Abstract
Ionotropic orphan delta receptors (GluD) are not gated by glutamate or any other endogenous ligand but are grouped with ionotropic glutamate receptors based on sequence similarity. GluD1 receptors play critical roles in synaptogenesis, synapse maintenance and have been implicated in neuronal disorders including schizophrenia, cognitive deficits, and cerebral ataxia. Here we report cryo-electron microscopy structures of the rat GluD1 receptor complexed with calcium and the ligand 7-chlorokynurenic acid, elucidating molecular architecture and principles of receptor assembly. The structures reveal a non-swapped architecture at the extracellular amino-terminal (ATD) and ligand-binding domain (LBD) interface. This is in contrast to other families of ionotropic glutamate receptors (iGluRs) where the dimer partners between the ATD and LBD layers are swapped. Our results demonstrate that principles of architecture and symmetry are not conserved between delta receptors and other iGluRs and provide a molecular blueprint for understanding the functions of the “orphan” class of iGluRs.
Keywords: Orphan delta receptors, cryo-electron microscopy, ion channel, GluD1, Synaptic plasticity, Neurotransmission, membrane protein
Introduction
Delta receptors belong to the ionotropic glutamate receptor (iGluR) family along with α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA), kainate (KA) and N-methyl-D-aspartate (NMDA) receptors. This enigmatic class of iGluRs is referred to as “orphan” because they are not activated by endogenous ligands. The family consists of two members, GluD1 and GluD2, that are expressed in multiple regions of the brain with GluD1 predominantly expressed in the inner ear1 and GluD2 in cerebellar Purkinje cells (PC)2. The two subtypes share ~50 % sequence similarity with each other and around 20-30 % with other iGluRs 3. While multiple structures of the intact iGluRs have been reported for AMPA, KA and NMDA receptors4–5, structural insights into a full-length Delta receptor is still lacking. Unlike other iGluRs, Delta receptors do not generate ligand-gated currents6–8 and are believed to exert their physiological functions via a non-canonical (metabotropic) pathway9–12. Dysfunction of GluD receptors is associated with social and cognitive deficits,9,13 cerebellar long-term depression (LTD),11–12, 14 cerebellar ataxia and atrophy15–16 and is also linked to retinal dystrophy,17 schizophrenia,18–19 and cocaine addiction20. The primary function of GluD receptors is believed to be their ability to act as a bidirectional synaptic organizer via trans-synaptic interactions at presynaptic termini mediated through neurexin and cerebellin,21–25 and at postsynaptic sites via direct interactions with shank scaffold proteins26–27. On the other hand, the ionotropic roles of GluD receptors are enigmatic because they possess a functional ion channel as demonstrated by electrophysiological assays on both recombinantly expressed GluD receptors harbouring the Lurcher point mutation28–30 and on chimeric receptors where the ligand-binding domains were swapped with those from AMPA or KA receptors31–32. Further, the ionotropic properties of Lurcher mutant receptors could be modulated by ligands like D-Ser, 7-Chlorokynurenic acid (7-CKA) and Ca2+ that bind to the ligand-binding domain (LBD)31,33–35. Moreover, recent reports suggest that GluD receptors not only interact directly with metabotropic glutamate receptors36–37 but are also gated by their activation19, 38–39.
While crystal structures of isolated amino-terminal domains (ATD), ligand-binding domains (LBD) and the intact extracellular region (ATD-LBD) have been reported for GluD receptors, the full-length structure of either member of this family is still elusive. In order to address this and to gain structural insight into the function of these orphan receptors, we have determined the structure of homotetrameric rat GluD1 receptors using single-particle cryo-electron microscopy (cryo-EM). The structure reveals a distinct architecture when compared to other iGluRs. We validated the observed receptor assembly via cysteine crosslinking experiments and whole-cell patch-clamp electrophysiology. Our results provide insights into architecture and assembly of orphan delta receptors and provide molecular blueprints for understanding their functions.
Results
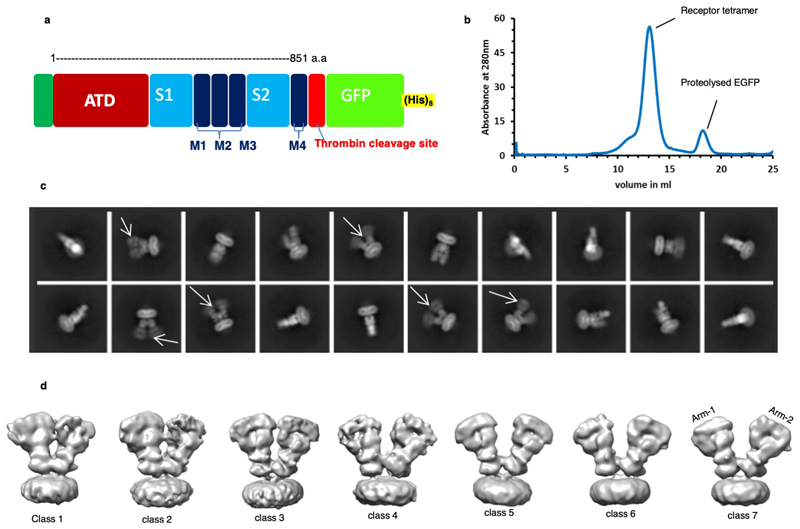
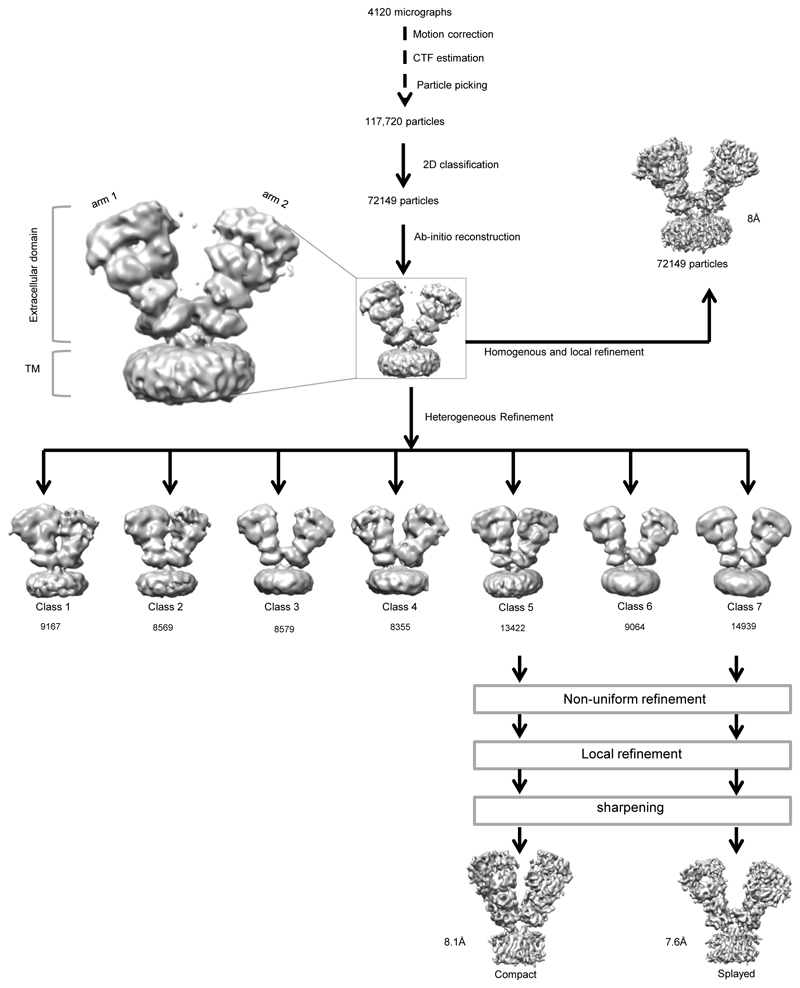
Screening of C-terminally truncated rat GluD1 via FSEC40 identified GluD1△851 as a promising construct for large-scale expression and purification from HEK293 GnTI- cells in suspension using established protocols41 (Extended Data Fig. 1 a, b; Supplementary Notes). Purified receptor (GluD1△851) was complexed with 1mM 7-chlorokyneurenic acid (7-CKA) to trap open cleft LBD conformation35, and 1mM Ca2+ to stabilize the LBD dimer assembly33 and subjected to cryo-EM analysis. 2D and 3D classification of the cleaned-up particle stack resulted in seven distinct classes displaying heterogeneity in the extracellular domains (Extended Data Fig. 1c, d) resulting primarily due to the movement of the two extracellular arms (Extended Data Fig. 2). Out of the seven 3D classes, we focussed on classes 5 and 7 representing a “compact” and a “splayed” conformation (Extended Data Fig. 2 and Supplementary Fig. 1) for 3D refinement resulting into final maps at 8.1 Å and 7.6 Å resolution respectively as estimated by the gold-standard FSC 0.143 criteria42 (Extended Data Fig. 3) into which protein co-ordinates were modelled and refined (Supplementary Fig. 2-3, Supplementary notes and Table 1).
Table 1. Cryo-EM data collection, refinement and validation statistics.
| Glud1Δ851 Compact EMD-0774 PDB- 6KSS | Glud1Δ851 Splayed EMD-0773 PDB-6KSP | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 130000x | 130000x |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 40.38 | 40.38 |
| Defocus range (μm) | -1.5 to -3.3 | -1.5 to -3.3 |
| Pixel size (Å) | 1.067 | 1.067 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 72149 | 72149 |
| Final particle images (no.) | 13422 | 14939 |
| Map resolution (Å) | 8.1 | 7.6 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 8-16 | 7.5-16 |
| Refinement | ||
| Initial model used (PDB code) | 5KC9(ATD),5CC2(LBD) 5KUF(TM) | 5KC9(ATD),5CC2(LBD) 5KUF(TM) |
| Model resolution (Å) | 8.4 | 8.2 |
| FSC threshold | 0.5 | 0.5 |
| Model resolution range (Å) | ||
| Map sharpening B factor (Å2) | -630 | -452 |
| Model composition | ||
| Nonhydrogen atoms | 23768 | 23768 |
| Protein residues | 3012 | 3012 |
| Ligands | - | - |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.005 | 0.005 |
| Bond angles (°) | 0.964 | 0.986 |
| Validation | ||
| MolProbity score | 2.03 | 2.2 |
| Clashscore | 10.24 | 10.20 |
| Poor rotamers (%) | 0.15 | 1.69 |
| Ramachandran plot | ||
| Favored (%) | 91.60 | 91.80 |
| Allowed (%) | 8.40 | 8.20 |
| Disallowed (%) | 0.00 | 0.00 |
A unprecedented architecture of the GluD1 receptor
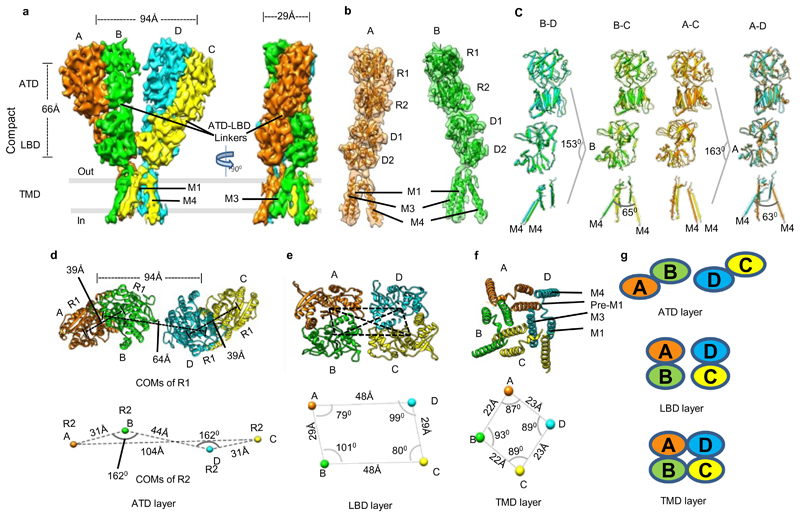
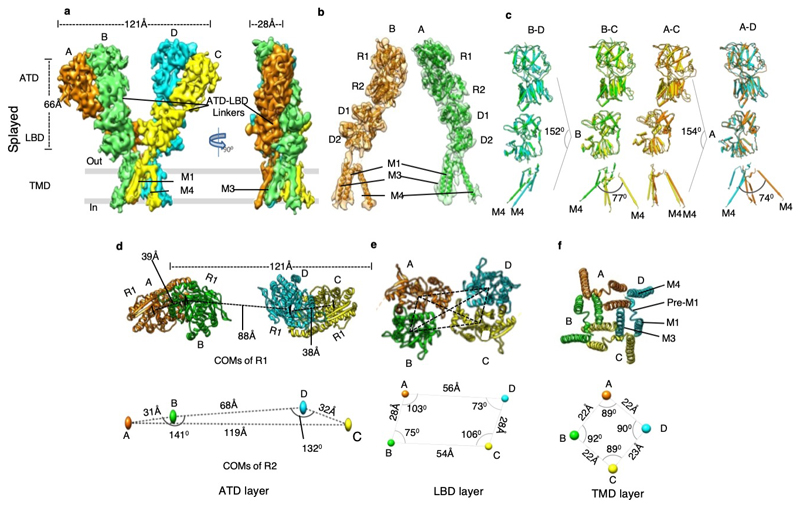
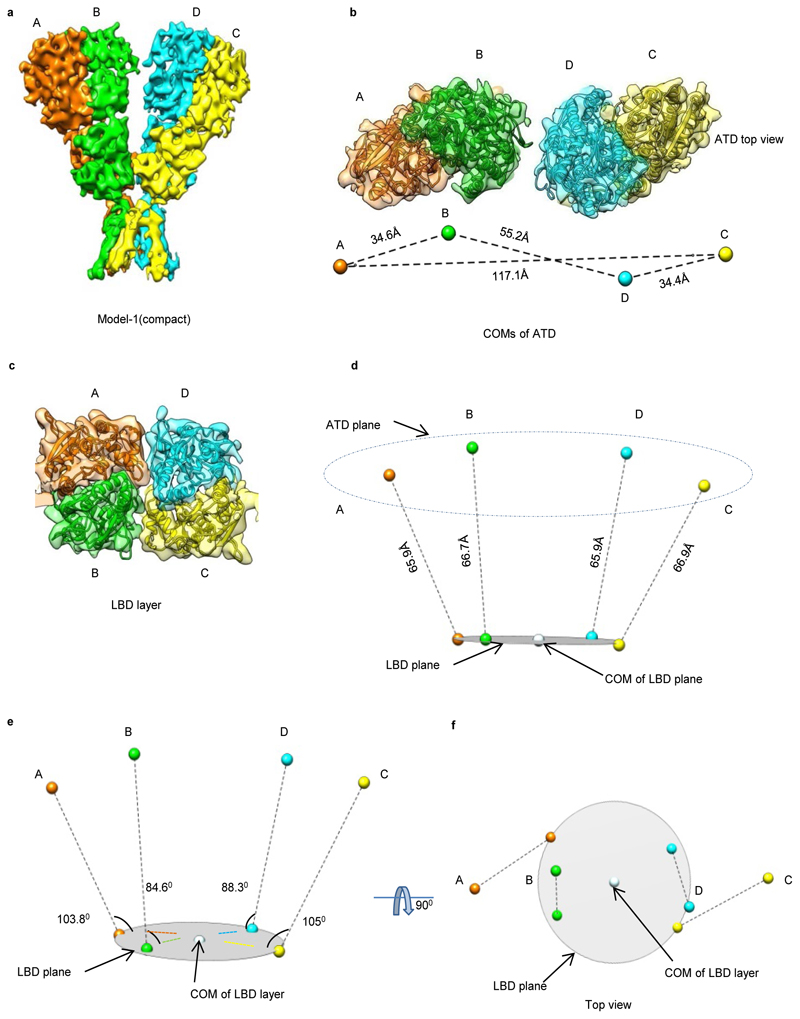
Our cryo-EM analysis revealed a Y-shaped GluD1 receptor tetramer with a three-layered arrangement of the ATD, LBD, and TM domains. The ATD and LBD are arranged in a 2-fold symmetric dimer-of-dimers configuration as observed for other iGluRs. However, remarkably, domain swapping is not observed at the ATD-LBD layer (Fig. 1, and Extended Data Fig. 4-6) between the proximal and distal subunits. It is noteworthy that all ionotropic glutamate receptor structures reported till-date for N-methyl-D-aspartate (NMDA), AMPA, and KA receptors exhibit domain swapping at the LBD layer (Extended Data Fig. 7). Due to this non-swapped architecture of GluD1 receptors, the two arms of the receptor tetramer are formed by subunits AB and DC with the domains of same subunits forming 2-fold symmetric dimers at the ATD and LBD layers (Fig. 1 a, d, e, g and Extended Data Fig. 4). Thus the conformations of subunits BD, proximal to the axis of tetramerization and subunits AC, that are distal to the axis of tetramerization are similar.
Fig. 1. GluD1 has an unprecedented non-swapped architecture.
Panels a-g show the architecture of compact conformation of GluD1 receptors in complex with 7-CKA and calcium. a, Side view highlighting the broadest face of the Y- shaped receptor and 90° rotated views of the sharpened 3D density map is shown. Each subunit is depicted in a different color. The EM reconstructions clearly show the non-swapped arrangement of the ATD and LBD layers. The distances between the centroids (R1-R1 of ATD domains) for AB and CD dimer pairs are shown above the model. The vertical separation between the COMs of ATD dimers and LBD dimers are also shown. Panel b shows the segmented density map for subunits A and B fitted with protein co-ordinates. c, Superimposition of subunits B/D, B/C, A/D and A/C are shown highlighting similar AB and BC conformations. Helices and sheets are represented as pipes and planks, respectively. Top views of ATD (d), LBD (e) and TM domains (f) are shown. The distances and the angles subtended between the COM (Centre of Mass) of various subunits were measured and are indicated below the top views. Panel g shows the schematic of the domain arrangement for the ATD, LBD, and TM layer. Also, see Extended Data Fig. 4, 5 and 6
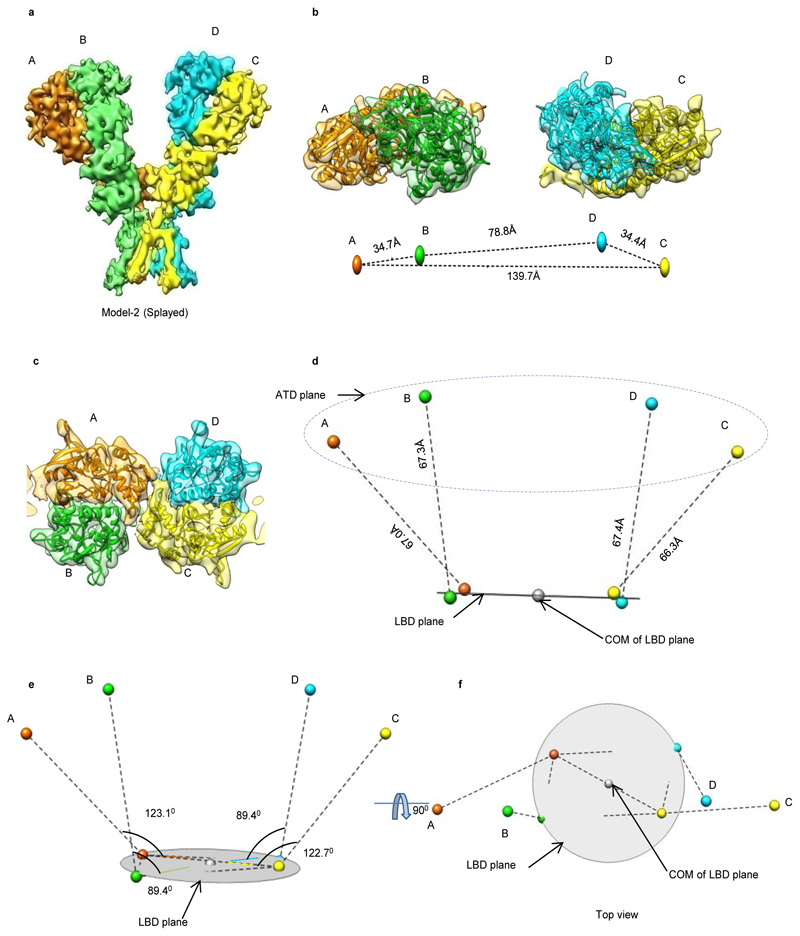
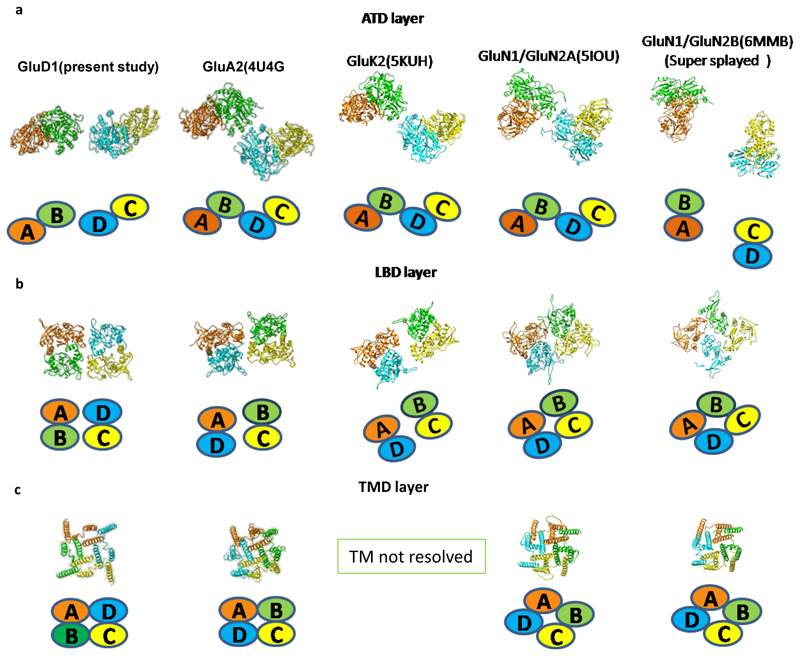
Further, the extracellular domains of subunits AD and BC could also be superimposed with an RMSD of ~2.6 Å but the TM domains do not superimpose such that the angle formed between M4 helices of the two subunits is ~ 63° (B-C) and 65° (A-D) respectively (Fig. 1 c). This assembly is distinct from the reported structures of AMPA, Kainate and NMDA receptors where the ATD dimer pairs are formed by AB and CD subunit pairs but due to domain swapping, the LBD dimers are formed between subunits AD and BC. This arrangement results in the formation of receptor tetramer by pairs of conformationally distinct AC and BD subunits (Extended Data Fig. 7 and Supplementary Fig. 4). The non-swapped arrangement of GluD1 receptors seemingly allows more conformational freedom for the movements of the two dimer arms (Fig. 1 a, d, and Extended Data Fig. 4 a, d) leading to the heterogeneity observed in our cryo-EM analysis.
Assembly of the Extracellular domains in GluD1
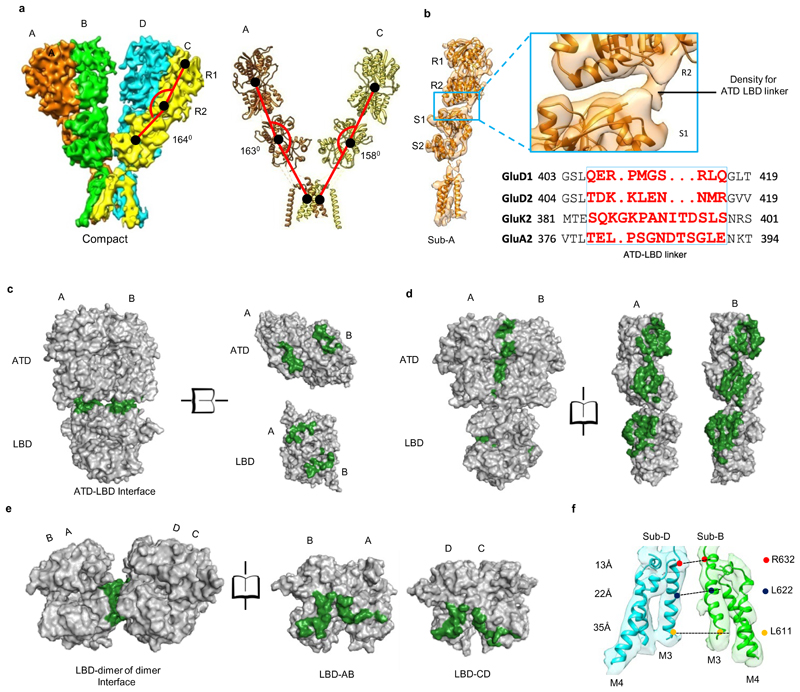
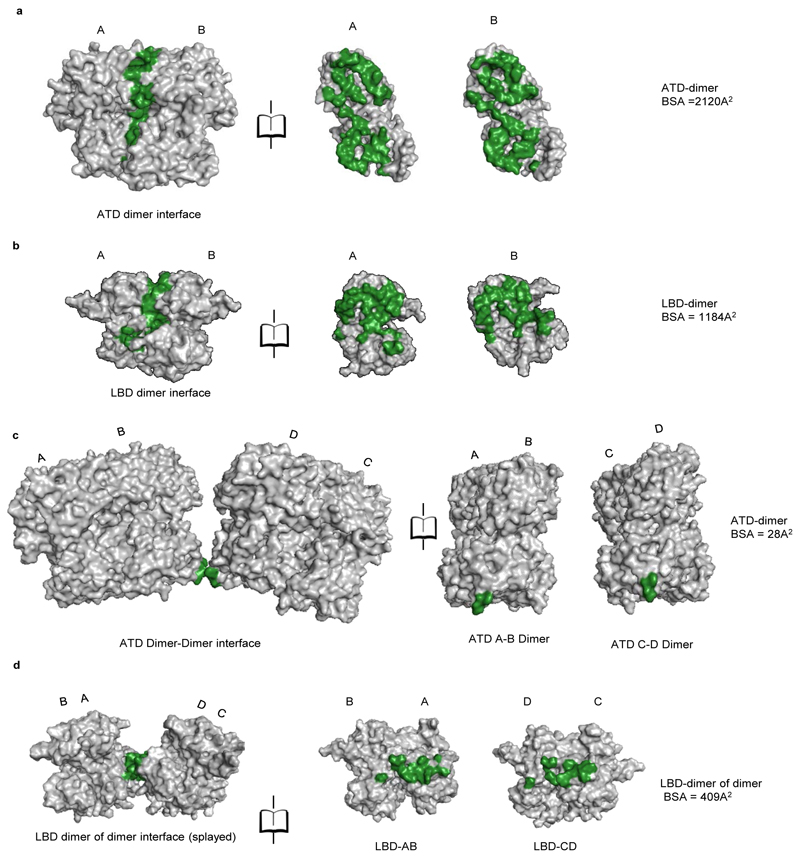
Each of the four ATDs has a clamshell-like structure formed by the upper (R1) and lower (R2) lobes, with a nearly identical conformation in each subunit. ATD dimers assemble via contacts mediated by both the upper R1 and lower R2 lobes, with a buried surface of ~2120 Å2 per subunit. Further, due to the movement of the two extracellular arms, the GluD1 ATD dimers form tetrameric (dimer-of-dimer) contacts only in the “compact” conformation, with a small buried surface of ~28 Å2 (Extended Data Fig. 8) unlike that in AMPA and kainate receptors where the buried surface is much larger at ~300 Å2. However, consistent with the nanomolar affinity for ATD homodimer formation observed for GluD2 receptors25, the ATD dimer interface is intact in both the compact and extended conformations (Fig. 2).
Fig. 2. Inter Subunit arrangement and solvent accessible surface.
a, Side view of the “compact” GluD1 model is shown along with the angles subtended between COMs (shown in black solid circles) of R1, R2 lobes of ATD, LBD and TM domain for different subunits are shown. b, Shows segmented density map of subunit B fitted with atomic co-ordinates along with a zoomed view of the ATD-LBD interface. The density for the linker region is visible, but not modeled. Panels c-e show solvent accessible surface area analysis for the ATD-LBD dimer interface (c), ATD-LBD interface (d) and LBD dimer of dimer interface (e). The buried surface is colored in green while the solvent accessible surface is represented in grey. It is important to note that we have not modeled the ATD-LBD linker due to limited resolution and hence are not included in the solvent-accessible surface calculation. The buried interface is likely to be more significant if the linker residues are placed and accounted. f, The distance between residues R611 (top) L622 (middle) and L632 (bottom) of the M3 helices of subunits B and D are shown. Also, see supplementary Fig. 4 and 6.
Further, due to the absence of subunit crossover, the two ATD–LBD arms of the receptor tetramer are in the same plane (Extended Data Fig. 5 and 6) while domain swapping and the dimer-of-dimers interface interactions in AMPA and kainate receptors leads to tilting of the ATD dimer pairs away from the overall axis of symmetry, such that subunits B and D lie proximal to the center of mass (COM), whereas subunits A and C form the distal edges of the tetramer assembly (Supplementary Fig. 4). Owing to this, the COMs of R2 lobes of ATD proximal subunits B-D in the "compact" conformation are at a distance of ~45 Å and an angle of ~162° is formed between COMs of subunits A, B and D, indicating that the two dimer are almost in the same plane. This is in contrast to AMPA receptor GluA2 where the B-D COMs are a distance of ~35 Å and subunit A, B and D COMs make an angle of ~119° (Fig. 1 d, and Extended Data Fig. 4 d).
Within the LBD layer, dimer pairs like those crystallized for the isolated domains for AMPA, kainate and delta receptors assemble via contacts mediated by the upper lobes, with a buried surface for each subunit of ~1184 Å2 (Fig. 2 d and Extended Data Fig. 8 b). The dimer-of-dimers interface in the compact conformation for the LBD is ~510 Å2 which is similar to that observed in AMPA and kainate receptors (Fig. 2 e). Further due to non-crossover, in GluD1, the ATD and LBD layers in subunits AB and CD pack on top of each other in an almost linear arrangement (Fig. 2 a). We also observe that the ATD and LBD layers pack closer with respect to each other than in AMPA and kainate receptors. Whether this assembly is due to non-crossover and the shorter ATD-LBD linker in GluD1 needs further exploration (Fig. 2 b). Our imaging conditions had 1mM 7-CKA and Ca2+, which helped in trapping the LBD in the dimeric state. The LBD adopts an open cleft similar to antagonist bound AMPA and kainate receptor LBDs with a cleft opening of ~ 25° compared to D-Ser bound GluD2 LBD35. (Supplementary Fig. 5). In overview, the LBD layer arrangement is similar to that observed in antagonist bound GluA2 or GluK2 receptors in a closed state with a classical 2-fold symmetric dimer and dimer-of-dimer assembly.
Transmembrane domain arrangement
In contrast to the two-fold symmetry of the LBD and ATD, the ion channel pore of GluD1 has 4-fold rotational symmetry such that the M4 segment of each subunit is packed against M1 and M3 from an adjacent subunit in a counter-clockwise rotation A-B-C-D when viewed from above (Fig. 1 f and g). While due to domain swap, the TM arrangement is clockwise A-B-C-D in the AMPA and kainate receptors (Extended Data Fig. 7 c). Similar to AMPA and kainate receptors, the pre-M1 cuff helix in GluD1 that lies parallel to the plane of the membrane, wraps around the exterior of the ion channel assembly. As observed in other iGluRs, the M3 helix bundle forms barrier to ion permeation. We also observe that although the TMs adopt a closed-pore, their assembly is more splayed similar to that observed in NMDA receptors and unlike AMPA or kainate receptors where the assembly is more compact. Consistent with this, the top of the M3 helix is more constricted and immediately separates below to form an expanded vestibule. Owing to this, the distances between residues R632 (top), L622 (middle) and L611 (bottom) of the M3 helices from two subunits D and B are ~13, 22 and 35 Å respectively (Fig. 2 f). In contrast, the TM packing is more compact in case of GluA2 receptors where the corresponding residues on M3 helix L620 (top), L610 (middle) and R599 (bottom) are separated by 15, 16 and 30 Å respectively (Supplementary Fig. 4 b). Similarly, the distance between M4 helix residues A812 (top), L823 (middle) and A836 (bottom) between subunits C and D are separated by ~33, 38 and 49 Å while those in GluA2 for corresponding residues are at a distance of ~29, 36 and 38 Å (Supplementary Fig. 4 c and Supplementary Fig. 6).
Validating receptor interfaces and assembly
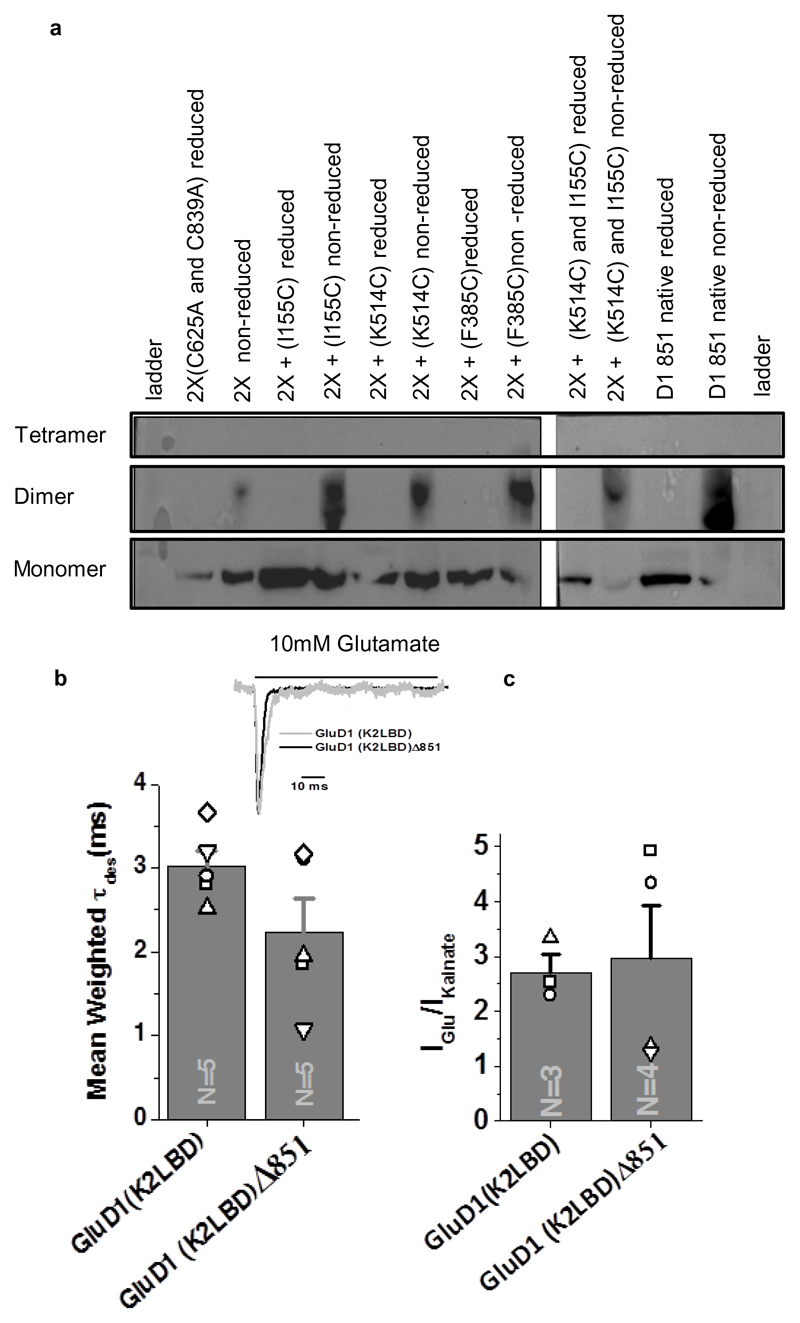
In order to validate that the unprecedented subunit arrangement and molecular symmetry of GluD1 receptor is physiological, we performed cysteine mutant crosslinking experiments. We introduced cysteine residues into the ATD dimer-of-dimers (F385C), ATD dimer (I155C) and LBD dimer (K514C) interfaces. These are the sites that should result in spontaneous disulfide bond formation due to the 2-fold symmetric assembly at these layers (Fig. 3 a; Supplementary Fig. 7). Our results show bands corresponding to GluD1 dimers in non-reducing conditions for I155C and K514C, indicating that the ATD and LBD indeed exist in a 2-fold symmetric dimeric arrangement (Fig. 3 a). Further, the F385C mutant also formed dimers suggesting that the two extracellular arms of the receptor tetramer can interact at the ATD dimer-of-dimers interface reminiscent to that observed in AMPA and kainate receptors.
Fig. 3. Probing GluD1 receptor interfaces.
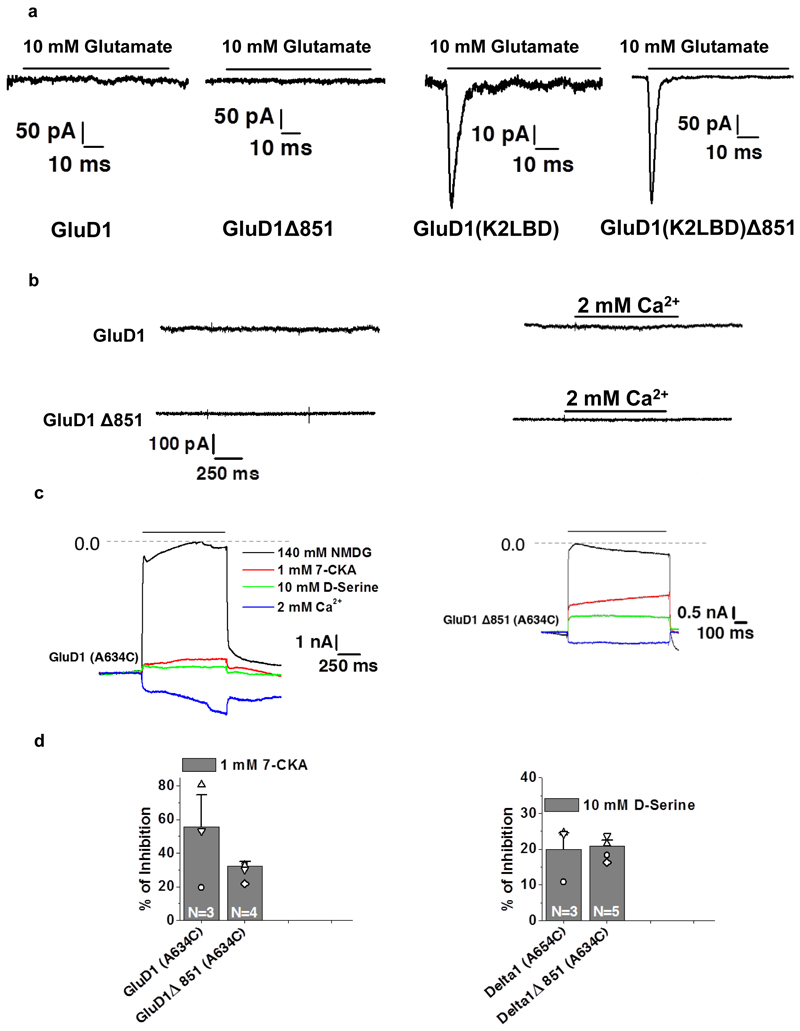
a, Western blot analysis for the cysteine crosslinking experiment is shown. GluD1 Δ851-2X construct (C625A/C839A) was used to generate cysteine mutants for the various domain interfaces due to low non-specific crosslinking. Dimer bands increase in non-reducing conditions for crosslinking of the ATD (2X-I155C), LBD (2X-K514C) and ATD-dimer-of-dimer interface (2X-F385C). For ATD and LBD crosslinking (2X-I155C/K514C) only dimer bands are observed, and bands corresponding to the tetramer are not seen. In all the cases, under reducing conditions, only bands corresponding to monomer were observed. Uncropped blot is provided with the paper online as Source Data. b, The comparison of mean weighted tau desensitization and (c) ratio of glutamate and kainate evoked currents for GluD1(K2LBD) and GluD1 (K2LBD) Δ851. The number of cells used for the recordings is shown. The error bars represent standard error from the mean. Representative normalized traces for GluD1Δ851 and GluD1(K2LBD) Δ851 overexpressed in HEK-293T on the application of 10 mM Glutamate is shown in the inset in b. Data for graphs in b-c are available as source data. Also, see Extended Data Fig. 9 and Supplementary notes.
We also made the double cysteine mutant I155C/K514C which would lock the receptor at both the ATD and LBD layers resulting into bands corresponding to tetramers on non-reducing SDS-PAGE gels in case of domain-swapped architecture and dimers for non-swapped assemblies. Consistent with our GluD1 structure, we observe bands corresponding to dimers and not tetramers indicating non-swapped architecture of GluD1 receptors in vivo, although we do observe higher oligomers (bigger than tetramers) probably due to non-specific linkages (Fig. 3 a).
In order to validate this assembly further, we carried out glutaraldehyde cross-linking of purified GluD1 receptors along with GluA2 as control. Due to its non-swapped architecture, the probability for getting dimers would be higher in GluD1 receptors compared to GluA2 where due to subunit cross-over, the probability for all protomers becoming cross-linked would be higher, leading to tetramers in SDS-PAGE gels. Consistent with this, crosslinking with 3mM glutaraldehyde for 2 and 5 minutes yielded dimers primarily on SDS-PAGE for GluD1 receptors, while tetramers were observed for GluA2 receptors (Supplementary Fig. 8).
C-terminal truncation does not affect the assembly of GluD1 receptors
We strived to use a minimally modified construct of GluD1 for protein expression and purification. The only modification we have made is C-terminal truncation at residue 851. In order to demonstrate that this CT deletion does not affect the receptor assembly and its functionality, we utilized a chimeric receptor approach, as native GluD1 receptors do not evoke ligand-gated currents. For this, we generated chimeric receptors where the LBD of GluD1 is swapped with that of GluK2, resulting in glutamate and kainate sensitive receptors32 (see Supplementary Notes). We carried out whole-cell recordings on wild type GluD1, GluD1△851, GluD1 (K2LBD) and GluD1(K2LBD) △851 receptors. While we got no measurable response on 10 mM glutamate application from wild type GluD1 and GluD1△851 receptors, robust currents were recorded from both GluD1 (K2LBD) and GluD1 (K2LBD) △851 suggesting that the CT deletion does not interfere with the assembly of GluD1 receptors (Fig. 3 b; Extended Data Fig. 9 a). Our observation is also consistent with the report of CT-deletion affecting synaptic trafficking but not the surface expression of GluD1 receptors43. In addition, we also generated A634C mutant receptors which result into formation of constitutively active GluD1 receptors30. Consistent with this, our whole-cell patch clamp recordings showed robust constitutively active receptors for both GluD1 A634C and GluD1△851-A634C (Extended Data Fig. 9 b-e). These constitutive currents were blocked when Na+ in extracellular solution was replaced with large cation NMDG demonstrating that these constitutive currents are mediated by GluD1 receptors (Extended Data Fig. 9 c-d). Further, as reported earlier for GluD1 and GluD2 receptors, these constitutive currents could also be modestly inhibited by application of 7-CKA and D-Ser and potentiated by calcium (Extended Data Fig. 9 c-e). Thus our electrophysiology experiments demonstrate that CT-deletion necessary for overexpression and purification of GluD1 does not affect receptors assembly.
Discussion
Using a minimally modified construct, we have purified and determined the structure of detergent-solubilized GluD1 receptors in-solution. Owing to the conformational heterogeneity of GluD1, the resolution of our EM maps is limited to ~8Å. However, by fitting crystal structures and models of various domains into the constraints of EM density map, we provide the first insight into the subunit arrangement for a homo-tetrameric GluD1 receptor. While all the previous models for GluD receptors depict domain swapping at the ATD-LBD interface, our study shows a unique non-swapped architecture for this enigmatic class of receptors. However, whether this non-crossover is the sole cause for the inactive ion channel needs to be addressed in the future.
Due to non-crossover, the two extracellular arms of the receptor seem to have a broader range of movement resulting in conformations where the receptor could adopt a "splayed" conformation. Recent data shows that other iGluRs, which unlike delta receptors have swapped architecture and show agonist gated ion channel activation, can also adopt splayed ATD conformations44–48 in detergent-solubilized states (Extended Data Fig. 7) albeit to a limited extent. It is to be noted that, the splayed conformations may not be physiological due to the higher concentration of proteins in membranes and interactions with other synaptic proteins,25, 49 auxiliary subunits50–51 that would likely limit movements.
Another important feature of the GluD1 receptor is the close packing of ATD and LBD domains. While, it’s not clear currently if this is driven by the shorter ATD-LBD linker when compared to other iGluRs, it has been reported that mutation of the linkers leads to loss of function in GluD2 receptors. Pertaining to this, D-serine application induced Parallel Fibre-long term depression (PF-LTD) in cerebellar slices in Purkinje cells (PCs) expressing wild-type receptors while it failed to do so in PCs expressing GluD2 in which a glycosylated linker was inserted between the two domains25. This could result likely from the uncoupling of the ATD-LBD interactions, which might lead to reduced transduction of forces generated by ligand binding to the transmembrane domains. However, the GluA2 receptor with shorter ATD-LBD linker still crystallized in a domain-swapped configuration and was gated by glutamate52. This along with other recent pieces of evidence suggest that not just the short linkers, but the ATD-LBD interface interactions, and contribution of the hinge region of the GluD2-LBD in the weak ligand affinity53 among other things might contribute to the inactivity of the GluD receptors. However, this needs to be established by future studies.
Our study also raises an important question of what drives the subunit cross-over in AMPA and kainate receptors and why this is not observed in GluD1, the answer to which is still not clear. For AMPA and kainate receptors, evidence from electron microscopy of intermediates of the biosynthesis process shows that GluA2 dimer synthesis precedes tetramer formation and that in the dimers, the ATD and TM segments are closely apposed, whereas the LBDs are too far apart to interact54. This arrangement is suitable for domain swapping on the assembly of a second dimer pair to form tetramers. By contrast, for the LBD dimer–stabilizing L483Y mutant, all three segments are closely apposed similar to that observed in our GluD1 structure. However, subsequent tetramer formation in GluA2 L483Y was observed to be strongly inhibited, as the subunit crossover observed in the full-length GluA2 structure cannot occur when the LBD dimer pairs cannot separate. This assembly mechanism is supported by nanomolar affinities of the ATD dimerization and low micromolar to the millimolar affinity for LBD dimerization respectively55–56. The GluD1 structure with closely apposed ATD-LBD domains and non-crossover, however, points towards a different mode of assembly. It is speculated that due to subunit crossover in the ATD and LBD layers, the tetramer in AMPA and kainate is unlikely to be assembled in a cooperative process involving all four subunits. While in case of GluD1 the assembly is likely to be straightforward with subunits assembling as two dimers and the tetramer formed by a simple assembly of the two dimers with no subunit crossover at the ATD-LBD layer. This is also supported by the nanomolar affinities of ATD dimerization in GluD receptors25 and the fact that isolated GluD2 LBDs in the apo state have been shown to crystallize as dimers in the presence of Ca2+ ions33 that can modulate the strength of dimer formation57. Further, the extracellular domains (ATD-LBD) of GluD2 also crystallized as dimers but with interactions only at the ATD layer and the apo-LBDs adopted an unusual “swing-out” conformation in the absence of calcium ions and antagonists58. This GluD2 ATD-LBD dimer could not be modelled into a receptor tetramer without significant reorganization and reorientation of the LBD domains. We believe that in the absence of the constraints exerted by linkers and transmembrane domain, and due to crystal lattice contacts, the LBDs likely adopt a non-physiological conformation in this study56.
Further, AMPA, kainate and NMDA are able to assemble into functional receptors even after ATD deletion. However, a recent study on ATD-deleted NMDA receptors revealed that a fraction of receptor population adopted LBD packing analogous to what we observe in the GluD1 structure highlighting the importance of ATDs in guiding the subunit arrangement of the LBD layer. The well resolved LBD-TM linkers in this △-ATD-NMDA receptor adopted a relaxed conformation likely rendering the receptor inactive59. Thus, aspects of our structures are seen in other iGluRs, but unique to GluD1 is the lack of subunit crossover.
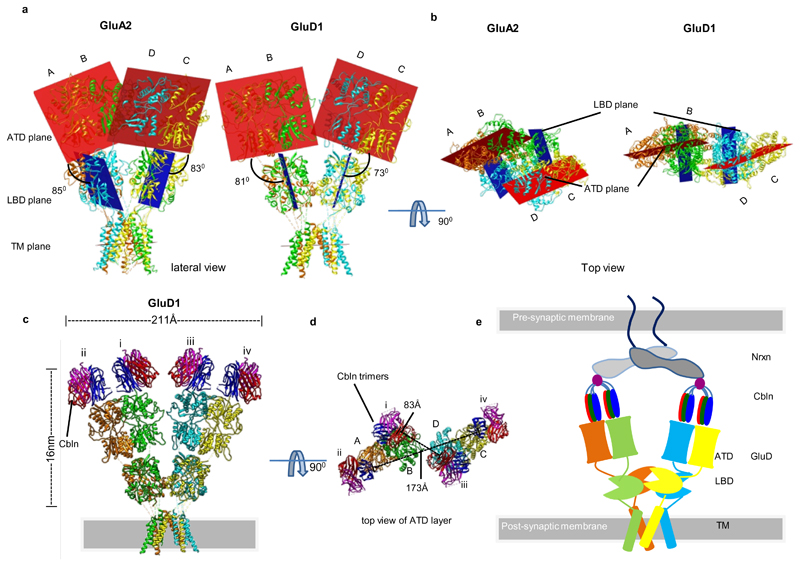
Multiple studies have shown that ATDs which are the most distal domains of GluD receptors directly interact with cerebelins, which in turn couples with neurexins forming the trans-synaptic complex. The arrangement and orientation of ATDs in GluD1 (Fig 4 a, b) does not occlude or restrict the cerebelin interaction surface. On superimposing ATD-cerebelin complex25 onto GluD1 ATD, the receptor ectodomain (ATD-LBD) and cerebelin (Cbln) has a length of ~16 nm similar to ~17 nm distance reported earlier25 (Fig 4 c, d). Based on this we propose a model (Fig. 4 e) for the tripartite complex formed between GluD receptors, cerebelin, and neurexin that mediate trans-synaptic interactions and is essential for maintaining the synaptic integrity of PF-PC synapses. Our model is similar to that proposed earlier25, 60 except for the non-crossover observed in GluD1 receptor. We also postulate that this anchoring of GluD receptors to the β-NRX1(+4)–Cbln1 complex will limit or prevent large-scale motions of the two extracellular arms.
Fig. 4. Orientation and arrangement of the ATD and LBD domains.
Panels a-b illustrate arrangement of the planes of ATD and LBD dimers in GluA2 and GluD1 in side a and top views b. The ATD planes are shown in brick red color while the LBD planes are depicted in blue. The angle formed between the ATD-LBD planes are measured and shown in a. c, Model of GluD1 and Cbln complex generated via superimposition of the GluD2 ATD-Cbln1 complex onto the ATD dimers of GluD1 (compact conformation). d, Top view of the GluD1-Cbln1 binary complex is shown along with the distances between the COMs of cerebelin1 trimers. Panel e shows schematic representation of the tripartite Neurexin, Cerebelin (Cbln) and GluD receptor trans-synaptic complex. The non-swapped architecture may allow for movements of the GluD receptor arms to accommodate the entire trans-synaptic complex in the 20-25 nm synaptic cleft.
In overview, our results provide a molecular framework to design future studies directed towards resolving the long-standing questions concerning this family of receptors. These results suggest that orphan delta receptors of the iGluR family likely have a different mode of assembly and provide a foundation for future studies directed towards understanding the functions of these receptors in light of this structural information.
Online Methods
Cell Lines
The HEK293S GnTI – (ATCC CRL-3022) were obtained and authenticated by ATCC and no further authentication or mycoplasma testing was performed.
Construct design
Rat GluD1 was cloned into pEGBacMam vector41 in frame with a C-terminal thrombin recognition site (GLVPRGSAAAA) and EGFP (A207K mutant) with a C-terminal octa-histidine (His8) tag. Full-length GluD1 had weak expression and stability as judged via FSEC40. Further screening of constructs identified GluD1△851 as a promising candidate for overexpression and purification.
For cysteine crosslinking experiments, mutants were generated by using site-directed mutagenesis. The cysteine knockout constructs; C625A and C839A were first generated in the GluD1△851 background (GluD1-2x) and then used for incorporating mutations I155C (ATD), F385C (ATD dimer-of-dimer interface) and K514C (LBD). Additionally, combination mutant GluD1-2x-I155C/K514C was also generated.
For electrophysiology experiments, wild type rat GluD1 and GluD1△851 was cloned into a pRK5 expression vector. GluK2 LBD chimeras were generated by exchanging GluD1 : S1, T418 - P528; S2, P644 – D792) with that of GluK2 : S1, S398-N515; S2, P636-E775 to generate constructs GluD1(K2LBD), GluD1(K2LBD) △851.
The constitutively active receptors were generated by site directed mutagenesis of residue alanine 634 in SYTANLAAF motif to cysteine30 (numbering as per mature polypeptide) in GluD1 wild type and GluD1△851 construct. All the constructs were verified by sequencing of the entire coding region.
Expression and Purification
HEK293GnTI- cells were adapted to grow in suspension cultures in freestyle 293 expression media supplemented with 2 % FBS (Gibco), 2 mM Glutamine (Gibco) and 1% Penstrep (Gibco) and were infected at a cell density of ~2.5*106 cells/ml with P2 baculovirus at a multiplicity of infection (MOI) of ~1. To boost the expression of the protein, 10mM sodium butyrate (Sigma) was added 20 hrs post-infection and cultures moved to 30ºC. The cells were harvested ~48-52 hrs later by centrifugation at 6000 rpm for 20 minutes. The cell pellet was collected and stored at -80ºC for further processing. The frozen cell pellet was thawed at room temperature for 10 minutes and was resuspended in a buffer (20 ml/L cell culture) containing 150 mM NaCl, 20mM Tris pH 8.0 along with protease inhibitor cocktail (Roche). The resuspended cells were disrupted by ultrasonication (QSonica sonicator, four cycles of 90 sec (15 sec on/ 15 sec off) with power level 7 using medium size probe) with constant stirring. Care was taken to keep the temperature below 12°C throughout the sonication. The lysate was first clarified by low-speed centrifugation and membranes were collected by ultracentrifugation at 40,000 rpm, 1 hour. Membrane pellets were homogenized and solubilized for 45 min in buffer containing 150 mM NaCl, 20 mM Tris (pH 8.0), 40 mM n-dodecyl-β-D-maltopyranoside, and 6 mM cholesterol hemisuccinate at 4°C. Detergent solubilized fraction was collected by centrifugation at 40,000 rpm, 1 hour and cobalt-charged TALON metal affinity resin (~4 ml bed volume) was added to the supernatant together with 10 mM imidazole to allow batch binding for 3 hours at 4°C. The resin was packed in a column, washed with 40 mM imidazole containing buffer (20 mM Tris, 150 mM NaCl, 0.75 mM n-dodecyl-β-D-maltopyranoside, 0.03 mM cholesterol hemisuccinate) till the baseline reached zero. Bound GluD1 receptors were eluted by 250 mM imidazole containing buffer. The fractions containing protein of interest were pooled and kept for thrombin (Millipore) digestion (1:100 wt/wt) overnight at 4 °C. The thrombin digested protein was further purified by size exclusion chromatography (Superose 6 10/300) equilibrated with 150 mM NaCl, 20 mM Tris pH 8.0 and 0.75 mM DDM, 0.03 mM cholesterol hemisuccinate. Eluted fractions were analyzed for homogeneity by SDS-PAGE and FSEC, only fractions containing tetrameric receptors were pooled and concentrated to ~0.9 mg/ml.
Cryo-electron microscopy data collection and analysis
A droplet of 3 μl each of purified GluD1△851 at a concentration of ~0.9 mg/ml was applied twice onto a glow discharged 1.2/1.3 300 mesh ultrafoil gold grid (Quantifoil). It was blotted for 8 seconds at a blot force of 0 using the FEI Vitrobot Mark IV at 4°C and 95 % humidity, and plunge-freezing the grid in liquid ethane.
Cryo-EM data collection was carried on a 300 kV Titan Krios microscope equipped with a K2 camera with a post-column energy filter61. Micrographs were recorded in super-resolution mode at a magnified nominal pixel size of 1.067 Å and defocus ranging from −1.5 to −3.5 μm. Imaged at a dose rate of 6.5 e− per pixel per s, each micrograph consisted of 40 dose-fractionated frames with a total exposure time of 6 s and total dose of 40.38 e− per Å2.
A total of 4120 movies were collected, aligned, and dose-weighted to correct for movement during imaging and account for radiation damage via Motioncor262. The CTF parameters for each micrograph were determined by Gctf63. Manual particle picking (~2000 particles) and reference-free 2D classification were carried out to generate templates for automated particle picking in CryoSPARC v264. A total of ~117k particles were autopicked and subjected to several rounds of reference-free 2D classification followed by manual inspection and selection of classes with iGluR like features. This process yielded a stack of 72149 cleaned particles that were subjected to per particle local motion correction followed with 2D classification and abinitio 3D reconstruction in cryoSPARC V2. Subsequent heterogenous 3D classification yielded seven classes, out which classes 5 (13422 particles) and 7 (14939 particles) representing “compact” and “splayed” conformations of GluD1 were taken for further refinement. Homogenous 3D refinement in C1 symmetry, followed with the non-uniform and local refinement as implemented in cryoSPARC V2 workflow yielded final maps to a resolution of ~ 8 Å and ~7.6 Å (0.143 FSC) for compact and splayed conformations respectively. 3D refinement by imposing C2 symmetry for the extracellular domains and masking out the TM domains didn’t improve the resolution significantly.
Model building and refinement
GluD1 tetramer model was built by the rigid-body fitting of individual domains into the EM map in UCSF Chimera65. Four copies each of GluD1 amino-terminal domain (PDB code, 5KC9) and ligand-binding domain modelled via I-TASSER using 5CC2 (GluD2 LBD complex with 7-CKA as a template) was used. Model for GluD1 TM domain was generated by threading its sequence onto the transmembrane domain of GluK2 (PDB code: 5KUF) and was used for fitting into EM density. The fits were improved by using a molecular dynamics-based flexible fitting simulation66 followed by multiple rounds of real-space refinement in Phenix67. After refinement, map CC between models and EM maps was 0.8 and 0.7 for the two models, indicative of a reasonable fit at the present resolution. Model-map FSC curve calculation yielded values of ~8.2 and ~8.4 that agreed well with the gold-standard FSCs generated during the 3D refinement (Supplementary Figure S4). The final model has good stereochemistry, as evaluated using MolProbity68 (Supplementary Data Table 1). All of the figures were prepared with Pymol69, UCSF Chimera65, and Prism 8.0.
Cysteine crosslinking and Western blots
For GluD1 cysteine crosslinking experiments, plasmid DNA encoding GluD1-2x (pEGBacMam), GluD1-2x with mutants I155C (ATD), F385C (ATD dimer of dimer interface), K514C (LBD) and GluD1-2x-I155C/K514C were transiently transfected for expression in HEK293T cells. Cells were harvested 24–48 h after transfection and resuspended in TBS buffer (20 mM Tris pH 8.0, 150 mM NaCl supplemented with protease inhibitor cocktail). Cells were sonicated, and membrane fractions were harvested following low-speed and ultra-speed centrifugation. Membranes were solubilized in TBS buffer supplemented with 40 mM DDM / 2mM CHS for 1 hour at 4 ºC, clarified by ultracentrifugation and then run on a 6 % SDS–PAGE gel either in the absence (non-reducing condition) or presence (reducing condition) of 100 mM DTT. Protein bands were electroblotted onto PVDF membranes (Amersham Biosciences) and were blocked for 1 hour at room temperature in TBST (150 mM NaCl, 10 mM Tris-HCl pH 7.6, 0.1 % Tween-20) containing 5% non-fat milk and then incubated for 1 h with monoclonal His5 antibodies (Sigma) raised in rabbit. After four 15-min washes with TBST (TBS +Tween-20 0.05%), the membranes were incubated for 1 h at room temperature with anti-rabbit goat antibodies conjugated to horseradish peroxidase. Then the membranes were rewashed two times for 15 min with TBST and twice with TBS and immunoreactivity was visualized using the ECL detection kit (Invitrogen).
Glutaraldehyde crosslinking
Purified and detergent solubilized rat GluD1△851, and GluA2cryst46 receptors in 20 mM HEPES, 150 mM NaCl, 1 mM DDM were incubated with 3 mM of glutaraldehyde for 2, 5 and 10 minutes at 37 °C to allow for crosslinking. Crosslinking reaction was stopped by quenching with 50 mM Tris following which, the protein was resolved on SDS-PAGE and stained with Coomassie blue for band visualization.
Electrophysiology
Rat GluD1 wild type, GluD1△851, GluD1(K2LBD), and GluD1(K2LBD) △851 receptors were tested for activity by whole-cell patch-clamp recordings. Assays were carried out 36–48 hours post-transfection in HEK293T cells. Pipettes were pulled (Sutter, P-1000) from borosilicate glass capillaries (1.5 OD x 1.17 x 100 L mm, Harvard Apparatus) and polished to 2–3 MΩ resistance, filled with internal solution containing 30 mM CsCl, 100 mM CsF, 4 mM NaCl, 10 mM HEPES, 5 mM EGTA, 2 mM Na2ATP and 0.5 mM CaCl2, pH 7.2 (osmolarity ranging between 290-300 mosmol/L). External solution (ECS) contained 150 mM NaCl, 2.8 mM KCl, 10 mM HEPES, and 0.5 mM CaCl2, pH 7.3 and osmolarity ranging between 295-305 mosmol/L. 10 mM glutamate or 1mM kainate dissolved in ECS was applied for 100 ms to measure the whole-cell desensitization kinetics. The whole-cell recordings were acquired using Patchmaster V2X90.2 (Heka Elektronick) 3 min after the establishment of the whole-cell configuration. Raw data files were exported into Igor pro (ITX) and converted into abf files, compatible for pClamp by using ABF Utility. The macroscopic rate of desensitization (τdes) was measured by the exponential fit to the decay of current from ~90 % of its peak amplitude (Ipeak) to baseline. The desensitization kinetics were fitted by using the single exponential, 2-term fitting (Levenberg-Marquardt). Mean weighted τdes values were calculated from our 2-term fitting using the equation TWeighted = (A1*T1+A2*T2)/ (A1+A2), where weight 1= A1, weight 2= A2, T1=Tau 1, T2=Tau2. Ratios of glutamate and kainate-evoked currents were determined in 3-4 independent experiments were subjected to statistical analysis using Prism 8.0.
For recording constitutively active currents, mutation A634C30 (numbering as per mature polypeptide) was introduced by site directed mutagenesis in both full-length GluD1 and GluD1△851. Recordings were performed between 16-28 hrs post-transfection from HEK293T cells. Pipettes were pulled as described above and were filled with internal solution containing 140 mM CsCl, 10 mM HEPES, 1 mM BAPTA, 2 mM Na2ATP with pH adjusted to 7.2 with CsOH (osmolarity ranging between 290-300 mosmol/L). The extracellular solution consisted of 135 mM NaCl, 5.4 mM KCl, 0.5 MgCl2, 5 mM HEPES, pH 7.2 adjusted by NaOH as described before28. For 7-CKA, D-Ser or calcium application, solutions were prepared by addition of either 2mM CaCl2 or 1mM 7-CKA, 10 mM D-Ser in the extracellular solution. The NMDG (N-Methyle-D-glucamine) solution consisted of 140 mM NMDG, 0.5 mM MgCl2 and 5mM HEPES. The seal resistance before entering into the whole-cell configuration was always at least 1 GΩ. Currents were recorded at room temperature using HEKA EPC10 with Patchmaster as described earlier.
Statistics
No statistical methods were used to predetermine the sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Extended Data
Extended Data Fig. 1. GluD1 purification and Cryo-EM data processing.
a, Schematic representation of the optimized GluD1 construct showing the C-terminal truncation at residue 851 and C-terminal thrombin cleavage site along with GFP and Octa histidine tag. Also, see Supplementary Fig.1. b, Size-exclusion profile of the final purified protein showing receptor stability in optimized buffer conditions. c, Selected 2D class averages from reference-free 2D classification of GluD1 in complex with 1mM 7-CKA and 1mM Ca2+. The white arrows mark a few classes that show conformational heterogeneity of the extracellular receptor domains. d, 3D classification of GluD1 into seven classes reveals heterogeneity due to the movement of the two extracellular arms. Also, see Extended Data Fig.2.
Extended Data Fig. 2. Cryo-EM data processing work flow.
A total of 72149 good particles were obtained by several cycles of 2D class averaging of particles from 4120 micrographs. The 3D map generated by ab-initio 3D reconstruction was further refined heterogeneously into seven conformationally distinct 3D classes. The 3D classes showed heterogeneity due to movement of the two extracellular arms. For the purpose of model building and analysis, a "compact" (Class 5) and a "splayed" (class 7) conformation maps were further refined to a resolution of 8.1 Å and 7.6 Å respectively.
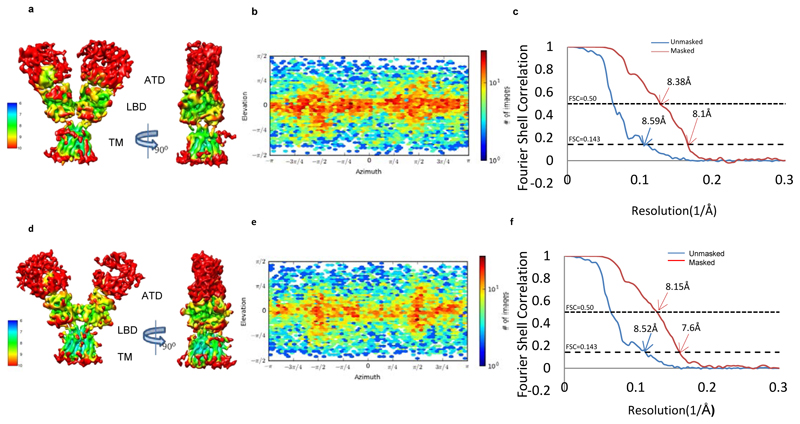
Extended Data Fig. 3. Local resolution estimates of the cryo-EM maps.
a and d, The sharpened Cryo-EM densities of GluD1Δ851 in 7-CKA and calcium bound form, colored based on local resolution. b and e, Euler angle distribution of particles for the two models is shown. c and f, Fourier shell correlation curves for the Cryo-EM maps with mask (red) without mask (blue). The resolution of map corresponding to FSC 0.5 and 0.143 is indicated.
Extended Data Fig. 4. Splayed conformation of GluD1 receptor.
Panels a-f show the architecture of splayed conformation of GluD1 receptors in complex with 7-CKA and calcium. a, Side view highlighting the broadest face of the Y- shaped receptor and 90° rotated views of the sharpened 3D density map is shown. Each subunit is depicted in a different color. The EM reconstructions clearly show the non-swapped arrangement of the ATD and LBD layers. The distances between the centroids (R1-R1 of ATD domains) for AB and CD dimer pairs are shown above the model. The vertical separation between the COMs of ATD dimers and LBD dimers are also shown. Panel b shows the segmented density map for subunits A and B fitted with protein co-ordinates. c, Superimposition of subunits B/D, B/C, A/D and A/C are shown highlighting similar AB and BC conformations. Helices and sheets are represented as pipes and planks, respectively. Top views of ATD (d), LBD (e) and TM domains (f) are shown. The distances and the angles subtended between the COM (Centre of Mass) of various subunits were measured and are indicated below the top views. Also, see Extended Data Fig. 5 and 6.
Extended Data Fig. 5. Architecture and domain arrangement in compact GluD1 model.
a, Cryo-EM density map of compact GluD1 model is shown in a view parallel to the membrane. The four subunits A, B, C, D are colored in orange, green, yellow and cyan respectively. The colored spheres represent the Centre of mass (COM) of ATD and LBD domains. b, Top view of ATD with segmented EM density map fitted with atomic models is shown. The distances between the COMs of ATDs are shown with dashed lines below the EM-density map, depicting the arrangement of ATDs in the plane. c, Densities corresponding to LBDs fitted with atomic models is shown. d, The distances from COMs of ATD and LBD are shown. The LBD plane is depicted as a circular disk and ATD plane is shown as dashed ellipse. Panels e and f, show side and top vies of angles subtended by COMs of ATD with COM of LBD layer. COM plane of the LBD layer is indicated by metallic disk.
Extended Data Fig. 6. Architecture and domain arrangement in splayed GluD1 model.
a, Cryo-EM density map of splayed GluD1 model is shown in a view parallel to the membrane. The four subunits A, B, C, D are colored in orange, green, yellow and cyan respectively. The colored spheres represent the Centre of mass (COM) of ATD and LBD domains. b, Top view of ATD with segmented EM density map fitted with atomic models is shown. The distances between the COMs of ATDs are shown with dashed lines below the EM-density map, depicting the arrangement of ATDs in the plane. c, Densities corresponding to LBDs fitted with atomic models is shown. d, The distances from COMs of ATD and LBD are shown. The LBD plane is depicted as a circular disk and ATD plane is shown as dashed ellipse. Panels e and f, show side and top vies of angles subtended by COMs of ATD with COM of LBD layer. COM plane of the LBD layer is indicated by metallic disk.
Extended Data Fig. 7. Domain arrangement in GluD1, GluA2, GluK2, GluN1/GluN2A and GluN1/GluN2B receptors.
a, Top views of the ATD (a), LBD (b) and TM domains (c) are shown for GluD1, GluA2, GluK2, GluN1/GluN2A and GluN1/GluN2B receptors highlighting the subunit arrangement. Each chain is uniquely colored and domain arrangement is also depicted in cartoon below each layer. Comparisons for "compact" and "super-splayed" conformations of NMDA receptors with that of GluD1 are shown highlighting the fact that in all the conformations of AMPA, KA and NMDA receptors the domain swapping between the ATD and LBD layers exists unlike that in GluD1.
Extended Data Fig. 8. Buried surface area between the sub domains.
Surface illustration of the isolated sub domains in grey with buried surface represented in green. The calculated buried surface area for the various domains is also shown. Panels a-d show the analysis for ATD dimer, LBD dimer, ATD dimer-of–dimer and LBD dimer-of-dimer interface for the compact GluD1 model.
Extended Data Fig. 9. C-terminal truncation does not affect assembly of GluD1 receptors.
a, Representative traces for the whole-cell recording of the GluD1, GluD1Δ851, GluD1(K2LBD) and GluD1(K2LBD)Δ851 expressed in HEK-293T cells are shown in response to 10 mM glutamate application. Panels b-e show whole-cell patch clamp recordings (holding potential = -60 mV) from constitutively active GluD1 A634C point mutant receptors. The seal resistance before entering into the whole-cell configuration was always at least 1 GΩ. b, shows that no spontaneous currents were observed for wild-type GluD1 or GluD1 Δ851 receptors and no effect was observed on 2mM Ca2+ application. Panels c and d show overlay of representative traces showing application of either NMDG solution or 1 mM 7-CKA (red), 10 mM D-Ser (green) or 2 mM CaCl2 (blue). Dashed line indicates zero current level achieved by application of impermeant NMDG which blocks the constitutive inward currents for both GluD1 A634C (c) and GluD1 Δ851-A634C receptors (d). The constitutive currents are also modestly inhibited by D-Ser or 7-CKA application and potentiated by Ca2+ (c, d) for both the full-length and CT truncated GluD1 receptors. e, shows percent inhibition of spontaneous currents by 7-CKA and D-Ser calculated with respect to NMDG inhibition. Data for graphs are available as source data. The number of cells used for the recordings is shown. The error bars represent standard error from the mean.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust DBT India Alliance fellowship (grant number IA/I/13/2/501023) awarded to J. Kumar. A. P. Burada thanks ICMR, India for senior research fellowship. R.V. thanks SERB for N-PDF fellowship (N-PDF/2016/002621). Dr. M. L. Mayer, NIH, Bethesda kindly gifted the various iGluR constructs that were subcloned and used for construct optimization and mutational studies. Dr. E. Gouaux, OHSU, Portland kindly provided the pEG BacMam vector. We acknowledge the European Synchrotron Radiation Facility for provision of microscope time on CM01, and we would like to thank M. Hons for their assistance in EM data collection. We thankfully acknowledge the kind help of Dr. M. Karuppasamy, EMBL in grid vitrification. Help with initial screening of conditions for grid vitrification from Dr. P. J. Peters, R. Ravelli at M4i, Maastricht University, Masstricht, Netherlands and Dr. V. K. Ragunath, National Electron cryo-microscopy facility at the Bangalore Life Sciences Cluster (DBT/PR12422/MED/31/287/2014), NCBS, Bangalore is gratefully acknowledged. We are also thankful to Dr. A. Kembhavi and Dr. K. Vaghmare, The Inter-University Centre for Astronomy and Astrophysics, Pune for helping with transfer and storage of raw EM data.
Footnotes
Author Contributions
A.P.B optimized construct and purified protein, did all the molecular biology, biochemical experiments, and processed EM data with assistance from J.K. Electrophysiology experiments were done by R.V. JK supervised the overall project design and its execution. All authors contributed to the analysis and preparation of the manuscript and approved the final draft.
Competing interests
The authors declare no competing interests.
Reporting Summary statement
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
The cryo-EM density reconstruction and final models were deposited in the Electron Microscopy DataBase (accession codes EMD-0744 for the compact conformation and EMD-0773 for the splayed conformation) and in the Protein Data Bank (accession codes 6KSS for the compact conformation and 6KSP for the splayed conformation). The raw movie data has been submitted to EMPIAR database. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files or from the corresponding author upon request. Source data for Fig. 3a, 3b, 3c and Extended Data Fig. 9 e are available with the paper online.
References
- 1.Gao J, et al. Orphan Glutamate Receptor δ 1 Subunit Required for High-Frequency Hearing. Mol Cell Biol. 2007;27:4500–4512. doi: 10.1128/MCB.02051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuzaki M. The delta2 glutamate receptor: a key molecule controlling synaptic plasticity and structure in Purkinje cells. Cerebellum. 2004;3:89–93. doi: 10.1080/14734220410028921. [DOI] [PubMed] [Google Scholar]
- 3.Yuzaki M, Aricescu AR. A GluD Coming-Of-Age Story. Trends in Neurosciences. 2017;40:138–150. doi: 10.1016/j.tins.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Twomey EC, Sobolevsky AI. Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry. 2018;57:267–276. doi: 10.1021/acs.biochem.7b00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greger IH, Mayer ML. Structural biology of glutamate receptor ion channels: towards an understanding of mechanism. Curr Opin Struct Biol. 2019;57:185–195. doi: 10.1016/j.sbi.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki M, Araki K, Shibata A, Mishina M. Molecular cloning of a cDNA encoding a novel member of the mouse glutamate receptor channel family. Biochem Biophys Res Commun. 1992;183:886–892. doi: 10.1016/0006-291x(92)90566-4. [DOI] [PubMed] [Google Scholar]
- 7.Lomeli H, et al. The rat delta-1 and delta-2 subunits extend the excitatory amino acid receptor family. FEBS Lett. 1993;315:318–322. doi: 10.1016/0014-5793(93)81186-4. [DOI] [PubMed] [Google Scholar]
- 8.Schmid SM, Hollmann M. To gate or not to gate: are the delta subunits in the glutamate receptor family functional ion channels? Mol Neurobiol. 2008;37:126–141. doi: 10.1007/s12035-008-8025-0. [DOI] [PubMed] [Google Scholar]
- 9.Hirai H, et al. Rescue of abnormal phenotypes of the delta2 glutamate receptor-null mice by mutant delta2 transgenes. EMBO Rep. 2005;6:90–95. doi: 10.1038/sj.embor.7400312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuzaki M. New insights into the structure and function of glutamate receptors: the orphan receptor delta2 reveals its family’s secrets. Keio J Med. 2003;52:92–99. doi: 10.2302/kjm.52.92. [DOI] [PubMed] [Google Scholar]
- 11.Kakegawa W, Kohda K, Yuzaki M. The delta2 ‘ionotropic’ glutamate receptor functions as a non-ionotropic receptor to control cerebellar synaptic plasticity. J Physiol Lond. 2007;584:89–96. doi: 10.1113/jphysiol.2007.141291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohda K, et al. The δ 2 glutamate receptor gates long-term depression by coordinating interactions between two AMPA receptor phosphorylation sites. Proc Natl Acad Sci USA. 2013;110:E948–957. doi: 10.1073/pnas.1218380110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav R, et al. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS ONE. 2012;7:e32969. doi: 10.1371/journal.pone.0032969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo T, Kakegawa W, Yuzaki M. Induction of long-term depression and phosphorylation of the delta2 glutamate receptor by protein kinase C in cerebellar slices. Eur J Neurosci. 2005;22:1817–1820. doi: 10.1111/j.1460-9568.2005.04319.x. [DOI] [PubMed] [Google Scholar]
- 15.Utine GE, et al. A homozygous deletion in GRID2 causes a human phenotype with cerebellar ataxia and atrophy. J Child Neurol. 2013;28:926–932. doi: 10.1177/0883073813484967. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi Y, et al. A new mouse allele of glutamate receptor delta 2 with cerebellar atrophy and progressive ataxia. PLoS ONE. 2014;9:e107867. doi: 10.1371/journal.pone.0107867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Schil K, et al. Early-onset autosomal recessive cerebellar ataxia associated with retinal dystrophy: new human hotfoot phenotype caused by homozygous GRID2 deletion. Genet Med. 2015;17:291–299. doi: 10.1038/gim.2014.95. [DOI] [PubMed] [Google Scholar]
- 18.Guo S-Z, et al. A case-control association study between the GRID1 gene and schizophrenia in the Chinese Northern Han population. Schizophr Res. 2007;93:385–390. doi: 10.1016/j.schres.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Benamer N, et al. GluD1, linked to schizophrenia, controls the burst firing of dopamine neurons. Mol Psychiatry. 2018;23:691–700. doi: 10.1038/mp.2017.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Gandhi PJ, Pavuluri R, Shelkar GP, Dravid SM. Glutamate delta-1 receptor regulates cocaine-induced plasticity in the nucleus accumbens. Transl Psychiatry. 2018;8:219. doi: 10.1038/s41398-018-0273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uemura T, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 22.Lee S-J, Uemura T, Yoshida T, Mishina M. GluRδ2 assembles four neurexins into trans-synaptic triad to trigger synapse formation. J Neurosci. 2012;32:4688–4701. doi: 10.1523/JNEUROSCI.5584-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao W, Díaz-Alonso J, Sheng N, Nicoll RA. Postsynaptic δ1 glutamate receptor assembles and maintains hippocampal synapses via Cbln2 and neurexin. Proc Natl Acad Sci USA. 2018;115:E5373–E5381. doi: 10.1073/pnas.1802737115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yasumura M, et al. Glutamate receptor δ1 induces preferentially inhibitory presynaptic differentiation of cortical neurons by interacting with neurexins through cerebellin precursor protein subtypes. J Neurochem. 2012;121:705–716. doi: 10.1111/j.1471-4159.2011.07631.x. [DOI] [PubMed] [Google Scholar]
- 25.Elegheert J, et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science. 2016;353:295–299. doi: 10.1126/science.aae0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uemura T, Mori H, Mishina M. Direct interaction of GluRdelta2 with Shank scaffold proteins in cerebellar Purkinje cells. Mol Cell Neurosci. 2004;26:330–341. doi: 10.1016/j.mcn.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi T, et al. Control of synaptic connection by glutamate receptor delta2 in the adult cerebellum. J Neurosci. 2005;25:2146–2156. doi: 10.1523/JNEUROSCI.4740-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wollmuth LP, et al. The Lurcher mutation identifies delta 2 as an AMPA/kainate receptor-like channel that is potentiated by Ca(2+) J Neurosci. 2000;20:5973–5980. doi: 10.1523/JNEUROSCI.20-16-05973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeno K, Yamakura T, Yamazaki M, Sakimura K. The Lurcher mutation reveals Ca(2+) permeability and PKC modification of the GluRdelta channels. Neurosci Res. 2001;41:193–200. doi: 10.1016/s0168-0102(01)00277-2. [DOI] [PubMed] [Google Scholar]
- 30.Yadav R, Rimerman R, Scofield MA, Dravid SM. Mutations in the transmembrane domain M3 generate spontaneously open orphan glutamate δ1 receptor. Brain Res. 2011;1382:1–8. doi: 10.1016/j.brainres.2010.12.086. [DOI] [PubMed] [Google Scholar]
- 31.Schmid SM, Kott S, Sager C, Huelsken T, Hollmann M. The glutamate receptor subunit delta2 is capable of gating its intrinsic ion channel as revealed by ligand binding domain transplantation. Proc Natl Acad Sci USA. 2009;106:10320–10325. doi: 10.1073/pnas.0900329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orth A, Tapken D, Hollmann M. The delta subfamily of glutamate receptors: characterization of receptor chimeras and mutants. Eur J Neurosci. 2013;37:1620–1630. doi: 10.1111/ejn.12193. [DOI] [PubMed] [Google Scholar]
- 33.Naur P, et al. Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci USA. 2007;104:14116–14121. doi: 10.1073/pnas.0703718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen KB, et al. Modulation of the dimer interface at ionotropic glutamate-like receptor delta2 by D-serine and extracellular calcium. J Neurosci. 2009;29:907–917. doi: 10.1523/JNEUROSCI.4081-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kristensen AS, et al. Pharmacology and Structural Analysis of Ligand Binding to the Orthosteric Site of Glutamate-Like GluD2 Receptors. Mol Pharmacol. 2016;89:253–262. doi: 10.1124/mol.115.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perroy J, et al. Direct Interaction Enables Cross-talk between Ionotropic and Group I Metabotropic Glutamate Receptors. J Biol Chem. 2008;283:6799–6805. doi: 10.1074/jbc.M705661200. [DOI] [PubMed] [Google Scholar]
- 37.Suryavanshi PS, et al. Glutamate delta-1 receptor regulates metabotropic glutamate receptor 5 signaling in the hippocampus. Mol Pharmacol. 2016 doi: 10.1124/mol.116.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ady V, et al. Type 1 metabotropic glutamate receptors (mGlu1) trigger the gating of GluD2 delta glutamate receptors. EMBO Rep. 2014;15:103–109. doi: 10.1002/embr.201337371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dadak S, et al. mGlu1 receptor canonical signaling pathway contributes to the opening of the orphan GluD2 receptor. Neuropharmacology. 2017;115:92–99. doi: 10.1016/j.neuropharm.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–81. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 41.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao W, et al. Mechanisms underlying the synaptic trafficking of the glutamate delta receptor GluD1. Molecular Psychiatry. 2019:1. doi: 10.1038/s41380-019-0378-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–9. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 45.Dürr KL, et al. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyerson JR, et al. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jalali-Yazdi F, Chowdhury S, Yoshioka C, Gouaux E. Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor. Cell. 2018;175:1520–1532.e15. doi: 10.1016/j.cell.2018.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y, Chen S, Swensen AC, Qian W-J, Gouaux E. Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science. 2019;364:355–362. doi: 10.1126/science.aaw8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuda K, et al. Transsynaptic Modulation of Kainate Receptor Functions by C1q-like Proteins. Neuron. 2016;90:752–767. doi: 10.1016/j.neuron.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Chen S, et al. Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell. 2017;170:1234–1246.e14. doi: 10.1016/j.cell.2017.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron. 2017;94:569–580.e5. doi: 10.1016/j.neuron.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tapken D, et al. The low binding affinity of D-serine at the ionotropic glutamate receptor GluD2 can be attributed to the hinge region. Sci Rep. 2017;7 doi: 10.1038/srep46145. 46145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shanks NF, Maruo T, Farina AN, Ellisman MH, Nakagawa T. Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci. 2010;30:2728–40. doi: 10.1523/JNEUROSCI.5146-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao H, et al. Preferential assembly of heteromeric kainate and AMPA receptor amino terminal domains. Elife. 2017;6 doi: 10.7554/eLife.32056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao H, et al. Analysis of high-affinity assembly for AMPA receptor amino-terminal domains. J Gen Physiol. 2013;141:747–749. doi: 10.1085/jgp.20121077004292013c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaudhry C, Plested AJ, Schuck P, Mayer ML. Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions. Proc Natl Acad Sci U S A. 2009;106:12329–34. doi: 10.1073/pnas.0904175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng S, Seven AB, Wang J, Skiniotis G, Özkan E. Conformational Plasticity in the Transsynaptic Neurexin-Cerebellin-Glutamate Receptor Adhesion Complex. Structure. 2016;24:2163–2173. doi: 10.1016/j.str.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song X, et al. Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature. 2018;556:515–519. doi: 10.1038/s41586-018-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuda K, et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- 61.Kandiah E, et al. CM01: a facility for cryo-electron microscopy at the European Synchrotron. Acta Crystallogr D Struct Biol. 2019;75:528–535. doi: 10.1107/S2059798319006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 65.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Kidmose RT, et al. Namdinator – automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ. 2019;6:526–531. doi: 10.1107/S2052252519007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schrödiner LLC. The PyMOL Molecular Graphics System, Version 1.3. DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.