Abstract
Background
KLF16, a member of the Kruppel-like factor (KLF) family, functions in the regulation of dopaminergic transmission, metabolism, and endocrinology. However, the role of KLF16 in prostate cancer (PCa) remains unknown.
Methods
We screened the expression of KLFs in PCa based on bioinformatics analysis. The protein levels of KLF16 in PCa specimens were confirmed by immunohistochemistry. Inhibiting KLF16 by RNA interference with shRNA was used to determine the effects of KLF16 on PCa cell growth in vitro and in vivo. RNA sequencing was used to investigate the signaling regulated by KLF16 in PCa. Bioinformatics analysis was also used to determine the possible correlations of KLF16 and signaling in PCa cohorts.
Results
Bioinformatics analysis showed that KLF16 may be required for PCa development. Notably, the expression of KLF16 was elevated in human PCa tissues. In vitro and in vivo experiments both demonstrated that depleting KLF16 significantly inhibited the growth of PCa cells. Downregulation of KLF16 significantly decreased the expression of MYC signaling in PCa cells. Furthermore, KLF16 expression was correlated with MYC signaling activity.
Conclusion
KLF16 was overexpressed in PCa tissues compared to normal tissues. KLF16 knockdown suppressed PCa cell growth in vitro and in vivo, and a deficiency of KLF16 inhibited activation of MYC signaling.
Keywords: Kruppel-like factor, KLF16, prostate cancer, MYC
Introduction
Prostate cancer (PCa) is a common cause of cancer deaths among males.1 When patients are diagnosed, PCa is usually androgen-dependent, so androgen deprivation treatment is the basic therapy for advanced PCa.2,3 During progression of PCa, part of the tumor becomes hormone refractory. Patients must then receive chemotherapy using docetaxel or cabazitaxel4,5 Many molecular mechanisms have been reported that take part in the development of PCa2,6 However, there is little known about the occurrence and progression of PCa. Hence, it is urgent to identify the mechanism of action, which could be used in individualized therapies.
Kruppel-like factors (KLFs) constitute a family of transcription factors that participate in diverse biological processes, including proliferation, migration, differentiation, inflammation, and pluripotency.7,8 It has been reported that some KLFs are involved in the initiation and development of PCa. For example, the expression of KLF4 is decreased in advanced PCa and regulates the epithelial-mesenchymal transition.9 KLF6 is mutated in a subset of PCa, and heterozygosity of KLF6 is lost in 77% of primary prostate tumors.10 In the following study, it has shown that wild type KLF6 is a key prostate cancer tumor suppressor gene, and caused cell‐cycle arrest through inhibiting cyclin‐dependent kinase.11,12 KLF8 is an androgen receptor transcriptional co-activator and is overexpressed in PCa.13 Our previous study showed that KLF9 and KLF13 functioned as tumor suppressers in PCa.14,15 However, the involvement of KLF16 in PCa, a member of the KLF family, is still unknown.
In this study, we found that KLF16 expression was upregulated in PCa tissues compared to normal tissues. Knockdown of KLF16 significantly suppressed tumor growth of PCa cells in vitro and in vivo. Furthermore, the MYC signature was inhibited following disruption of KLF16 in PCa cells. Overall, our results strongly suggest that elevated KLF16 contributes to the progression of PCa.
Materials and Methods
Cell Culture
The 22RV1 and LNCaP cell lines were kindly provided by the Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China). The PC3 cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). The 22RV1, LNCaP and PC3 cell lines were grown in RPMI-1640 medium (Corning, Inc., Corning, NY, USA) containing 10% fetal bovine serum (Gemini, Woodland Hills, CA, USA), 1% penicillin/streptomycin (Gibco, Grand Island, NY, USA), and 1% HEPES (Corning, Inc.). All cultures were maintained at 37°C in a 5% CO2 incubator.
Reagents, Plasmids and Lentivirus
Puromycin was purchased from MedChem Express (Shanghai, China). The short hairpin RNA (shRNA) expression sequence (5′-CTCGCACCTAAAGTCGCACCT-3′) and the control shRNA sequence (5′-GCTCCGTGAACGGCCACGAGT-3′) were cloned into the pLKO.1 vector. Lentivirus were produced by cotransfection of the psPAX2 (Addgene 12260) and the pCMV-VSVG plasmid (Addgene 8454) into 293T cells using Lipofectamine 2000 (Invitrogen). The culture medium was changed and collected in 48h and 72h, and supernatants were filtered through a 0.45-μm filter syringe to obtain the products containing lentivirus. Then, the products were used for transduction of the indicated cells with of 5 μg mL−1 of polybrene (H9268; Sigma-Aldrich, St. Louis, MO, USA) and selected on 5 μg mL−1 puromycin to establish the stable line.
Cell Growth and Colony Formation Assay
Cell growth was detected by seeding 2000 cells per well in 96-well plates. The cells were cultured in 200 μL of complete medium for 6 days. The MTT assay was used to measure cell growth. The detailed procedures have been described in a previous study.16 For colony formation assay, the indicated cells were planted in 6-well plates (2000–5000 cells/well) and cultured in complete medium for 1–2 weeks until the macroscopic colonies appearing. Then, the colonies were fixed in methanol for 10 min and stained with 0.5% crystal violet for more than 1 hr.
Western Blotting and Immunohistochemistry
Western blotting (WB) and immunohistochemistry (IHC) were performed as previously described.14 The following antibodies were used: KLF16 (HPA052481; Sigma-Aldrich, St. Louis, MO, USA; WB: 1:1000, IHC: 1:100), MYC (E5Q6W; Cell Signaling Technology, Danvers, MA, USA; WB:1:1000), MAX (D154193; Sangon Biotech, Shanghai, China; WB:1:1000) and β-actin (sc-47778; Santa Cruz Biotechnology, Santa Cruz, CA, USA; WB: 1:5000). IHC scores were calculated using the following formula: IHC score = intensity score × percentage score. The intensity score was measured according to the intensity of staining (0: negative, 1: weak, 2: moderate, and 3: strong); the percentage score was determined using the percentage of stained area (0: 0%, 1: 1–25%, 2: 26–50%, 3: 51–75%, and 4: 76–100%). The prostate cancer and normal tissues were collected by The Fifth People’s Hospital of Shanghai, Fudan University with written informed consent. This study was approved by Medical Ethics Committee of The Fifth People’s Hospital of Shanghai, Fudan University (2018-LL-076B). All experiments were performed following the guidelines and regulations of Medical Ethics Committee of The Fifth People’s Hospital of Shanghai, Fudan University.
Animal Experiments
PC3 cells were infected with a lentivirus target control or KLF16. Then, 1 × 106 PC3 cells were injected with Matrigel (volume 1:1; Corning, Inc.) into the flanks of 6-week-old male nude mice (n = 6). After 6 weeks, all mice were sacrificed, and the tumor xenografts were dissected and weighed. All animal experiments were subject to approval by the Department of Laboratory Animal Science, Fudan University (2018-WY-JS038), and conducted according to the regulations for Department of Laboratory Animal Science, Fudan University.
RNA Sequencing and Analysis
Total RNA extracted from indicated groups of PC3 cells was subjected to RNA sequencing (RNA-seq) performed by Majorbio Biopharm Technology (Shanghai, China). The sequencing reads were analyzed using the free online platform of Majorbio Cloud Platform (www.majorbio.com) to obtain the expression profiles. The sequence data have been submitted to Gene Expression Omnibus (GEO, GSE138286). The gene set enrichment analysis (GSEA) used the GSEA software provided by the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp) following the website’s instructions. We used the curated gene sets (C6 oncogenic gene sets) within MSigDB.
Published Datasets and Analysis
The Cancer Genome Atlas (TCGA)17 and Memorial Sloan Kettering Cancer Center (MSKCC)18 expression profiles were downloaded from UCSC Xena (http://xena.ucsc.edu/). The cell cycle progression (CCP) activity was calculated by the z-score of the sum of the expression levels of each CCP gene. When the published datasets were performed by GSEA, the analyses included the following: the metric for ranking genes was “Pearson,” the permutation type was “phenotype,” and the other sets were default.
Statistics
All statistical analyses were calculated by Prism software, version 8 (GraphPad Software, San Diego, CA, USA). A value of p < 0.05 was considered statistically significant. The semi-quantitation of Western blotting was made by gray scale measurement using ImageJ (https://imagej.nih.gov/ij/).
Results
Identification of KLF16 as a Regulator in PCa
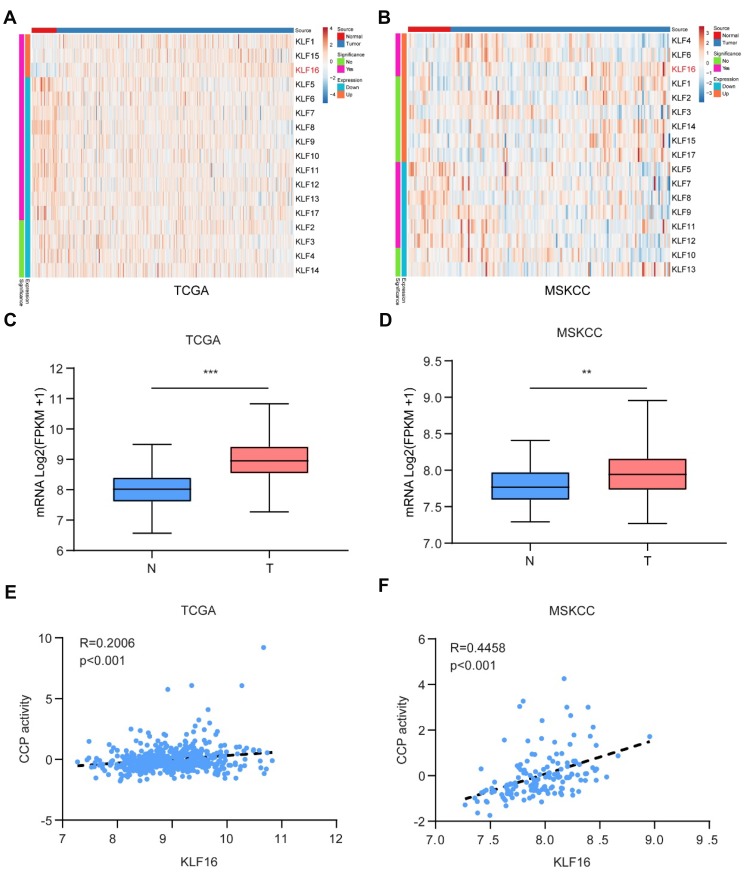
To identify possible oncogenes in KLFs, we screened the expression levels of KLFs in prostatic normal or tumor tissues by in silico analysis (Figure 1A and B). In two published datasets (TCGA and MSKCC), KLF16 expression was found to be increased in tumor tissues (Figure 1C and D). Thirty-one genes involved in CCP were shown to be correlated with the mortality of primary PCa.19 We also confirmed that CCP activity was correlated with cell growth in PCa.20 We analyzed the correlation between KLF16 expression levels and CCP activities in published datasets for human PCa. As expected, the level of KLF16 was positively correlated with CCP activity in the two cohorts (Figure 1E and F). It has been shown that KLF16 may influence the proliferation of PCa cells, and all analyses of published datasets indicate that KLF16 plays an important role in the development of PCa.
Figure 1.
KLF16 is a potential oncogene in prostate cancer (PCa). (A, B) Expression of Kruppel-like factors (KLFs) of normal prostate tissues and PCa tissues in The Cancer Genome Atlas (TCGA) and Memorial Sloan Kettering Cancer Center (MSKCC) is shown by a heat map using a classification of differential expression. (C, D) RNA expression of PCa tissues (T) showing KLF16 expression was significantly higher than normal prostate tissues in the TCGA and MSKCC cohorts (t-test; **p < 0.01; ***p < 0.001). (E, F) The plots show significant positive correlations between KLF16 and cell cycle progression in the TCGA or MSKCC datasets (Pearson’s correlation).
KLF16 Expression Is Upregulated in PCa Tissues
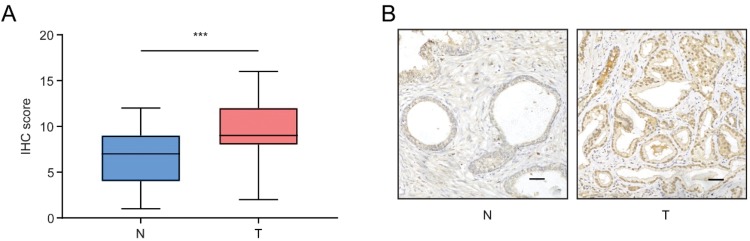
To confirm the expression of KLF16 in clinical specimens, we determined the expression levels of KLF16 in normal (n = 32) and PCa (n = 56) tissues using IHC. The stain scores of tumor tissues were significantly higher than normal tissues (Mann–Whitney test; p < 0.001; Figure 2A). The staining of normal prostate tissues was weaker than PCa tissues (Figure 2B). Consistent with the results of published datasets, KLF16 expression was elevated in PCa tissues. Taken together, these results strongly suggest that KLF16 plays an important role in the tumorigenesis of PCa.
Figure 2.
KLF16 expression is upregulated in prostate cancer (PCa). (A) Expression of KLF16 in normal prostate tissues and PCa tissues examined by immunohistochemistry (Mann–Whitney test; ***p < 0.001). (B) Representative photographs of normal prostate tissues and PCa tissues (scale bar: 200 μm).
KLF16 Knockdown Decreases the Growth of PCa Cells in vitro and in vivo
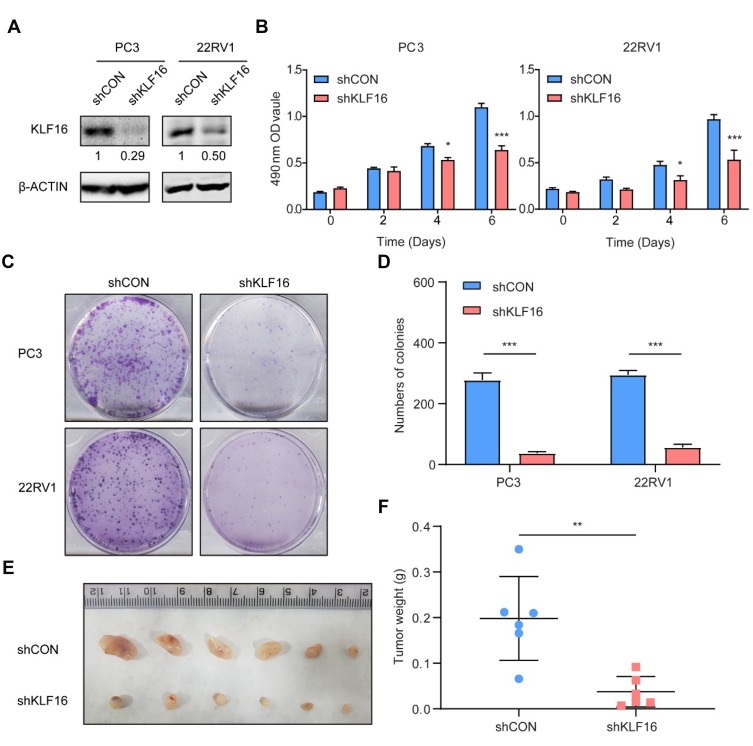
To test the function of KLF16 on PCa cell proliferation in vitro, we interrupted KLF16 by shRNA targeting KLF16. The knockdown efficiency of KLF16 was confirmed by Western blotting (Figure 3A, Supplemental Fig. A). Using in vitro experiments, we found that deletion of KLF16 in PC3 or 22RV1 cells dramatically affected cell growth and colony formation (Figure 3B–D) Activity of KLF16 in prostate cancer was confirmed in another cell line, LNCaP cells (Supplemental Fig. B-D). Next, we subcutaneously injected PC3 cells with or without downregulated KLF16 into nude mice. After 6 weeks, the mice were sacrificed. Xenografts of the control group were significantly larger (Figure 3E) and weighed more than those in the KLF16 knockdown group (Figure 3F). These results show that inhibiting KLF16 significantly suppressed PCa cell growth both in vitro and in vivo.
Figure 3.
Knockdown of KLF16 inhibits prostate cancer (PCa) cell growth in vitro and in vivo. PC3 and 22RV1 cells were infected by a lentivirus with a short hairpin RNA (shRNA) targeting KLF16 or a control sequence. (A) The efficiency of the shRNA in PCa cells was confirmed by Western blotting. The semi-quantitation of Western blotting was shown under the pictures. (B) PC3 and 22RV1 cells expressing shRNA targeting KLF16 or a control sequence were cultured in 96-well plates for 6 days. The MTT assay was conducted to determine viability of the indicated cells (t-test; *p < 0.05; ***p < 0.001). (C) PC3 and 22RV1 cells expressing their respective shRNAs were examined using a colony formation assay. (D) The numbers of colonies for each group were counted and the results are shown (t-test; ***p < 0.001). (E) The mice with xenograft were shown after implanting PC3 cells 6 weeks later. (F) The weight of the xenograft tumors was calculated. Error bars represent mean ± SD (Mann–Whitney, ** p < 0.01, n = 6).
Downregulation of KLF16 Influences MYC Signaling
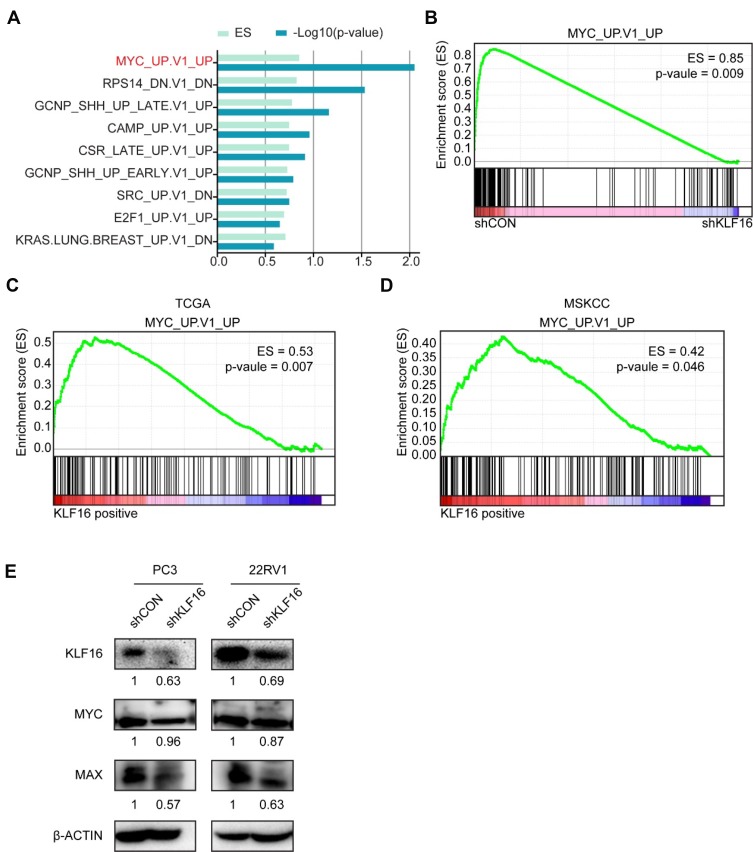
To evaluate the possible oncogenic signaling responsible for the suppressive effects of KLF16 deletion on PCa cell growth, we performed RNA-seq analyses of PC3 cells with or without KLF16 knockdown. Then, the gene expression profiles of PC3 cells followed by GSEA were shown in rank order (Figure 4A). The set of genes, which was upregulated in human mammary epithelial cells overexpressing the MYC gene, was significantly enriched in control PC3 cells (Figure 4B). In clinical specimens, the gene set reflecting MYC signaling activity was also significantly increased in cohorts with high KLF16 expression (Figure 4C and D). Although the expression of MYC did not change remarkably after KLF16 knockdown, we have found that the important MYC transcriptional partner, MAX, has significant downregulated (Figure 4E).
Figure 4.
Loss of KLF16 results in inhibition of MYC signaling. (A) Gene set enrichment analysis (GSEA) was performed based on oncogenic signatures that disrupt KLF16 in PC3 cells. The results are shown in rank order of p-value and enrichment scores. (B) GSEA of the profile data inhibiting expression of KLF16 in PC3 cells based on the gene set of MYC signaling in the control (left) and shKLF16 (right). (C, D) GSEA of the TCGA and MSKCC datasets based on the MYC signaling gene set and KLF16 transcription levels. (E) PC3 and 22RV1 cells were infected with the indicated shRNAs virus, and the indicated proteins were detected by Western blot. The semi-quantitation of Western blotting was shown under the pictures.
Discussion
The roles that KLFs play in cancer have been reported by an increasing number of investigators.8 In particular, KLFs have been found to be crucial for the development of PCa. In our previous studies, we showed that two KLFs (KLF914 and KLF1315) functioned as tumor suppressers in PCa. Nevertheless, much remains to be understood regarding the roles of KLFs in PCa. In this study, using in silico analyses, we found that KLF16 might promote the progression of PCa.
KLF16 was first reported as a dopamine receptor regulating factor, which modulated dopaminergic transmission in the brain.21 One study revealed that KLF16 is involved in the regulation of metabolism and endocrinology.22 However, there have been few studies regarding its roles in different cancers. In glioma cells, KLF16 suppresses cell proliferation, which negatively regulates the expression of TFAM.23 However, another study showed that the deletion of KLF16 inhibited the growth of gastric cancer cells by decreasing the expression of p21 and CDK4.24 In the present study, our findings indicate that KLF16 may promote the proliferation and occurrence of PCa. Overall, these studies have shown a diverse role of KLF16 in tumors.
Our study is the first to investigate the expression of KLF16 in PCa tissues, and to determine potential biological roles in PCa cells. We found that KLF16 was highly expressed in PCa tissues compared to normal prostate tissues, both in our specimens and in published cohorts. Moreover, the mRNA expression of KLF16 was positively correlated with CCP activity, which reflected the prognosis of PCa in two individual datasets. We then confirmed that loss of KLF16 attenuated the proliferation of PCa cells in vitro and in vivo. The RNA-seq data using PCa cells and published datasets both showed that disrupting KLF16 inhibited MYC signaling. In the present study, we did not observe significant changes in MYC protein levels following KLF16 knockdown, indicating that KLF16 might influence MYC function rather than its expression. The downregulation of MAX after KLF16 knockdown had confirmed it. We then found that MYC activity was inhibited following disruption of KLF16. Because MYC plays important roles in cell cycle transitions,25,26 our findings are consistent with other studies, which have reported that deletion of KLF16 causes cell cycle arrest.
Conclusions
In both datasets and clinical specimens, KLF16 expression was evaluated in PCa and normal tissues. Expression of KLF16 dramatically inhibited PCa cell growth in vitro and in vivo. A deficiency of KLF16 inhibited MYC signaling in PCa cells. KLF16 expression was also positively correlated with MYC signaling in PCa cohorts. Our findings indicate that KLF16 may play important roles in the development of PCa.
Funding Statement
The project was supported by Natural Science Research Funds of Minhang District, Shanghai (Grant No.2018MHZ016), Scientific Research Project funded by The Fifth People’s Hospital of Shanghai, Fudan University (Grant No.2018WYZT06) and the Shanghai Key Medical Specialty Program (Grant No.ZK2019A03).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Attard G, Parker C, Eeles RA, et al. Prostate cancer. Lancet. 2016;387(10013):70–82. doi: 10.1016/S0140-6736(14)61947-4 [DOI] [PubMed] [Google Scholar]
- 3.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701–711. doi: 10.1038/nrc4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrylak DP, Tangen CM, Hussain MH, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351(15):1513–1520. doi: 10.1056/NEJMoa041318 [DOI] [PubMed] [Google Scholar]
- 5.Tannock IF, de Wit R, Berry WR, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351(15):1502–1512. doi: 10.1056/NEJMoa040720 [DOI] [PubMed] [Google Scholar]
- 6.Karantanos T, Evans CP, Tombal B, Thompson TC, Montironi R, Isaacs WB. Understanding the mechanisms of androgen deprivation resistance in prostate cancer at the molecular level. Eur Urol. 2015;67(3):470–479. doi: 10.1016/j.eururo.2014.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90(4):1337–1381. doi: 10.1152/physrev.00058.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tetreault MP, Yang Y, Katz JP. Kruppel-like factors in cancer. Nat Rev Cancer. 2013;13(10):701–713. doi: 10.1038/nrc3582 [DOI] [PubMed] [Google Scholar]
- 9.Liu YN, Abou-Kheir W, Yin JJ, et al. Critical and reciprocal regulation of KLF4 and SLUG in transforming growth factor beta-initiated prostate cancer epithelial-mesenchymal transition. Mol Cell Biol. 2012;32(5):941–953. doi: 10.1128/MCB.06306-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narla G, Heath KE, Reeves HL, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294(5551):2563–2566. doi: 10.1126/science.1066326 [DOI] [PubMed] [Google Scholar]
- 11.Narla G, DiFeo A, Yao S, et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005;65(13):5761–5768. doi: 10.1158/0008-5472.CAN-05-0217 [DOI] [PubMed] [Google Scholar]
- 12.Benzeno S, Narla G, Allina J, et al. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64(11):3885–3891. doi: 10.1158/0008-5472.CAN-03-2818 [DOI] [PubMed] [Google Scholar]
- 13.He HJ, Gu XF, Xu WH, Yang DJ, Wang XM, Su Y. Kruppel-like factor 8 is a novel androgen receptor co-activator in human prostate cancer. Acta Pharmacol Sin. 2013;34(2):282–288. doi: 10.1038/aps.2012.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen P, Sun J, Xu G, et al. KLF9, a transcription factor induced in flutamide-caused cell apoptosis, inhibits AKT activation and suppresses tumor growth of prostate cancer cells. Prostate. 2014;74(9):946–958. doi: 10.1002/pros.v74.9 [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Peng R, Wang B, et al. Transcription factor KLF13 inhibits AKT activation and suppresses the growth of prostate carcinoma cells. Cancer Biomark. 2018;22(3):533–541. doi: 10.3233/CBM-181196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu W, Li J, Wang Q, et al. Targeting POH1 inhibits prostate cancer cell growth and enhances the suppressive efficacy of androgen deprivation and docetaxel. Prostate. 2019;79(11):1304–1315. doi: 10.1002/pros.v79.11 [DOI] [PubMed] [Google Scholar]
- 17.Abeshouse A, Ahn J, Akbani R; Cancer Genome Atlas Research N. The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12(3):245–255. doi: 10.1016/S1470-2045(10)70295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A, Coleman I, Morrissey C, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22(4):369–378. doi: 10.1038/nm.4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang CK, D’Souza UM, Eisch AJ, et al. Dopamine receptor regulating factor, DRRF: a zinc finger transcription factor. Proc Natl Acad Sci USA. 2001;98(13):7558–7563. doi: 10.1073/pnas.121635798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daftary GS, Lomberk GA, Buttar NS, et al. Detailed structural-functional analysis of the Kruppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. J Biol Chem. 2012;287(10):7010–7025. doi: 10.1074/jbc.M111.266007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Li S, Ke Y, et al. KLF16 suppresses human glioma cell proliferation and tumourigenicity by targeting TFAM. Artif Cells Nanomed Biotechnol. 2018;46(sup1):608–615. doi: 10.1080/21691401.2018.1431654 [DOI] [PubMed] [Google Scholar]
- 24.Ma P, Sun CQ, Wang YF, et al. KLF16 promotes proliferation in gastric cancer cells via regulating p21 and CDK4. Am J Transl Res. 2017;9(6):3027–3036. [PMC free article] [PubMed] [Google Scholar]
- 25.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849(5):506–516. doi: 10.1016/j.bbagrm.2014.03.013 [DOI] [PubMed] [Google Scholar]