Abstract
The complex relationship between metabolic disease risk and body fat distribution in humans involves cellular characteristics which are specific to body fat compartments. Here we show depot-specific differences in the stromal vascual fraction of visceral and subcutaneous adipose tissue by performing single-cell RNA sequencing of tissue specimen from obese individuals. We characterize multiple immune cells, endothelial cells, fibroblasts, adipose and hematopoietic stem cell progenitors. Subpopulations of adipose-resident immune cells are metabolically active and associated with metabolic disease status and those include a population of potential dysfunctional CD8+ T cells expressing metallothioneins. We identify multiple types of adipocyte progenitors that are common across depots, including a subtype enriched in individuals with type 2 diabetes. Depot-specific analysis reveals a class of adipocyte progenitors unique to visceral adipose tissue, which shares common features with beige preadipocytes. Our human single-cell transcriptome atlas across fat depots provides a resource to dissect functional genomics of metabolic disease.
Background
White adipose tissue (WAT) and its endocrine activities are known to be implicated in the development of obesity and associated metabolic disorders. Specifically, the risk increases with increase in abdominal obesity contributed by excessive visceral adipose tissue (VAT)1 – a linear relationship that is not seen with abdominal subcutaneous adipose tissue (SAT)2. Susceptibility to obesity-related cardiovascular and metabolic disorders has also been linked with the increase in adipose volume resulting from enlargement of tissue resident adipocytes (i.e. hypertrophy)3. On the other hand, adipocyte expansion by recruiting new progenitors (hyperplasia) is often considered as a protective mechanism from the metabolic standpoint4. Studies have also shown that adipose tissue dysfunction leading to insulin resistant type 2 diabetes (T2D) is marked by inflammation, fibrosis and / or lipodystrophy5 which emphasizes the importance of adipose-infiltrating immune cell populations in modulating and developing metabolic disorders. For instance, M1 macrophages, mast cells, B-2 cells, CD8+ T cells and IFN-γ+ Th1 cells were seen to be increased in adipose tissue of individuals with obesity compared with those who were normal weight and the reverse pattern was observed in M2 macrophages, eosinophils, Treg, iNKT, B1 and γδ T cells6. These adipose tissue resident immune cells have also been shown to create a microenvironment that can inhibit adipocyte progenitor differentiation to lipid-storing adipocytes7. However, despite extensive work on characterizing various cell subpopulation in adipose tissue, the complete human non-adipocyte fraction also known as the stromal vascular fraction (SVF) has not been profiled across depots in an unbiased manner. Given the multitude of factors affecting adipose tissue function, a thorough understanding of the cell types involved, and their specific gene expression pattern is essential. The advent of single-cell transcriptomic approaches in the past years have made it possible to use these technologies to determine cellular heterogeneity and functional states at the single-cell level with high reproducibility and sensitivity8. Current high-throughput microfluidics techniques are capturing thousands of cells from each sample simultaneously for gene expression profiling and together with new algorithms for clustering, visualization, and modeling this allows for high-powered analysis of disease-targeted tissue samples for efficient cataloging of cellular composition and the role in disease risk. Recent studies utilizing single-cell RNA sequencing (scRNA-Seq) in adipose tissue from mouse models have identified a subset of adipocyte progenitors that regulates adipocyte differentiation9 as well as the presence of a novel type of inflammatory progenitors residing in the visceral fat depot of the mice10. Similar strategies in human adipose samples have not been applied to date.
We present a high-throughput single-cell expression profiling study of human adipose tissue including 25 samples derived from multiple depots of individuals with obesity. We provide a rich catalog of cell types residing in adipose tissue including both latent and common cell populations. We characterize and validate distinct cell types that are metabolically active, specific to each depot or correlate with metabolic disease status.
Results
Characterization of SVF across multiple adipose depots
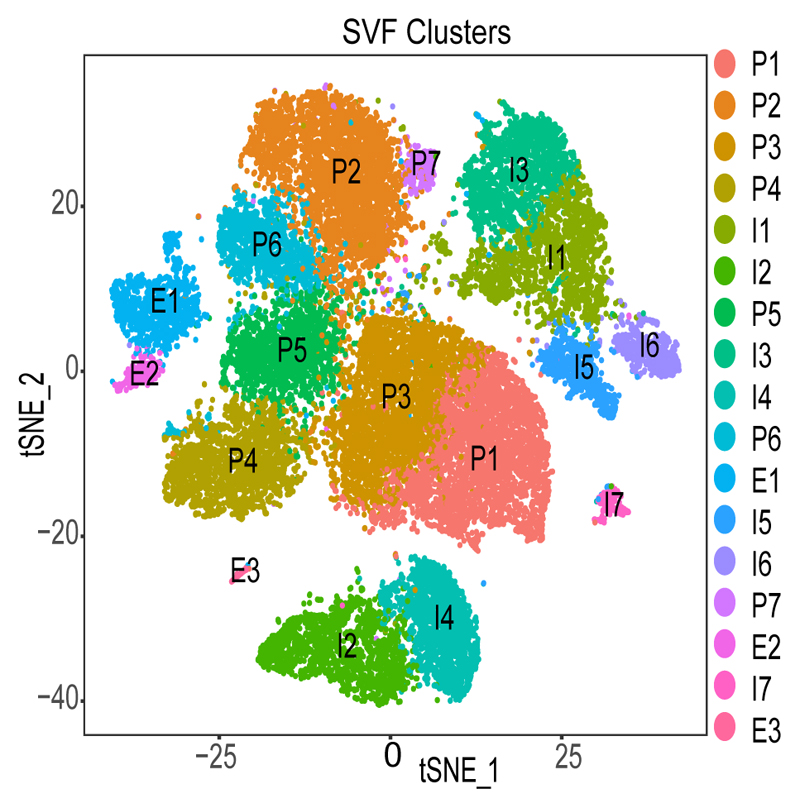
We generated scRNA-Seq data from 25 adipose samples (12 VAT and 13 SAT) derived from 14 individuals undergoing bariatric surgery (Supplementary Table 1, Supplementary Figure 1, Methods). All samples were matched for age and BMI but differed based on fasting glycemia as an indication of T2D (Table 1). We annotated the clusters using marker genes (Supplementary Table 2-3) which resulted in three groups of cells: adipocyte progenitors and stem cells (P1-P7), immune cells (I1-I7) and endothelial cells (E1-E3) (Figue 1). The proportion of the cell types based on individual average was 55%, 37% and 8% for progenitors, immune and endothelial cells, respectively. This distribution was similar for the merged sample average and corresponded to 60%, 34% and 6% which are in line with classical fluorescence-activated cell sorting (FACS) experiments 11. To avoid subjectivity and to add strength to analyses, we also performed reference-based single-cell annotation (Methods) which confirmed our broad clusters of cell populations (Supplementary Table 4). Finally, as the distribution of female and male samples was slightly skewed (Table 1) we performed in-depth sex-specific expression analysis of the marker genes in 245 (N=165 male and N=80 female) VAT and SAT samples, respectively, from the GTEx Consortium (Supplementary Table 5).
Table 1. Characteristics of the study subjects. P values are calculated used two sided ttest.
| T2D | non-T2D | ||
|---|---|---|---|
| N | 5 | 9 | |
| Male:Female (N) | 2:03 | 2:07 | |
| mean ± SD | mean ± SD | P-value | |
| Age (years) | 52.8±12.5 | 43.3±11.4 | 0.2 |
| BMI (kg/m2) | 41.0±6.0 | 43.5±7.2 | 0.49 |
| Fasting glycemia (mmol/L) | 10.7±1.3 | 5.57±0.5 | 4.97E-04 |
| Percentage of glycated hemoglobin | 0.08±0.005 | 0.05±0.002 | 1.61E-04 |
Figure 1. Identified cell populations in the non-adipocyte fraction of adipose tissue.
Clustering results of 26,350 cells from the stromal vascular fraction (SVF) derived from 25 adipose samples that underwent single-cell RNA sequencing identifying 17 clusters. Cell populations were classified as Progenitors (P), Immune cells (I) and Endothelial cells (E) and labelled accordingly.
Multiple types of endothelial cells reside in adipose tissue
We identified three types of endothelial cells (E1-E3, Figure 1) all showing selective expression for GNG11 and SEPW1. E1 and E2, were found to be relatively similar sharing nearly half of the top expressed genes including SPARCL1, FABP4 and IFI27. However, comparative analysis between E1 and E2 cells showed the latter group expressing classical endothelial markers such as ACKR1, SELE, TM4SF1, VCAM1, TMEM173, PLVAP, ICAM1, PECAM1, VWF, ADAMTS9 and TFPI. On the other hand, E1 cells had pronounced expression of FABP4, LGALS1, RBP7, GPX3 and CD36 which are known to be expressed in microvascular endothelial cells of adipose tissue involved in the fatty acid handling machinery12. To characterize E3, we performed differential gene expression analysis between E3 and the combined population of E1 and E2 cells, respectively, (Supplementary Table 6) and found LYVE1 among the top expressed genes. LYVE1 is a marker of lymphatic endothelial cells 13 and thus these results indicate the presence of lymphatic vasculature in SVF with 78% of its cell population from VAT samples.
Characterization of Immune cells and their link to adipose tissue inflammation
We found that 34% of cells from our merged clusters (Figure 1, I1-I7; Supplementary Table 4) expressed markers of different immune cell populations. To facilitate the classification of the cell types we subset the immune cell clusters and identified 14 new clusters, labelled as IS1 - IS14, where all groups were present across depots (Figure 2a-b).
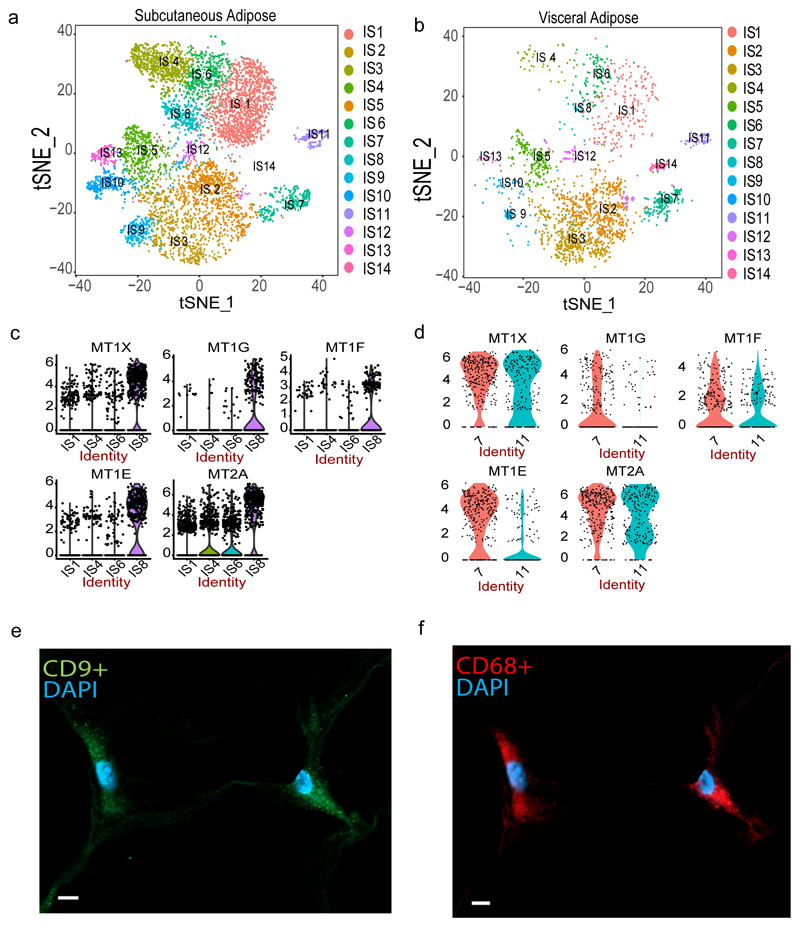
Figure 2. SVF-derived immune cells.
Re-clustering of 9,025 CD34- cells representing immune cells in the stromal vascular fraction (SVF) of adipose tissue identified 14 cell types including T/NK cells (IS1, IS4, IS6, IS8), macrophages (IS2, IS3, IS7, IS9, IS12), dendritic cells (IS5, IS13), monocyte (IS10) and B-cells (IS11) in SAT (a) and VAT (b). Violin plots of expression density of Metallothionein genes across T / NK cell clusters in discovery (c) and CD34- validation samples (d) comprising 25 and 3 samples respectively. The y axis indicate log transformed expression values and the width indicate number of cells expressing the particular gene. Immunohistochemistry in subcutaneous adipose derived from a 60 year-old woman with a BMI of 52.2 kg/m2 showing co-expression of CD9 (e) and CD68 (f) cells. The scale bar indicates 5 μm. The staining was done on 4 independent individuals to confirm.
A large proportion (40%) of the immune cells (IS1, IS4, IS6, IS8) were clustered closely together and showed gene expression signatures of NK / T cells. IS1 exhibited selective expression of IL7R which is known to be expressed in naive T cells14. However, unsupervised cell type definition pointed towards a mix of naïve and memory CD4+ and CD8+ T cells (Supplementary Table 7). Manual annotation of IS4 cells identified specific expression of GNLY, NKG7, FGFBP2, GZMB, GZMH and CTSW which points toward these cells being NK cells (Supplementary Table 8). Indeed, this was supported by unsupervised annotation which showed 68% of IS4 cells being correlated with the NK cell reference. A recent report found that obesity is associated with reprogramming of blood-derived NK cells with upregulation of genes involved in lipid metabolism including CD3615. However, the IS4 NK cells identified here did not express any genes involved in lipid metabolism pointing towards different obesity-related effect on NK cells dependent on resident tissue.
IS6 showed increased expression of CCL5 and IL32, indicating that these cells may be activated T cells 16 which was confirmed by the unsupervised annotation revealing 68% of the cells being associated with memory CD8+ T cells. While large proportion (59%) of the IS8 cells were, similar to IS6 cells, mapped as memory CD8+ T cells in our unsupervised annotation we noted in the manual curation that they showed a unique pattern of high expression of metallothionein genes – MT1E, MT1F, MT1G, MT1X and MT2A (Supplementary Table 8, Figure 2c). We followed-up this finding by performing additional scRNA-Seq profiling on CD34- sorted SVF cells and found evidence of the presence of metallothionein-rich T cells also in the validation sample (Cluster 7; Supplementary Table 9; Figure 2d) where both discovery and validation samples specifically showed MT1F and MT1G to be uniquely expressed by this T-cell subpopulation (Figure 2c-d). Similar to what was recently shown for circulating immune cells15, these potential adipose-resident dysfunctional T cells may be induced by the obesity and thus enriched in our scRNA study. To test this, we used bulk gene expression data from two large SAT population-based collections including 1) 776 female samples from the MuTHER resource and 2) 770 male samples from the METSIM Study where both studies have in addition to tissue collection, study subjects deeply phenotyped for obesity-related traits 17,18. Here, we used BMI and DXA-derived % Trunk fat (PTF) in association with the expression of MT1E, MT1F, MT1G, MT1X and MT2A (Supplementary Table 10). Encouragingly, we found consistent strong positive associations between the obesity-traits and expression of MT1F, MT1G and MT2A whereas MT1E and MT1X showed weaker or absence of significant association in line with our observations of cell-specific expression of these two genes. Finally, we found that all metallothionein genes are expressed in adipose tissue irrespective of tissue depot with no sex-specific expression pattern (Supplementary Figure 2, Supplementary Table 5).
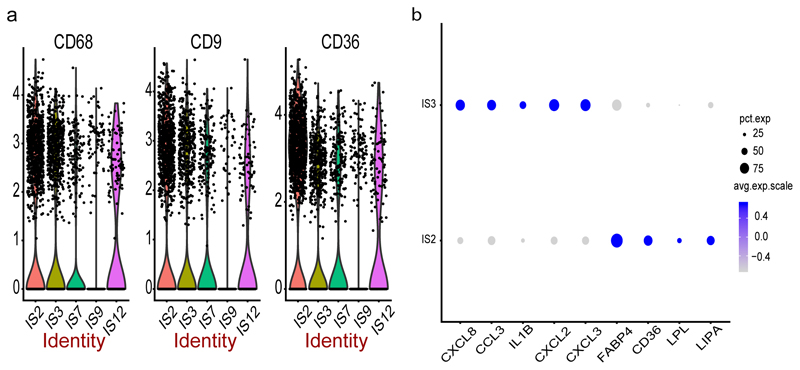
Our manual and/or unsupervised annotation mapped clusters IS2,IS3, IS7, IS9 and IS12 to adipose tissue macrophages (ATM) with the proportion of cells expressing the classical macrophage marker CD68 ranging from 19 to 52% (Extended Data 1a) with IS2 having the highest (52%) and IS9 the lowest (19%). IS2 cells were also enriched with genes that are involved in lipid metabolism during obesity (Supplementary Table 8) including LIPA, LPL, CD36 and FABP4 – findings in line with the notion that obesity activates a non-classical inflammatory phenotype of ATM involving lipid accumulation and trafficking19. In addition, we find these metabolically active ATMs (i.e. IS2 cells) to express CD9 more pronounced than the other ATMs (Extended Data 1a). We performed cellular phenotyping by immunohistochemistry and validated the co-expression of CD68 and CD9 in obese adipose tissue (Figure 2e-f) as well as confirmed 17,18 the linear relationship of CD9 expression and BMI in the MuTHER and METSIM SAT cohorts, respectively (MuTHER BMI: p-value=2.2E-13, beta=0.011; METSIM BMI: p-value=9.4E-28, beta=0.379).
IS12 and IS3 clusters showed a similar signature as IS2 but where IS3 cells had increased expression for inflammatory genes CXCL3, CXCL2, IL1B, CCL3 and CXCL8 (Extended Data 1b). On the other hand, IS9 cells are marked by the expression of FOLR2 and KLF4 which are known signatures of M2 macrophages 20,21. Finally, although our unsupervised annotation indicated IS12 cells as macrophages we could not distinguish them further.
Our unsupervised annotation identified IS5 and IS10 as classical CD16- monocytes with high expression of S100A8, S100A9 and S100A12 in IS10. However, further literature mining showed that IS5 cells are rather dendritic cells due to their high expression of HLA genes including HLA-DPB1, HLA-DQA1, HLA-DPA1, HLA-DRA and HLA-DQB1. IS13 cells possessed signatures of a newly identified subtype of dendritic cells 22 with high expression of LST1, SERPINA1, AIF1 and FCGR3A.
Finally, we were able to link the IS11 cluster to B cells due to the expression of IGKC, JCHAIN, CD79A and CD37. Remaining cluster (IS14) showed mixed signatures in both annotation strategies and thus was not explored further.
Identification of adipocyte progenitors derived from different adipose tissue depots
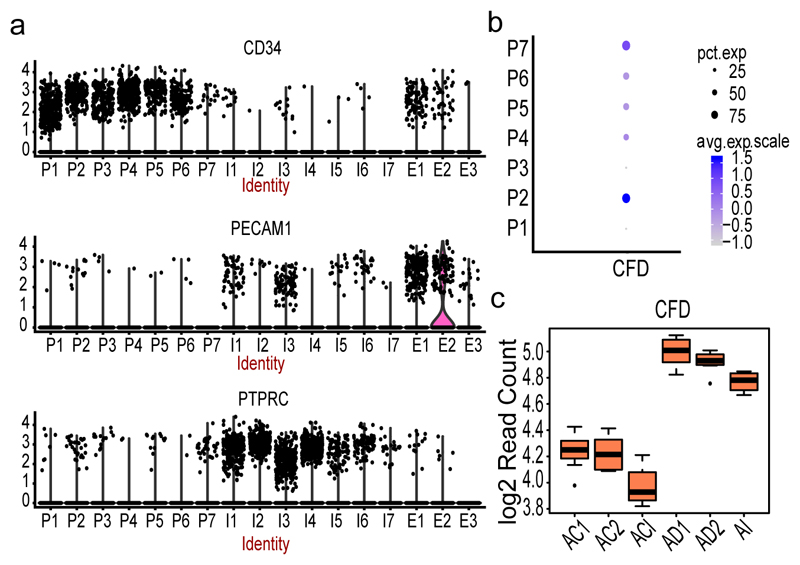
Next, we characterized our progenitor clusters (P1-P7) by studying the expression pattern of cell surface makers used in FACS experiment of SVF progenitors23 and noted that all clusters appeared CD45− CD34+ CD31− (Figure 3a) with a profound signature in P1-P6. We also noted that five of the clusters (P2, P4-P7) were identified to express CFD24 encoding the adipokine Adipsin which is known as a marker of adipocyte differentiation (Figure 3b) and thus the range of CFD expression observed across the clusters may be linked to different stages of adipogenesis. To validate this, we used in vitro differentiation studies of human mesenchymal stem cells (MSC) and identified a 5.6 log2 fold induction between undifferentiated MSCs and the first stage of adipogenic induction (ACI vs AI, Adjusted p-value=1.19E-26, Figure 3c). We further noted that CFD expression is additionally increased at later stages of adipogenesis (AI vs AD1, log2fold=1.3, Adjusted p-value=0.012; AI vs AD2, log2fold=1.2, Adjusted p-value=0.036). Incorporating this information with our progenitor signature indicated that P2 and P7 include more mature preadipocytes compared to cells within clusters P4-P6. We could further validate this by unsupervised annotation which linked on average 88% of the cells in P2 and P7 to Adipocytes compared to only on average 70% of cells in P4-P6 (Supplementary Table 11).
Figure 3. SVF-derived progenitor clusters.
(a) Violin plots of log transformed expression density of CD34 (upper panel), CD31/PECAM1 (middle panel) and CD45/PTPRC (bottom panel) across all SVF clusters from 25 samples. The width of the violin plot indicate number of cells expressing the particular gene. (b) Dot plot of the expression of CFD across progenitor (P) clusters. The size of the dot corresponds to the percentage of cells expressing CFD in each cluster and the color represents the average CFD expression level (c) Box plot of CFD expression at 4 different time points of adipocyte differentiation from mesenchymal stem cells (MSCs, n= 8 individuals). AI corresponds to the first time point three days after culturing MSCs in the induction media. AD1 and AD2 are 1 and 2 weeks of differentiation in adipogenic media after the AI time point. ACI, AC1 and AC2 are corresponding control sets for AI, AD1 and AD2 without any adipocyte differentiation treatments. Each time point with corresponding control includes three independent MSC cultures and shown by average log 2 read count. The black line inside the boxplot represent and median value and the size of the box is determined by the 25th and 75th percentile of the data. The wiskers of box plot represents the maximum and minimum values of the data shown.
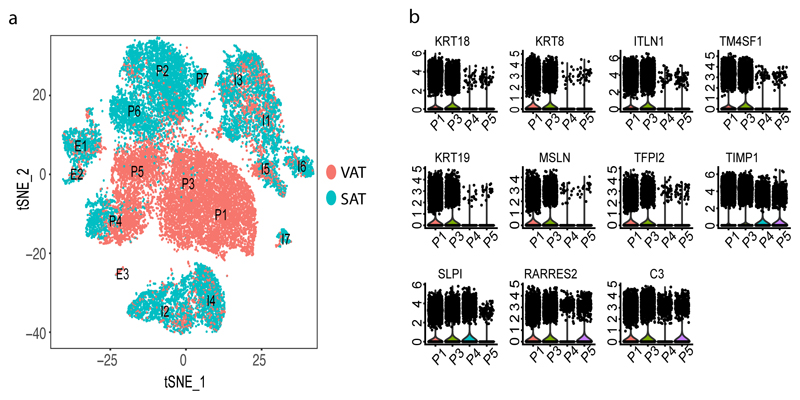
We then compared the seven progenitor clusters based on tissue type and found a striking depot-specific pattern (Figure 4a) where three of the clusters (P2, P6, P7) were mainly composed of cells from SAT whereas P1, P3, P5 included mostly cells from VAT and only one cluster of progenitors (P4) included a mixed population of SAT- and VAT-derived cells. For instance, the VAT-specific clusters P1 and P3 exhibited pronounced expression (average log fold change > 1.5) of omentin (ITLN1)25 and mesothelin (MSLN). Omentin is a VAT-specific adipocytokine serving as a biomarker for metabolic diseases26 and together with MSLN also a marker of mesothelial cells27 in line with findings that VAT develops from the mesothelium28. Additionally, we identified 16 other genes characterizing P1 and P3 (average log fold change >1, Supplementary Table 2) which we followed-up with bulk RNA-Seq and found 11/16 (68%) genes being differentially expressed (Adjusted p-value < 0.05 and log fold change >= 1) (Supplementary Table 12, Figure 4b).
Figure 4. Main cell clusters in SVF based on depot.
(a) Clustering results of all SVF samples that underwent single-cell RNA sequencing which identified 17 clusters from 26,350 cells. Clusters were classified as Progenitors (P), Immune cells (I) and Endothelial cells (E) and labelled accordingly. Cells are labelled as VAT (red)- or SAT (blue)-derived, respectively. (b)Violin plots of expression density of 11 genes that are specific to VAT progenitor clusters. The y axis indicate log transformed expression values and the width indicate number of cells expressing the particular gene.
To study these cell populations in more detail, we created two subsets for re-clustering including 1) SAT- (N=5,458) and VAT-derived (N=9,847) progenitor cells, respectively.
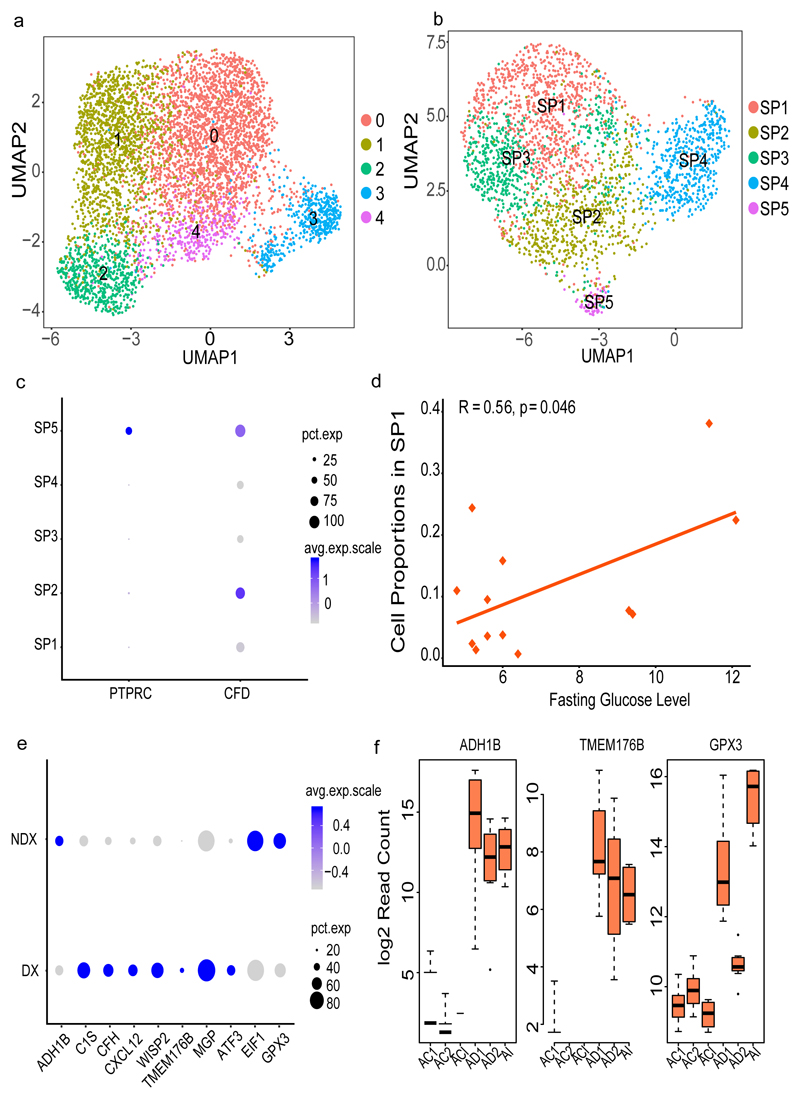
Subcutaneous adipocyte progenitors and their role in metabolic conditions
The clustering of the adipocyte progenitors identified five groups (SP1-SP5, Figure 5a-5b) all expressing CFD. SP1 and SP3 cells expressed pre-adipocyte/adipose stem cell markers (e.g. MGP, APOD, CXCL14, WISP2) whereas SP2 showed signatures of a more mature adipocyte progenitor cell (e.g. APOE, FABP4, CEBPB and CD36). The SP4 group contained cells expressing genes involved in fibrosis and extra cellular matrix accumulation including COL3A1, COL6A3, COL1A1 and COL6A1. This fibroblast signature was validated by unsupervised annotation where 69% of the cells were mapped as fibroblasts compared to SP1-SP3 with over 80% of the cells mapped as adipocytes (Supplementary Table 13). SP5 cells expressed high levels of inflammatory markers (e.g. CCL5, CD3E, IL7R and IL32) including CD45/PTPRC (Figure 5c) indicating that SP5 cells represent hematopoietic stem cell (HSC) progenitors.
Figure 5. Progenitor clusters specific to SAT.
(a) Re-clustering of SAT-specific progenitors from the complete sample set of 26,350 cells (b) Re-clustering of SAT-specific progenitors (2,705 cells) by subsampling the high coverage library to 1500 cells (c) Dot plot of the expression of PTPRC (CD45) and CFD across SAT progenitors (SP1-SP5). (d) Pearson correlation of fasting glucose levels (mmol/L) and SP1 proportion across all samples obtained from 13 individuals (e) Dot plot of differentially expressed SP1 genes in cells derived from T2D (DX) versus non-T2D (NDX) samples previously validated in the MuTHER study (f) Box plot of T2D-associated SP1 gene expression at different time points of adipocyte differentiation from mesenchymal stem cells (MSCs, n= 8 individuals). AI corresponds to the first time point three days after culturing MSCs in the induction media. AD1 and AD2 are 1 and 2 weeks of differentiation in adipogenic media after the AI timepoint. ACI, AC1 and AC2 are corresponding control sets for AI, AD1 and AD2 without any adipocyte differentiation treatments. Each timepoint with corresponding control includes three independent MSC cultures and shown by average log 2 read count and error bars corresponding to standard deviation. The black line inside the boxplot represent and median value and the size of the box is determined by the 25th and 75th percentile of the data. The wiskers of box plot represents the maximum and minimum values of the data shown. The size of the dot in (c) and (e) corresponds to the percentage of cells expressing the genes in each cluster and the color represents the average expression level.
We then investigated whether the proportion of adipocyte progenitor clusters specific to SAT in each individual sample was associated with T2D status. We correlated fasting glucose levels with all the SP cell proportions and found the abundance of SP1 to be significantly correlated with glucose levels (Pearson r=0.56, p-value=0.046, Figure 5d, Supplementary Table 14). We followed-up this finding in multiple ways and provide further evidence of the link between SP1 cells and T2D. First, we used top differently expressed genes in this group (SP1, N=43, Adjusted p-value < 0.01) compared to all other cell types (SP2-SP5; Supplementary Table 15) for disease annotation (Methods) and found T2D as the top annotated disease (Supplementary Tables S16, p-value=3.21E-6). Secondly, we used the MuTHER resource 17 followed by the METSIM Study18 for transcription-wide association analysis of multiple T2D-related traits. We found the SP1 gene set (N=43) to be significantly enriched for association to the T2D traits in both studies (Supplementary Table 17) corresponding to fold changes between 2.5 and 9.4 dependent on the trait and study (Fisher’s p-value = 0.06 – 7.96E-08, Supplementary Table 18). Specifically, the female MuTHER Study included enriched association for 44% of the SP1 genes using strict Bonferroni corrected study-wise p-value threshold of p<2.53E-6. Replication of these genes in the male METSIM Study showed strong correlation of effect sizes across T2D traits (r=0.92, Supplementary Figure 3).
Finally, we went back to our single-cell data set and performed differential expression analysis between SP cells from T2D vs non-T2D samples as defined by their fasting glycemia restricting to the 19 validated SP1-specific genes (Supplementary Table 17-18). Here, we found 53% of the genes (10/19) being also differently expressed (Bonferroni p-value < 0.05/19, Figure 5e) in cells based on T2D status with the same direction of effect as seen in MuTHER (Supplementary Table 19). These genes include GPX3, WISP2, ATF3, EIF1, MGP, TMEM176B, CXCL12, CFH, C1S and ADH1B29. The top differentially expressed gene was GPX3 with significantly higher proportion of cells expressing the gene in non-T2D samples (73%) versus in T2D samples (66%) (Seurat DGE, p-value=1.5.9E-14). In line with these findings, we found GPX3 to be negatively correlated with Homeostatic Model Assessment for Insulin Resistance (HOMA-IR) in both the MuTHER (beta=-0.084, p-value=1.03E-11) and METSIM cohort (beta=-0.147, p-value=4.46E-5), respectively.
We also noted WISP2 among those top 10 genes with differential expression between T2D and non-T2D cells (p-value=3.1E-5) but as opposed to GPX3, WISP2 seems to have a negative effect on T2D status. We found 72% of T2D cells expressing the genes versus only 52% of the non-T2D cells. Again, we validated this pattern in the two large MuTHER and METSIM adipose tissue cohorts with a positive correlation with HOMA-IR (beta=0.115, p-value=2.2E-20 and beta=0.132, p-value=2.3E-4). Similar to WISP2, we found that T2D cells expressed ATF3 to a larger extent (51%) compared to non-T2D cells (29%) and ATF3 was positively correlated to HOMA-IR in MuTHER (beta=0.079, p-value=2.9E-21) and METSIM (beta=0.102, p-value=4.4E-3),respectively.
Validation of depot independent adipocyte progenitors in CD34+ sorted cells
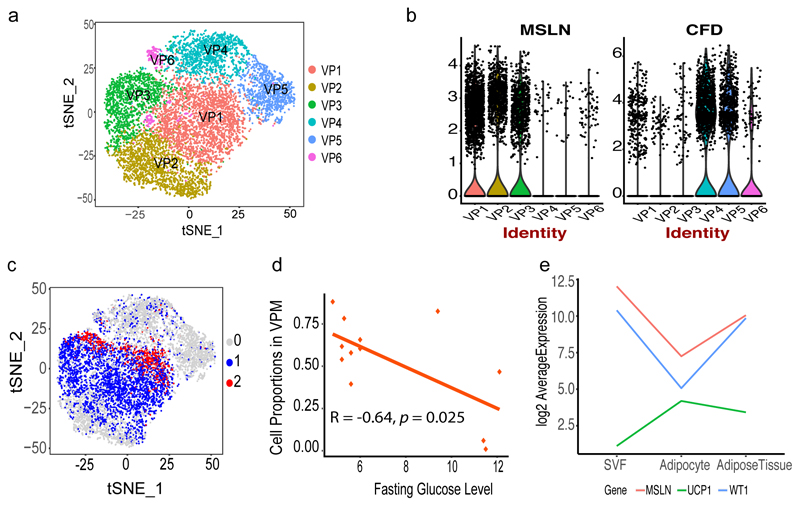
Clustering of the VAT-derived progenitor cells resulted in six clusters (Figure 6a, Supplementary Table 20) with cells expressing CFD (referred to as VPC and correspond to VP4-6) versus MSLN (referred to as VPM and correspond to VP1-3) were clustered separately (Figure 6b). The VPCs (VP4-6) resembled the progenitor types identified in SAT and thus may represent adipocyte progenitors that are present across depots. Specifically, VP4 included cells enriched for genes such as APOD, CXCL14, DPT, GPX3, MGP, EIF1, C1S and ADH1B and thus with a similar expression pattern as the T2D-linked SP1 cells. VP5 cells show an enrichment for MFAP5 which in turn is involved in adipose tissue remodeling in individuals with obesity 30. Another notable gene expressed in cluster VP5 was S100A4, a biomarker for inhibition of adipogenesis, associated with reduced obesity and inflammation in rodents 31. In all, the expression signature of VP5 was similar to the fibrotic signatures of the cells grouped as SP4. Finally, VP6 is marked by the expression of TYROBP, HLA-DRA and CD74 and in general have the inflammatory signature identified for the HSC progenitors in SP5. Taken together, the combined results from our SAT and VAT single-cell data indicate presence of three depot-independent cell types: T2D-assocaited adipocyte progenitors, fibroblasts and HPC, respectively. To provide further support to these observations, we generated scRNA-Seq profiles of CD34+ sorted SVF cells (Methods). The clustering of these cells resulted in thirteen groups (0-12, Supplementary Figure 4, Supplementary Table 21) with clear evidence for overlapping fibrotic and HPC properties of two of the clusters. Specifically, we linked cluster 10 (134 cells) to HPC progenitors which corresponded to 2% of the complete cell population concurring with the proportion of cells in the SP5 cluster (3%) from the deep sequenced sample when restricting to progenitors only. The expression signature of this cluster also resembles that of SP5 with PTPRC, CCL5 and IL32 among the top genes distinguishing these CD34+ progenitors from the other. Similarly, we found clusters 5 and 7 in the validation sample to have similar properties as the fibrotic SP4 cluster including expression of FBN1, PI16 and IGFBP6.
Figure 6. Progenitor clusters specific to VAT derived from individuals with obesity.
(a) Re-clustering of VAT-specific progenitors (9,847 cells) from the complete sample set (b) Violin plots showing expression density of MSLN and CFD in 6 VP clusters. The y axis indicate log transformed expression values and the width indicate number of cells expressing the particular gene (c) VAT progenitor cells labelled based on mitochondrial gene distribution: group 0 (grey, 5,018 cells) shows cells with mitochondrial gene expression <=5%, group 1 (blue, 4,100 cells) represents 6% to 14% and group 2 (red, 729 cells) represents 15% to 24% expression. See also supplementary figure 5 (d) Pearson correlation of fasting glucose levels (mmol/L) and VPM cell proportion across all samples from 12 individuals (e) Expression pattern of UCP1, MSLN wnd WT1 using bulk RNASeq showing change in expression in mature adipocytes.
Visceral specific progenitor cells have different origin and mitochondrial activity
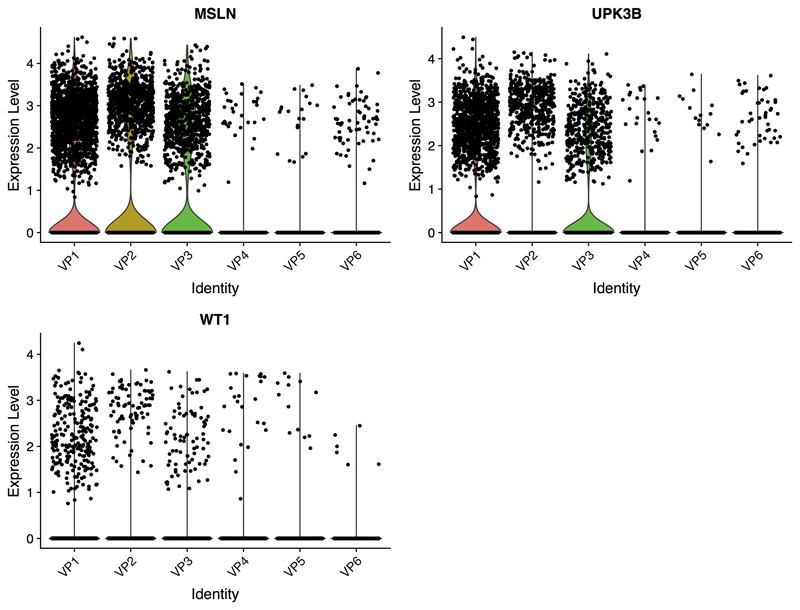
In contrast to VPC, VPM appeared to have unique properties specific to the visceral depot with signature of a mesothelial origin as discussed above. Apart from specific expression of MSLN, the VPM progenitor cells also expressed the mesothelial adipocyte markers UPK3B and WT1 (Extended Data 2). Comparing VPC and VPM cells, we also noted clear differences in mitochondrial gene expression. Specifically, we found that the VPM had comparatively high mitochondrial expression (i.e. 6-20%) whereas VPC showed a consistent pattern of cells with low (i.e. <=5%) expression of mitochondrial genes (Figure 6c). As it is well known that brown adipocytes contain higher number of mitochondria than white adipocytes, this intriguing finding prompted us to test whether the VPM cells constitute progenitors with differentiation potential towards beige adipocytes and thus potentially have a more protective function against obesity and insulin resistance. To test this, we first correlated VPM proportion with fasting glucose levels and found a significant negative correlation (Pearson r=-0.64, p-value=0.025, Figure 6d). We also noted that VPM expressed PLA2G2A (VP1; Adjusted p-value= 5.84E-109) which was recently shown to activate mitochondrial uncoupling in brown adipose, and in line with our observations here, provided protection from the deleterious effects of high fat diets in mice32. In addition, the mitochondrial marker gene SOD2 (VP3; Adjusted p-value=2.72E-245) known to be upregulated in beige and brown AT33 compared to white AT was found to be specific to the VPM34 cells.
Next, we investigated whether this beige adipose signature found among a subset of the VAT progenitor cells (i.e. VPM) could be validated in independent datasets of purified VAT and SAT - derived mature adipocytes from the same clinical study population (Supplementary Table 22). Indeed, we noted striking overrepresentation of not only the mesothelial markers MSLN (log2fold=1.48, Adjusted p-value=1.7E-3), WT1 (log2fold=2.73, Adjusted p-value=8.1E-10) and UPK3B (log2fold=2.54, Adjusted p-value=8.3E-9) but also the classical markers for beige and brown adipocytes EBF2 (log2fold=2.39, Adjusted p-value=2.1E-20), PRDM16 (log2fold=1.44, Adjusted p-value=9.9E-5) and UCP1 (log2fold=2.20, Adjusted p-value=2.3E-7). However, progenitor or mature brown adipocyte markers MYF5 (log2fold=0.04, Adjusted p-value=0.96) or ZIC1 (log2fold=-0.01, Adjusted p-value=0.98) were not differentially expressed, suggesting that the VPM cells are not of the brown adipocyte lineage. We then used bulk SVF and whole tissue from VAT and SAT to validate the induction of UCP1 during differentiation to mature adipocytes whereas the expression of MSLN and WT1 peaks in progenitors (Figure 6e). In addition, we noted a strong negative correlation in VAT between UCP1 and both mesothelial markers (MSLN vs UCP1, r=-0.72, p-value=0.018 and WT1 vs UCP1, r=-0.72, p-value=0.019; Supplementary Figure 6). We also noted a significant positive correlation between UCP1 and IRX3 in VAT (r=0.85, p-value=1.71E-3) in line with recent reports showing the induction of IRX3 in the browning process of adipocytes35 (Supplementary Figure 6). Lastly, in line with our observations of marker genes characterizing VPM, we observed a trend of SOD2 expression to be positively correlated with the expression of UCP1 (r=0.56, p=0.09) further indicating browning signatures of a subset of VPM-derived adipocytes residing in the visceral depot only.
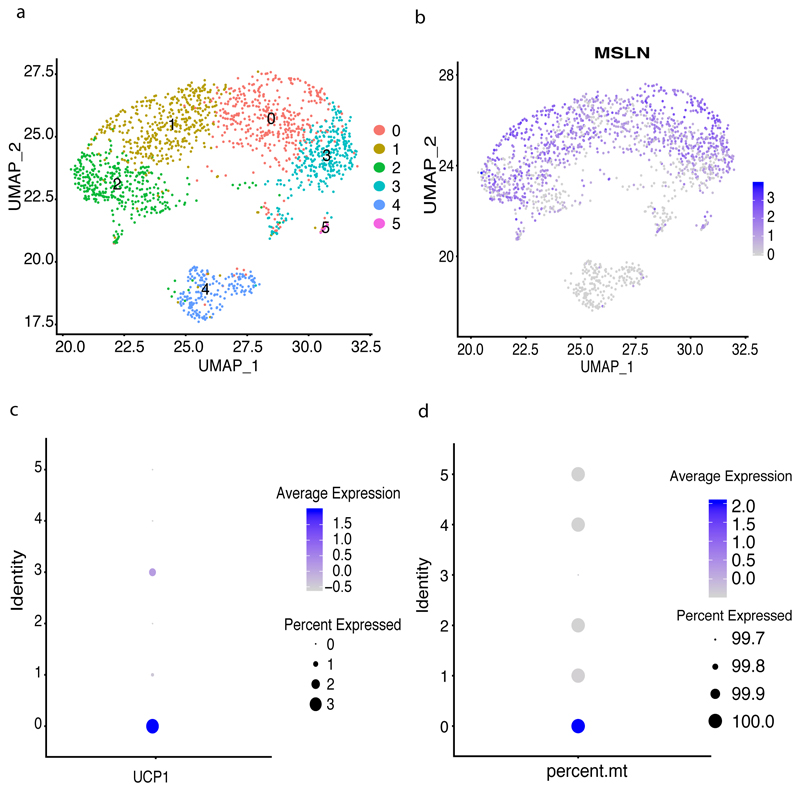
These several lines of evidence indicate that there are UCP1+ cells originating from MSLN / WT1 expressing progenitors in VAT only where the expression of UCP1 is peaking in mature adipocytes that underwent browning. This may explain why we failed to detect any UCP1 expressing cells in our large single-cell data of VPM progenitors from individuals with obesity. However, as we observed a negative correlation between proportions of VPM cells and metabolic disease status (Figure 6d) we hypothesized that UCP1-expressing preadipocytes derived from VPM progenitors may be more pronounced in healthy tissue early in development. To address this, we accessed VAT from a healthy young donor. We subset CD34+ cells for clustering (Figure 7) and confirmed the high abundance of MSLN / WT1 progenitors in SVF derived from VAT. Encouragingly, we also identified a distinct cluster of cells among the VPM that not only expressed UCP1 (Figure 7c) but also had the highest mitochondrial content (Figure 7d). Finally, we also noted SOD2 among the top genes with differential expression between UCP1-expressing versus non-UCP1 VPMs (Adjusted p-value=7.02E-93) with 95% of UCP1+ cells expressing the gene versus only 75% of the UCP1- VPMs. On the contrary, as shown above, UCP1+ cells had significantly lower expression of WT1 (Adjusted p-value= 4.24E-08) with only 22% of UCP1+ cells expressing the gene versus 56% of the UCP1- VPMs (Supplementary Table 23).
Figure 7. Progenitor clusters specific to VAT derived from a healthy individual.
(a) Re-clustering of progenitors identified 6 clusters comprising 1,781 cells(b) Umap highlighting expression of MSLN in all progenitor clusters(c) UCP1 expression was detected in cluster 0 which had high mitochondrial gene expression (d)
Discussion
We have performed a large, unbiased assessment of the cellular landscape in the non-adipocyte fraction of human adipose tissue (known as the SVF). The corresponding mature adipocyte fraction per depot was studied in parallel by bulk sequencing approaches due to current technical limitations in profiling large and lipid-rich cells at single-cell resolution. Study subjects were selected based on obesity-related metabolic phenotypes including presence or absence of type 2 diabetes (T2D), which allowed us to incorporate differences in cell populations not only dependent on depot but also based on disease status.
Our initial classification resulted in three broad categories of cell types including progenitors or stem cells, immune cells and endothelial cells. The latter cell type represented ~8% of all cells but with distinctive signatures dividing the group further into subpopulations. Although in minority, it is well established that endothelial cells play an important role in adipose tissue inflammation where obesity-induced T2D has been associated with profound dysfunction of endothelial cells36. Our largest population of endothelial cells were characterized as fatty acid handling microvascular endothelial cells due to pronounced expression of genes encoding fatty acid transport and binding proteins, respectively. These results are in line with recent work showing the importance of endothelial fatty acid uptake in adipose tissue especially during obesity-induced tissue inflammation12. Another subpopulation was identified as lymphatic-derived due to expression of the lymphatic vessel endothelial hyaluronan receptor (LYVE-1) gene. This is of interest as recent efforts have shown the importance of a crosstalk between lymphatic vessels and adipose tissue, and that lymphatic dysfunction are linked to metabolic diseases 37. We extend these efforts and show that the lymphatic vasculature is depot-specific as the majority of these cells were derived from visceral samples.
Our characterization of adipose-resident immune cells, representing ~30% of the cells, confirmed the complex nature of obesity-induced adipose tissue inflammation. We identified 14 different immune cell types including T cells, B cells, NK cells, dendritic cells, monocytes and macrophages. T cells, and particular the CD8+ subset, are known to play an important role in adipose inflammation where the obese adipose tissue activates these cells which then recruit and activate adipose-resident macrophages38. We identified activated memory CD8+ T cells expressing CCL5 which is known to be positively correlated with obesity39 where increased expression associate with increased BMI. Interestingly, we found a subpopulation of these CD8+ T cells expressing metallothionein with unique clustering pattern compared to the activated memory CD8+ cells and with indication of an association to obesity-related traits. Specifically, using two large population-based adipose tissue resources we found a strong association of the expression of the cell-specific metallothionein genes MT2A, MT1F and MT1G and obesity phenotypes indicating a role of these cells in adipose dysfunction. This T cell expression profile of metallothionein genes was recently linked to a novel type of dysfunctional CD8+ T cell population identified in single-cell studies of tumor-infiltrating lymphocytes40 where targeted deletion of the metallothionein resulted in loss of T cell dysfunction. Similar to Singer et al, we also found that these metallothionein genes are expressed only in the potential dysfunctional adipose-resident T cells but not in the activated memory CD8+ T cells, and as such break new grounds in possibly defining the role of T cells in adipose inflammation and potentially insulin resistance.
As mentioned above, macrophages are activated by T cells and infiltrate the adipose tissue contributing to the obesity-related tissue inflammation and subsequently insulin resistance. Adipose resident macrophages play different key role in energy metabolism, clearance of dead adipocytes and removing extracellular lipids 41 which are found to be altered during the development of obesity and insulin resistance. We observed that with high expression of genes involved in lipid metabolism during obesity, our largest macrophage population likely represent the so called metabolically active ATMs which have been associated with a beneficial role in obesity including promoting dead adipocyte clearance42. We further showed that they have selective expression of CD9 which points towards them representing a distinct type of obesity-related ATMs.
We identified multiple clusters of progenitor cells with clear expression signatures dependent on cell source (i.e. subcutaneous or visceral). They were further characterized to be at different stages of adipogenesis which was marked with varying degree of CFD expression. CFD has been identified as a key player in adipogenesis where knockdown of the gene inhibits lipid accumulation and expression of adipocyte markers during adipocyte differentiation43. On the other hand, overexpression of CFD promotes adipocyte differentiation. We confirmed the association of CFD expression and adipocyte differentiation with validation experiments using mesenchymal stem cells that were monitored during adipogenesis. These stem cells were derived from bone-marrow and cultured in vitro and thus may possess different properties than adipose-derived stem cells, however it was recently shown that they do have similar adipogenic differential potential44. We also show clear distinction of the subpopulations beyond what has been shown before45. We identified a subtype of premature adipocytes that associated with T2D where individuals with high glucose levels had higher abundance of this cell type than those with normal glucose levels. We noted that this T2D-linked cell type was characterized by genes within the PPARγ pathway, some with protective role against insulin resistance. One of those key protective genes was GPX3 where significantly higher expression was identified in cells derived from nondiabetics. It is known that T2D-associated oxidative stress can be reversed by PPARγ-mediated antioxidant regulation mediated through the expression of GPX346. Furthermore, PPARγ agonists such as Thiazolidinediones (TZDs), commonly used to treat T2D, although not used in our samples, induce these antioxidant effect with increased expression of GPX3 as a result. On the contrary, we also found genes expressed by these T2D-associated cells with seemingly negative effect on T2D status including WISP2 and ATF3. The expression of these genes was significantly higher in T2D cells as well as a larger proportion of T2D cells expressing the genes compared to non-T2D cells. WISP2 is an adipokine and while one of its key function is to regulate adipogenic commitment and PPARγ activation, our results confirms previous efforts showing it can also induce insulin resistance likely through the inhibition of adipocyte recruitment and promotion of hypertrophic obesity47. ATF3 inhibits PPARγ-mediated transactivation48 and hepatic ATF3 expression is associated with T2D49 and as such ATF3, similar to GPX3, may serve as a potential therapeutic target for the reduction of insulin resistance.
Finally, while our depot-specific analysis confirmed earlier efforts showing that visceral-derived adipocyte progenitors originate from the mesothelium 28, we provide additional support to the notion that a subset of these are also likely inducible by responding to environmental stimuli and may be differentiating into beige preadipocytes. We noted that for instance the visceral-specific VPM cells, while being CD34+/WT1+ they did not express MYF5, suggesting that rather than being classical brown preadipocytes, they are more likely induced and could be consistent with so called beige adipocyte progenitors50. Although the physiological significance of such beige adipocyte progenitors in visceral adipose of individuals with morbid obesity is uncertain, they may possibly represent a compensatory mechanism to reduce the adverse effects of obesity. This hypothesis is supported by our finding of overrepresentation of this particular cell population in non-insulin resistant as well as healthy subjects.
We acknowledge that a limitation of our study is the focus of individuals with obesity where further studies are needed to contrast cellular architechture in adipose tissue derived from healty individuals. However, as there are clear depot and region-specific differences in cellular distributions, single-cell profiling spanning healthy and obese individuals would require a surgical procedure common in the general population where tissue resection can be obtained for research purpose.
In conclusion, we show the richness of the cellular contribution in human adipose tissue with multiple types of subpopulations of immune cells, endothelial cells, fibroblasts and progenitors. Interrogation of the non-adipocyte fraction of adipose tissue at single cell level suggests that development of obesity and T2D could be a joint action of subpopulation of different cell types residing in adipose tissue with altered function.
Methods
Study Subjects
Participants were selected through an institutionally approved biobank infrastructure with ongoing recruitment at the Quebec Heart and Lung Institute (IUCPQ; Université Laval, Quebec City, Canada). Inclusion and exclusion criteria were those related to bariatric surgery. Specifically, men and women who had a BMI≥40kg/m2 or ≥35kg/m2 with major comorbidities, who required surgery and who met the NIH Guidelines for bariatric surgery were invited to participate. Exclusion criteria were general contra-indications for bariatric surgery, a BMI<35 kg/m2, age under 18 or over 60 years, respectively, abnormal bowel habits including irritable bowel syndrome, pregnancy, cirrhosis, inflammatory bowel disease and previous bariatric surgery. Participants were further selected based on i) the presence of a signed consent form and ii) availability of type 2 diabetes (T2D) status where subgroups (i.e. with or without T2D) were matched for BMI, age as well as males:females ratio as much as possible.
For the discovery cohort used for single-cell RNA sequencing, we identified 14 individuals (4 males and 10 females) using the above described inclusion criteria where five individuals were diagnosed with type 2 diabetes and nine individuals without type 2 diabetes. Detailed clinical information is provided in Table 1. For single- cell expression validation, two additional individuals were identified using the same selection criteria including one 35-year-old male without type 2 diabetes and with BMI of 59 kg/m2 and one 42-year-old female with type 2 diabetes with BMI 44.8 kg/m2
For validation studies using bulk SVF and adipocyte samples, we identified 13 independent individuals from the same study population. This sample set included 12 females and one male with a BMI range from 39.7 to 59.2 kg/m2 and age values ranging from 29 to 60 years. Six of these individuals were diagnosed with type 2 diabetes and six were not. Similarly, for validation studies using bulk adipose tissue samples, we identified another set of 10 independent individuals with BMI 35.5 to 71.8 kg/m2, age between 42 and 49 years and the male-to-female ratio as 1:1.
For all sample sets, adipose tissue was collected during the bariatric surgery from two different fat compartments: greater omentum corresponding to VAT and abdominal subcutaneous fat compartment (SAT). Informed consent was obtained from each participant through the management of the framework of the Quebec Heart and Lung Institute Obesity Biobank. The protocol was approved by the Research Ethics Committee of the IUCPQ (Protocol # 21320).
Sample Preparation
SVF and mature adipocytes were obtained as follows: adipose tissue was digested within 30 min of collection with collagenase according to a modification of the Robdell method51. Briefly, adipose tissue samples were digested with collagenase type 1 in Krebs-Ringer-Henseleit (KRH) buffer for 45 minutes at 37°C. Cell suspensions containing mature adipocytes and SVF were then filtered with a nylon mesh and washed 3 times with KRH buffer. The nature of the buoyancy adipocytes allows them to float to the surface. Mature adipocytes were aliquoted and the remaining solution containing the SVF was centrifuged 1500 rpm for 5 minutes. The pellet was washed with pre-adipocyte growth medium (PGM) (DMEM-F12 supplemented with 10% calf serum, 1% penicillin-streptomycin, 17μM pantothenic acid, 33μM biotin, 100μM ascorbic acid and 2,5 μm/ml amphoB) followed by a second centrifugation. SVF cells were then cryopreserved using freezing medium (PGM supplemented with 40% FBS and 10% DMSO). The medium was added to the pellet and was frozen with a temperature gradient (-1 °C/ minutes) and stored in liquid nitrogen until analysis. Whole adipose tissue samples were following collection quickly frozen in liquid nitrogen and stored until analysis.
Isolation of CD34+/CD34- SVF cells by fluorescence activated cell sorting
Cells from the SVF were thawed and suspended in PBS-0.1% BSA. They were centrifuged at 1500 rpm for 5 minutes. Erythrocyte lysis buffer was added for 5 minutes to the suspension to get rid of red blood cells. The suspension was centrifuged again at 1500 rpm for 5 minutes. Primary antibody (CD34, PE, eBioscience) was added to the samples and incubated for 60 minutes at 4°C in the dark. PBS-0.1% BSA was added to wash the suspension followed by 80 μm filtration with nylon mesh to remove debris. The suspension was then transfer to a 10ml polypropylene tube, centrifuged at 1500 rpm for 5 minutes and the pellet was suspended with PBS-0.1% BSA. Viability staining solution has been added 15 minutes before the sorting process. Viability control was performed with live/dead cells to ascertain that the staining solution worked well and for gate adjustment. OneComp Ebeads control was also used to provide positive/negative control for the antibody. The samples were sorted with the BD FACSAria II (BD, San Diego, California, USA) with the following parameters: Cell size, graininess, viability, singlet cell and CD34+/CD34-.
Dead and non-singlet cells were automatically discarded. Cells that were positive for CD34 and negative for CD34 were collected in two distinct tubes (Supplementary Figure 7). After sorting, cells were centrifuged at 1500 rpm for 5 minutes and were conserved in freezing medium as described above and stored at -80 °C until further analysis.
MSC culture and differentiation
Bone-marrow derived mesenchymal stromal cells (LY-MSCs) from eight healthy 19-27 years old female donors were provided by the GMP facility of the Advanced Cell Therapy Centre, Finnish Red Cross Blood Service (FRCBS), Helsinki, Finland. The cells were thawed, expanded and differentiated into adipocytes following the FRCBS protocol previously published52. Briefly, in preparation for adipogenic differentiation cells at passage #3 were plated in 10 cm cell culture plates and grown in expansion media containing low glucose DMEM (Thermo Fisher Scientific), 100 U/ml penicillin /100 μg/ml streptomycin (Thermo Fisher Scientific) and 10% Stemulate (Pooled Human Platelet Lysate culture media supplement, Cook Regentec) until they reached 70%-90% confluence (Timepoint 1), at which point the media was changed to adipogenic basal medium containing alpha-MEM Glutamax, 10 % FBS, 20 mM HEPES, 100 U/ml penicillin and 100 μg/ml streptomycin (Thermo Fisher Scientific). Half of the culture plates were used as controls and maintained in the adipogenic basal medium until harvesting (AC samples). The other half was subjected to adipogenic differentiation (AD samples). More specifically the cells were first incubated for 3-4 days in adipogenic basal media supplemented with the induction cocktail of 0.1 mM indomethacin (Sigma), 0.5 μg/ml insulin, 0.2 mM 3-isobutyl-1-methylxanthine (IBMX), and 0.4 μg/ml dexamethasone (Preadipocyte Differentiation Medium Supplement Pack, PromoCell) (Timepoint 2), then the media was changed to the terminal differentiation cocktail containing 0.1 mM indomethacin (Sigma), 0.5 μg/ml insulin and 3 μg/ml Ciglitazone (PromoCell). The media of all plates was changed twice a week and the differentiation was allowed to proceed for 3 weeks for Timepoint 3 or 4 weeks for Timepoint 4. For each donor samples were harvested at Timepoint 1 (ND – non-differentiated cells), Timepoint 2 (AIC and AID: Adipocyte induced control without drugs in the media or differentiated with induction drugs), Timepoint 3 (AC1, AD1: Adipocyte control without drugs in the media or differentiated with drugs) and Timepoint 4 (AC2, AD2: Adipocyte control without drugs in the media or differentiated with drugs).
Single cell RNA sequencing
Cells from the SVF were thawed and serially diluted in DMEM/F12 supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin in 15ml conical tube to a volume of 14ml. They were centrifuged at 300 rcf for 10 minutes and the supernatant was removed and discarded. The pelleted cells were suspended in an adequate amount of DMEM/F12 supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin and transferred to a 1.5ml microfuge tube for counting. 10 μl of the suspension was used for a cell count and viability assay on a Countess II Automated Cell Counter (Invitrogen) to determine the volume of suspension to be used in the 10x Chromium Single Cell Library protocol. The desired number of viable cells to be loaded onto the Chromium Single Cell A Chip (10x Genomics) was 12,000 cells. After cell capture, the remaining cells were centrifuged at 300 rcf for 10 minutes and cryopreserved using freezing medium (Recovery Cell Culture Freezing Medium, Thermo Fisher Scientific). The cells were suspended in freezing medium and frozen with a temperature gradient of -1°C per minute. Cell capture, cDNA amplification, and library preparation were performed using the Chromium Single Cell 3' Library & Gel Bead Kit v2 (10x Genomics), according to the manufacturer’s protocol. The final libraries were analyzed using a TapeStation to determine library size and a Qubit Broad Range dsDNA assay to determine library concentration and sequenced using Illumina HiSeq at high depth (mean 171,560 reads/cell) capturing on average 1,527 cells/sample.
Bulk RNA Sequencing
In total of 0.5 to 3 million cells were re-suspended in 500 uL TRIzol Reagent and total RNA was extracted using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA library preparations were carried out on 500 ng of RNA with RNA integrity number (RIN)>7 using the Illumina TruSeq Stranded Total RNA Sample preparation kit, according to manufacturer's protocol. Final libraries were analyzed on a Bioanalyzer and sequenced on the Illumina HiSeq 2500 (pair-ended 100 - 150 bp sequences). On average 61,222,621, 49,175,966, 66,301,281 and 84,454,561 paired-end reads were sequenced in adipocytes, SVF, adipose tissue and MSCs respectively.
QC and clustering of single cell data from SVF
Initial analysis of the single cell libraries was done using cellranger v.2.1.0 from 10X Genomics. GRCh38 was used as the genome reference. The analysis was started with 26 samples. All sequenced samples were aggregated using cellranger aggr pipeline where read depth difference between libraries was normalized by subsampling higher depth libraries. Seurat53 was used for further analysis of clustering, dimensionality reduction and differential gene expression analysis. For filtering, we used only cells with a minimum gene count of 200 and a maximum of 2500 genes/cell based on their distribution in the sample. We also removed cells with unique molecular identifiers (UMIs) less than 200 and greater than 13,750 per cell. Cells with percentage of mitochondrial gene expression greater than 20 was also removed. However, clustering showed that the majority of the cells (90%) from one sample were clustered together apart from the clusters of all the other samples. The sample was removed the sample and analysis was repeated.
The final input data to Seurat comprised of 38,170 cells and after filtering we obtained 26,350 cells. 11,111 cells were removed because of low gene / UMI count. We regressed out the variability due to UMI distribution, mitochondrial gene expression and the difference between G2M and S phase scores based on the gene expression of cell cycle genes 54. Top 23 dimensions were used to generate final clusters using principal component analysis (PCA) and graph based clustering. Top 100 genes in each cluster was identified in comparison with all other cells using the function – ‘FindAllMarkers’ in Seurat while keeping a cutoff of padj <= 0.01. Genes that are expressed to a minimum of 25% of cells in either of the test population is considered for analysis. Clusters were annotated using overlapping known marker genes among the cluster specific genes. Differential gene expression analysis between cell population was done using Wilcoxon rank sum test in ‘FindMarkers’ function. The raw expression data of the filtered cells used in clustering of Seurat was used for generating annotation using SingleR with default parameters55. The annotation for each cell is then overlapped with Seurat cluster identities to identify the cell type of each cluster. The subset for the VAT progenitors and immune clusters were created from the clustering results using ‘SubsetData’ function of Seurat. Top 10 and 15 dimensions were used for clustering VAT progenitors and Immune cells respectively using highly variable genes in the corresponding data.
The sample with highest number of cells captured (n=6,672) is randomly subsampled to 1,500 cells to match the cell counts of remaining samples. The clustering is repeated using the same settings as that of initial analysis. A subset of SAT progenitors (P2, P4, P6, P7) was created. Cells from VAT that were included in the subset was removed and we obtained 2,705 that are from SAT progenitors. Top variable genes were identified and clustering was done using top 10 dimensions. The cluster specific genes were used for functional annotation using DAVID Bioinformatics Resources 6.8 56,57.
Differential gene expression analysis using bulk RNA
Raw reads were trimmed for quality (phred33 ≥30) and length (n≥32), and Illumina adapters were clipped off using Trimmomatic v.0.3558. Filtered reads were then aligned to the GRCh38 human reference using STAR v.2.5.3a59. Raw read counts of genes were obtained using htseq-count v.0.6.060. Differential gene expression analysis was done using DeSeq2 v.1.18.161.
The gene raw count table (V6p) of RNAseq data was downloaded from the GTEx web portal. The subset of data is then created selecting only tissues of interest based on the sample attribute file provided in the GTEx portal. Differential expression gene analysis is done using DeSeq2 v.1.18.1.
Detection of CD9+/CD68+ macrophages by histological immunofluorescence
Visceral and subcutaneous adipose tissue samples from four patients undergoing bariatric surgery were identified of which two were diagnosed with type 2 diabetes. The mean age and BMI were 47.3±11.7 years and 49.9±5.2 kg/m2 respectively. Adipose tissue slides were disembedded by xylene (2 x 10 min) followed by rehydration step using ethanol (2 x 5 min 100% and 2 x 5min 95%). Antigen retrieval was performed using citrate buffer 0.01M (0.1M citric acid with 0.1 sodium citrate mixed with water) for 5 min at 65°C and 10 min at 95°C. The final rehydration step was performed by immersion of the slides for 5 minutes in water and for 5 minutes in PBS. Non-specific sites were blocked for at least 45 minutes using PBS 1X, 0.1% BSA, 0.4% Triton-X100 and 10% goat serum. Primary antibodies, rabbit anti-human CD9 (PA5-11556, Invitrogen, California, USA) and mouse anti-human CD68 (ab955, Abcam, Cambridge, United Kingdom) were incubated overnight at 4°C in a humidified chamber. The slides were washed 3 times in PBS 1X for 5 minutes. Secondary antibodies, goat anti-rabbit Alexa488 (A11008, Invitrogen, California, USA) and goat anti-mouse Alexa594 (A11005, Invitrogen, California, USA) were incubated for 2 hours at room temperature in the dark. The slides were then washed 3 times with PBS 1X for 5 minutes. Vectashield mounting medium with DAPI (H-1200, Vectors Laboratories, California, USA) was used with coverslip and sealed with nail polish. Digital images were taken using Zeiss LSM800 confocal system (Zeiss, Oberkochen, Germany). Image were taken at 20x and 63x magnification. The slides were stored at 4°C in the dark and imaged less than 4 days after staining.
Validation of beige adipocyte signatures by single cell sequencing
Adipose tissue was collected from a 15-month-old boy undergoing inguinal hernia repair at Children’s Mercy Kansas City. Consent was obtained through the management of the study protocol approved by the Institutional Review Board at Children’s Mercy Kansas City (Protocol # 17110653). An easily accessible portion of the greater omentum at the lower end of the omental drape was identified and a portion was pulled out and a 2cm segment was amputed to allow for areas of cautery changes to be discarded. Specimen was passed off the field in a jar containing saline. Adipose tissue was then prepared as described above to obtain SVF. The SVF pellet was treated with ACK lysis buffer at RT for 5 minutes, followed by another centrifugation step. The ACK lysis buffer was removed from the pellet and the cells were washed once more with HBSS+200uM adenosine+2%FBS and centrifuged one final time. Cells were counted to determine the total number of cells in suspension and viability. Cells were then cryopreserved using freezing medium (Gibco™ Recovery™ Cell Culture Freezing Medium, 10% DMSO). The medium was added to the pellet and was frozen with a temperature gradient (-1 °C/ minutes) and stored in liquid nitrogen until analysis. At the time of single-cell analysis, cells from the SVF were thawed and prepared as described above including cell count and viability assay on a Countess II Automated Cell Counter (Invitrogen) to determine the volume of suspension to be used in the 10x Chromium Single Cell Library protocol. The desired number of viable cells to be loaded onto two wells of a Chromium Single Cell A Chip (10x Genomics) was 8000 cells for a total of two captures with 4000 cells/capture. Cell capture, cDNA amplification, and library preparation were performed using the Chromium Single Cell 3' Library & Gel Bead Kit v2 (10x Genomics), according to the manufacturer’s protocol. The final two libraries were analyzed using a TapeStation to determine library size and a Qubit Broad Range dsDNA assay to determine library concentration and sequenced on an Illumina NovaSeq6000 instrument. For QC and clustering, we followed same procedure used for the discovery samples to analyze the validation sample. However, for filtering, we used only cells with a minimum gene count of 200 and a maximum of 6000 genes/cell based on their distribution in the sample. We also removed cells with unique molecular identifiers (UMIs) greater than 50,000 per cell. Cells with percentage of mitochondrial gene expression greater than 40 was also removed. After filtering we used 2,779 cells for further clustering and analysis. Top 55 dimensions were used to generate final clusters using principal component analysis (PCA) and graph-based clustering. The subset for the progenitors were created from the clustering results using ‘SubsetData’ function of Seurat. Top 50 dimensions were used for clustering VAT progenitors using highly variable genes in the corresponding data.
Gene expression analysis in population based cohorts
Associations between gene expression levels (IlluminaHT12) and phenotypes within the MuTHER cohort were modeled using a linear mixed effects model as described previously17. Briefly, the lmer function in the lme4 package, was fitted by maximum-likelihood. The linear mixed effects model was adjusted for age and experimental batch (fixed effects) and family relationship (twin-pairing) and zygosity (random effects). A likelihood ratio test was used to assess the significance of the phenotype effect. The p-value of the phenotype effect in each model was calculated from the Chi-square distribution with 1 degree of freedom using -2log(likelihood ratio) as the test statistic. Summary statistics from associations between gene expression levels and phenotypes within the METSIM cohort were obtained from METSIM Study18.
Extended Data
Extended Data 1.
Multiple macrophage clusters were identified in SVF from both SAT and VAT depots (a) 4 distinct macrophage clusters showing varying expression of CD68 (19 - 52% of cells), CD9 (10 – 51% of cells) and CD36 (25 – 72% of cells). The y axis of the violin plot indicate log transformed expression values and the width indicate number of cells expressing the particular gene. (b)Genes involves in lipid metabolism is found expressed in macrophage cluster – IS2, whereas IS3 is rich in inflammatory markers.
Extended Data 2.
Gene expression of marker genes in 6 visceral specific progenitor clusters. The y axis of the violin plot indicate log transformed expression values and the width indicate number of cells expressing the particular gene.
Supplementary Material
Acknowledgements
This work was supported by a Canadian Institute of Health Research (CIHR) team grant awarded to E.G (EGM141898) and the CIHR funded Epigenome Mapping Centre at McGill University (EP1-120608) awarded to T.P. E.G. holds the Roberta D. Harding & William F. Bradley, Jr. Endowed Chair in Genomic Research and T.P. holds the Dee Lyons/Missouri Endowed Chair in Pediatric Genomic Medicine. A.T. is the director of a Research Chair in Bariatric and Metabolic Surgery. M.C.V. is the recipient of the Canada Research Chair in Genomics Applied to Nutrition and Metabolic Health (Tier 1). The MuTHER Study was funded by a program grant from the Wellcome Trust (081917/Z/07/Z) and core funding for the Wellcome Trust Centre for Human Genetics (090532). The TwinsUK study was funded by the Wellcome Trust and European Community’s Seventh Framework Programme (FP7/2007-2013). The TwinsUK study also receives support from the National Institute for Health Research (NIHR)- funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London. We further thank additional members of the MuTHER consortium for providing valuable data for this study; Kourosh R. Ahmadi, Chrysanthi Ainali, Amy Barrett, Veronique Bataille, Jordana T. Bell, Alfonso Buil, Emmanouil T. Dermitzakis, Antigone S. Dimas, Richard Durbin, Daniel Glass, Neelam Hassanali, Catherine Ingle, David Knowles, Maria Krestyaninova, Cecilia M. Lindgren, Christopher E. Lowe, Eshwar Meduri, Paola di Meglio, Josine L. Min, Stephen B. Montgomery, Frank O. Nestle, Alexandra C. Nica, James Nisbet, Stephen O'Rahilly, Leopold Parts, Simon Potter, Johanna Sandling, Magdalena Sekowska, So-Youn Shin, Kerrin S. Small, Nicole Soranzo, Gabriela Surdulescu, Mary E. Travers, Loukia Tsaprouni, Sophia Tsoka, Alicja Wilk, Tsun-Po Yang & Krina T. Zondervan. The authors also thank Matti Korhonen and Marja Ahti for valuable help in this study. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the dbGaP accession number phs000424.v7.p2 on 07/10/2019.
Footnotes
Author Contributions
E.G. conceived the study. E.G., A.T., T.P. and M.C.V. designed experiments. A.T., J.F. and M.F.G prepared and/or provided the clinical samples. L.B. and B.B. managed clinical aspects of the study. A.P., R.B., M.G., D.A.L. and M.M.S performed the experiments. W.A.C, J.J., H. D., A. S. and G.B. provided bioinformatics support. A.L and J.N. provided MSC cell lines and culture protocols. J.V. and E.G. analyzed the data and drafted the manuscript. All authors reviewed and contributed feedback on the final manuscript.
Declaration of Interests
A.T. receives funding from Johnson & Johnson Medical Companies and Medtronic for research unrelated to the present manuscript.
Data Availability
Raw data files from single-cell and bulk RNA-Sequencing used in this study are uploaded in GEO in a SuperSeries with accession number: GSE136230. The expression data from the MuTHER cohort have been deposited in the ArrayExpress, (https://www.ebi.ac.uk/arrayexpress/) with accession number E-TABM-1140.
References
- 1.Tchernof A, Despres JP. Pathophysiology of human visceral obesity: an update. Physiol Rev. 2013;93:359–404. doi: 10.1152/physrev.00033.2011. [DOI] [PubMed] [Google Scholar]
- 2.Porter SA, et al. Abdominal subcutaneous adipose tissue: a protective fat depot? Diabetes Care. 2009;32:1068–1075. doi: 10.2337/dc08-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laforest S, Labrecque J, Michaud A, Cianflone K, Tchernof A. Adipocyte size as a determinant of metabolic disease and adipose tissue dysfunction. Crit Rev Clin Lab Sci. 2015;52:301–313. doi: 10.3109/10408363.2015.1041582. [DOI] [PubMed] [Google Scholar]
- 4.Denis GV, Obin MS. 'Metabolically healthy obesity': origins and implications. Mol Aspects Med. 2013;34:59–70. doi: 10.1016/j.mam.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michaud A, et al. Relevance of omental pericellular adipose tissue collagen in the pathophysiology of human abdominal obesity and related cardiometabolic risk. Int J Obes (Lond) 2016;40:1823–1831. doi: 10.1038/ijo.2016.173. [DOI] [PubMed] [Google Scholar]
- 6.Schipper HS, Prakken B, Kalkhoven E, Boes M. Adipose tissue-resident immune cells: key players in immunometabolism. Trends Endocrinol Metab. 2012;23:407–415. doi: 10.1016/j.tem.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Nawaz A, et al. CD206(+) M2-like macrophages regulate systemic glucose metabolism by inhibiting proliferation of adipocyte progenitors. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00231-1. 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen TK, Baryawno N. Introduction to Single-Cell RNA Sequencing. Curr Protoc Mol Biol. 2018;122:e57. doi: 10.1002/cpmb.57. [DOI] [PubMed] [Google Scholar]
- 9.Schwalie PC, et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018;559:103–108. doi: 10.1038/s41586-018-0226-8. [DOI] [PubMed] [Google Scholar]
- 10.Hepler C, et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife. 2018;7 doi: 10.7554/eLife.39636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrlund A, et al. The cell-type specific transcriptome in human adipose tissue and influence of obesity on adipocyte progenitors. Sci Data. 2017;4 doi: 10.1038/sdata.2017.164. 170164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briot A, et al. Senescence Alters PPARgamma (Peroxisome Proliferator-Activated Receptor Gamma)-Dependent Fatty Acid Handling in Human Adipose Tissue Microvascular Endothelial Cells and Favors Inflammation. Arterioscler Thromb Vasc Biol. 2018;38:1134–1146. doi: 10.1161/ATVBAHA.118.310797. [DOI] [PubMed] [Google Scholar]
- 13.Banerji S, et al. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J Cell Biol. 1999;144:789–801. doi: 10.1083/jcb.144.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schluns KS, Kieper WC, Jameson SC, Lefrancois L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol. 2000;1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- 15.Michelet X, et al. Metabolic reprogramming of natural killer cells in obesity limits antitumor responses. Nat Immunol. 2018;19:1330–1340. doi: 10.1038/s41590-018-0251-7. [DOI] [PubMed] [Google Scholar]
- 16.Schall TJ, et al. A human T cell-specific molecule is a member of a new gene family. J Immunol. 1988;141:1018–1025. [PubMed] [Google Scholar]
- 17.Small KS, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Civelek M, et al. Genetic Regulation of Adipose Gene Expression and Cardio-Metabolic Traits. Am J Hum Genet. 2017;100:428–443. doi: 10.1016/j.ajhg.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kratz M, et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014;20:614–625. doi: 10.1016/j.cmet.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jager NA, et al. Folate receptor-beta imaging using 99mTc-folate to explore distribution of polarized macrophage populations in human atherosclerotic plaque. J Nucl Med. 2014;55:1945–1951. doi: 10.2967/jnumed.114.143180. [DOI] [PubMed] [Google Scholar]
- 21.Liao X, et al. Kruppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villani AC, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356 doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acosta JR, et al. Increased fat cell size: a major phenotype of subcutaneous white adipose tissue in non-obese individuals with type 2 diabetes. Diabetologia. 2016;59:560–570. doi: 10.1007/s00125-015-3810-6. [DOI] [PubMed] [Google Scholar]
- 24.Ribeiro R, et al. Human periprostatic white adipose tissue is rich in stromal progenitor cells and a potential source of prostate tumor stroma. Exp Biol Med (Maywood) 2012;237:1155–1162. doi: 10.1258/ebm.2012.012131. [DOI] [PubMed] [Google Scholar]
- 25.Yang RZ, et al. Identification of omentin as a novel depot-specific adipokine in human adipose tissue: possible role in modulating insulin action. Am J Physiol Endocrinol Metab. 2006;290:E1253–1261. doi: 10.1152/ajpendo.00572.2004. [DOI] [PubMed] [Google Scholar]
- 26.de Souza Batista CM, et al. Omentin plasma levels and gene expression are decreased in obesity. Diabetes. 2007;56:1655–1661. doi: 10.2337/db06-1506. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe T, Watanabe-Kominato K, Takahashi Y, Kojima M, Watanabe R. Adipose Tissue-Derived Omentin-1 Function and Regulation. Compr Physiol. 2017;7:765–781. doi: 10.1002/cphy.c160043. [DOI] [PubMed] [Google Scholar]
- 28.Chau YY, et al. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat Cell Biol. 2014;16:367–375. doi: 10.1038/ncb2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winnier DA, et al. Transcriptomic identification of ADH1B as a novel candidate gene for obesity and insulin resistance in human adipose tissue in Mexican Americans from the Veterans Administration Genetic Epidemiology Study (VAGES) PLoS One. 2015;10:e0119941. doi: 10.1371/journal.pone.0119941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaittinen M, et al. MFAP5 is related to obesity-associated adipose tissue and extracellular matrix remodeling and inflammation. Obesity (Silver Spring) 2015;23:1371–1378. doi: 10.1002/oby.21103. [DOI] [PubMed] [Google Scholar]
- 31.Hou S, et al. S100A4 protects mice from high-fat diet-induced obesity and inflammation. Lab Invest. 2018;98:1025–1038. doi: 10.1038/s41374-018-0067-y. [DOI] [PubMed] [Google Scholar]
- 32.Kuefner MS, et al. Secretory phospholipase A2 group IIA modulates insulin sensitivity and metabolism. J Lipid Res. 2017;58:1822–1833. doi: 10.1194/jlr.M076141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perdikari A, et al. BATLAS: Deconvoluting Brown Adipose Tissue. Cell Rep. 2018;25:784–797 e784. doi: 10.1016/j.celrep.2018.09.044. [DOI] [PubMed] [Google Scholar]
- 34.Wankhade UD, et al. TGF-beta receptor 1 regulates progenitors that promote browning of white fat. Mol Metab. 2018;16:160–171. doi: 10.1016/j.molmet.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou Y, et al. IRX3 Promotes the Browning of White Adipocytes and Its Rare Variants are Associated with Human Obesity Risk. EBioMedicine. 2017;24:64–75. doi: 10.1016/j.ebiom.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts AC, Porter KE. Cellular and molecular mechanisms of endothelial dysfunction in diabetes. Diab Vasc Dis Res. 2013;10:472–482. doi: 10.1177/1479164113500680. [DOI] [PubMed] [Google Scholar]
- 37.Escobedo N, Oliver G. The Lymphatic Vasculature: Its Role in Adipose Metabolism and Obesity. Cell Metab. 2017;26:598–609. doi: 10.1016/j.cmet.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimura S, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 39.Wu H, et al. T-cell accumulation and regulated on activation, normal T cell expressed and secreted upregulation in adipose tissue in obesity. Circulation. 2007;115:1029–1038. doi: 10.1161/CIRCULATIONAHA.106.638379. [DOI] [PubMed] [Google Scholar]
- 40.Singer M, et al. A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell. 2016;166:1500–1511 e1509. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russo L, Lumeng CN. Properties and functions of adipose tissue macrophages in obesity. Immunology. 2018;155:407–417. doi: 10.1111/imm.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coats BR, et al. Metabolically Activated Adipose Tissue Macrophages Perform Detrimental and Beneficial Functions during Diet-Induced Obesity. Cell Rep. 2017;20:3149–3161. doi: 10.1016/j.celrep.2017.08.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song NJ, et al. Small Molecule-Induced Complement Factor D (Adipsin) Promotes Lipid Accumulation and Adipocyte Differentiation. PLoS One. 2016;11:e0162228. doi: 10.1371/journal.pone.0162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li CY, et al. Comparative analysis of human mesenchymal stem cells from bone marrow and adipose tissue under xeno-free conditions for cell therapy. Stem Cell Res Ther. 2015;6:55. doi: 10.1186/s13287-015-0066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Acosta JR, et al. Single cell transcriptomics suggest that human adipocyte progenitor cells constitute a homogeneous cell population. Stem Cell Res Ther. 2017;8:250. doi: 10.1186/s13287-017-0701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung SS, et al. Glutathione peroxidase 3 mediates the antioxidant effect of peroxisome proliferator-activated receptor gamma in human skeletal muscle cells. Mol Cell Biol. 2009;29:20–30. doi: 10.1128/MCB.00544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammarstedt A, et al. WISP2 regulates preadipocyte commitment and PPARgamma activation by BMP4. Proc Natl Acad Sci U S A. 2013;110:2563–2568. doi: 10.1073/pnas.1211255110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang MK, Jung MH. ATF3 inhibits PPARgamma-stimulated transactivation in adipocyte cells. Biochem Biophys Res Commun. 2015;456:80–85. doi: 10.1016/j.bbrc.2014.11.037. [DOI] [PubMed] [Google Scholar]
- 49.Kim JY, et al. Activating transcription factor 3 is a target molecule linking hepatic steatosis to impaired glucose homeostasis. J Hepatol. 2017;67:349–359. doi: 10.1016/j.jhep.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 50.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tchernof A, et al. Regional differences in adipose tissue metabolism in women: minor effect of obesity and body fat distribution. Diabetes. 2006;55:1353–1360. doi: 10.2337/db05-1439. [DOI] [PubMed] [Google Scholar]
- 52.Laitinen A, et al. A robust and reproducible animal serum-free culture method for clinical-grade bone marrow-derived mesenchymal stromal cells. Cytotechnology. 2016;68:891–906. doi: 10.1007/s10616-014-9841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aran D, et al. Reference-based annotation of single-cell transcriptomes identifies a profibrotic macrophage niche after tissue injury. bioRxiv. 2018 doi: 10.1101/284604. 284604. [DOI] [Google Scholar]
- 56.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 57.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.