Abstract
Objective
Pathogenic variants in KCNB1, encoding the voltage-gated potassium channel KV2.1, are associated with developmental and epileptic encephalopathies (DEE). Previous functional studies on a limited number of KCNB1 variants indicated a range of molecular mechanisms by which variants affect channel function, including loss of voltage sensitivity, loss of ion selectivity, and reduced cell-surface expression.
Methods
We evaluated a series of 17 KCNB1 variants associated with DEE or neurodevelopmental disorder (NDD) to rapidly ascertain channel dysfunction using high-throughput functional assays. Specifically, we investigated the biophysical properties and cell-surface expression of variant KV2.1 channels expressed in heterologous cells using high-throughput automated electrophysiology and immunocytochemistry-flow cytometry.
Results
Pathogenic variants exhibited diverse functional defects, including altered current density and shifts in the voltage-dependence of activation and/or inactivation, as homotetramers or when co-expressed with wild-type KV2.1. Quantification of protein expression also identified variants with reduced total KV2.1 expression or deficient cell-surface expression.
Interpretation
Our study establishes a platform for rapid screening of KV2.1 functional defects caused by KCNB1 variants associated with DEE and other NDDs. This will aid in establishing KCNB1 variant pathogenicity and the mechanism of dysfunction, which will enable targeted strategies for therapeutic intervention based on molecular phenotype.
Keywords: Voltage-gated K+ channel, KCNB1, KV2.1, epilepsy, neurodevelopmental disorder
INTRODUCTION
De novo variants in a diverse range of genes constitute a significant cause of developmental and epileptic encephalopathies (DEE) and other neurodevelopmental disorders (NDD).1,2 Despite an increasing number of genes identified for DEE and increased clinical genetic testing, many variants are classified as variants of uncertain significance (VUS) that are difficult to interpret and/or act on. Obtaining functional data is a powerful approach to bridge the knowledge gap between genetics and molecular pathology3, which can provide additional evidence for interpretation of variant pathogenicity and offer functional information to guide treatment strategies. However, due to the low throughput of many functional assays, functional annotation of variants is a major bottleneck in the field. The development of high-throughput assays is a necessary step for experimental evaluation of the large volume of novel variants being identified by increased clinical genetic testing.
KCNB1 [NM004975] encodes the KV2.1 voltage-gated potassium (K+) channel α-subunit that conducts delayed rectifier K+ current4, a key modulator of membrane repolarization in electrically excitable cells, including various neuron subtypes.5 Kcnb1-deficient mice display enhanced seizure susceptibility and behavioral hyperexcitability, indicating that KV2.1 acts as a homeostatic suppressor of heightened neuronal activity.6 De novo missense variants in KCNB1 are associated with DEE7–13, which includes multiple seizure types, developmental delay (DD), and neuropsychiatric sequelae. Correlational analyses based solely on the substituted amino acid type and position have proven insufficient for genotype-phenotype association11, highlighting the need for experimental studies to determine functional effects of additional variants. To date, seven KCNB1 variants associated with DEE, all located in the pore (S347R, T374I, V378A, G379R, G401R) and voltage-sensor (I199F, R306C) domains, were shown to exhibit altered potassium current density, voltage-dependence, and/or ion selectivity.7–9,14 However, determining the functional consequences of a larger series of variants is necessary to define the range of dysfunction and broaden the associated clinical phenotypes studied.
In this study, we performed high-throughput functional studies of 19 KCNB1 variants (17 missense, 1 frameshift, 1 nonsense) to determine their effect on protein function and provide functional evidence for weighing potential pathogenicity. Several broad categories of channel dysfunction (i.e. altered peak current density, voltage-dependence, protein expression) were identified, and most variants conferred some degree of loss-of-function. Our results provide insight into functional pathogenicity for a large series of KCNB1 variants. This will aid in prioritization for development of more complex experimental models (i.e. knock-in mouse or iPSC) by identifying a subset of representative variants based on underlying pathophysiologic mechanisms. Uncovering a range of KCNB1 functional defects will help define genotype-phenotype relationships by adding a molecular phenotype to the genotypes, and may ultimately enable development of targeted treatment strategies for individuals with KCNB1 DEE.
MATERIALS AND METHODS
Information was collected for individuals with DEE or NDD and a potentially pathogenic KCNB1 variant (identified as part of routine care) under approval of the Ann & Robert H. Lurie Children’s Hospital of Chicago IRB or through collection of de-identified information deemed non-human subjects research by the Northwestern University IRB. Consent for release of de-identified information was obtained in accordance with local institutional policies. In addition, 11 KCNB1 variants were obtained from the literature and variant databases.8–13,15–19 No functional studies were available for 17 of 19 variants.
Plasmids
Full-length human KCNB1 cDNA in plasmid pIRES2-DsRed-MST-KV2.1-WT (Addgene #131707) was described.7,14 For co-transfections, pIRES2-smGFP-KV2.1-WT (Addgene #131709), was engineered by replacing DsRed-MST with smGFP, allowing detection of co-transfection of WT+variant cells (Addgene #131709). For cell-surface labeling, a 2xHA-tag was inserted between a.a.220–221 and the IRES2-DsRed-MST cassette was removed, resulting in pCMV-KV2.1-HA-WT. Variants were introduced using QuikChange mutagenesis (Agilent). Clones were sequenced to confirm desired modifications and absence of unwanted mutations.
Heterologous Expression
Transient expression of WT and variant KCNB1 in CHO-K1 cells (CRL 9618, ATCC) was achieved by electroporation using the Maxcyte STX system (MaxCyte Inc.) with the preset CHO or CHO-PE protocol (7.5–10μg KCNB1 cDNA per 1e7 cells). Following electroporation, cells were grown for 36 hours, harvested, and frozen in liquid N2. Co-expression of WT and variant was achieved by co-electroporation of pIRES2-DsRed-MST-KV2.1-WT or -variant (5 μg) plus pIRES2-smGFP-KV2.1-WT (5 μg).
Cell Preparation for Automated Patch Clamp Recording
Cells were thawed, incubated for 24 hours, and harvested using 0.25% trypsin-EDTA in F-12K media (GIBCO/Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals). Cell number and viability were determined (Vi-Cell, Beckman-Coulter). Cells were diluted to 200,000 cells/mL with external solution (see below) and allowed to recover for 40 minutes at 15°C, shaking at 200 rpm.
Automated Patch Clamp Recording
Automated patch clamp recording was performed using the SyncroPatch 768PE system (Nanion Technologies). External solution contained (in mM): 140 NaCl, 4 KCl, 1 MgCl2, 2 CaCl2, 5 glucose, and 10 HEPES, pH 7.4. Internal solution contained (in mM): 60 KF, 50 KCl, 10 NaCl, 20 EGTA, 10 HEPES, pH 7.2. Pulse generation and data collection used PatchController 384 V1.3.0 and DataController384 V1.2.1 software (Nanion). Whole-cell currents were acquired at 5 kHz and filtered at 1 kHz. Currents were not leak subtracted. Access resistance and apparent membrane capacitance were estimated using built-in protocols. Whole-cell currents were measured at room temperature from a holding potential of −80 mV and elicited with depolarizing steps (500 ms) from −100 to +60 mV (10 mV steps) followed by a 250 ms step to −30 mV or 0 mV (tail-currents). Background currents were removed by digital subtraction of whole-cell currents recorded from non-transfected CHO-K1 cells off-line. Current amplitudes were analyzed from all cells with seal resistance ≥0.5GΩ, series resistance ≤20MΩ and cell capacitance >2pF. Electrically unstable cells with loss of seal or voltage control were excluded. The peak current (Ipeak) was normalized for cell capacitance and plotted against voltage to generate current density–voltage relationships. Voltage-dependence of activation was determined by plotting tail currents normalized to the largest tail current amplitude against the depolarizing test potential (−100 to +60 mV). Normalized G-V curves were fit with the Boltzmann function: G=1/(1+exp[(V-V½)/k] to determine voltage for half-maximal channel activation (V½) and slope factor (k). Voltage-dependence of inactivation was assessed following a 5 second pre-pulse from −100 to +40 mV (10 mV steps) followed by a 250 ms step to +60 mV. Normalized current measured at the +60 mV test potential were plotted against the pre-pulse voltage and fit with the Boltzmann function (I/Imax=1/(1+exp[(V-V½)/k]) to determine voltage for half-maximal channel inactivation (V½) and slope factor (k). Voltage-dependence of channel activation or inactivation were determined only from cells with positive outward current values following background subtraction. Statistical outliers (≥±3 standard deviations) were excluded.
Immunocytochemistry-Flow Cytometry
Cells were electroporated as described above and incubated for 24-h prior to labeling. For analysis of total and cell-surface expression, cells were incubated with αHA-Alexa488 (16B12, Thermo Fisher; 1:250 in FACS buffer (1% FBS, 0.05% NaN3 in PBS, pH 7.4)). Cells were then harvested, and fixed/permeabilized using Fix&Perm buffer (BD Biosciences). Following permeabilization, KV2.1 was detected using Alexa647-conjugated-anti-KV2.1 mouse monoclonal antibody (K89/34, Antibodies Inc.; A20186 ThermoFisher; 1:200 in Perm buffer with 3% NGS). Fluorescence signals were measured by flow cytometry (CytoFlex, Beckman-Coulter) and analyzed with CytExpert (Beckman-Coulter) and FlowJo software (FlowJo LLC). Non-transfected CHO-K1 cells processed identically served as negative controls. Light-scatter based gates (FSC/SSC) were used to exclude non-viable cells and cell debris. A total of 10,000 events were analyzed for each variant per experiment.
For independent analyses of KV2.1 total expression (Fig. 2E&G), frozen aliquots of electroporated cells were thawed, cultured for 24 hours, and then directly harvested with Accutase, without cell-surface labeling. Percent-positive variant-expressing cells were normalized to WT-expressing cells tested on the same day. Histograms were aligned to correct for between-run variability in fluorescence intensity; y-axes were uniformly set across conditions (Fig. 2E&G).
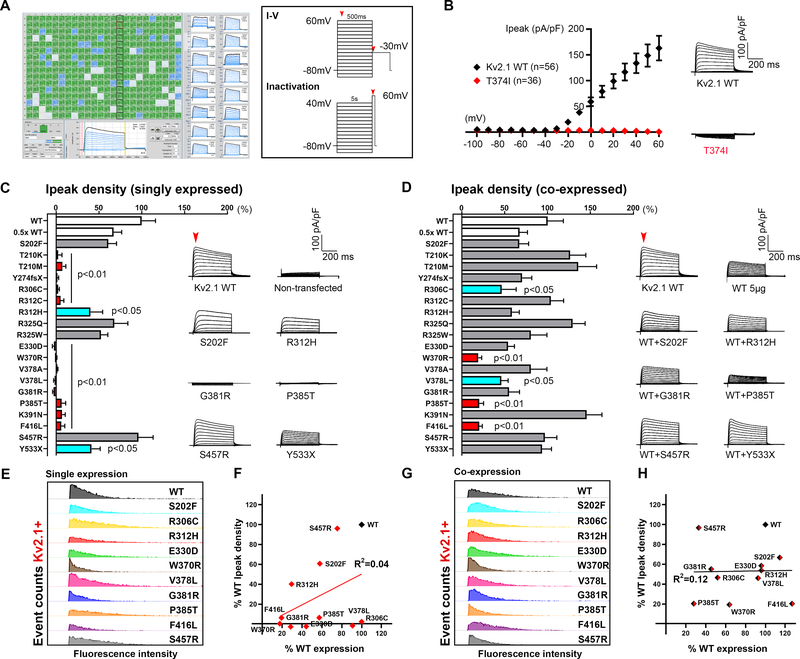
Figure 2. High-throughput functional screening of KV2.1 variants.
A. Screenshot of automated whole-cell current recordings from CHO-K1 cells transiently expressing KV2.1 channels, with 16 individual cell recordings shown on the right blue traces. Voltage protocols used for functional studies are depicted in the box; red arrows indicate time-points of current measurement. B. Validation of automated patch clamp recordings of KV2.1 channels. Averaged KV2.1 WT and T374I variant, (previously published by our group using manual patch clamp) current-voltage relationships and whole-cell current density traces show a similar loss-of-function phenotype for the T374I variant. C-D. Comparison of Ipeak density of KV2.1 variants in homomeric and co-expression (with WT) configuration (red bars p<0.001 vs WT, p<0.05 vs 0.5x WT; turquoise bars p<0.05 vs WT; One-way ANOVA Dunnett’s multiple comparisons). Averaged whole-cell current traces of selected variants are shown on the right; endogenous current recorded from non-transfected CHO-K1 cells were subtracted off-line. Red arrow denotes the time of Ipeak measurement. For co-expression configuration, KV2.1 variants were co-expressed with WT by electroporation of equimolar amount of DNA. E&G. Protein expression analyses of variants by flow cytometry in the homomeric (left) and co-expression (right) configuration confirm KV2.1 protein expression in variant-expressing cells, in particular for the ones with severely reduced current density, albeit with differing degrees of severity. Data are shown in histograms aligned to correct for between-run variability in fluorescence intensity; Y-axes were uniformly set at a maximum of 250 and 350 event counts (for C and E respectively) across conditions. F&H. Correlation of Ipeak density and protein expression by percent-KV2.1 positive cells, in either homomeric or co-expression configuration, did not reach statistical significance (p=0.22 and 0.96 in order; Pearson’s correlation).
Cell-Surface Biotinylation and Immunoblotting
Frozen aliquots of CHO-K1 cells electroporated with WT or variant KCNB1 channels were thawed and grown under the same conditions used for automated patch clamp recording. Cell-surface biotinylation and immunoblotting were performed as previously described7,14, 60–72h post-electroporation, using mouse anti-KV2.1 (1:250; K89/34, Antibodies Inc), mouse anti-transferrin receptor (1:500; H68.4, ThermoFisher), and rabbit anti-calnexin (1:250; H70, Santa Cruz Biotechnology), Alexa Fluor 680-goat anti-rabbit IgG (1:20,000, Jackson ImmunoResearch) and Alexa Fluor 790-goat anti-mouse IgG (1:20,000, Jackson ImmunoResearch) antibodies.
Normalized total, surface, and surface/total protein ratio results were derived from 3 independent experiments. Calnexin immunoreactivity was present in total protein lysates and absent from the cell-surface fraction, confirming the selectivity of biotin labeling for cell-surface protein.
Data Analysis
Data were analyzed using DataController384 V1.2.1 (Nanion), SigmaPlot 2000 (Systat Software, Inc.), GraphPad Prism (GraphPad Software), and OriginPro 2018 (OriginLab). Variants were compared to WT using one-way ANOVA with Dunnett’s post-hoc comparisons or unpaired t-tests. Normalizations and statistical comparisons to WT were conducted per plate (for electrophysiology) or per experiment (for protein expression) to account for potential batch effects. For voltage-dependence in co-expression experiments, obtained parameters (V1/2 or k) could not be subject to percentile normalization (multiple batches); thus, average values of WT and each variant were compared using Brown-Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparisons. Data fits with Boltzmann’s equation were done for each individual cell using non-linear curve fitting in OriginPro2018; solid fit lines represent fits of averaged data (Fig. 5). Whole-cell currents normalized for membrane capacitance are expressed as mean±S.E.M.
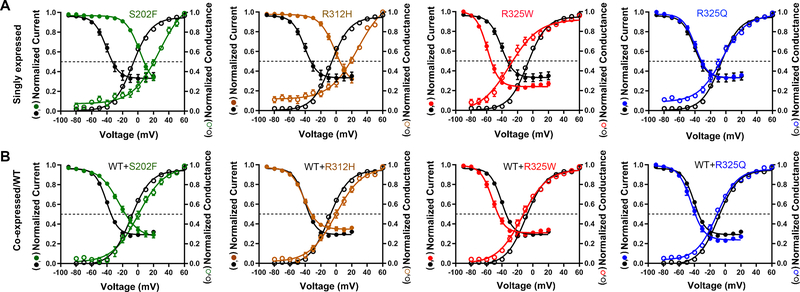
Figure 5. Voltage-dependence of activation/inactivation of KV2.1 variants.
A-B. Voltage-dependent channel activation (open circles) and inactivation (solid circles) curves obtained from selected homomeric and co-expression KV2.1 variants, compared to WT (circles in black). Solid lines represent averaged data fits with the Boltzmann equation. Numerical values of V1/2 and slope factor (k) for the other variants are listed in Table 1.
Principal component analysis (PCA) was performed using ClustVis20, with principal components (PC) calculated using the Nipals algorithm. Six variables (homomeric) were compared: Ipeak density, V1/2 and k for activation and inactivation, cell-surface/total expression ratio; all normalized with respect to WT by percentile-value or Z-score. Spearman non-parametric correlational analysis was performed using the leading variables from PCA with number of seizure types and age of onset.
RESULTS
KCNB1 Variant In Silico Analysis and Clinical Phenotypes
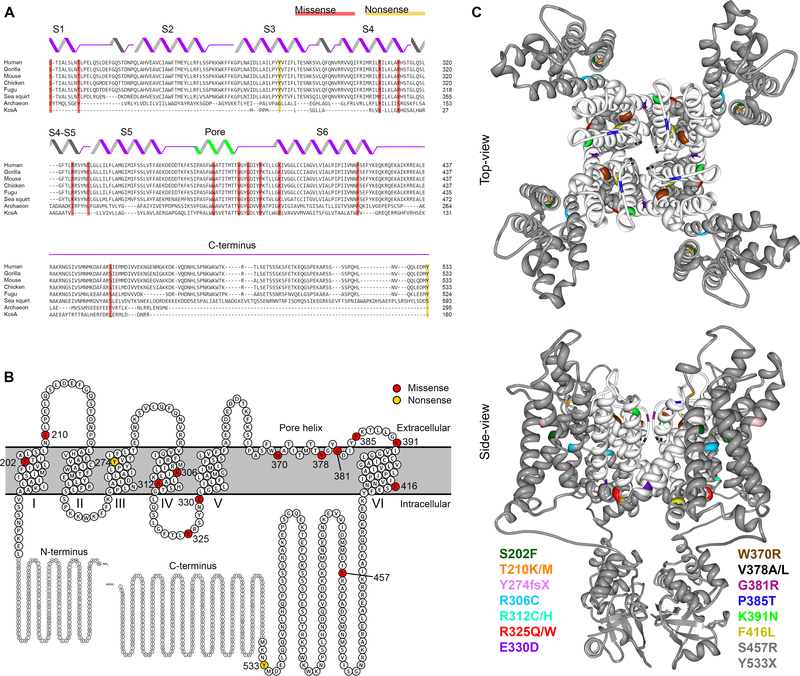
KCNB1 variant residues reported here all show a high degree of evolutionary conservation (Fig. 1A) and are located mainly in the transmembrane domains of KV2.1 (Fig. 1B–C). S202, T210, R306 and R312 are located in the voltage-sensing domain.21 R325 and E330 are in the S4-S5 linker, critical for electromechanical coupling between the voltage-sensing and pore domains.22 W370, P385, K391, and F416 are in the pore domain, with V378 and G381 located within the selectivity filter.23 Truncation variants at Y274 and Y533 are not predicted to undergo nonsense-mediated mRNA decay, as the premature stop codons are located in the last exon, and are likely generate truncated proteins. All disease-associated (pathogenic/likely-pathogenic) variants had a scaled CADD score of >22, indicating they rank among the top 1% of deleterious variants with respect to the all possible SNVs in the reference genome (Supplementary Table 1).24
Figure 1. KCNB1 variants identified in individuals with DEE.
A. Evolutionary conservation of KV2.1 shown by multiple sequence alignment of KV2.1 species orthologues (Clustal Omega 39); secondary structural elements are illustrated above the sequences. KV2.1 variants are shaded in red (missense) and yellow (nonsense). B. Schematic view of the entire KV2.1 subunit and the variants in the membrane: modified from Protter plot of Q14721 (KCNB1_Human)40. C. Locations of variants (color-coded) mapped onto crystal structure of KV2.1/Kv1.2 chimera (PDB2R9R)21: a top-view and a side-view across lipid bilayer. Pore domain is highlighted in white; two variants (S457R and Y533X) are not depicted, because their location in the distal C-terminus is not available in crystal structure.
Clinical phenotypes summarized in Supplementary Table 1 include DEE in 28 patients and NDD in four patients. Among those with NDD, one had a single seizure and three had no history of seizure at last reported follow-up at ≤6 years of age. Phenotypes for novel variants reported in Supplementary Table 1 are consistent with previous reports.7–14 Supplementary Table 1 also includes variants from previous clinically focused reports11,16, summarized here for completeness, with updates when available. In total, there are 17 pathogenic/likely-pathogenic variants from 27 individuals, including several recurrent variants (T210M x3; R306C x6; R312C x2; R312H x2; E330D x 2; V378A x2; P385T x2; F416L x3).
Consequences of KV2.1 variants on current density
To screen for functional consequences of the KCNB1 variants, we recorded whole-cell currents in voltage-clamp mode of CHO-K1 cells transiently expressing wild-type (WT) or variant KV2.1 channels using the SyncroPatch 768PE automated patch clamp platform (Fig. 2A). Previous studies showed feasibility of this approach to study channel properties of KV2.1 and the closely related KV7.1 channel encoded by KCNQ1.14,25,26 In addition, we also tested on this platform the T374I variant that we previously characterized by manual patch clamp recording7, and reproduced the loss-of-function phenotype (Fig. 2B; p<0.001, unpaired t-test @ +60mV vs WT). This further validates use of this platform to screen for KV2.1 dysfunction.
KV2.1 channels are a tetrameric assembly of four alpha subunits encoded by KCNB1. When singly-expressed in CHO cells, the KV2.1 channels form a homomeric configuration of 4 WT or 4 variant subunits. In the homomeric configuration, the majority of the KV2.1 variants (13/19) exhibited low K+ conductance (loss-of-function)(Fig. 2C, red), while the remaining variants displayed moderate K+ conductance (partial loss-of-function)(blue), or K+ conductance similar to WT (grey). The degree of loss-of-function was considered ‘partial’ if peak current (Ipeak) density was reduced 50–70% relative to WT and ‘severe’ if >75%. Averaged whole-cell current density traces for KV2.1 WT and representative variants are displayed to show diversity in current density (Fig. 2C right). Averaged whole-cell Ipeak density traces for all KV2.1 variants are in Figure 4.
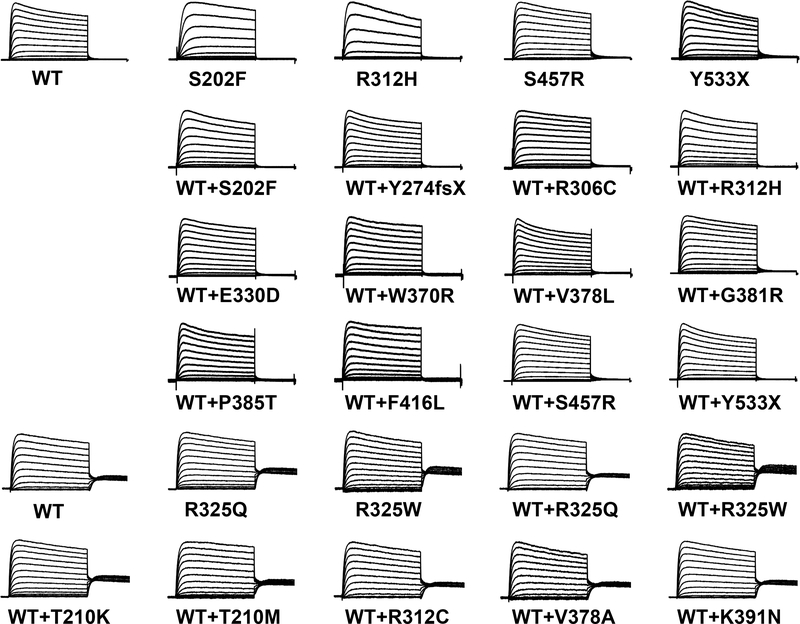
Figure 4. Averaged current density traces normalized to the maximal peak.
Averaged whole-cell currents recorded from CHO-K1 cells transiently expressing homomeric WT or variant KV2.1, or co-expressing WT+variant KV2.1 and normalized to the maximum peak current at +60 mV. Traces were normalized to peak current to enable comparison of gating behaviors among variants with divergent current density. Horizontal scale bar represents 200 ms.
KCNB1 DEE is associated with de novo heterozygous variants. Thus, an individual has KV2.1 subunits produced from both WT and variant alleles. To determine the effect of the KCNB1 variants in a condition resembling heterozygosity in the patient, we co-expressed each variant with WT channels in a 1:1 equimolar ratio. In the co-expression configuration (WT+variant), many variants exhibited Ipeak density similar to cells transfected with WT+WT channels, suggesting these variants can be rescued by WT co-expression. In contrast, W370R, P385T and F416L exhibited significantly lower Ipeak density compared to WT+WT cells (Fig. 2D, bars in red). These variants also showed lower Ipeak density when compared to cells transfected with half the amount of WT plasmid (0.5x WT), suggesting a dominant-negative interaction of the variant with WT subunits. R306C and V378L exhibited partial loss-of-function (bars in turquoise) similar to 0.5x WT. Averaged current traces for a subset of variants are displayed (Fig. 2D, right). Complete statistical information including number of cells and p-values are shown in Supplementary Table 2, and normalized traces are in Fig. 4.
Total protein expression of KV2.1 variants
To test whether loss-of-function phenotypes with impaired current density were due to less channel protein, we evaluated total expression levels for KV2.1 variants by immunoblotting and/or immunocytochemistry-flow cytometry using frozen aliquots of the same cells tested by electrophysiology. In the homomeric configuration, population analyses of KV2.1-positive cells confirmed that KCNB1 variants with severe loss-of-function phenotypes had less KV2.1 protein compared to WT (Fig. 2E&G). The same variants co-expressed 1:1 with WT showed similar results. However, correlational analyses of current density and total protein expression showed no relationship between parameters in either homomeric (R2=0.16, p=0.22, Pearson’s) or co-expression conditions (R2=0.0003, p=0.96)(Fig. 2F&H). Variants not shown in Fig. 2E&G were tested by immunoblotting (Figure 3E–G).
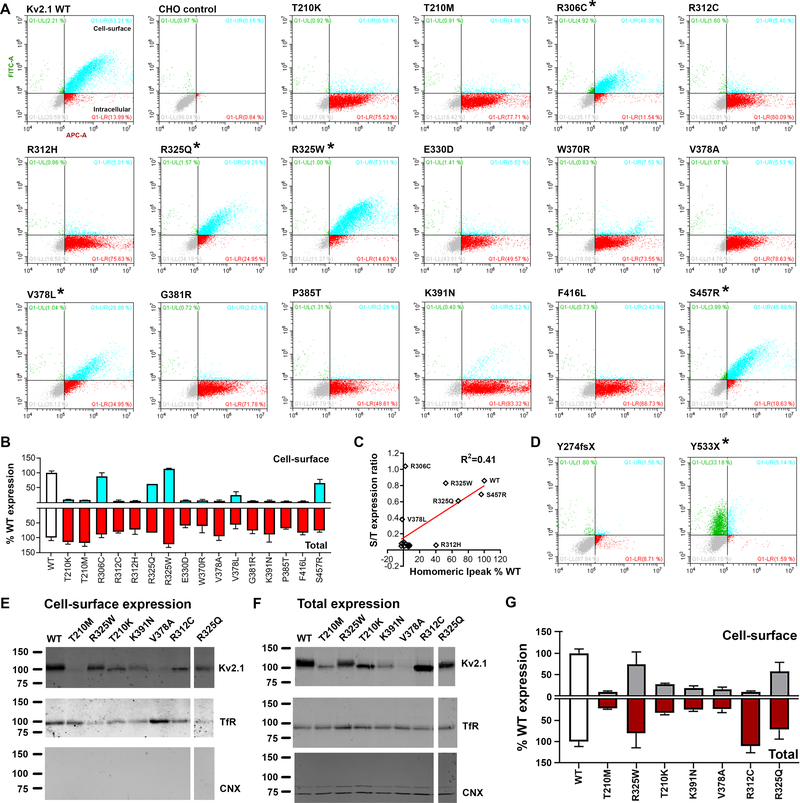
Figure 3. Total and cell-surface protein expression levels of KV2.1 variants.
A. Representative dot-plots of flow cytometry analyses of CHO cells expressing KV2.1 tagged with HA-epitope at the extracellular site between S1-S2 linker regions; anti-HA-Alexa488 and anti-KV2.1-Alexa647 antibodies were used to probe the cell-surface and intracellular KV2.1 respectively. Population in turquoise (HA+/KV2.1+) denote KV2.1 located at the cell-surface, while the dots in red (HA+/KV2.1-) indicate expressed KV2.1 protein located intracellular. WT and non-transfected CHO cells (negative control) are shown at the top left for comparison. Non-transfected CHO cells underwent the entire immunocytochemistry labeling protocol. B. Expression level comparisons of KV2.1 variants to that of WT (white bars) revealed reduced cell-surface trafficking of variants (* denote variants with intact cell-surface trafficking, i.e. S/T ratio > 0.3): cell-surface expression in turquoise and the total expression in red. C. Correlational analyses of surface-to-total expression level ratio and homomeric Ipeak density (values in % WT) result in R2=0.41 and p=0.006: Pearson’s. D. Analyses of two truncation variants, devoid of the epitope site for anti-KV2.1-Alexa647, further validate the assay. The Y274fsX36 variant fails to reach the cell-surface. In contrast, for the Y533X, the population in green (HA+/KV2.1-) denote KV2.1 expressed and reaching the cell-surface, yet unable to be detected by the anti-KV2.1-Alexa647 antibody due to the absent epitope. E-F. Cross-validation analyses of select KV2.1variants for cell-surface and total protein expression; Transferrin receptor (TfR), calnexin (CNX). G. Quantification of western blot analyses confirms similar expression pattern as ICC-FC assay.
Expression analysis of KV2.1 variants: cell-surface trafficking
Presence at the cell-surface is critical for voltage-gated ion channel function, and some variants may interfere with KV2.1 cell-surface expression. Cell-surface and total expression were simultaneously examined using immunocytochemistry-flow cytometry (ICC-FC). An extracellular HA-epitope tag enabled discrimination of cell-surface and total KV2.1 fractions, allowing rapid determination of cell-surface expression. A substantial population of WT KV2.1 reached the cell-surface, with a mean surface/total expression ratio >0.8 (Fig. 3A–B), indicating highly efficient cell-surface trafficking as previously shown.29,30 The majority of the variants (13/19) exhibited deficits in cell-surface expression of KV2.1, potentially explaining loss of K+ conductance observed by electrophysiology (Fig. 2C). The non-pathogenic variants (R325Q, S457R) exhibited cell-surface and total expression indistinguishable from WT (Fig. 3A–C). Cross-validation of select variants (T210K/M, R312C, R325Q/W, V378A, K391N) with cell-surface biotinylation followed by immunoblotting produced largely congruent results (Fig. 3E–G). The observed reduction in protein expression for some variants (Fig. 3E–G) that was not observed by ICC-FC result is likely due to the time difference at which the proteins were analyzed. It is possible that during the extended time (extra 24–36h compared to ICC-FC experiment), variants may have triggered misfolded protein responses, which may slow down protein translation or induce cell death.
S202F located near the HA-epitope site (a.a.220) likely had altered immunoreactivity considering the lack of anti-HA signal, but intact current density of S202F. The substituted phenylalanine may result in steric hindrance, preventing HA antigen-antibody interaction. For the T210K/M variants, also located nearby the HA-tag site, immunoblotting of non-tagged T210K/M confirmed lower cell-surface expression (Fig. 3A–B, Fig. 3E–G), consistent with loss-of-function phenotypes in homomeric configuration (Fig. 2C).
The two truncation variants, Y274fsX36 and Y533X, were also tested for expression, despite lacking the epitope site for anti-Kv2.1-Alexa647 (a.a.837–853). The Y274fsX variant had no cell-surface HA signal (Fig. 3D). However, in a subsequent experiment, permeabilization prior to anti-HA incubation revealed abundant total expression, suggesting a trafficking defect for Y274fsX36. Y533X exhibited cell-surface HA immunoreactivity (Fig. 3D), indicating that truncated Y533X proteins reach the plasma membrane.
Voltage-dependence of channel activation and inactivation
Voltage-dependence of channel activation and inactivation was assessed for KV2.1 variants with sufficient K+ conductance to reliably measure these parameters. In the homomeric configuration, several variants exhibited altered voltage-dependence of channel activation and inactivation when compared WT (Table 1). S202F and R312H exhibited depolarizing shifts in their V1/2 for both channel activation and inactivation (Fig. 5A) and larger slope factors, suggesting a decrease in voltage-sensitivity for channel inactivation. Y533X induced a depolarizing shift in V1/2 of activation without affecting channel inactivation (Table 1). In contrast, R325W caused hyperpolarizing shifts in the V1/2 values for both channel activation and inactivation (Fig. 5A).
Table 1. Biophysical properties of Kv2.1 variants singly or co-expressed with WT.
Singly expressed variants with at least 10pA/pF are included. Values that differ from the respective WT condition are shown in red (Dunnett’s T3 multiple comparisons), and n = the number of cells recorded.
| Configurations or variants | Voltage-dependence of activation | Voltage-dependence of inactivation | ||
|---|---|---|---|---|
| V½ | k | V½ | k | |
| WT | −4.6 ± 1.0 | 13.0 ± 0.4 | −35.5 ± 2.6 | 7.5 ± 0.6 |
| S202F |
24.3 ± 3.8 p<0.001 (n=14) |
16.4 ± 1.7 p>0.189 (n=14) |
2.7 ± 1.2 p<0.001 (n=30) |
8.8 ± 0.4 p<0.001 (n=30) |
| R312H |
28.1 ± 1.5 p<0.001 (n=21) |
18.2 ± 1.0 p<0.001 (n=21) |
−0.1 ± 2.3 p<0.001 (n=18) |
8.5 ± 0.7 p<0.001 (n=18) |
| R325Q | −5.0 ± 2.5 p>0.884 (n=18) |
14.8 ± 1.2 p>0.804 (n=18) |
−39.6 ± 1.8 p> 0.704 (n=17) |
7.1 ± 0.4 p>0.083 (n=17) |
| R325W |
−23.4 ± 3.9 p<0.001 (n=22) |
17.7 ± 1.2 p>0.033 (n=22) |
−58.0 ± 1.4 p<0.001 (n=25) |
5.7 ± 0.2 p<0.001 (n=25) |
| S457R | −13.3 ± 1.4 p>0.050 (n=25) |
12.0 ± 0.7 p>0.952 (n=25) |
−44.2 ± 1.7 p>0.543 (n=38) |
6.5 ± 0.2 p>0.418 (n=38) |
| Y533X |
13.0 ± 3.6 p<0.001 (n=17) |
18.3 ± 1.4 p<0.001 (n=17) |
−45.7 ± 2.6 p>0.313 (n=26) |
6.8 ± 0.6 p>0.200 (n=26) |
| WT+WT | −7.3 ± 0.9 | 12.9 ± 0.9 | −39.6 ± 0.6 | 6.3 ± 0.1 |
| WT+S202F | 1.3 ± 3.1 p>0.184 (n=25) |
22.2 ± 0.8 p<0.001 (n=25) |
−29.6 ± 1.7 p<0.001 (n=54) |
10.4 ± 0.4 p<0.001 (n=54) |
| WT+T210K | −10.8 ± 1.3 p>0.395 (n=44) |
13.1 ± 0.5 p>0.999 (n=44) |
−46.2 ± 1.3 p<0.001 (n=34) |
6.3 ± 0.2 p>0.999 (n=34) |
| WT+T210M | −6.9 ± 1.7 p>0.999 (n=29) |
12.9 ± 0.9 p>0.999 (n=29) |
−43.3 ± 2.4 p>0.917 (n=25) |
6.5 ± 0.3 p>0.996 (n=25) |
| WT+Y274fsX | −7.1 ± 4.0 p>0.999 (n=18) |
14.5 ± 1.1 p>0.949 (n=18) |
−40.2 ±1.6 p>0.999 (n=65) |
5.7 ± 0.2 p>0.380 (n=65) |
| WT+R306C | −15.9 ± 3.7 p>0.435 (n=11) |
15.3 ± 2.2 p>0.989 (n=11) |
−44.9 ± 3.2 p>0.847 (n=22) |
9.0 ± 0.6 p<0.003 (n=22) |
| WT+R312C | −9.1 ± 1.3 p>0.989 (n=35) |
13.4 ± 0.5 p>0.999 (n=35) |
−42.0 ± 1.3 p>0.850 (n=27) |
6.0 ± 0.1 p>0.946 (n=27) |
| WT+R312H | 0.1 ± 2.7 p>0.198 (n=31) |
14.3 ± 0.7 p>0.776 (n=31) |
−38.7 ± 1.5 p>0.999 (n=78) |
7.7 ± 0.4 p>0.010 (n=78) |
| WT+R325Q | −10.7 ± 1.4 p>0.527 (n=43) |
12.8 ± 0.6 p>0.999 (n=43) |
−43.4 ± 1.7 p>0.531 (n=32) |
5.9 ± 0.2 p>0.957 (n=32) |
| WT+R325W |
−14.3 ± 1.7 p<0.011 (n=39) |
16.1 ± 1.0 p>0.077 (n=39) |
−51.9 ± 1.5 p<0.001 (n=35) |
5.9 ± 0.2 p>0.957 (n=35) |
| WT+E330D | −12.4 ± 2.3 p>0.529 (n=18) |
15.3 ± 1.4 p>0.825 (n=18) |
−40.4 ± 2.3 p>0.999 (n=52) |
7.4 ± 1.0 p>0.987 (n=52) |
| WT+W370R | −3.5 ± 3.3 p>0.986 (n=12) |
16.4 ± 1.8 p>0.663 (n=12) |
−38.9 ± 2.9 p>0.999 (n=26) |
6.9 ± 0.4 p>0.813 (n=26) |
| WT+V378A | −7.5 ± 1.4 p>0.999 (n=29) |
13.3 ± 0.5 p>0.999 (n=29) |
−41.4 ± 1.2 p>0.976 (n=32) |
5.6 ± 0.2 p>0.159 (n=32) |
| WT+V378L | −5.8 ± 4.8 p>0.999 (n=20) |
16.0 ± 1.2 p>0.298 (n=20) |
−46.4 ± 1.9 p>0.022 (n=61) |
7.5 ± 0.3 p<0.003 (n=61) |
| WT+G381R | −9.4 ± 2.7 p>0.999 (n=27) |
14.2 ± 1.0 p>0.916 (n=27) |
−41.2 ±1.4 p>0.995 (n=62) |
6.3 ± 0.5 p>0.999 (n=62) |
| WT+P385T | −6.7 ± 4.1 p>0.999 (n=11) |
14.9 ± 1.2 p>0.850 (n=11) |
−40.6 ± 2.1 p>0.999 (n=43) |
6.9 ± 0.5 p>0.958 (n=43) |
| WT+K391N | −9.4 ± 1.2 p>0.947 (n=44) |
12.6 ± 0.4 p>0.999 (n=44) |
−41.4 ± 1.2 p>0.977 (n=39) |
5.9 ± 0.1 p>0.492 (n=39) |
| WT+F416L | −3.1 ± 3.7 p>0.988 (n=16) |
15.6 ± 1.1 p>0.388 (n=16) |
−39.5 ± 1.6 p>0.999 (n=48) |
7.3 ± 0.4 p>0.175 (n=48) |
| WT+S457R |
−18.9 ± 3.0 p<0.019 (n=22) |
14.1 ± 1.0 p>0.989 (n=22) |
−42.0 ± 1.6 p>0.960 (n=42) |
5.9 ± 0.2 p>0.959 (n=42) |
| WT+Y533X | −11.6 ± 1.7 p>0.384 (n=42) |
14.8 ± 0.8 p>0.468 (n=42) |
−43.0 ± 1.4 p>0.432 (n=65) |
6.0 ± 0.2 p>0.998 (n=65) |
When co-expressed with WT, the effects of R312H on channel activation and inactivation were abolished, suggesting rescue by WT subunits. Co-expression with WT channels prevented the depolarizing shift in activation due to S202F, but did not alter the effects on inactivation (Fig. 5B, Table 1). Hyperpolarizing shifts in V1/2 for activation and inactivation caused by R325W were still evident in the presence of WT, albeit with smaller magnitude. Although T210K was non-functional as homotetramer, it caused a hyperpolarizing shift in inactivation V1/2 when co-expressed with WT. A complete summary is presented in Table 1.
Functional effects of non-pathogenic variants
Two likely non-pathogenic variants (R325Q, S457R) were included in functional assays, with experimenters blinded to this status. R325Q was present as a singleton variant (MAF 3.983e−06) in the genome aggregation database (gnomAD) that is devoid of individuals with severe pediatric disease.27 R325Q was indistinguishable from WT in contrast to the disease-associated R325W variant that induced changes in both channel activation and inactivation, (Fig. 5A–B).
S457R was classified as non-pathogenic based on inheritance from an unaffected parent and presence of S457R as a singleton variant (MAF 3.983e−06) in gnomAD.27 In the homomeric configuration, S457R was indistinguishable from WT. However, there was a hyperpolarizing shift in voltage-dependence of activation when S457R was co-expressed with WT. S457 is subject to post-translational phosphorylation, and dephosphorylation of KV2.1 is associated with hyperpolarizing shifts in voltage-dependent gating.28 The observed difference in voltage-dependence of activation may reflect differences in phosphorylation state between the variant (non-phosphorylated residue 457) and WT subunits (phosphorylated residue 457) under our experimental conditions, which require intracellular sodium fluoride, a phosphatase inhibitor, for gigaohm seal formation.
Genotype-Phenotype analyses of KV2.1 variants
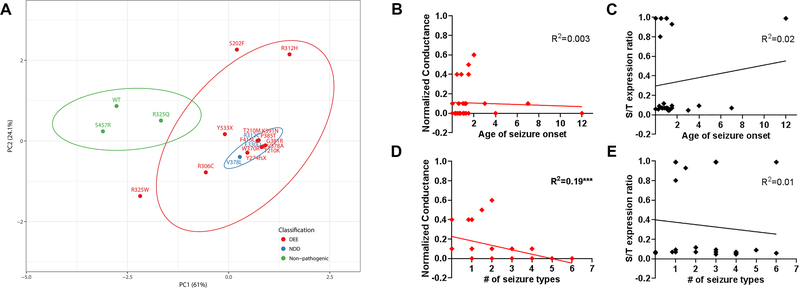
To visualize the relationship among various parameters, we performed principal component analyses (PCA) using 6 variables measured in the homomeric configuration: Ipeak density, V1/2 and k for activation and inactivation, and cell-surface/total expression ratio (Fig. 6). The top-3 leading dimensions for PC1 were Ipeak density (0.61), cell-surface/total expression ratio (0.62), and V1/2 for activation (−0.28); and for PC2 were Ipeak density (0.62), and inactivation V1/2 (0.47) and k (0.49). Plotting PC1 vs. PC2 separated nonpathogenic (R325Q, S457R, WT) from pathogenic variants with 95% confidence. However, it was not possible to separate the pathogenic variants by clinical classification.
Figure 6. Principal component analyses of KV2.1 variants.
A. Experimental data for biophysical and protein expression analyses were transformed with respect to WT, to generate eigenvector-based multivariate analyses, and plotted using ClustVis 20. Nipals PCA is used to calculate principal components, with ellipses depicting 95% confidence interval. Each axes explains 61% and 24.1% of the total variance (X and Y, in order): N = 20 data points. Total of 6 variables measured in the homomeric condition were used: Ipeak density, V1/2 and k for activation and inactivation, protein expression cell-surface/total ratio. The top 3 leading dimensions (variable) for PC1 were Ipeak density (0.61), cell-surface/total expression ratio (0.62), V1/2 for activation (−0.28); and for PC2 were Ipeak density (0.62), and inactivation V1/2 (0.47) and k (0.49). Nonpathogenic variants and WT (in green) are separable from the pathogenic variants (red, blue). DEE, developmental epileptic encephalopathy; NDD, neurodevelopmental disorder. B-E. Correlational analysis of quantifiable clinical parameters and the leading functional parameters identified in this study. Conductance values normalized to that of WT and surface-to-total protein expression ratio were used for analyses. R2 values are noted for each plot (***p<0.02, Spearman’s).
In addition, we performed correlational analyses with quantifiable clinical phenotypes and the leading variables identified by PCA: conductance and cell-surface/total expression ratio (Fig. 6B–E). Among these parameters, we identified a single correlation between the number of seizure types and conductance (R2=0.19; p<0.02, Spearman’s), with low/no conductance associated with a more complex clinical presentation that include multiple seizure types. Conversely, variants with some level of preserved conductance (>0.2 of WT) had only 1–2 seizures types, indicative of a less complex clinical presentation.
DISCUSSION
KV2.1 loss-of-function in KCNB1 DEE
Here we report functional studies of 19 KCNB1 variants, including 17 likely-pathogenic variants identified in individuals with DEE or NDD, and two presumed non-pathogenic variants present in the gnomAD database that excludes severe pediatric disease.27 High-throughput electrophysiological and biochemical analysis of KV2.1 variants expressed in CHO-K1 cells revealed partial or near-complete loss-of-function as a common mechanism, consistent with prior manual electrophysiology.7–9 The molecular mechanisms underlying loss-of-function phenotypes included lower Ipeak density due to low total and/or cell-surface KV2.1 expression, or shifts in voltage-dependence of activation and inactivation.
The majority of variants exhibited significantly lower Ipeak density when expressed as homotetramers. However, co-expression with WT subunits normalized Ipeak density for many variants. Rescue by WT subunits suggest that variant subunits traffic to the cell-surface as part of a heteromeric assembly, raising the possibility for rescue by molecular chaperones. In contrast, co-expression of some variants (R306C, V378L) with WT resulted in lower Ipeak density approximating 0.5x WT, suggesting that while the mutant subunits cannot be rescued, they do not interfere with cell-surface expression of WT. Co-expression of other variants (W370R, P385T, F416L) with WT resulted in Ipeak density less than 0.5x WT, suggesting a dominant-negative effect on cell-surface expression of WT subunits.
Several variants encoded functional KV2.1 channels with Ipeak density indistinguishable from WT, but with defects in voltage-dependence. Depolarizing shifts in V1/2 for both activation and inactivation were observed for S202F and R312H, resulting in loss-of-function due to impaired channel opening in a physiological voltage range. Conversely, R325W had hyperpolarized shifts in V1/2 for activation and inactivation. Although this likely results in enhanced channel opening at physiological voltages, the corresponding shift in the voltage-dependence of inactivation limits channel availability, suggesting overall loss-of-function. Co-expression of WT channels to approximate heterozygosity resulted in attenuated or altered voltage-dependence. This may be explained by variable populations of WT+WT, WT+var, and var+var channels. The presence of significant voltage-dependence phenotypes specific to co-expression conditions (activation k for WT+S202F or activation V1/2 for WT+S457R) suggests that some variants co-assemble with WT subunits as heterotetramers.
Comparison of same-site substitutions
The importance of experimental validation is highlighted by pairwise comparison of substitutions of different amino acids at the same position (T210K/M, R312C/H, R325Q/W and V378A/L). The T210K/M variants resulted in loss of K+ conductance and cell-surface KV2.1 expression. T210 is a critical residue forming the S1-pore interface.31,32 A hydroxyl group is required at this position for channel function.32 Thus, substitution with either lysine or methionine would be incompatible with channel function. In contrast, R312H and R312C resulted in partial and severe loss-of-conductance, respectively, which was opposite of the in silico consequence prediction that suggested R312H would be more deleterious than R312C (Supplementary Table 1). Retaining a positive charge at this position in the S4 segment by substituting histidine may lessen severity relative to a charge-neutralization cysteine substitution. Similarly, the R325Q/W variants differed in their voltage-dependence, with R325Q retaining WT-like behavior, consistent with non-pathogenicity. This suggests that substitution of R325 with a bulky hydrophobic tryptophan may alter electrochemical coupling between S4-S5 and the S6 segment33, while glutamine is tolerated.
Both the V378A/L variants resulted in loss of K+ conductance in the absence of WT subunits. V378L exhibited modest cell-surface expression, while the V378A variant was absent, suggesting discrete mechanisms underlying loss of K+ conductance for these variants. Leucine is present at the equivalent position in KV3 and KV4 channels, indicating this substitution is structurally tolerable and maintains K+ selectivity, although it likely alters conductance.34,35 Conversely, substitution with alanine has been shown to disrupt ion selectivity and affect channel stability.35 Previous studies of the V378A variant in CHO-K1 and COS-7 cells showed impaired expression that could be rescued by the KV2 inhibitor GxTx, suggesting enhanced internalization and degradation as a possible mechanism.9 Consistent with this, we observed robust total expression of V378A at 24 hours post-transfection (Fig. 3), while there was little V378A expression at 72 hours (Fig. 3E–G).
High-throughput variant characterization pipeline
One goal of this study was to optimize electroporation and automated planar patch clamp technologies for high-throughput characterization of KCNB1 variants identified by clinical genetic testing, making it feasible to rapidly characterize new variants. Moreover, the immunocytochemistry-flow-cytometry assay enables high-throughput evaluation of cell-surface and total protein expression for further delineation of pathogenic mechanisms. Efficient functional characterization of ion channel variants is critical to realize the precision medicine goal of rapid, definitive precision diagnosis and targeted treatment based on the specific genetic variant.
Utility of functional characterization for clinical genetic testing
Based on ACMG guidelines, evidence of a deleterious effect on protein function can be considered as ‘strong’ evidence for pathogenicity, given the functional assay is robust and validated.3 Electrophysiology is the gold standard for characterizing the effects of ion channel variants, and is a well-validated, robust approach. In contrast, in silico evidence based on pathogenicity prediction algorithms are only considered ‘supporting’ evidence, thus insufficient when considered alone. Functional variant characterization, both in channelopathies and other disorders, has untapped potential to provide diagnostic benefit for weighing variant pathogenicity. Functional characterization of a series of KCNB1 variants can serve as a reference dataset for genetic test interpretation in newly diagnosed patients, and provide additional evidence for re-classification of variants previously classified as variants of uncertain significance.
The KCNB1 variants R325Q and S457R provide examples of how functional variant characterization can provide useful information. R325Q is a rare population variant that exhibited similar functional characteristics to WT channels, demonstrating that glutamine substitution at R325 is tolerated. However, R325W had significant functional defects as a homotetramer and when co-expressed with WT (Fig. 2C&D), supporting this variant as potentially pathogenic. S457R was initially identified by clinical genetic testing in a child with DEE, but was later found to be inherited from an unaffected parent. Prior to parental testing, we performed electrophysiological characterization of S457R and found no deleterious effect on protein function, consistent with non-pathogenicity. Principal component analysis allowed reliable separation of the non-pathogenic group (R325Q, S457R, WT) from the other 17 variants studied, highlighting the viability of functional screening to differentiate nonpathogenic versus pathogenic variants with high confidence (95%).
Genotype-Phenotype Correlations
A goal of our study was to assess potential correlations between KV2.1 channel dysfunction and clinical severity. Although the clinical phenotypes of DEE and NDD were not distinguished by PCA, it is possible that the NDD cases (3–6 years-of-age at the time of reporting) will go on to develop epilepsy, as there are at least two DEE cases with seizure onset in adolescence (Supplementary Table 1). We identified a correlation between conductance and the number of seizure types, indicating that variants with preserved functionality are associated with less complex epilepsy. However, due to the higher likelihood of genetic testing in complex epilepsy cases, our cohort is biased toward the more severe end of the spectrum. Characterization of additional variants across a broad clinical spectrum may help establish clearer genotype-phenotype correlations. Alternatively, the underlying pathogenic mechanisms may require more complex experimental systems, and/or final expression of the clinical phenotype can be influenced by other genetic, epigenetic or environmental modifiers. The variable severity observed among patients with the same recurrent variant is consistent with other factors contributing to clinical presentation.
Limitations of high-throughput functional screening
Despite the aforementioned strengths of functional evaluation in a heterologous expression model, there are some limitations. Some KV2.1 variant effects detected in our study may become more or less pronounced in neurons. For example, variant and WT alleles are co-expressed in individuals with KCNB1 DEE, which is associated with heterozygous variants. However, in CHO-K1 cells co-expression with WT sometimes masked variant effects. This highlights a difference between heterologous expression versus the nervous system and suggests that homomeric expression provides a more robust assay. In neurons, a population of non-conducting KV2.1 clustered at plasma membrane-endoplasmic reticulum junctions operate as a docking site for membrane proteins30, a function that is not assessed in our system. In addition, the effect of variants that alter binding site affinity for interacting proteins like KChAP, AMIGO, and KCNE1–336–38 may not be detected if interacting proteins are not present at relevant levels in CHO-K1 cells. Hence, additional characterization of variants in more complex experimental systems will be necessary for complete understanding of disease mechanisms in neurons and other specialized cells where KV2.1 is a significant contributor.
Conclusions
Efficient characterization of KCNB1 variants can provide the precision diagnoses required for precision medicine. Many of the disease-associated KCNB1 variants (14/17) exhibited phenotypes that ultimately result in a channel hypofunction, through 1) decreased K+ conductance, 2) altered voltage-dependence, 3) reduced protein expression, or 4) altered cell-surface trafficking. Classification of KCNB1 variants into categories of defects suggests that molecular chaperones to increase cell-surface expression may be a suitable therapeutic strategy for some, while subtype-selective activators of KV2.1 may be a viable therapy for others. Alternatively, antisense oligonucleotide or gene therapy approaches that increase expression may hold promise for treatment of KCNB1 DEE.
Supplementary Material
1) Supplementary Table 1. Clinical features of individuals with KCNB1 missense variants. Heterozygous de novo variants were reported as pathogenic or likely pathogenic, with the exception of a single variant determined to be non-pathogenic following parental testing (grey shading).
2) Supplementary Table 2. Number of cells and p-values for the variant channel peak current density in comparison to that of WT.
ACKNOWLEDGEMENTS
This work was supported by funding from NIH/NINDS grants R01 NS053792 (J.A.K.), U54 NS108874 (A.L.G.), K08 NS097633 (E.M.G.), the Ann & Robert H. Lurie Children’s Hospital of Chicago Precision Medicine Initiative (J.J.M., J.B.O., S.N.M.) and Pediatric Physician-Scientist Research Award (S.N.M.). AT is supported by The Scripps Translational Science, an NIH-NCATS Clinical and Translational Science Award (CTSA; 5 UL1 TR001114). AP was supported by the Translational Research Program, Boston Children’s Hospital. SKK is supported by a pre-doctoral fellowship from the American Epilepsy Society. We thank the patients and their families for their cooperation, and Tatiana Abramova, Reshma Desai, Nicole Hawkins, Tyler Thaxton, and Alex Huffman for technical assistance.
Footnotes
POTENTIAL CONFLICTS OF INTEREST
Dianalee McKnight is employed by GeneDx, Inc., a wholly-owned subsidiary of OPKO Health, Inc., that provides genetic testing that includes KCNB1 on epilepsy and ASD/ID panels. The other authors report no competing interests related to this study.
REFERENCES
- 1.Allen AS, Berkovic SF, Cossette P, et al. De novo mutations in epileptic encephalopathies. Nature. 2013. September 12;501(7466):217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvill GL, Mefford HC. Next-Generation Sequencing in Intellectual Disability. J Pediatr Genet. 2015. September;4(3):128–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015. May;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murakoshi H, Trimmer JS. Identification of the Kv2.1 K+ channel as a major component of the delayed rectifier K+ current in rat hippocampal neurons. J Neurosci. 1999. March 01;19(5):1728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop HI, Guan D, Bocksteins E, et al. Distinct Cell- and Layer-Specific Expression Patterns and Independent Regulation of Kv2 Channel Subtypes in Cortical Pyramidal Neurons. J Neurosci. 2015. November 04;35(44):14922–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speca DJ, Ogata G, Mandikian D, et al. Deletion of the Kv2.1 delayed rectifier potassium channel leads to neuronal and behavioral hyperexcitability. Genes Brain Behav. 2014. April;13(4):394–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torkamani A, Bersell K, Jorge BS, et al. De novo KCNB1 mutations in epileptic encephalopathy. Ann Neurol. 2014. 10/;76(4):529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saitsu H, Akita T, Tohyama J, et al. De novo KCNB1 mutations in infantile epilepsy inhibit repetitive neuronal firing. Sci Rep. 2015. October 19;5:15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiffault I, Speca DJ, Austin DC, et al. A novel epileptic encephalopathy mutation in KCNB1 disrupts Kv2.1 ion selectivity, expression, and localization. J Gen Physiol. 2015. November;146(5):399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latypova X, Matsumoto N, Vinceslas-Muller C, Bezieau S, Isidor B, Miyake N. Novel KCNB1 mutation associated with non-syndromic intellectual disability. J Hum Genet. 2017. April;62(5):569–73. [DOI] [PubMed] [Google Scholar]
- 11.de Kovel CGF, Syrbe S, Brilstra EH, et al. Neurodevelopmental Disorders Caused by De Novo Variants in KCNB1 Genotypes and Phenotypes. JAMA Neurol. 2017. August 14;74(10):1228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen NM, Conroy J, Shahwan A, et al. Unexplained early onset epileptic encephalopathy: Exome screening and phenotype expansion. Epilepsia. 2016. January;57(1):e12–7. [DOI] [PubMed] [Google Scholar]
- 13.Miao P, Peng J, Chen C, Gai N, Yin F. [A novel mutation in KCNB1 gene in a child with neuropsychiatric comorbidities with both intellectual disability and epilepsy and review of literature]. Zhonghua Er Ke Za Zhi. 2017. February 2;55(2):115–9. [DOI] [PubMed] [Google Scholar]
- 14.Calhoun JD, Vanoye CG, Kok F, George AL Jr., Kearney JA. Characterization of a KCNB1 variant associated with autism, intellectual disability, and epilepsy. Neurol Genet. 2017. December;3(6):e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald TW, Gerety SS, Jones WD, et al. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015. March 12;519(7542):223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marini C, Romoli M, Parrini E, et al. Clinical features and outcome of 6 new patients carrying de novo KCNB1 gene mutations. Neurol Genet. 2017. December;3(6):e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018. January 4;46(D1):D1062–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016. August 18;536(7616):285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaminathan GJ, Bragin E, Chatzimichali EA, et al. DECIPHER: web-based, community resource for clinical interpretation of rare variants in developmental disorders. Human molecular genetics. 2012. October 15;21(R1):R37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015. July 1;43(W1):W566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007. November 15;450(7168):376–82. [DOI] [PubMed] [Google Scholar]
- 22.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science (New York, NY). 2005. August 5;309(5736):903–8. [DOI] [PubMed] [Google Scholar]
- 23.Taglialatela M, Drewe JA, Brown AM. Barium blockade of a clonal potassium channel and its regulation by a critical pore residue. Molecular pharmacology. 1993. July;44(1):180–90. [PubMed] [Google Scholar]
- 24.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014. March;46(3):310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanoye CG, Desai RR, Fabre KL, et al. High-Throughput Functional Evaluation of KCNQ1 Decrypts Variants of Unknown Significance. Circ Genom Precis Med. 2018. November;11(11):e002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, Kuenze G, Smith JA, et al. Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations. Sci Adv. 2018. March;4(3):eaar2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karczewski KJ, Francioli LC, Tiao G, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019:531210. [Google Scholar]
- 28.Cerda O, Baek JH, Trimmer JS. Mining recent brain proteomic databases for ion channel phosphosite nuggets. J Gen Physiol. 2011. January;137(1):3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox PD, Loftus RJ, Tamkun MM. Regulation of Kv2.1 K(+) conductance by cell surface channel density. J Neurosci. 2013. January 16;33(3):1259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deutsch E, Weigel AV, Akin EJ, et al. Kv2.1 cell surface clusters are insertion platforms for ion channel delivery to the plasma membrane. Mol Biol Cell. 2012. August;23(15):2917–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Full Y, Seebohm G, Lerche H, Maljevic S. A conserved threonine in the S1-S2 loop of KV7.2 and K V7.3 channels regulates voltage-dependent activation. Pflugers Archiv : European journal of physiology. 2013. June;465(6):797–804. [DOI] [PubMed] [Google Scholar]
- 32.Lee SY, Banerjee A, MacKinnon R. Two separate interfaces between the voltage sensor and pore are required for the function of voltage-dependent K(+) channels. PLoS biology. 2009. March 3;7(3):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betts MJRR. Amino acid properties and consequences of substitutions In: Gray MRBaIC, editor. Bioinformatics for Geneticists: John Wiley & Sons, Ltd; 2003. [Google Scholar]
- 34.Brown AM, Drewe JA, Hartmann HA, et al. The potassium pore and its regulation. Annals of the New York Academy of Sciences. 1993. December 20;707:74–80. [DOI] [PubMed] [Google Scholar]
- 35.Heginbotham L, Lu Z, Abramson T, MacKinnon R. Mutations in the K+ channel signature sequence. Biophysical journal. 1994. April;66(4):1061–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peltola MA, Kuja-Panula J, Lauri SE, Taira T, Rauvala H. AMIGO is an auxiliary subunit of the Kv2.1 potassium channel. EMBO reports. 2011. December 1;12(12):1293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuryshev YA, Gudz TI, Brown AM, Wible BA. KChAP as a chaperone for specific K(+) channels. Am J Physiol Cell Physiol. 2000. May;278(5):C931–41. [DOI] [PubMed] [Google Scholar]
- 38.McCrossan ZA, Roepke TK, Lewis A, Panaghie G, Abbott GW. Regulation of the Kv2.1 potassium channel by MinK and MiRP1. J Membr Biol. 2009. March;228(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011. October 11;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omasits U, Ahrens CH, Muller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014. March 15;30(6):884–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1) Supplementary Table 1. Clinical features of individuals with KCNB1 missense variants. Heterozygous de novo variants were reported as pathogenic or likely pathogenic, with the exception of a single variant determined to be non-pathogenic following parental testing (grey shading).
2) Supplementary Table 2. Number of cells and p-values for the variant channel peak current density in comparison to that of WT.