SUMMARY
The study of genetic factors associated with BD can be safely said to have entered a mature phase, with the appropriate capacity to begin to provide the long awaited insights into the pathophysiology of BD. These advances have been made possible by technological innovations such as large-scale microarray genotyping and next generation sequencing, which have been applied in a comprehensive genome-wide manner to large samples. Several genes have now been robustly associated with BD and in 2016 analyses by the PGC are expected to yield many more similar findings. The initial fruits of such efforts will be to identify molecular targets and pathways that provide insights into the underlying biology of BD and thereby facilitate novel drug development. Eventually, as the field progresses and a greater proportion of heritability is accounted for, more immediate clinical applications such as risk stratification and biologically informed drug selection may also become feasible. However, to keep pace with the technological advances, there is a pressing need now for clinical care to “partner” with research and provide both the samples sizes and the relevant phenotypes necessary to fulfill the promise of precision medicine.
Keywords: Bipolar disorder, Genetics, GWAS, Sequencing, Copy number variation (CNV), Family study, Personalized medicine, Pharmacogenetics
INTRODUCTION
Bipolar disorder (BD) is a lifelong disorder marked by periodic disturbances in mood, cognition and behavior. When strictly defined, it is characterized by the presence of manic episodes with marked changes in mood, energy, and cognition that are often associated with psychotic symptoms. The magnitude or severity of manic episodes can be variable, and milder (hypo)manic episodes can be seen in up to 2% to 3% of the general population.1 Despite the nosologic emphasis on elevated mood episodes, most of the cumulative morbidity arises from depressive episodes, which occur more often and last longer than the briefer periods of mood elevation. Although descriptions of syndromic alteration in mood states have been present since antiquity, it was Emil Kraepelin who provided an initial “synthesis” of a bewildering variety of pathologic mood states into a single nosologic entity that still forms the basic conception of the modern definitions of BD. Kraepelin’s notion of “manic-depressive insanity” included a broad variety of mood states, of variable severity that were often marked by instability (manifesting as mixed states), and were linked by a “uniform prognosis” in their tendency to remit and a “hereditary taint.”2 The latter observation has spawned more than a century of genetic research into BD, although it has only been in last several years that clearly replicated findings have emerged, in large part owing to the application of reliable, high-throughput technologies to samples of sufficient size made possible by worldwide collaboration. This review highlights recent findings in the field of BD genetics and looks forward to the potential role that future genetic findings will have on clinical care.
FAMILY STUDIES: MEASURING HERITABILITY FROM THE TOP DOWN
A long line of family studies, beginning from the early 20th century work of Kraepelin’s student Ernst Rudin, to the more recent studies based on the Diagnostic and Statistical Manual of Mental Disorders in the early 21st century, have found consistent evidence for strong familial aggregation of BD type I, ranging from concordance rates of 40% to 70% in monozygotic twins, to approximately 8% to 10% in first-degree relatives.3 More recent family studies have made use of nationwide registry data in the Scandinavian countries, which provide essentially the only method to ascertain a comprehensive sample despite the disadvantage of only including subjects with the disorder who have sought clinical care and have been accurately diagnosed.4,5 A recent study of BD using Swedish registry data has been particularly informative for its consideration of the familial aggregation of BD with all the major psychiatric disorders.5 Consistent with prior interview-based family studies, Song and colleagues5 found strong evidence for familial aggregation of BD-I with first-degree relative risks (RR) of 5.8 to 7.9. However, the large sample size provided the statistical power to detect coaggregation of BD and schizophrenia (RR, 2.8), major depression (RR, 2.1), anxiety disorders (RR, 1.8), attention deficit hyperactivity disorder (RR, 2.4), personality disorders (RR, 2.2), autism (RR, 2.6), as well as drug abuse disorders (RR, 1.7). These findings are consistent with the familial transmission of a more generalized increased risk for a broad array of psychopathology, which has also gained support from genome-wide association studies (GWAS).
In contrast, 2 recent interview-based family studies of BD that attempted to ascertain the full spectrum of mood disorders have highlighted the relative independence of BD-I and psychotic disorders from the more common unipolar and anxiety diagnoses.6,7 Although these studies distinguish themselves from prior interview-based studies by their broader ascertainment, and their use of nonhierarchical diagnoses, they may have failed to detect a modest coaggregation with psychotic disorders because of their more limited sample size. Indeed, prior family studies of BD often showed a modest increase in rates of schizophrenia, but because of the difficulties in collecting large numbers of families, they lacked the power to detect a statistically significant difference. However, a recent metaanalysis of 38 family showed evidence for a consistent but modest increase in rates of schizophrenia in first degree family members of subjects with BD (odds ratio [OR], 2.10).8 Nevertheless, these studies raise the important point that ascertainment and diagnostic assessment (including the emphasis placed on diagnostic hierarchy) is an important factor influencing patterns of familial transmission. Despite highlighting differing conclusions, the overall pattern seen within the broader context of published BD literature is generally consistent: BD-I has the highest RRs among first degree family members (RR ~ 8–10), with far lower familial risks for “other” disorders (RR ~ 2) that range from subthreshold mood and anxiety disorders to psychotic disorders.
What is the relevance of such family studies in an era of increasing molecular sophistication? By relying almost entirely on clinical phenotypes, family studies are subject to the usual criticisms of clinically defined studies based on descriptive syndromes: overlapping symptoms, variable reliability, and uncertain mapping onto an underlying biology. However, although clinical phenotypes remain broad proxies for an elusive underlying biology, they reflect the clinical reality that is most pertinent to patients and clinicians and still provide the most discriminative “index” relevant for clinical decision making. Indeed, because family history is a broad proxy encompassing all transmitted genetic variation as well as shared environment, it remains the single strongest risk factor for development of BD.
MOLECULAR STUDIES: MEASURING HERITABILITY FROM THE BOTTOM UP
Heritability is an estimate of what proportion of a disorder can be attributed to genetic variation, but it provides no guidance on whether the underlying genetic causes are few or many, common or rare, or somewhere in between.9 The relative contribution of common versus rare variants for psychiatric phenotypes has been a subject of much debate. Fortunately, empirical data are beginning to settle this debate.
Initial approaches to mapping genes for BD were limited by sparse, low-resolution linkage technologies, which could point to a broad area of a chromosome if a disorder was strongly linked to 1 or only few specific regions. Although the genome-wide linkage approach worked well with Mendelian diseases, it has been generally unsuccessful for common diseases that likely consist of more diverse and nuanced genetic causes. Numerous genome-wide linkage studies of BD were performed in the last 2 decades, although the failure to identify a consistent linkage signal is an indication that the more moderately penetrant variants seen in certain complex disorders (such as the MHC locus in autoimmune disorders) are unlikely to be found for BD.10
ASSOCIATION STUDIES: SUCCESS OF GENOME-WIDE APPROACHES AT SCALE
After the sequencing of the human genome it became apparent that most genetic variation is composed of single nucleotide polymorphisms, which represent single base-pair changes that occur regularly throughout the genome at approximately every 1000 bases for a total of approximately 3.5 million in a genome.11 Technological advances led to accurate and cost-effective methods to genotype single nucleotide polymorphisms throughout out the genome in a highly automated assay that has become known as a GWAS. Before GWAS, association studies in BD were limited to 1 or a few genes (“candidate gene studies”) and suffered from poor reproducibility owing to small sample sizes and the challenges of selecting candidate genes given the limited knowledge of the pathophysiology of BD. Fortunately, the last decade has seen a transition to genome-wide approaches, which provide an agnostic yet comprehensive approach.
GWAS have now been performed successfully on almost all medically relevant phenotypes and, despite a few exceptions,12 has led to replicable associations that can provide “entry points” into disease related biology. Importantly, GWAS data are easily compared or metaanalyzed across studies through a process known as imputation, in which common variants in correlation with a known genotype can be probabilistically deduced (“imputed”) based on known patterns within the human population. A second important consideration has been the establishment of clear guidelines for considering a finding to be genome-wide significant.13 Simulation and empirical approaches have demonstrated that the genome has approximately 1 million common independent markers, leading to a corrected association P value threshold of 5 × 10−8 (derived by dividing the conventional P<.05 value by approximately 1 million tests).13 Typically findings that cross this threshold in a primary analysis have a high rate of replication and go on to show evidence of association in subsequent metaanalyses.
In most disorders, including psychiatric disorders, initial GWAS samples (consisting of a few thousand cases and controls) have been underpowered to detect genome-wide significant associations. This has led to the formation of consortia, such as the Psychiatric GWAS Consortium (PGC), in which large-scale metaanalyses have been performed in sample sizes that now include tens of thousands cases and controls. As sample sizes have increased and initial GWAS findings begin to emerge, an “inflection point” is reached where further addition of samples leads to a regular, linear increase in genome-wide significant findings. For example, in schizophrenia, the latest published analysis of 36,989 cases and 113,075 controls is now beyond the “inflection point,” with a yield of 108 genome-wide significant loci.14
Progress in BD seems to be following a similar pattern, although sample sizes are smaller and the “inflection point” has not yet been reached. The first GWAS of BD consisted of 1868 cases and 2938 controls and was conducted by the Wellcome Trust consortium in 2007.15 This was followed by several similarly sized studies, with no individual study large enough to identify genome-wide significant findings.16–19 Yet, as samples began to be metaanalyzed a number of replicated findings have emerged. In 2011 the PGC published its first BD metaanalysis consisting of 11,974 cases and 51,792 controls,20 finding genome-wide significant evidence in 2 loci within the L-type calcium channel subunit gene CACNA1C and the cell surface receptor protein ODZ4. Several additional metaanalyses have been published subsequently,21–24 and their genome-wide significant results are summarized in Table 1.
Table 1.
Genome-wide significant loci in bipolar disorder
| Nearest Gene | Chr | Marker | Risk Allele | P-value | Odds Ratio | Cases (N) | Controls (N) | Study |
|---|---|---|---|---|---|---|---|---|
| Loci with initial genome-wide significant evidence (P<5 × 10−8) and at least one additional replication | ||||||||
| TRANK1 | 3p22 | rs9834970 | C | 2.4 × 10−11 | 1.18 | 6,658 | 7,155 | Chen et al,22 2011 |
| 4.8 × 10−8 | 1.12 | 9,747 | 14,278 | Muhleisen et al,24 2014 | ||||
| ANK3 | 10q21 | rs10994415 | C | 6.9 × 10−11 | 1.27 | 9,747 | 14,278 | Muhleisen et al,24 2014 |
| ODZ4 | 11q14 | rs12576775 | A | 4.4 × 10−8 | 0.88 | 11,974 | 51,792 | PGC-BD et al,20 2011 |
| 6.2 × 10−9 | 0.88 | 13,192 | 54,705 | Green et al,23 2013 | ||||
| 4.5 × 10−9 | 0.85 | 9,747 | 14,278 | Muhleisen et al,24 2014 | ||||
| CACNA1C | 12p13 | rs4765913 | A | 1.5 × 10−8 | 1.14 | 11,974 | 51,792 | PGC-BD et al,20 2011 |
| 9.8 × 10−10 | 1.14 | 13,192 | 54,705 | Green et al,23 2013 | ||||
| NCAN | 19p13 | rs1064395 | A | 2.1 × 10−9 | 1.17 | 7,759 | 34,062 | Cichon et al,21 2011 |
| Loci with initial genome-wide significant evidence (P < 5 × 10−8) | ||||||||
| RHEBL1/DHH | 12q13 | rs7296288 | A | 9.0 × 10−9 | 0.90 | 13,192 | 54,705 | Green et al,23 2013 |
| TRPC4AP | 20q11 | rs3818253 | A | 3.9 × 10−8 | 1.16 | 8,699 | 12,163 | Green et al,23 2013 |
| ADCY2 | 5p15 | rs17826816 | G | 9.9 × 10−9 | 1.14 | 9,747 | 14,278 | Muhleisen et al,24 2014 |
| MIR2113; POU3F2 | 6q16 | rs12202969 | A | 1.1 × 10−8 | 1.12 | 9,747 | 14,278 | Muhleisen et al,24 2014 |
| PTGFR | 1p31 | rs4650608 | T | 8.4 × 10−9 | 0.88 | 7,773 | 9,883 | Chen et al,22 2011 |
| LMAN2L | 2q11 | rs2271893 | G | 5.2 × 10−9 | 0.86 | 7,773 | 9,883 | Chen et al,22 2011 |
| Many genes | 3p21 | rs7618915 | G | 1.6 × 10−9 | 0.87 | 7,773 | 9,883 | Chen et al,22 2011 |
| SYNE1 | 6q25 | rs9371601 | T | 2.9 × 10−8 | 1.10 | 9,368 | 10.929 | Green et al,74 2013 |
Loci with subsequent evidence for replication in an independent sample are shown in the top part of the table, while loci with initial evidence for genome-wide significance only are shown in the bottom of the table. Although studies are listed independently, there is substantial overlap among the cases and controls between the studies.
The results shown in Table 1 show typical characteristic of GWAS findings: (1) most highly associated markers are intronic (ie, within or next to genes but outside of the protein coding region), (2) their effect sizes (OR) are small and all are less than 1.2, and (3) associations are independent of each other, with no statistical evidence for gene-gene interactions. Importantly, owing to the close correlation of markers in nearby regions of the chromosomes, GWAS associations highlight broad genomic regions, known as loci, that can span up to several hundred thousand base pairs. The actual marker that led to the association is generally unlikely to be “causal” marker within a locus and additional investigation is necessary to uncover the actual genetic variants that have a functional effect.
FROM GENOME-WIDE ASSOCIATION STUDIES TO FUNCTION: ELUCIDATING MECHANISM
Once a genome-wide association is confirmed and replicated in subsequent metaanalysis, the arguably more challenging task of elucidating the molecular mechanisms behind the association lies ahead.25 As an example of the type of further evaluation required to link a GWAS loci with its interacting gene(s), Roussos and colleagues26 performed a comprehensive evaluation of findings from a recently published schizophrenia GWAS that included the same CACNA1C locus identified in GWAS metaanalyses of BD (see Table 1). The authors identified a number of additional markers in strong correlation with the index marker in regions known as enhancers that regulate the transcription of a nearby gene. The marker associated with BD and schizophrenia was shown subsequently to be associated with decreased enhancer binding to the promoter of CACNA1C and lower levels of transcription.26 These experiments involved the use of post mortem brain tissue, neural progenitor cells, and nonneural cell lines, reflecting the need for a comprehensive approach to functional validation of a GWAS loci, particularly for psychiatric disorders where not all tissues or cell lines may be equally relevant.
Historically, the main source of human neural tissue has been post mortem brain samples, which are limited by well known confounds such as differential environmental exposures and post mortem related artifacts. As a result, there has been a widespread embracement of novel cellular reprograming technologies that can transform somatic cells (obtained from skin or blood) into neural progenitor cell lines or line-age specific neural cell lines.27 Although this remains a nascent field, particularly in its application to complex brain disorders, the use of transformed neural cells is likely to provide a feasible and relevant model system to study important aspects of neural physiology. Only a few initial stem cell studies have been performed with BD,28–30 with preliminary results of the most comprehensive study indicating that BD may be associated with a hyperexcitable neuronal phenotype that is differentially responsive to lithium in subjects successfully treated with lithium.30 If confirmed, such cellular phenotypes may show promise as potential biomarkers of clinical response and may provide a potentially means for the high-throughput testing the biological effects of newly discovered genetic variants and novel compounds.
Of course, gene expression is the most initial “downstream” phenotype and establishing a causal chain from genome to the ultimate (clinical) phenotype will require studying the effects of an associated locus on protein, cell, and ultimately physiologic function. Elucidating how a putative disease related biological effect is “transmitted” across such increasingly complex hierarchies of biological processes will be a major focus of research for the conceivable future.31 Imaging modalities will no doubt play an increasingly important role, although most current studies are hindered by small sample size and limited interstudy comparability.32 Fortunately, the technological innovations and large-scale consortia that were integral to success of genetic studies are also now occurring in the field of imaging and imaging genetics.33,34
Polygenic Risk Scores
In addition to the few associations that meet the stringent level for genome-wide significance, a GWAS study will have a much greater number of associated markers that do not meet genome-wide significance. Although the confidence that any 1 such marker is associated with the studied phenotype is low, these subthreshold markers can be grouped to create a polygenic “signature” that is highly reproducible across GWAS datasets and can account for a much higher proportion of phenotypic variation compared with any individual marker.35 The magnitude of the polygenic association generally depends on the power of the initial “training” dataset from which the subthreshold associations are derived. Because power in a GWAS study correlates strongly with sample size, increasing the sample size of disorder-specific GWAS will lead to predictable improvements in the ability of polygenic scores to discriminate cases from controls. For example, in the schizophrenia PGC2 study, the polygenic score was associated with approximately 10% to 20% of the phenotypic variance in an independent sample.14 The smaller BD training dataset so far yields a more modest, but still highly significant, association that accounts for approximately 3% of the phenotypic variance.20
Another important use of polygenic scores is that they can be used to model the proportion of the overall heritability that can be attributed to common variants. The development of analytical methods that measure the overall proportion of shared common variation throughout the genome in cases versus controls has provided estimates that at least 25% of the overall heritability of BD can be explained by common variants.36 This estimate provides a strong impetus to increase GWAS sample sizes, because the currently discovered genome-wide significant findings explain no more than a few percent of the overall heritability.
A more elusive goal is the application of polygenic scores for phenotype prediction. Currently available scores, for either schizophrenia or BD, explain far too little of the phenotypic variance to be clinically meaningful by themselves, with prediction area under the curve estimates no greater than approximately 0.6.37 However, under “optimal” conditions, when polygenic scores can be estimated from extremely large discovery samples (at least an order of magnitude greater than currently available samples), clinical utility may be achievable with potential under the curve values in the 0.8 to 0.9 range.38 One important caveat is to remember that even the perfect genetic test will be constrained by the overall heritability of BD (≈0.7). Hence, the most informative type of test will also likely need to incorporate other markers or “proxies” of pathophysiological or environmental states that provide further predictive capacity.
Although the prospects for clinical prediction lie in the future, polygenic scores are currently of research interest as an index of genetic liability to a specific disorder. In a recent study of schizophrenia, for example, family history was found to interact with polygenic scores to increase the risk of schizophrenia,39 pointing to the need for further studies to characterize important elements of family history that are both dependent and independent of genetic liability. More broadly, this type of analysis can also be used to better characterize the genetic effects on clinically relevant phenotypes, such as illness course and drug response, which are likely to be better predicted by polygenic rather than single marker models.
Rare Variants: The Next Frontier
Genetic variation can be arbitrarily defined as rare if it is present in less than 1% of a studied sample or population. Rare variation may be particularly relevant for the understanding of disease related biology, because rare variants are evolutionarily more recent and have had less time to be selected against or removed by evolution.40 Hence, rare variants may be more likely to be pathogenic compared with common variants and could therefore provide a more direct and actionable insight into disease pathophysiology.41 Highly penetrant rare variants are responsible for most Mendelian disorders and, although such strongly associated variants have not been found for most psychiatric disorders, the identification of variants with penetrance in between those responsible for Mendelian disorders and those found by GWAS is currently a major focus of research.
Copy Number Variants
A recently described type of variation that arises from subtle misalignment errors during DNA repair and replication is known as a copy number variation (CNV) and consists of large deletions or duplications of several thousand to a million base pairs. For much the same reasons described previously, rare CNVs tend to be more pathogenic than common CNVs, especially larger CNVs that are more likely to disrupt a single gene or a group of genes. Rare CNVs were first found to be overrepresented in cases with early onset neurodevelopmental disorders such as autism and intellectual disability.42 Subsequent studies have expanded the range of disorders affected by CNVs to include adult onset disorders such as schizophrenia43 and, to a lesser extent, BD.44,45
In a recent study that also collated findings from the literature, Georgieva and colleagues44 reported significantly increased risk of de novo CNVs in BD, when defining a CNV as any deletion or duplication greater than 10 kilobases (crude OR, 2.2; P = .0003). Although they also found a trend toward increased risk for larger (greater than 100 kilobases) and more pathogenic de novo CNVs, this difference was not statistically significant (crude OR, 1.46; P = .18). Compared with schizophrenia, the association of de novo CNV variants with BD seems to be more modest in both the magnitude of the association and the actual size of the CNV. A similar pattern is seen in case-control studies of CNVs where most of the discovered CNVs consist of inherited, rather than de novo CNVs.45 Table 2 highlights the findings of a recent large scale study and meta-analysis of CNVs in BD, which found the most consistent evidence for the association of 3 recurrent CNVs with BD, with the strongest findings seen with the 16p11.2 duplication. The CNV associations shown in Table 2 share a number of important characteristics: (1) The effect size of the associations are much higher compared with common variants; (2) All CNVs have been found in controls, albeit at a much lower frequency; (3) Each CNV is associated with multiple genes; (4) Each CNV is individually rare and collectively affect only approximately 0.3% of all cases; and (5) All highlighted CNVs have also been associated more strongly with schizophrenia.
Table 2.
Copy number variations (CNV) nominal significance (P<.05) for association with BD
| CNV Locus | Odds Ratioa | P-Value | Frequency Cases | Frequency Controls |
|---|---|---|---|---|
| 1q21.1 duplication | 2.6 | .02 | 0.099% (8/8084) | 0.037% (24/64,046) |
| 3q29 deletion | 17.3 | .03 | 0.025% (2/8084) | 0.0014% (1/69,965) |
| 16p11.2 duplication | 4.4 | 2.3 ×10−4 | 0.13% (12/9129) | 0.03% (19/63,068) |
Abbreviation: CNV, copy number variation.
Effect estimates and raw numbers obtained from the meta-analysis performed by Green and colleagues.
Data from Green EK, Rees E, Walters JT et al. Copy number variation in bipolar disorder. Mol Psychiatry 2015;[Epub ahead of print].
Next-Generation Sequencing
Most rare variation, like most common variation, is composed of single nucleotide changes or deletions. Yet, because rare variation is poorly represented in GWAS arrays, the only comprehensive means of assessing its role in disease susceptibility is by DNA sequencing. Fortunately, recent technological advances have changed the feasibility of large scale DNA sequencing, with whole genomes now routinely sequenced in several days for approximately $1000. Two major assays are used to measure rare variants in clinical and research settings: whole genome sequencing and a more targeted form of sequencing known as whole exome sequencing, which is limited to the approximately 1.5% of the genome that is transcribed into messenger RNA and translated into protein. Whole exome sequencing has the advantage of lower cost, a lower bioinformatic “footprint,” and a focus on the more easily interpretable part of the genome where most Mendelian disorders have been identified to date.46 However, whole exome sequencing misses variation in the noncoding (including regulatory) regions and is less accurate in detecting important types of variations such as insertions, deletions, or CNVs. Although whole exome sequencing is currently the most widely used genetic assay of rare variants, it will ultimately be supplanted by whole genome sequencing as sequencing costs decrease.
The role of rare sequence variation in psychiatric disorders is an active area of research, with particular emphasis on the study of more heritable syndromes such as intellectual disability, autism, schizophrenia, and BD. In a pattern similar to that seen in the CNV studies, the effect of rare variants—particularly de novo rare variants—seem to be more prominent in neurodevelopmental disorders, such as intellectual disability or certain forms of autism, where gene-disrupting mutations have been found to cluster in pathways involved in transcriptional regulation and synaptic function.47,48 In schizophrenia, a recent exome sequencing study of 623 trios did not implicate a specific gene, but found enrichment of rare de novo variants in pathways associated with the postsynaptic density.49 A complementary exome case-control study of schizophrenia49 showed similar enrichment for very rare mutations in postsynaptic gene sets (in particular, the activity-regulated cytoskeleton-associated protein and W-methyl-D-aspartate receptor gene sets), as well as in calcium channel subunits, which had been previously implicated in the PGC2 GWAS study.
A number of BD sequencing studies are ongoing, but only a few family-based sequencing studies have been published so far.50–54 Two of these studies have included Amish samples, including the exceptionally large pedigree first described by Egeland and colleagues55 in 1987. However, the investigators were unable to find rare pathogenic variants that segregated in most affected family members, leading them to conclude that genetic heterogeneity may be present even within a family.51 The results of these initial studies have so far not shown clear convergence on a specific gene, although larger and more comprehensive analyses are currently under way within the Bipolar Sequencing Consortium. At the same time, a number of research groups are also performing exome or whole genome sequencing of large case-control samples, with results expected in early 2016. Together, these case-control studies should have sufficient sample size (several thousand cases and controls) to identify what may be considered “low-hanging fruit.” Ongoing studies of other complex disorders with similar samples sizes have identified significant enrichment of rare-variants within genes in certain disorders like amyotrophic lateral sclerosis56; however, the study of most other complex disorders has not yet led to significant findings, indicating the need for larger samples sizes likely in the range of 10 to 20,000 cases.57
Genetic Architecture of Bipolar Disease
The overall number of risk alleles, their individual frequency, and their effect sizes is collectively referred to as the genetic architecture of a particular phenotype or trait. Although significant heritability implies that genetics plays a causal role in disease, the actual genetic architecture underlying the heritability cannot be predicted and needs to be determined empirically. At 1 extreme are many of the classically Mendelian disorders, where only 1 very rare and penetrant type of mutation can account for the entire disease phenotype. At the other extreme are disorders with prominent polygenicity, marked by a very large number (several hundreds or thousands) of risk alleles that individually increases risk modestly but collectively account for most of the heritability. The gap between heritability predicted from twin and family studies and the heritability explained by identified variants has been termed the “missing heritability.” Yet, as Witte and colleagues37 have commented, much of it should be termed “hidden heritability” because increasing samples for both GWAS and rare variants studies should in the near future explain a large amount of this hidden heritability, as is occurring in other complex phenotypes such as height, Crohn’s disease, and body mass index.
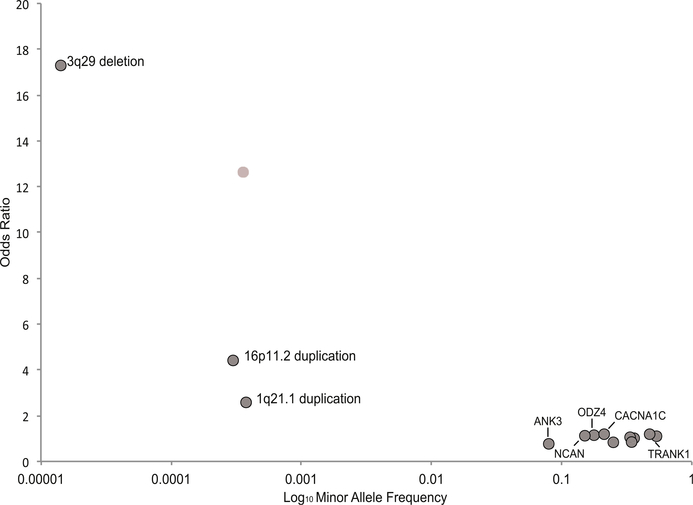
Fig. 1 shows where the current findings described in this article fall on the genomic architecture space, with relatively high effect sizes seen for the rare CNV mutations, compared with the much lower effects sizes seen for the more common GWAS associations. Although the number of robustly confirmed loci remains relatively small, sample sizes in BD have only just reached the numbers where most complex disorders begin to yield findings in a predictable incremental manner. By early 2016, the PGC metaanalysis is expected to include more than 35,000 BD cases, which is equivalent to the sample size that yielded more than 100 genome-wide significant findings for schizophrenia. Consequently, it is reasonable to assume that the number of common variants associated with BD will increase rapidly over the next year.
Fig. 1.
Emerging genetic architecture of bipolar disorder. Risk loci are show in relationship to their effect size (y axis) and their frequency in control populations (x axis). Loci represent those shown in Tables 1 and 2. For the common variants, only those with genome-wide significant evidence for replication and additional replication are labeled with text. (Data fromRefs.20–24,45,74)
The near term results for rare variants is perhaps more uncertain, because the methodology is newer and the number of samples with whole exome or genome sequencing is still much less than that available for GWAS. Available data from other complex disorders along with theoretically based simulations suggest that, like GWAS, sample sizes will likely need to be in the tens of thousands to begin yielding significant findings at realistic effect sizes.58 The next several years should reveal whether more penetrant mutations exist in BD, as has been found in autism and intellectual disability. Although there are evolutionary reasons to believe that rare variants of large effect are more likely to be found in early onset disorders with low fecundity,59 this does not diminish the potential importance of identifying even a relatively small number of rare but highly penetrant variants, because such variants can yield almost immediate insights into disease biology and therapeutic targets. There are now several successful examples of very rare variants of high effect that have identified therapeutic targets, including the recently approved PCSK-9 inhibitors for hyperlipidemia.60
Blurring of Diagnostic Boundaries: Cross-Disorder Associations
A major feature of many of the discovered genetic associations with BD has been their concurrent associations with other phenotypes, particularly schizophrenia. For example, 4 of the genome-wide significant variants shown in Table 1 (within or near the genes TRANK1, CACNA1C, NCAN, and the multigenic 3p21 locus) were also found to be genome-wide significant in the PCG2 GWAS of schizophrenia.14 In addition, all the rare CNVs shown in Table 2 have also been found in CNV studies of schizophrenia.42 Interestingly, the rare CNV variants also show overlap with studies of autism, a phenotype for which the rare variants have thus far been studied more intensively than common variants. Prominent overlap is also present among the many subthreshold common associations that drive significant polygenic risk associations across most major psychiatric disorders, with a particularly strong genetic correlation (GWAS based correlation of 0.68) between BD and schizophrenia.36
At a genome-wide level, these correlations reflect a widely seen property of individual or aggregated variants, both common and rare, known as pleiotropy, which in its simplest definition is the association of a genetic risk factor with more than 1 trait.61 Pleiotropy is a phenomenon widely seen across species and is likely to be mechanistically diverse, because there are many potential ways to affect a phenotype in the long complex causal chain between gene and the phenotype of interest. At a broad level, potential factors involved in pleiotropy may include differing gene–gene interactions (including the effect of the overall genomic background), developmental effects, and differential environmental effects, which are all areas of active scientific investigation.
These correlations mirror the results seen from the more recent population-based studies,4,5 and have important implications for psychiatry nosology, largely confirming long-held suspicions that the diagnostic borders across phenotypes may not reflect biological boundaries.62 However, it must also be emphasized that their specific relevance for clinical care is still uncertain, because the relationship of polygenic risk scoresto clinically relevant phenotypes such as prognosis and treatment response remains unknown.
Pharmacogenetics
Potentially the most immediately relevant application of genetics to patients with BD may arise from the field of pharmacogenetics, which investigates how genetic variation affects both the efficacy and metabolism of therapeutic drugs. Because it is less likely that variants associated with drug response or drug tolerability have been under negative evolutionary pressure, there are theoretic reasons to expect that pharmacogenetics variants may have larger effect sizes and be more clinically actionable. Indeed, in other fields of medicine, a number of high-effect variants have been identified in a wide variety of drug response studies that range from efficacy of interferon treatment in hepatitis C to the dosing of anticoagulants.63 Most identified variants have been related to drug metabolism and adverse side effects rather than therapeutic effect, presumably reflecting the more complex nature of the latter.
In BD, initial investigations have focused on the response to mood stabilizers, particularly lithium.64 One difficulty of such studies is the appropriate measure of the drug response phenotype, which usually requires a substantial longitudinal component and careful attention to common confounders like the use of additional medications, illicit drug use, and variable medication adherence. Not surprisingly, most published studies have been limited by small sample sizes and prominent interstudy heterogeneity. A number of candidate gene studies have been published and, although some have shown evidence for replication, the overall level of evidence remains insufficient and below that of the previously described disease-specific GWAS findings.65
To date, there have been 3 GWAS of lithium response and no such studies of response to other BD medications. The first lithium response GWAS was a secondary analysis using longitudinal data from the STEP-BD trial (up to 2 years of follow-up) to characterize the association between common variants and relapse in 458 subjects on lithium treatment.66 The authors did not find any genome-wide significant findings, but attempted to replicate their top results in an independent case-control sample using a retrospective measure of lithium response. Five markers showed modest evidence for replication, which included the GluR2 glutamate receptor and ODZ4, a gene previously found to be a BD risk gene (see Table 1). A more recent study from Taiwan, however, reported an unusually strong association with a marker in glutamate decarboxylase-like protein 1 (GADL1), with an effect size (OR, 73.9) that is almost 2 orders of magnitude greater than those seen in any BD case-control GWAS.67 Although the original study authors also presented consistent replication in an additional sample, a number of failed replications have also been published.68,69 Hence, the relevance of this finding remains uncertain and in need for further confirmation. The largest and most recent study of lithium response included 2,563 patients collected by the Consortium for Lithium Genetics and phenotyped using a uniform retrospective lithium rating scale. 70 This study found a genome-wide significant association on a locus on chromosome 21 that encompasses two long non-coding RNAs (lncRNAs), which are sequences of DNA that are actively transcribed and may have important roles in the regulation of gene-expression. However, further replication is needed and additional work is required to determine whether any potential causal relationship between the associated markers and the lncRNAs.70 With the exception of the unusually large effect finding from the Taiwanese study, these initial GWAS show the more typical modest effect sizes that require large samples sizes and also point to the likelihood that lithium response may also be a complex, polygenic phenotype.
Toward Personalized Medicine
As a disorder defined at the level of syndrome, with prominent heterogeneity at the level of clinical symptoms, long-term outcome, and response to treatment, BD is long due some element of personalization. The goals of personalized medicine are not new, but the recent widespread availability of genomic and other “-omic” based technologies, along with availability of large-scale cohorts, has renewed focus on identifying novel ways to classify patients and help identify treatments targeted to the specific disease pathophysiology.
For clinicians treating patients with BD, there are a number of well-known clinical dilemmas to be (re)addressed in this new era of personalized or precision medicine. First, there is a need for improved prediction of BD in the symptomatic years when children and adolescents suffer primarily from a broad range of subthreshold mood, anxiety, and behavioral disorders before the emergence of (hypo)mania.71 Second, the course of BD can differ widely between patients, with marked heterogeneity in terms of episode polarity, recurrence, association with comorbid syndromes, and cognitive impairment.72 Finally, the most desired outcome of the precision medicine initiative will be to help tailor treatment to patients based on their individual risk factors.
What role will genetics play in these precision medicine goals? To quote a prominent geneticist, “Genetics alone cannot tell us why we are the way we are, but it has a seat at the table.”73 How direct the influence of genetic risk factors will be on clinically relevant phenotypes will be determined by the underlying genetic architecture. If psychiatric disorders are primarily polygenic, then genetics will play a more probabilistic rather than deterministic role in prediction of clinical outcomes, much in the way that certain laboratory tests aid physicians in “risk stratification.” For the small minority of patients or families for whom a “simpler” genetic architecture is responsible for their disorder (eg, a rare patient who carries a penetrant CNV mutation), genetics is likely to play a more strongly informative role, but it is currently unclear for what proportion of patients with BD this will be applicable. However, even in such scenarios, because the heritability of BD is less than 100%, nongenetic factors will also play an important role in the manifestation of clinically relevant phenotypes. Elucidation of such factors, which fall under the overly vague label of “environmental” causes, is perhaps a greater challenge than genomics, but it is one that must be addressed to fulfill the promises of personalized medicine.
KEY POINTS.
Bipolar disorder (BD) is a disabling, often lifelong, disorder that is among the most highly heritable of all common disorders.
Ongoing genome-wide association studies (GWAS) are yielding more robustly associated markers; next-generation sequencing technologies are poised to identify of rare and potentially more penetrant variants.
The modest effect sizes seen among GWAS associations are consistent with a polygenic genetic architecture, with risk being distributed across a large number of loci.
Future progress in common (GWAS) and rare variant studies will provide novel insights into the biology of BD, and help pave the way for personalized medicine and improved, targeted therapies.
ACKNOWLEDGMENTS
The author thanks the valuable assistance of David Liebers, MPhil, in performing the literature review.
Disclosures: The author has received research support from the NIMH (R00MH86049). The author has no relevant conflict of interests.
REFERENCES
- 1.Merikangas KR, Jin R, He JP, et al. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch Gen Psychiatry 2011; 68(3):241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraepelin E Manic-depressive insanity and paranoia Translated by R. Mary Barclay from the eighth German edition of the ‘Textbook of Psychiatry’. Edinburgh: E. & S. Livingstone; 1921. [Google Scholar]
- 3.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet 2003;123C(1):48–58. [DOI] [PubMed] [Google Scholar]
- 4.Mortensen PB, Pedersen CB, Melbye M, et al. Individual and familial risk factors for bipolar affective disorders in Denmark. Arch Gen Psychiatry 2003;60(12): 1209–15. [DOI] [PubMed] [Google Scholar]
- 5.Song J, Bergen SE, Kuja-Halkola R, et al. Bipolar disorder and its relation to major psychiatric disorders: a family-based study in the Swedish population. Bipolar Disord 2014;17(2):184–93. [DOI] [PubMed] [Google Scholar]
- 6.Merikangas KR, Cui L, Heaton L, et al. Independence of familial transmission of mania and depression: results of the NIMH family study of affective spectrum disorders. Mol Psychiatry 2014;19(2):214–9. [DOI] [PubMed] [Google Scholar]
- 7.Vandeleur CL, Merikangas KR, Strippoli MP, et al. Specificity of psychosis, mania and major depression in a contemporary family study. Mol Psychiatry 2014;19(2): 209–13. [DOI] [PubMed] [Google Scholar]
- 8.Van Snellenberg JX, deCandia T. Meta-analytic evidence for familial coaggregation of schizophrenia and bipolar disorder. Arch Gen Psychiatry 2009;66(7):748–55. [DOI] [PubMed] [Google Scholar]
- 9.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era–concepts and misconceptions. Nat Rev Genet 2008;9(4):255–66. [DOI] [PubMed] [Google Scholar]
- 10.Badner JA, Koller D, Foroud T, et al. Genome-wide linkage analysis of 972 bipolar pedigrees using single-nucleotide polymorphisms. Mol Psychiatry 2012;17(8): 818–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levinson DF, Mostafavi S, Milaneschi Y, et al. Genetic studies of major depressive disorder: why are there no genome-wide association study findings and what can we do about it? Biol Psychiatry 2014;76(7):510–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008; 9(5):356–69. [DOI] [PubMed] [Google Scholar]
- 14.Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S, Neale BM, Corvin A, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511(7510):421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wellcome Trust Case Control Consortium, Burton PR, Clayton DG, Cardon LR, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447(7145):661–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baum AE, Akula N, Cabanero M, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry 2008;13(2):197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sklar P, Smoller JW, Fan J, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry 2008;13(6):558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith EN, Bloss CS, Badner JA, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry 2009;14(8):755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott LJ, Muglia P, Kong XQ, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A 2009;106(18):7501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Psychiatric GWAS Consortium Bipolar Disorder Working Group, Sklar P, Ripke S, Scott LJ, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43(10):977–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cichon S, Muhleisen TW, Degenhardt FA, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet 2011;88(3):372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen DT, Jiang X, Akula N, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry 2011;18(2):195–205. [DOI] [PubMed] [Google Scholar]
- 23.Green EK, Hamshere M, Forty L, et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry 2013;18(12):1302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muhleisen TW, Leber M, Schulze TG, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun 2014;5:3339. [DOI] [PubMed] [Google Scholar]
- 25.Chakravarti A, Clark AG, Mootha VK. Distilling pathophysiology from complex disease genetics. Cell 2013;155(1):21–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roussos P, Mitchell AC, Voloudakis G, et al. A role for noncoding variation in schizophrenia. Cell Rep 2014;9(4):1417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennand KJ, Simone A, Tran N, et al. Modeling psychiatric disorders at the cellular and network levels. Mol Psychiatry 2012;17(12):1239–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen HM, DeLong CJ, Bame M, et al. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl Psychiatry 2014;4:e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madison JM, Zhou F, Nigam A, et al. Characterization of bipolar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and mRNA expression abnormalities. Mol Psychiatry 2015;20(6):703–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mertens J, Wang QW, Kim Y, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 2015;527(7576):95–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Insel TR, Cuthbert BN. Medicine. Brain disorders? Precisely. Science 2015; 348(6234):499–500. [DOI] [PubMed] [Google Scholar]
- 32.Savitz JB, Rauch SL, Drevets WC. Clinical application of brain imaging for the diagnosis of mood disorders: the current state of play. Mol Psychiatry 2013; 18(5):528–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jorgenson LA, Newsome WT, Anderson DJ, et al. The BRAIN Initiative: developing technology to catalyse neuroscience discovery. Philos Trans R Soc Lond B Biol Sci 2015;370(1668). 10.1098/rstb.2014.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hibar DP, Stein JL, Renteria ME, et al. Common genetic variants influence human subcortical brain structures. Nature 2015;520(7546):224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wray NR, Lee SH, Mehta D, et al. Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry 2014;55(10):1068–87. [DOI] [PubMed] [Google Scholar]
- 36.Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee SH, Ripke S, Neale BM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013;45(9):984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witte JS, Visscher PM, Wray NR. The contribution of genetic variants to disease depends on the ruler. Nat Rev Genet 2014;15(11):765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudbridge F Power and predictive accuracy of polygenic risk scores. PLoS Genet 2013;9(3):e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agerbo E, Sullivan PF, Vilhjalmsson BJ, et al. Polygenic risk score, parental socioeconomic status, family history of psychiatric disorders, and the risk for schizophrenia: a Danish population-based study and meta-analysis. JAMA Psychiatry 2015;72(7):635–41. [DOI] [PubMed] [Google Scholar]
- 40.Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 2012; 337(6090):64–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lupski JR, Belmont JW, Boerwinkle E, et al. Clan genomics and the complex architecture of human disease. Cell 2011;147(1):32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 2012;148(6):1223–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirov G, Pocklington AJ, Holmans P, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 2012;17(2):142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georgieva L, Rees E, Moran JL, et al. De novo CNVs in bipolar affective disorder and schizophrenia. Hum Mol Genet 2014;23(24):6677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green EK, Rees E, Walters JT, et al. Copy number variation in bipolar disorder. Mol Psychiatry 2015. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 2010;11(6):415–25. [DOI] [PubMed] [Google Scholar]
- 47.lossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014;515(7526):216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014;515(7526):209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fromer M, Pocklington AJ, Kavanagh DH, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014;506(7487):179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collins AL, Kim Y, Szatkiewicz JP, et al. Identifying bipolar disorder susceptibility loci in a densely affected pedigree. Mol Psychiatry 2012;18(12):1245–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Georgi B, Craig D, Kember RL, et al. Genomic view of bipolar disorder revealed by whole genome sequencing in a genetic isolate. PLoS Genet 2014;10(3): e1004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strauss KA, Markx S, Georgi B, et al. A population-based study of KCNH7 p.Arg394His and bipolar spectrum disorder. Hum Mol Genet 2014;23(23): 6395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cruceanu C, Ambalavanan A, Spiegelman D, et al. Family-based exome-sequencing approach identifies rare susceptibility variants for lithium-responsive bipolar disorder. Genome 2013;56(10):634–40. [DOI] [PubMed] [Google Scholar]
- 54.Ament SA, Szelinger S, Glusman G, et al. Rare variants in neuronal excitability genes influence risk for bipolar disorder. Proc Natl Acad Sci U S A 2015; 112(11):3576–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Egeland JA, Gerhard DS, Pauls DL, et al. Bipolar affective disorders linked to DNA markers on chromosome 11. Nature 1987;325(6107):783–7. [DOI] [PubMed] [Google Scholar]
- 56.Cirulli ET, Lasseigne BN, Petrovski S, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 2015;347(6229): 1436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuk O, Schaffner SF, Samocha K, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A 2014; 111(4):E455–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiezun A, Garimella K, Do R, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet 2012;44(6):623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agarwala V, Flannick J, Sunyaev S, et al. Evaluating empirical bounds on complex disease genetic architecture. Nat Genet 2013;45(12):1418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov 2013;12(8):581–94. [DOI] [PubMed] [Google Scholar]
- 61.Solovieff N, Cotsapas C, Lee PH, et al. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet 2013;14(7):483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kendell RE. Diagnosis and classification of functional psychoses. Br Med Bull 1987;43(3):499–513. [DOI] [PubMed] [Google Scholar]
- 63.Sala R, Strober MA, Axelson DA, et al. Effects of comorbid anxiety disorders on the longitudinal course of pediatric bipolar disorders. J Am Acad Child Adolesc Psychiatry 2014;53(1):72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alda M Lithium in the treatment of bipolar disorder: pharmacology and pharmacogenetics. Mol Psychiatry 2015;20(6):661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salloum NC, McCarthy MJ, Leckband SG, et al. Towards the clinical implementation of pharmacogenetics in bipolar disorder. BMC Med 2014;12:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perlis RH, Smoller JW, Ferreira MA, et al. A genomewide association study of response to lithium for prevention of recurrence in bipolar disorder. Am J Psychiatry 2009;166(6):718–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen CH, Lee CS, Lee MT, et al. Variant GADL1 and response to lithium therapy in bipolar I disorder. N Engl J Med 2014;370(2):119–28. [DOI] [PubMed] [Google Scholar]
- 68.Consortium on Lithium Genetics, Hou L, Heilbronner U, Rietschel M, et al. Variant GADL1 and response to lithium in bipolar I disorder. N Engl J Med 2014;370(19): 1857–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ikeda M, Kondo K, Iwata N. Variant GADL1 and response to lithium in bipolar I disorder. N Engl J Med 2014;370(19):1856–7. [DOI] [PubMed] [Google Scholar]
- 70.Hou L, Heilbronner U, Degenhardt F, et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. The Lancet, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Axelson D, Goldstein B, Goldstein T, et al. Diagnostic precursors to bipolar disorder in offspring of parents with bipolar disorder: a longitudinal study. Am J Psychiatry 2015;172(7):638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frank E, Nimgaonkar VL, Phillips ML, et al. All the world’s a (clinical) stage: rethinking bipolar disorder from a longitudinal perspective. Mol Psychiatry 2015. 10.1176/appi.ajp.2014.14010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olson MV. Human genetic individuality. Annu Rev Genomics Hum Genet 2012;13: 1–27. [DOI] [PubMed] [Google Scholar]
- 74.Green EK, Grozeva D, Forty L, et al. Association at SYNE1 in both bipolar disorder and recurrent major depression. Mol Psychiatry 2013;18(5):614–7. [DOI] [PubMed] [Google Scholar]