on drug regimens is often stored in an unstructured format, such as free-text clinical notes, requiring natural language processing (NLP) methodologies to extract medication information. Dosing information such as the strength or amount of a drug as well as how often it is taken are needed to compute quantities such as dose given intake and daily dose. To use this information to understand patients’ drug response and improve treatment, care must be taken to build research datasets of the highest quality possible. This requires careful extraction of medication information from unstructured EHR databases, including validation of each step in this process. Such processes may prove particularly beneficial in cases that rely on real-world data rather than on randomized studies (eg, population pharmacokinetic or pharmacodynamic analyses in pediatric populations).3
We describe medExtractR, an NLP algorithm we developed using the R programming language version 3.5.3 (R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org) to extract medication information such as strength, dose amount, and frequency from clinical notes. Our system provides a targeted approach to identify medication entities that facilitate computation of clinical dose-related quantities. This system is also easy to tune and customize, which allows it to achieve high performance in a variety of contexts. We first compare medExtractR with 3 existing NLP systems that can be used for general-purpose medication extraction: MedEx, MedXN, and CLAMP (Clinical Language Annotation, Modeling, and Processing).4–6 Then, we describe how to apply medExtractR on a new dataset, demonstrating and evaluating this procedure on the MIMIC-III (Medical Information Mart for Intensive Care III) Clinical Care Database.7
NLP for clinical texts
As EHR databases became more common in large health systems, the need to implement information extraction tasks to harvest data stored in unstructured formats grew. Although research related to EHRs has been growing exponentially, publications about NLP have increased only marginally, suggesting that NLP systems may currently be underutilized in EHR-based research.8 A recent literature review identified 71 mentions of NLP systems in papers published between 2006 and 2016, a majority of which were rule-based. Alternative approaches include hybrid systems combining rule-based and machine learning methodologies as well as ensemble methods incorporating multiple NLP systems.9,10
Clinical information extraction encompasses a variety of applications. Many NLP tools have previously been developed for various purposes, including MetaMap,11 cTAKES,12 MedLEE,13 and MedTagger,14 among others.15–18 A common application is phenotyping disease areas, for example by extracting diagnoses from clinical text.19,20 Information extraction has also been used to improve clinical workflows by detecting adverse events during treatment or to improve patient management by identifying care coordination activities after hospital discharge.21,22 Another area of application, and the focus of this article, is extracting medication information from clinical text in EHRs to perform drug-related studies.
NLP for medication extraction
Medication information, including characteristics of a dosing regimen, is important to understand and improve patient treatment. Medication extraction is a variant of information extraction, or the process of creating structured data from an unstructured format within a text source.23 Most medication extraction systems combine lexicon and rule-based approaches, though some incorporate machine learning methods.24,25
In the context of medication extraction, a major focus of some clinical NLP systems has been on the identification of drug names alone. While early systems were successful at extracting drug names, they often struggled to extract other drug entities such as strength or frequency with which the drug is taken. Jagannathan et al26 compared 4 commercial NLP systems and found that the systems were able to achieve above 90% F-measure for drug names but not for strength, route, and frequency, with F-measures as low as 48.3% on frequency. The extraction of drug information beyond the medication name was the focus of the Third i2b2 Workshop on NLP Challenges for Clinical Records. Overall F-measure scores for the top 10 teams ranged from 0.764 to 0.857, indicating room for improvement when extracting this type of information.27
In the past decade or so, NLP systems with a focus on extracting medication entities have emerged. MedEx, developed at Vanderbilt University, combines a semantic tagger and chart parser to identify drug names and entities.4 MedXN is a rule-based system developed at Mayo Clinic to extract drug information and normalize it to RxNorm concept unique identifiers, particularly for use in medication reconciliation contexts.5 CLAMP uses a pipeline architecture to break its process into several steps, including part-of-speech tagging and section header identification. It has flexible capabilities to identify concepts such as treatments and laboratory tests in addition to identifying drug-related information.6 These NLP systems are more general-purpose medication extraction algorithms, intended to be generally well performing across all drugs. Such systems may face difficulty when trying to optimize accuracy of extracted information with respect to a specific drug, study site, or patient cohort.
In contrast to other existing medication extraction NLP systems, medExtractR was developed to extract dosing information for a particular drug of interest to perform medication-specific studies such as pharmacokinetic, pharmacodynamic, or pharmacogenomic studies. This system is written in R, a programming language developed and widely used for statistical analysis, and is available as an R package (“medExtractR”) for download from the Comprehensive R Archive Network (CRAN).
MATERIALS AND METHODS
Data
Two primary drugs of interest were selected as candidates for developing medExtractR: tacrolimus and lamotrigine. Both share a wide dosing range that is titrated to achieve a clinical effect, making them ideal targets for pharmacokinetic studies. Tacrolimus, an immunosuppressive drug commonly used for transplant patients to prevent organ rejection, tends to have a simple prescribing pattern, typically involving the same dose given twice a day. Lamotrigine, an antiepileptic medication, has much more complex and variable prescribing patterns. Patients may take the drug 2 or 3 times daily, often with different morning, midday, or evening dosages. A third drug, allopurinol, provided an additional test drug but was not used for developing medExtractR. Allopurinol is a commonly used uric acid–lowering drug to treat gout, and often has a simple prescription pattern with a single dose being given once daily.
Clinical notes were randomly sampled from the Synthetic Derivative, a de-identified copy of Vanderbilt University Medical Center (VUMC) EHRs,28 and Research Derivative, an identified repository of clinical data drawn from VUMC EHRs. First, we defined a cohort for each medication. For tacrolimus, we used the same cohort of patients used in a previous study.2 For lamotrigine and allopurinol, we created new cohorts. For lamotrigine, we identified Synthetic Derivative records containing the keywords lamotrigine or Lamictal and selected subjects with ICD-9-CM (International Classification of Diseases–Ninth Revision–Clinical Modification) and ICD-10-CM (International Classification of Diseases–Tenth Revision–Clinical Modification) billing code for epilepsy and their first lamotrigine level measured between 18 and 70 years of age. Of those, we retained subjects with at least 3 lamotrigine levels and 3 records of dose information within 5 years of data, yielding 305 subjects. For allopurinol, we selected Research Derivative records with the keywords allopurinol or Zyloprim. We then refined the cohort who had clinical laboratory measures for uric acid and had “gout” mentioned in a problem list or had received either an ICD-9-CM or ICD-10-CM billing code for gout. After removing patients with an ICD-9-CM or ICD-10-CM code for malignant neoplasms, we identified a final cohort of 6264 patients. For subjects in each cohort of the 3 medications, we pulled all clinical notes, from which 60 training notes and 50 test notes were randomly sampled for tacrolimus and lamotrigine, while 110 test notes were randomly sampled for allopurinol.
Description of the medExtractR system
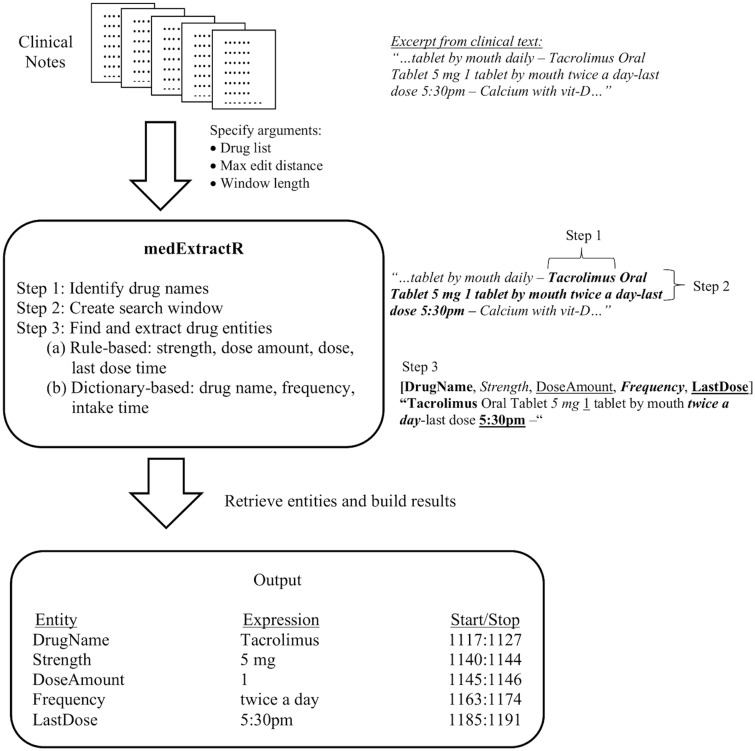
The medExtractR system relies on a combination of lexicon dictionaries and regular expression patterns to identify relevant medication information. A schematic of the system is shown in Figure 1 and examples of its application to clinical note excerpts are illustrated in Figure 2. Once a drug mention is found within a note, medExtractR searches in a surrounding window for entities including drug name, strength, dose amount, dose, intake time, frequency, and last dose time. The term drug mention refers to an appearance of a drug name within a clinical note along with its associated entities. Drug mentions are only returned when either key dosing information (strength, dose amount, or dose) or last dose time is found. Thus, phrases with only a drug name present are not extracted.
Figure 1.
Conceptual representation of the medExtractR system.
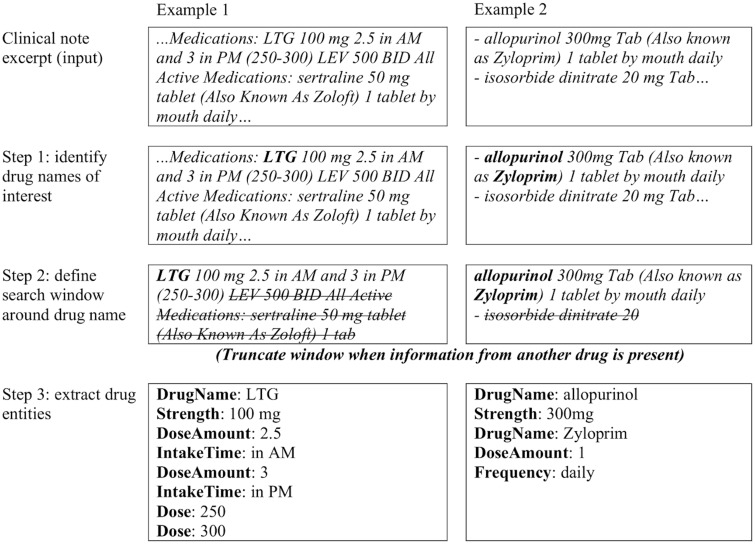
Figure 2.
Examples of medExtractR applied to excerpts from clinical notes.
To determine rules for how medExtractR should identify various entities, we manually reviewed the training notes to observe common patterns in how each entity is represented. For some, dictionaries were built based on expressions observed in the training sets while for others, regular expression rules were initially constructed by hand to capture the most commonly observed patterns. We then iteratively modified the dictionaries and rules/regular expressions to maximize performance (F-measure) in the training notes. The following steps outline how medExtractR operates.
Step 1: Identify drug names
In addition to the clinical note, medExtractR takes as an argument a vector of names for medications it should extract. Each drug under consideration had its own curated list of names, which consists of variations such as generic, brand, and abbreviated names. It then searches within the clinical note to identify all text matching a name from this drug list. For drug names with more than 5 characters, we allow approximate string matching with an edit distance specified by a function argument.
Step 2: Create search window
Once a drug name has been identified, medExtractR then identifies a search window around the mention from which to extract related drug entities. The length of the window (in number of characters) is another function argument. The ideal window length was chosen to optimize F-measure performance on the training set, resulting in 60 characters for tacrolimus and 130 for the more complicated lamotrigine. We used 60 characters for allopurinol since its prescribing patterns are closer to tacrolimus in simplicity. The search window is truncated at the first occurrence of an unrelated drug name. The list of unrelated drug names was extracted from RxNorm (ingredient or brand name),29 from which we removed words that could be confused with regular English words (eg, today or tomorrow) and supplemented with drug abbreviations observed in the training notes.
Step 3: Find and extract drug entities
Within the search window, medExtractR finds and extracts entities of interest (Table 1). For the dose change entity, we have a default dictionary of possible words (eg, reduce or switch) observed within the training set.
Table 1.
Drug entities identified by medExtractR
| Entity Name | Description | Examples |
|---|---|---|
| Drug name | Name of the drug for which dose information should be extracted | “Prograf,” “tacrolimus,” “FK-506,” “tac,” “fk” |
| Strength | Strength of an individual unit of the medication | “5 mg,” “100 mg” |
| Dose amount | Number of units taken at each intake | “2,” “one,” “0.5” |
| Dose | Total dose given intake | “10 mg,” “200 mg” |
| Frequency | Number of times per day the drug is taken | “once daily,” “2x/day,” “each day” |
| Intake time | Relative time period during the day when a drug is taken | “morning,” “bedtime,” “QPM,” “breakfast” |
| Dose change | Keyword indicating a change in the dosing regimen | “reduce,” “increase,” “switch,” “change” |
| Last dose time | Time expression indicating the time at which the last dose of the drug was taken | “8: 30 pm,” “2030,” “830 last night” |
The remaining entities are identified either using manually curated dictionaries or a combination of regular expressions and rule-based approaches. Last dose time is an optional entity identified using time expressions in various formats, including am/pm (eg, “9 pm”), military time (eg, “2100”), or a qualifying expression (eg, “9 last night”), which we identify with regular expressions. To be extracted as a last dose time, the search window also must contain a keyword such as last or taken to reduce false positives.
For both frequency and intake time, we developed dictionaries based on expressions observed within clinical notes and confirmed with physicians when the expressions were ambiguous (eg, “x1”: dosed 1 time). Strength, dose amount, and dose primarily rely on regular expressions to identify numeric expressions within the window (eg, “2” or “two”). Pattern-based rules are then used to label the expression as 1 of these 3 entities. Strength requires a number followed by a unit that is specified as a function argument (eg, “mg”). Examples of rules for dose amount include a number preceding a word like tablet or capsule or “take|takes|taking” followed by a number. Dose (ie, dose given intake) is mathematically equivalent to strength multiplied by dose amount. Dose is often identical in appearance to strength when no dose amount is present within that search window. The code for extracting the drug entities is provided as a function that could be easily customized by the user depending on medication of interest or institution.
Evaluation
To create gold standard datasets for each drug, we used the brat rapid annotation tool (BRAT) to manually annotate drug entities in clinical notes.30 We developed a set of annotation guidelines for each entity by examining how drug information was written within the training notes. Two reviewers familiar with the chosen medications independently annotated a set of 20 notes for each of tacrolimus and lamotrigine. We assessed the interannotator agreement using Cohen’s kappa separately for each drug. Cases where the 2 reviewers disagreed were resolved by review from a third expert. The annotation guidelines were then revised to clarify instances that resulted in disagreements.
We annotated a set of 60 training and 50 test clinical notes for each of tacrolimus and lamotrigine. Additionally, we annotated 110 clinical notes for an additional drug, allopurinol, to assess performance on an independent drug not used in medExtractR development. We evaluated medExtractR independently and compared it to 3 existing clinical NLP systems: MedXN, MedEx, and CLAMP. Performance was assessed using
where TP, FP, and FN represent true positives, false positives, and false negatives, respectively. To provide uncertainty in our estimates of precision, recall, and F1 score, we computed 95% bootstrapped percentile confidence intervals, with 5000 bootstrap samples at the clinical note level. For independent evaluation of medExtractR, we assessed performance for all drug-entity pairings with the exception of last dose time, which was only evaluated for tacrolimus.
When comparing medExtractR with the existing NLP systems, we standardized the raw output from both the existing systems and medExtractR to ensure compatibility during evaluation. This removed entities not extracted by all systems (eg, “duration”) and recoded entities having different meaning across systems, for example consolidating medExtractR “dose” and “strength” extractions to all be “strength.” Output generated by the existing NLP systems was manually compared with the gold standard. In cases where another NLP system extracted the same information in the gold standard but represented it differently, we created a validated output field, which conformed the extraction to the gold standard representation. For example, in the expression “tacrolimus 1mg 2 capsules twice daily,” MedEx extracts “2 capsules” as dose amount, while the gold standard only has “2.” This manual review ensured that evaluation and comparison across algorithms was fair. The validated entities used in comparing NLP systems were drug name, strength, dose amount, and frequency.
Application to a new dataset
A major goal of developing medExtractR is to make it possible for researchers to easily customize the medication extraction algorithm to their study of interest. We describe the steps one should take to implement medExtractR in a new dataset using example data obtained from the MIMIC-III Clinical Care Database (v1.4).7
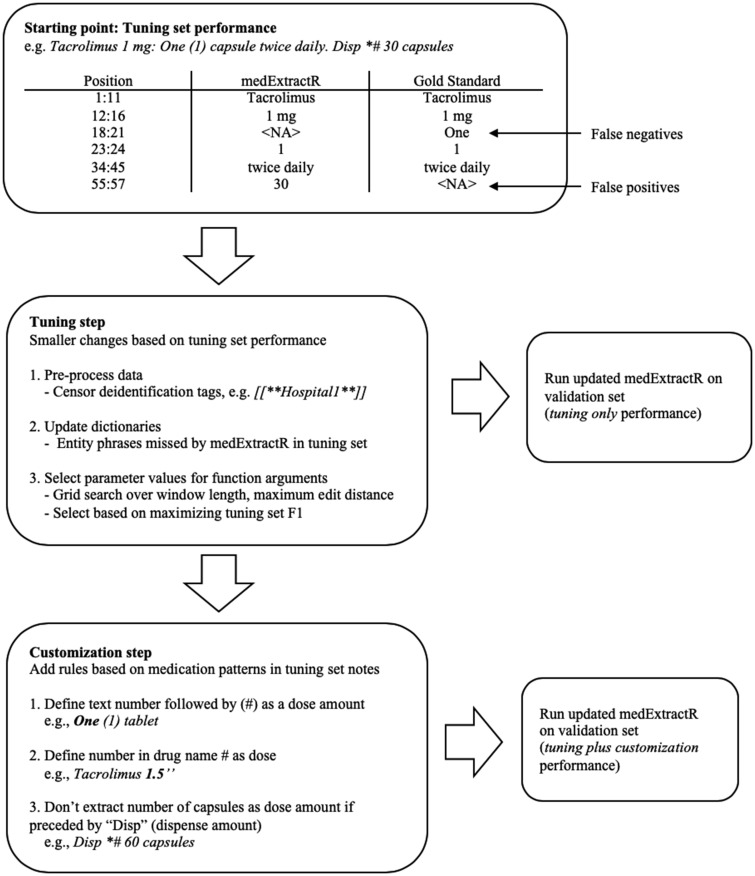
We selected tacrolimus and lamotrigine to directly compare with performance on the VUMC notes as well as oxcarbazepine, an antiepileptic drug not used in our VUMC evaluation to further assess medExtractR’s ability to adapt to a new drug. From MIMIC-III clinical notes associated with intensive care unit admissions, we identified all notes containing a variation of one of these drug names, including brand names or abbreviations. Each drug required the sampling of 2 different sets: a tuning set and a validation set. The tuning set was used for medExtractR customization on the MIMIC-III notes, which consisted of a tuning step and customization step. The tuning step involves easy-to-implement fixes, such as identifying whether preprocessing of notes is required, updating entity dictionaries with additional phrases, and selecting values for function arguments (eg, window length or maximum edit distance) by optimizing performance on the tuning set. The customization step involves changing or adding rules to identify drug entities within the source code. This step is considered separately, as it requires a higher level of coding ability to implement.
To create the tuning sets, notes were sampled at random for each drug and read by a researcher one at a time. If the note contained information about the dosage of the drug of interest, then that note was included in that drug’s tuning set until 10 notes were acquired. This method of sampling helped ensure there would be enough dosing information to adequately tune medExtractR. Validation sets were sampled to contain 50 discharge summaries, which were more likely to contain dosing information, and 50 notes from all other note types. This resulted in oversampling of discharge summaries, as 20% of tacrolimus and 30% of lamotrigine and oxcarbazepine notes were discharge summaries. For both the tuning and validation sets for each drug, we developed gold standard datasets using the same procedure as for VUMC notes: reviewers manually annotated dosing entities according to the previously created annotation guidelines.
To obtain an initial evaluation on the MIMIC-III notes, we ran medExtractR on the tuning set using the same parameter values as those used in VUMC. Cases where medExtractR missed frequency or intake time expressions were added to the dictionary. We also censored all MIMIC-III de-identification patterns to avoid false positives (eg, medExtractR occasionally identifies numbers in patterns like “Telephone/Fax(3)” as a dose amount). Once these changes were made, we performed a grid search to determine parameter values of function arguments like search window length and maximum edit distance. We selected the parameter values for each drug that maximized F-measure performance on the tuning set.
For the customization step, we manually analyzed the false positives and false negatives generated by medExtractR on the tuning set and implemented additional rules to better identify drug entities (Figure 3). As a result, we created rules based on 3 patterns within the MIMIC-III notes that medExtractR wasn’t able to capture. For example, we excluded numbers in expressions like “Disp #*45 tablets” because they refer to dispense amounts and not dose amounts. These reflected common ways that medications in general were represented in MIMIC-III, not necessarily those specific to each individual drug.
Figure 3.
Flow chart of implementation of medExtractR in MIMIC-III (Medical Information Mart for Intensive Care III) Clinical Care Database.
We evaluated medExtractR’s performance on the MIMIC-III validation sets under 3 different scenarios: (1) no tuning or customization, (2) tuning but no customization, and (3) both tuning and customization. For scenario 1, we used the same function arguments that were selected for the VUMC notes to demonstrate what the expected “out-of-the-box” performance on a new dataset would be. Lamotrigine parameter values were used for oxcarbazepine in scenario 1 because their prescribing patterns are more closely aligned.
RESULTS
Evaluation and comparison in the Synthetic Derivative
On 20 notes used for double annotation, the Cohen’s kappa for interannotator agreement was 0.970 for tacrolimus and 0.837 for lamotrigine on 84 and 101 annotations, respectively, of labeled medication entities. The lower agreement for lamotrigine was primarily due to titration schedules, or periods of time during which a dose is gradually increased over the course of several days or weeks until it reaches a maintenance dose. In these cases, the reviewers did not initially agree on whether drug entities throughout the entire titration schedule should be annotated for the gold standard. Annotation guidelines were updated and clarified to resolve any instances of disagreement before annotating the training and test sets.
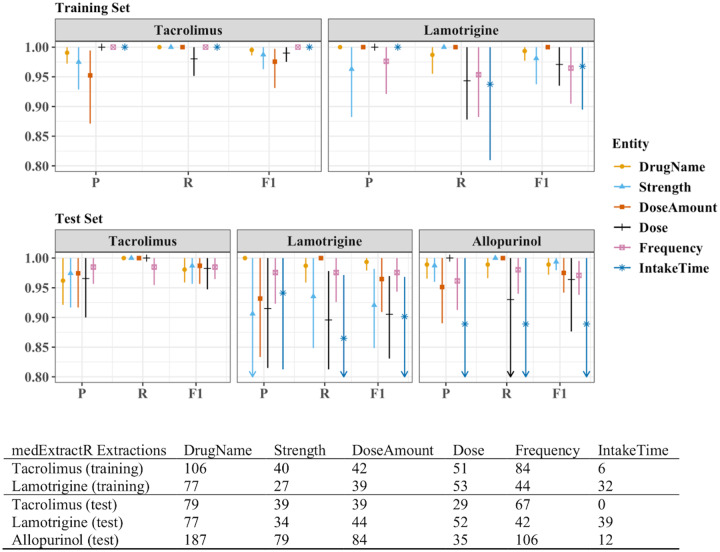
The training sets for tacrolimus and lamotrigine contained 60 notes each, with 105 and 102 drug mentions, respectively. The test sets for tacrolimus, lamotrigine, and allopurinol contained 50, 50, and 110 notes with 88, 76, and 191 drug mentions, respectively. All training set performance measures for medExtractR were >0.95 for all drug-entity combinations except recall for lamotrigine-dose and lamotrigine-intake time (Figure 4, Supplementary Table 1). The medExtractR system performance remained high on the test set. For most entities, medExtractR achieved precision, recall, and F1 score above 0.90 for tacrolimus and lamotrigine as well as allopurinol, the drug on which it was not trained. Lower performance for intake time may be partially due to relatively low occurrence with only 12 mentions. For lamotrigine, dose syntax was highly variable and thus more complicated to extract, resulting in slightly lower performance. Results are not presented for the dose change entity because there were 10 or fewer mentions for each of tacrolimus, lamotrigine, and allopurinol. High performance measures (>0.95) were also observed for last dose time in tacrolimus notes (Table 2).
Figure 4.
Entity-level precision, recall, and F1 performance measures for medExtractR. The training sets consisted of 60 notes each for tacrolimus and lamotrigine. The test sets consisted of 50 notes each for tacrolimus and lamotrigine and 110 notes for allopurinol. P, R, and F1 represent precision, recall, and F-measure (F1 score), respectively. Symbols and lines represent estimates and 95% bootstrapped confidence intervals, respectively. Arrows along the bottom x-axis indicate that either part or all of the confidence interval is below 0.80. Note that there were no annotations or extractions of intake time for tacrolimus in the test set of clinical notes. Dose change is not shown here because there were 10 or fewer mentions for each drug. Numeric results for all entities in this figure and dose change can be found in Supplementary Table 1.
Table 2.
MedExtractR system performance for extraction of last dose time on tacrolimus notes
| Training Set (n = 63 Extractions) | Test Set (n = 57 Extractions) | |
|---|---|---|
| Precision | 0.97 (0.92-1.00) | 0.98 (0.94-1.00) |
| Recall | 0.97 (0.92-1.00) | 0.96 (0.91-1.00) |
| F-measure | 0.97 (0.92-1.00) | 0.97 (0.94-1.00) |
Values are presented as estimate (95% bootstrap confidence interval). Based on 60 training notes and 50 test notes.
When comparing medExtractR with the other NLP systems, we considered both an overall and entity-level comparison. Here, overall means the combined performance aggregating across drug name, strength, dose amount, and frequency entities for each NLP system. MedExtractR achieved high overall recall, precision, and F1 score (>0.95) for tacrolimus and lamotrigine on both the training and test sets as well as for the allopurinol test set. This performance exceeded or matched that of all 3 existing NLP with respect to F1 score for tacrolimus, lamotrigine, and allopurinol (Table 3).
Table 3.
Performance measures for medExtractR and 3 existing natural language processing systems across standardized and combined drug name, strength, dose amount, and frequency
| Tacrolimus |
Lamotrigine |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F-measure | Precision | Recall | F-measure | |||||||
| Training set performancea | ||||||||||||
| medExtractR | 0.99 (0.98, 1.00) | 1.00 (1.00, 1.00) | 0.99 (0.99, 1.00) | 1.00 (0.99, 1.00) | 0.97 (0.94, 1.00) | 0.98 (0.97, 1.00) | ||||||
| MedEx | 0.79 (0.74-0.84) | 0.74 (0.69-0.79) | 0.76 (0.72-0.81) | 0.91 (0.87-0.95) | 0.73 (0.63-0.84) | 0.81 (0.74-0.87) | ||||||
| MedXN | 0.96 (0.93-0.99) | 0.90 (0.84-0.95) | 0.93 (0.89-0.96) | 0.96 (0.93-0.99) | 0.76 (0.64-0.87) | 0.85 (0.76-0.92) | ||||||
| CLAMP | 0.83 (0.77-0.88) | 0.60 (0.54-0.65) | 0.70 (0.64-0.74) | 0.94 (0.90-0.97) | 0.57 (0.46-0.68) | 0.71 (0.62-0.79) | ||||||
| Test set performance | ||||||||||||
| medExtractR | 0.97 (0.95-0.99) | 1.00 (0.99, 1.00) | 0.98 (0.97, 1.00) | 0.97 (0.94, 1.00) | 0.96 (0.91-0.99) | 0.96 (0.94-0.98) | 0.96 (0.92-0.99) | 0.98 (0.96, 1.00) | 0.97 (0.95-0.99) | |||
| MedEx | 0.77 (0.71-0.84) | 0.76 (0.71-0.82) | 0.77 (0.71-0.82) | 0.92 (0.87-0.96) | 0.82 (0.74-0.89) | 0.87 (0.81-0.91) | 0.94 (0.90-0.97) | 0.94 (0.86-0.98) | 0.94 (0.89-0.97) | |||
| MedXN | 0.96 (0.92-0.98) | 0.96 (0.92-0.99) | 0.96 (0.93-0.98) | 0.93 (0.89-0.96) | 0.83 (0.74-0.90) | 0.88 (0.82-0.92) | 0.97 (0.94-0.99) | 0.97 (0.93, 1.00) | 0.97 (0.94-0.99) | |||
| CLAMP | 0.84 (0.78-0.91) | 0.65 (0.58-0.71) | 0.73 (0.68-0.79) | 0.94 (0.90-0.97) | 0.66 (0.57-0.74) | 0.78 (0.70-0.84) | 0.96 (0.93-0.99) | 0.81 (0.75-0.87) | 0.88 (0.84-0.92) | |||
Values are presented as estimate (95% bootstrap confidence interval). Results are based on 60 training notes and 50 test notes each for tacrolimus and lamotrigine, and 110 test notes for allopurinol. These are overall results, combining performance across the entities drug name, strength, dose amount, and frequency, which were standardized across systems to ensure comparability.
aThe training set is for medExtractR, and served as another test set for the 3 existing natural language processing systems.
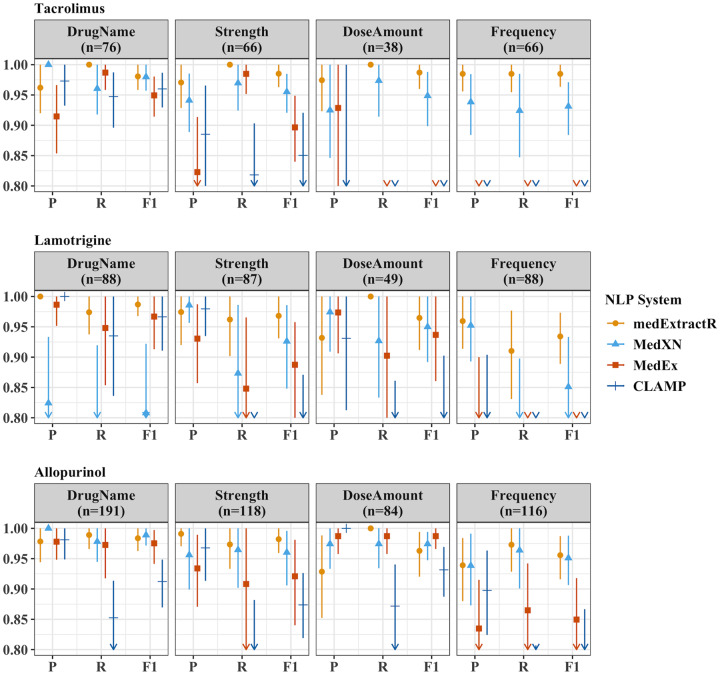
When comparing entity-level extraction, medExtractR often performed as well as or better than the existing NLP systems (Figure 5, Supplementary Table 2). The 2 cases in which medExtractR had a lower F-measure than the existing systems were for drug name and dose amount with allopurinol. Additionally, low performance on frequency for both tacrolimus and lamotrigine was observed with both MedEx and CLAMP (F1 score <0.8). An error analysis of medExtractR is provided in the Supplementary Appendix.
Figure 5.
Comparison of medication extraction natural language processing (NLP) systems for entity-level precision, recall, and F1 performance measures on test sets. The test sets consisted of 50 notes each for tacrolimus and lamotrigine, and 110 notes for allopurinol. Here, n refers to the number of annotations for that drug-entity combination in the gold standard dataset. P, R, and F1 represent precision, recall, and F-measure (F1 score), respectively. The drug entities presented here reflect a restricted list of entities that have been standardized across all 4 NLP systems to ensure comparability. Symbols and lines represent estimates and 95% bootstrapped confidence intervals, respectively. Arrows along the bottom x-axis indicate that either part or all of the confidence interval is below 0.80. Numeric results for this figure can be found in Supplementary Table 2.
Application to MIMIC-III
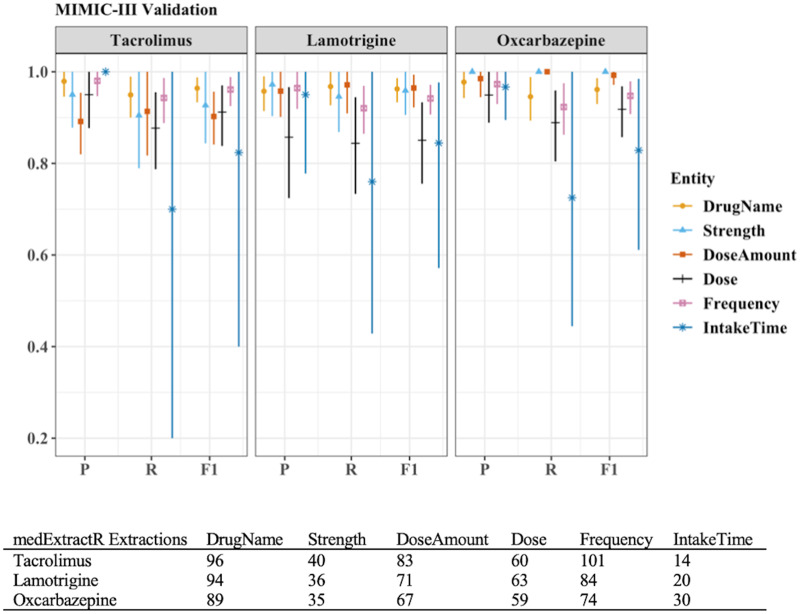
To demonstrate implementation of medExtractR on an external dataset, we compare results across the no tuning or customization, tuning but no customization, and tuning plus customization scenarios in Table 4. Using the parameter values optimized on the VUMC EHRs (ie, no tuning or customization), the system achieved F-measure values in the 0.81-0.85 range. Recall was typically lower than precision in these cases. With tuning but no customization, performance improved marginally for each drug (F-measures 0.86-0.90). After implementing the tuning plus customization, the largest improvements in performance were obtained, with the F-measure increasing by 0.07, 0.09, and 0.14 over no tuning for tacrolimus, lamotrigine, and oxcarbazepine, respectively. Upon further investigation, almost all improvement at the customization step was from a single new rule: a text number followed by a digit in parentheses indicates a dose amount, eg, “five (5).” When restricting customization to only this rule, the F-measures for tacrolimus, lamotrigine, and oxcarbazepine were 0.91, 0.92, and 0.95, respectively. Entity-level medExtractR performance for the combined tuning plus customization approach is presented in Figure 6.
Table 4.
Performance measures for medExtractR on MIMIC-III validation sets
| No Tuning or Customization | Tuning Without Customization | Tuning Plus Customization | |
|---|---|---|---|
| Tacrolimus (n = 423 Annotations) | |||
| Precision | 0.96 (0.92-0.99) | 0.93 (0.89-0.96) | 0.95 (0.91-0.98) |
| Recall | 0.77 (0.71-0.83) | 0.81 (0.76-0.85) | 0.89 (0.84-0.94) |
| F-measure | 0.85 (0.81-0.89) | 0.86 (0.83-0.90) | 0.92 (0.88-0.95) |
| Lamotrigine (n = 381 Annotations) | |||
| Precision | 0.87 (0.82-0.92) | 0.93 (0.89-0.97) | 0.94 (0.90-0.98) |
| Recall | 0.81 (0.77-0.85) | 0.83 (0.78-0.87) | 0.92 (0.87-0.96) |
| F-measure | 0.84 (0.81-0.87) | 0.88 (0.84-0.91) | 0.93 (0.89-0.96) |
| Oxcarbazepine (n = 375 Annotations) | |||
| Precision | 0.79 (0.72-0.86) | 0.97 (0.94-0.99) | 0.97 (0.95-0.99) |
| Recall | 0.83 (0.79-0.87) | 0.85 (0.80-0.89) | 0.92 (0.88-0.96) |
| F-measure | 0.81 (0.76-0.85) | 0.90 (0.87-0.93) | 0.95 (0.92-0.97) |
Values are presented as estimate (95% bootstrap confidence interval). Results are based on 100 validation notes for each drug. These are overall results, combining performance across the entities. “No tuning” results were obtained by running medExtractR on MIMIC-III (Medical Information Mart for Intensive Care III) notes using the same parameter values used for Vanderbilt University Medical Center notes, with lamotrigine parameter values used for oxcarbazepine. Tuning includes note preprocessing, dictionary updates, and optimizing parameter values. Customization refers to encoding different rules within medExtractR. Tuning and customization were developed on a separate tuning set of 10 notes for each drug.
Figure 6.
Entity-level performance of medExtractR after tuning and customization on (Medical Information Mart for Intensive Care III) validation sets. Results are based on 100 validation notes after tuning and customization for each drug. P, R, and F1 represent precision, recall, and F-measure (F1 score), respectively. Tuning includes note preprocessing, dictionary updates, and optimizing parameter values. Customization refers to encoding different rules within medExtractR. Tuning and customization were developed on a separate tuning set of 10 notes for each drug. Dose change results are not presented, as there were <5 mentions within each drug.
DISCUSSION
Medication information can be critical data to perform many studies using EHRs. Detailed medication dosing information allows for estimation of drug exposure more accurately, which is foundational in pharmacokinetic studies or assessment of drug exposure-response relationships in pharmacodynamic studies. Informative models require high quality data, and thus accurate extraction of medication information from EHRs is a pivotal step in advancing research in the medical field. Our results showed high performance for medExtractR both independently and in comparison with 3 existing NLP systems (MedEx, MedXN, and CLAMP).
MedExtractR operates on a much smaller scale than existing medication extraction systems by targeting particular drugs of interest. In some applications, this narrow scope may be considered a limitation because it is not optimized for more general medication extraction. However, in cases such as building research datasets to study particular drugs of interest, a system like medExtractR that can achieve higher performance for a narrower scope of medications is desirable. This is analogous to general phenotype exercises, which often employ customized NLP solutions instead of using general-purpose NLP systems (eg, MetaMap).1,10
The medExtractR system was able to generalize on drugs not used in development (allopurinol and oxcarbazepine), as well as on an entirely different source of data (MIMIC-III) with tuning and customization. The higher performance seen with medExtractR is in part owing to its ability to better capture such variations through arguments such as the window length parameter, which is easily customizable. Different medications are not only subject to variations in prescribing patterns, but also in writing styles between providers. When applying the VUMC-tuned medExtractR to an outside source of notes from MIMIC-III, performance was initially suboptimal (overall F1 score ≈ 0.85). However, after performing tuning and customization on a small set of tuning notes, which can be done with a little effort, all precision, recall, and F-measures increased to above 0.90 for all drugs (except tacrolimus recall, which was 0.89). While the combination of tuning and customization will ultimately produce the highest performance for medExtractR, we strongly recommend at least performing tuning as a necessary step when applying medExtractR to a new dataset. The medExtractR system is not intended to be implemented without some degree of tailoring to the user’s dataset, helping to ensure high-quality extraction of medication information across a variety of contexts. In an updated version of the “medExtractR” R package, we plan to incorporate customization rules that are useful for outside databases in addition to better documentation, so that users can easily follow an example using these tuning and customization steps.
A limitation with our initial VUMC evaluation is the choice of allopurinol as a test drug, which has fairly simple prescription patterns. Applying medExtractR to a drug with even more complex patterns, for example, different dosages on different days of the week, would likely require more expansive dictionaries or the addition of new rules within the system. Another limitation of this study would be the size of our VUMC training set (60 notes). While this might be sufficient to capture the variability in prescribing patterns for some medications, a larger annotated set may be needed for drugs with more complex patterns (eg, with different doses on different days of the week). MedExtractR is currently developed for use with oral tablet or capsule medications. A future aim is to determine changes that should be made for extraction of alternative administration routes or dosage forms (eg, intravenous or oral solution medicine).
We also acknowledge that some existing medication extraction NLP systems, such as MedEx, incorporate an additional “postprocessing” step in which individual entities are paired up to provide a dosing regimen given intake. We are currently developing a tool of this nature for both medExtractR and MedXN to construct patient dosing schedules from their entity-level extraction. By separately developing these 2 procedures (ie, entity-level dose extraction and pairing of the extracted entities), each of which is optimized and validated, we can generate more robust clinical datasets. We noticed that sometimes a note may describe both the patient’s current dose and a new dose being prescribed at that visit, from which a correct dose should be identified. This competing dose issue is also an important problem to be solved to build more accurate medication-based research data.
CONCLUSION
We presented medExtractR, a medication extraction algorithm written in R designed to focus on individual drugs for creating research datasets. We demonstrated that medExtractR achieved high performance for 3 different drugs and outperformed the existing medication extraction systems MedEx, MedXN, and CLAMP. The flexible and easily customizable nature of the medExtractR system allows the system to generalize across other drugs or datasets from different institutions, as long as researchers are committed to tuning the model appropriately. The medExtractR system offers an alternative for users who prefer the trade-off of putting in some extra effort to tailor medication extraction to their dataset in order to improve the quality of results.
The ultimate goal of using medExtractR is to develop datasets from EHRs for various medication-related studies requiring more detailed dosing information such as pharmacokinetic or pharmacodynamic or pharmacogenomic analyses. Future work will validate medExtractR’s ability to correctly identify quantities such as dose given intake and daily dose from extracted entities. This information is critical to accurately estimate drug exposure, which can serve as an outcome or an exposure of interest in many drug-related studies.
FUNDING
This work was supported by the National Institute of General Medical Sciences under award R01-GM123109 (principal investigator: LC).
AUTHOR CONTRIBUTIONS
As the principal investigator, LC conceived the research and managed the project. HLW developed the medExtractR software, performed analyses, and drafted the manuscript. CB revised the software and implemented as an R package. EM provided quality assurance of data and entity dictionaries. MLW annotated gold standards for evaluation datasets. CAB and JCD contributed to methods for software development and evaluation. CAB also provided data infrastructure management. All authors provided feedback to improve medExtractR throughout development, and also reviewed and edited the final manuscript.
Supplementary Material
Acknowledgments
The software in this article uses publicly available data courtesy of the U.S. National Library of Medicine, National Institutes of Health, Department of Health and Human Services; the National Library of Medicine is not responsible for the product and does not endorse or recommend this or any other product.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23 (6): 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Birdwell KA, Grady B, Choi L, et al. Use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenet Genomics 2012; 22 (1): 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van SD, Choi L.. Real-world data for pediatric pharmacometrics: can we upcycle clinical data for research use? Clin Pharmacol Ther 2019; 106 (1): 84–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010; 17 (1): 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sohn S, Clark C, Halgrim SR, et al. MedXN: an open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc 2014; 21 (5): 858–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Soysal E, Wang J, Jiang M, et al. CLAMP–a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc 2018; 25 (3): 331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: a literature review. J Biomed Inform 2018; 77: 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform 2017; 73: 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuo TT, Rao P, Maehara C, et al. Ensembles of nlp tools for data element extraction from clinical notes. AMIA Annu Symp Proc 2016; 2016: 1880–9. [PMC free article] [PubMed] [Google Scholar]
- 11. Aronson AR, Lang FM.. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010; 17 (3): 229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994; 1 (2): 161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Torii M, Wagholikar K, Liu H.. Using machine learning for concept extraction on clinical documents from multiple data sources. J Am Med Inform Assoc 2011; 18 (5): 580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Denny JC, Irani PR, Wehbe FH, et al. The KnowledgeMap project: development of a concept-based medical school curriculum database. AMIA Annu Symp Proc 2003; 2003: 195–9. [PMC free article] [PubMed] [Google Scholar]
- 16. Goryachev S, Sordo M, Zeng QT.. A suite of natural language processing tools developed for the I2B2 project In: AMIA Annu Symp Proc 2006; 2006: 931. [PMC free article] [PubMed] [Google Scholar]
- 17. Nguyen AN, Lawley MJ, Hansen DP, et al. A simple pipeline application for identifying and negating SNOMED clinical terminology in free text. In: HIC 2009: Proceedings, Frontiers of Health Informatics – Redefining Healthcare; 2009: 188–93.
- 18. Gold S, Elhadad N, Zhu X, et al. Extracting structured medication event information from discharge summaries. AMIA Annu Symp Proc 2008; 2008: 237–41. [PMC free article] [PubMed] [Google Scholar]
- 19. Sada Y, Hou J, Richardson P, et al. Validation of case finding algorithms for hepatocellular cancer from administrative data and electronic health records using natural language processing. Med Care 2016; 54 (2): e9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang M, Cyhaniuk A, Cooper DL, et al. Identification of people with acquired hemophilia in a large electronic health record database. J Blood Med 2017; 8: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rochefort CM, Buckeridge DL, Forster AJ.. Accuracy of using automated methods for detecting adverse events from electronic health record data: a research protocol. Implement Sci 2015; 10 (1): 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruud KL, Johnson MG, Liesinger JT, et al. Automated detection of follow-up appointments using text mining of discharge records. Int J Qual Health Care 2010; 22 (3): 229–35. [DOI] [PubMed] [Google Scholar]
- 23. Martin JH, Jurafsky D.. Speech and Language Processing: An Introduction to Natural Language Processing, Computational Linguistics, and Speech Recognition. Upper Saddle River, NJ: Pearson/Prentice Hall; 2009. [Google Scholar]
- 24. Patrick J, Li M.. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc 2010; 17 (5): 524–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z, Liu F, Antieau L, et al. Lancet: a high precision medication event extraction system for clinical text. J Am Med Inform Assoc 2010; 17 (5): 563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jagannathan V, Mullett CJ, Arbogast JG, et al. Assessment of commercial NLP engines for medication information extraction from dictated clinical notes. Int J Med Inform 2009; 78 (4): 284–91. [DOI] [PubMed] [Google Scholar]
- 27. Uzuner Ö, Solti I, Cadag E.. Extracting medication information from clinical text. J Am Med Inform Assoc 2010; 17 (5): 514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roden DM, Pulley JM, Basford MA, et al. Development of a large‐scale de‐identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84 (3): 362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nelson SJ, Zeng K, Kilbourne J, et al. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 2011; 18 (4): 441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stenetorp P, Pyysalo S, Topić G, et al. BRAT: a web-based tool for NLP-assisted text annotation. In: proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics; 2012: 102–7.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.