Abstract
Background
RP11-334E6.12 is a dysregulated long noncoding RNA (lncRNA) that has never been studied in breast cancer. The biological function and potential mechanism of RNA RP11-334E6.12 in tumorigenesis are still unknown.
Methods
We scanned the Cancer Genome Atlas (TCGA) database and identified RP11-334E6.12 as one of the most dysregulated lncRNAs. The level of RP11-334E6.12 was assessed in breast cancer (BC) tissue samples and BC cell lines. The survival and RP11-334E6.12 expression of patients were analysed. The biological influence of RP11-334E6.12 on BC cell lines was studied using proliferation, Transwell migration, and invasion assays.
Results
RP11-334E6.12 was upregulated in both the TCGA database and our own database. Moreover, survival analyses indicated that RP11-334E6.12 was related to poor overall survival. Moreover, RP11-334E6.12 promoted the proliferation, migration and invasion of BC cells. RP11-334E6.12 promotes the epithelial mesenchymal transition of BC by activating the STAT3 pathway.
Conclusion
Taken together, our results demonstrate that RP11-334E6.12 is associated with the progression of breast cancer. Our findings indicate that long noncoding RNA RP11-334E6.12 promotes the proliferation, migration and invasion of breast cancer cells by activating the STAT3 pathway.
Keywords: breast cancer, RP11-334E6.12, survival, proliferation, migration, invasion
Background
Breast cancer (BC) is one of the most common cancers worldwide and is the second leading cause of cancer-related mortality in women.1 Although advances in the therapy have been made, many patients still suffer from metastasis and recurrence. Immunotherapeutic strategies, such as CAR T cells and PDL1, have been applied, but immunotherapy was not suitable for all patients.2 Hence, more therapeutic targets are urgently needed.
Long noncoding RNAs (lncRNAs) are involved in many physiological and pathological processes in humans, such as cell division and differentiation.3,4 LncRNAs are reported to be involved in the progression of multiple kinds of carcinomas.5,6 Many studies have also shown that lncRNAs contribute to the progression of breast carcinoma through processes such as proliferation, angiogenesis and tissue invasion.7,8
In the present study, we scanned the TCGA database and identified RP11-334E6.12 as a significantly upregulated lncRNA in BC. We next detected the expression difference between normal tissue and BC. We next uncovered the biological function of RP11-334E6.12 through a series of experiments and found that RP11-334E6.12 promotes the proliferation, migration and invasion of BC by activating STAT3.
Methods
Tissue Samples
All human tissues were obtained from the surgical suite in the Department of Thyroid and Breast Surgery, Shandong Provincial Hospital affiliated with Shandong University after confirmation by a pathologist. Tissues were obtained with the patients’ written consent under a protocol approved by the institution’s Institutional Review Board.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, NY, USA) according to the manufacturer’s instructions. qRT-PCR was performed with the SYBR green detection RT-PCR system (Takara, Japan). Actin was used for normalization. All conditions were repeated in triplicate. The 2−ΔΔCt method was used to calculate the relative RNA expression.
Cell Culture and Cell Transfection Assay
The human mammary cancer cell lines MCF-10A, MDA-453, BT-549, BT-474, MCF-7 and MDA-231 were gifts from Professor Tian of the Department of Thyroid and Breast Surgery, Shandong Provincial Hospital affiliated with Shandong University and was approved by institutional review board or ethics committee of Shandong Provincial Hospital affiliated with Shandong University. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM), Minimum Essential Medium (MEM) and RPMI 1640 supplemented with 10% foetal bovine serum (Life Technologies, Grand Island, NY, USA), 1% penicillin G, and streptomycin in the incubator at 37°C with 5% CO2.
Cell Counting Kit-8 Assay
The Cell Counting Kit-8 (CCK-8, Dojondo, Tabaru, Japan) assay was applied to measure the proliferation ability. Cells (500) were seeded into 96-well plates. The absorption values were measured at 24, 48, and 72 hrs after primary seeding. The experiments were repeated three times, and the data are shown as the mean±standard deviation (SD).
Colony Formation Assay
Stable cell lines were seeded in a 6-well plate at a density of 500 cells per well. After being cultured for 2 weeks, cell colonies were fixed with 4% paraformaldehyde and stained with 1% crystal violet. The colonies were examined and counted under a microscope.
Transwell Migration and Invasion Assay
A Transwell assay was applied to detect migration and invasion ability. For the invasion assay, Matrigel chambers (BD Biosciences, San Jose, CA, USA) were used to determine the effect of cells on invasion according to the manufacturer’s instructions. Cells (7.5×104 cells/well) were resuspended in 250 μL of medium in the upper chamber (8-μm pore size, Costar, Corning, NY, USA) of a Transwell system, whereas the lower chamber was filled with 0.5 mL of medium supplemented with 10% FBS. After incubation for 24 hrs at 37°C, the invasive cells were fixed with 100% methanol and stained with 0.5% crystal violet before counting under an inverted microscope. For the migration assay, the indicated cells were plated on uncoated Matrigel upper chambers. The number of migrated cells was estimated under a microscope (Nikon, Tokyo, Japan) at 200× magnification.
Lentivirus Production and Stable Cell Line Construction
Lentiviral vectors expressing shRNA and control were cotransfected with packaging vectors psPAX2 and pMD2G (Addgene) into HEK293FT cells for lentivirus production using Lipofectamine 3000 in accordance with the manufacturer’s instructions. To establish stable cell lines, cells were transduced by using the above lentiviruses and polybrene (8 mg/mL, Sigma). After incubating for 72 hrs, cells were selected with 2 mg/mL puromycin for 3 days.
Statistical Analysis
All data analyses were performed with SPSS 20.0 statistical software. The χ2 test was used to analyse the relationships between categorical variables. The differences between groups were compared by Student’s t-test. Cox regression and Kaplan-Meier methods were used to analyse OS, and p<0.05 was considered to be a significant difference from the control.
Results
RP11-334E6.12 Is Upregulated in BC Tissues and Associated with Poor Prognosis
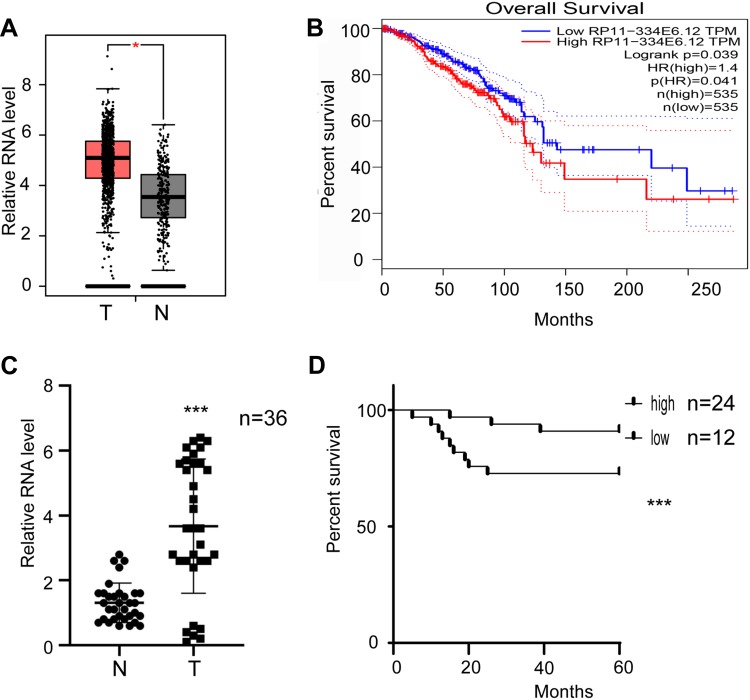
After TCGA screening, the long noncoding RNA RP11-334E6.12 was identified as one of the lncRNAs contributing to the progression of BC (Figure 1A, p<0.05, 1B, p <0.05). To further verify this hypothesis, we analysed the expression pattern in 36 paired normal tissues and tumour tissues. RP11-334E6.12 was significantly upregulated in tumour tissue compared to normal tissue (p <0.001) (Figure 1C). Moreover, we analysed the correlation between RP11-334E6.12 and the overall survival of patients. Our own database showed that RP11-334E6.12 was negatively associated with prognosis (p <0.001) (Figure 1D), and patients with a high level of RP11-334E6.12 had a shorter overall survival time than those with a low level. Taken together, RP11-334E6.12 was negatively associated with the tumorigenesis and progression of breast cancer.
Figure 1.
RP11-334E6.12 was upregulated in BC and was negatively corelated with prognosis.
Notes: (A) The relative level of RP11-480I12.5 in TCGA database (student‘s two tailed paired test *p<0.05). (B) Survive analysis of patients in TCGA database (Log rank test). (C) The relative level of RP11-480I12.5 in our own database (student‘s two tailed paired test ***p<0.001). (D) Survive analysis of patients in our own database (Log rank test p<0.001).
RP11-334E6.12 Promotes the Proliferation of BC
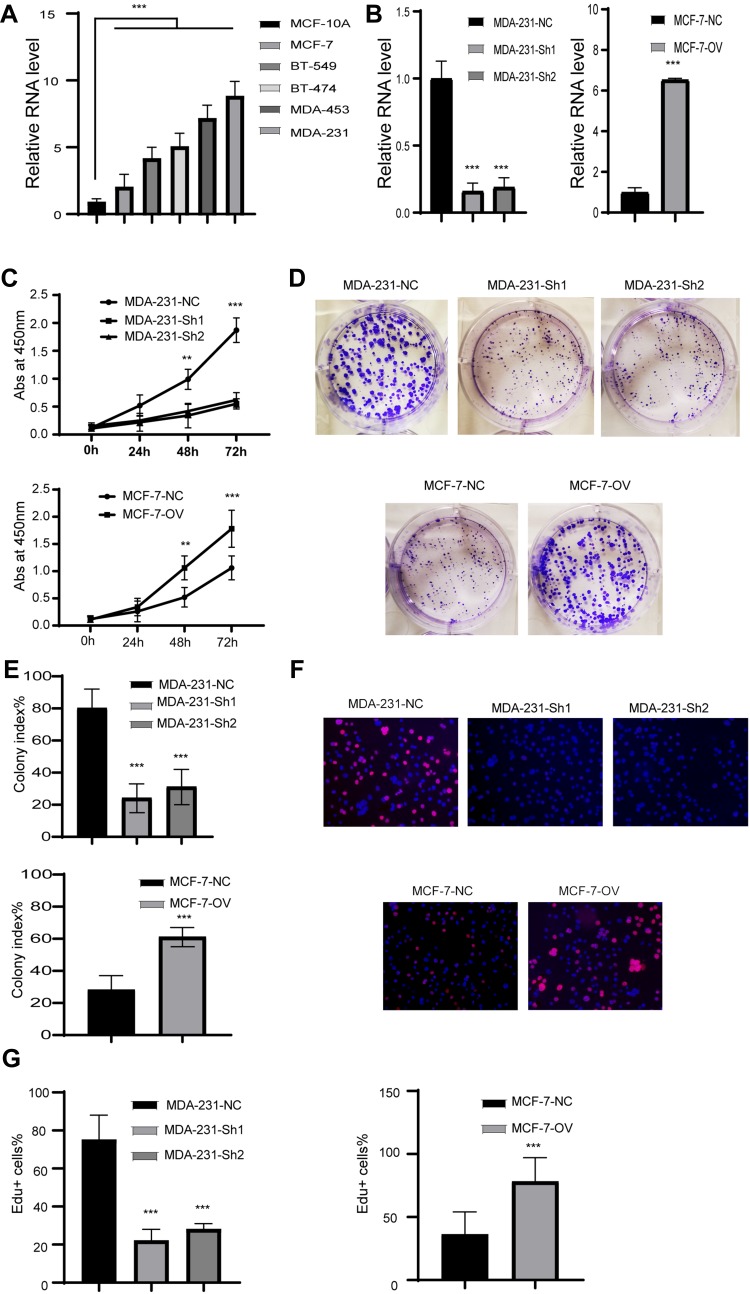
We showed that RP11-334E6.12 is upregulated in BC and negatively associated with prognosis. To further uncover the biological function of RP11-480I12.5, we next detected the expression pattern in BC cell lines. The level of RP11-334E6.12 was higher in BC cell lines than in the control normal cell line MCF-10A (p <0.001) (Figure 2A). We next established a stable RP11-334E6.12 knockdown MDA-231 cell line and an RP11-334E6.12-overexpressing MCF-7 cell line. The relative RNA level is shown in Figure 2B (p <0.001). We applied a series of experiments to measure the proliferation ability of the cell lines described above. Cells with higher RP11-334E6.12 developed an improved proliferation ability in both MDA-231-NC and MCF-7-OV cell lines. Cells with a higher level of RP11-334E6.12 harboured a higher absorbance at 450 nm (p <0.01, p <0.001) (Figure 2C) and a higher colony index (Figure 2D and E, p <0.001). We next detected the DNA synthesis rate in the cell lines described above through an EdU assay. Both MDA-231-NC and MCF-7-OV cells harboured a higher percentage of Edu+ cells. These results indicate that RP11-334E6.12 promotes DNA synthesis (Figure 2F and G, p <0.001).
Figure 2.
RP11-334E6.12 promotes the proliferation in BC.
Notes: (A) The relative level of RP11-334E6.12 in BC cell line (***p<0.001). (B) The relative level of RP11-334E6.12 in stable cell line (***p<0.001). (C) The abs at 450nm of different cell line (**p<0.01, ***p<0.001). (D) The representative image of colony formation assay. (E) The statistical analysis of colony formation assay (***p<0.001). (F) The representative image of Edu assay. (G) The statistical analysis of Edu assay (***p<0.001).
RP11-334E6.12 Promotes the Migration and Invasion of BC Cells
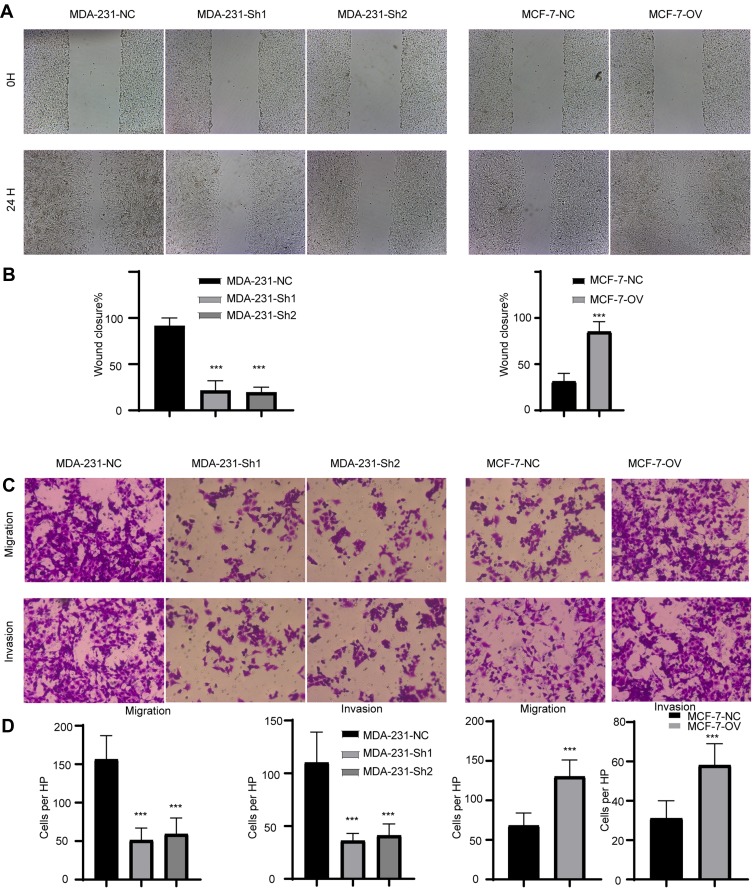
We have previously demonstrated that RP11-334E6.12 promotes proliferation in BC cell lines. Recurrence and metastasis are the major reasons for cancer-related death. To uncover the biofunction of RP11-334E6.12 in migration and invasion, we applied wound healing, Transwell and invasion assays. Cells with higher levels of RP11-334E6.12 developed improved migration and invasion abilities in wound healing (Figure 3A and 3B, p <0.001) and transwell and invasion chamber assays (Figure 3C and 3D p <0.001).
Figure 3.
RP11-334E6.12 promotes the migration and invasion of BC.
Notes: (A) The representative image of wound healing. (B) The statistical analysis of wound healing (***p<0.001). (C) The representative image of transwell and invasion chamber. (D) The statistical analysis of transwell and invasion chamber (***p<0.001).
RP11-334E6.12 Promotes the Epithelial Mesenchymal Transition of BC Cell Lines
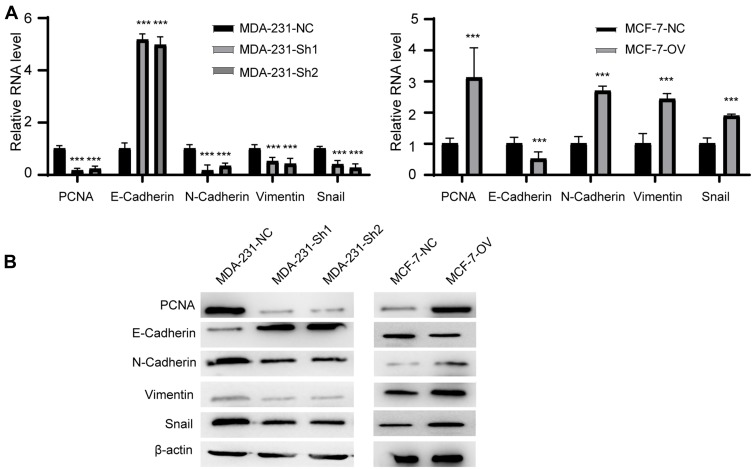
We proved that RP11-334E6.12 promotes proliferation, migration and invasion in BC cell lines. Epithelial mesenchymal transition is one of the most popular pathways reflecting the status of BC cells. We next examined EMT markers and proliferation markers in the different cell lines. The results showed that mesenchymal markers and proliferation markers decreased in stable knockdown cell lines compared to control cells, while epithelial markers increased at both the RNA level (Figure 4A, p <0.001) and protein level (Figure 4B).
Figure 4.
RP11-334E6.12 promotes EMT of BC.
Notes: (A) The relative RNA level of EMT markers and PCNA of different cell line (***p<0.001). (B) The Western blot of EMT markers and PCNA of different cell line.
RP11-334E6.12 Activates the STAT3 Pathway in BC
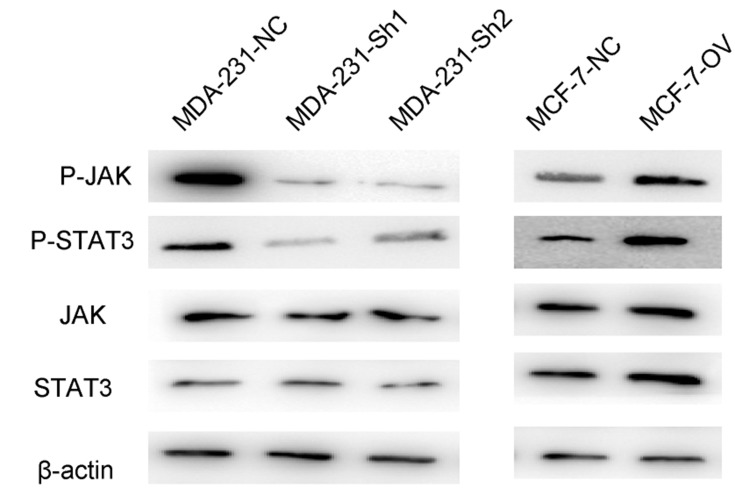
We next tried to determine the potential mechanism of promoting EMT. We examined some critical pathways, such as the β-catenin, STAT3 and NOTCH pathways (data not shown). Finally, we found that the STAT3 pathway was the most dysregulated pathway. P-JAK and P-STAT3 decreased in stable knocking cell lines (Figure 5A). We hypothesized that RP11-334E6.12 may exert its function through the phosphorylation of the JAK family.
Figure 5.
RP11-334E6.12 promotes the phosphorylation of STAT3 pathway.
Discussion
Breast carcinoma is the most common malignancy in women worldwide. Although many advancements have been made, many patients still suffer from recurrence and metastasis. However, the mechanism of the tumorigenesis of breast carcinoma is still unknown. Recent studies have shown that many lncRNAs participate in the tumorigenesis and progression of BC through multiple aspects. Dong proved that FBXL19-AS1 promotes cell proliferation and inhibits cell apoptosis via the miR-876-5p/FOXM1 axis in breast cancer.9 Tang found that LncCCAT1 promotes breast cancer stem cell function by activating WNT/β-catenin signalling.10 lncRNA GAS5 was recently reported to promote apoptosis in triple-negative breast cancer by targeting miR-378a-5p/SUFU signalling.7 Rossi found that lncRNAs interact with proteins to further facilitate breast progression.11
STAT3 is one of the key pathways sustaining the stemness of stem cells, and it has been reported to be overactivated in multiple kinds of cancers.12 After activation by inducers, STAT3 is phosphorylated by the JAK family and thus enters the nucleus to exert its function as a transcription factor.13 STAT3 was reported to be overactivated in the breast and to promote proliferation and migration. However, STAT3 is one key transcription factor shared in both normal cells and malignant cells. Therapies directly targeting STAT3 are not available.
In the present research, we found that RP11-334E6.12 is commonly overexpressed in BC. RP11-334E6.12 promotes proliferation, migration and invasion through the EMT pathway by activating STAT3 pathways.
Conclusion
Taken together, our results indicate that RP11-334E6.12 is associated with tumour progression in breast cancers. Our findings indicate that the long noncoding RNA RP11-334E6.12 promotes the proliferation, migration and invasion of breast cancer cells through the EMT pathway by activating STAT3 pathways.
Abbreviations
TCGA, The Cancer Genome Atlas; BC, breast cancer; LncRNA, long noncoding RNA.
Ethics and Consent
Tissues were obtained with the patients’ written and informed consent under a protocol approved by the institution’s Institutional Review Board in accordance with the Declaration of Helsinki. Tissues were obtained with the patients’ written consent for publication.
Data Sharing Statement
The data are available in this article.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 2.Keren L, Bosse M, Marquez D, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174(6):1373–1387. doi: 10.1016/j.cell.2018.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo ZH, Walid AA, Xie Y, et al. Construction and analysis of a dysregulated lncRNA-associated ceRNA network in a rat model of temporal lobe epilepsy. Seizure. 2019;69:105–114. doi: 10.1016/j.seizure.2019.04.010 [DOI] [PubMed] [Google Scholar]
- 4.Huang T, Wang J, Zhou Y, Zhao Y, Hang D, Cao Y. LncRNA CASC2 is upregulated in osteoarthritis and participates in the regulation of IL-17 expression and chondrocyte proliferation and apoptosis. Biosci Rep. 2019;39. doi: 10.1042/BSR20182454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taheri M, Ghafouri-Fard S. Long non-coding RNA signature in cervical cancer. Klin Onkol. 2018;31(6):403–408. [DOI] [PubMed] [Google Scholar]
- 6.Xu YD, Shang J, Li M, Zhang YY. LncRNA DANCR accelerates the development of multidrug resistance of gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23(7):2794–2802. doi: 10.26355/eurrev_201904_17554 [DOI] [PubMed] [Google Scholar]
- 7.Zheng S, Li M, Miao K, Xu H. lncRNA GAS5-promoted apoptosis in triple-negative breast cancer by targeting miR-378a-5p/SUFU signaling. J Cell Biochem. 2019. [DOI] [PubMed] [Google Scholar]
- 8.Wang N, Hou M, Zhan Y, Sheng X. LncRNA PTCSC3 inhibits triple-negative breast cancer cell proliferation by downregulating lncRNA H19. J Cell Biochem. 2019;120(9):15083–15088. [DOI] [PubMed] [Google Scholar]
- 9.Dong G, Pan T, Zhou D, Li C, Liu J, Zhang J. FBXL19-AS1 promotes cell proliferation and inhibits cell apoptosis via miR-876-5p/FOXM1 axis in breast cancer. Acta Biochim Biophys Sin (Shanghai). 2019. doi: 10.1093/abbs/gmz110 [DOI] [PubMed] [Google Scholar]
- 10.Tang T, Guo C, Xia T, et al. LncCCAT1 promotes breast cancer stem cell function through activating WNT/beta-catenin signaling. Theranostics. 2019;9(24):7384–7402. doi: 10.7150/thno.37892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi T, Pistoni M, Sancisi V, et al. RAIN is a novel enhancer-associated lncRNA that controls RUNX2 expression and promotes breast and thyroid cancer. Mol Cancer Res. 2019;18(1):140–152. [DOI] [PubMed] [Google Scholar]
- 12.Palmitoylation alters STAT3 activity and tumor response to high-fat diet. Cancer Discov. 2019. [DOI] [PubMed] [Google Scholar]
- 13.Kang S, Tanaka T, Narazaki M, Kishimoto T. Targeting interleukin-6 signaling in clinic. Immunity. 2019;50(4):1007–1023. doi: 10.1016/j.immuni.2019.03.026 [DOI] [PubMed] [Google Scholar]