Abstract
Herein we describe, to our knowledge for the first time the use of the clustered regularly interspaced short palindromic repeats/CRISPR-associated gene 9 (CRISPR/Cas9) system for genome editing of Neospora caninum, an apicomplexan parasite considered one of the main causes of abortion in cattle worldwide. By using plasmids containing the CRISPR/Cas9 components adapted to the closely related parasite Toxoplasma gondii, we successfully knocked out a green fluorescent protein (GFP) in an Nc-1 GFP-expressing strain, and efficiently disrupted the NcGRA7 gene in the Nc-Spain7 isolate by insertion of a pyrimethamine resistance cassette. The successful use of this technology in N. caninum lays the foundation for an efficient, targeted gene modification tool in this parasite.
Keywords: Neospora caninum, CRISPR/Cas9, Transfection, Gene disruption, NcGRA7, Nc-Spain7
Neospora caninum is a protozoan parasite from the phylum Apicomplexa, and is closely related to Toxoplasma gondii. As the causative agent of bovine neosporosis, it is considered one of the main causes of abortion and neonatal mortality in cattle worldwide, entailing significant economic losses (Dubey et al., 2007; Dubey and Schares, 2011; Reichel et al., 2013). At present, there is an urgent need to develop control measures that protect cattle from abortion and vertical transmission. In order to find new vaccines and drug targets, it is imperative to increase knowledge of the molecular factors and genes involved in the parasite’s biology. Thus, developing specific tools for genetic manipulation of Neospora will help us understand host-parasite interactions, and possibly aid in the development of vaccine and treatment strategies. While a wide array of tools has been developed for the closely related parasite T. gondii–including an assortment of promoters, fluorescent proteins, selectable markers, and genome editing tools–few such methods are available for N. caninum (reviewed in Suarez et al., 2017). Indeed, the genetic manipulation of N. caninum is limited to a few studies such as the introduction of the LacZ gene, the expression of bradyzoite (the semi-dormant stage of the parasite that is present in tissue cysts) antigens such as NcSAG4, the expression of antigenic and secreted Toxoplasma proteins (e.g. TgSAG1, TgGRA2, TgGRA15 or TgROP16), the mutation of dihydrofolate reductase-thymidylate synthase (DHFR-TS; conferring resistance to pyrimethamine) or the insertion of drug selectable markers such as chloramphenicol acetyl transferase (CAT; resistance to chloramphenicol) or Ble (resistance to phleomycin) (Howe et al., 1997; Beckers et al., 1997; Howe and Sibley, 1997; Zhang et al., 2010; Marugán-Hernández et al., 2011; Pereira et al., 2014; Pereira and Yatsuda, 2014; Mota et al., 2017).
Recently, the adaptation of the clustered regularly interspaced short palindromic repeats/CRISPR-associated gene 9 (CRISPR/Cas9) technology–a naturally occurring DNA recognition system used as a defense mechanism in bacteria and archaea–has led to extremely efficient gene editing in a variety of organisms (Jinek et al., 2012; Mali et al., 2013; Shen et al., 2014; Sidik et al., 2014). Briefly, this system relies on the introduction of site-specific double-strand DNA breaks (DSBs) by the endonuclease Cas9 in a target sequence that is homologous to the single guide RNA (sgRNA, or gRNA hereafter). These DSBs are subsequently repaired by the organism either by the non-homologous end joining (NHEJ) pathway, leading to insertion and deletion mutations in the targeted genes (“indels”), or by homologous direct repair (HDR) in the presence of a DNA donor template. A gRNA complex will be able to guide the Cas9 only if an appropriate protospacer-adjacent motif (PAM) is located immediately after the target sequence. The utility of using gRNAs designed to target specific genes, combined with the high efficiency of disruption by Cas9, has proven useful for gene manipulation in many model organisms (Cong et al., 2013). In apicomplexan parasites, to date, this has only been achieved in Plasmodium, Cryptosporidium and Toxoplasma (Suarez et al., 2017). In the latter, the CRISPR/Cas9 system was recently adapted to produce efficient targeted gene disruption and site-specific insertion of selectable markers (Shen et al., 2014; Sidik et al., 2014). Since then, it has been widely employed as an efficient and powerful means of testing the role of specific genes in diverse genetic backgrounds and even to perform a genome-wide screen of Toxoplasma genes conferring fitness in human foreskin fibroblasts (HFFs) (Sidik et al., 2016).
In the present work we describe, to our knowledge, the first successful use of the CRISPR/Cas9 system in N. caninum by transfecting plasmids developed for Toxoplasma. We disrupted two different genes, the green fluorescence protein (GFP) expressed in Nc-1, and the NcGRA7 in the wild-type Nc-Spain7 isolate, demonstrating the utility of the Toxoplasma-adapted plasmids and CRISPR/Cas9 technology in N. caninum. To disrupt the genes, we used the universal pU6 plasmid (Addgene plasmid # 52694), which contains Cas9 with a nuclear localisation sequence driven by the TgTUBl promoter and a gRNA expression site driven by the T. gondii U6 promoter (Sidik et al., 2014). Using the BsaI-specific sites in the vector, we generated different plasmids containing 20 nucleotide gRNAs that targeted two regions of the GFP gene (GenBank accession number U87973) close to the ATG site and the 5’ and 3’ ends of the coding sequence of the NcGRA7 gene (Fig. 1A and 2A). A complete description of the primers and gRNAs used in this work is provided in Table 1. Parasite culture and transfections were performed as described elsewhere (e.g. Mota et al., 2017).
Fig. 1.
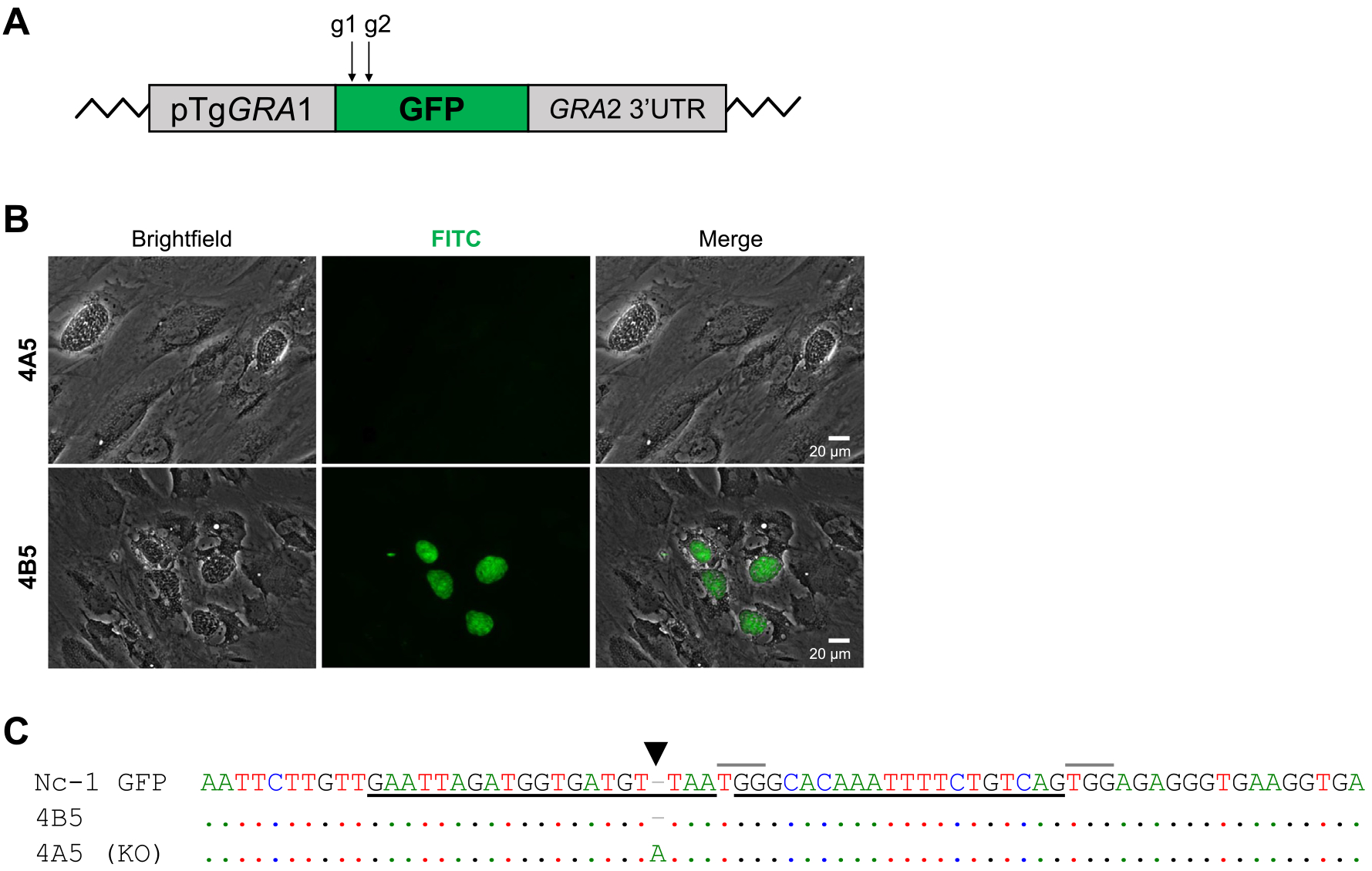
Analysis of the Neospora caninum Nc-1 GFP knockout (KO) generated with clustered regularly interspaced short palindromic repeats/CRISPR-associated gene 9 (CRISPR/Cas9). (A) Schematic representation of the GFP gene disruption achieved by employing two guide RNAs (gRNAs; g1 and g2) in the Nc-1 GFP strain. (B) Images (20×) from cell culture flasks containing human foreskin fibroblast cells infected with two different clones obtained after transfection and limiting dilution (4A5 and 4B5) showing green fluorescence. Note the absence of green fluorescence in clone 4A5. White bars represent scale bars (20 μm). (C) Sequence alignment of the parental strain (Nc-1 GFP) and two clones, 4B5 and 4A5 (KO). Identical nucleotides are presented as dots. The two gRNA sites are underlined in black. Protospacer adjacent motif (PAM) sites are indicated by grey lines above the sequence. The cutting site is indicated by an arrowhead. UTR, untranslated region. pTgGRA1, promoter region of the Toxoplasma dense granule 1 gene.
Fig. 2.

Analysis of the Neospora caninum Nc-GRA7 knockout generated with clustered regularly interspaced short palindromic repeats/CRISPR-associated gene 9 (CRISPR/Cas9). (A) Schematic representation of the GRA7 locus and the repair template used in the transfection. Disruption was achieved by employing two guide RNAs (gRNAs; g1 and g2). The positions and directions of primers (P1–4) used for diagnostic PCRs are indicated by arrows. The position of the NotI restriction enzyme used to linearize the donor template is indicated. (B) Schematic representation of the mutant clones that incorporated the donor template in forward (Fo) and reverse (Ro) orientation. g1 and g2 indicate gRNAs, and positions and directions of primers used for diagnostic PCRs (P1–4) are indicated by arrows. Sequence alignments of regions surrounding the cutting site are shown, comparing a representative of Fo (clone #50) and Ro (clone #16) with the NcGRA7 locus and the loxP-DHFR-mCherry plasmid. gRNA sites and protospacer adjacent motif (PAM) sequences are indicated by red and blue lines above the sequences. Slashes within sequences are used to visually separate distant unconnected regions. The cutting sites are indicated by arrowheads. (C) Diagnostic PCR demonstrating the loss of GRA7 (P1 + P2) and the integration of the donor template in forward (P2 + P4 and P1 + P3) and reverse (P1 + P4 and P2 + P3) orientations. WT, wild type; UTR, untranslated region; DHFR-TS, dihydrofolate reductase-thymidylate synthase (DHFR-TS); LoxP: loxP sites. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Guide RNAs (gRNAs) and primers used in the present work.
| Region | Primer | Sequence | Used for |
|---|---|---|---|
| GFP | GFP gRNA1 | GAATTAGATGGTGATGTTAA | Disrupt GFP gene in the N-terminal end |
| GFP gRNA2 | GGGCACAAATTTTCTGTCAG | ||
| GFP-Fw | TGGCCAAATCAAAGGCTATT | Amplify and sequence the gRNA targeted region of the GFP gene | |
| GFP-Rv | TGGGTATCTTGAAAAGCATTGA | ||
| GRA7 | GRA7 gRNA1 | GTTTGTGGACTGGCAATCGC | Disrupt GRA7 gene in both N- and C-terminal ends |
| GRA7 gRNA2 | GCAGTGATCCGGAACAGGAG | ||
| GRA7-Fw (P1) | TCGCTGTTCCTGTAGGCTTT | Amplify and sequence the gRNA targeted region of the GRA7 gene together with repair template | |
| GRA7-Rv (P2) | CTGTCATCTGGGACACGAAA | ||
| Loxp-DHFR-mCherry plasmid | Plasmid-Fw (P4) | GGCGTGAAGATCTGGGACAA | Amplify and sequence the gRNA targeted region of the GRA7 gene together with repair template |
| Plasmid-Rv (P3) | ACGACCTACACCGAACTGAGAT |
First, ~1 × 103 tachyzoites of the Nc-1 GFP strain were transfected with a combination of 35 μg of each of the two universal pU6 plasmids (70 μg in total) containing the two gRNAs described above. Immediately after transfection, a limiting dilution of the pool of transfected parasites was performed in 96-well plates to obtain individual clones (Sidik et al., 2016). After 7 days, 16 individual clones were obtained. From these, one clone (4A5) showed parasites that were not green, implying a disruption of the GFP gene (Fig. 1B). To further characterise this clone, we sequenced a 285 bp PCR-amplified region containing the gRNA homology site (Fig. 1C). Results showed that the loss of GFP expression was due to a frame shift caused by the insertion of a single nucleotide (A) 4 bp upstream of the PAM site, which has been reported to be the usual position where Cas9 cleaves DNA (e.g. Sidik et al., 2014).
After confirming that CRISPR/Cas9 can be used for gene disruption in Neospora, we wanted to test whether this system could also produce gene disruptions or deletions by insertion of a selectable marker following site-specific targeting as previously described for Toxoplasma (Shen et al., 2014; Sidik et al.,2014). For that, we aimed to disrupt the NcGRA7 gene (GenBank accession number JQ410455; ToxoDB ID NCLIV_021640) with two gRNAs targeting the 5’ and 3’ ends of the coding sequence, and produce the deletion of most of the NcGRA7 open reading frame (ORF) and insertion of a donor DNA template bearing a pyrimethamine resistance cassette (Fig. 2A). In order to do that, we used as a repair template the LoxP-mCherry-DHFR plasmid (Addgene Plasmid #70147) (Long et al., 2016), which contains the Toxoplasma DHFR-TS pyrimethamine-resistant allele marker (Donald and Roos, 1993) fused to the fluorescent protein mCherry flanked by two loxP sites that allows the excision of this cassette in the presence of Cre recombinase. After transfecting ~107 tachyzoites of the Nc-Spain7 isolate with a combination of the two gRNAs (6 μg each) and the linearised donor vector (Notl cut, 2 μg) described above, parasites were returned to culture. Selection with 10 iM pyrimethamine was initiated 24 h later and continued for three passages. To confirm the insertion of the resistance cassette into the GRA7 locus, single clones of pyrimethamine-resistant parasites were isolated as described above and diagnostic PCR was performed to check for integration (Fig. 2B and C). Unlike the wild-type strain (WT), we failed to amplify the NcGRA7-specific band from any of the 59 clones obtained, suggesting the insertion of a large piece of DNA in the GRA7 locus. To rule out the possibility of low DNA yields or DNA degradation, we performed a GRA6 PCR (Walsh et al., 2001). As expected, all clones amplified the specific GRA6 band (Fig. 2C). Moreover, since all clones were pyrimethamine-resistant (in contrast to WT), the plasmid, or at least a portion containing the DHFR-TS resistance cassette, was inserted either in the GRA7 locus or randomly elsewhere in the genome. To identify clones that specifically inserted the plasmid in the GRA7 locus, further PCRs were carried out. From these 59 GRA7-negative clones, nine were negative for all the plasmid insertion PCRs and 31 showed band sizes larger or smaller than the expected amplicon size, which suggests that only a portion or altered parts of the plasmid were integrated in these clones. On the other hand, a total of 19 clones demonstrated integration of the complete repair template into the GRA7 locus, 17 of which incorporated the plasmid in forward orientation (Fo) and two in reverse orientation (Ro). To further characterise the exact position of the insertion in the GRA7 locus, we selected eight Fo clones and the two Ro clones, and sequenced the regions upstream of the first gRNA and downstream of the second gRNA. For all the Fo clones, gene disruption was produced by insertion of the resistance cassette at the site where Cas9 is predicted to cut after hybridisation of the second gRNA, which was close to the stop codon (Fig. 2B). Thus, it would be expected that these Fo clones still express a minimally truncated, but otherwise functional, GRA7 protein, since the gene was only disrupted towards the 3’ end. In contrast, a deletion of most of the NcGRA7 ORF was produced in the two Ro clones, integrating the repair template between the two gRNAs. Furthermore, the DSB was produced at a position corresponding to 3–4 bp upstream of the PAM in both types of clones (Fig. 2B), similar to the cutting site mentioned above for the GFP mutant.
It is worth mentioning that PCR analysis by itself cannot rule out the possibility that other copies of our donor template were inserted elsewhere in the genome. Furthermore, it is possible that gRNAs have off-target effects. Therefore, it is recommended to always complement knockout (KO) clones with the WT copy of the gene, for example by insertion into a neutral locus such as the uracil phosphoribosyltransferase (UPRT) gene and subsequent selection with 5-fluorodeoxyuridine (FUDR) (Shen et al., 2014). The disruption of the Neospora UPRT gene and repair with a WT copy of the gene of interest should be straightforward using the CRISPR/Cas9 approach we describe in this paper.
Finally, the present work shows that N. caninum can acquire resistance to pyrimethamine by expression of the DHFR-TS resistance cassette described for Toxoplasma. It was already known that Neospora is extremely sensitive to pyrimethamine and that resistance to pyrimethamine can be achieved by expression of a mutated version of the NcDHFR-TS gene, employing the same mutations described for Toxoplasma, which occur in highly conserved regions (approximately 80% with the DHFR-TS of Toxoplasma) (Pereira et al., 2014). Herein, we show that Neospora can also become resistant to pyrimethamine by expression of the Toxoplasma mutated DHFR-TS gene.
In conclusion, our results demonstrate that (i) effective gene disruption can be achieved in Neospora by using CRISPR/Cas9; (ii) the ability to introduce a selectable marker at the site of the DSB induced by the gRNA appears not to require homology arms, as previously described for Toxoplasma (Shen et al., 2014); and (iii) production of N. caninum mutants by CRISPR/Cas9 can be achieved by using plasmids already available for the closely related parasite Toxoplasma. The use of CRISPR/Cas9 in N. caninum will contribute to the identification of critical parasite-encoded pathways which, in turn, could lead to the design of novel methods for its control, particularly vaccines and drugs.
Acknowledgements
This research was partially funded by the Spanish Ministry of Economy and Competitiveness (AGL2016-75935-C2-1-R). We gratefully acknowledge all members of the Saeij laboratory group for their help with this work.
References
- Beckers CJ, Wakefield T,Joiner KA, 1997. The expression of Toxoplasma proteins in Neospora caninum and the identification of a gene encoding a novel rhoptry protein. Mol. Biochem. Parasitol 89, 209–223. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, 2013. Multiplex genome engineering using CRISPR/cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RG, Roos DS, 1993. Stable molecular transformation of Toxoplasma gondii: A selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc. Natl. Acad. Sci. U.S.A 90, 11703–11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP, Schares G, Ortega-Mora LM, 2007. Epidemiology and control of neosporosis and Neospora caninum. Clin. Microbiol. Rev 20, 323–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP, Schares G, 2011. Neosporosis in animals–the last five years. Vet. Parasitol 180, 90–108. [DOI] [PubMed] [Google Scholar]
- Howe DK, Mercier C, Messina M, Sibley LD, 1997. Expression of Toxoplasma gondii genes in the closely-related apicomplexan parasite Neospora caninum. Mol. Biochem. Parasitol 86, 29–36. [PubMed] [Google Scholar]
- Howe DK, Sibley LD, 1997. Development of molecular genetics for Neospora caninum: A complementary system to Toxoplasma gondii. Methods 13, 123–133. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S, Wang Q, Sibley LD, 2016. Analysis of noncanonical calcium-dependent protein kinases in Toxoplasma gondii by targeted gene deletion using CRISPR/Cas9. Infect. Immun 84, 1262–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM, 2013. Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marugán-Hernández V, Ortega-Mora LM, Aguado-Martfnez A, Alvarez-Garcfa G, 2011. Genetic manipulation of Neospora caninum to express the bradyzoite-specific protein NcSAG4 in tachyzoites. Parasitology 138, 472–480. [DOI] [PubMed] [Google Scholar]
- Mota CM, Chen AL, Wang K, Nadipuram S, Vashisht AA, Wohlschlegel JA, Mineo TWP, Bradley PJ, 2017. New molecular tools in Neospora caninum for studying apicomplexan parasite proteins. Sci. Rep 7, 3768 10.1038/s41598-017-03978-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pereira LM, Baroni L, Yatsuda AP, 2014. A transgenic Neospora caninum strain based on mutations of the dihydrofolate reductase-thymidylate synthase gene. Exp. Parasitol 138, 40–47. [DOI] [PubMed] [Google Scholar]
- Pereira LM, Yatsuda AP, 2014. The chloramphenicol acetyltransferase vector as a tool for stable tagging of Neospora caninum. Mol. Biochem. Parasitol 196, 75–81. [DOI] [PubMed] [Google Scholar]
- Reichel MP, Alejandra Ayanegui-Alcerreca M, Gondim LF, Ellis JT, 2013. What is the global economic impact of Neospora caninum in cattle - The billion dollar question. Int. J. Parasitol 43, 133–142. [DOI] [PubMed] [Google Scholar]
- Shen B, Brown KM, Lee TD, Sibley LD, 2014. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio 5, e01114–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S, 2014. Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One 9, e100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik SM, Huet D, Ganesan SM, Huynh MH, Wang T, Nasamu AS, Thiru P, Saeij JP, Carruthers VB, Niles JC, Lourido S, 2016. A genome-wide CRISPR screen in Toxoplasma identifies essential apicomplexan genes. Cell 166, 1423–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez CE, Bishop RP, Alzan HF, Poole WA, Cooke BM, 2017. Advances in the application of genetic manipulation methods to apicomplexan parasites. Int. J. Parasitol 47, 701–710. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Vemulapalli R, Sriranganathan N, Zajac AM, Jenkins MC, Lindsay DS, 2001. Molecular comparison of the dense granule proteins GRA6 and GRA7 of Neospora hughesi and Neospora caninum. Int. J. Parasitol 31, 253–258. [DOI] [PubMed] [Google Scholar]
- Zhang G, Huang X, Boldbaatar D, Battur B, Battsetseg B, Zhang H, Yu L, Li Y, Luo Y, Cao S, Goo YK, Yamagishi J, Zhou J, Zhang S, Suzuki H, Igarashi I, Mikami T, Nishikawa Y, Xuan X, 2010. Construction of Neospora caninum stably expressing TgSAG1 and evaluation of its protective effects against Toxoplasma gondii infection in mice. Vaccine 28, 7243–7247. [DOI] [PubMed] [Google Scholar]


