Abstract
Transcription is a highly regulated process that supplies living cells with coding and non-coding RNA molecules. Failure to properly regulate transcription is associated with human pathologies, including cancers. RNA polymerase II is the enzyme complex that synthesizes messenger RNAs that are then translated into proteins. In spite of its complexity, RNA polymerase requires a plethora of general transcription factors to be recruited to the transcription start site as part of a large transcription pre-initiation complex, and to help it gain access to the transcribed strand of the DNA. This chapter reviews the structure and function of these eukaryotic transcription pre-initiation complexes, with a particular emphasis on two of its constituents, the multisubunit complexes TFIID and TFIIH. We also compare the overall architecture of the RNA polymerase II pre-initiation complex with those of RNA polymerases I and III, involved in transcription of ribosomal RNA and non-coding RNAs such as tRNAs and snRNAs, and discuss the general, conserved features that are applicable to all eukaryotic RNA polymerase systems.
Keywords: Transcription, Initiation, General transcription factors, TFIIH, TFIID, RNA polymerase, Gene expression, Structural biology, Cryo-electron microscopy
Introduction to Transcription in Eukaryotes
Transcription is the synthesis of RNA based on a DNA template. While phage, viral, and organellar gene expression systems generally use simple single-subunit RNA polymerase enzymes, bacterial, archaeal, and eukaryotic RNA polymerases are increasingly complex multisubunit enzymes. A 5-subunit core is conserved from bacteria to humans, though archaeal and eukaryotic RNA polymerases harbor up to 12 additional subunits (Fig. 5.1a–e). Eukaryotes use three structurally and functionally distinct RNA polymerases (Roeder and Rutter 1969, 1970; Sklar et al. 1975), abbreviated Pol I, Pol II, and Pol III, each of them specialized for the synthesis of certain classes of RNAs. Pol I synthesizes long ribosomal RNA (rRNA) precursors in the nucleolus that are later processed into 25S/28S, 18S, and 5.8S rRNAs, Pol II synthesizes mostly messenger RNAs (mRNAs) that are later translated into proteins, and Pol III synthesizes tRNAs, 5S rRNA, and other small non-coding RNAs (Roeder and Rutter 1970; Weinmann and Roeder 1974).
Fig. 5.1.
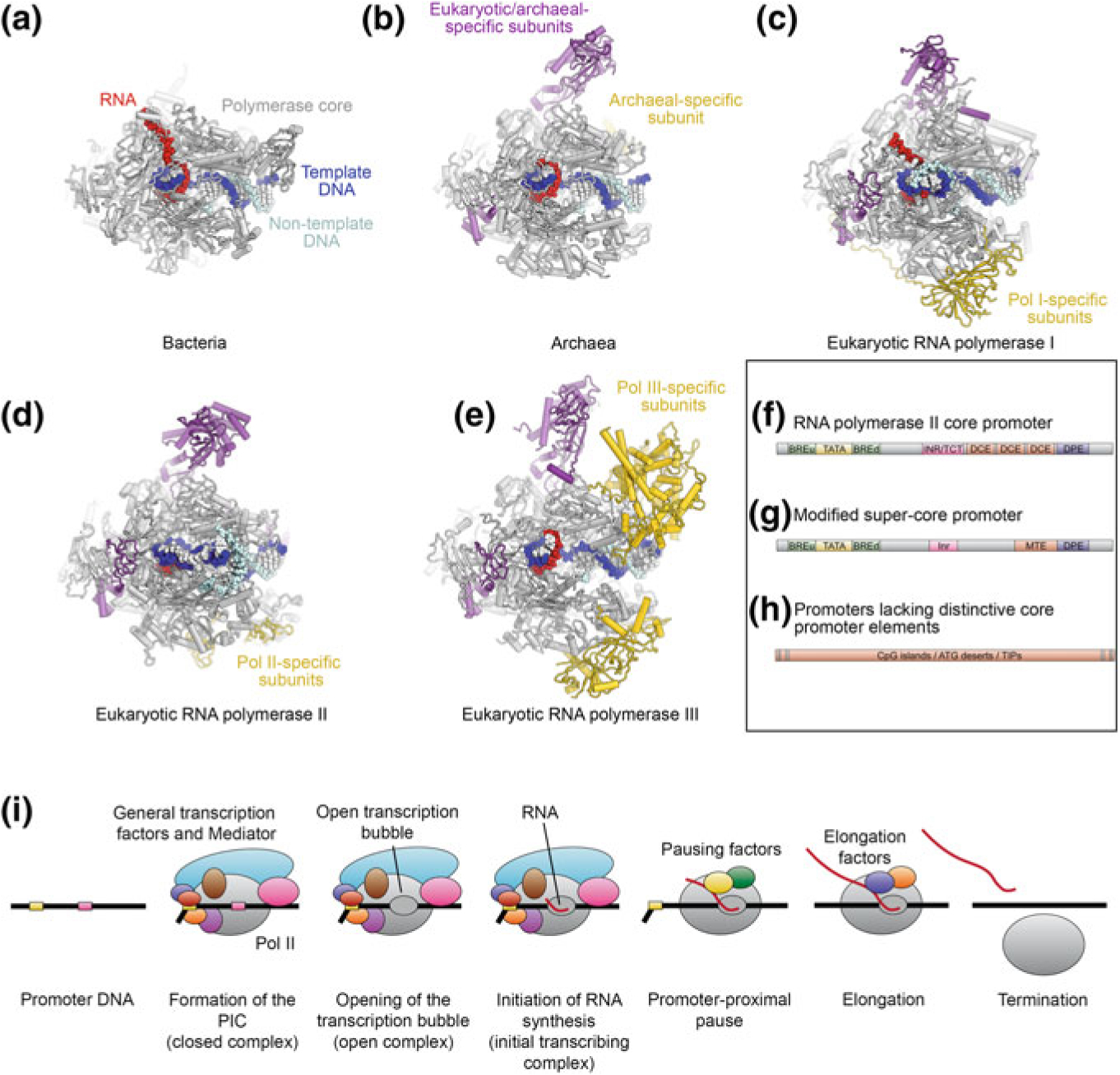
Architecture of multisubunit RNA polymerases across kingdoms and organization of eukaryotic Pol II promoters. a–e Depictions of transcribing multisubunit RNA polymerases. Subunits occurring only in eukaryotes or archaea are colored purple, subunits specific to polymerase type are colored yellow. a Elongating bacterial RNA polymerase (Vassylyev et al. 2007). b Archaeal RNA polymerase (Korkhin et al. 2009), DNA superposed from (Vassylyev et al. 2007). c Elongating yeast Pol I (Tafur et al. 2016). d Elongating fungal Pol II (Ehara et al. 2017). d Elongating yeast Pol III (Hoffmann et al. 2015). f–h Organization of eukaryotic Pol II core promoters. f Idealized Pol II core promoter elements. Abbreviations: TATA, TATA-box; INR, initiator; TCT, initiator like element present in ribosomal protein coding genes; DCE, downstream core element; DPE, downstream promoter element (Lenhard et al. 2012; Roy and Singer 2015). g Modified super core promoter used for structural studies of the human Pol II-PIC and TFIID. The upstream and downstream TFIIB-response elements BREu (Lagrange et al. 1998) and BREd (Deng and Roberts 2005) were added to the super core promoter (Juven-Gershon et al. 2006), which comprises TATA-box (Gannon et al. 1979), Inr (Chen and Struhl 1985), MTE (Lim et al. 2004), and DPE (Burke and Kadonaga 1996, 1997). h Many eukaryotic Pol II promoters lack these distinct promoter elements and contain longer DNA segments harbouring CpG islands, ATG deserts, or transcription initiation platforms (TIPs) instead (Roy and Singer 2015). i Simplified schematic of the Pol II transcription cycle from PIC assembly to transcription termination
The DNA core promoters that control expression of their corresponding genes also differ substantially between the three eukaryotic polymerase systems. In the Pol II system, several core promoter elements have been identified (Burke and Kadonaga 1996, 1997; Chen and Struhl 1985; Deng and Roberts 2005; Gannon et al. 1979; Juven-Gershon and Kadonaga 2010; Lagrange et al. 1998; Lenhard et al. 2012; Lim et al. 2004; Roy and Singer 2015), and a synthetic promoter combining a TATA-box, initiator (Inr), downstream promoter element (DPE), and motif ten element (MTE), which provides a highly specific and high-affinity binding site for the transcription machinery (Juven-Gershon et al. 2006) has been used in structural analysis of promoter-bound human Pol II pre-initiation complexes. However, not all of these elements are typically present in any natural promoter. For example, most Pol II promoters lack a TATA box (Carninci et al. 2006; Sandelin et al. 2007), the canonical binding site of TATA-box binding protein (TBP), which is a subunit of the general transcription factor IID (TFIID) and also ubiquitously used in the Pol I and Pol III systems (see below). Nevertheless, TBP still participates in assembly of the transcription machinery on Pol II promoters lacking TATA boxes because the recognition of the other core promoter motifs by TFIID allows TBP loading on the DNA (Burke and Kadonaga 1997; Kutach and Kadonaga 2000; Lim et al. 2004; Pugh and Tjian 1991). Panels f–h of Fig. 5.1 provide examples of the promoter elements typically found in eukaryotic genes.
Transcription is commonly subdivided into three phases (Fig. 5.1i). After the step-wise assembly of a pre-initiation complex (PIC) on the DNA (Buratowski et al. 1989), whereby general transcription factors and RNA polymerase are recruited to the promoter, the transcription bubble opens, allowing the synthesis of the first phosphodiester bond in the active site of the RNA polymerase. This step is referred to as initiation of transcription. Subsequently, RNA polymerase needs to clear the promoter and, in the case of Pol II, overcome a promoter-proximal pause, allowing it to enter the elongation phase. After synthesis of the product RNA, transcription needs to be terminated during the termination phase of the transcription reaction.
The initiation of transcription is a highly regulated process. Together with the subsequent regulation of RNA processing, RNA stability, and regulated translation of mRNAs into proteins, transcription plays a major role in the control of gene expression as a whole. Deregulation of transcription is implicated in human pathologies, and elevated transcription levels by each of the three eukaryotic RNA polymerases have been associated with cancer (Cabarcas and Schramm 2011; Johnson et al. 2008; Lockwood et al. 2010).
The Pol II Pre-initiation Complex
Pol II and the General Transcription Factors
The enzymatic core of Pol II shows extensive structural homology to the 5-subunit α2ββ’ω core of bacterial RNA polymerases (Cramer et al. 2000; Zhang et al. 1999) (Fig. 5.1a, d) and the fundamental mechanism of phosphodiester bond synthesis is highly conserved (Cramer et al. 2001; Gnatt et al. 2001; Kaplan 2013; Svetlov and Nudler 2013; Wang et al. 2006; Westover et al. 2004). However, Pol II includes 7 additional subunits to form a 12-subunit complex (Figs. 5.1a, d and 5.2a, b). Structural landmarks of Pol II (Fig. 5.2c, d) include the stalk, two jaws that interact with the incoming DNA substrate, the flexible clamp domain that closes over the bound nucleic acids in initiation and elongating complexes, and two pores that allow substrates to enter and product mRNAs to exit the catalytically active complex (Fig. 5.2c, d) (Cramer et al. 2000, 2001; Gnatt et al. 2001). Behind the unwound transcription bubble in the active site, a protein wall forces the DNA to adopt a bend before exiting the polymerase (Fig. 5.2c). In the transcribing complex (Fig. 5.2e, f), two fork loops from the Rpb2 subunit maintain the open transcription bubble (Cramer et al. 2001; Gnatt et al. 2001). Motions of the bridge helix that contacts the DNA-RNA hybrid in the active site are coupled to substrate translocation, while opening and closing of the trigger loop is associated with catalysis, which is mediated by two metal ions bound in the active site (Fig. 5.2f) (Gnatt et al. 2001; Wang et al. 2006; Westover et al. 2004).
Fig. 5.2.
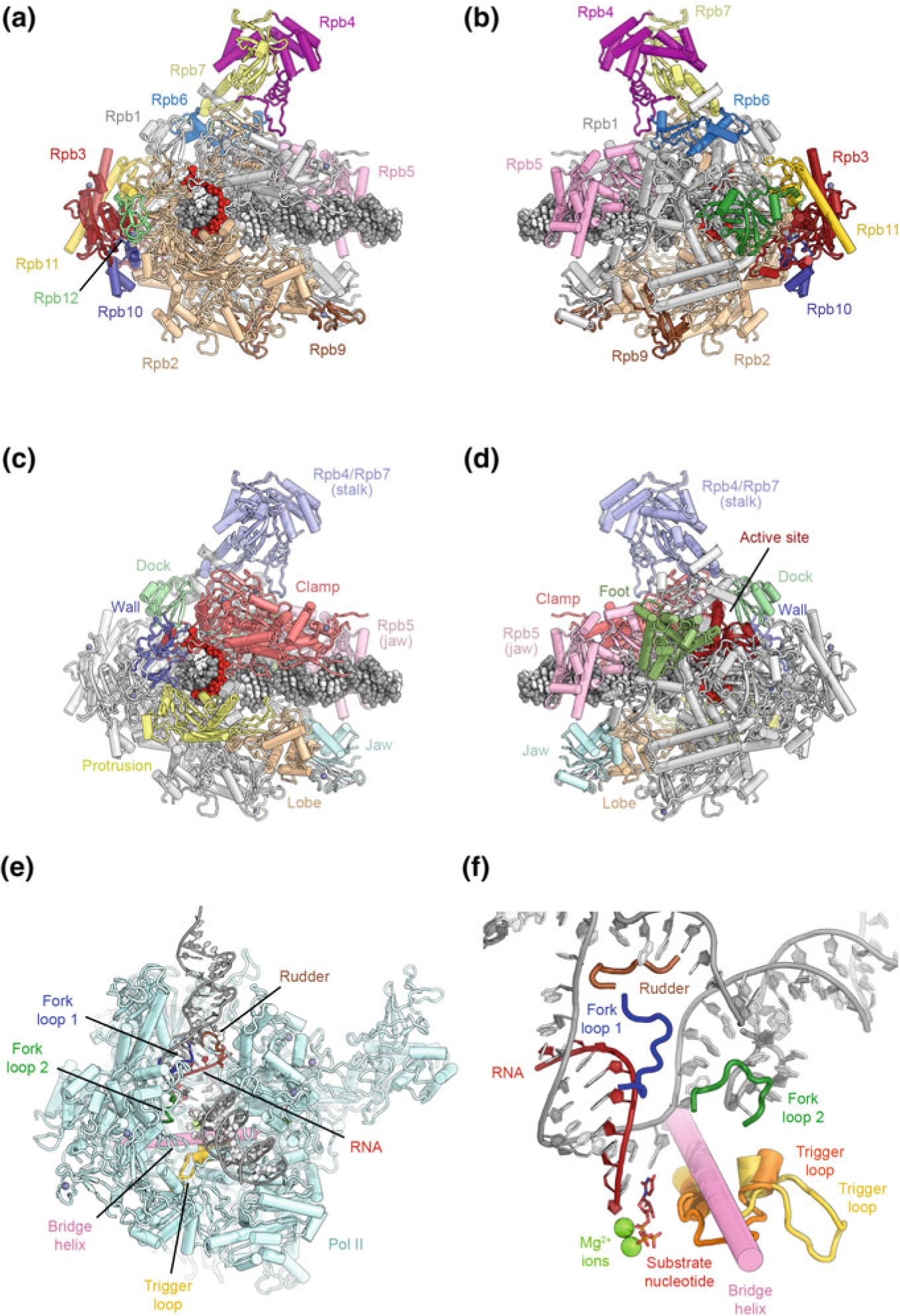
Architecture of eukaryotic RNA polymerase II. a, b Depiction of transcribing Pol II extracted from the structure of an elongation complex (Ehara et al. 2017), shown from the front and back and colored by protein subunits. c, d Structural domains of the Pol II subunits Rpb1 and Rpb2 are highlighted in color according to (Cramer et al. 2001). Additional structural landmarks mentioned in the text are highlighted as well (protein names are indicated). e View of the transcription bubble at the Pol II active site (Barnes et al. 2015). Key structural and functional elements near the active site that are mentioned in the text are indicated. f Detailed view of the Pol II active site (Barnes et al. 2015; Wang et al. 2006). Two conformations of the trigger loop are shown in orange and yellow
The initiation of mRNA synthesis requires Pol II and the general transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (Sainsbury et al. 2015) to form a pre-initiation complex (PIC) on the promoter. Activated transcription additionally depends on Mediator, a 1.2 MDa multiprotein complex, and the activity of a number of cofactors and gene-specific activators and repressors. In contrast to the smaller, oligomeric general transcription factors TFIIA, TFIIB, TFIIE, and TFIIF, two general transcription factors are large multiprotein assemblies: TFIIH is a heterodecameric protein complex with a molecular weight of 0.5 MDa, and TFIID is a 1.3 MDa multiprotein assembly of TATA-box binding protein (TBP) and 13 different TBP-associated factors (TAFs) in humans. We will discuss the structures and functional mechanisms of these two general transcription factors in more detail in the following sections, while the remaining general transcription factors will be discussed in the context of the Pol II-PIC.
TFIID
Function of TFIID
The assembly of the PIC begins with the recognition of the core promoter DNA by TFIID, with the help of TFIIA (Burke and Kadonaga 1997; Chalkley and Verrijzer 1999; Lee et al. 2005; Verrijzer et al. 1995). TBP then interacts with TFIIB, which in turn interacts with Pol II, thereby nucleating the assembly of the full PIC (Buratowski et al. 1989). In addition to TBP, direct interactions between TFIID TAFs and other components of the general transcription machinery have also been proposed (Dubrovskaya et al. 1996; Ruppert and Tjian 1995). Furthermore, TFIID, together with Mediator and the multifunctional SAGA complex, integrates signals from transcriptional activators and has been shown to be required for activated transcription (Baek et al. 2002; Chen et al. 1993, 1994; Horikoshi et al. 1988; Pugh and Tjian 1990; Stringer et al. 1990; Wu and Chiang 2001). Additionally, a number of domains within the complex recognize specific histone modifications associated with active genes (Jacobson et al. 2000; Vermeulen et al. 2007). Thus, TFIID is important for nucleating PIC assembly at the promoter and also serves as a hub for the integration of regulatory cues that are important for regulated transcription.
Conformational Complexity of TFIID
Due to the size and complexity of TFIID, structural efforts to characterize the full complex have involved electron microscopy (EM) of natively purified samples. Early studies of negatively stained human and budding yeast TFIID showed it to have an overall horseshoe-shaped architecture of three major lobes, termed lobes A, B and C (Andel et al. 1999; Brand et al. 1999; Leurent et al. 2002). Improving the resolution by cryo-EM proved to be extremely challenging due to conformational heterogeneity (Elmlund et al. 2009; Grob et al. 2006; Papai et al. 2009). It was ultimately shown that lobes B and C form a relatively stable core (the BC core), while lobe A is flexibly attached and can undergo conformational rearrangements on the scale of 100 Å, from interacting with lobe C to interacting with lobe B (Fig. 5.3a) (Cianfrocco et al. 2013). Free TFIID was found to occupy two preferred conformational states, termed the canonical and rearranged states, with addition of TFIIA and promoter DNA shifting this equilibrium towards the rearranged state (Fig. 5.3a). In this conformation, lobe A interacts with lobe B and is positioned near the TATA box, suggesting the presence of TBP within lobe A (Cianfrocco et al. 2013) (Fig. 5.3a).
Fig. 5.3.
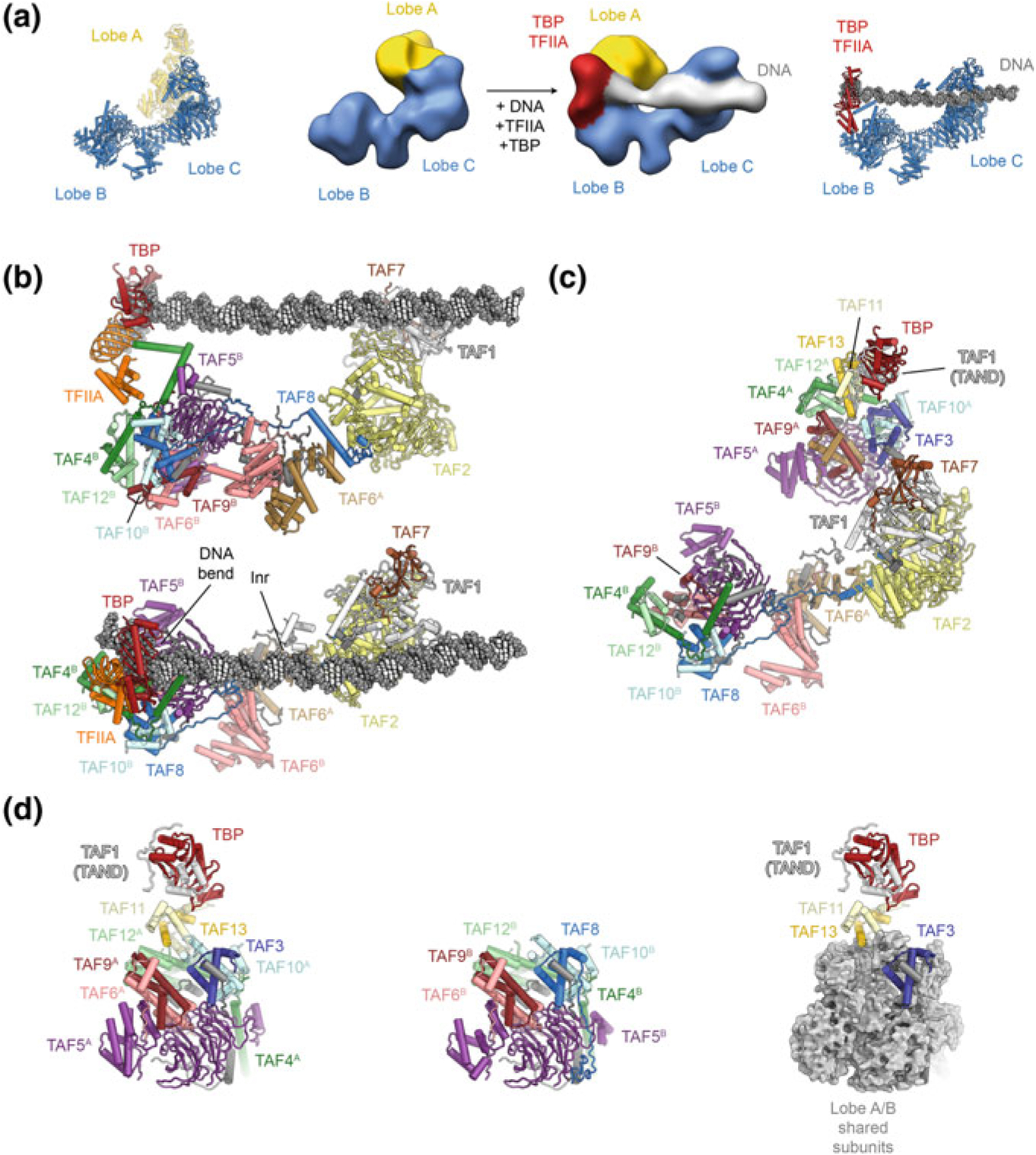
The structure of human TFIID. a Organization of human TFIID into three lobes. Early cryo-EM maps (Cianfrocco et al. 2013) that revealed the tri-lobal overall structure of TFIID are shown in the center, with recent coordinate models (Patel et al. 2018) shown next to them for comparison. b Structure of TFIID bound to DNA and TFIIA (Patel et al. 2018) in side view (top) and top view (bottom). Protein subunits are shown in colour and labelled; the TBP-induced DNA bend and the location of the transcription start site (Inr) are labeled. c Structure of TFIID in the canonical state (Patel et al. 2018) shown in the same orientation as bottom panel in (b). d Composition of lobes A and B. Left: lobe A. Center: lobe B. Right: Subunits shared between lobes A and B are shown as grey surface to highlight the subunits in lobe A that distinguish between lobes A and B
The Structure of TFIID
Subsequent structural studies of human TFIID aimed to provide higher-resolution analysis of both free and DNA-bound TFIID (Louder et al. 2016; Patel et al. 2018) (Fig. 5.3b, c). While we will focus on the structure of human TFIID below, similar work in yeast has also been reported recently, although promoter DNA could not be clearly visualized in the cryo-EM structure of promoter-bound yeast TFIID (Kolesnikova et al. 2018).
The structure of stably DNA-bound human TFIID (Fig. 5.3b), using the super core promoter, and in complex with TFIIA, showed that lobe C includes TAFs 1, 2, and 7, with a TAF1-TAF7 module binding the DPE and MTE downstream promoter elements, while a small domain within TAF1 contacts the Inr motif (indicated in Fig. 5.3b) that includes the transcription start site (TSS) (Louder et al. 2016). Lobe C is connected to lobe B via a long helix in TAF8 that also interacts with a centrally positioned dimer of HEAT repeats from the two copies of TAF6. The TAF6 dimer connects to lobe B, which in turn contacts the TBP-TFIIA module, as well as promoter sequences near the TATA box (Louder et al. 2016; Nogales et al. 2017a; Nogales et al. 2017c). The TATA box DNA in this structure is bent (Fig. 5.3b, bottom), mirroring the structure of the isolated TBP-DNA complex (Nikolov et al. 1996). By virtue of this arrangement, lobes B and C act as a molecular ruler around the TSS, providing a platform for transcription pre-initiation complex assembly (Louder et al. 2016; Nogales et al. 2017b).
The complete molecular architecture of all three lobes of human TFIID was derived from the cryo-EM analysis of free TFIID (Patel et al. 2018) (Fig. 5.3c). Due to the scarcity, structural heterogeneity, fragility, and preferred orientation of this specimen on the cryo-EM grid, determination of this structure was a tour-de-force. A striking structural feature of TFIID is that lobes A and B share a common core architecture (Fig. 5.3d), with six TAFs being present in two copies, one in each lobe (Bieniossek et al. 2013; Leurent et al. 2002; Patel et al. 2018; Sanders et al. 2002). In this common module, TAF5, which contains a helical N-terminal domain and a WD40 domain, binds to three dimers of TAFs that contain histone-fold domains (HFDs) and themselves form a hexameric assembly that resembles the overall architecture of the nucleosome core particle (Patel et al. 2018), in agreement with earlier biochemical and structural data (Hoffmann et al. 1996; Xie et al. 1996). TAF5 and five of the histone fold-containing TAFs, TAF10, and the pairs TAF4-TAF12 and TAF6-TAF9, are present in both lobes A and B (Fig. 5.3d). The TAF6 HFDs present in both lobes, A and B, are flexibly connected to their corresponding C-terminal HEAT repeat domains that exist as a dimer at the center of the complex, within lobe C (Fig. 5.3b). In spite of their shared core architecture, lobes A and B are functionally and structurally unique because their common TAF10 subunit associates with the HFD of TAF3 in lobe A but with the HFD of TAF8 in lobe B (Fig. 5.3d). TAF8 contributes to the lobe B-lobe C interaction, while TAF3 recruits the TAF11-TAF13 HFD dimer to lobe A. Together with N-terminal regions of TAF1 (Anandapadamanaban et al. 2013; Kokubo et al. 1993, 1994), the TAF11-TAF13 pair binds TBP, which consequently hangs peripherally on this mobile lobe A within TFIID.
Mechanism of TBP Loading onto the Promoter
The extreme mobility of lobe A, along with its ability to carry TBP, suggests a role for the dynamics of this lobe in the deposition of TBP onto the promoter (Fig. 5.4a). An exhaustive classification of a cryo-EM dataset where TFIID, TFIIA and core promoter DNA were mixed together revealed five distinct states, including previously observed as well as new conformational and functional states of lobe A. These states were termed canonical, extended, scanning, rearranged and engaged states (Fig. 5.4a), with the first corresponding to lobe A positioned proximal to lobe C, and consequent states being further from lobe C and ultimately contacting lobe B (Patel et al. 2018). The model derived from the co-existence of these states proposes that in the canonical state, where lobe A is far removed from the DNA, TBP is bound by the TAF11-TAF13 HFD dimer, as well as the TAF1 TAND1 and TAND2 motifs (Figs. 5.3d, 5.4b), which have been implicated in inhibition of DNA and TFIIA binding by TBP (Fig. 5.4c) (Anandapadamanaban et al. 2013; Kokubo et al. 1993, 1994). These inhibitory interactions are sequentially broken in the scanning and rearranged states as they are replaced by interactions of TBP with DNA and TFIIA (Fig. 5.4d), facilitated by the scaffolding role of TFIID and by lobe A-lobe B interactions. Specifically, we have proposed that in the scanning state TAND1 is displaced as TBP loosely associates with the DNA, while in the rearranged state TAND2 is displaced as TBP binds to TFIIA that also interacts with lobe B. In the final, engaged state, TBP binds tightly and bends the DNA, and it releases from TAF11-TAF13. Displacement of TAF11-TAF13 opens up the binding site for TFIIB, enabling PIC assembly (Patel et al. 2018).
Fig. 5.4.
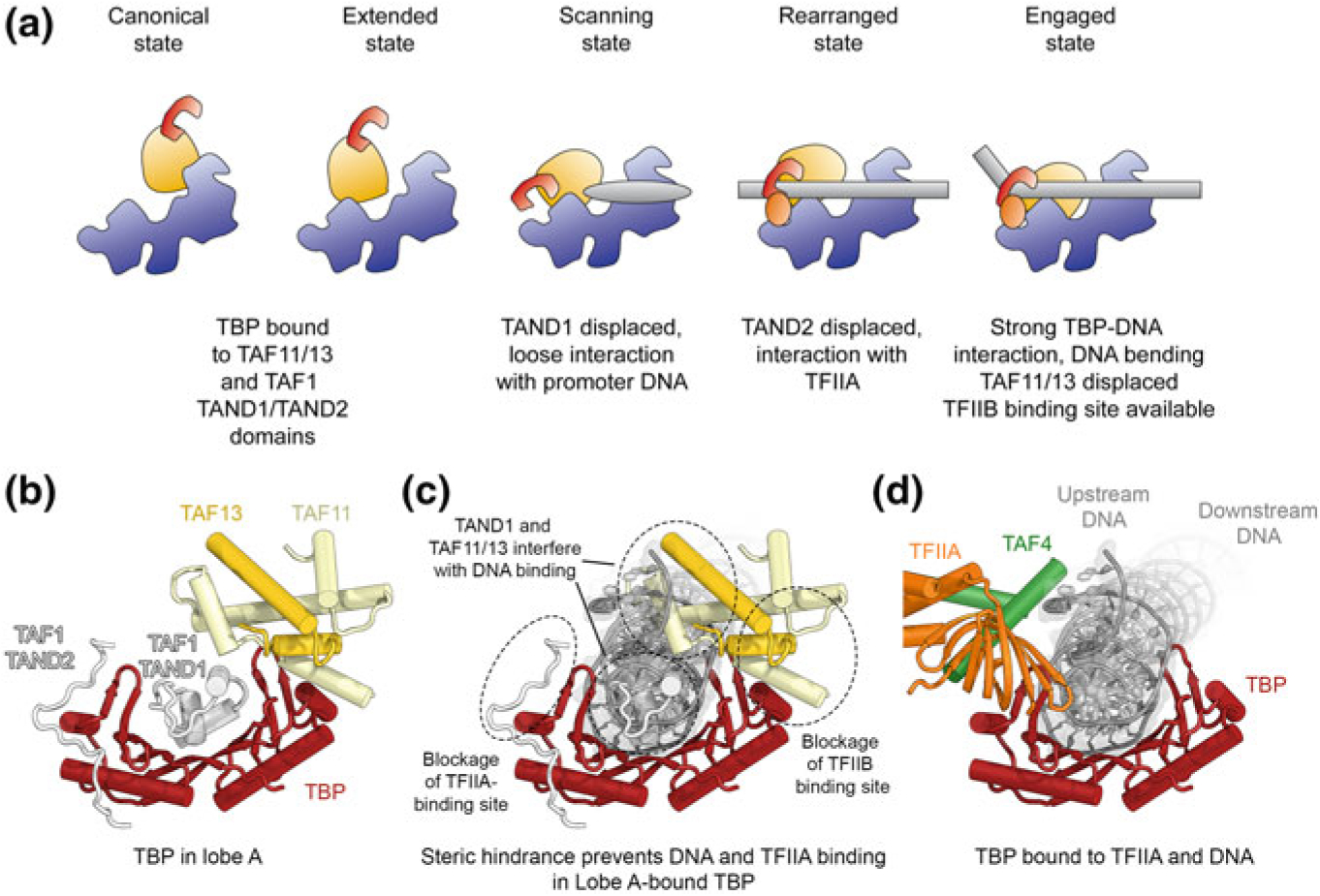
Proposed conformational transitions of TFIID during DNA binding and mechanism of TBP deposition. a Five different conformational states (canonical, extended, scanning, rearranged, and engaged state) of TFIID as identified by cryo-EM analysis of a mixed dataset (Patel et al. 2018). For each state, a schematic depiction and the interaction environment of TBP, which is initially bound to TFIID lobe A and eventually deposited onto upstream promoter DNA in a stably bound manner, is provided. b TBP in lobe A is contacted by the TAF11-TAF13 dimer and the TAND1 and TAND2 domains of TAF1 (Anandapadamanaban et al. 2013; Patel et al. 2018). c TAF11-TAF13 and the TAF1 TAND1 and TAND2-domains block DNA- and transcription factor-binding sites on TBP. d TBP bound to and bending the DNA in the context of the engaged state of DNA-bound TFIID (Patel et al. 2018)
Notably, current data indicate that the overall architecture of TFIID is conserved between promoters that do and do not contain TATA boxes (Cianfrocco et al. 2013; Patel et al. 2018), in agreement with the observation that TBP also participates in initiation on TATA-less promoters (Pugh and Tjian 1991). The intricate mechanism of TBP loading onto the upstream promoter may ensure that even in the absence of a TATA box, TBP engages with DNA in a defined, controlled, and productive fashion, rather than binding DNA nonspecifically. Completion of TBP loading onto the core promoter in such cases (reaching the engaged state) may depend on chromatin marks, transcriptional activators, or co-activator complexes, because both structural and biochemical analysis shows that TBP only weakly binds TATA-free promoters in reconstituted systems that contain TFIID and TFIIA (Cianfrocco et al. 2013) even though transcription assays show transcription from such promoters (Juven-Gershon et al. 2006; Lim et al. 2004).
TFIIH
Functional Roles of TFIIH in Transcription
TFIIH is a protein complex of ten different subunits organized into a core and a CdK activating kinase (CAK) subcomplex (Fig. 5.5a). Both the core and the CAK are required for TFIIH to function in transcription initiation, while only the TFIIH core complex is required in DNA repair (Svejstrup et al. 1995). The TFIIH core complex includes seven protein subunits: XPB, XPD, p62, p52, p44, p34, and p8 (Fig. 5.5a). Two of these, XPB and XPD, are SF2-family DNA-dependent ATPases/helicases (Guzder et al. 1994; Sung et al. 1993; Weeda et al. 1990). While the enzymatic function of XPD is dedicated to DNA repair (Kuper et al. 2014), only XPB activity is required to help promoter opening during transcription initiation (Fishburn et al. 2015; Grünberg et al. 2012; Kim et al. 2000). Originally thought to act like a wrench to introduce torsion into downstream promoter DNA to enable opening of the transcription bubble (Kim et al. 2000), it was later proposed that XPB acts as a DNA translocase that promotes the movement of downstream promoter DNA towards the Pol II active site, thereby facilitating bubble opening (Fig. 5.5b) (Fishburn et al. 2015; Grünberg et al. 2012; He et al. 2013).
Fig. 5.5.

Structure and function of TFIIH. a Schematic of TFIIH architecture. Main enzymatic subunits are shown in color (XPD and XPB, SF2-family DNA-dependent ATPases; CDK7, Cyclin-dependent kinase). Core and CAK subcomplexes are indicated. b Model for the activity of the TFIIH subunit XPB during promoter opening; depiction based on the structures in (He et al. 2016). c Cryo-EM reconstruction of the human Pol II-PIC (He et al. 2016) with fitted coordinate models. TFIIH subunits are highlighted in color. d, e Relocalization of the CAK kinase-cyclin module in the Mediator-bound Pol II-PIC (Schilbach et al. 2017): The cryo-EM volume corresponding to the CAK in the Mediator-containing PIC is shown in orange. The presumed location of the CAK in the absence of Mediator is indicated by an orange ellipse. f Structure of the TFIIH core complex with bound MAT1 (Greber et al. 2019). g Interactions of p62 with the ATP-and DNA-binding sites of XPD. h Mapping of human disease mutations onto the structure of TFIIH (Cleaver et al. 1999; Greber et al. 2019)
The CAK subcomplex includes the CDK7, Cyclin H, and MAT1 subunits (Devault et al. 1995; Fisher et al. 1995; Fisher and Morgan 1994; Shiekhattar et al. 1995) (Fig. 5.5a). CDK7 is a cyclin dependent kinase that requires the presence of Cyclin H for full activity, while MAT1 aids formation of the CDK-cyclin complex (Devault et al. 1995; Fisher et al. 1995) and anchors the CAK subcomplex to the core complex via interactions with XPD and XPB (Abdulrahman et al. 2013; Busso et al. 2000; Greber et al. 2017; Rossignol et al. 1997). The role of XPD in recruitment of the CAK subcomplex (which is required only for transcription) explains why its presence in TFIIH is important for transcription initiation, while its enzymatic activity is not (Dubaele et al. 2003; Kuper et al. 2014). The CAK is able to phosphorylate a number of targets, which include the C-terminal low-complexity region of the Pol II subunit Rpb1, referred to as the Pol II CTD here, as well as transcriptional activators, general transcription factors, and, in humans, cell-cycle CDKs (Fisher and Morgan 1994; Shiekhattar et al. 1995).
TFIIH as a DNA Repair Complex
In addition to its role as a Pol II general transcription factor, TFIIH also serves as a helicase complex in the nucleotide excision DNA repair pathway. This pathway is responsible for removing a large number of bulky base alterations that result in distortion of the DNA backbone, including 6–4 photoproducts, cyclobutane pyrimidine dimers, and other large base adducts (Koch et al. 2016). In these pathways, TFIIH serves to open a DNA bubble around damaged nucleotides, which can then be excised by the structure-specific endonucleases XPF-ERCC1 and XPG (Marteijn et al. 2014). Initial recognition of the damage occurs independently of TFIIH, either by a global genome surveillance pathway that depends on the DDB1-DDB2 and XPC complexes, or via transcription-coupled repair (TCR), in which Pol II stalled by DNA lesions acts as the initial damage sensor, leading to the recruitment of specialized TCR factors such as CSA and CSB, and ultimaltely, TFIIH (Compe and Egly 2012). Unlike in transcription initiation, the enzymatic activity of both XPD and XPB is required during NER. XPD acts as a bona-fide helicase to unwind the DNA double helix and scan for the DNA lesion (Kuper et al. 2014; Li et al. 2015), while the role of XPB has been described either as an ATP-dependent conformational switch that anchors TFIIH to DNA (Coin et al. 2007), or as a weak helicase and DNA translocase that supports scanning of the DNA for the lesion that originally triggered TFIIH recruitment (Li et al. 2015; Sugasawa et al. 2009). Upon lesion recognition in a binding pocket in XPD (Buechner et al. 2014; Mathieu et al. 2013), both helicase motors stall (Li et al. 2015; Naegeli et al. 1993), enabling assembly of the repair bubble, double incision, and ultimately repair of the DNA lesion by DNA polymerases Pol δ and Pol κ (Compe and Egly 2012; Marteijn et al. 2014).
The Structure of TFIIH
Due to its importance for two critical cellular processes, the structure of TFIIH has been intensively investigated for almost two decades. Initial studies resulted in low-resolution EM reconstructions that showed a ring- or horseshoe-shaped complex (Chang and Kornberg 2000; Schultz et al. 2000) and allowed the approximate assignment of the XPB and XPD ATPase subunits to the open ends of a horseshoe (Gibbons et al. 2012). An integrative modelling approach provided a tentative overall architecture for the complex, which was, however, complicated by the horseshoe-shaped overall architecture of the complex that puts many subunits within crosslinking distance of each other, and the high flexibility of the CAK subcomplex (Luo et al. 2015). Sub-nanometer resolution structures of TFIIH in the context of both human and yeast PICs (Fig. 5.5c) confirmed the assignment of the ATPase subunits to density at the open end of the horseshoe-shaped TFIIH complex and revealed the locations of p44 near XPD and of the p52-p8 dimer near XPB (Fig. 5.5c) (He et al. 2013, 2016; Murakami et al. 2015), in agreement with regulatory roles of these subunits on the two ATPase subunits (Coin et al. 2007; Dubaele et al. 2003; Jawhari et al. 2002; Kim et al. 2015). The interpretation of these cryo-EM maps relied heavily on docking of crystal structures or homology models. Structures of bacterial and archaeal homologs of XPB (Fan et al. 2006) and XPD (Bienstock et al. 2003; Fan et al. 2008; Liu et al. 2008; Wolski et al. 2008) provided structural templates for interpretation of the low-resolution cryo-EM maps, and for modelling of human disease mutations.
The TFIIH-containing PIC structures (He et al. 2013, 2016; Tsai et al. 2017) also suggested that in the absence of Mediator, the CAK subcomplex localizes close to XPD and the C-terminal region of Rpb1, thus in proximity to its major substrate (Greber et al. 2017; He et al. 2013; Tsai et al. 2017). However, later Mediator-bound structures showed that in this context, which likely better resembles the in vivo situation, CAK is found in a different location (Robinson et al. 2016; Schilbach et al. 2017) (Fig. 5.5d, e). Both in the absence and in the presence of Mediator, the CAK subcomplex is poorly resolved, and in spite of the availability of high-resolution crystal structures for both CDK7 and Cyclin H (Andersen et al. 1997; Kim et al. 1996; Lolli et al. 2004), the interaction with the third CAK subunit, MAT1, and the mechanism of CAK regulation remain unresolved.
A more detailed and complete molecular model of the TFIIH core complex resulted from the improved cryo-EM reconstructions of apo-TFIIH (Greber et al. 2017) and of the yeast Pol II-PIC (Schilbach et al. 2017). These studies revealed the overall structure of additional TFIIH subunits, including p62, and the positioning of the zinc-binding domains of p34, p44, and the CAK subunit MAT1. The latest breakthrough came from the 3.7 Å-resolution cryo-EM structure of human apo-TFIIH (Greber et al. 2019). This structure revealed the complete network of interactions that governs the assembly of the TFIIH core complex (Fig. 5.5f, g), allowed the mapping of numerous disease mutations in XPD and XPB (Fig. 5.5h and see below), and showed that XPD is probably tightly regulated, not only by MAT1 and p44, as demonstrated by previous biochemical and mutagenesis experiments (Abdulrahman et al. 2013; Dubaele et al. 2003; Kim et al. 2015; Sandrock and Egly 2001), but also by XPB and p62, which block access to known XPD DNA- and ATP-binding sites (Fig. 5.5g) (Greber et al. 2019).
TFIIH in Health and Disease
Defects in TFIIH subunits are associated with xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy (Fig. 5.5h), human disease syndromes characterized by high incidence of cancers, premature ageing, or transcriptional defects (Cleaver et al. 1999; Rapin 2013). Mutations that cause xeroderma pigmentosum, characterized by extreme sensitivity to sunlight and a high incidence of cancers, interfere with the global genome NER pathway by impairing (i) detection of lesions by the specialized damage recognition sensors (e.g. XPC), (ii) repair bubble opening by interfering with the function of the TFIIH helicases XPB and XPD, or (iii) excision of the damaged nucleotide by the XPF-ERCC1 or XPG endonucleases. Cockayne syndrome is a premature ageing condition, which in the case of mutations in the XPD subunit of TFIIH, occurs in combination with xeroderma pigmentosum (Cleaver et al. 1999; Rapin 2013). Cockayne syndrome is thought to be a disease induced by defects in TCR. However, recent data suggest that CS mutations in XPD may lead to stalling of the helicase after initiation of the repair process, which may interfere with completion of the repair reaction and with the resumption of transcription (Herrera-Moyano et al. 2014; Moriel-Carretero et al. 2015; Theron et al. 2005). Mapping of XP and XP-CS mutations onto the structure of human XPD (Greber et al. 2017, 2019) (Fig. 5.5h) or its archaeal and bacterial homologs (Bienstock et al. 2003; Fan et al. 2008; Liu et al. 2008; Wolski et al. 2008) shows that they localize primarily to the enzymatic core of the helicase, affecting either nucleotide binding and hydrolysis, or DNA binding and translocation, in agreement with the enzymatic function of XPD in NER (Kuper et al. 2014), and the observation that these mutations primarily lead to NER defects.
In contrast, trichothiodystrophy (TTD) mutations in TFIIH mostly affect peripheral regions of XPD (Fig. 5.5h) (Bienstock et al. 2003; Fan et al. 2008; Greber et al. 2017; Liu et al. 2008; Wolski et al. 2008), where they structurally destabilize the protein or its interactions within TFIIH, thereby disrupting TFIIH assembly and impairing proper placement of the CAK during transcription initiation (Coin et al. 1998; Dubaele et al. 2003). Additional TTD mutations in XPB and p8 have been shown to destabilize these proteins and reduce the overall levels of properly assembled TFIIH (Botta et al. 2002; Dubaele et al. 2003; Giglia-Mari et al. 2004; Kainov et al. 2008). This lack of structurally intact TFIIH, either due to reduced protein levels or disrupted protein-protein interfaces, is what leads to a general transcription defect in addition to DNA repair defects.
In addition to its association with these inherited disorders, TFIIH has also been implicated in promoting cancer cell growth due to the transcription-promoting activity of its CAK module and the requirement for elevated transcription in cancer cells. Therefore, TFIIH is a possible drug target in cancer chemotherapy (Berico and Coin 2017; Fisher 2018; Gervais et al. 2018).
Structural Insight into the Pol II-PIC
Visualization of the Step-Wise Assembly of the Human Pol II-PIC
Early crystallographic studies revealed the structures of promoter-bound TBP, alone and in complex with TFIIA and TFIIB (Fig. 5.6a, b), as well as TFIIB bound to Pol II (Pol II-ITC with bound TFIIB shown in (Fig. 5.6c). These studies showed that TBP binding to the TATA-box induces an almost 90° bend in the overall trajectory of the upstream promoter DNA (Fig. 5.6a), and how TFIIB interacts with TBP and with the BRE promoter elements near the TATA-box (Fig. 5.6b) (Bleichenbacher et al. 2003; Nikolov et al. 1992, 1995, 1996; Tan et al. 1996; Tsai and Sigler 2000). Structures of TFIIB bound to Pol II (Bushnell et al. 2004; Kostrewa et al. 2009; Liu et al. 2010; Sainsbury et al. 2013) showed how the cyclin domains of TFIIB bind above the Pol II wall, while the N-terminal B-ribbon binds to the Pol II dock (Fig. 5.6c). The segment between the B-ribbon and the cyclin folds is seen inserted into the Pol II cleft, approaching the active site, which led to the suggestion that this B-reader aids in start site selection (Kostrewa et al. 2009; Liu et al. 2010). A combination of these structures with the previously mentioned structure of the TPB-TFIIB-DNA complex (Nikolov et al. 1995; Tsai and Sigler 2000) and the structure of the Pol II elongation complex (Gnatt et al. 2001) suggested approximate models for the closed and open Pol II-PIC, and the subsequent structure of the TFIIB-bound Pol II-ITC (Fig. 5.6c), complete with substrate DNA and an RNA product mimic, revealed that the TFIIB B-reader contacts the single stranded DNA in the polymerase active site, leads to binding of one of the catalytic metals, and stimulates RNA synthesis (Sainsbury et al. 2013).
Fig. 5.6.
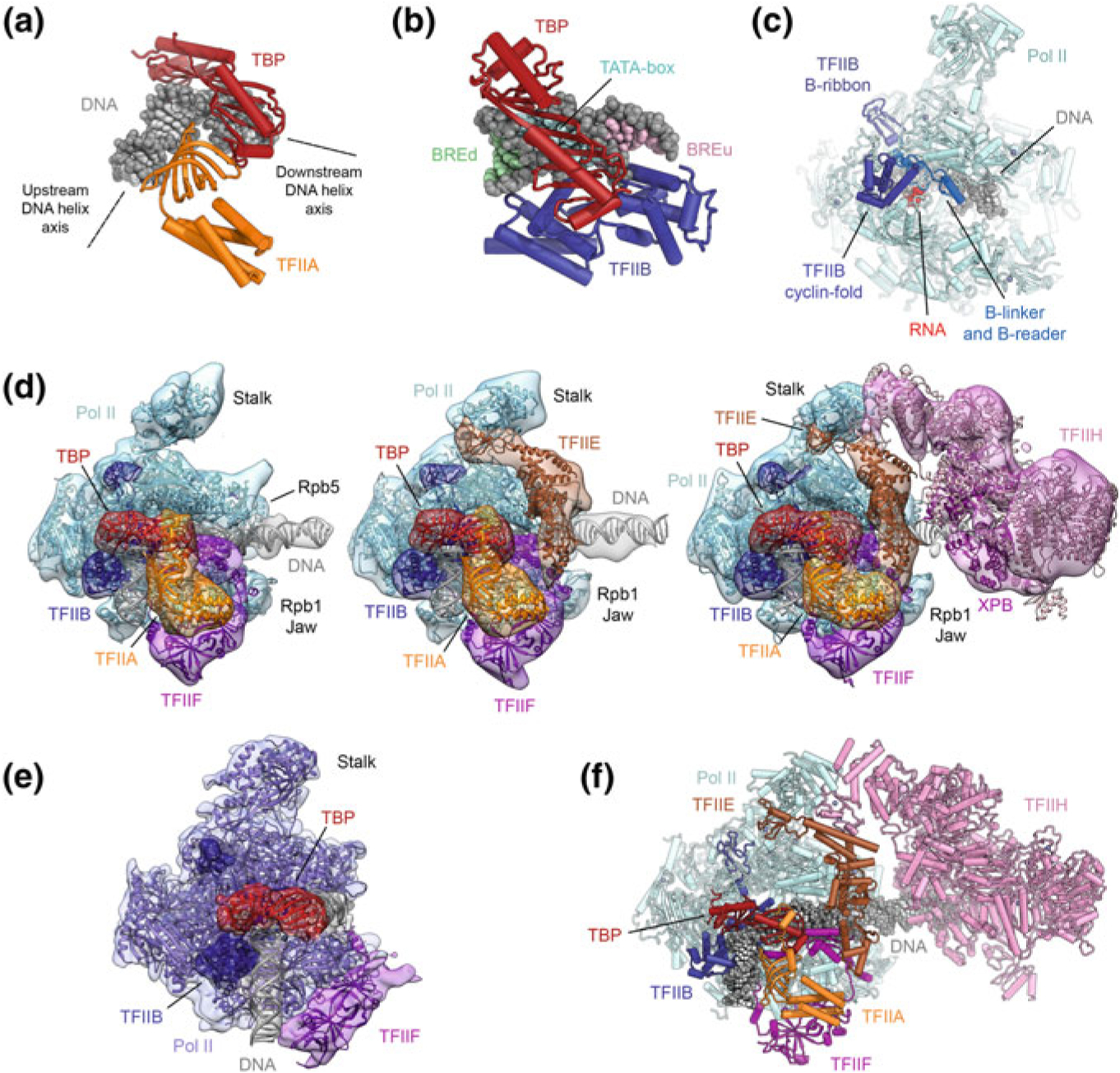
The structure of the Pol II-PIC and its components. a Structure of promoter DNA bound to TBP and TFIIA (Bleichenbacher et al. 2003). TBP introduces a pronounced bend into the TATA box (indicated by dotted black lines representing the upstream and downstream DNA helix axes). b Structure of promoter DNA bound to TBP and TFIIB (Tsai and Sigler 2000). The TATA box and regions corresponding to the BREd and BREu regions recognized by TFIIB are indicated by light blue, green, and pink shading of the DNA bases. c Structure of initially transcribing Pol II bound to TFIIB (Sainsbury et al. 2013). d Stepwise assembly of the human Pol II-PIC: Pol II-DNA-TBP-TFIIA-TFIIB-TFIIF (left), Pol II-DNA-TBP-TFIIA-TFIIB-TFIIF-TFIIE (middle), and Pol II-DNA-TBP-TFIIA-TFIIB-TFIIF-TFIIE-TFIIH (right), the latter showing a bilobal architecture. Depiction based on (He et al. 2013) with fitted coordinates from (Greber et al. 2019; He et al. 2016). e The structure of the yeast Pol II-ITC lacking TFIIH, TFIIA, and TFIIE (Plaschka et al. 2015) is in good agreement with the human Pol II-PIC core (compare e.g. to leftmost complex in panel d). f Complete molecular structure of the human Pol II-PIC with TFIIH, assembled from coordinates for the human core Pol II-PIC and the structure of human TFIIH (Greber et al. 2019; He et al. 2016)
The architecture of the complete Pol II-PIC and a structural characterization of its assembly pathway have remained refractory to crystallographic approaches and were finally obtained using cryo-EM analysis of in vitro reconstituted complexes (Fig. 5.6d). In order to arrive at a more complete understanding of the complete Pol II-PIC, and to map the location of all the individual general transcription factors, the human Pol II-PIC was assembled in a step-wise fashion, following the generally accepted model for PIC assembly (Buratowski et al. 1989), and analyzed by cryo-EM (He et al. 2013). The system was simplified by substituting TFIID with just TBP (a consensus TATA box was included in the promoter DNA), and Mediator was not included in the reconstitution. Additionally, although TFIIF is thought to join the growing PIC pre-bound to Pol II (Killeen and Greenblatt 1992; Rani et al. 2004; Roeder 1996), the structure was determined for complexes with and without TFIIF to allow mapping of the density corresponding to this transcription factor.
The cryo-EM density of the TBP-TFIIA-TFIIB-Pol II complex resulting from these studies (He et al. 2013) could be fully accounted for by the crystallographic structures of isolated sub-complexes (see above) and showed the general transcription factors clustered on the upstream promoter DNA near the TATA box, with density for the DNA approaching the Pol II cleft. The otherwise poorly ordered downstream promoter DNA became stabilized upon addition of TFIIF, which binds to the cluster of general transcription factors near the TATA box (Fig. 5.6d, left), Pol II, and the upstream promoter DNA itself (He et al. 2013). The stabilization of the DNA was induced by interactions of TFIIF with DNA near the BREd element and the opening of the Pol II clamp, allowing DNA to access the Pol II cleft and form interactions with the clamp head and Rpb5. These interactions are consistent with roles of TFIIF in start site selection and transcription bubble opening (Ghazy et al. 2004; Yan et al. 1999).
Addition of TFIIE, which spans from TFIIF to the Pol II stalk (Fig. 5.6d, middle), led to further stabilization of the core PIC (He et al. 2013). Winged-helix domains in TFIIE connect the Pol II stalk region to the TFIIF subunit Rap30, topologically trapping the DNA on the Pol II-PIC. The DNA in this complex is slightly bent and contacted at either side of the initiator element by Pol II elements near the clamp head and at the Rpb5 jaw (He et al. 2013, 2016).
Finally, TFIIH was seen to bind to this core-PIC (Fig. 5.6d, right) such that its XPB subunit is in contact with the downstream promoter DNA near Rpb5 (Figs. 5.5c, 5.6d, right) (He et al. 2013, 2016), in support of its proposed function as a double-stranded DNA translocase during promoter opening (Fishburn et al. 2015; Grünberg et al. 2012; Kim et al. 2000). During the closed-to-open transition, mimicked in the studies of the human system by synthetic constructs that provide single-stranded DNA at the side of the transcription bubble, the downstream promoter DNA is bent around the Rpb5 contact for insertion of the template strand into the active site, while the Pol II clamp domain closes and assumes a conformation similar to that observed in elongation complexes (He et al. 2013, 2016).
The TFIIH-containing Pol II-PIC exhibits a bi-lobed overall appearance (Figs. 5.5c, 5.6d), with TFIIH forming one lobe and Pol II and the remaining GTFs forming the other lobe (He et al. 2013, 2016). In addition to the interaction between XPB and the downstream promoter DNA, TFIIH and the core PIC are connected by two main protein-protein interfaces: (i) Mobile regions in the C-terminal half of TFIIEα interact with the N-terminal PH domain of p62 (He et al. 2016; Okuda et al. 2008; Schilbach et al. 2017) and form additional contacts with the C-terminal region of p62, and with XPB (Schilbach et al. 2017), with the latter interaction explaining previous biochemical data that suggest a role of TFIIE in XPB regulation (Drapkin et al. 1994; Ohkuma and Roeder 1994; Maxon et al. 1994); and (ii) the N-terminal RING domain of MAT1 interacts with the Pol II stalk and TFIIE (Schilbach et al. 2017).
In spite of initial claims to the contrary (Murakami et al. 2013), the model derived from the sequential assembly of human proteins on promoter DNA (He et al. 2013) is in good agreement with subsequent structures from both the human (He et al. 2016) and the yeast Pol II system (Fig. 5.6d, e) (Murakami et al. 2015; Plaschka et al. 2015, 2016; Schilbach et al. 2017). However, slight differences between the human and yeast systems were visualized in the higher-resolution structures. For example, the promoter DNA in the human closed complex is contacted by the Rpb1 clamp and Rbp5, while the DNA is suspended further away from Pol II in the yeast complexes, where such contacts are not present (Nogales et al. 2017b) (for further discussion see next section).
High-Resolution Analysis of the Pol II Core-PIC and Mechanism of Open Complex Formation
High-resolution cryo-EM analysis of human and yeast Pol II core-PICs (the latter excluding TFIIH) (Figs. 5.6f, 5.7a–c) have revealed the intricate architecture of closed, open, and initially transcribing complexes in near-atomic detail (He et al. 2016; Plaschka et al. 2016). In the human open Pol II-PIC, the B-reader and B-linker elements of TFIIB contact the single stranded DNA of the open transcription bubble (Fig. 5.7d–f), and all of these elements are resolved in the map (He et al. 2016). In the yeast Pol II-PIC reconstructions (Plaschka et al. 2016), both the single-stranded DNA and the interacting TFIIB elements are not visualized or poorly resolved. The better definition of the open DNA bubble in the human system might be linked to the scanning mode of start site selection in yeast (Nogales et al. 2017b).
Fig. 5.7.

Structure of the open and initially transcribing human Pol II-PICs and comparison with their yeast counterparts. a Structure of the open human Pol-II PIC (He et al. 2016). b Structure of the human Pol II-ITC (He et al. 2016). RNA near the Pol II active site is shown in red. c Comparison to the structure of the yeast open complex (Plaschka et al. 2016) reveals overall conservation of PIC architecture. d–f DNA interactions of TFIIB in the human Pol II open complex (e) and ITC (f) (He et al. 2016). g Interaction between the TFIIE E-wing domain and bound promoter DNA in the yeast closed PIC (Plaschka et al. 2016). h In the human Pol II-PIC (He et al. 2016), the closed promoter DNA is inserted more deeply into the Pol II cleft, and the TFIIE-DNA interaction visualized in yeast is not present. i Human Pol II open PIC (He et al. 2016)
The reverse is true for interactions of TFIIE (Fig. 5.7g–i), where the human PIC lacks density for part of the TFIIE E-wing domain near the upstream end of the open bubble, while an interaction between this domain and the DNA is visible in the yeast structure (He et al. 2016; Plaschka et al. 2016). This might account for the propensity of the yeast Pol II-PIC system to spontaneously open the transcription bubble (Plaschka et al. 2016), while this has not been observed in the reconstituted human complexes. Nearby, the WH1 domain of TFIIEβ (Tfa2 in yeast) that interacts with the TFIIFβ (Rap30 in humans, Tfg2 in yeast) WH domain and in particular with the additional helix of this domain in yeast is, accordingly, also in a different position between the human and yeast systems. The WH2 domain of TFIIEb (Tfa2) also has a slightly different position in the two systems.
The structure of the human Pol II-ITC is very similar to the open complex (Fig. 5.7a, b, e, f), with the exception of an enlarged DNA bubble and the presence of RNA, which had been introduced with the substrate DNA template (He et al. 2016).
The Mediator-Bound Pol II PIC
Mediator is a co-activator complex composed of more than 20 subunits and with a molecular weight of approximately 1.2 MDa. Mediator integrates the signals from transcriptional activators and repressors for regulated transcription initiation (Borggrefe and Yue 2011; Brzovic et al. 2011; Flanagan et al. 1991; Jeronimo et al. 2016; Lee et al. 1999; Natarajan et al. 1999; Soutourina 2018), a prerequisite for cellular development and differentiation. Additionally, Mediator stimulates the activity of CDK7 within the TFIIH CAK module towards the C-terminal, low-complexity region of the Pol II subunit Rpb1 (Kim et al. 1994). Mediator itself includes the CDK8-Cyclin C pair, which together with the Med12 and Med13 subunits form a dissociable module (Borggrefe et al. 2002; Tsai et al. 2013) that phosphorylates a range of targets, with broad implications for the mechanism of transcription regulation and for cellular development and disease (e.g. (Bancerek et al. 2013; Donner et al. 2010; Firestein et al. 2008; Knuesel et al. 2009)).
The overall structure of Mediator can be subdivided into four modules, termed head, middle, tail, and arm (Cai et al. 2009), as well as the dissociable kinase module (Borggrefe et al. 2002; Tsai et al. 2013). Early low-resolution electron microscopy analysis indicated that an open conformation of Mediator wraps around one side of Pol II to form the holoenzyme (Asturias et al. 1999; Davis et al. 2002; Dotson et al. 2000). Because of its size, complexity, conformational variability, and low abundance in cells, structure determination of Mediator has been pursued by X-ray crystal structures of subcomplexes, such as structures of its head and kinase modules (Imasaki et al. 2011; Lariviere et al. 2012a; Robinson et al. 2012; Schneider et al. 2011). These structures were then combined with chemical crosslinking-mass spectrometry distance restraints and low-resolution cryo-EM maps to arrive at structural models for the full complex (Lariviere et al. 2012b, 2013; Robinson et al. 2015, 2016; Tsai et al. 2014; Verger et al. 2019).
The most complete atomic model for any Mediator complex to date is that of the Schizosaccharomyces pombe Mediator head and middle modules (or core Mediator), determined by X-ray crystallography at 3.4 Å resolution (Fig. 5.8a) (Nozawa et al. 2017) and cryo-EM at 4.4 Å resolution (Tsai et al. 2017). Even though the architecture of the complex in these two structures is in overall agreement, the higher-resolution X-ray crystallographic analysis resulted in a more accurate and more complete model. The structure shows that the Med14 subunit spans the entire middle module and serves as a scaffold for the assembly of the remaining subunits of this module (Fig. 5.8a). The head and middle modules are bridged by the Med17 and Med6 subunits (Fig. 5.8a), each of which contributes folded domains to both modules, with flexible linkers connecting them (Nozawa et al. 2017). The head module had been visualized previously in isolation (Imasaki et al. 2011; Lariviere et al. 2012a; Robinson et al. 2012). Interestingly, one of these structures had been solved in complex with a CTD peptide bound in a groove formed by the Med6, Med8, and Med17 subunits (Fig. 5.8b) (Robinson et al. 2012). The Mediator tail module is critical for interactions with transcriptional activators, but in spite of the availability of partial X-ray crystal structures (e.g. for the human Med23 and Med25 subunits (Monté et al. 2018; Vojnic et al. 2011)), it has not been visualized in its entirety at high resolution. However, an approximate subunit mapping has been obtained from an integrative modeling approach (Robinson et al. 2015).
Fig. 5.8.
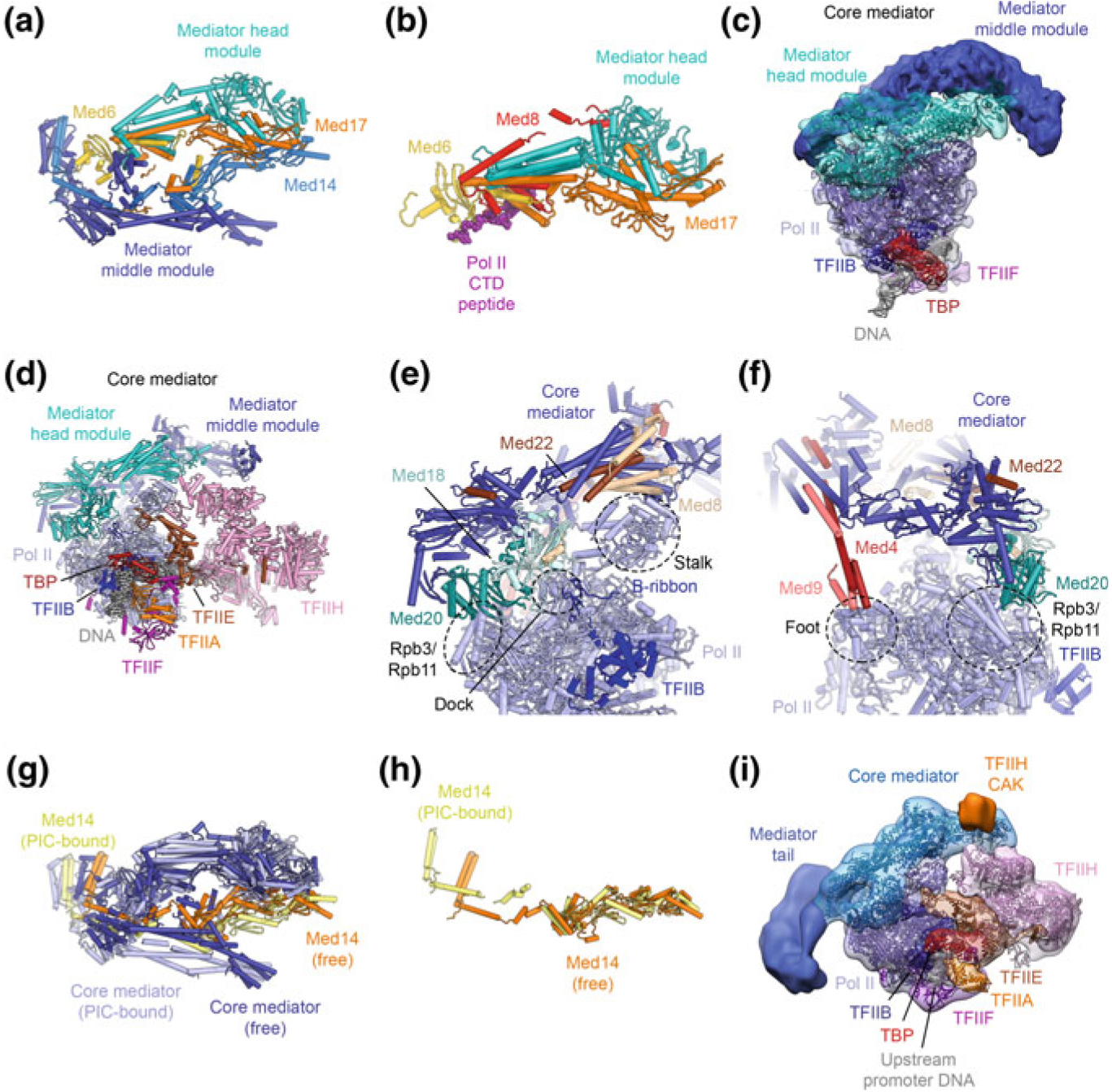
The Mediator-bound Pol II-PIC. a Crystal structure of the Mediator head and middle modules (core Mediator) (Nozawa et al. 2017). b Crystal structure of the Mediator head module bound to a Pol II CTD peptide (Robinson et al. 2012). c Low-resolution cryo-EM structure of the core-Pol II-ITC bound to core Mediator (Plaschka et al. 2015). d Cryo-EM structure of the yeast Pol II-PIC, including TFIIH, bound to core Mediator (Schilbach et al. 2017). e, f Contacts sites between core Mediator and Pol II, according to the structure shown in (d). g, h Conformational changes in Mediator upon binding to the PIC (g) and associated with conformational changes in Med14 (Nozawa et al. 2017; Schilbach et al. 2017). h Low-resolution cryo-EM map of the yeast Pol-II PIC with Mediator (Robinson et al. 2016), including the Mediator tail module, fitted with the coordinates of the most recent yeast Pol II-core Mediator structure (Schilbach et al. 2017)
Detailed insight into the architecture of the Mediator-bound Pol II-PIC came from cryo-EM analysis of reconstituted complexes of yeast core-PIC (without TFIIH) or ITC complexes (Plaschka et al. 2015; Tsai et al. 2017) bound to a core-Mediator complex (Fig. 5.8c). These cryo-EM reconstructions revealed extensive density for the Mediator middle and head modules, and initially allowed fitting of several sub-modules of the head module, aided by distance restraints from chemical crosslinking-mass spectrometry (Plaschka et al. 2015). Subsequently, with higher resolution cryo-EM structures and with the determination of the X-ray crystal structure of the head and middle modules (Nozawa et al. 2017), the entire core Mediator complex could be placed in the density of a TFIIH-containing and Mediator-bound Pol II-PIC (Schilbach et al. 2017) (Fig. 5.8d). Core Mediator binds to the PIC at three contact sites that are arranged near the Pol II Rpb4-Rpb7 stalk (Fig. 5.8e, f). First, the so-called movable jaw (Med18-Med20) forms contacts with the TFIIB B-ribbon, the Pol II dock, and the Pol II subunits Rpb3 and Rpb11 (Fig. 5.8e). Second, the Mediator head module subunits Med8 and Med22 directly contact the Rpb4-Rpb7 stalk (Fig. 5.8e). Finally, the plank domain of the middle module forms transient contacts to the Pol II foot (Fig. 5.8f), as deduced from different conformations observed in cryo-EM particle sub-classes (Plaschka et al. 2015; Schilbach et al. 2017). While the Mediator head and middle modules themselves assume a relatively constant conformation when associating with Pol II—which also does not undergo any major conformational rearrangements upon complex formation—the relative orientation of the two Mediator modules is changed upon core-Mediator-Pol II complex formation due to conformational rearrangements in the Med14 subunit (Schilbach et al. 2017; Tsai et al. 2017) (Fig. 5.8g, h).
This structural model just described, together with a low-resolution cryo-EM reconstruction of a complete Pol II-PIC, including TFIIH and the Mediator tail module (Robinson et al. 2016), showed the Mediator tail module pointing towards the upstream promoter DNA, where it may interact with bound transcriptional activators, consistent with their function in Mediator recruitment (Fig. 5.8i) (Borggrefe and Yue 2011; Brzovic et al. 2011; Jeronimo et al. 2016; Lee et al. 1999; Natarajan et al. 1999). The structures of TFIIH-containing PICs in complex with Mediator or core Mediator suggest that the hook motif of the Mediator middle module aids in positioning of the TFIIH CAK subcomplex, such that it is optimally placed to phosphorylate the Pol II CTD, which runs inside a cradle formed by the inner surface of the remainder of the Mediator middle module (Robinson et al. 2012, 2016; Schilbach et al. 2017) (Figs. 5.5d, e, 5.8b).
Possible Organization of a Complete Pol II-PIC Containing TFIID and Mediator
The structure of the Pol II-PIC in the presence of full TFIID has not yet been obtained. An approximate architecture can be deduced by superimposing the components that are shared between the high-resolution Pol II-PIC reconstructions and the structure of promoter-bound TFIID, specifically TBP, TFIIA, and the upstream promoter DNA (Louder et al. 2016; Patel et al. 2018). Similarly, by superposition of Pol II in the Mediator-containing Pol II-PIC reconstructions (Plaschka et al. 2015; Robinson et al. 2016; Schilbach et al. 2017; Tsai et al. 2017) allows approximate positioning of Mediator in this architectural model (Fig. 5.9).
Fig. 5.9.
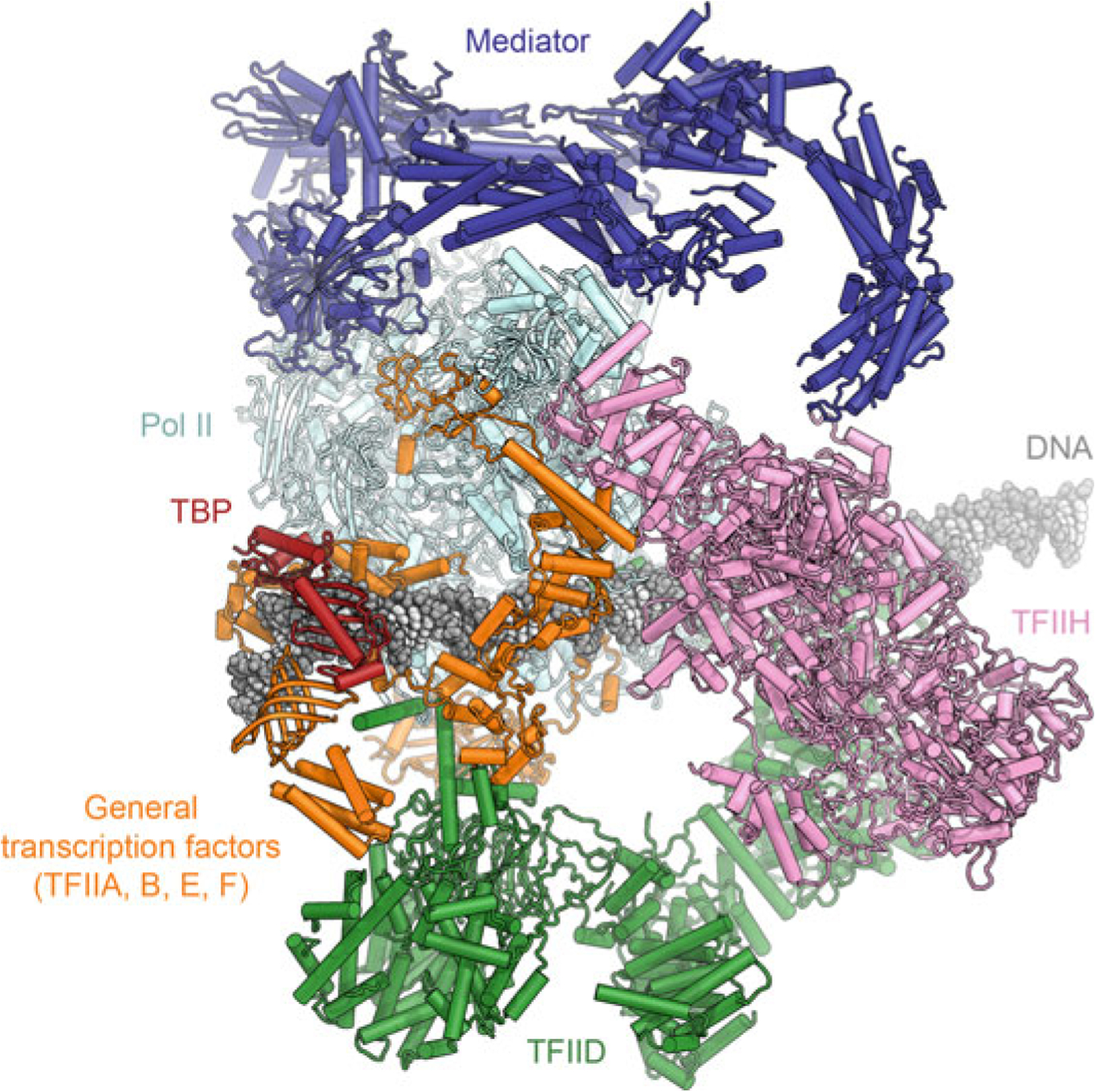
Architectural model of the holo-Pol II-PIC with TFIID and Mediator. Proposed model of an entire Pol II-PIC, including Mediator and TFIID, generated by superimposing component structures (Greber et al. 2019; He et al. 2016; Patel et al. 2018; Schilbach et al. 2017)
This analysis shows that Mediator and TFIID would occupy opposite sides of Pol II (Fig. 5.9), and thus would not clash with each other. However, structural rearrangements may be necessary concerning the position of TAF contacts with the DNA during PIC assembly. The region of TAF4 that, within lobe B, approaches TFIIA and the upstream promoter DNA partially overlaps with the Rap30 winged-helix domain within TFIIF in the model (Louder et al. 2016; Patel et al. 2018). Furthermore, the downstream promoter contacts formed by TFIID lobe C partially overlap with regions of Pol II and TFIIH that contact the DNA in the PIC, and would, in any case, need to be released for Pol II to transcribe through the downstream promoter region. Indeed, it has been proposed that an isomerization of these contacts and release of TAF7, localized in the immediate vicinity of the downstream promoter DNA, is required for transcription initiation (Gegonne et al. 2006; Zhang et al. 2015). Because this isomerization is apparently required only for the first round of initiation, but not for subsequent re-initiation events (Zhang et al. 2015), it is possible that TFIID does not re-form these downstream promoter contacts after it has functioned as a molecular ruler to load TBP for PIC recruitment.
In spite of the release of downstream promoter contacts, TFIID may remain bound to the promoter if the upstream TFIID architecture can rearrange such that the sterical hindrance between the TFIID TAFs and the GTFs of the core Pol II-PIC can be mitigated. It has been shown that transcription re-initiation can be up to fourfold faster than the first round of initiation (Jiang and Gralla 1993) and activator-independent (Joo et al. 2017), suggesting that a set of general transcription factors remains bound at the promoter and allows incoming Pol II complexes to re-initiate rapidly. Transcription re-initiation depends on ATP hydrolysis, implicating the XPB or CDK7 subunits of TFIIH in the process (Yudkovsky et al. 2000). Even though much remains to be elucidated to gain a full understanding of the molecular details that distinguish the processes of initiation and re-initiation in Pol II transcription, the recent detailed structures of the Pol II-PIC and its components provide an architectural framework for future studies.
The Pol I and Pol III Pre-initiation Complexes
Structure of the Pol I-PIC and Mechanism of Initiation of rRNA Transcription
Pol I and Its General Transcription Factors
Pol I is responsible for synthesis of long rRNA precursors in the nucleolus, contributing up to 60% of total RNA synthesis in yeast (Warner 1999). The X-ray crystal structure of the 14-subunit Pol I was that of a transcription-incompetent dimer (Fig. 5.10a) with a widened cleft that was blocked by inserted protein segments (Engel et al. 2013; Fernández-Tornero et al. 2013). In solution, Pol I exists in an equilibrium of inactive dimers and functional monomers (Milkereit et al. 1997), a situation that may parallel the physiological response to nutrient starvation (Torreira et al. 2017). Binding of Rrn3, one of the Pol I transcription initiation factors, renders Pol I monomeric (Fig. 5.10b) and with a widened cleft that can be accessed by substrate DNA (Engel et al. 2016; Pilsl et al. 2016).
Fig. 5.10.
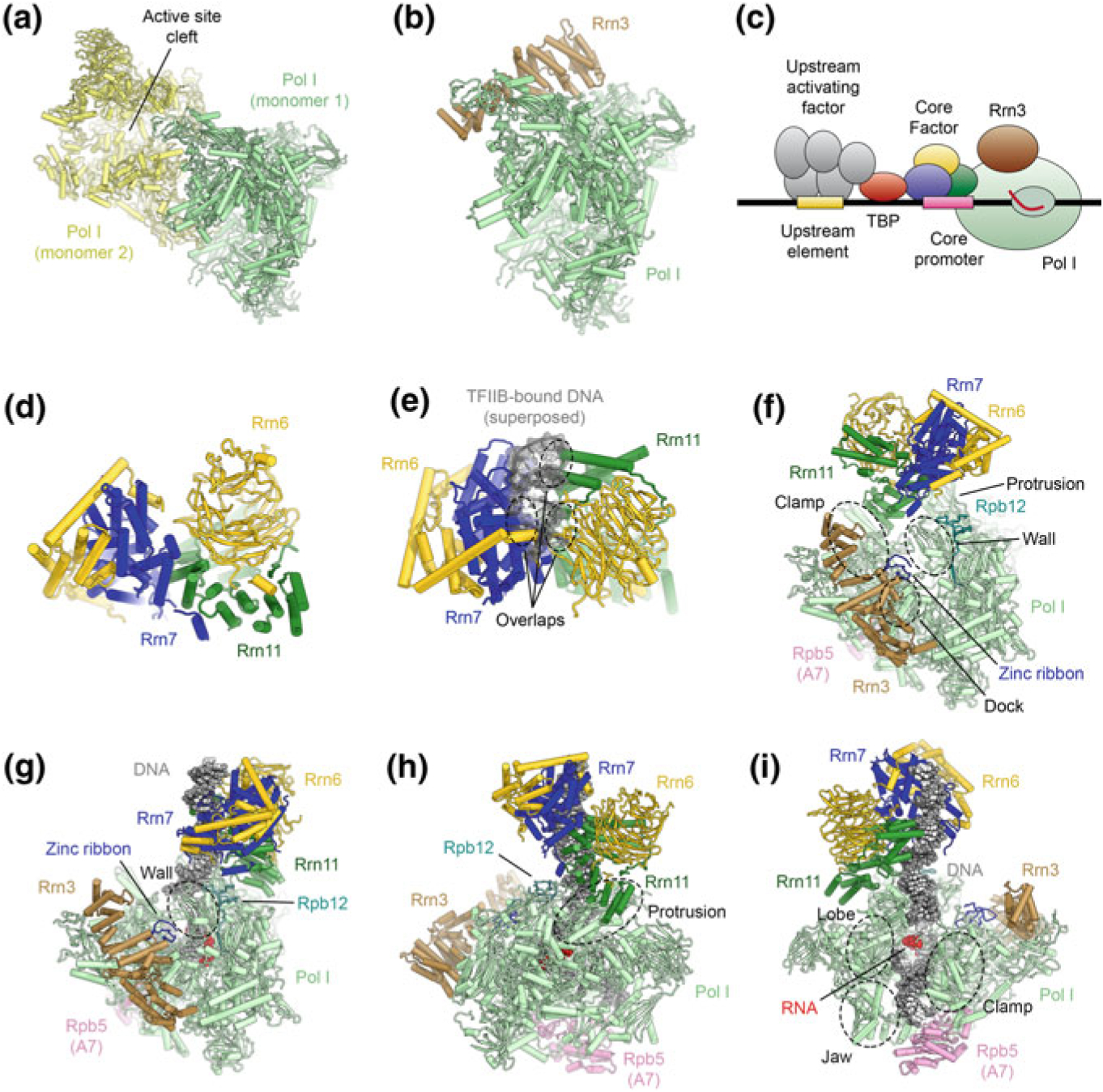
The architecture of the RNA polymerase I PIC and ITC. a Structure of the Pol I-dimer (Engel et al. 2013), with a widened active site cleft, as observed in Pol I crystals. b Structure of Pol I bound with Rrn3 (Engel et al. 2016), which blocks the Pol I dimerization interface. c Schematic of the Pol I core promoter and general transcription factors in yeast. d Structure of yeast core factor (CF) (Engel et al. 2017). e Structure of yeast core factor shown with DNA superposed from a TFIIB-DNA structure (Tsai and Sigler 2000). The superposed DNA overlaps with CF, indicating that Rrn7 in CF cannot bind to DNA by the same mechanism as TFIIB (also see (Fig. 5.15). f Structure of the Pol I-Rrn3-CF complex (Engel et al. 2017). Regions of Pol I that are in contact with CF are indicated. g–i Depiction of the Pol I-ITC, with bound DNA, Rrn3, and CF (Engel et al. 2017; Han et al. 2017). Contacts differ from the DNA-free complex (shown in f) and corresponding regions on Pol I are indicated. Downstream promoter DNA is stabilized by contacts from the Pol I clamp, lobe, and jaw domains and Rpb5
The Pol I initiation system differs substantially from that of Pol II. In yeast, the Pol I-PIC comprises the general transcription factors TBP, Rrn3, upstream activating factor (UAF), and the heterotrimeric core factor (CF) (Fig. 5.10c) (Bedwell et al. 2012; Hontz et al. 2008; Keener et al. 1998; Keys et al. 1996; Schneider 2012; Steffan et al. 1998). Rrn3 and CF support basal Pol I initiation, while binding of UAF and TBP precedes CF recruitment in vivo and is required for full activity (Bordi et al. 2001; Hontz et al. 2008; Keys et al. 1996; Steffan et al. 1998). Yeast CF is formed by Rrn6, Rrn7, and Rrn11 (Keys et al. 1994; Lalo et al. 1996; Lin et al. 1996). Its mammalian counterpart, selectivity factor 1 (SL1), harbors three subunits homologous to the yeast system and two additional mammalian-specific factors (Comai et al. 1992; Denissov et al. 2007; Friedrich et al. 2005; Gorski et al. 2007; Learned et al. 1985; Naidu et al. 2011), suggesting a conserved core architecture with certain mammalian-specific features. The structure of CF (Fig. 5.10d), determined independently by both X-ray crystallography and cryo-EM, shows a bi-lobal assembly, with Rrn11 and Rrn7 each forming the core of one lobe, and with Rrn6 spanning across both lobes (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017). In spite of sequence and structural homology between Rrn7 and TFIIB (Knutson and Hahn 2011), these two initiation factors are not functionally equivalent because, due to its position in the Pol I-PIC, Rrn7 cannot bind to DNA in the way TFIIB does (Fig. 5.10e) (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017).
Structural Insight into RNA Polymerase I Pre-initiation Complexes
Three independent cryo-EM studies have provided detailed insights into the structures of the Pol I-PIC and the promoter-free Pol I-Rrn3-CF complex to elucidate the mechanism of transcription initiation by Pol I (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017). In the Pol I-Rrn3-CF complex, lacking promoter DNA (Fig. 5.10f), CF binds to the upstream end of the Pol I cleft, which is partially widened (Engel et al. 2017). There are three contact sites between CF and the PIC core (Fig. 5.10f): (i) The N-terminal zinc-ribbon domain of Rrn7 contacts the Pol I dock domain and a loop in Rrn3; (ii) the Rrn7 insertion domain contacts the Pol I wall; and (iii) Rrn11 contacts the Pol I clamp and protrusion via its TPR domain (Engel et al. 2017). Notably, the N-terminal cyclin domain of Rrn7 is embedded in CF and therefore unable to form contacts equivalent to the TFIIB-Pol II wall interactions present in the Pol II system (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017).
Upon binding of DNA and formation of an ITC, mimicked experimentally by introduction of a mismatch bubble with bound RNA, the Pol I active site cleft assumes a contracted conformation (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017), similar to that observed for the elongation complex structures (Neyer et al. 2016; Tafur et al. 2016). The promoter DNA in the active site cleft of Pol I is stabilized by the clamp head, cleft, and jaw of Pol I subunit A190, the lobe domain of A135, and Rpb5 (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017). Upon DNA binding, CF is relocated to a different binding site on Pol I (Fig. 5.10g) (Engel et al. 2017). Though CF uses the same binding motifs, its footprint on Pol I moves to the protrusion, Rpb12, and a Pol I-specific structural element of the wall (Engel et al. 2017) (Fig. 5.10g–i). Density for Rrn3 was absent in one of the Pol I-ITC cryo-EM maps, indicating that it may dissociate under conditions that might mimic a late initiation intermediate (Han et al. 2017), consistent with Rrn3 dissociation after Pol I initiation (Bier et al. 2004; Milkereit and Tschochner 1998).
In all cryo-EM reconstructions of Pol I-ITCs, the upstream promoter DNA appears bent (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017). In one case, two kinks, by 35° and 45°, are observed near nucleotide −16 (Han et al. 2017); in the second, the DNA bends by approximately 30° between the CF binding site, where it runs along Rrn11 (DNA nucleotides −35 to −25), and the entry point into the Pol I cleft between the protrusion and the wall (DNA nucleotides −20 to −12) (Engel et al. 2017); and in the third, the bend localizes to approximately nucleotide −30 (Sadian et al. 2017). The ability of the DNA to bend and form these contacts is likely important for promoter recognition, along with the ability of the region around the transcription start site to melt and form the transcription bubble.
Interestingly, the DNA is suspended above the cleft in the closed Pol II-PIC (He et al. 2013, 2016; Louder et al. 2016; Murakami et al. 2015; Plaschka et al. 2016), while it is already sandwiched between the wall and the protrusion in the CF-bound PIC within the Pol I system (Fig. 5.11a, b) (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017). This difference indicates that Pol I-DNA interactions in the Pol I-PIC already assume an elongation complex-like state, while the Pol II-PIC requires the DNA to shift by approx. 20 Å between the PIC and the elongation complex to reach this state (Fig. 5.11c).
Fig. 5.11.
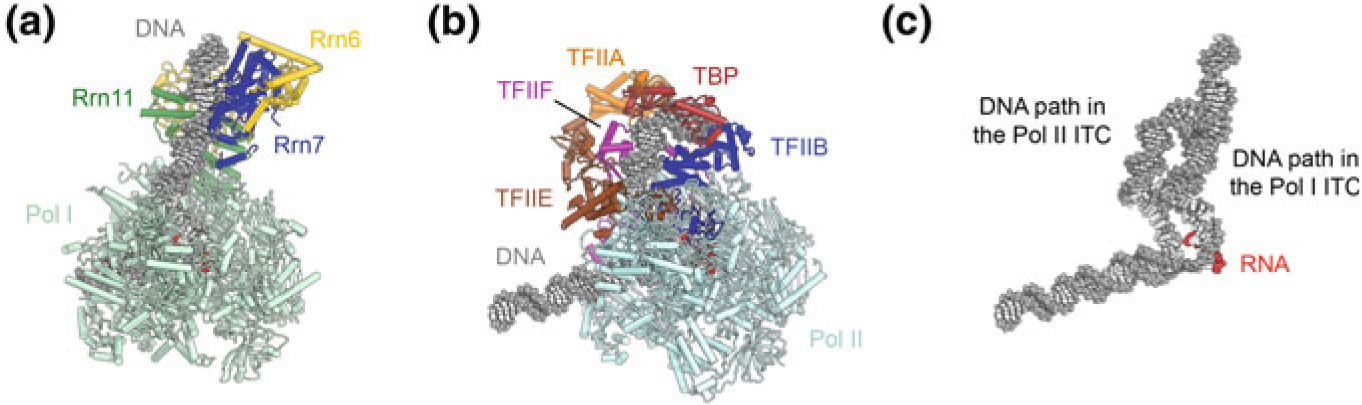
Comparison of DNA paths in the Pol I and Pol II ITCs. a Structure of the Pol I-ITC (Han et al. 2017). b Structure of the Pol II-ITC (He et al. 2016). c The path of the upstream promoter DNA is different between the Pol I and Pol II ITCs because of differences in the mode of binding of Rrn7 and TFIIB to the DNA. See also Fig. 5.15
Model for Initiation by RNA Polymerase I
The model for Pol I initiation that emerges from the structural studies just described is that Rrn3 prepares Pol I for initiation by stabilizing it in a monomeric, open-cleft conformation, while CF binds the upstream promoter DNA, and, after docking to Pol I, loads the DNA into the Pol I cleft. Contacts between proximal promoter regions and Pol I specific structural elements likely contribute to promoter recognition at this stage. DNA unwinding and open complex formation leads to initiation of RNA synthesis and eventual displacement of Rrn7 and CF once the RNA reaches a suitable length (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017).
Structure of the Pol III-PIC and Similarities to the Pol II System
Pol III and Its Redox-Sensing General Transcription Factor TFIIIB
Pol III contains 17 subunits, 10 of which form the conserved core shared across multisubunit RNA polymerases, whereas 2 form a stalk reminiscent of that in Pol II (Fig. 5.1d, e). 5 Pol III-specific subunits, the C82-C34-C31 trimer and the C53-C37 dimer, appear to be homologous to general transcription factors in the Pol II system (Fig. 5.12), but have become stably incorporated into Pol III (Khatter et al. 2017; Vannini and Cramer 2012). Specifically, there is strong structural homology between TFIIF and the Pol III C53-C37 dimer, which is involved in both transcription initiation and termination (Arimbasseri and Maraia 2015; Kassavetis et al. 2010; Rijal and Maraia 2013; Wu et al. 2011) while the C82-C34-C31 trimer, located on the Pol III C160 clamp, shows homology to TFIIE (Vannini and Cramer 2012) and appears to be a functional fusion of TFIIE and TFIIF (Vorländer et al. 2018; Wu et al. 2012). C82 and C34 contain multiple winged-helix domains that participate in interactions with the Pol III general transcription factors and facilitate Pol III-PIC formation (Khoo et al. 2014, 2018).
Fig. 5.12.
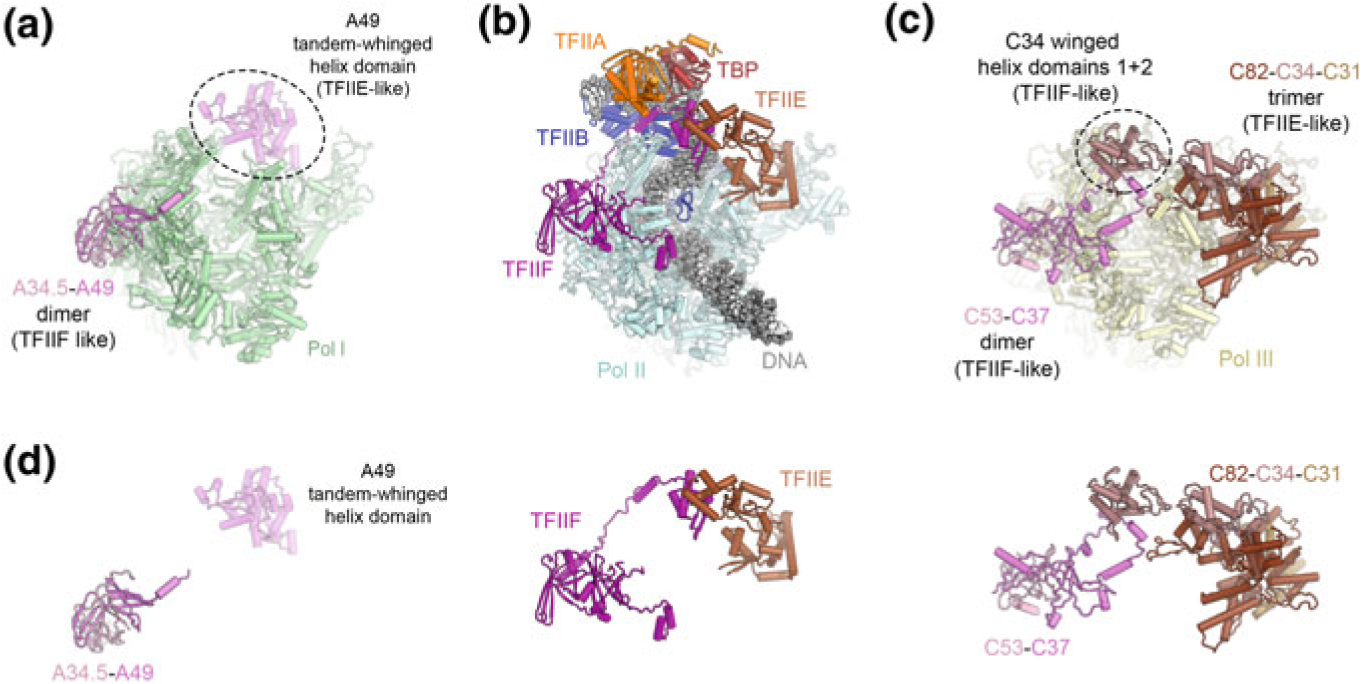
Transcription factor-like subunits of Pol III. a Structure of Pol I extracted from the structure of a Pol I elongation complex in which the tandem winged helix domain of A49 is resolved (Tafur et al. 2016). b Structure of the Pol II-PIC shown in the same orientation for comparison (He et al. 2016). c Structure of Pol III extracted from the structure of an open Pol III-PIC (Abascal-Palacios et al. 2018) showing the locations of the C82-C34-C31 trimer and the C53-C37 dimer. d Comparison of the overall architecture of the A34.5-A49 dimer (Pol I system, left), TFIIE and TFIIF (Pol II system, middle), and the C82-C34-C31 trimer and C53-C37 dimer (Pol III system, right). The Pol I A34.5-A49 and Pol III C53-C37 dimers are positioned similarly to TFIIF, except that the A49 tandem-winged helix domain shows similarities with TFIIE, though the binding site on the polymerase is not identical. Most of the Pol III C82-C34-C31 trimer is positioned similarly to TFIIE, with the C82 cleft loop and TFIIE E-wing domain occupying similar positions, juxtaposed to bound promoter DNA in closed PICs. The C34 winged-helix 1 domain is positioned similarly to the winged-helix domain of the TFIIF subunit Rap30
Transcription initiation by Pol III requires the trimeric general transcription factor TFIIIB (Fig. 5.13a–c), which is sufficient for Pol III transcription initiation in vitro (Kassavetis et al. 1990, 1999). TFIIIB is formed by TBP, TFIIB-related factor 1 (Brf1), and the SANT-domain containing protein B-double prime 1 (Bdp1) (Colbert and Hahn 1992; Ishiguro et al. 2002; Kassavetis et al. 1995; Lobo et al. 1992; López-De-León et al. 1992; Schramm and Hernandez 2002; Schramm et al. 2000; Wang and Roeder 1995). In addition to Brf1, vertebrates encode a second TFIIB-related subunit, termed Brf2 (Cabart and Murphy 2001; Mital et al. 1996; Schramm et al. 2000; Teichmann et al. 2000), which acts at certain promoters, including the U6 small nuclear RNA (snRNA) promoter, characterized by a strong TATA-box and an upstream proximal sequence element (PSE) that is recognized by the SNAPc complex (Henry et al. 1995; Sadowski et al. 1993). TFIIIB not only functions in recruitment of Pol III to the promoter, but also supports initial promoter melting and extension of the open bubble via its Bdp1 and Brf1 subunits, respectively (Kassavetis et al. 2001).
Fig. 5.13.
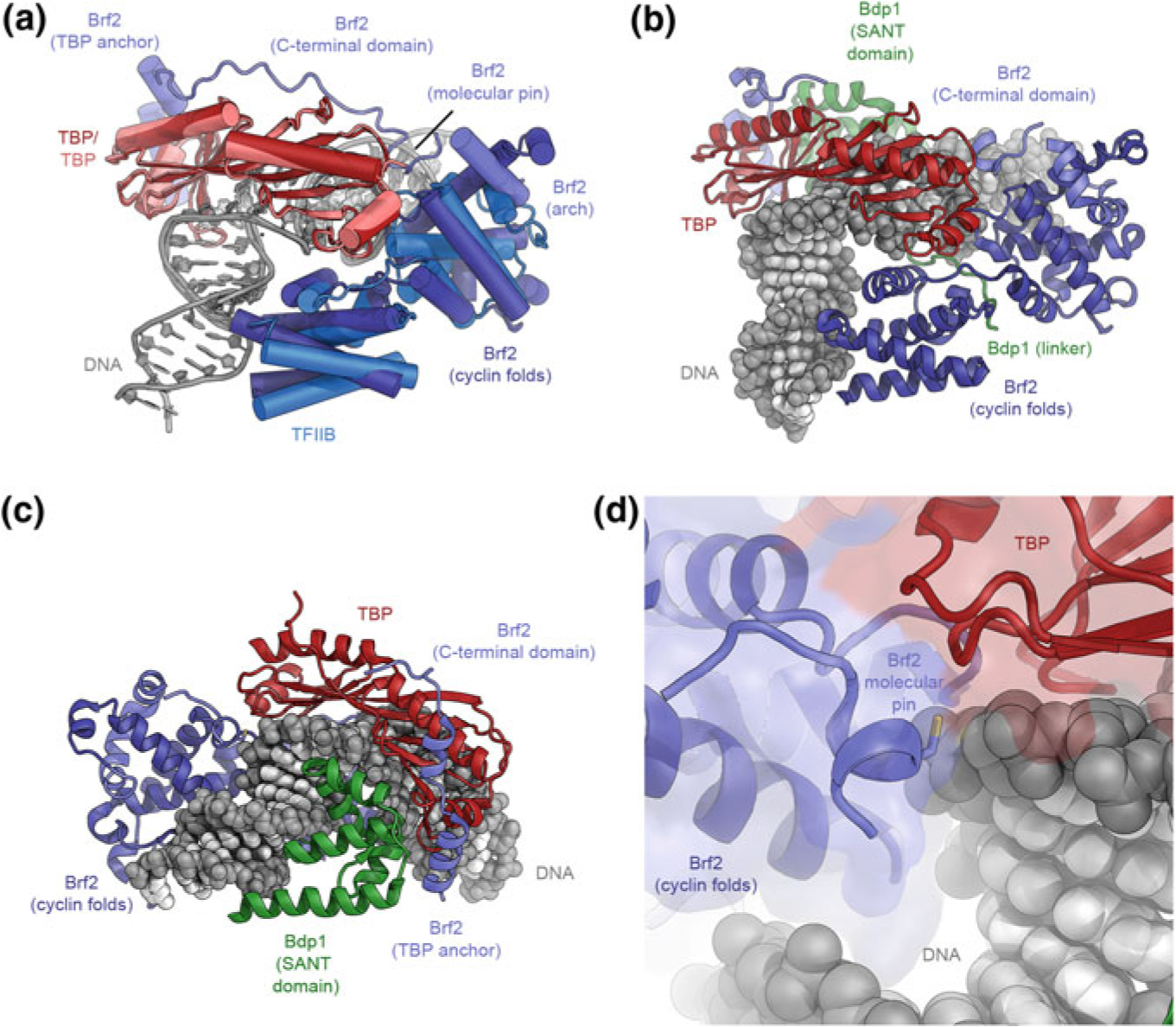
The structure of TFIIIB. a Superposition of the structures of the DNA-bound TBP-Brf2 complex (Gouge et al. 2015) and the TBP-TFIIB-DNA complex (Tsai and Sigler 2000). TBP and the Brf2 cyclin folds in TFIIIB occupy very similar positions as their Pol II-system counterparts (TBP and TFIIB, respectively). The C-terminal domain of Brf2 comprises arch, molecular pin, and TBP anchor regions (as indicated). b, c Structure of DNA-bound TFIIIB (Gouge et al. 2017). The Bdp1 SANT domain binds to DNA at the side opposite to the Brf2 cyclin folds. d Structure of the redox-sensitive molecular pin in the context of bound DNA and TBP (Gouge et al. 2017)
The N-terminal halves of Brf1 and Brf2 are structurally homologous to TFIIB (Fig. 5.13a), contributing to the conservation of the overall architecture of the Pol II and Pol III-PICs (López-De-León et al. 1992; Vannini and Cramer 2012; Wang and Roeder 1995), while the C-terminal half includes a Brf-specific domain that forms tight and specific interactions with TBP (Juo et al. 2003; Khoo et al. 1994). The crystal structure of a Brf2-TBP-DNA complex (Gouge et al. 2015) shows that TBP engages DNA in the same way as in the TFIIB-TBP-DNA complex (Nikolov et al. 1995; Tsai and Sigler 2000) and induces the same strong bend at the TATA box (Gouge et al. 2015; Nikolov et al. 1996) (Fig. 5.13a). Even though details differ, the interactions between DNA and the regions conserved between Brf2 and TFIIB are similar overall, including contacts of the cyclin domains with the promoter DNA immediately upstream and downstream of the TATA box (Gouge et al. 2015; Nikolov et al. 1995; Tsai and Sigler 2000). The Brf2 C-terminal region, however, is unique, and contains three architectural regions, termed arch, anchor, and molecular pin (Gouge et al. 2015) (Fig. 5.13a). The molecular pin harbors a conserved LPPC-motif and binds to a ternary interface between the Brf2-cyclin domain, TBP, and the DNA (Fig. 5.13d). Interestingly, the cysteine in the molecular pin allows Brf2 to serve as a redox-sensing transcription factor that links oxidative stress to cellular responses, including apoptosis, because oxidation of the cysteine impairs complex assembly and leads to down-regulation of survival-promoting genes (Gouge et al. 2015).
The third component of TFIIIB, Bdp1, is specific to the Pol III system and contributes to an extremely tight interaction of TFIIIB with promoter DNA (Kassavetis et al. 1990, 2005; Shah et al. 1999). The crystal structure of a TFIIIB (TBP-Brf2-Bdp1)-DNA complex (Fig. 5.13b, c) shows that the Bdp1 SANT domain plays a key role in association of the protein with the remainder of the complex and binds to a location similar to that of TFIIA in the Pol II system (Bleichenbacher et al. 2003; Gouge et al. 2017; Tan et al. 1996).
Once assembled on the promoter, TFIIIB binds very stably (Kassavetis et al. 1990) and remains bound even after Pol III starts elongating, allowing for efficient Pol III recycling and high rates of transcription initiation on Pol III promoters (Dieci et al. 2013; Dieci and Sentenac 1996). Indeed, the TFIIIB association is so stable that it serves as a roadblock that impedes pervasive Pol II transcription and leads to dissociation of lagging strand replicative polymerases (Roy et al. 2016; Smith and Whitehouse 2012).
As detailed above, TFIIIB is the key transcription factor in the Pol III system. Two additional transcription factors, TFIIIA and TFIIIC are assembly and specificity factors that aid in positioning of TFIIIB on the upstream promoter DNA (Kassavetis et al. 1990; Roberts et al. 1995). TFIIIC a hexameric 0.5 MDa protein complex that binds to intragenic promoter elements (Conesa et al. 1993; Ducrot et al. 2006; Male et al. 2015; Stillman and Geiduschek 1984), but is dispensable after TFIIIB binding and is displaced by transcribing Pol III (Bardeleben et al. 1994).
The Structure of the RNA Polymerase III Pre-initiation Complex
Three independent cryo-EM studies (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018) have revealed the mechanism of Pol III-PIC assembly and promoter opening at high resolution. Two of these structures were determined on a U6 snRNA promoter (Abascal-Palacios et al. 2018; Vorländer et al. 2018), one of them on the asparagine-tRNA promoter (Han et al. 2018). These studies used promoter DNA substrates mimicking closed and open Pol III-PICs as well as Pol III-ITCs (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018). Interestingly, it was found that the open Pol III-PIC spontaneously forms even from fully base-paired templates (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018) and even in presence of mutants deficient in promoter melting (Abascal-Palacios et al. 2018; Han et al. 2018). The RNA substrate was lost from the Pol III-ITC under some conditions, either due to washing or nucleolytic cleavage aided by the Pol III subunit C11 (Han et al. 2018; Vorländer et al. 2018), and the Pol III-OC and ITC complexes exhibited similar architecture (Han et al. 2018).
The cryo-EM reconstructions comprise Pol III, TFIIIB, and promoter DNA, and show TFIIIB tightly bound to and wrapped around the upstream DNA (Fig. 5.14a). Similarly to the Brf2-containing structure (Gouge et al. 2015, 2017), the cyclin folds of Brf1, present in the cryo-EM sample as a Brf1-TBP fusion protein (Kassavetis et al. 2005), bind to TBP and promoter DNA upstream and downstream of TBP (Figs. 5.13a, 5.14b). The SANT domain of Bdp1 binds TBP, DNA, and Brf1, leading to a highly stable assembly of TFIIIB on the upstream promoter DNA, suitable for supporting multiple rounds of initiation by Pol III (Dieci et al. 2013; Dieci and Sentenac 1996).
Fig. 5.14.
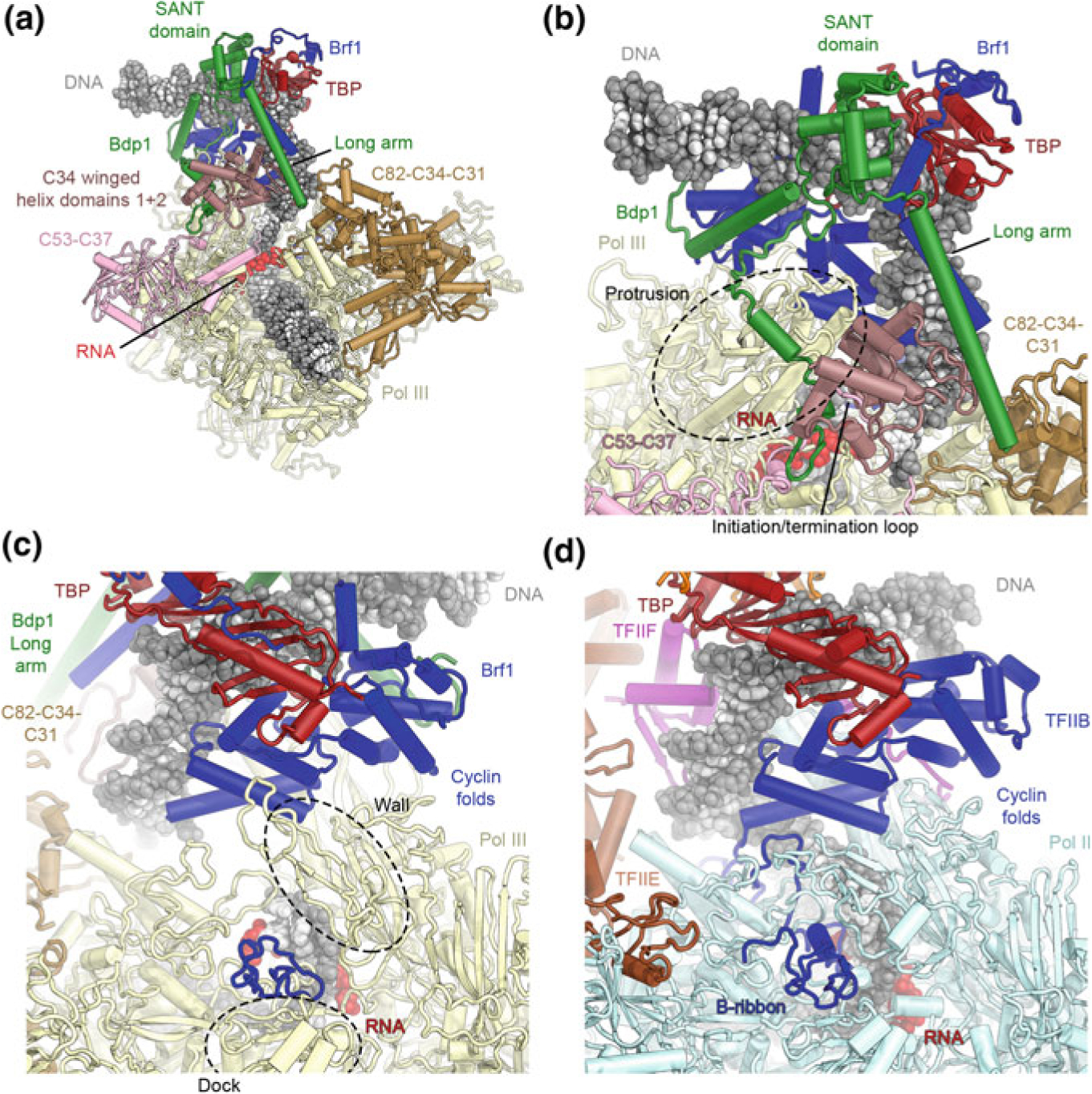
The structure of the Pol III-ITC. a Overview of the structure of the yeast Pol III-ITC (Vorländer et al. 2018). Compared to the structure of isolated TFIIIB, additional elements reaching towards the polymerase are visualized in the complete PIC and ITC structures. b TFIIIB encloses the upstream promoter. Structural elements of Bdp1 (green) contact the transcription-factor like subunits of Pol III (pink and brown), thereby contributing to a stable platform for ordering of some of these factors. c, d The B-ribbon domains of Brf1 (c) and TFIIB (d) occupy similar positions and interact with nucleic acids
In the open Pol III-PIC, Pol III itself exhibits a closed clamp conformation and tightly stabilizes the open DNA bubble (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018). Pol III subunits C37, C34 and C31, all of them part of the “built-in transcription factors” of Pol III are flexible in the structure of transcribing Pol III (Hoffmann et al. 2015), but ordered in the open PIC (Fig. 5.14a). Bound to the upstream promoter DNA, TFIIIB sits above the Pol III cleft, where its Bdp1 and Brf1 subunits form several contacts with Pol III. Due to these contacts, parts of the general transcription factor subunits that are not visualized in the crystal structures (Gouge et al. 2015, 2017) are visible in the cryo-EM reconstructions (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018).
In Brf1, the N-terminal zinc-ribbon and the cyclin folds contact the Pol III dock, wall, and protrusion, similar to the TFIIB contacts observed in the Pol II system (Fig. 5.14c, d) (He et al. 2016; Murakami et al. 2015; Nikolov et al. 1995; Plaschka et al. 2016; Tsai and Sigler 2000). The zinc-ribbon is inserted through the active site cleft and contacts the Pol III dock domain (Fig. 5.14c), and an adjacent linker region interacts with the template DNA strand (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018). The zinc-ribbon interaction is also found in the Pol I system (Engel et al. 2017; Han et al. 2017; Sadian et al. 2017), while the interaction of the N-terminal cyclin fold with the wall and protrusion (Fig. 5.14c, d) is restricted to Pol II and Pol III.
Notably, Brf1 includes a structural element termed the helical pin, structurally homologous to the Brf2 molecular pin and occupying the same site at the interface between TBP, DNA, and the Brf1 cyclin folds (Gouge et al. 2017), but without the redox-sensing activity of its Brf2 counterpart (Abascal-Palacios et al. 2018; Han et al. 2018).
The extended SANT domain and a linker of Bdp1 bind to the major and minor grooves of the DNA, as visualized previously (Gouge et al. 2017). Additionally, several long helices of Bdp1 are seen in the cryo-EM maps (Fig. 5.14a), reaching across the Pol III C34 subunit (part of the TFIIE/TFIIF-like Pol III subunits), and together with C37 (part of the TFIIF-like Pol III subunits) form a platform that stabilizes C34 (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018). Notably, these interactions between Bdp1 and C34-C37 also lead to the stabilization of the initiation/termination loop of C37 (C37 residues 211–224) (Kassavetis et al. 2010; Rijal and Maraia 2013; Wu et al. 2011) by interactions with part of a Bdp1 region termed the “tether” (Bdp1 residues 360–398). After stabilization by the C37-Bdp1 platform, winged-helix domain 2 in C34 contacts the open transcription bubble near its upstream edge (Fig. 5.14b), implicating C34 in promoter melting or stabilization of the open bubble (Abascal-Palacios et al. 2018; Brun et al. 1997; Han et al. 2018; Vorländer et al. 2018). Bdp1 mutants that are defective in open complex formation (Kassavetis et al. 2001) map to this platform region, underscoring the importance of the molecular arrangement in this region for promoter opening (Abascal-Palacios et al. 2018).
The cryo-EM maps visualized the interactions that contribute to melting and stabilization of the transcription bubble at the active site. In the Pol III-PIC, the active site assumes a conformation that is reminiscent of elongating Pol III (Hoffmann et al. 2015), with a disordered trigger loop and rudder, and a bent bridge helix. Upon transition to the ITC, the upstream bubble edge is stabilized mostly by contacts from Pol III subunits, including the C34 and C82 winged helix domains and the C160 clamp, as well as the N-terminal Brf1 cyclin fold (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018). The arrangement of TFIIIB elements, with the Brf1 N-terminal zinc-ribbon, the adjacent linker, and the N-terminal cyclin fold close to the transcription bubble, and with Bdp1 stabilizing C34 (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018), explains the dual function of TFIIIB in promoter melting (Kassavetis et al. 2001).
Model for Transcription Initiation by RNA Polymerase III and Transition to Elongation
The structural results on the Pol III-PIC described in the previous section, combined with structures of apo-Pol III and transcribing Pol III (Hoffmann et al. 2015), suggest a model for Pol III transcription initiation. Initially, Pol III has a closed clamp, as visualized in the closed Pol III-PIC complex, with disordered downstream DNA projecting away from the polymerase (Vorländer et al. 2018). This is in contrast to e.g. the closed Pol II-PIC, where the DNA runs along the length of the Pol II active site cleft and interacts with the jaws at the downstream end (He et al. 2013, 2016; Murakami et al. 2015). The transcription factor-like modules of Pol III that later become stabilized, as well as TFIIIB elements interacting with them, are initially partially disordered in closed Pol III-PIC (Vorländer et al. 2018). Upon insertion of promoter DNA into the cleft by TFIIIB and the consequent Pol III cleft closure, TFIIIB stabilizes the winged-helix domains of C34 above the cleft, leading to entrapment of the promoter DNA and initial promoter melting. Subsequently, the Brf1 zinc-ribbon and an adjacent linker region become stabilized, with the linker region contacting the opening bubble and facilitating bubble extension, leading to an ITC and eventually an elongating state (Abascal-Palacios et al. 2018; Han et al. 2018; Vorländer et al. 2018).
As detailed above, TFIIIB interacts with promoter DNA very stably and may participate in the Pol III transcription re-initiation pathway (Dieci et al. 2013; Dieci and Sentenac 1996; Kassavetis et al. 1990). An interesting and unexpected finding was the visualization of a transcribing Pol III complex that is still attached to promoter-bound TFIIIB, with up to 33 bp of melted DNA accommodated by Pol III (Han et al. 2018). Based on these findings, and the fact that most Pol III-transcribed genes are short, it was proposed that Pol III may stay associated with TFIIIB throughout the entire transcription cycle (Han et al. 2018).
Comparison of PIC Architectures
Conserved and Divergent Features of Eukaryotic Pre-initiation Complex Architectures
Now that detailed structures of pre-initiation complexes of Pol I, II, and III have been determined, it is possibly to compare the shared and unique features of these three polymerase systems on an architectural level.
Universality of TFIIB-like Factors and Positioning of the Promoter DNA in the PIC
TFIIB-like factors play a critical role in all three eukaryotic RNA polymerase systems discussed in this chapter (see e.g. Figures 5.6, 5.7, 5.10, 5.11, 5.13, 5.14). While Pol II employs TFIIB itself, the Pol I core factor harbours the TFIIB homolog Rrn7, and the Pol III initiation factor TFIIIB contains the TFIIB-related factors Brf1 or Brf2 (Fig. 5.15a–c). The structures of the Pol II and Pol III-PICs (Abascal-Palacios et al. 2018; Han et al. 2018; He et al. 2016; Plaschka et al. 2016; Vorländer et al. 2018) have revealed that in spite of the presence of additional domains in the Pol III initiation factors Brf1/Brf2, the general architecture and role of TFIIB and Brf1/Brf2 are conserved, with all of them forming similar interactions with the upstream promoter DNA and the polymerase, including interactions of the cyclin domains with the polymerase wall and the positioning of the zinc-ribbon domain at the polymerase dock and the adjacent linker in proximity of the transcription bubble (Figs. 5.14c, d, 5.15b, c). The structure of the Pol I-PIC revealed that the zinc-ribbon of Rrn7 assumes a similar position as its TFIIB and Brf1/Brf2 counterparts. However, in spite of the conservation of the two cyclin folds that lie at the core of all TFIIB-related initiation factors, the Rrn7 cyclin folds interact differently with both the upstream promoter DNA (Fig. 5.15a–c) and the RNA polymerase, as compared to TFIIB and Brf1/Brf2 (Fig. 5.15d–f).
Fig. 5.15.
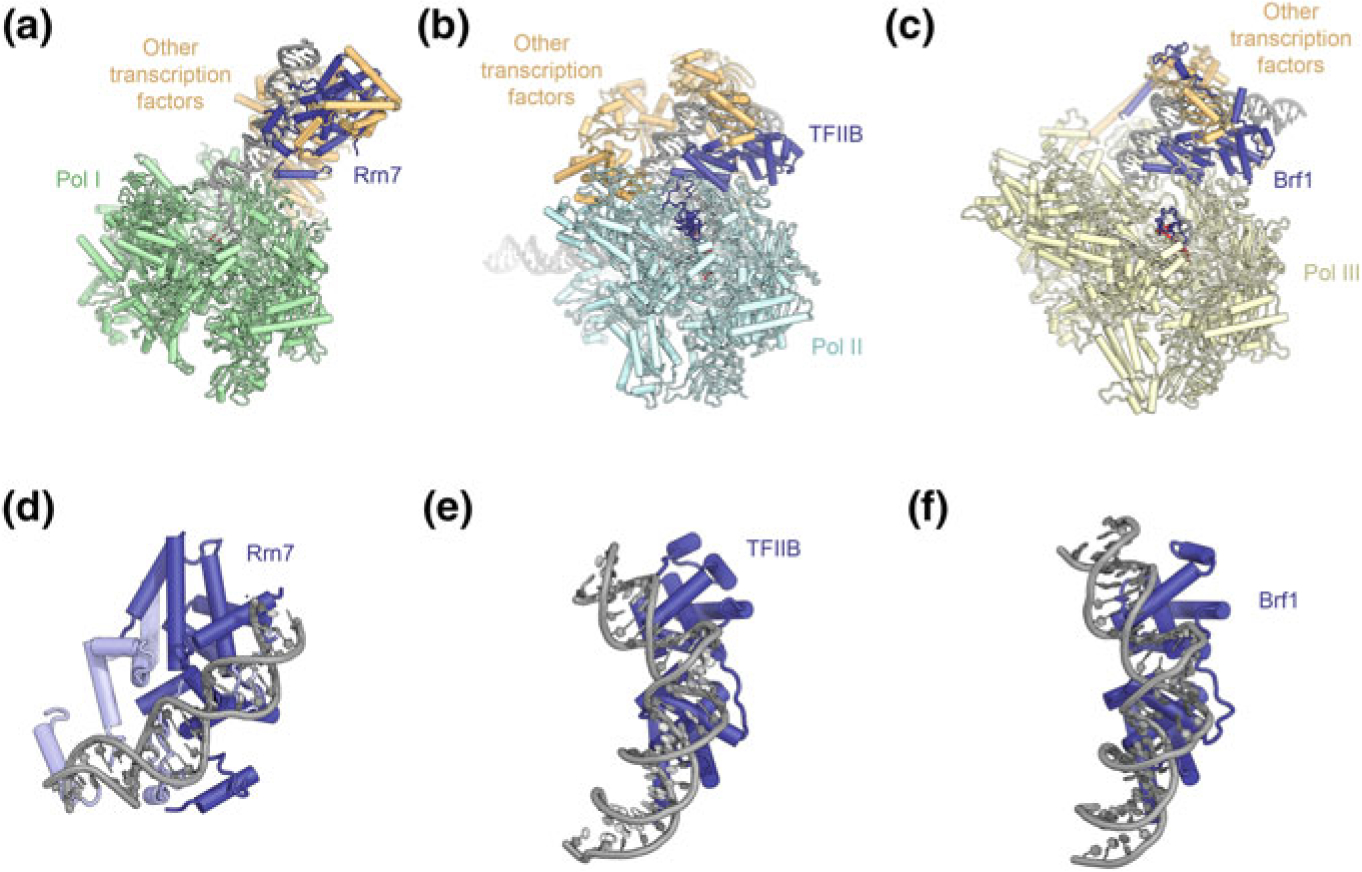
Comparison of DNA-bound structures of TFIIB-like initiation factors. a–c Structure of the Pol I (a), Pol II (b), and Pol III (c) ITCs (Han et al. 2017; He et al. 2016; Vorländer et al. 2018). TFIIB-related factors are colored dark blue, other transcription factors orange. d–f Conformation of upstream promoter DNA bound to the cyclin folds of Rrn7 (d), TFIIB (e), and Brf1 (f)
As a consequence of this difference in PIC architecture, the path of the DNA differs between the Pol I-PIC on the one hand and the Pol II- and Pol III-PICs on the other (Engel et al. 2017, 2018; Han et al. 2017; Sadian et al. 2017). In the Pol I-ITC, the promoter DNA is accommodated in the active site cleft very similarly as in the Pol I elongation complex, likely assuming such a conformation as early as during closed complex formation. In contrast, in both the Pol II- and Pol III-PICs, the upstream promoter DNA in the closed complex is suspended high above the active site cleft, remains in this conformation during ITC formation, and rearranges to its final trajectory only in the elongation complex (Fig. 5.11c) (Abascal-Palacios et al. 2018; Han et al. 2018; He et al. 2016; Plaschka et al. 2016; Vorländer et al. 2018).
Built-in General Transcription Factor-like Subunits in Pol I and Pol III
Pol II is a 12-subunit enzyme, while Pol I has 14 subunits, and Pol III comprises 17 subunits (see Fig. 5.1). As noted above, the supernumerary subunits in Pol I and Pol III share structural and functional similarity to general transcription factors of the Pol II system. Specifically, the location of the Pol I A49-A34.5 and Pol III C53-C37 heterodimers is highly similar to the binding site of TFIIF on the Rpb2 lobe in the Pol II-PIC (Fig. 5.12). This suggests structural and functional overlap with TFIIF, even though A49 in the Pol I system also contains a tandem winged helix domain reminiscent of TFIIE in terms of its DNA interactions (Khatter et al. 2017; Tafur et al. 2016; Vannini and Cramer 2012). Notably, TFIIF is thought to arrive to the Pol II-PIC pre-bound to Pol II (Killeen and Greenblatt 1992; Rani et al. 2004; Roeder 1996), suggesting a relatively straightforward evolutionary path to how its Pol I and Pol III equivalents became incorporated into the multisubunit polymerase (Khatter et al. 2017). In spite of these similarities, there are also differences. The C53-C37 dimer in Pol III is involved not only in initiation, but also in termination, enabling Pol III to terminate autonomously upon encountering the poly-thymidine stretch that serves as the termination signal in this system (Khatter et al. 2017; Landrieux et al. 2006; Rijal and Maraia 2013).
The second polymerase subassembly that shows similarity to Pol II transcription factors is the Pol III C82-C34-C31 trimer. Similarly to TFIIE, the C82-C34-C31 trimer binds to the clamp head (Fig. 5.12b–d) and may contact and stabilize the transcription bubble (Vorländer et al. 2018). At the same time, the C34 winged helix domains 1 and 2 bind in a location that is highly similar to the TFIIFβ (yeast Tfg2) winged helix domain (Fig. 5.7g, h), indicating that the C82-C34-C31 may be a functional fusion of TFIIE and TFIIF (Vorländer et al. 2018).
Pausing of elongating Pol II can lead to backtracking, at which stage the 3′-end of the nascent mRNA leaves the active site and is extruded through a tunnel in the enzyme (Cramer et al. 2000; Gnatt et al. 2001). At this point, critical elements of the Pol II active centre are trapped in inactive conformations and the nascent RNA can no longer be elongated (Cheung and Cramer 2011; Kettenberger et al. 2003), unless the polymerase either advances to place the 3′-end in the active site again, or the transcript is cleaved. In the Pol II-system, TFIIS aids in transcript cleavage and permits resumption of elongation (Izban and Luse 1992; Reines 1992; Wang and Hawley 1993). While the intrinsic Pol II nuclease activity is weak and requires enhancement by TFIIS, the Pol I and Pol III systems harbour strong intrinsic nuclease activity, residing in a TFIIS-like extension domain of their A12.2 and C11 subunits (Ruan et al. 2011; Vannini and Cramer 2012), which for the Pol III system has also been implicated in the Pol III re-initiation pathway (Iben et al. 2011; Khatter et al. 2017). Lacking or unexpectedly weak density for the product RNA in some cryo-EM reconstructions of Pol I and Pol III complexes that were formed with a synthetic DNA bubble scaffold and bound RNA may be due to cleavage of initially bound RNA by intrinsic nuclease activity (Han et al. 2018; Sadian et al. 2017; Vorländer et al. 2018).
Opening of the Transcription Bubble
The Pol II system is unique in its ability to exploit the activity of a DNA-dependent ATPase, XPB within TFIIH, for opening of the transcription bubble. Bacterial RNA polymerases and eukaryotic Pol I and Pol III are able to independently melt the promoter to form open PICs, as strikingly illustrated by structural studies in which attempted structure determination of closed complexes resulted in the formation of open complexes (Abascal-Palacios et al. 2018; Engel et al. 2017; Han et al. 2018; Vorländer et al. 2018). Recent structural, biochemical, and computational analyses (Alekseev et al. 2017; Dienemann et al. 2018; Plaschka et al. 2016) suggest that at least under some circumstances, Pol II is also able to spontaneously melt the promoter, suggesting a universally conserved mechanism of promoter opening in multi-subunit RNA polymerases.
Open Complex Formation in Yeast
For the yeast Pol II transcription initiation system, a reconstitution of the yeast Pol II core-PIC using double stranded DNA for cryo-EM analysis resulted in an open complex (Plaschka et al. 2016). A subsequent study showed that in yeast, there are two distinct promoter classes concerning the requirement for TFIIH; one promoter class can be melted spontaneously without contribution from TFIIH, while the second class benefits more strongly from the presence of TFIIH (Dienemann et al. 2018). The two promoter classes seem to be distinguished by subtle differences in their free energy of melting. Interestingly, the HIS4 promoter used for the structural analysis that resulted in spontaneous open-complex formation (Plaschka et al. 2016) belongs to the first, spontaneously melting class. A follow-up experiment using the more highly TFIIH-responsive GAT1 promoter resulted in structure determination of both open and closed core-PICs, indicating that indeed, promoter melting is less energetically favorable compared to HIS4 (Dienemann et al. 2018). The DNA in one of these closed complexes assumes a different, more highly distorted and more deeply inserted conformation compared to previously determined closed PICs (Dienemann et al. 2018) and may represent an initiation intermediate. This DNA conformation is observed both in the presence and absence of TFIIH.
The Mammalian RNA Polymerase System
Whether these findings can be applied to the human Pol II system is not fully established. Critically, the closed Pol II-PIC could be successfully reconstituted and structurally characterized using human proteins (He et al. 2016). However, experiments in human cells where XPB was depleted using spironolactone indicate that XPB depletion predominantly affects DNA repair, while global transcription is less affected. In contrast, when XPB is present, but its enzymatic activity is inhibited by triptolide, global transcription is reduced (Alekseev et al. 2017). A hypothesis that is consistent with these observation is that XPB may, at least in some cases, act in a checkpoint-like manner. According to this model, inactive XPB binds to the downstream promoter DNA and prevents its insertion into the Pol II active site. Activation of the enzymatic DNA translocase activity of XPB—or alternatively, its depletion—would allow the downstream promoter DNA to reach the active site of Pol II and promoter melting would occur driven by binding energy alone (Alekseev et al. 2017).
Further analysis will be required to establish whether spontaneous melting of Pol II promoters is a general feature and occurs in vivo, consistent with a universally applicable mechanism for melting of promoters by multisubunit polymerases, or if higher eukaryotes have indeed evolved a unique, TFIIH-dependent solution, possibly to allow for more precise regulation of gene expression.
Conclusion
Recent breakthroughs in the structure determination of transcriptional assemblies have revealed the detailed structures of the PICs of Pol I, Pol II, and Pol III. These structures uncovered the intricate architecture of these molecular machines in atomic detail, and have provided valuable mechanistic insight into the process of PIC assembly, transcription bubble opening, and initiation of transcription. This progress has been critically enabled by technical advances in cryo-EM, which allowed the study of highly complex but low-abundance and conformationally heterogeneous molecular assemblies at ever-higher resolution.
Future progress will likely involve the reconstitution and visualization of even larger complexes, in the presence of gene-specific transcription factors, and in the context of chromatin.
Acknowledgements
Molecular depictions were created using UCSF Chimera (Pettersen et al. 2004) and PyMol (The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC.). This work was funded through NIGMS grant R35-GM127018 to E. N; B. J. G. was supported by the Swiss National Science Foundation (projects P300PA-160983 and P300PA-174355). E. N. is a Howard Hughes Medical Investigator.
References
- Abascal-Palacios G, Ramsay EP, Beuron F, Morris E, Vannini A (2018) Structural basis of RNA polymerase III transcription initiation. Nature 553:301–306 [DOI] [PubMed] [Google Scholar]
- Abdulrahman W et al. (2013) ARCH domain of XPD, an anchoring platform for CAK that conditions TFIIH DNA repair and transcription activities. Proc Natl Acad Sci USA 110:E633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseev S, Nagy Z, Sandoz J, Weiss A, Egly J-M, Le May N, Coin F (2017) Transcription without XPB establishes a unified helicase-independent mechanism of promoter opening in eukaryotic gene expression. Mol Cell 65:504–513.e505 [DOI] [PubMed] [Google Scholar]
- Anandapadamanaban M et al. (2013) High-resolution structure of TBP with TAF1 reveals anchoring patterns in transcriptional regulation. Nat Struct Mol Biol 20:1008–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andel F, Ladurner AG, Inouye C, Tjian R, Nogales E (1999) Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286:2153–2156 [DOI] [PubMed] [Google Scholar]
- Andersen G et al. (1997) The structure of cyclin H: common mode of kinase activation and specific features. EMBO J 16:958–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Maraia RJ (2015) Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol Cell 58:1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD (1999) Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283:985–987 [DOI] [PubMed] [Google Scholar]
- Baek HJ, Malik S, Qin J, Roeder RG (2002) Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol 22:2842–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancerek J et al. (2013) CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38:250–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeleben C, Kassavetis GA, Geiduschek EP (1994) Encounters of Saccharomyces cerevisiae RNA polymerase III with its transcription factors during RNA chain elongation. J Mol Biol 235:1193–1205 [DOI] [PubMed] [Google Scholar]
- Barnes CO et al. (2015) Crystal structure of a transcribing RNA polymerase II complex reveals a complete transcription bubble. Mol Cell 59:258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedwell GJ, Appling FD, Anderson SJ, Schneider DA (2012) Efficient transcription by RNA polymerase I using recombinant core factor. Gene 492:94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berico P, Coin F (2017) Is TFIIH the new Achilles heel of cancer cells? Transcription 9:47–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C et al. (2013) The architecture of human general transcription factor TFIID core complex. Nature 493:699–702 [DOI] [PubMed] [Google Scholar]
- Bienstock RJ, Skorvaga M, Mandavilli BS, Van Houten B (2003) Structural and functional characterization of the human DNA repair helicase XPD by comparative molecular modeling and site-directed mutagenesis of the bacterial repair protein UvrB. J Biol Chem 278:5309–5316 [DOI] [PubMed] [Google Scholar]
- Bier M, Fath S, Tschochner H (2004) The composition of the RNA polymerase I transcription machinery switches from initiation to elongation mode. FEBS Lett 564:41–46 [DOI] [PubMed] [Google Scholar]
- Bleichenbacher M, Tan S, Richmond TJ (2003) Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J Mol Biol 332:783–793 [DOI] [PubMed] [Google Scholar]
- Bordi L, Cioci F, Camilloni G (2001) In vivo binding and hierarchy of assembly of the yeast RNA polymerase I transcription factors. Mol Biol Cell 12:753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Yue X (2011) Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell Dev Biol 22:759–768 [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD (2002) A complex of the Srb8, −9, −10, and −11 transcriptional regulatory proteins from yeast. J Biol Chem 277:44202–44207 [DOI] [PubMed] [Google Scholar]
- Botta E, Nardo T, Lehmann AR, Egly J-M, Pedrini AM, Stefanini M (2002) Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum Mol Genet 11:2919–2928 [DOI] [PubMed] [Google Scholar]
- Brand M, Leurent C, Mallouh V, Tora L, Schultz P (1999) Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286:2151–2153 [DOI] [PubMed] [Google Scholar]
- Brun I, Sentenac A, Werner M (1997) Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J 16:5730–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS et al. (2011) The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell 44:942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechner CN, Heil K, Michels G, Carell T, Kisker C, Tessmer I (2014) Strand-specific recognition of DNA damages by XPD provides insights into nucleotide excision repair substrate versatility. J Biol Chem 289:3613–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S, Hahn S, Guarente L, Sharp PA (1989) Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56:549–561 [DOI] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT (1996) Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev 10:711–724 [DOI] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT (1997) The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev 11:3020–3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell DA, Westover KD, Davis RE, Kornberg RD (2004) Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science 303:983–988 [DOI] [PubMed] [Google Scholar]
- Busso D, Keriel A, Sandrock B, Poterszman A, Gileadi O, Egly JM (2000) Distinct regions of MAT1 regulate cdk7 kinase and TFIIH transcription activities. J Biol Chem 275:22815–22823 [DOI] [PubMed] [Google Scholar]
- Cabarcas S, Schramm L (2011) RNA polymerase III transcription in cancer: the BRF2 connection. Mol Cancer 10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabart P, Murphy S (2001) BRFU, a TFIIB-like factor, is directly recruited to the TATA-box of polymerase III small nuclear RNA gene promoters through its interaction with TATA-binding protein. J Biol Chem 276:43056–43064 [DOI] [PubMed] [Google Scholar]
- Cai G, Imasaki T, Takagi Y, Asturias FJ (2009) Mediator structural conservation and implications for the regulation mechanism. Structure 17:559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P et al. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38:626–635 [DOI] [PubMed] [Google Scholar]
- Chalkley GE, Verrijzer CP (1999) DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J 18:4835–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WH, Kornberg RD (2000) Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell 102:609–613 [DOI] [PubMed] [Google Scholar]
- Chen W, Struhl K (1985) Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J 4:3273–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Farmer G, Zhu H, Prywes R, Prives C (1993) Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev 7:1837–1849 [DOI] [PubMed] [Google Scholar]
- Chen JL, Attardi LD, Verrijzer CP, Yokomori K, Tjian R (1994) Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79:93–105 [DOI] [PubMed] [Google Scholar]
- Cheung ACM, Cramer P (2011) Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471:249–253 [DOI] [PubMed] [Google Scholar]
- Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E (2013) Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 152:120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE, Thompson LH, Richardson AS, States JC (1999) A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum Mutat 14:9–22 [DOI] [PubMed] [Google Scholar]
- Coin F, Marinoni JC, Rodolfo C, Fribourg S, Pedrini AM, Egly JM (1998) Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet 20:184–188 [DOI] [PubMed] [Google Scholar]
- Coin F, Oksenych V, Egly J-M (2007) Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell 26:245–256 [DOI] [PubMed] [Google Scholar]
- Colbert T, Hahn S (1992) A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev 6:1940–1949 [DOI] [PubMed] [Google Scholar]
- Comai L, Tanese N, Tjian R (1992) The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68:965–976 [DOI] [PubMed] [Google Scholar]
- Compe E, Egly J-M (2012) TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol 13:343–354 [DOI] [PubMed] [Google Scholar]
- Conesa C, Swanson RN, Schultz P, Oudet P, Sentenac A (1993) On the subunit composition, stoichiometry, and phosphorylation of the yeast transcription factor TFIIIC/tau. J Biol Chem 268:18047–18052 [PubMed] [Google Scholar]
- Cramer P et al. (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640–649 [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863–1876 [DOI] [PubMed] [Google Scholar]
- Davis JA, Takagi Y, Kornberg RD, Asturias FA (2002) Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol Cell 10:409–415 [DOI] [PubMed] [Google Scholar]
- Deng W, Roberts SGE (2005) A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev 19:2418–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S et al. (2007) Identification of novel functional TBP-binding sites and general factor repertoires. EMBO J 26:944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devault A et al. (1995) MAT1 (‘menage à trois’) a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J 14:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Sentenac A (1996) Facilitated recycling pathway for RNA polymerase III. Cell 84:245–252 [DOI] [PubMed] [Google Scholar]
- Dieci G, Bosio MC, Fermi B, Ferrari R (2013) Transcription reinitiation by RNA polymerase III. Biochim Biophys Acta 1829:331–341 [DOI] [PubMed] [Google Scholar]
- Dienemann C, Schwalb B, Schilbach S, Cramer P (2018) Promoter distortion and opening in the RNA polymerase II cleft. Mol Cell 73:97–106.e4 [DOI] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM (2010) CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol 17:194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson MR et al. (2000) Structural organization of yeast and mammalian mediator complexes. Proc Natl Acad Sci USA 97:14307–14310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin R, Reardon JT, Ansari A, Huang J-C, Zawel L, Ahn KJ, Sancar A, Reinberg D (1994) Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 368:769–772 [DOI] [PubMed] [Google Scholar]
- Dubaele S, Proietti De Santis L, Bienstock RJ, Keriel A, Stefanini M, Van Houten B, Egly J-M (2003) Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell 11:1635–1646 [DOI] [PubMed] [Google Scholar]
- Dubrovskaya V, Lavigne AC, Davidson I, Acker J, Staub A, Tora L (1996) Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J 15:3702–3712 [PMC free article] [PubMed] [Google Scholar]
- Ducrot C, Lefebvre O, Landrieux E, Guirouilh-Barbat J, Sentenac A, Acker J (2006) Reconstitution of the yeast RNA polymerase III transcription system with all recombinant factors. J Biol Chem 281:11685–11692 [DOI] [PubMed] [Google Scholar]
- Ehara H, Yokoyama T, Shigematsu H, Yokoyama S, Shirouzu M, Sekine S-I (2017) Structure of the complete elongation complex of RNA polymerase II with basal factors. Science 357:921–924 [DOI] [PubMed] [Google Scholar]
- Elmlund H et al. (2009) Cryo-EM reveals promoter DNA binding and conformational flexibility of the general transcription factor TFIID. Structure 17:1442–1452 [DOI] [PubMed] [Google Scholar]
- Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P (2013) RNA polymerase I structure and transcription regulation. Nature 502:650–655 [DOI] [PubMed] [Google Scholar]
- Engel C, Plitzko J, Cramer P (2016) RNA polymerase I-Rrn3 complex at 4.8 Å resolution. Nat Commun 7:12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel C et al. (2017) Structural basis of RNA polymerase I transcription initiation. Cell 169:120–131.e22 [DOI] [PubMed] [Google Scholar]
- Engel C, Neyer S, Cramer P (2018) Distinct mechanisms of transcription initiation by RNA polymerases I and II. Annu Rev Biophys 47:425–446 [DOI] [PubMed] [Google Scholar]
- Fan L, Arvai AS, Cooper PK, Iwai S, Hanaoka F, Tainer JA (2006) Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol Cell 22:27–37 [DOI] [PubMed] [Google Scholar]
- Fan L et al. (2008) XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell 133:789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Tornero C et al. (2013) Crystal structure of the 14-subunit RNA polymerase I. Nature 502:644–649 [DOI] [PubMed] [Google Scholar]
- Firestein R et al. (2008) CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 455:547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J, Tomko E, Galburt E, Hahn S (2015) Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci USA 112:3961–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP (2018) Cdk7: a kinase at the core of transcription and in the crosshairs of cancer drug discovery. Transcription 10:47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP, Morgan DO (1994) A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell 78:713–724 [DOI] [PubMed] [Google Scholar]
- Fisher RP, Jin P, Chamberlin HM, Morgan DO (1995) Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell 83:47–57 [DOI] [PubMed] [Google Scholar]
- Flanagan PM, Kelleher RJ, Sayre MH, Tschochner H, Kornberg RD (1991) A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350:436–438 [DOI] [PubMed] [Google Scholar]
- Friedrich JK, Panov KI, Cabart P, Russell J, Zomerdijk JCBM(2005) TBP-TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J Biol Chem 280:29551–29558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon F et al. (1979) Organisation and sequences at the 5′ end of a cloned complete ovalbumin gene. Nature 278:428–434 [DOI] [PubMed] [Google Scholar]
- Gegonne A, Weissman JD, Zhou M, Brady JN, Singer DS (2006) TAF7: a possible transcription initiation check-point regulator. Proc Natl Acad Sci USA 103:602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais V et al. (2018) Small molecule-based targeting of TTD-A dimerization to control TFIIH transcriptional activity represents a potential strategy for anticancer therapy. J Biol Chem 293:14974–14988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazy MA, Brodie SA, Ammerman ML, Ziegler LM, Ponticelli AS (2004) Amino acid substitutions in yeast TFIIF confer upstream shifts in transcription initiation and altered interaction with RNA polymerase II. Mol Cell Biol 24:10975–10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons BJ et al. (2012) Subunit architecture of general transcription factor TFIIH. Proc Natl Acad Sci USA 109:1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G et al. (2004) A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet 36:714–719 [DOI] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292:1876–1882 [DOI] [PubMed] [Google Scholar]
- Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, nameZomerdijk JCBM (2007) A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J 26:1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouge J et al. (2015) Redox signaling by the RNA polymerase III TFIIB-related factor Brf2. Cell 163:1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouge J et al. (2017) Molecular mechanisms of Bdp1 in TFIIIB assembly and RNA polymerase III transcription initiation. Nat Commun 8:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Nguyen THD, Fang J, Afonine PV, Adams PD, Nogales E (2017) The cryo-electron microscopy structure of human transcription factor IIH. Nature 549:414–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Toso D, Fang J, Nogales E (2019) The complete structure of the human TFIIH core complex. eLife 8:e44771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob P, Cruse MJ, Inouye C, Peris M, Penczek PA, Tjian R, Nogales E (2006) Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure 14:511–520 [DOI] [PubMed] [Google Scholar]
- Grünberg S, Warfield L, Hahn S (2012) Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol 19:788–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder SN, Sung P, Bailly V, Prakash L, Prakash S (1994) RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature 369:578–581 [DOI] [PubMed] [Google Scholar]
- Han Y, Yan C, Nguyen THD, Jackobel AJ, Ivanov I, Knutson BA, He Y (2017) Structural mechanism of ATP-independent transcription initiation by RNA polymerase I. Elife 6:e27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Yan C, Fishbain S, Ivanov I, He Y (2018) Structural visualization of RNA polymerase III transcription machineries. Cell Discov 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fang J, Taatjes DJ, Nogales E (2013) Structural visualization of key steps in human transcription initiation. Nature 495:481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E (2016) Near-atomic resolution visualization of human transcription promoter opening. Nature 533:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RW, Sadowski CL, Kobayashi R, Hernandez N (1995) A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerase II and III. Nature 374:653–656 [DOI] [PubMed] [Google Scholar]
- Herrera-Moyano E, Moriel-Carretero M, Montelone BA, Aguilera A (2014) The rem mutations in the ATP-binding groove of the Rad3/XPD helicase lead to Xeroderma pigmentosum-Cockayne syndrome-like phenotypes. PLoS Genet 10:e1004859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Chiang CM, Oelgeschläger T, Xie X, Burley SK, Nakatani Y, Roeder RG (1996) A histone octamer-like structure within TFIID. Nature 380:356–359 [DOI] [PubMed] [Google Scholar]
- Hoffmann NA et al. (2015) Molecular structures of unbound and transcribing RNA polymerase III. Nature 528:231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontz RD, French SL, Oakes ML, Tongaonkar P, Nomura M, Beyer AL, Smith JS (2008) Transcription of multiple yeast ribosomal DNA genes requires targeting of UAF to the promoter by Uaf30. Mol Cell Biol 28:6709–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi M, Carey MF, Kakidani H, Roeder RG (1988) Mechanism of action of a yeast activator: direct effect of GAL4 derivatives on mammalian TFIID-promoter interactions. Cell 54:665–669 [DOI] [PubMed] [Google Scholar]
- Iben JR, Mazeika JK, Hasson S, Rijal K, Arimbasseri AG, Russo AN, Maraia RJ (2011) Point mutations in the Rpb9-homologous domain of Rpc11 that impair transcription termination by RNA polymerase III. Nucleic Acids Res 39:6100–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imasaki T et al. (2011) Architecture of the Mediator head module. Nature 475:240–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro A, Kassavetis GA, Geiduschek EP (2002) Essential roles of Bdp1, a subunit of RNA polymerase III initiation factor TFIIIB, in transcription and tRNA processing. Mol Cell Biol 22:3264–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS (1992) The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′–5′ direction in the presence of elongation factor SII. Genes Dev 6:1342–1356 [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R (2000) Structure and function of a human TAFII250 double bromodomain module. Science 288:1422–1425 [DOI] [PubMed] [Google Scholar]
- Jawhari A et al. (2002) p52 Mediates XPB function within the transcription/repair factor TFIIH. J Biol Chem 277:31761–31767 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Langelier M-F, Bataille AR, Pascal JM, Pugh BF, Robert F (2016) Tail and kinase modules differently regulate core mediator recruitment and function in vivo. Mol Cell 64:455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Gralla JD (1993) Uncoupling of initiation and reinitiation rates during HeLa RNA polymerase II transcription in vitro. Mol Cell Biol 13:4572–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SAS, Dubeau L, Johnson DL (2008) Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem 283:19184–19191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo YJ, Ficarro SB, Soares LM, Chun Y, Marto JA, Buratowski S (2017) Downstream promoter interactions of TFIID TAFs facilitate transcription reinitiation. Genes Dev 31:2162–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo ZS, Kassavetis GA, Wang J, Geiduschek EP, Sigler PB (2003) Crystal structure of a transcription factor IIIB core interface ternary complex. Nature 422:534–539 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT (2010) Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339:225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Cheng S, Kadonaga JT (2006) Rational design of a super core promoter that enhances gene expression. Nat Meth 3:917–922 [DOI] [PubMed] [Google Scholar]
- Kainov DE, Vitorino M, Cavarelli J, Poterszman A, Egly J-M (2008) Structural basis for group A trichothiodystrophy. Nat Struct Mol Biol 15:980–984 [DOI] [PubMed] [Google Scholar]
- Kaplan CD (2013) Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochim Biophys Acta 1829:39–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP (1990) S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60:235–245 [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Nguyen ST, Kobayashi R, Kumar A, Geiduschek EP, Pisano M (1995) Cloning, expression, and function of TFC5, the gene encoding the B″ component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci USA 92:9786–9790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Letts GA, Geiduschek EP (1999) A minimal RNA polymerase III transcription system. EMBO J 18:5042–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Letts GA, Geiduschek EP (2001) The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. EMBO J 20:2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Soragni E, Driscoll R, Geiduschek EP (2005) Reconfiguring the connectivity of a multiprotein complex: fusions of yeast TATA-binding protein with Brf1, and the function of transcription factor IIIB. Proc Natl Acad Sci USA 102:15406–15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Prakash P, Shim E (2010) The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J Biol Chem 285:2695–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Josaitis CA, Dodd JA, Nomura M (1998) Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J Biol Chem 273:33795–33802 [DOI] [PubMed] [Google Scholar]
- Kettenberger H, Armache K-J, Cramer P (2003) Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114:347–357 [DOI] [PubMed] [Google Scholar]
- Keys DA, Vu L, Steffan JS, Dodd JA, Yamamoto RT, Nogi Y, Nomura M (1994) RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev 8:2349–2362 [DOI] [PubMed] [Google Scholar]
- Keys DA et al. (1996) Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev 10:887–903 [DOI] [PubMed] [Google Scholar]
- Khatter H, Vorländer MK, Müller CW (2017) RNA polymerase I and III: similar yet unique. Cur Opin Struct Biol 47:88–94 [DOI] [PubMed] [Google Scholar]
- Khoo B, Brophy B, Jackson SP (1994) Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev 8:2879–2890 [DOI] [PubMed] [Google Scholar]
- Khoo S-K, Wu C-C, Lin Y-C, Lee J-C, Chen H-T (2014) Mapping the protein interaction network for TFIIB-related factor Brf1 in the RNA polymerase III preinitiation complex. Mol Cell Biol 34:551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo S-K, Wu C-C, Lin Y-C, Chen H-T (2018) The TFIIE-related Rpc82 subunit of RNA polymerase III interacts with the TFIIB-related transcription factor Brf1 and the polymerase cleft for transcription initiation. Nucleic Acids Res 46:1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen MT, Greenblatt JF (1992) The general transcription factor RAP30 binds to RNA polymerase II and prevents it from binding nonspecifically to DNA. Mol Cell Biol 12:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599–608 [DOI] [PubMed] [Google Scholar]
- Kim KK, Chamberlin HM, Morgan DO, Kim SH (1996) Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase. Nat Struct Biol 3:849–855 [DOI] [PubMed] [Google Scholar]
- Kim TK, Ebright RH, Reinberg D (2000) Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288:1418–1422 [DOI] [PubMed] [Google Scholar]
- Kim JS, Saint-André C, Lim HS, Hwang C-S, Egly J-M, Cho Y (2015) Crystal structure of the Rad3/XPD regulatory domain of Ssl1/p44. J Biol Chem 290:8321–8330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ (2009) The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev 23:439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson BA, Hahn S (2011) Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science 333:1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch SC, Simon N, Ebert C, Carell T (2016) Molecular mechanisms of xeroderma pigmentosum (XP) proteins. Q Rev Biophys 49:e5. [DOI] [PubMed] [Google Scholar]
- Kokubo T, Gong DW, Yamashita S, Horikoshi M, Roeder RG, Nakatani Y (1993) Drosophila 230-kD TFIID subunit, a functional homolog of the human cell cycle gene product, negatively regulates DNA binding of the TATA box-binding subunit of TFIID. Genes Dev 7:1033–1046 [DOI] [PubMed] [Google Scholar]
- Kokubo T, Yamashita S, Horikoshi M, Roeder RG, Nakatani Y (1994) Interaction between the N-terminal domain of the 230-kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA-box binding. Proc Natl Acad Sci USA 91:3520–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova O, Ben-Shem A, Luo J, Ranish J, Schultz P, Papai G (2018) Molecular structure of promoter-bound yeast TFIID. Nat Commun 9:4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhin Y et al. (2009) Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol 7:e1000102–1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrewa D, Zeller ME, Armache K-J, Seizl M, Leike K, Thomm M, Cramer P (2009) RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462:323–330 [DOI] [PubMed] [Google Scholar]
- Kuper J et al. (2014) In TFIIH, XPD helicase is exclusively devoted to DNA repair. PLoS Biol 12: e1001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach AK, Kadonaga JT (2000) The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol 20:4754–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH (1998) New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev 12:34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalo D, Steffan JS, Dodd JA, Nomura M (1996) RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J Biol Chem 271:21062–21067 [DOI] [PubMed] [Google Scholar]
- Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C (2006) A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J 25:118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P (2012a) Structure of the Mediator head module. Nature 492:448–451 [DOI] [PubMed] [Google Scholar]
- Lariviere L, Seizl M, Cramer P (2012b) A structural perspective on Mediator function. Curr Opin Cell Biol 24:305–313 [DOI] [PubMed] [Google Scholar]
- Lariviere L, Plaschka C, Seizl M, Petrotchenko EV, Wenzeck L, Borchers CH, Cramer P (2013) Model of the Mediator middle module based on protein cross-linking. Nucleic Acids Res 41:9266–9273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned RM, Cordes S, Tjian R (1985) Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol Cell Biol 5:1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Park JM, Min S, Han SJ, Kim YJ (1999) An activator binding module of yeast RNA polymerase II holoenzyme. Mol Cell Biol 19:2967–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-H, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA (2005) Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol Cell Biol 25:9674–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B, Sandelin A, Carninci P (2012) Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet 13:233–245 [DOI] [PubMed] [Google Scholar]
- Leurent C et al. (2002) Mapping histone fold TAFs within yeast TFIID. EMBO J 21:3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-L, Golebiowski FM, Onishi Y, Samara NL, Sugasawa K, Yang W (2015) Tripartite DNA lesion recognition and verification by XPC, TFIIH, and XPA in nucleotide excision repair. Mol Cell 59:1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT (2004) The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev 18:1606–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder RH (1996) A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol Cell Biol 16:6436–6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H et al. (2008) Structure of the DNA repair helicase XPD. Cell 133:801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD (2010) Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 327:206–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo SM, Tanaka M, Sullivan ML, Hernandez N (1992) A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell 71:1029–1040 [DOI] [PubMed] [Google Scholar]
- Lockwood WW et al. (2010) Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Med 7:e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolli G, Lowe ED, Brown NR, Johnson LN (2004) The crystal structure of human CDK7 and its protein recognition properties. Structure 12:2067–2079 [DOI] [PubMed] [Google Scholar]
- López-De-León A, Librizzi M, Puglia K, Willis IM (1992) PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell 71:211–220 [DOI] [PubMed] [Google Scholar]
- Louder RK, He Y, López-Blanco JR, Fang J, Chacón P, Nogales E (2016) Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531:604–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J et al. (2015) Architecture of the human and yeast general transcription and DNA repair factor TFIIH. Mol Cell 59:794–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Male G et al. (2015) Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat Commun 6:7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JHJ (2014) Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 15:465–481 [DOI] [PubMed] [Google Scholar]
- Mathieu N, Kaczmarek N, Rüthemann P, Luch A, Naegeli H (2013) DNA quality control by a lesion sensor pocket of the xeroderma pigmentosum group D helicase subunit of TFIIH. Curr Biol 23:204–212 [DOI] [PubMed] [Google Scholar]
- Maxon ME, Goodrich JA, Tjian R (1994) Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev 8:515–524 [DOI] [PubMed] [Google Scholar]
- Milkereit P, Tschochner H (1998) A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J 17:3692–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Schultz P, Tschochner H (1997) Resolution of RNA polymerase I into dimers and monomers and their function in transcription. Biol Chem 378:1433–1443 [DOI] [PubMed] [Google Scholar]
- Mital R, Kobayashi R, Hernandez N (1996) RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol 16:7031–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monté D et al. (2018) Crystal structure of human Mediator subunit MED23. Nat Commun 9:3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel-Carretero M, Herrera-Moyano E, Aguilera A (2015) A unified model for the molecular basis of Xeroderma pigmentosum-Cockayne Syndrome. Rare Dis 3:e1079362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K et al. (2013) Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342:1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Tsai K-L, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD (2015) Structure of an RNA polymerase II preinitiation complex. Proc Natl Acad Sci USA 112:13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli H, Bardwell L, Friedberg EC (1993) Inhibition of Rad3 DNA helicase activity by DNA adducts and abasic sites: implications for the role of a DNA helicase in damage-specific incision of DNA. Biochemistry 32:613–621 [DOI] [PubMed] [Google Scholar]
- Naidu S, Friedrich JK, Russell J, Zomerdijk JCBM(2011) TAF1B is a TFIIB-like component of the basal transcription machinery for RNA polymerase I. Science 333:1640–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Zhou H, Winston F, Hinnebusch AG (1999) Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol Cell 4:657–664 [DOI] [PubMed] [Google Scholar]
- Neyer S et al. (2016) Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature 540:607–610 [DOI] [PubMed] [Google Scholar]
- Nikolov DB et al. (1992) Crystal structure of TFIID TATA-box binding protein. Nature 360:40–46 [DOI] [PubMed] [Google Scholar]
- Nikolov DB et al. (1995) Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377:119–128 [DOI] [PubMed] [Google Scholar]
- Nikolov DB, Chen H, Halay ED, Hoffman A, Roeder RG, Burley SK (1996) Crystal structure of a human TATA box-binding protein/TATA element complex. Proc Natl Acad Sci USA 93:4862–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Fang J, Louder RK (2017a) Structural dynamics and DNA interaction of human TFIID. Transcription 8:55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Louder RK, He Y (2017b) Structural insights into the eukaryotic transcription initiation machinery. Annu Rev Biophys 46:59–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Patel AB, Louder RK (2017c) Towards a mechanistic understanding of core promoter recognition from cryo-EM studies of human TFIID. Curr Opin Struct Biol 47:60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K, Schneider TR, Cramer P (2017) Core Mediator structure at 3.4 Å extends model of transcription initiation complex. Nature 545:248–251 [DOI] [PubMed] [Google Scholar]
- Ohkuma Y, Roeder RG (1994) Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature 368:160–163 [DOI] [PubMed] [Google Scholar]
- Okuda M, Tanaka A, Satoh M, Mizuta S, Takazawa M, Ohkuma Y, Nishimura Y (2008) Structural insight into the TFIIE-TFIIH interaction: TFIIE and p53 share the binding region on TFIIH. EMBO J 27:1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papai G, Tripathi MK, Ruhlmann C, Werten S, Crucifix C, Weil PA, Schultz P (2009) Mapping the initiator binding Taf2 subunit in the structure of hydrated yeast TFIID. Structure 17:363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AB et al. (2018) Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362(6421):eaau8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- Pilsl M et al. (2016) Structure of the initiation-competent RNA polymerase I and its implication for transcription. Nat Commun 7:12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C et al. (2015) Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518:376–380 [DOI] [PubMed] [Google Scholar]
- Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P (2016) Transcription initiation complex structures elucidate DNA opening. Nature 533:353–358 [DOI] [PubMed] [Google Scholar]
- Pugh BF, Tjian R (1990) Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell 61:1187–1197 [DOI] [PubMed] [Google Scholar]
- Pugh BF, Tjian R (1991) Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev 5:1935–1945 [DOI] [PubMed] [Google Scholar]
- Rani PG, Ranish JA, Hahn S (2004) RNA polymerase II (Pol II)-TFIIF and Pol II-mediator complexes: the major stable Pol II complexes and their activity in transcription initiation and reinitiation. Mol Cell Biol 24:1709–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapin I (2013) Disorders of nucleotide excision repair. Handb Clin Neurol 113:1637–1650 [DOI] [PubMed] [Google Scholar]
- Reines D (1992) Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem 267:3795–3800 [PMC free article] [PubMed] [Google Scholar]
- Rijal K, Maraia RJ (2013) RNA polymerase III mutants in TFIIFα-like C37 that cause terminator readthrough with no decrease in transcription output. Nucleic Acids Res 41:139–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S, Colbert T, Hahn S (1995) TFIIIC determines RNA polymerase III specificity at the TATA-containing yeast U6 promoter. Genes Dev 9:832–842 [DOI] [PubMed] [Google Scholar]
- Robinson PJJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD (2012) Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA 109:17931–17935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ et al. (2015) Molecular architecture of the yeast Mediator complex. Elife 4:e08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei P-J, Burlingame AL, Kornberg RD (2016) Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell 166:1411–1422.e1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 21:327–335 [PubMed] [Google Scholar]
- Roeder RG, Rutter WJ (1969) Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature 224:234–237 [DOI] [PubMed] [Google Scholar]
- Roeder RG, Rutter WJ (1970) Specific nucleolar and nucleoplasmic RNA polymerases. Proc Natl Acad Sci USA 65:675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol M, Kolb-Cheynel I, Egly JM (1997) Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J 16:1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AL, Singer DS (2015) Core promoters in transcription: old problem, new insights. Trends Biochem Sci 40:165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Gabunilas J, Gillespie A, Ngo D, Chanfreau GF (2016) Common genomic elements promote transcriptional and DNA replication roadblocks. Genome Res 26:1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P (2011) Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J Biol Chem 286:18701–18707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert S, Tjian R (1995) Human TAFII250 interacts with RAP74: implications for RNA polymerase II initiation. Genes Dev 9:2747–2755 [DOI] [PubMed] [Google Scholar]
- Sadian Y et al. (2017) Structural insights into transcription initiation by yeast RNA polymerase I. EMBO J 36(18):2698–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski CL, Henry RW, Lobo SM, Hernandez N (1993) Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev 7:1535–1548 [DOI] [PubMed] [Google Scholar]
- Sainsbury S, Niesser J, Cramer P (2013) Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 493:437–440 [DOI] [PubMed] [Google Scholar]
- Sainsbury S, Bernecky C, Cramer P (2015) Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 16:129–143 [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA (2007) Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 8:424–436 [DOI] [PubMed] [Google Scholar]
- Sanders SL, Garbett KA, Weil PA (2002) Molecular characterization of Saccharomyces cerevisiae TFIID. Mol Cell Biol 22:6000–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrock B, Egly JM (2001) A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. J Biol Chem 276:35328–35333 [DOI] [PubMed] [Google Scholar]
- Schilbach S, Hantsche M, Tegunov D, Dienemann C, Wigge C, Urlaub H, Cramer P (2017) Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551:204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA (2012) RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene 493:176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider EV, Böttcher J, Blaesse M, Neumann L, Huber R, Maskos K (2011) The structure of CDK8/CycC implicates specificity in the CDK/cyclin family and reveals interaction with a deep pocket binder. J Mol Biol 412:251–266 [DOI] [PubMed] [Google Scholar]
- Schramm L, Hernandez N (2002) Recruitment of RNA polymerase III to its target promoters. Genes Dev 16:2593–2620 [DOI] [PubMed] [Google Scholar]
- Schramm L, Pendergrast PS, Sun Y, Hernandez N (2000) Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev 14:2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz P, Fribourg S, Poterszman A, Mallouh V, Moras D, Egly JM (2000) Molecular structure of human TFIIH. Cell 102:599–607 [DOI] [PubMed] [Google Scholar]
- Shah SM, Kumar A, Geiduschek EP, Kassavetis GA (1999) Alignment of the B″ subunit of RNA polymerase III transcription factor IIIB in its promoter complex. J Biol Chem 274:28736–28744 [DOI] [PubMed] [Google Scholar]
- Shiekhattar R et al. (1995) Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature 374:283–287 [DOI] [PubMed] [Google Scholar]
- Sklar VE, Schwartz LB, Roeder RG (1975) Distinct molecular structures of nuclear class I, II, and III DNA-dependent RNA polymerases. Proc Natl Acad Sci USA 72:348–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Whitehouse I (2012) Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483:434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J (2018) Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 19:262–274 [DOI] [PubMed] [Google Scholar]
- Steffan JS, Keys DA, Vu L, Nomura M (1998) Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Mol Cell Biol 18:3752–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman DJ, Geiduschek EP (1984) Differential binding of a S. cerevisiae RNA polymerase III transcription factor to two promoter segments of a tRNA gene. EMBO J 3:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer KF, Ingles CJ, Greenblatt J (1990) Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature 345:783–786 [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Akagi J-i, Nishi R, Iwai S, Hanaoka F (2009) Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell 36:642–653 [DOI] [PubMed] [Google Scholar]
- Sung P, Bailly V, Weber C, Thompson LH, Prakash L, Prakash S (1993) Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature 365:852–855 [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ et al. (1995) Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell 80:21–28 [DOI] [PubMed] [Google Scholar]
- Svetlov V, Nudler E (2013) Basic mechanism of transcription by RNA polymerase II. Biochim Biophys Acta 1829:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafur L et al. (2016) Molecular structures of transcribing RNA polymerase I. Mol Cell 64:1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Hunziker Y, Sargent DF, Richmond TJ (1996) Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381:127–151 [DOI] [PubMed] [Google Scholar]
- Teichmann M, Wang Z, Roeder RG (2000) A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc Natl Acad Sci USA 97:14200–14205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theron T et al. (2005) Transcription-associated breaks in xeroderma pigmentosum group D cells from patients with combined features of xeroderma pigmentosum and Cockayne syndrome. Mol Cell Biol 25:8368–8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torreira E et al. (2017) The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. Elife 6:e20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K-L et al. (2017) Mediator structure and rearrangements required for holoenzyme formation. Nature 544:196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FT, Sigler PB (2000) Structural basis of preinitiation complex assembly on human pol II promoters. EMBO J 19:25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K-L, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ (2013) A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol 20:611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K-L, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ (2014) Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157:1430–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A, Cramer P (2012) Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 45:439–446 [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I (2007) Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448:157–162 [DOI] [PubMed] [Google Scholar]
- Verger A, Monté D, Villeret V (2019) Twenty years of Mediator complex structural studies. Biochem Soc Trans 47:399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M et al. (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131:58–69 [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Chen JL, Yokomori K, Tjian R (1995) Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 81:1115–1125 [DOI] [PubMed] [Google Scholar]
- Vojnic E et al. (2011) Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nat Struct Mol Biol 18:404–409 [DOI] [PubMed] [Google Scholar]
- Vorländer MK, Khatter H, Wetzel R, Hagen WJH, Müller CW (2018) Molecular mechanism of promoter opening by RNA polymerase III. Nature 553:295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hawley DK (1993) Identification of a 3′–5′ exonuclease activity associated with human RNA polymerase II. Proc Natl Acad Sci USA 90:843–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Roeder RG (1995) Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA 92:7026–7030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD (2006) Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127:941–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24:437–440 [DOI] [PubMed] [Google Scholar]
- Weeda G, van Ham RC, Vermeulen W, Bootsma D, van der Eb AJ, Hoeijmakers JH (1990) A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne’s syndrome. Cell 62:777–791 [DOI] [PubMed] [Google Scholar]
- Weinmann R, Roeder RG (1974) Role of DNA-dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc Natl Acad Sci USA 71:1790–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover KD, Bushnell DA, Kornberg RD (2004) Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 119:481–489 [DOI] [PubMed] [Google Scholar]
- Wolski SC, Kuper J, Hänzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C (2008) Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol 6:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM (2001) TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J Biol Chem 276:34235–34243 [DOI] [PubMed] [Google Scholar]
- Wu C-C, Lin Y-C, Chen H-T (2011) The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol 31:2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-C et al. (2012) RNA polymerase III subunit architecture and implications for open promoter complex formation. Proc Natl Acad Sci USA 109:19232–19237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X et al. (1996) Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380:316–322 [DOI] [PubMed] [Google Scholar]
- Yan Q, Moreland RJ, Conaway JW, Conaway RC (1999) Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J Biol Chem 274:35668–35675 [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S (2000) A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225–229 [DOI] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98:811–824 [DOI] [PubMed] [Google Scholar]
- Zhang Z et al. (2015) Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. Elife 4:e07777. [DOI] [PMC free article] [PubMed] [Google Scholar]


