Summary
Chimeric antigen receptor (CAR) therapy targeting CD19 has yielded remarkable outcomes in patients with acute lymphoblastic leukemia. To identify potential CAR targets in acute myeloid leukemia (AML), we probed the AML surfaceome for over-expressed molecules with tolerable systemic expression. We integrated large transcriptomics and proteomics data sets from malignant and normal tissues, and developed an algorithm to identify potential targets expressed in leukemia stem cells, but not in normal CD34+CD38− hematopoietic cells, T cells or vital tissues. As these investigations did not uncover candidate targets with a profile as favorable as CD19, we developed a generalizable combinatorial targeting strategy fulfilling stringent efficacy and safety criteria. Our findings indicate that several target pairings hold great promise for CAR therapy of AML.
Keywords: Immunotherapy, CAR T cell, surfaceome, target discovery, high-throughput annotation, algorithm, acute myeloid leukemia (AML), proteomics, combinatorial strategies, leukemia
Introduction
Acute myeloid leukemia (AML) is the most common acute leukemia in adults and the second most common in children. The standard induction chemotherapy regimens have not substantially changed over the past 40 years (Pulte et al., 2008). Most patients initially respond to chemotherapy and achieve a complete remission (CR), but only a minority achieves long-term survival because of relapse with chemoresistant disease. Patients may achieve a second CR in 30–45% of cases, but only in 10–15% of patients whose first CR lasted less than a year (Becker et al., 2011). Patients who never achieve remission (Estey et al., 1996) or relapse within 6 months of a CR are less likely to respond to any treatment (Breems et al., 2005).
Immunotherapy is emerging as an approach to treat cancer, based on the successes of antibody-mediated checkpoint blockade and engineered T cells (Couzin-Frankel, 2013). Checkpoint blockade has so far proven to be especially effective against cancers with high mutation burdens (McGranahan et al., 2016; Rizvi et al., 2015; Snyder et al., 2014). AML however presents with a low mutation burden relative to other cancers (Alexandrov et al., 2013), averaging only 13 genic mutations in newly diagnosed disease (Cancer Genome Atlas Research, 2013). Immune recognition of potential neoantigens arising from mutations in coding sequences is therefore less probable (Le et al., 2015; Van Allen et al., 2015) and less likely to respond to immune checkpoint blockade (Rizvi et al., 2015; Snyder et al., 2014). Chimeric antigen receptor (CAR) therapy targeting up-regulated normal cell-surface gene products represents a potential alternative immunotherapeutic approach (Sadelain et al., 2017). CARs are synthetic receptors that retarget and reprogram T cells (Jensen and Riddell, 2015; Sadelain, 2016). Unlike the physiological T cell receptor, which engages HLA-peptide complexes, CARs bind to native cell surface molecules that do not require antigen processing or HLA expression for tumor recognition. CAR T cells therefore can recognize target antigens on any HLA background or in tumor cells that have down-regulated HLA expression or proteasomal antigen processing, two mechanisms known to promote tumor immune escape (Zhou and Levitsky, 2012). The frequency of AML mutations affecting epigenetic modifiers, including DNA methylation-related genes such as DNMT3A and IDH1/2 (Cancer Genome Atlas Research, 2013), may promote dysregulation of non-mutated cell surface proteins that could serve as effective CAR targets. The degree to which such global genomic effects may shape a targetable AML cell surfaceome is unknown. We hypothesized that AML cells display cell-surface protein alterations that provide targets for CAR therapy.
CAR therapy targeting CD19 has been particularly effective in acute lymphoblastic leukemia (ALL) (Sadelain, 2015), resulting in high complete remission rates in adults and children with chemorefractory disease (Davila et al., 2014; Lee et al., 2015; Maude et al., 2014a; Qasim et al., 2017; Turtle et al., 2016a). CD19 is found in B lineage lymphomas and leukemias, where it is expressed in most if not all cancer cells, while absent from hematopoietic stem cells as well as all normal tissues outside the B cell lineage (LeBien and Tedder, 2008). CD19 CAR T cells induce a B cell aplasia in murine models (Davila et al., 2013; Pegram et al., 2012) and in leukemia and lymphoma patients (Brentjens et al., 2013; Davila et al., 2014; Kochenderfer et al., 2013; Kochenderfer et al., 2015; Lee et al., 2015; Maude et al., 2014b; Qasim et al., 2017; Turtle et al., 2016a; Turtle et al., 2016b). This B cell aplasia is reversible when CAR T cells wane or are depleted (Paszkiewicz et al., 2016). Other “on-target/off-tumor” activities of CAR T cells can induce a range of toxicities, which may be tolerable, as is the case for transient B cell aplasias induced by CD19 CAR T cells, or severe and even lethal (Curran et al., 2012). Thus, CAR T cells specific for carbonic anhydrase-IX, which is overexpressed in renal cell carcinoma but is also present in normal biliary epithelium, can induce cholestasis (Lamers et al., 2013; Lamers et al., 2006). T cells targeting carcino-embryonic antigen (CEA) in patients with colon cancer caused severe hemorrhagic colitis due to CEA expression in normal colonic tissue (Parkhurst et al., 2011). A fatal outcome occurred in a patient infused with CAR T cells specific for HER2, which led to rapid respiratory failure and multi-organ dysfunction, attributed to reactivity against HER2 in normal lung cells (Morgan et al., 2010). Inversely, loss of antigen expression in a fraction of tumor cells will result in tumor escape and relapse, as seen in more than 30% of B-ALL patients after CD19 CAR therapy(Wang et al., 2017). Loss or down-regulation of CD19 on malignant B cells may occur via different mechanisms including splicing, missense mutation or lineage switch(Gardner et al., 2016; Jacoby et al., 2016; Maude et al., 2014a; Perna F, 2016; Sotillo et al., 2015; Yu et al., 2017). Effective CAR targets must therefore fulfill a number of criteria to enable complete tumor eradication while sparing normal tissues from intolerable damage.
An ideal CAR target should be expressed at high density, in most if not all tumor cells including cancer stem cells, and in a large fraction of patients (Table 1). Unlike native T cells, which are known to signal through the TCR in response to very low antigen density, CAR T cells require higher antigen densities to fully activate effector functions (Drent et al., 2017; Turatti et al., 2007; Walker et al., 2017; Weijtens et al., 2000). High absolute antigen expression that is easily detected by FACS analysis is thus much preferred for CAR target selection. Clonal heterogeneity creates complex tumors that are prone to escape targeted therapies. Expression of the target in normal tissues may be tolerable (transient or partial elimination of non-vital cell types) or unacceptable (destruction of vital tissues, hematopoietic stem cell depletion). To prevent undue toxicity, the ideal tumor target should not be expressed on any normal tissue/organ of, or at least not in vital tissues (heart, liver, CNS, lung and other tissues that cannot withstand transient damage) nor in closely related normal cellular counterparts, i.e., CD34+ hematopoietic stem/progenitor cells (HSPCs) in the case of AML. The target antigen should also not be expressed in CAR T cells to obviate fratricide elimination (Table 1). It is therefore imperative to carefully evaluate candidate targets not just in tumor cells, but across all normal tissues. Consequently, this task requires comprehensive sources of antigen annotation, as well as analytical tools specifically designed to identify potential CAR target antigens.
Table 1.
Features of an ideal CAR target
| Goal | Activity | Expression |
|---|---|---|
| Efficient recognition and targeting by CAR T cells | High on-tumor | - in all tumor cells - at high level - in many patients |
| Safe discrimination of target cells by CAR T cells | Low off-tumor | NOT in: - any normal tissue, especially vital tissues - normal counterparts (eg HSPCs for AML) - resting/activated T cells |
To date, searches for CAR targets have largely relied on transcriptome analyses, under the assumption that there exists a direct correspondence between mRNA transcripts and protein expression. The correlation between mRNA and protein expression is complex and potentially unrepresentative of the cell proteome, due to multiple factors including variable half-lives and post transcriptional regulatory mechanisms (Hack, 2004). An integrated and comprehensive analysis of transcriptomic and proteomic data in both malignant and normal cells is therefore needed to capture information lacking from indirect analyses of mRNA or limited protein expression assays (Haider and Pal, 2013). Moreover, advanced proteomic technologies combined with enrichment strategies for identifying plasma cell membrane proteins can provide direct measurements of the surface proteome. The goal of this study is to define a CAR target discovery strategy by integrating both proteomic and genomic datasets from AML and normal cell populations and compiling a comprehensive dataset of antigen annotations for the identification of therapeutic targets.
Results
Assembling a comprehensive dataset of AML surface molecule annotations
To search for potential CAR targets, we first performed surface-specific proteomic studies in a diverse panel of AML cell lines (THP1, Mono-mac, Kasumi, Molm13, OCI/AML3 and TF-1). After biotinylating the cell surface (Figure S1A), we performed mass-spectrometric analysis and identified 4,942 proteins. In order to generate the largest possible inclusive data set, we further added to this list the findings of surface-specific proteomic studies conducted in other human myeloid leukemia lines (NB4, HL60, THP1, PLB985, K562) (Strassberger et al., 2014) and all previously reported surface proteins, such as CD123 (Jordan et al., 2000), CLL1 (Bakker et al., 2004), CD33 (Taussig et al., 2005), CD44 (Jin et al., 2006), CD96 (Hosen et al., 2007), CD47 (Majeti et al., 2009), CD32 and CD25 (Saito et al., 2010), TIM3 (Kikushige et al., 2010), CD99 (Chung et al., 2017), adding another 80 proteins (Figure 1 - orange boxes and Figure S1B).
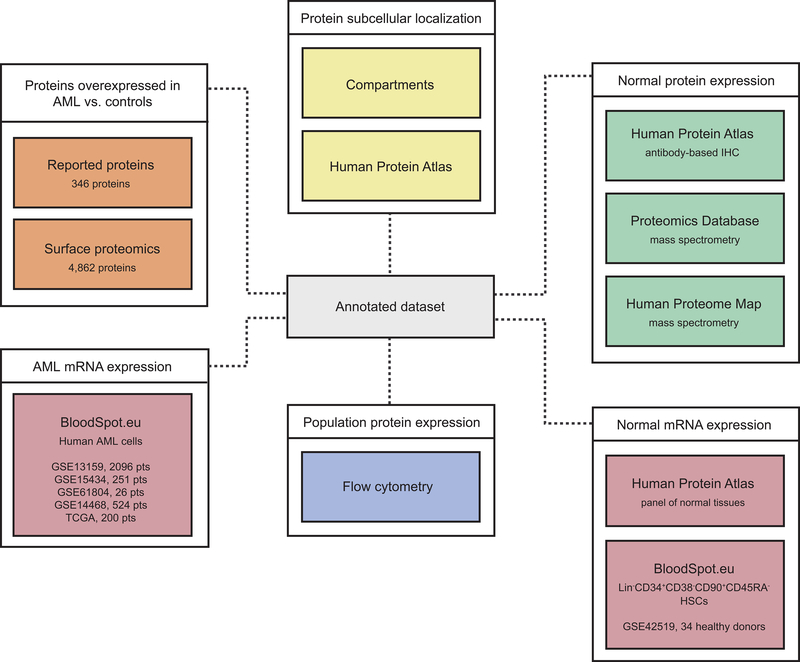
Figure 1. Generation of a comprehensive dataset of AML surface molecule annotations.
Orange boxes represent the information derived from proteomics studies in AML. Yellow boxes represent data sources providing information on subcellular localization. Green boxes represent three distinct published repositories of protein expression in several normal tissues and the platforms in which those data was generated. Pink boxes represent RNA data from AML (left) or normal cells (right). The blue box represents expression data obtained by flow cytometry in multiple distinct subsets of hematopoietic cells. The grey box at the center represents the combined annotation repository. See also Figure S1 and Table S1.
To annotate the expression of these molecules in normal tissues, we integrated data from the Human Protein Atlas (HPA) (Uhlen et al., 2015), the Human Proteome Map (HPM) (Kim et al., 2014) and the Proteomics Database (PD) (Wilhelm et al., 2014). These data sources provided protein expression information for several normal tissues/organs, including liver, gallbladder, pancreas, stomach, gut, rectum, testis, epididymis, prostate, breast, vagina, uterus, ovary, skin, skeletal and smooth muscle, brain, thyroid, bronchus, lung, heart, eye, tonsil, synovial fluid and others, listed in Table S1. Data in the atlases were obtained by antibody-based immunohistochemistry (HPA) or protein Mass Spectrometry (HPM and PD) (Figure 1 - green boxes). To focus our study on molecules specifically annotated to the membrane, we relied on two subcellular localization data sources, the HPA’s subcellular annotation and the Jensen Lab’s Compartments repository (Figure 1 - yellow boxes).
To remove surface molecules that are not over-expressed in AML cells compared to normal counterparts, we utilized publicly available gene expression analyses in normal BM CD34+CD38−CD90+CD45RA− HSCs and Lin−CD34+CD38−CD90−CD45RA− multipotent progenitors (MPP), Lin−CD34+CD38+CD45RA−CD123+ common myeloid progenitor cells (CMP), Lin−CD34+CD38+CD45RA+CD123+ granulocyte monocyte progenitors (GMP), Lin−CD34+CD38+CD45RA− CD123− megakaryocyte-erythroid progenitor cells (MEP) from healthy donors (GSE42519) and 3,097 primary AML patient samples clustered in 26 distinct subtypes based on specific cytogenetics such as del5q, t(8;21), t(11q23)/MLL, inv(16)/t(16;16) etc (GSE13159, GSE15434, GSE61804, GSE14468 and the Cancer Genome Atlas) (Bagger et al., 2016) (Figure 1 - pink boxes). In the final stage of our study, we characterized the expression of candidate targets selected by our algorithms, described below, in a panel of 30 primary AML patient samples based on flow cytometric analyses (Figure 1 - blue box). This compilation (Figure 1 - grey box) represents a comprehensive repository of AML surface protein annotation.
Design of an algorithm to identify CAR targets
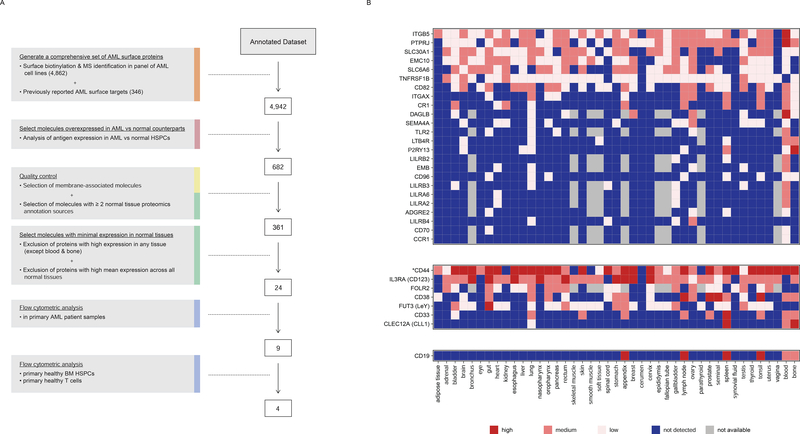
Starting from an annotated dataset of 23,118 Ensembl gene entries (19,876 unique HUGO gene identities) including 4,942 encoding surface molecules, we designed an algorithm to select CAR targets (Figure 2A). Given that an ideal target should be over-expressed in tumor cells compared to normal tissue counterparts, we first computed a log10 expression ratio between AML cells and normal HSPCs per molecule by comparing RNA expression levels in 26 genetically defined subtypes of AML, to normal BM HSCs, MPP, CMP, GMP, and MEP progenitor cells. A mean expression value for each molecule in both malignant and normal groups was calculated and a normal distribution fit to the AML/normal HSPCs ratios. A threshold of two standard deviations above the distribution peak maximum was applied, leaving 823 Ensembl gene entries corresponding to 682 unique HUGO entries. We then prioritized antigens with a membrane-associated sub-localization and redundant protein expression data in at least 2 of the 3 databases (HPA, HPM and PDB) that annotate expression levels in normal tissues. This “quality control” step further removed 321 molecules, leaving us with 361 candidates.
Figure 2. Algorithm for candidate AML CAR target discovery.
(A) The algorithm shows the steps used to identify surface molecules overexpressed in AML relative to normal HSPCs, the quality control, the assessment of minimal expression in a large panel of normal tissues, and the flow cytometric analyses. Step descriptions are color-coded to match data sources in Figure 1. The number of molecules remaining after each analytical step is indicated to the right of each box. (B) Expression profile of 24 selected AML target candidates (top panel), previously reported AML CAR targets (middle panel), and CD19 (bottom panel) in normal tissues. *Aggregate of CD44 isoforms (only PDB distinguishes between CD44 and CD44v6). See also Figure S2.
To eliminate any molecule highly expressed in normal tissues, we merged protein expression data in normal tissues from HPA, HPM and PDB, ranging from 0 (below the level of detection) to 3 (high) (Figure S2A–B). Notably, our integrated database confirmed the presence of normal tissues adversely affected in earlier trials targeting CAIX (Lamers et al., 2013; Lamers et al., 2006), CEA (Parkhurst et al., 2011) and HER2 (Morgan et al., 2010), including gallbladder, gut and lung, respectively (Figure S2C). Conversely, CD19, whose only reported on-target/off-tumor toxicity is the induction of B cell aplasia, exhibited a profile of expression limited to the expected lymphoid-rich tissues (Figure 2B). We excluded molecules exhibiting high average expression (>2) across all normal tissues as well as molecules exhibiting high expression (3) in any normal tissue, except for blood and bone/bone marrow. With this algorithm, we identified 24 molecules overexpressed in AML vs. their normal counterparts and with no high expression in clusters of normal tissues, except for blood and bone marrow (Figure 2B). Further exclusion of the spleen would additionally include CD33 and CLEC12A amongst the top 24 CAR candidate targets (Figure S2D).
Expression analyses in primary AML samples and normal hematopoietic cells
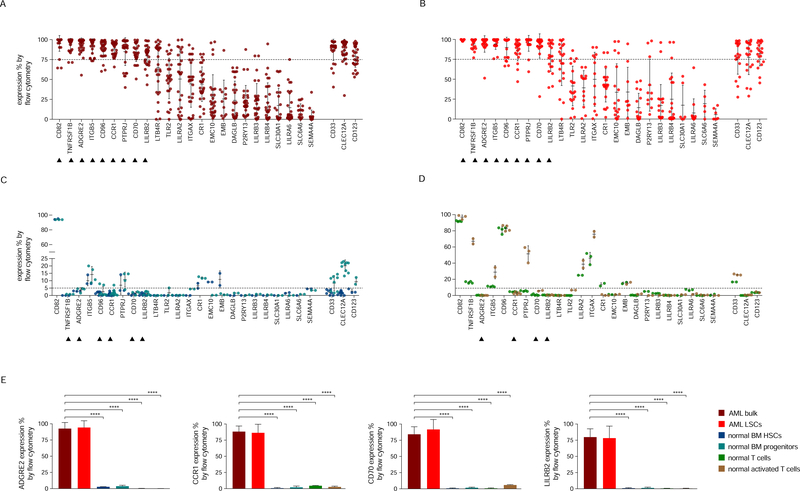
We analyzed expression levels of our 24 candidates by flow cytometry in 30 primary specimens of AML, enriched for AML harboring genetic abnormalities predisposing to clinical relapse (Table S2). These samples bear frequently recurring genetic abnormalities, including mutations in DNMT3A (14), CEBPA (12), IDH2 (11), FLT3 (9), NPM1 (7), IDH1 (7), WT1 (4), RUNX1 (4), ASXL1 (4), SUZ12 (3), KRAS (2), TET2 (2), TP53 (1) and CBL (1). Nine of the 24 candidate targets were present in all analyzed patient specimens and detected therein in >75% cells: CD82, TNFRSF1B (aka CD120b), ADGRE2 (aka EMR2 or CD312), ITGB5, CCR1 (aka CD191), CD96, PTPRJ (aka CD148), CD70, and LILRB2 (aka CD85d). We included in our analyses CD123 (IL3RA), CLEC12A (CLL1) and CD33, as these molecules are targets in current AML clinical trials. These antigens were also found to be expressed in >75% of AML cells in all patients (Figure 3A).
Figure 3. Flow cytometric analyses in primary AML samples and normal hematopoietic cells.
(A-D) Expression (% positive) of the 24 candidate antigens and the three CAR targets in current clinical investigation (most right three) in bulk AML population (A), in leukemic CD34+CD38− cells (B), in normal BM CD34+CD38−CD45RA−CD90+ HSCs (blue) and CD34+CD38+ progenitor cells (light blue) (C), or in freshly purified (green) or activated (brown) normal CD3+ peripheral blood T cells (D). Data are represented as mean ± standard deviation. (E) Summary of expression levels (mean ± SEM) of four top targets in indicated cell populations. ****, p value <0.0001 (Student’s t-test). See also Table S2.
As an ideal CAR target should be expressed in leukemic stem cells (LSCs) (Table 1), we further examined the expression of our selected markers in AML CD34+CD38− cells using flow cytometry. We found that all nine targets were also highly expressed (>75%) in this essential AML cell subset (Figure 3B). Their mean expression levels (range 78–99%) were comparable to that of CD123 (mean 82%) and slightly higher than that of CLEC12A (mean 77%) and CD33 (mean 77%) in LSCs. Flow cytometric analyses in normal BM CD34+CD38−CD90+CD45RA− HSCs and CD34+CD38+ progenitor cells showed that six out of nine molecules (TNFRSF1B, ADGRE2, CCR1, CD96, CD70 and LILRB2) were expressed at low levels (<5%) in normal HSPCs (Figure 3C). CD123, CLEC12A and CD33 were present at higher levels in these normal progenitor cells (9%, 20% and 8%, respectively) (Figure 3C).
As CAR therapy requires the sustained activity of functional CAR T cells, we investigated expression of these antigens in freshly purified and activated T cells from healthy donors. Four of the latter six candidates (ADGRE2, CCR1, CD70 and LILRB2) showed low-level expression (<5%) in T cells (Figure 3D). TNFRSF1B and CD96 were more abundant (up to 67% and 83%, respectively), which may complicate the generation or activity of CAR T cells and would require adapted strategies (e.g., target gene ablation) if one were to pursue these antigens in a CAR therapy.
In summary, our selection process identified four potential CAR targets with high expression in AML bulk cells and AML LSCs (Figure 3A–B) and low expression in normal tissues (Figure 2B), normal HSPCs (Figure 3C) and resting/activated T cells (Figure 3D), as depicted in Figure 3E. These expression profiles compare favorably with CD123, which is highly expressed in AML, especially LSCs (Figure 3A–B), but is also abundant in multiple normal tissues (Figure 2B). CD33 and CLEC12A are also highly expressed in AML (Figure 3A), although they exhibit a high degree of expression in normal hematopoietic progenitor cells (Figure 3C), consistent with their RNA expression levels (Bagger et al., 2016). Integrated systemic proteomics data indicate that CD33 is more abundant in the lung, prostate and skin (Figure 2B).
It is noteworthy that none of these molecules showed a profile comparable to that of CD19, which is expressed at high levels in virtually all B cell leukemia cells, completely absent from HSPCs and T cells, and undetectable systemically outside B cell areas. The absence of a target expression profile similarly favorable to CD19 thus prompted us to leverage our annotated database to explore combinatorial targeting strategies.
Combinatorial pairing of candidate targets
Combinatorial strategies fall in two major categories (Figure 4). One is based on cumulative CAR targeting through the generation of bi-specific T cells that co-express two CARs or a dual-specific CAR (Duong et al., 2011; Grada et al., 2013; Wilkie et al., 2012; Zah et al., 2016). The other takes advantage of split signaling (Alvarez-Vallina and Hawkins, 1996; Krause et al., 1998) to target two antigens, using one antigen to direct costimulation to enhance or rescue the function of a CAR or TCR specific for the other antigen (Kloss et al., 2013; Krause et al., 1998). In the former approach (CAR/CAR, Figure 4), T cells recognize target cells that express any of two given antigens and will thus engage tissues expressing either antigen alone. Some low or moderate expression in normal tissues, albeit not optimal, may be tolerable depending on the tissues in question. In the latter approach (CAR/CCR, Figure 4), T cells are more restricted to dual-antigen positive tumor cells, thus relaxing the expression criteria for at least one of the paired antigens. This approach however requires pan-tumor expression of the CAR target to avert antigen escape. In both instances, target pairings depend on the systemic expression and co-expression of the two prospective matches to minimize cumulative expression in normal tissues.
Figure 4. Principles of combinatorial targeting for CAR therapy.
The top panel represents single (CD19, left) and combinatorial (CAR/CAR, middle and CAR/CCR, right) targeting approaches. The combinatorial strategies require that the paired targets (Antigen A and Antigen B) meet stringent criteria that we defined in 6 principles illustrate below. The top three address safety and the bottom three therapeutic efficacy. Heatmaps indicate the expression level for the respective antigens in different tissues (not detected, low, medium and high, color-coded as indicated in the last panel). To minimize systemic on-target/off-tumor toxicity, an ideal pair should not present overlapping expression in normal tissues. Albeit suboptimal, some low or moderate expression in normal tissues may be tolerable in the CAR/CAR approach, depending on the tissues in question. This criterion may be relaxed for the CCR target in the CAR/CCR approach. To minimize HSC toxicity, the expression of both paired targets should be very low in CD34+CD38− HSCs. To minimize T-cell reactivity, the expression of two targets in a pair should be very low in normal resting and activated (r/a) T cells. To overcome clonal heterogeneity, the combined targets should mark all tumor cells. The CAR/CCR approach requires pan-tumor expression of the CAR target. To prioritize leukemic stem cell targeting, both antigens should be expressed in LSCs, but not obligatorily in all leukemic cells for one of the antigens in the CAR/CAR approach. To prevent antigen-negative relapse, both antigens should be co-expressed in tumor cells as much as possible (t0 and t1 represent pre-treatment and relapse time points).
To improve the targeting of AML without aggravating off-target toxicity, we wrote a software package to pair antigens minimizing cumulative target expression in normal organ/tissues. Two other related safety considerations are the avoidance of normal HSC and T cell recognition. The requirements for increasing therapeutic efficacy entail additional, overlaid selection criteria. The first principle is to maximize the number of targetable tumor cells, addressing the challenge of clonal heterogeneity. Another priority is to target LSCs, without which a CAR therapy could not be potentially curative. Finally, pairing choices should favor redundant expression of the two targets in the tumor in order to minimize the risk of antigen escape. We applied these principles to a pool of twelve molecules, including the nine top single targets defined in Figure 3, to which we added CD123, CD33 and CLEC12A, which represent 66 possible combinations.
The script we generated pairs targets with non-overlapping expression in normal tissues, wherein expression levels in vital and non-vital tissues were weighted. Pairing in vital tissues required that at least one of two antigens be “not-detected” (0) in each tissue. Pairing in non-vital tissues allowed both or one of the two antigens to exhibit “low” (1) expression. Our top four targets (ADGRE2, CCR1, CD70 and LILRB2), did not present overlapping expression in normal tissues (other than myeloid-rich tissues-bone, blood, spleen, appendix) when paired with CD33, CLEC12A or CD96. CD96 was removed from further study because of its high expression in T cells (Figure 3D), thus failing to meet one of the Figure 4 principles. The preferred pairings should recognize virtually all AML cells in a given clinical specimen, prioritize LSC recognition and favor redundancy (Figure 4).
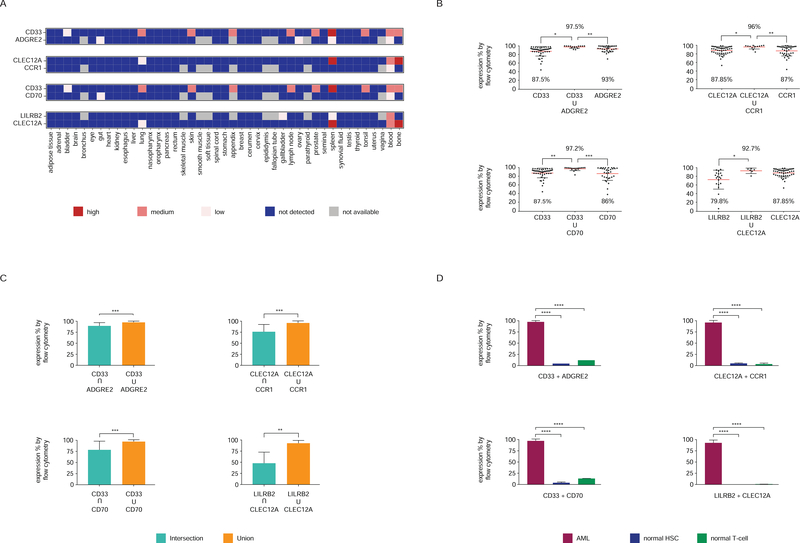
The following four combinations were analyzed in primary AML samples: ADGRE2+CD33, CCR1+CLEC12A, CD70+CD33 and LILRB2+CLEC12A (Figure 5A). All four combinations increased the rate of targeting, reaching near 100% FACS positivity in all AML specimens. For each one of these pairs, the dual targeting exceeds the targeting of either antigen alone: (ADGRE2+CD33 = 97.5%) vs ADGRE2 (93%) and CD33 (87.5%); (CLEC12A+CCR1 = 96%) vs CLEC12A (87.8%) and CCR1 (87%); (CD70+CD33= 97.2%) vs CD70 (86%) and CD33 (87.5%); (LILRB2+CLEC12A = 92.7%) vs LILRB2 (79.8%) (Figure 5B). We further analyzed the co-expression of two given targets in comparison to the sum of each one’s expression. Most AML cells expressed both antigens (Figure 5C), but total positivity (union) was significantly higher than dual-positivity (intersection) (Figure 5C), suggesting the presence of cells expressing one antigen only (Figure S3). This finding is consistent with AML clonal heterogeneity and favors using these antigen pairs in the dual-targeting approach (CAR/CAR) rather than the CAR/CCR approach (Figure 4). Finally, expression levels of these four combinations in normal bone marrow HSPCs and peripheral blood T cells were low (Figure 5D), confirming that one may maximize AML recognition without increasing toxicity towards normal hematopoietic cells.
Figure 5. Paired expression of potential CAR targets in AML.
(A) Co-expression of antigen pairs in normal tissues (see text for algorithm criteria). (B) Expression of each and both of indicated antigen pair in primary AML samples (% positive by flow cytometry). Data are represented as mean ± standard deviation. (C) Levels of co-expression (intersection) and additive expression (union) of antigen pairs in primary AML samples. Data are represented as mean ± standard deviation. (D) Additive expression of antigen pairs in AML cells (mean ± SEM) compared to normal BM HSCs and T cells. See also Figure S3.
Discussion
CAR therapy has demonstrated great potential against relapsed B-cell malignancies, in particular ALL. One may hope for a comparable outcome in AML, if targets as effective as CD19 are identified. We have generated a discovery platform that relies on large data sets of protein and RNA expression in malignant and normal tissues to search for optimal CAR targets. As we did not identify single targets with an ideal expression profile, we focused on combinatorial pairings designed to target nearly all AML cells within a tumor sample, including LSCs, without increasing potential off-target toxicity.
We assembled an extensive AML surfaceome dataset, combining public protein repositories and our own cell-surface proteomics performed in six AML cell lines, thus generating an inclusive list of AML-associated cell surface proteins. Studies on candidate targets typically focus on one molecule, comparing expression in cancer cells to their normal counterparts, rarely taking into account the systemic expression of the candidate target, thus underestimating the toxicity risks across normal organs. To address body-wide protein expression, we combined three extensive proteomics databases that map the human proteome. Both immuno-histochemical assays and mass spectrometry data were combined, increasing confidence in their dependability, especially for low levels of expression. This database included annotations of every candidate molecule in a large panel of normal tissues, in addition to AML and normal HSP cells.
Starting from over 5,000 Ensembl gene IDs (4,942 HGNC), our algorithm identified 24 candidates with features potentially suitable for CAR targeting. Four of these targets, all present in our cell surface proteomics, were G-protein coupled receptors (G-PCRs): ADGRE2, CCR1, LTB4R and P2RY13. Prior studies based on RNA-seq have found these G-PCRs to be amongst the most highly expressed G-PCRs in AML cells (Maiga et al., 2016). A possible role for G-PCRs in leukemia has been suggested for chemokine receptors (such as CCR1), adhesion receptors (such as ADGRE2) and purine receptors (including P2RY13) (Wilhelm et al., 2011). Our FACS analyses, conducted for all 24 candidates in a panel of 30 primary AML samples and AML LSCs, further shortened our candidate list to nine molecules, based on positive detection by FACS analysis in most patients and in >75% cells per clinical specimen. Six of these molecules exhibited low levels of expression in normal bone marrow CD34+CD38−CD45RA−CD90+ HSCs: TNFRSF1B, ADGRE2, CCR1, CD96, CD70 and LILRB2. TNFRSF1B and CD96 were found to be expressed at high levels in T cells, which may result in CAR T cell self-elimination. The expression of TNFRSF1B and CD96 in T cells could be eliminated by gene editing, requiring additional CAR T cell manufacturing steps (Riviere and Sadelain, 2017).
The remaining four candidate targets identified by our algorithm were ADGRE2, CCR1, CD70 and LILRB2. ADGRE2 (aka EMR2) is a member of the epidermal growth factor (EGF)-TM7 family of proteins, along with EMR1(Baud et al., 1995), F4/80 and CD97(Lin et al., 1997). Like CD97, ADGRE2/EMR2 possesses calcium-binding EGF domains (Downing et al., 1996), but unlike CD97, which is ubiquitously expressed in many cell types, EMR2 expression is restricted to monocytes/macrophages and granulocytes and is not up-regulated in activated T and B cells (Lin et al., 2000). We found ADGRE2 to be expressed at low levels in the gut, ovary and spleen and our FACS analyses detected ADGRE2 in ~93% of cells in all patient specimens.
CCR1 (aka CD191) is a G-PCR that binds to members of the C-C chemokine family. An immunohistochemical analysis of 944 hematolymphoid neoplasias previously identified CCR1 expression in a subset of AML, B and T cell lymphomas, plasma cell myeloma, and Hodgkin lymphoma (Anderson et al., 2010). The majority of our AML cases showed strong CCR1 expression, averaging 88% positivity by FACS in all specimens.
CD70 is a member of the TNF-family and the ligand of the CD27 T cell costimulatory receptor (Bowman et al., 1994; Goodwin et al., 1993). It is expressed in multiple tumor types and serves as a target for antibody and drug-conjugated antibody depletion in both renal cell carcinoma and non-Hodgkin lymphoma (Law et al., 2006; McEarchern et al., 2007; McEarchern et al., 2008; Ryan et al., 2010). CD70-specific CARs have been shown to induce sustained regression of established Raji Burkitt lymphoma xenografts (Shaffer et al., 2011). We found CD70 in ~86% of cells in all patient specimens.
LILRB2 (aka CD85d) is a member of the leukocyte immunoglobulin-like receptor (LIR) family, expressed in myeloid and B cells, acting to suppress the immune response. It is also expressed in non-small cell lung cancer (Sun et al., 2008), and in the gallbladder and spleen at low levels. We found LILRB2 in ~76% of cells in most patient specimens. This finding supports the notion that HSCs can express immune inhibitors of innate and adaptive immunity to evade potential immune surveillance (Zheng et al., 2012).
Seven targets have been previously reported as potential AML CAR targets. None of these meet our criteria for optimal single CAR targeting, however they may nonetheless prove effective with acceptable levels of toxicity. CD33 is a myeloid-specific sialic acid-binding receptor, targeted by gentuzumab ozogamicin (GO) (Administration, 2010) with demonstrated survival benefit in AML patients (Hills et al., 2014; Ravandi et al., 2012). Vadastuximab talirine, a CD33-directed antibody-drug conjugated in phase III clinical development (2016), was recently halted following 5 serious adverse events in patients with AML. Preclinical studies evaluating CD33 CARs have shown reduction of myeloid progenitors (Kenderian et al., 2015; Pizzitola et al., 2014). Two clinical trials targeting CD33 are currently active ( and ). One AML patient was treated with CD33 CAR T cells at the Chinese PLA General Hospital, showing transient efficacy and mild fluctuations in bilirubin (Wang et al., 2015). We found CD33 to be detected systemically at higher levels than other myeloid markers, in particular in lung, skin, and prostate. Our FACS analyses detected CD33 in ~87% of cells from all patient specimens. CLEC12A (aka CLL1, CD371), a type II transmembrane receptor family containing a C-type lectin/C-type lectin-like domain, is over-expressed in LSCs (van Rhenen et al., 2007). CLEC12A was more prominent in the lung and detected in ~87% of cells in most patient specimens, as well as in committed progenitor cells, consistent with RNA expression levels (Bakker et al., 2004). CLEC12A plays a role as a negative regulator of granulocyte and monocyte function. CLEC12A CAR T cells have been shown to be effective against HL60 (Tashiro et al., 2017), but exhibited modest activity against primary AML xenografts (Kenderian et al., 2016). Lewis (Le)-Y, a difucosylated carbohydrate antigen, has been targeted in four patients with relapsed AML. Infusion of second-generation CD28-based CAR T cells resulted in stable/transient remission of three patients, all of whom ultimately progressed despite T cell persistence (Ritchie et al., 2013), suggesting possible antigen escape. We found Le-Y to be highly expressed in the gut. Two trials for CARs targeting CD123 ( and ), the high-affinity interleukin-3 receptor a-chain, have been conducted, one of which is still in progress. In one instance, partial remission was induced in a patient with FLT3-ITD+ AML treated with a third generation CD123-specific CAR (Luo et al., 2015). Preclinical studies have revealed significant myeloablation in one study (Gill et al., 2014) but not another (Pizzitola et al., 2014). CD123 is however expressed at high levels in several normal tissues, which resulted in its elimination by our algorithm. Low affinity CARs may mitigate some of the on-target/off-tumor toxicity (Arcangeli et al., 2017). Two other myeloid-lineage markers, folate receptor β and CD44v6, the isoform variant 6 of the adhesive receptor CD44 (Bendall et al., 2000; Legras et al., 1998; Lynn et al., 2016; Lynn et al., 2015), were also found to be expressed in multiple normal tissues. CD44v6 is found in AML stem cells (Casucci et al., 2013) and some epithelial tissues, particularly skin keratinocytes (Heider et al., 2004). Reports of CD44v6 expression are conflicting, depending on antibody usage(Bendall et al., 2000). CD38 is a non-lineage-restricted, type II transmembrane glycoprotein targeted by Daratumumab, the first U.S. Food and Drug Administration-approved anti-CD38 antibody (Dimopoulos et al., 2016; Lonial et al., 2016) the activity of which on AML is limited without ATRA (Yoshida et al., 2016). It is expressed in all normal hematopoietic progenitor cells, T cell and NK cells. Drent et al. generated ~124 antibodies specific for CD38 spanning over 2 logs of affinity and demonstrated that CAR T cells bearing scFvs with reduced affinity can strongly lyse CD38++ myeloma cells (on-target/on-tumor effect), while sparing CD38+ normal hematopoietic cells (on-target/off-tumor effect) (Drent et al., 2017). These findings extend previous reports linking scFv affinity to CAR activity (Caruso et al., 2015; Hudecek et al., 2013; Liu et al., 2015).
While several of the above targets have therapeutic potential, we did not find any with an expression profile comparable to CD19. The lesser homogeneity of their expression raises the concern of antigen escape and relapse. This prompted us to explore combinatorial targeting strategies. Combinatorial strategies differ in both intent and approach (Sun and Sadelain, 2015; Wu et al., 2015). Some combine activating receptors (CAR/CAR), enabling T cells to recognize target cells that express any of two given antigens. This approach broadens T cell reactivity and likely decreases the risk of antigen escape, but at the cost of cumulating toxicities associated with each target. In contrast, combinatorial approaches that restrict T cell reactivity to dual-positive tumor cells will reduce toxicity towards normal tissues that express either target alone (Alvarez-Vallina and Hawkins, 1996; Kloss et al., 2013). These, however, require pan-expression of the CAR target in AML cells. Our analyses did not identify optimal targets for this strategy.
We laid out 6 principles to guide combinatorial CAR pairing with the purpose of enhancing therapeutic efficacy without increasing off-tumor toxicity. Several suitable pairs emerged from analyses in a pool of twelve promising molecules and 66 combinatorial pairings. We studied four of these, CD33+ADGRE2, CLEC12A+CCR1, CD33+CD70 and LILRB2+CLEC12A, in greater depth, examining their expression in primary AML specimens. Three of these pairings positively stained >97% of cells in AML samples, (LILRB2+CLEC12A scored slightly lower, averaging 93%), while all stained <5% of normal HSCs and T cells. Thus, the aggregate staining of ADGRE2 and CD33 increased the rate of FACS recognition to 97%. CD33 is more abundant in lung, skin, and prostate than other myeloid markers, but this combinatorial pairing is not predicted to compound this on-target/off-tumor activity. Similarly, the combined targeting of CCR1 and CLEC12A is not predicted to increase off-tumor targeting in the lung. In pairing targets with non-overlapping expression in normal tissues, one may leverage a co-targeting strategy with minimal cumulative antigen expression in non-tumor cells.
This present study is a new approach to discovering of CAR targets. It rests on two central concepts. First is the use of a composite high-throughput annotation database based on proteomics and transcriptomics to evaluate many candidates simultaneously. Second is the application of a series of principles to guide combinatorial pairings in an algorithm as we used here. Future CAR T cell studies, in relevant tumor models and more likely in phase I clinical trials, are needed to fully validate our concepts and approach. We hope this paradigm will accelerate the development of CAR therapy for AML and other cancers including solid tumors.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michel Sadelain (m-sadelain@ski.mskcc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Primary AML specimens were obtained from the Hematology Oncology Tissue Bank (HOTB) of MSKCC (IRB protocol Y2017P026 was reviewed and approved by the Institutional Review Board/Privacy Board-A). Informed consent was obtained from all subjects. Patient characteristics are illustrated in Table S2.
Primary human bone marrow CD34+ cells were purchased from Stem Cell Technologies (70002.2, 70002.3).
Human T cells isolation and activation
De-identified buffy coats from healthy volunteer donors were purchased from the New York Blood Center (IRB exempted). Peripheral blood mononuclear cells were isolated by density gradient centrifugation, and T lymphocytes were then purified using the Pan T cell isolation kit (Miltenyi Biotech). Cells were activated with Dynabeads (1:1 beads:cell) Human T-Activator CD3/CD28 (ThermoFisher) in X-vivo 15 medium (Lonza) supplemented with 5% human serum (Gemini Bioproducts) with 100 U/ml IL-2 (Miltenyi Biotech) at a density of 106 cells/ml. The beads were removed by magnetic separation 48 hr after activation. The medium was changed every 2 days, and cells were replated at 106 cells/ml.
AML cell lines
THP1, Mono-mac, Kasumi, Molm13, OCI/AML3 and TF-1 cells were maintained in RPMI1640/l-Glutamine (Life Technologies, Inc., Carlsbad, CA), supplemented with 10% FBS (20% for HL60) (Life Technologies) at 37 °C. THP1 and Mono-mac lines bear MLL-AF9 translocation, the THP1 line also bears deletion of TP16, TP53, UTX and rearrangement of RB1; Kasumi bears an AML1-ETO translocation; Molm13 FLT3, OCI/AML3 a NPM mutation and TF-1 a highly rearranged hyperdiploid karyotype with TP53 mutation.
METHOD DETAILS
Normal Tissue Proteomics Compilation and Data Retrieval
Expression data for normal tissues was retrieved from three data repositories, the Human Protein Atlas (HPA) (http://www.proteinatlas.org/about/download, normal_tissue.csv.zip, accessed 10/15/16), the Human Proteome Map (HPM) (RRID:SCR_015560, http://www.humanproteomemap.org/download.php,HPM_gene_level_epxression_matrix_Kim_et_al_052914.csv accessed 10/13/16), and the Proteomics Database (PDB) (RRID:SCR_015562, Accessed via the PDB API, available at https://www.proteomicsdb.org/proteomicsdb/#api, 10/13/16). Additionally, subcellular localization data was obtained from the HPA (RRID:SCR_006710, http://www.proteinatlas.org/about/download, subcellular_localization.csv.zip, accessed 10/15/16) and COMPARTMENTS (RRID:SCR_015561, http://compartments.jensenlab.org/Downloads, LOCATE_human_v6_20081121.xml, accessed 10/15/15) repositories, and transcriptomic data retrieved from the HPA (http://www.proteinatlas.org/about/download, rna_tissue.csv.zip, accessed 10/15/15) and Bloodspot.eu data archives (RRID:SCR_015563).
Correcting Tissue and Organelle Nomenclature
Due to differing tissue nomenclature among source repositories, each data set was mapped to a set of consensus tissue labels. In cases where multiple tissues from one repository mapped to a single label from another source, the maximum expression value was taken, for instance the PDB’s “retina” and “vitreous humor” tissues were collapsed into a single tissue category, “eye.” For consistency, fetal and placental tissues were also discarded, resulting in 43 distinct tissue categories (Table S1): adipose tissue, adrenal, appendix, bladder, blood, bone, brain, breast, bronchus, cerumen, cervix, epididymis, eye, fallopian tube, gallbladder, gut, heart, kidney, esophagus, liver, lung, lymph node, nasopharynx, oropharynx, ovary, pancreas, parathyroid, prostate, rectum, seminal, skeletal muscle, skin, smooth muscle, soft tissue, spinal cord, spleen, stomach, synovial fluid, testis, thyroid, tonsil, uterus, vagina.
Similarly, maps of subcellular localization labels from the HPA and Compartments databases were generated by manually classifying organelle labels as either cell membrane-associated or otherwise unaffiliated, and then applying the resulting dictionary to proteins in both repositories.
Calculation of Distribution Metrics
In the interest of facilitating subsequent selection steps, expression entries were classified into three categories: “not detected”, “low”, “medium”, and “high”, the native format in which HPA protein and RNA data was made available. To accomplish the binning, both the HPM and PDB datasets were first log10 transformed, after HPM data was then temporarily corrected for the purpose of abundance distribution estimation so as to minimize artifactual cases in which LC-MS/MS peptide fragment masses were underdetermined, resulting in multiple gene assignments. This was accomplished by collapsing protein entries originating from the same experiment and occurring with precisely the same spectral abundance measure, into a single entry during the curve fitting process (after which all entries were restored). To fit normal curves of best fit to the observed distributions, the Broyden–Fletcher–Goldfarb–Shanno algorithm was applied(Team, 2016), after which the peak maximum and standard deviation measure was recorded for each curve.
Gene Nomenclature Conversion
Gene identification labels for all data sources not natively provided in Ensembl gene format were then converted using the biomaRt and mygene R packages, as well as the Proteomics Database API to ensure comprehensive mapping between differing nomenclatures(Adam Mark, 2014; Durinck et al., 2009). In cases where multiple entries from a given data source mapped to the same gene ID only the highest expression value for each tissue was retained. And in cases where entries mapped to more than one Ensembl Gene ID, duplicate entries for each ID were made.
For each unique (protein, tissue, repository) entry, the maximum expression value was retained and the remaining expression values, which arose from native IDs mapped to two Ensembl Gene IDs, were discarded.
Generation of Repository Metrics
The number of available data sources for every unique (protein, tissue) entry was then recorded and the maximum binned expression abundance for each unique (protein, tissue) entry was then computed.
Expression and Binning
Expression values within one standard deviation and above normal peaks were considered to be of “medium” (2) abundance, expression values above this threshold were considered of “high” (3) abundance. Similarly, expression values within one standard deviation and below the normal peak were considered of “low” (1) abundance, and abundance values falling below one standard deviation considered to have an expression level low enough to consider “not detected” (0), for the purpose of epitope selection.
AML/normal HSPCs ratio analysis
The experiment-normalized RNA data obtained from Bloodspot.eu was exponentiated using a base of two as the data was natively provided in log2 format, after which the maximum values taken for each unique (gene-cell type) entry. Data was then divided into two groups, AML and normal HSPCs, with the AML group consisting of the following subtypes: −5/7(Q), −9Q, 7, 8, ABN(3Q), COMPLEX, COMPLEX_ DEL(5Q), COMPLEX_ UNTYPICAL, DEL(5Q), DEL(7Q)/7Q-, DEL(9Q), INV(16), INV(3), nan, NORMAL, OTHER, T(1;3), T(11Q23)/MLL, T(6;9), T(8;16), T(8;21), T(9;11), T(9;22), TRISOMY 11, TRISOMY 13, and TRISOMY 8. The normal HSPCs group consisting of the following cell types: HSC, MPP, CMP, GMP, and MEP. A mean expression value for each gene in the AML group was then calculated by averaging across the five normal cell types. The resulting mean values were then taken as devisors for expression ratios in which the dividends were each cell type’s RNA abundance. The base ten logarithm was then taken for each expression ratio and normal curves were fit to the observed distribution using the Broyden–Fletcher–Goldfarb–Shanno algorithm, after which the peak maximum and standard deviation measure was recorded for each curve. A threshold of two standard deviations above the distribution maximum was then applied, and any protein candidate with a ratio above this threshold recorded for later use.
Target Selection
Step 0 - Surface proteomics
Only proteins found during surface-biotinylation proteomics assays performed on six AML cell lines, THP1, Mono-mac, Kasumi, Molm13, OCI/AML3 and TF-1 and those reported by Strassberger et al., or in a select list of 346 previously reported molecules, including CLEC12A, IL3RA, FOLR2, FUT3, CD33, and CD38 were kept for further study.
Step 1 - RNA
Further, exclusively molecules whose ratio of RNA expression in AML samples versus normal HSPCs cells was greater than or equal to 2 SD above the mean were retained.
Step 2 - QC
Only protein entries reported only in two or more normal tissue proteomics databases were retained for further study. Additionally, proteins were discarded from further consideration if the locations reported in either the HPA or Compartments databases were not on the cell membrane.
Step 3 - Exclude High Expressors
Any protein entry whose mean expression across all normal tissues exceeded the classification threshold as a “medium” (2) expressor was excluded from further study. Further, all proteins whose expression was classified as high (3) for any tissue type, in any dataset, apart from those originating from blood and bone were excluded from further study.
Combinatorial Selection
Pairwise Exclusion
9 candidates, in addition to CLEC12A, IL3RA and CD33, were then assessed as combinatorial pairs by evaluating their expression each tissue sites at once. Vital and non-vital tissues were then assessed using distinct criteria, which all tissues possessing a given protein were required to pass. Due to their high concentration of hematopoietic cells, the appendix, bone, blood, and spleen tissues were not considered for the purposes of selection. Criteria for vital tissues (adipose tissue, adrenal, bladder, brain, bronchus, eye, gut, heart, kidney, esophagus, liver, lung, nasopharynx, oropharynx, pancreas, rectum, skeletal muscle, skin, smooth muscle, soft tissue, spinal cord, and stomach) required at least one of the tissue pairs to possess no detectable expression. Criteria for non-vital tissues (breast, cerumen, cervix, epididymis, fallopian tube, gallbladder, lymph node, ovary, parathyroid, prostate, seminal, spleen, synovial fluid, testis, thyroid, tonsil, uterus, and vagina) permit tissue expression in both antigens in a pair to exhibit “low” expression, or to possess no detectable expression, as qualified above.
Surface Proteomics
We performed cell surface biotinylation of six (THP1, Mono-mac, Kasumi, Molm13, OCI/AML3 and TF-1) human AML cell lines, which was used for mass spectrometric analysis.
For the isolation and collection of surface proteins, we used the Pierce® Cell Surface Protein Isolation Kit #89881 (Thermo Scientific 89881). 6×106 cells were cultured in 75 cm2 flasks. Prior to surface protein biotinylation, all reagents were cooled to 4°C. The cells were washed four times with ice-cold phosphate buffered saline (PBS) followed by incubation with 0.25 mg/mL Sulfo-NHS-SS-Biotin in 10 mL ice-cold PBS per flask on a rocking platform for 30 min at 4°C. The biotinylation reaction was quenched by adding 500 μL of the provided quenching solution (Pierce). Centrifuge cells at 500xg for 5 min and discard supernatant. Cells were washed with ice-cold PBS, harvested by gentle scraping and pelleted by centrifugation. The cells were lysed using the provided lysis buffer (Pierce) containing a protease inhibitor cocktail (Sigma) for 30 min on ice with intermittent vortexing. Lysates were centrifuged at 16,000xg for 2 min at 4°C. The clarified supernatant was used for purification of biotinylated proteins on NeutrAvidin Agarose. Before use, 500 μL of NeutrAvidin Agarose slurry was washed three times with Pierce wash buffer in a provided column (Pierce). The clarified supernatant was added to the slurry and incubated for 2 hr at room temperature in the closed column using an end-over-end tumbler to mix vigorously and allow the biotinylated proteins to bind to the NeutrAvidin Agarose slurry. Unbound proteins were removed by repetitive washing; three times with 500 μL Pierce Wash Buffer in a provided column (Pierce), three times with 500μL (50mM Ammonium bicarbonate) and eight times with 500μL digestion buffer (50mM Tric-Cl, pH 7.5, 1mM CaCl2). Finally bounded proteins on biotin-NeutrAvidin Agarose were digested with 4μg of trypsin (prepared in digestion buffer) over night at 37°C in shaking incubator (~750 rpm). The next day, digested peptides were filtered through column and protease reaction was stopped by of 0.5% TFA. Samples were cleared by centrifuging 10 min at 14,000xg, 15°C and desalted by stage tips. Desalted peptides were dry down by speed vac and re-suspended in 10μl of 3% acetonitrile/0.1% formic acid for LC-MS/MS analysis.
LC-MS/MS analysis
Desalted peptides were dissolved in 3% acetonitrile/0.1% formic acid and injected onto a C18 capillary column on a nano ACQUITY UPLC system (Water) which was coupled to the Q Exactive mass spectrometer (Thermo Scientific). Peptides were eluted with a non-linear 200 min gradient of 2–35% buffer B (0.1% (v/v) formic acid, 100% acetonitrile) at a flow rate of 300 nl/min. After each gradient, the column was washed with 90% buffer B for 5 min and re-equilibrated with 98% buffer A (0.1% formic acid, 100% HPLC-grade water) for 4 min. MS data were acquired with an automatic switch between a full scan and 10 scan data-dependent MS/MS scan (TopN method). Target value for the full scan MS spectra was 3 X 106 charges in the 380–1800 m/z range with a maximum injection time of 30 ms and resolution of 70,000 at 200 m/z in profile mode. Isolation of precursors was performed with 2.0 m/z. Precursors were fragmented by higher-energy C-trap dissociation (HCD) with a normalized collision energy of 27 eV. MS/MS scans were acquired at a resolution of 17,500 at 200 m/z with an ion target value of 5 X 104 maximum injection time of 60 ms and dynamic exclusion for 60 s in centroid mode.
Protein identification
MS raw files were converted into MGF by Proteome Discover (Thermo Scientific) and processed using Mascot 2.4 (Matrix Science, U.K.) by searching against the Uniprot human Database (version 2014 with 20209 protein entries) supplemented with common contaminant proteins. Search criteria included 10 ppm mass tolerance for MS spectra, 0.8 Da mass tolerance for MS/MS spectra, a maximum of two allowed missed cleavages, fixed carbamidomethylation of cysteine modifications, variable methionine oxidation and N-terminal protein acetylation, Mascot significance threshold of 0.05, and a false discovery rate of <0.01. Mascot data were assembled by Scaffold and X!-Tandem software and search criteria for identification 2 minimum peptides and 1% FDR at the peptide, and protein level.
Flow-cytometric analysis
The following antibodies were used to define antigen expression by flow-cytometry:
CD82-PE (Biolegend, 342103); CD120b (TNF-RII)-PE (Miltenyi, 130–107-740); EMR2-FITC, -APC (Miltenyi, 130–104-654, 130–104-656); ITGB5-PE (Biolegend, 345203); CCR1-PE, -APC (Miltenyi, 130–100-367, 130–100-358); CD96-APC (Miltenyi, 130–101-031); PTPRJ (CD148)-PE (life technologies, A15799); CD70-PE, -FITC (Biolegend, 355104, 355106); CD85d (ILT4)-PE -APC (Miltenyi, 130–100-567, 130–100-559); LTB4R1-AF700 (Novus Biologicals, FAB099N); CD85h (ILT1)-APC (Miltenyi, 130–100-920); TLR2-APC (Miltenyi, 130–099-020); CR1 (aka CD35)-APC (Miltenyi, 130–099-923); ITGAX (CD11c)-APC (Biolegend, 301613); EMB (abcam, 179801); EMC10 (abcam PA5–25112); LILRB3-PE (Miltenyi, 130–101-662); LILRB4-APC (R&D, FAB24251A); DAGLB (abcam, PA5–26331); P2RY13 (Novus Biologicals, NBP2–37382); LILRA6-APC (Miltenyi, 130–101-665); SLC30A1 (Alomone labs, AZT-011); SLC6A6 (LSBio, LS-C179237); SEMA4A (R&D, FAB4694A); CD123-PE (BD Biosciences, 555644); CLEC12A-PE (Miltenyi, 130–106-482); CD33-APC (Miltenyi, 130–098-864); CD38-BV421 (BD Biosciences, 562444); CD34-PE/Cy7 (Biolegend, 343515); CD45RA-BV640 (Biolegend, 304135); CD90-FITC (BD Biosciences, 555595).
QUANTIFICATION AND STATISTICAL ANALYSIS
Student’s t-test was used for significance testing in the bar graphs using a two-sample, normally distributed equal-variance model. P values less than 0.05 were considered to be significant. Graphs and error bars reflect means and standard deviations. All statistical analyses were carried out using GraphPad Prism 4.0 and the R statistical environment. ****, p value <0.0001.
Allowing for a 20% margin, a sufficient single-tailed estimate of arbitrarily large population size can be assessed at 95% confidence with 23 patients. We chose a sample size of 30 to further narrow the window of uncertainty.
DATA AND SOFTWARE AVAILABILITY
Proteomic data were submitted (ProteomeXchange Submission) to PRIDE database under the project accession: PXD007552.
Supplementary Material
Highlights.
We generated and annotated an extensive dataset of AML cell surface proteins
We designed an algorithm to identify candidate CAR targets
We defined six criteria for combinatorial pairing of CAR targets
We identified target combinations fulfilling stringent criteria for CAR therapy
Perna et al. try to identify potential ideal chimeric antigen receptor therapy targets in acute myeloid leukemia but fail to identify any. They then develop a generalizable combinatorial targeting strategy that fulfils stringent efficacy and safety criteria, which identifies several promising target pairings.
Significance.
The foremost challenge in extending the success of CD19 CAR therapy to AML is to identify targets as favorable as CD19 is for ALL. We outline a framework defining ideal features of CAR targets and establish a methodology for mining composite high-throughput surfaceome expression data. Given the intrinsic complexity of AML and the risks of antigen escape and off-tumor CAR T cell toxicity, we optimized combinatorial target selection based on expression profiles in normal and malignant tissues. The approach reported herein provides the basis for rational design of CAR therapies for AML and a template for combinatorial targeting in general.
Acknowledgments
This work was supported by a Technology Development Fund from Memorial Sloan Kettering Cancer Center to F.P. We thank Ahmet Dogan and Mariko Yake for assistance with pathology of AML samples; Jessica Schulman, Minal Patel, Akshar Patel, Erin McGovern and Amber Turner for assistance with the HOTB samples; Ly Vu for assistance with cell lines; Rupa Juthani for assistance with selection of public datasets; Sjoukje van der Stegen for assistance with flow cytometric data analysis; all members of the Sadelain laboratory for their critical comments. The authors have no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- (2016). ADCs Show Promise in Leukemias. Cancer Discov 6, 939. [DOI] [PubMed] [Google Scholar]
- Adam Mark RT, Afrasiabi Cyrus and Wu Chunlei (2014).
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Vallina L, and Hawkins RE (1996). Antigen-specific targeting of CD28-mediated T cell costimulation using chimeric single-chain antibody variable fragment-CD28 receptors. Eur J Immunol 26, 2304–2309. [DOI] [PubMed] [Google Scholar]
- Anderson MW, Zhao S, Ai WZ, Tibshirani R, Levy R, Lossos IS, and Natkunam Y (2010). C-C chemokine receptor 1 expression in human hematolymphoid neoplasia. Am J Clin Pathol 133, 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangeli S, Rotiroti MC, Bardelli M, Simonelli L, Magnani CF, Biondi A, Biagi E, Tettamanti S, and Varani L (2017). Balance of Anti-CD123 Chimeric Antigen Receptor Binding Affinity and Density for the Targeting of Acute Myeloid Leukemia. Mol Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagger FO, Sasivarevic D, Sohi SH, Laursen LG, Pundhir S, Sonderby CK, Winther O, Rapin N, and Porse BT (2016). BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res 44, D917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker AB, van den Oudenrijn S, Bakker AQ, Feller N, van Meijer M, Bia JA, Jongeneelen MA, Visser TJ, Bijl N, Geuijen CA, et al. (2004). C-type lectin-like molecule-1: a novel myeloid cell surface marker associated with acute myeloid leukemia. Cancer Res 64, 8443–8450. [DOI] [PubMed] [Google Scholar]
- Baud V, Chissoe SL, Viegas-Pequignot E, Diriong S, N’Guyen VC, Roe BA, and Lipinski M (1995). EMR1, an unusual member in the family of hormone receptors with seven transmembrane segments. Genomics 26, 334–344. [DOI] [PubMed] [Google Scholar]
- Becker PS, Kantarjian HM, Appelbaum FR, Petersdorf SH, Storer B, Pierce S, Shan J, Hendrie PC, Pagel JM, Shustov AR, et al. (2011). Clofarabine with high dose cytarabine and granulocyte colony-stimulating factor (G-CSF) priming for relapsed and refractory acute myeloid leukaemia. Br J Haematol 155, 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall LJ, Bradstock KF, and Gottlieb DJ (2000). Expression of CD44 variant exons in acute myeloid leukemia is more common and more complex than that observed in normal blood, bone marrow or CD34+ cells. Leukemia 14, 1239–1246. [DOI] [PubMed] [Google Scholar]
- Bowman MR, Crimmins MA, Yetz-Aldape J, Kriz R, Kelleher K, and Herrmann S (1994). The cloning of CD70 and its identification as the ligand for CD27. J Immunol 152, 1756–1761. [PubMed] [Google Scholar]
- Breems DA, Van Putten WL, Huijgens PC, Ossenkoppele GJ, Verhoef GE, Verdonck LF, Vellenga E, De Greef GE, Jacky E, Van der Lelie J, et al. (2005). Prognostic index for adult patients with acute myeloid leukemia in first relapse. J Clin Oncol 23, 1969–1978. [DOI] [PubMed] [Google Scholar]
- Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, Bartido S, Stefanski J, Taylor C, Olszewska M, et al. (2013). CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med 5, 177ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2013). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368, 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso HG, Hurton LV, Najjar A, Rushworth D, Ang S, Olivares S, Mi T, Switzer K, Singh H, Huls H, et al. (2015). Tuning Sensitivity of CAR to EGFR Density Limits Recognition of Normal Tissue While Maintaining Potent Antitumor Activity. Cancer Res 75, 3505–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casucci M, Nicolis di Robilant B, Falcone L, Camisa B, Norelli M, Genovese P, Gentner B, Gullotta F, Ponzoni M, Bernardi M, et al. (2013). CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood 122, 3461–3472. [DOI] [PubMed] [Google Scholar]
- Chung SS, Eng WS, Hu W, Khalaj M, Garrett-Bakelman FE, Tavakkoli M, Levine RL, Carroll M, Klimek VM, Melnick AM, and Park CY (2017). CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzin-Frankel J (2013). Breakthrough of the year 2013. Cancer immunotherapy. Science 342, 1432–1433. [DOI] [PubMed] [Google Scholar]
- Curran KJ, Pegram HJ, and Brentjens RJ (2012). Chimeric antigen receptors for T cell immunotherapy: current understanding and future directions. J Gene Med 14, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila ML, Kloss CC, Gunset G, and Sadelain M (2013). CD19 CAR-targeted T cells induce long-term remission and B Cell Aplasia in an immunocompetent mouse model of B cell acute lymphoblastic leukemia. PLoS One 8, e61338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila ML, Riviere I, Wang X, Bartido S, Park J, Curran K, Chung SS, Stefanski J, Borquez-Ojeda O, Olszewska M, et al. (2014). Efficacy and toxicity management of 19–28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med 6, 224ra225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos MA, Oriol A, Nahi H, San-Miguel J, Bahlis NJ, Usmani SZ, Rabin N, Orlowski RZ, Komarnicki M, Suzuki K, et al. (2016). Daratumumab, Lenalidomide, and Dexamethasone for Multiple Myeloma. N Engl J Med 375, 1319–1331. [DOI] [PubMed] [Google Scholar]
- Downing AK, Knott V, Werner JM, Cardy CM, Campbell ID, and Handford PA (1996). Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell 85, 597–605. [DOI] [PubMed] [Google Scholar]
- Drent E, Themeli M, Poels R, de Jong-Korlaar R, Yuan H, de Bruijn J, Martens ACM, Zweegman S, van de Donk N, Groen RWJ, et al. (2017). A Rational Strategy for Reducing On-Target Off-Tumor Effects of CD38-Chimeric Antigen Receptors by Affinity Optimization. Mol Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong CP, Westwood JA, Berry LJ, Darcy PK, and Kershaw MH (2011). Enhancing the specificity of T-cell cultures for adoptive immunotherapy of cancer. Immunotherapy 3, 33–48. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey E, Kornblau S, Pierce S, Kantarjian H, Beran M, and Keating M (1996). A stratification system for evaluating and selecting therapies in patients with relapsed or primary refractory acute myelogenous leukemia. Blood 88, 756. [PubMed] [Google Scholar]
- Gardner R, Wu D, Cherian S, Fang M, Hanafi LA, Finney O, Smithers H, Jensen MC, Riddell SR, Maloney DG, and Turtle CJ (2016). Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 127, 2406–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Tasian SK, Ruella M, Shestova O, Li Y, Porter DL, Carroll M, Danet-Desnoyers G, Scholler J, Grupp SA, et al. (2014). Preclinical targeting of human acute myeloid leukemia and myeloablation using chimeric antigen receptor-modified T cells. Blood 123, 2343–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin RG, Alderson MR, Smith CA, Armitage RJ, VandenBos T, Jerzy R, Tough TW, Schoenborn MA, Davis-Smith T, Hennen K, and et al. (1993). Molecular and biological characterization of a ligand for CD27 defines a new family of cytokines with homology to tumor necrosis factor. Cell 73, 447–456. [DOI] [PubMed] [Google Scholar]
- Grada Z, Hegde M, Byrd T, Shaffer DR, Ghazi A, Brawley VS, Corder A, Schonfeld K, Koch J, Dotti G, et al. (2013). TanCAR: A Novel Bispecific Chimeric Antigen Receptor for Cancer Immunotherapy. Mol Ther Nucleic Acids 2, e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack CJ (2004). Integrated transcriptome and proteome data: the challenges ahead. Brief Funct Genomic Proteomic 3, 212–219. [DOI] [PubMed] [Google Scholar]
- Haider S, and Pal R (2013). Integrated analysis of transcriptomic and proteomic data. Curr Genomics 14, 91–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider KH, Kuthan H, Stehle G, and Munzert G (2004). CD44v6: a target for antibody-based cancer therapy. Cancer Immunol Immunother 53, 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills RK, Castaigne S, Appelbaum FR, Delaunay J, Petersdorf S, Othus M, Estey EH, Dombret H, Chevret S, Ifrah N, et al. (2014). Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: a meta-analysis of individual patient data from randomised controlled trials. Lancet Oncol 15, 986–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosen N, Park CY, Tatsumi N, Oji Y, Sugiyama H, Gramatzki M, Krensky AM, and Weissman IL (2007). CD96 is a leukemic stem cell-specific marker in human acute myeloid leukemia. Proc Natl Acad Sci U S A 104, 11008–11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudecek M, Lupo-Stanghellini MT, Kosasih PL, Sommermeyer D, Jensen MC, Rader C, and Riddell SR (2013). Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin Cancer Res 19, 3153–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby E, Nguyen SM, Fountaine TJ, Welp K, Gryder B, Qin H, Yang Y, Chien CD, Seif AE, Lei H, et al. (2016). CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat Commun 7, 12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MC, and Riddell SR (2015). Designing chimeric antigen receptors to effectively and safely target tumors. Curr Opin Immunol 33, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, and Dick JE (2006). Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med 12, 1167–1174. [DOI] [PubMed] [Google Scholar]
- Jordan CT, Upchurch D, Szilvassy SJ, Guzman ML, Howard DS, Pettigrew AL, Meyerrose T, Rossi R, Grimes B, Rizzieri DA, et al. (2000). The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia 14, 1777–1784. [DOI] [PubMed] [Google Scholar]
- Kenderian SS, Ruella M, Shestova O, Klichinsky M, Aikawa V, Morrissette JJ, Scholler J, Song D, Porter DL, Carroll M, et al. (2015). CD33-specific chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia 29, 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikushige Y, Shima T, Takayanagi S, Urata S, Miyamoto T, Iwasaki H, Takenaka K, Teshima T, Tanaka T, Inagaki Y, and Akashi K (2010). TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell 7, 708–717. [DOI] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. (2014). A draft map of the human proteome. Nature 509, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss CC, Condomines M, Cartellieri M, Bachmann M, and Sadelain M (2013). Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat Biotechnol 31, 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenderfer JN, Dudley ME, Carpenter RO, Kassim SH, Rose JJ, Telford WG, Hakim FT, Halverson DC, Fowler DH, Hardy NM, et al. (2013). Donor-derived CD19-targeted T cells cause regression of malignancy persisting after allogeneic hematopoietic stem cell transplantation. Blood 122, 4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenderfer JN, Dudley ME, Kassim SH, Somerville RP, Carpenter RO, Stetler-Stevenson M, Yang JC, Phan GQ, Hughes MS, Sherry RM, et al. (2015). Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J Clin Oncol 33, 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause A, Guo HF, Latouche JB, Tan C, Cheung NK, and Sadelain M (1998). Antigen-dependent CD28 signaling selectively enhances survival and proliferation in genetically modified activated human primary T lymphocytes. J Exp Med 188, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers CH, Sleijfer S, van Steenbergen S, van Elzakker P, van Krimpen B, Groot C, Vulto A, den Bakker M, Oosterwijk E, Debets R, and Gratama JW (2013). Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: clinical evaluation and management of on-target toxicity. Mol Ther 21, 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers CH, Sleijfer S, Vulto AG, Kruit WH, Kliffen M, Debets R, Gratama JW, Stoter G, and Oosterwijk E (2006). Treatment of metastatic renal cell carcinoma with autologous T-lymphocytes genetically retargeted against carbonic anhydrase IX: first clinical experience. J Clin Oncol 24, e20–22. [DOI] [PubMed] [Google Scholar]
- Law CL, Gordon KA, Toki BE, Yamane AK, Hering MA, Cerveny CG, Petroziello JM, Ryan MC, Smith L, Simon R, et al. (2006). Lymphocyte activation antigen CD70 expressed by renal cell carcinoma is a potential therapeutic target for anti-CD70 antibody-drug conjugates. Cancer Res 66, 2328–2337. [DOI] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. (2015). PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372, 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBien TW, and Tedder TF (2008). B lymphocytes: how they develop and function. Blood 112, 1570–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M, Shah NN, et al. (2015). T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legras S, Gunthert U, Stauder R, Curt F, Oliferenko S, Kluin-Nelemans HC, Marie JP, Proctor S, Jasmin C, and Smadja-Joffe F (1998). A strong expression of CD44–6v correlates with shorter survival of patients with acute myeloid leukemia. Blood 91, 3401–3413. [PubMed] [Google Scholar]
- Lin HH, Stacey M, Hamann J, Gordon S, and McKnight AJ (2000). Human EMR2, a novel EGF-TM7 molecule on chromosome 19p13.1, is closely related to CD97. Genomics 67, 188–200. [DOI] [PubMed] [Google Scholar]
- Lin HH, Stubbs LJ, and Mucenski ML (1997). Identification and characterization of a seven transmembrane hormone receptor using differential display. Genomics 41, 301–308. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang S, Fang C, Yang S, Olalere D, Pequignot EC, Cogdill AP, Li N, Ramones M, Granda B, et al. (2015). Affinity-Tuned ErbB2 or EGFR Chimeric Antigen Receptor T Cells Exhibit an Increased Therapeutic Index against Tumors in Mice. Cancer Res 75, 3596–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonial S, Weiss BM, Usmani SZ, Singhal S, Chari A, Bahlis NJ, Belch A, Krishnan A, Vescio RA, Mateos MV, et al. (2016). Daratumumab monotherapy in patients with treatment-refractory multiple myeloma (SIRIUS): an open-label, randomised, phase 2 trial. Lancet 387, 1551–1560. [DOI] [PubMed] [Google Scholar]
- Lynn RC, Feng Y, Schutsky K, Poussin M, Kalota A, Dimitrov DS, and Powell DJ Jr. (2016). High-affinity FRbeta-specific CAR T cells eradicate AML and normal myeloid lineage without HSC toxicity. Leukemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn RC, Poussin M, Kalota A, Feng Y, Low PS, Dimitrov DS, and Powell DJ Jr. (2015). Targeting of folate receptor beta on acute myeloid leukemia blasts with chimeric antigen receptor-expressing T cells. Blood 125, 3466–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiga A, Lemieux S, Pabst C, Lavallee VP, Bouvier M, Sauvageau G, and Hebert J (2016). Transcriptome analysis of G protein-coupled receptors in distinct genetic subgroups of acute myeloid leukemia: identification of potential disease-specific targets. Blood Cancer J 6, e431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD Jr., van Rooijen N, and Weissman IL (2009). CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138, 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, et al. (2014a). Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 371, 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Shpall EJ, and Grupp SA (2014b). Chimeric antigen receptor T-cell therapy for ALL. Hematology Am Soc Hematol Educ Program 2014, 559–564. [DOI] [PubMed] [Google Scholar]
- McEarchern JA, Oflazoglu E, Francisco L, McDonagh CF, Gordon KA, Stone I, Klussman K, Turcott E, van Rooijen N, Carter P, et al. (2007). Engineered anti-CD70 antibody with multiple effector functions exhibits in vitro and in vivo antitumor activities. Blood 109, 1185–1192. [DOI] [PubMed] [Google Scholar]
- McEarchern JA, Smith LM, McDonagh CF, Klussman K, Gordon KA, Morris-Tilden CA, Duniho S, Ryan M, Boursalian TE, Carter PJ, et al. (2008). Preclinical characterization of SGN-70, a humanized antibody directed against CD70. Clin Cancer Res 14, 7763–7772. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, et al. (2016). Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, and Rosenberg SA (2010). Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther 18, 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst MR, Yang JC, Langan RC, Dudley ME, Nathan DA, Feldman SA, Davis JL, Morgan RA, Merino MJ, Sherry RM, et al. (2011). T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther 19, 620–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkiewicz PJ, Frassle SP, Srivastava S, Sommermeyer D, Hudecek M, Drexler I, Sadelain M, Liu L, Jensen MC, Riddell SR, and Busch DH (2016). Targeted antibody-mediated depletion of murine CD19 CAR T cells permanently reverses B cell aplasia. J Clin Invest 126, 4262–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegram HJ, Lee JC, Hayman EG, Imperato GH, Tedder TF, Sadelain M, and Brentjens RJ (2012). Tumor-targeted T cells modified to secrete IL-12 eradicate systemic tumors without need for prior conditioning. Blood 119, 4133–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna F SM (2016). Myeloid leukemia switch as immune escape from CD19 chimeric antigen receptor (CAR) therapy. Transl Cancer Res 5, S221–S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzitola I, Anjos-Afonso F, Rouault-Pierre K, Lassailly F, Tettamanti S, Spinelli O, Biondi A, Biagi E, and Bonnet D (2014). Chimeric antigen receptors against CD33/CD123 antigens efficiently target primary acute myeloid leukemia cells in vivo. Leukemia 28, 1596–1605. [DOI] [PubMed] [Google Scholar]
- Pulte D, Gondos A, and Brenner H (2008). Improvements in survival of adults diagnosed with acute myeloblastic leukemia in the early 21st century. Haematologica 93, 594–600. [DOI] [PubMed] [Google Scholar]
- Qasim W, Zhan H, Samarasinghe S, Adams S, Amrolia P, Stafford S, Butler K, Rivat C, Wright G, Somana K, et al. (2017). Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med 9. [DOI] [PubMed] [Google Scholar]
- Ravandi F, Estey EH, Appelbaum FR, Lo-Coco F, Schiffer CA, Larson RA, Burnett AK, and Kantarjian HM (2012). Gemtuzumab ozogamicin: time to resurrect? J Clin Oncol 30, 3921–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DS, Neeson PJ, Khot A, Peinert S, Tai T, Tainton K, Chen K, Shin M, Wall DM, Honemann D, et al. (2013). Persistence and efficacy of second generation CAR T cell against the LeY antigen in acute myeloid leukemia. Mol Ther 21, 2122–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere I, and Sadelain M (2017). Chimeric Antigen Receptors: A Cell and Gene Therapy Perspective. Mol Ther 25, 1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. (2015). Cancer immunology Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MC, Kostner H, Gordon KA, Duniho S, Sutherland MK, Yu C, Kim KM, Nesterova A, Anderson M, McEarchern JA, et al. (2010). Targeting pancreatic and ovarian carcinomas using the auristatin-based anti-CD70 antibody-drug conjugate SGN-75. Br J Cancer 103, 676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadelain M (2015). CAR therapy: the CD19 paradigm. J Clin Invest 125, 3392–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadelain M (2016). Chimeric antigen receptors: driving immunology towards synthetic biology. Curr Opin Immunol 41, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadelain M, Riviere I, and Riddell S (2017). Therapeutic T cell engineering. Nature 545, 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Kitamura H, Hijikata A, Tomizawa-Murasawa M, Tanaka S, Takagi S, Uchida N, Suzuki N, Sone A, Najima Y, et al. (2010). Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med 2, 17ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer DR, Savoldo B, Yi Z, Chow KK, Kakarla S, Spencer DM, Dotti G, Wu MF, Liu H, Kenney S, and Gottschalk S (2011). T cells redirected against CD70 for the immunotherapy of CD70-positive malignancies. Blood 117, 4304–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. (2014). Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371, 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR, et al. (2015). Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer Discov 5, 1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassberger V, Gutbrodt KL, Krall N, Roesli C, Takizawa H, Manz MG, Fugmann T, and Neri D (2014). A comprehensive surface proteome analysis of myeloid leukemia cell lines for therapeutic antibody development. J Proteomics 99, 138–151. [DOI] [PubMed] [Google Scholar]
- Sun J, and Sadelain M (2015). The quest for spatio-temporal control of CAR T cells. Cell Res 25, 1281–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liu J, Gao P, Wang Y, and Liu C (2008). Expression of Ig-like transcript 4 inhibitory receptor in human non-small cell lung cancer. Chest 134, 783–788. [DOI] [PubMed] [Google Scholar]
- Tashiro H, Sauer T, Shum T, Parikh K, Mamonkin M, Omer B, Rouce RH, Lulla P, Rooney CM, Gottschalk S, and Brenner MK (2017). Treatment of Acute Myeloid Leukemia with T Cells Expressing Chimeric Antigen Receptors Directed to C-type Lectin-like Molecule 1. Mol Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig DC, Pearce DJ, Simpson C, Rohatiner AZ, Lister TA, Kelly G, Luongo JL, Danet-Desnoyers GA, and Bonnet D (2005). Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood 106, 4086–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, B. B. a. R. D. C. (2016). Tools for General Maximum Likelihood Estimation.
- Turatti F, Figini M, Balladore E, Alberti P, Casalini P, Marks JD, Canevari S, and Mezzanzanica D (2007). Redirected activity of human antitumor chimeric immune receptors is governed by antigen and receptor expression levels and affinity of interaction. J Immunother 30, 684–693. [DOI] [PubMed] [Google Scholar]
- Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, Sommermeyer D, Melville K, Pender B, Budiarto TM, et al. (2016a). CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest 126, 2123–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtle CJ, Hanafi LA, Berger C, Hudecek M, Pender B, Robinson E, Hawkins R, Chaney C, Cherian S, Chen X, et al. (2016b). Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor-modified T cells. Sci Transl Med 8, 355ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Geukes Foppen MH, Goldinger SM, et al. (2015). Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rhenen A, van Dongen GA, Kelder A, Rombouts EJ, Feller N, Moshaver B, Stigter-van Walsum M, Zweegman S, Ossenkoppele GJ, and Jan Schuurhuis G (2007). The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood 110, 2659–2666. [DOI] [PubMed] [Google Scholar]
- Walker AJ, Majzner RG, Zhang L, Wanhainen K, Long AH, Nguyen SM, Lopomo P, Vigny M, Fry TJ, Orentas RJ, and Mackall CL (2017). Tumor Antigen and Receptor Densities Regulate Efficacy of a Chimeric Antigen Receptor Targeting Anaplastic Lymphoma Kinase. Mol Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QS, Wang Y, Lv HY, Han QW, Fan H, Guo B, Wang LL, and Han WD (2015). Treatment of CD33-directed chimeric antigen receptor-modified T cells in one patient with relapsed and refractory acute myeloid leukemia. Mol Ther 23, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wu Z, Liu Y, and Han W (2017). New development in CAR-T cell therapy. J Hematol Oncol 10, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijtens ME, Hart EH, and Bolhuis RL (2000). Functional balance between T cell chimeric receptor density and tumor associated antigen density: CTL mediated cytolysis and lymphokine production. Gene Ther 7, 35–42. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Briau M, Austin P, Faubert A, Boucher G, Chagnon P, Hope K, Girard S, Mayotte N, Landry JR, et al. (2011). RNA-seq analysis of 2 closely related leukemia clones that differ in their self-renewal capacity. Blood 117, e27–38. [DOI] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. (2014). Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587. [DOI] [PubMed] [Google Scholar]
- Wilkie S, van Schalkwyk MC, Hobbs S, Davies DM, van der Stegen SJ, Pereira AC, Burbridge SE, Box C, Eccles SA, and Maher J (2012). Dual targeting of ErbB2 and MUC1 in breast cancer using chimeric antigen receptors engineered to provide complementary signaling. J Clin Immunol 32, 1059–1070. [DOI] [PubMed] [Google Scholar]
- Wu CY, Roybal KT, Puchner EM, Onuffer J, and Lim WA (2015). Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Mihara K, Takei Y, Yanagihara K, Kubo T, Bhattacharyya J, Imai C, Mino T, Takihara Y, and Ichinohe T (2016). All-trans retinoic acid enhances cytotoxic effect of T cells with an anti-CD38 chimeric antigen receptor in acute myeloid leukemia. Clin Transl Immunology 5, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Sotillo E, Harrington C, Wertheim G, Paessler M, Maude SL, Rheingold SR, Grupp SA, Thomas-Tikhonenko A, and Pillai V (2017). Repeated loss of target surface antigen after immunotherapy in primary mediastinal large B cell lymphoma. Am J Hematol 92, E11–E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zah E, Lin MY, Silva-Benedict A, Jensen MC, and Chen YY (2016). T Cells Expressing CD19/CD20 Bispecific Chimeric Antigen Receptors Prevent Antigen Escape by Malignant B Cells. Cancer Immunol Res 4, 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Umikawa M, Cui C, Li J, Chen X, Zhang C, Huynh H, Kang X, Silvany R, Wan X, et al. (2012). Inhibitory receptors bind ANGPTLs and support blood stem cells and leukaemia development. Nature 485, 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, and Levitsky H (2012). Towards curative cancer immunotherapy: overcoming posttherapy tumor escape. Clin Dev Immunol 2012, 124187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic data were submitted (ProteomeXchange Submission) to PRIDE database under the project accession: PXD007552.