Abstract
Ubiquitous across all domains of life, tRNAs constitute an essential component of cellular physiology, carry out an indispensable role in protein synthesis, and have been historically the subject of a wide range of biochemical and biophysical studies as prototypical folded RNA molecules. Although conformational flexibility is a well-established characteristic of tRNA structure, it is typically regarded as an adaptive property exhibited in response to an inducing event, such as the binding of a tRNA synthetase or the accommodation of an aminoacyl-tRNA into the ribosome. In this study, we present crystallographic data of a tRNA molecule to expand on this paradigm by showing that structural flexibility and plasticity are intrinsic properties of tRNAs, apparent even in the absence of other factors. Based on two closely related conformations observed within the same crystal, we posit that unbound tRNAs by themselves are flexible and dynamic molecules. Furthermore, we demonstrate that the formation of the T-loop conformation by the tRNA TΨC stem–loop, a well-characterized and classic RNA structural motif, is possible even in the absence of important interactions observed in fully folded tRNAs.
Keywords: tRNA, structure, X-ray crystallography, flexibility, T-loop
INTRODUCTION
Ribosomes in all living systems depend universally on transfer RNA (tRNA) molecules to incorporate the amino acid sequence encoded by messenger RNA transcripts during translation and this decoding process forms the physical basis of the genetic code (Ramakrishnan 2002). Given their vital role in cellular metabolism, tRNAs are not only highly abundant in cells, but also tightly regulated throughout their life cycle (El Yacoubi et al. 2012; Kirchner and Ignatova 2015; Roundtree et al. 2017). The biogenesis of tRNAs is a complex process whose details are still not fully understood; however, across all domains of life conserved mechanisms dictate that tRNAs are synthesized as precursor molecules that undergo a series of activating post-transcriptional modifications for subsequent aminoacylation and thus recruitment by the ribosome (Phizicky and Hopper 2010).
The physiological importance of tRNAs has prompted numerous studies in recent decades to examine closely the structure-function relationships exhibited by tRNAs both in isolation and in complex with other macromolecular entities (Korostelev and Noller 2007; Fei et al. 2011; Agirrezabala and Valle 2015; Marín et al. 2017). Although a substantial amount of structural data have been obtained for many larger and seemingly more complex non-coding RNAs ever since the elucidation of the first tertiary structures of tRNA in the 1970s (Kim et al. 1974; Robertus et al. 1974), tRNAs remain as prototypical molecules for RNA structural studies (Shi and Moore 2000). In fact, a significant amount of information relating to base pairing, base stacking, base modifications, and additional tertiary elements that facilitate long-range intramolecular interactions in folded RNAs were first identified and analyzed in detail in tRNA crystal structures (Batey et al. 1999; Moore 1999; Shi and Moore 2000; Hendrix et al. 2005).
In this study, we present crystallographic data derived from an unmodified Escherichia coli tRNAAsp produced by in vitro transcription that further support the notion that structural flexibility and plasticity are inherent properties of its structure. Although conformational flexibility is by now a well-established and assumed trait of tRNA tertiary structure, prior observations of this characteristic have been made typically in the context of an inducing agent or event, such as the binding of a tRNA synthetase or the accommodation of an aminoacyl-tRNA into the ribosome (e.g., Agirrezabala and Valle 2015; Kuhn 2016). Even when comparative analyses were conducted on free, full-length tRNAs, apparent differences in conformation were attributed generally to sequence variation, solvent conditions, or nucleobase modifications (Byrne et al. 2010). In the current paradigm of RNA structural biology, tRNAs are generally regarded as rigid, two-domain, L-shaped molecules that adaptively undergo conformational change principally in response to the action of other molecular entities (Dethoff et al. 2012). Here we propose that tRNAs are intrinsically flexible and dynamic molecules, which likely sample, without provocation, a wider range of conformational states than previously thought. Although crystal structures depict the conformational average of the molecules constituting the crystalline lattice, a crystal structure of tRNAAsp shown here consists of an unmodified tRNA molecule exhibiting two slightly different conformations within the asymmetric unit. This finding helps not only to further refine our understanding of the general structural properties of tRNAs obtained primarily through crystallographic data, but also to reconcile these static snapshots with solution-based and computational data that have offered more dynamic views of tRNA behavior (Giegé and Frugier 2000–2013; Fulle and Gohlke 2008; Zhang et al. 2014; Sutton and Pollack 2015; Bao et al. 2017). In addition, we determined the crystal structure of an RNA hairpin construct mimicking the tRNAAsp acceptor stem stacked onto the TΨC stem–loop (tRNAAsp-AS/TSL), which revealed that the TΨC loop assumes a tertiary conformation resembling a canonical T-loop, albeit not as well ordered as in tRNA and missing some key interactions that are normally present in tRNAs.
RESULTS
Model refinement against crystallographic data
Since we were able to obtain relatively large crystals of E. coli tRNAAsp, with the largest crystals exceeding 1 mm in the longest dimension, we merged several X-ray diffraction data sweeps collected from a single crystal to push the resolution limit of the data. In addition, as there was clear diffraction anisotropy, a data set was processed using the Staraniso server (Tickle et al. 2018) to produce an anisotropic data set. As the crystals are twinned and show anisotropic diffraction, care was taken not to over-interpret the data and to be cautious in the interpretation. We performed paired refinement calculations with increasingly small incremental changes in resolution near 2 Å (approximately where we expected the limit to be based on outer shell I/σ and CC1/2 values) to determine objectively the best resolution limit of the data to use in the refinement. By applying paired refinement analysis of Rwork and Rfree to these data (Karplus and Diederichs 2012), we found that there were meaningful data to as far as 1.99 Å in the isotropic data set that would otherwise be processed to 2.3 Å by previous conventional standards based on Rmerge or Rmeasure. A similar analysis showed that the anisotropic resolution limit was 1.95 Å in the best direction (CC1/2 = 0.488 in the outermost resolution shell). Consistently, model refinement against the isotropic 1.99 Å data set (CC1/2 = 0.404 in the outermost resolution shell) versus a previous 2.4 Å data set demonstrated clear improvements in the 2Fo–Fc electron density map and in the Rfree value of the refined model.
The crystals of the stacked acceptor stem and TΨC stem–loop had different c-axis lengths depending on the age of the crystals; older crystals had a significantly longer c-axis (281.8 Å vs. 259.3 Å) and a better ordered T-loop. Older crystals appear to have become dehydrated, but contrary to what is usually observed, the cell axes did not shrink due to dehydration. The apparent reason for the growth of the c-axis is ordering of the loop in the hairpin. In the crystals, the stems align almost parallel to the c-axis and six symmetry related loops face each other at the origin, where threefold and twofold axes intersect. In the long cell crystals, twofold symmetry related bases of C15 in the loop stack on each other as part of the lattice contacts. In the short cell crystals, the loops are disordered as they would interdigitate due to the crystal symmetry. This suggests that growing of the c-axis over time is due to the formation of more stable lattice contacts that allow the loops to separate and form a stable stacking interaction. Thus, in this case, better packing results in cell axis growth. In addition, crystal of both c-axis lengths showed anisotropic diffraction and were processed using the Staraniso server (Tickle et al. 2018) where the cut-off for the mean (I)/σ(I) of the accepted data was determined by a similar procedure as for paired refinement. Overall, for both the tRNAAsp and acceptor stem/TΨC stem–loop crystals, extending the resolution limit of the included data resulted in better maps and models, consistent with what has been reported earlier for other RNA crystals (Wang 2010).
Structural differences between the two molecules of E. coli tRNAAsp constituting the crystallographic asymmetric unit
The overall tertiary structures of the two molecules comprising the crystallographic asymmetric unit of the unmodified E. coli tRNAAsp crystal exhibit the characteristic L-shaped structure of tRNA molecules, which consists of two helical stacks, corresponding to the acceptor stem/TΨC stem–loop and the anticodon/D stem–loop, oriented near-perpendicularly to each other (Fig. 1). The electron density corresponding to nearly every residue in both molecules was surprisingly well-ordered, which allowed us to build complete models for both tRNAs. Nevertheless, a comparison between the two structures in the asymmetric unit of E. coli tRNAAsp reveals that there are both local and global conformational differences between them.
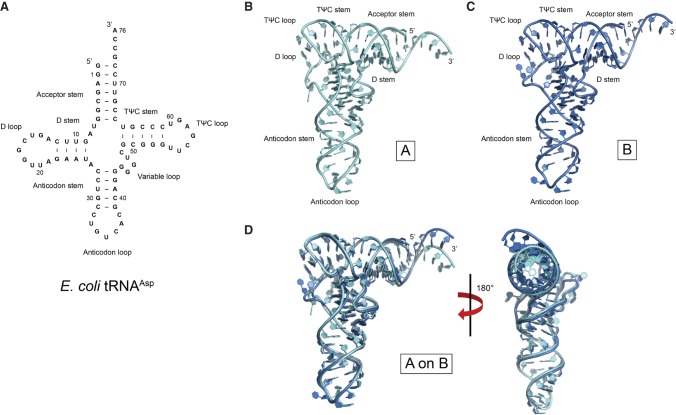
FIGURE 1.
Secondary and overall tertiary structure of E. coli tRNAAsp. (A) Secondary structure diagram of E. coli tRNAAsp. (B,C) Overall tertiary structure of the two molecules of E. coli tRNAAsp (denoted as A,B) that comprise the crystallographic asymmetric unit. (D) Juxtaposition of the two structures reveal several conformational differences between the two models.
Superposition of the two non-crystallographic symmetry-related molecules of E. coli tRNAAsp (referred to as tRNAAsp A and B hereafter) demonstrates that there are clear structural differences between the nucleotides at the 3′ end (Fig. 1B,C), the anticodon loops (Fig. 2A,B), the D loops (Fig. 2C,D), and the angle of their “elbow” region (Fig. 3). Superposition of the two complete structures show a root mean square deviation (rmsd) of 2.52 Å for all atoms, but the rmsd goes down to 1.84 Å when the last four nucleotides are excluded. If the bases are excluded in the latter comparison, the rmsd is 1.53 Å, emphasizing that most nucleobases have almost identical conformation, but that globally the differences are significant, likely reflecting a change in overall structure (see below). In addition, analysis of the contacts mediating the crystal packing shows that although there are significant differences between the two monomers in some regions, in general, the packing contacts do not promote these changes. Supplemental Figure S1 illustrates some of the contacts involving the regions mentioned above.
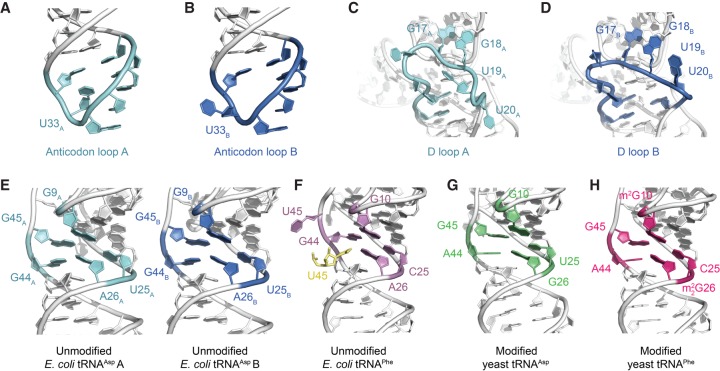
FIGURE 2.
Comparative structural analyses between the crystal structure of E. coli tRNAAsp with other free tRNA molecules. (A,B) Conformational differences in the anticodon loop of the two molecules of E. coli tRNAAsp that comprise the crystallographic asymmetric unit. (C,D) Conformational differences in the D loop of the two molecules of E. coli tRNAAsp that comprise the crystallographic asymmetric unit. (E–H) The core region of the two molecules of unmodified E. coli tRNAAsp that comprise the crystallographic asymmetric unit is very similar in conformation to the core region of both modified yeast tRNAAsp (PDB ID: 1VTQ) (Comarmond et al. 1986) and yeast tRNAPhe (PDB ID: 1EHZ) (Shi and Moore 2000), whereas the core region of an unmodified E. coli tRNAPhe (PDB ID: 3L0U) (Byrne et al. 2010) is comparatively different (m2G and m22G represent 2-methylguanosine and N2,N2-dimethylguanosine, respectively). In the latter structure, the extruded U45 forms a crystal lattice contact and is replaced by U45 from a symmetry related molecule (yellow).
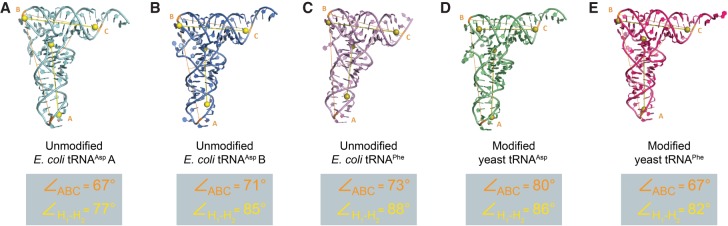
FIGURE 3.
Variation in angle of the “elbow” region of tRNAs. The angle of the “elbow” region (made up of the TΨC and D loops) varies from tRNA to tRNA without any obvious correlation to the nucleobase modification status of the entire tRNA molecule, as was previously proposed (Byrne et al. 2010). The figure shows the “elbow” angles calculated from three reference points (ABC) and from the angle formed by the best axes of the two stems (H1–H2; see Materials and Methods).
A previous analysis of the tRNA anticodon loop revealed that several of its defining structural features involve a conserved uridine at position 33; specifically, it was proposed that the atomic interactions between the ribose moiety of U33 and the nucleobase (purine or pyrimidine) of residue 35 and between the uracil moiety of U33 and the backbone phosphates of residues 35 and 36 help to establish the characteristic anticodon loop structure (Auffinger and Westhof 2001). The sugar-phosphate backbone conformations of the anticodon loops of both tRNAAsp A and B are indeed very similar; however, whereas U33A is oriented in the canonical position, the nucleobase of U33B is flipped out from the anticodon loop and faces the D loop of a symmetry-related tRNAAsp A molecule (Fig. 2A,B). In the crystal structure, G34A–U35A–C36A face C36B–U35B–G34B from a non-crystallographic related molecule, with G34A–C36B and C36A–G34B forming base pairs, and U35A–U36B facing each other, but too far apart to form hydrogen bonds. More importantly, this juxtaposition suggests that despite the conservation of U33 and its key role in the anticodon loop, the canonical position of U33 appears not to be essential for maintaining the backbone conformation of the anticodon loop. In the case of tRNAAsp B, the surrounding context of U33B has allowed for this particular residue to swivel out toward the solvent space without having any marked structural effect on the rest of the anticodon loop except for the creation of a void in the loop. In both cases, the region surrounding U33 is mainly solvent, thereby providing no apparent reason for the swiveling out of the nucleobase, although there is a minor interaction between U33B and a neighboring molecule.
A comparison of the D loops in tRNAAsp A and B reveals that there is significant conformational variation in this backbone region across the two molecules (Fig. 2C,D), consistent with a previous observation that the D loop exhibits relatively greater flexibility than other tRNA regions (Byrne et al. 2010). For example, U19A and U20A assume markedly different tertiary positions than U19B and U20B. In both tRNAAsp A and B, two consecutive and conserved guanidine residues, G17 and G18 (Juhling et al. 2009), insert into the TΨC T-loop structure as an intercalated base (Chan et al. 2013) and a hydrogen-bonded pair with the apex (C56) residue of the T-loop, respectively. Thus, G17 and G18 act to dock the proximal region of an otherwise flexible D loop into the TΨC loop and may help stabilize the T-loop. Despite the apparent importance of these docking interactions mediated by both G17 and G18, the crystal structure of a modified yeast tRNAAsp (PDB ID: 1VTQ) (Comarmond et al. 1986) reveals a D loop conformation in which G18 does not engage the TΨC loop. In a scenario in which both D loop G17 and G18 residues are undocked from the TΨC loop, the flexible D loop would be completely solvent accessible and thus primed to be peeled away from the tRNA core. Of note, a large conformational change has been observed in an archaeal tRNAVal when bound by the archaeosine tRNA-guanine transglycosylase; this alternate but distinct tRNA conformation was termed the λ-form structure and has a key feature of a disrupted D loop (Ishitani et al. 2003). However, intrinsic flexibility and structural variability of the D loop offer the possibility that the λ-form structure likely falls within a spectrum of conformations sampled independently by tRNAs and, in this particular case, the role of protein binding is to stabilize rather than to induce this specific conformation.
Given the overall L-shaped tertiary structure of canonical tRNAs, an important measure of global conformation is the angle of their “elbow” region, which is the inter-helical angle between the coaxial stacks forming the acceptor stem/TΨC stem–loop and the anticodon/D stem–loop. By measuring this “elbow” angle using the phosphorus positions of three residues (A35, C56, and C72 or their equivalent nucleotides) in unmodified E. coli tRNAPhe, and the angle in several other tRNA structures that are both unbound or bound to protein, it has been proposed previously that nucleotide sequence, modification status, as well as protein binding, all affect the global conformation of tRNA (Byrne et al. 2010). Since the crystal of E. coli tRNAAsp consists of two non-crystallographic-symmetry related molecules, a comparison of the “elbow” angle of the two non-crystallographic-symmetry related molecules of tRNAAsp provides a direct assessment of the intrinsic flexibility of this tRNA in the absence of protein binding, when the nucleotide sequences are identical, and when there are no nucleotide modifications. In comparison to unmodified E. coli tRNAPhe, which exhibits an “elbow” angle of 73°, tRNAAsp A and B exhibit more acute “elbow” angles of 67° and 71°, respectively, (Fig. 3). The difference between the angles measured for tRNAAsp A and B alone suggests that there is an intrinsic flexibility in the unmodified E. coli tRNAAsp tertiary structure. In this particular instance, the clear difference in global conformation of the tRNAs is likely a mere effect of their surrounding crystalline environment, which may cause the tRNAs to distort in different regions to allow for crystal packing. Of note, a model of yeast tRNAPhe based on SAXS data reveals a significantly more obtuse “elbow” angle than is present in its crystal structure (Hammond et al. 2009). Furthermore, comparison of the solution structure of a viral tRNA mimic (Hammond et al. 2009) and its crystal structure (Colussi et al. 2014), which shows a more conventional tRNA-like “elbow” angle, also reveals a significant degree of flexibility in the “elbow” region between the solution and crystal structures that is in turn modulated by an upstream structural element (Hammond et al. 2010).
Comparative structural analysis of representative free tRNA crystal structures
Despite a previous suggestion that RNA nucleotide sequence differences and nucleobase modifications exert global conformational changes in the overall structure of tRNA (Byrne et al. 2010; Lorenz et al. 2017), we found that crystal packing and crystal-to-crystal variation can account for many of the structural differences observed in the currently available crystal structures of free tRNAs (Figs. 1–3). For instance, in the crystal structure of an unmodified E. coli tRNAPhe (Byrne et al. 2010), U45 is rotated out from the tRNA core toward solvent space, where it is inserted into the core region of a symmetry-related molecule. In canonical tRNA, U45 would be expected to form a base triple with G10 and C25, but the extrusion of U45 precludes the formation of this triple. Instead, a small rearrangement of the backbone brings G44 to the expected location of U45, creating a void in the canonical position of G44 and preventing the formation of the expected A26–G44 base pair. The space left open by the movement of G44 to form the triple is taken over by U45 of a symmetry related molecule, creating an U45sym–A26 base pair. Thus, the core structure is rearranged relative to that of the prototypic tRNA core (Fig. 2), but the overall structure remains fairly similar due to the rearrangements and the movement of a symmetry related nucleotide.
The general structure of the tRNA core appears to be conserved across different tRNA molecules, irrespective of nucleobase modifications and, to some degree, sequence variation (Fig. 2E–H). A comparison of the cores of unmodified E. coli tRNAPhe and modified yeast tRNAPhe shows close similarity between their central base triple, G10–C25–G44/G10–C25–G45 (even though in the latter molecule G10 is N2-methylated) and between the adjacent base pair (U45sym–A26 and A44·G26 in the unmodified and modified molecules, respectively, where G26 is N2-dimethylated in the latter case). The U45sym–A26 pair does not form a canonical base pair as the symmetry related molecule forms a highly buckled pair with only N1 of A26 and N3 of U45 within hydrogen bonding distance, whereas N6 of A26 and O4 of U45 are too far apart due to the absence of coplanarity. Furthermore, a comparison of the cores of the unmodified E. coli tRNAAsp and the modified yeast tRNAAsp reveals that this adjacent base pair in the two molecules are interchangeably G·A or A·G, respectively. Finally, a comparison between modified tRNAAsp and modified tRNAPhe shows that either U or C can occur at position 25, as long as the base triple configuration is maintained. These comparisons reveal that despite the general importance ascribed to specific residues or modifications in tRNA function and metabolism, there exists a clear degree of plasticity in the canonical structure of the tRNA core that acts to buffer against the tertiary level effects of sequence variation and nucleobase modifications.
By measuring the “elbow” angle of the modified yeast tRNA structures and comparing them with the angles measured for their unmodified E. coli counterparts, we found that there was no obvious correlation between the measured angles and whether a tRNA molecule is modified or unmodified. This observation suggests that nucleobase modifications do not result in a global change in tRNA structure, at least to an extent that is discernable by changes in the “elbow” angle. In fact, we found that there were greater differences in the measured “elbow” angles simply due to how the angles were measured (i.e., by using three reference phosphorus atoms in the acceptor stem, the TΨC loop, and the anticodon loop [Byrne et al. 2010], or by defining coaxial vectors for the acceptor stem/TΨC stem–loop and the anticodon/D stem–loop stacks) than there were within each set of measurements (Fig. 3); however, we also note that some of the discrepancy between the two methods is that they do not equally reflect twisting at the “elbow.” Taken together, we propose an alternative explanation for the observed variation in the “elbow” angles of these tRNA structures: tRNAs are intrinsically flexible molecules whose “elbow” region can conform to both bending and twisting imposed by crystallization.
Crystal structure of a fragment of E. coli tRNAAsp consisting of its acceptor stem/TΨC stem–loop
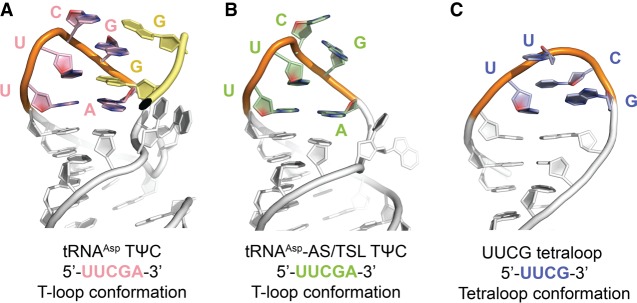
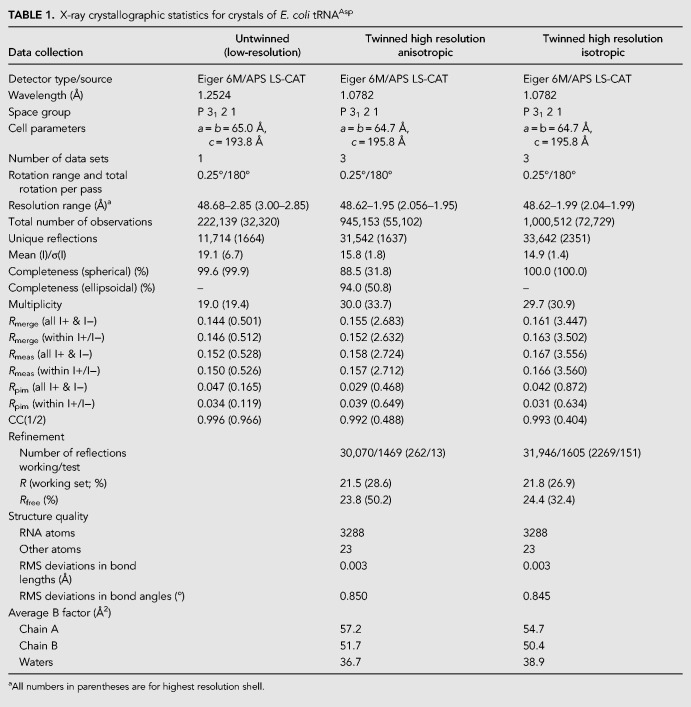
As a part of this study, we crystallized an RNA hairpin molecule equivalent to the E. coli tRNAAsp TΨC stem–loop stacked on its acceptor stem (Fig. 4). In the absence of its native tertiary context, the TΨC loop assumed a conformation that resembles a T-loop (Fig. 5), although the nucleotides forming the loop are not as well ordered as in tRNA. RNA structural motifs are frequently catalogued with an underlying assumption that there exists a sequence consensus for each motif and that they all share the same structure (Fernández-Millán et al. 2016). As noted previously (Chan et al. 2013), the T-loop motif does not appear to have as strong a sequence consensus as other motifs and, unlike many other motifs, T-loops can only be identified confidently based on tertiary structure. It is thus surprising that the T-loop is formed in the absence of important stabilizing interactions, such as the ones with the D-loop nucleotides. This suggests that the propensity to form a T-loop is in fact encoded in the E. coli tRNAAsp TΨC loop sequence and that T-loop formation can occur even in the absence of the D-loop, which is consistent with the modified tRNAAsp structure (Comarmond et al. 1986), which shows a T-loop even though some of the interactions are not present.
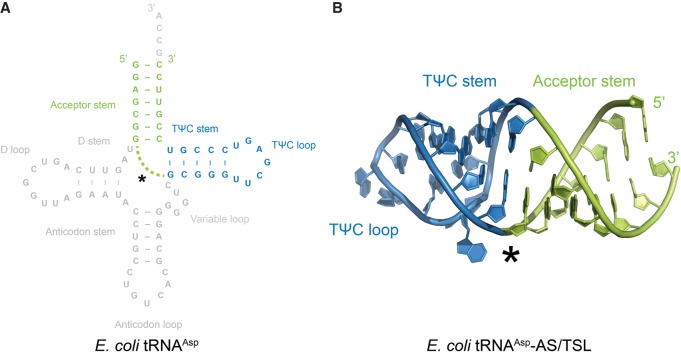
FIGURE 4.
Secondary and overall tertiary structure of E. coli tRNAAsp-AS/TSL. (A) Secondary structure diagram of the crystallized RNA stem–loop construct, which consists of the acceptor stem (AS, colored in green) and the TΨC stem–loop (TSL, colored in blue) of E. coli tRNAAsp. (B) Overall tertiary structure of the E. coli tRNAAsp-AS/TSL (with coloring as in A), in which the TΨC loop exhibits a T-loop conformation. The asterisk shows the region where the TΨC stem and acceptor stems were linked to bypass the anticodon and D stems.
FIGURE 5.
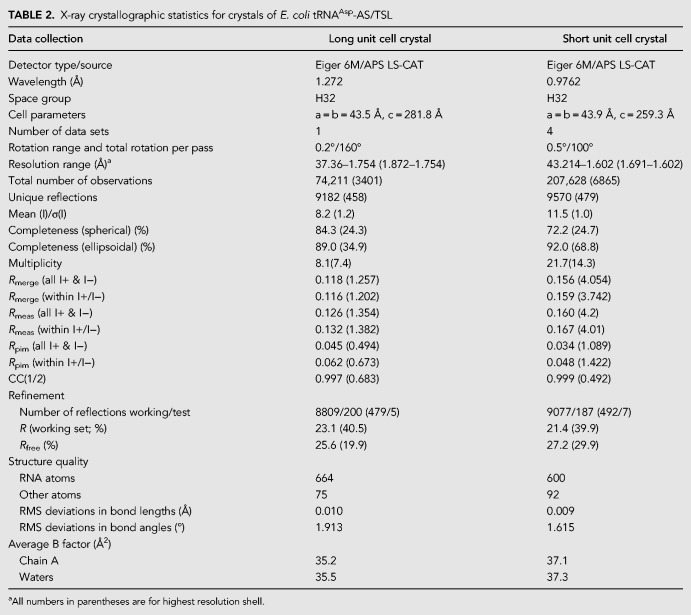
Residues comprising the E. coli tRNAAsp TΨC loop exhibit a T-loop structural fold in a context-independent manner. (A) TΨC loop in the crystal structure of full-length E. coli tRNAAsp assumes the canonical T-loop conformation while engaging in tertiary interactions with D loop residues (yellow) acting as intercalating and stacked nucleobases. (B) TΨC loop in the crystal structure of E. coli tRNAAsp-AS/TSL reveals a T-loop conformation, despite the absence of the D loop interactions, and not a UUCG tetraloop conformation (illustrated in C), as could be expected from sequence analysis.
DISCUSSION
The crystal structure of E. coli tRNAAsp demonstrates that structural flexibility is an intrinsic property of tRNAs. In the absence of external modifying factors, such as binding by proteins or other RNAs, the surrounding crystalline conditions are sufficient to generate conformational changes that in previous structures were attributed to the action of proteins or other factors, such as nucleotide sequence differences and nucleobase modifications. This is further illustrated in Supplemental Figure S2, which shows superpositions of both E. coli tRNAAsp molecules with other tRNAs found in complex with proteins, other RNAs, or the ribosome. The comparisons demonstrate that, in general, the structural variability found between both E. coli tRNAAsp molecules is within the range of changes induced by tRNA binding to another macromolecule. Similarly, prior suggestion that RNA nucleobase modifications exert a global effect on an entire RNA molecule were based on the observation that an unmodified tRNA molecule exhibits a different structural conformation than their modified counterparts (Byrne et al. 2010). Between the two molecules of E. coli tRNAAsp that comprise its crystallographic asymmetric unit, we found a similar degree of structural variation attributed previously to the absence versus presence of nucleobase modifications, thereby suggesting that it is rather the flexible nature of tRNAs that is being exemplified by these crystal structures.
We also demonstrated that the E. coli tRNAAsp TΨC loop can form even in the absence of all stabilizing interactions. Since the E. coli tRNAAsp TΨC loop consists of a sequence that fits the consensus of both a T-loop and a UUCG tetraloop, it could be assumed that the loop has an intrinsic propensity to assume either structural conformation. In the context of a full-length tRNA molecule, the 5′-UUCGA-3′ segment of the TΨC loop forms a T-loop motif with an intercalated nucleobase (G17) supplied by the D loop. In the absence of this tertiary interaction, the isolated TΨC loop still forms a loop that is closer in structure to a T-loop than to a UUCG tetraloop, although the apex of the loop is partially disordered. This is direct evidence showing that there is a hierarchy in structure formation when two distinct motifs can be formed in a single RNA sequence. In this case, the T-loop seems to take precedence over the tetraloop, even though not all the expected interactions are present.
Given that there are far fewer available crystal structures of RNAs than of proteins, it is interesting to note which RNA–RNA interactions mediate crystal lattice contacts in new RNA structures. In the crystal of E. coli tRNAAsp, which contains two non-crystallographic symmetry-related molecules, there are surprisingly few crystal lattice contacts. Furthermore, these intermolecular RNA–RNA interactions consist primarily of coaxial stacking, base pairing, and base stacking (Supplemental Fig. S3), which are also the primary and familiar mediators of intramolecular interactions in folded RNA molecules. This observation supports the notion that folded RNA molecules utilize the same RNA–RNA interactions for both intramolecular stabilization and assembly on the quaternary level.
Finally, an evolutionary implication of tRNA structural flexibility is that it helps to reduce selective pressures on both natural tRNA structural mimics and tRNA modifiers since the structures that they adopt or need to act on, respectively, do not necessarily need to assume a very specific and rigid conformation. This is not to say that tRNA tertiary structures are not significantly affected by nucleotide sequence or modification, but merely that their intrinsic flexibility alone allows for greater functional versatility. From a general functional perspective, inherent flexibility of tRNAs (and, by extension, folded RNA molecules) also suggests that they are perhaps more active participants in biological processes than previously thought, particularly since the current paradigm for RNA-based biological processes is that non-coding RNA molecules are passive and rigid structural entities that function primarily by being acted upon by proteins or other RNA–protein complexes.
In conclusion, our crystallographic studies of E. coli tRNAAsp provide further evidence that structural flexibility is an intrinsic property of tRNAs and of folded RNA molecules in general. In a broader context, this finding challenges a widespread view that folded tRNA molecules are structurally static unless chemically modified or physically acted upon. Furthermore, we also provide data demonstrating that there is structural hierarchy underlying RNA structural motifs, the basic structural units of folded RNA, such that a complete bottom-up approach to understanding RNA folding must take into account that sequences conforming to more than one structural motif may prefer one over the other in a manner that may not be easy to predict.
MATERIALS AND METHODS
Transcription and RNA purification
Full-length Escherichia coli tRNAAsp and a truncated RNA hairpin construct (tRNAAsp-AS/TSL, equivalent to the coaxial stack formed by the acceptor stem/TΨC stem–loop) were produced in vitro on the milligram scale by run-off transcription using recombinant T7 RNA polymerase and standard reaction conditions (Milligan et al. 1987). In brief, the DNA template encoding E. coli tRNAAsp for transcription was cloned into the multiple cloning site of pUC19 with an upstream T7 RNA polymerase promoter element and a BsmAI restriction site immediately 3′ to the coding region. This plasmid was transformed into and thus replicated in a culture of E. coli DH5α grown in Terrific Broth media and purified by conventional alkaline lysis methods, followed by phenol–chloroform extraction and selective PEG 6000 precipitation, to remove contaminating cellular host proteins and RNAs, respectively. The purified plasmid DNA was subsequently digested with BsmAI (New England BioLabs) and purified further by phenol–chloroform extraction and ethanol precipitation. In contrast, the DNA template encoding E. coli tRNAAsp-AS/TSL for transcription was generated by annealing complementary oligonucleotides purchased from Integrated DNA Technologies. The sequences of the two oligonucleotides are as follows: 5′-CGACGTAATACGACTCACTATAGGAGCGGGCGGGTTCGAGTCCCGTCCGTTCC-3′ and 5′-GGAACGGACGGGACTCGAACCCGCCCGCTCCTATAGTGAGTCGTATTACGTCG-3′; the annealed product contains the T7 RNA polymerase promoter element upstream of the coding region. For annealing, the two oligonucleotides were incubated for 2 min at 90°C at an equal molar stoichiometric ratio, cooled subsequently to room temperature on a benchtop, and used directly for in vitro transcription.
Following in vitro transcription, E. coli tRNAAsp and tRNAAsp-AS/TSL were separated from short transcripts and other reaction components by electrophoresis in a native 10% acrylamide gel ran in 89 mM Tris-HCl, pH 7.6, 89 mM boric acid (1× TB buffer). The principal gel band corresponding to each transcription product was located by UV shadowing and extracted into 50 mM potassium acetate, pH 7.0, 200 mM potassium chloride by passive diffusion at 4°C. The extracted RNAs were both precipitated with ethanol and stored at −20°C.
The gel-purified and precipitated E. coli tRNAAsp (Supplemental Fig. S4A) was resuspended in 50 mM Tris-HCl, pH 7.5, 200 mM sodium chloride, 5 mM magnesium chloride and further purified by size exclusion chromatography on an FPLC instrument (ÄKTA, GE Healthcare) using a Superdex 200 10/300 GL column ran with 50 mM Tris-HCl, pH 7.5, 200 mM sodium chloride, 5 mM magnesium chloride (Supplemental Fig. S4B). The FPLC-purified E. coli tRNAAsp was then both buffered-exchanged into 50 mM Tris-HCl, pH 7.5, 200 mM sodium chloride, 40 mM potassium chloride, 10 mM magnesium chloride and concentrated using 10 kDa MWCO spin concentrators containing either a cellulose acetate (Millipore) or polyethersulfone (GE Healthcare) membrane to a final concentration of ∼20 mg/mL. In contrast, the gel-purified and precipitated E. coli tRNAAsp-AS/TSL was resuspended in 50 mM Tris-HCl, pH 7.5, 200 mM sodium chloride, 5 mM magnesium chloride, filtered using a 0.22 µm spin column with a cellulose acetate membrane, and used directly for crystallization trials. The concentration of E. coli tRNAAsp-AS/TSL used for crystallization ranged between 1 to 4 mg/mL.
Crystallization and structure determination of E. coli tRNAAsp
Crystals of E. coli tRNAAsp were grown at 287 K by vapor diffusion in a hanging drop format over a range of equilibration solutions optimized around 50 mM sodium cacodylate, pH 7.0, 30% v/v 2-methyl-2,4-pentanediol (MPD). Although we found that the presence of various salts (up to 300 mM sodium chloride and 300 mM potassium chloride) and polyamines (1 to 10 mM spermine or spermine tetrahydrochloride), the coverslip material (siliconized glass, unsiliconized glass, and plastic coverslips), and different sample concentrations (ranging from 5 to 20 mg/mL of E. coli tRNAAsp) all had varying effects on the morphology of the crystals, which often grew in large clusters (Supplemental Fig. S4C), there was no obvious correlation between these factors and the eventual size or diffraction limit of the crystals. However, since the crystals tended to grow in clusters, manual manipulation was often needed to excise fragments suitable for data collection. Our best diffracting, and also the largest, single crystal of E. coli tRNAAsp (Supplemental Fig. S4C) was grown from a mixture of 2 µL of 20 mg/mL E. coli tRNAAsp and 2 µL of 50 mM sodium cacodylate, pH 7.5, 30% MPD, 10 mM spermine tetrahydrochloride, 300 mM sodium chloride, 300 mM potassium chloride (equilibration solution), and 0.5 µL of Hampton Research Silver Bullet Additive No. 23, which consists of 0.25% w/v p-coumaric acid, 0.25% w/v phenylurea, 0.25% w/v poly(3-hydroxybutyric acid), 0.25% w/v sulfaguanidine, 0.02 M sodium HEPES, pH 6.8.
X-ray diffraction data (Supplemental Fig. S4C) were collected at 100 K at the Life Sciences Collaborative Access Team (LS-CAT) beamlines at the Advanced Photon Source (APS), Argonne National Laboratory, using a Dectris Eiger 9M detector. Crystals of E. coli tRNAAsp were flash frozen directly in mother liquor with liquid nitrogen for data collection since the MPD content of the mother liquor was sufficient for cryo-protection. Processing of diffraction data was carried out using XDS (Kabsch 2010) for indexing and integration and AIMLESS (Evans and Murshudov 2013) for merging and scaling. The crystal structure of E. coli tRNAAsp was solved by molecular replacement using a homology model generated by Chainsaw (Stein 2008) in CCP4 (Winn et al. 2011) using the crystal structure of a modified yeast tRNAPhe (PDB ID: 1EHZ) (Shi and Moore 2000) with the primary sequence of E. coli tRNAAsp threaded through. Although the vast majority of E. coli tRNAAsp crystals were twinned, we were able to collect a data set from an untwinned crystal, which allowed us to solve its structure and generate an initial model (R/Rfree = 0.20/0.26 at 2.85 Å). Twinned crystals were subsequently used for refinement to high resolution. As the crystals showed anisotropic diffraction, data were re-processed using the Staraniso server (Tickle et al. 2018) to produce the final data set. All iterative rounds of model building and refinement were performed with Coot (Emsley and Cowtan 2004) and Refmac5 (Murshudov et al. 1997). The final models (isotropic to 1.99 Å R/Rfree = 0.218/0.244 and anisotropic to 1.95 Å R/Rfree = 0.215/0.238) were determined from the highest resolution data set collected. Additional X-ray crystallographic statistics for E. coli tRNAAsp are summarized in Table 1. The 2mFO–DFC map, calculated with coefficients output by Refmac (Murshudov et al. 1997), and simulated annealing omit maps calculated with Phenix (Afonine et al. 2012) for two different regions of the crystal are shown in Supplemental Figure S5 to illustrate the quality of the maps. Only the coordinates for the model refined against the anisotropic data were deposited in the PDB as the two sets are virtually identical.
TABLE 1.
X-ray crystallographic statistics for crystals of E. coli tRNAAsp
Crystallization and structure determination of an RNA hairpin consisting of the acceptor stem and TΨC stem–loop of E. coli tRNAAsp
Crystals of E. coli tRNAAsp-AS/TSL were also grown at 287 K by vapor diffusion in a hanging drop format over a range of equilibration solutions optimized around 2 M ammonium sulfate (Supplemental Fig. S6A). Crystals of E. coli tRNAAsp-AS/TSL were flash frozen with liquid nitrogen in crystallization well solution supplemented with either increased ammonium sulfate concentration or with 20% (v/v) glycerol for cryo-protection. For data collection, X-ray diffraction data (Supplemental Fig. S6B) were collected at 100 K at LS-CAT at the APS, Argonne National Laboratory, using a Dectris Eiger 9M detector. Processing of diffraction data was carried out using XDS (Kabsch 2010) for indexing and integration and AIMLESS (Evans and Murshudov 2013) for merging and scaling. Final anisotropic data sets were obtained using the Staraniso server (Tickle et al. 2018). The crystal structure of E. coli tRNAAsp-AS/TSL was solved by molecular replacement using coordinates for an idealized 5 bp A-form RNA helix (Robertson and Scott 2008) generated in Coot (Emsley and Cowtan 2004). Additional electron density for the rest of the molecule appeared subsequently in iterative rounds of model building and refinement, which were performed with Coot (Emsley and Cowtan 2004) and Refmac5 (Murshudov et al. 1997), respectively. The length of the c-axis was different depending on the age of the crystals such that older crystals had a longer c-axis and a better ordered T-loop, whereas fresher crystals had a shorter c-axis and a disordered T-loop. The final models for E. coli tRNAAsp-AS/TSL had an R/Rfree = 0.214/0.272 to 1.6 Å resolution and R/Rfree = 0.231/0.256 to 1.75 Å for the short and long axis crystals respectively. Additional X-ray crystallographic statistics are summarized in Table 2. The 2mFO–DFC map, calculated with coefficients output by Refmac (Murshudov et al. 1997), and simulated annealing omit maps calculated with Phenix (Afonine et al. 2012) are shown in Supplemental Figures S7, S8 to illustrate the quality of the maps. Coordinates for the long and short cell models refined against their respective anisotropic data were deposited in the PDB.
TABLE 2.
X-ray crystallographic statistics for crystals of E. coli tRNAAsp-AS/TSL
Visualization and comparative analysis of structural models
The crystal structures of the tRNA molecules were aligned by least squares fitting in Coot (Emsley and Cowtan 2004) using the atomic coordinates of their TΨC loop (residues 54 to 58 in E. coli tRNAAsp). PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC) was used to depict all structural models and to measure the “elbow” angle of the tRNA molecules based on the position of three reference atoms, as done in a previous analysis (Byrne et al. 2010). To calculate an alternate and more representative measure of the inter-helical angle between the acceptor stem/TΨC stem and the anticodon/D stem helical stacks of each tRNA molecule, we applied the law of cosines to stem axial vectors determined by the DSSR software (Lu et al. 2015) of the 3DNA program suite (Lu and Olson 2003, 2008a,b).
DATA DEPOSITION
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 6UGG (E. coli tRNAAsp model), 6UGI (E. coli tRNAAsp-AS/TSL, long cell model), and 6UGJ (E. coli tRNAAsp-AS/TSL, short cell model).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Lei Huang for providing us with a plasmid encoding the E. coli tRNAAsp and the beamline scientists at LS-CAT/Sector 21 at the Advanced Photon Source, Argonne National Laboratory, for their assistance. We also acknowledge support received from the Structural Biology Facility and the High Throughput Analysis Laboratory at Northwestern University. Funding was provided by the National Institutes of Health (NIH) grants R01 GM058443 and R35 GM118108 to A.M. C.W.C. was supported by the NIH Medical Scientist Training grant 4T32 GM008152, the Molecular Biophysics Training grant NIH 5T32 GM008382, and an Achievement Rewards for College Scientists Foundation Fellowship. LS-CAT/Sector 21 at the Advanced Photon Source, Argonne National Laboratory was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Structural Biology Facility is acknowledged.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.073478.119.
REFERENCES
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68: 352–367. 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Valle M. 2015. Structural insights into tRNA dynamics on the ribosome. Int J Mol Sci 16: 9866–9895. 10.3390/ijms16059866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P, Westhof E. 2001. An extended structural signature for the tRNA anticodon loop. RNA 7: 334–341. 10.1017/S1355838201002382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Zhang X, Shi YZ, Wu YY, Tan ZJ. 2017. Understanding the relative flexibility of RNA and DNA duplexes: stretching and twist-stretch coupling. Biophys J 112: 1094–1104. 10.1016/j.bpj.2017.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Rambo RP, Doudna JA. 1999. Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl 38: 2326–2343. [DOI] [PubMed] [Google Scholar]
- Byrne RT, Konevega AL, Rodnina MV, Antson AA. 2010. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res 38: 4154–4162. 10.1093/nar/gkq133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CW, Chetnani B, Mondragón A. 2013. Structure and function of the T-loop structural motif in noncoding RNAs. Wiley Interdiscip Rev RNA 4: 507–522. 10.1002/wrna.1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, Kieft JS. 2014. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature 511: 366–369. 10.1038/nature13378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comarmond MB, Giegé R, Thierry JC, Moras D, Fischer J. 1986. Three-dimensional structure of yeast tRNAAsp. I. Structure determination. Acta Cryst 42: 272–280. 10.1107/S0108768186098233 [DOI] [Google Scholar]
- Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. 2012. Functional complexity and regulation through RNA dynamics. Nature 482: 322–330. 10.1038/nature10885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bailly M, de Crécy-Lagard V. 2012. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 46: 69–95. 10.1146/annurev-genet-110711-155641 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69: 1204–1214. 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Richard AC, Bronson JE, Gonzalez RL Jr. 2011. Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol 18: 1043–1051. 10.1038/nsmb.2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Millán P, Schelcher C, Chihade J, Masquida B, Giege P, Sauter C. 2016. Transfer RNA: from pioneering crystallographic studies to contemporary tRNA biology. Arch Biochem Biophys 602: 95–105. 10.1016/j.abb.2016.03.005 [DOI] [PubMed] [Google Scholar]
- Fulle S, Gohlke H. 2008. Analyzing the flexibility of RNA structures by constraint counting. Biophys J 94: 4202–4219. 10.1529/biophysj.107.113415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R, Frugier M. 2000–2013. Transfer RNA structure and identity. In Madame Curie Bioscience Database [Internet]. Landes Bioscience, Austin, TX: https://www.ncbi.nlm.nih.gov/books/NBK6236/ [Google Scholar]
- Hammond JA, Rambo RP, Filbin ME, Kieft JS. 2009. Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA 15: 294–307. 10.1261/rna.1360709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JA, Rambo RP, Kieft JS. 2010. Multi-domain packing in the aminoacylatable 3′ end of a plant viral RNA. J Mol Biol 399: 450–463. 10.1016/j.jmb.2010.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DK, Brenner SE, Holbrook SR. 2005. RNA structural motifs: building blocks of a modular biomolecule. Q Rev Biophys 38: 221–243. 10.1017/S0033583506004215 [DOI] [PubMed] [Google Scholar]
- Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. 2003. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 113: 383–394. 10.1016/S0092-8674(03)00280-0 [DOI] [PubMed] [Google Scholar]
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162. 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. 2012. Linking crystallographic model and data quality. Science 336: 1030–1033. 10.1126/science.1218231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, Rich A. 1974. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185: 435–440. 10.1126/science.185.4149.435 [DOI] [PubMed] [Google Scholar]
- Kirchner S, Ignatova Z. 2015. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 16: 98–112. 10.1038/nrg3861 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Noller HF. 2007. The ribosome in focus: new structures bring new insights. Trends Biochem Sci 32: 434–441. 10.1016/j.tibs.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Kuhn CD. 2016. RNA versatility governs tRNA function: why tRNA flexibility is essential beyond the translation cycle. Bioessays 38: 465–473. 10.1002/bies.201500190 [DOI] [PubMed] [Google Scholar]
- Lorenz C, Lünse CE, Mörl M. 2017. tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7: E35 10.3390/biom7020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Olson WK. 2003. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res 31: 5108–5121. 10.1093/nar/gkg680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Olson WK. 2008a. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc 3: 1213–1227. 10.1038/nprot.2008.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Olson WK. 2008b. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc 3: 1213–1227. 10.1038/nprot.2008.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Bussemaker HJ, Olson WK. 2015. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res 43: e142 10.1093/nar/gkv716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M, Fernández-Calero T, Ehrlich R. 2017. Protein folding and tRNA biology. Biophys Rev 9: 573–588. 10.1007/s12551-017-0322-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8798. 10.1093/nar/15.21.8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PB. 1999. Structural motifs in RNA. Annu Rev Biochem 68: 287–300. 10.1146/annurev.biochem.68.1.287 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. 2002. Ribosome structure and the mechanism of translation. Cell 108: 557–572. 10.1016/S0092-8674(02)00619-0 [DOI] [PubMed] [Google Scholar]
- Robertson MP, Scott WG. 2008. A general method for phasing novel complex RNA crystal structures without heavy-atom derivatives. Acta Crystallogr D Biol Crystallogr D64: 738–744. 10.1107/S0907444908011578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. 1974. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 250: 546–551. 10.1038/250546a0 [DOI] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moore PB. 2000. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA 6: 1091–1105. 10.1017/S1355838200000364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N. 2008. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J Appl Crystallogr 41: 641–643. 10.1107/S0021889808006985 [DOI] [Google Scholar]
- Sutton JL, Pollack L. 2015. Tuning RNA flexibility with helix length and junction sequence. Biophys J 109: 2644–2653. 10.1016/j.bpj.2015.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickle IJ, Flensburg C, Keller P, Paciorek W, Sharff A, Vornhein C, Bricogne G. 2018. STARANISO. Global Phasing Ltd., Cambridge, UK. [Google Scholar]
- Wang J. 2010. Inclusion of weak high-resolution X-ray data for improvement of a group II intron structure. Acta Crystallogr D Biol Crystallogr 66: 988–1000. 10.1107/S0907444910029938 [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D 67: 235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Walker RC, Phizicky EM, Mathews DH. 2014. Influence of sequence and covalent modifications on yeast tRNA dynamics. J Chem Theory Comput 10: 3473–3483. 10.1021/ct500107y [DOI] [PMC free article] [PubMed] [Google Scholar]