Abstract
Cancer cells are able to evade immune surveillance through the expression of inhibitory ligands that bind their cognate receptors on immune effector cells. Expression of Programmed Death-Ligand 1 (PD-L1) in tumor micro-environments is a major immune checkpoint for tumor-specific T cell responses, by binding to Programmed Cell Death protein-1 (PD-1) on activated and dysfunctional T cells1. The activity of myeloid cells, such as macrophages and neutrophils, is likewise regulated by a balance between stimulatory and inhibitory signals. In particular, cell surface expression of the CD47 protein has been shown to form a “don’t eat me” signal on tumor cells, by binding to SIRPα expressed on myeloid cells2–5. Using a haploid genetic screen, we here identify glutaminyl-peptide cyclotransferase-like (QPCTL) as a major component of the CD47-SIRPα checkpoint. Biochemical analysis demonstrates that QPCTL is critical for pyroglutamate formation on CD47 at the SIRPα binding site shortly after biosynthesis. Both genetic and pharmacological interference with QPCTL activity enhances antibody-dependent cellular phagocytosis and cellular cytotoxicity of tumor cells. Furthermore, interference with QPCTL expression leads to a major increase in neutrophil-mediated tumor cell killing in vivo. These data identify QPCTL as a novel target to interfere with the CD47 pathway, and thereby augment antibody therapy of cancer.
CD47 is a broadly expressed inhibitory ligand for myeloid cells4,6. However, CD47 expression on tumor cells can be higher than on surrounding healthy tissue cells, providing a first rationale for the clinical targeting of this myeloid cell checkpoint3,7. In addition, pro-phagocytic signals such as calreticulin and SLAMF7 can be expressed on the surface of cancer cells8,9, leading to increased phagocytosis and clearance of these cells upon CD47-SIRPα blockade. Finally, pro-phagocytic signals can also be provided by therapy, in particular by administration of opsonizing tumor-specific antibodies, such as the anti-CD20 antibody rituximab, anti-Her2 antibody trastuzumab and anti-EGFR antibody cetuximab. In support of this strategy, preclinical data have demonstrated that targeting of the CD47-SIRPα axis in combination with tumor-opsonizing antibodies leads to increased tumor control10–12. Recent work has provided the first clinical validation of the CD47-SIRPα axis as a myeloid checkpoint, by demonstrating a 50% objective response rate of the combination of rituximab plus anti-CD47 in rituximab-refractory non-Hodgkin's Lymphoma13.
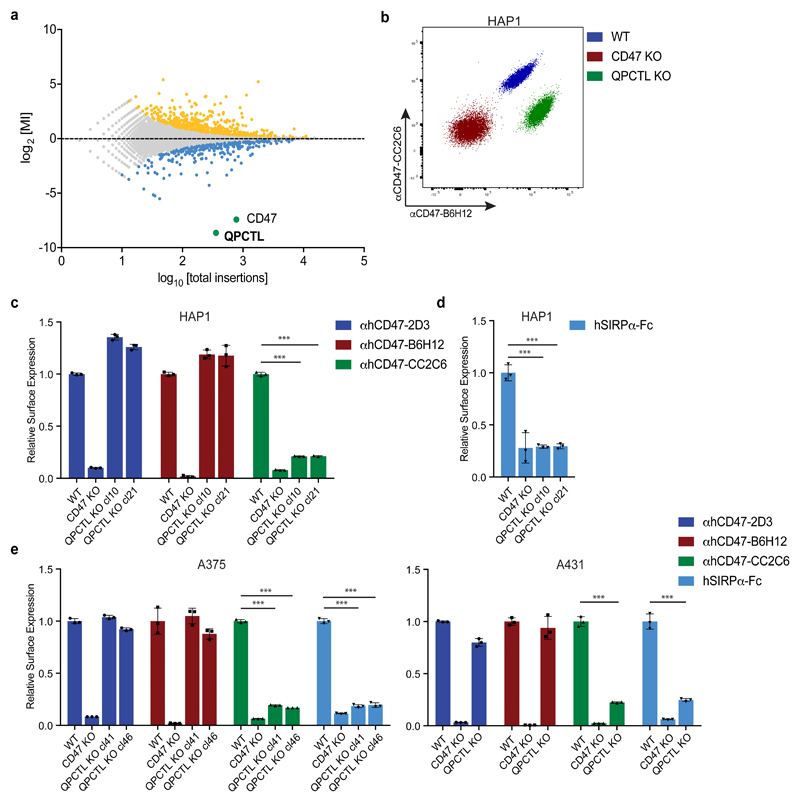
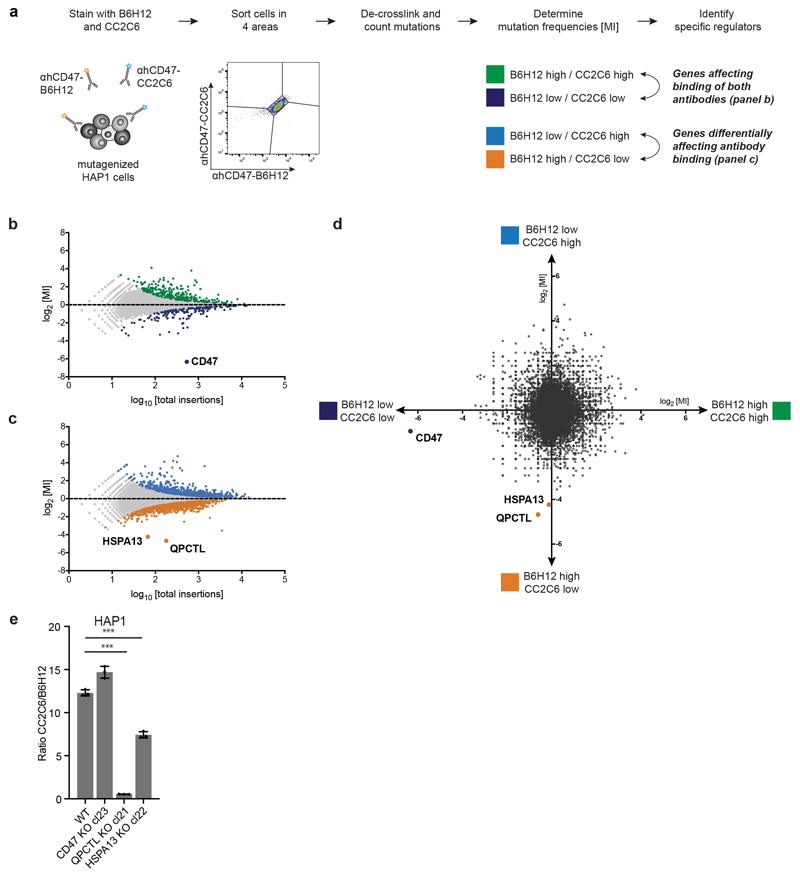
In spite of the potential value of the CD47 checkpoint molecule as a therapeutic target, transcriptional control by the Myc oncogene thus far forms the only identified regulatory mechanism of protein expression or function14. To reveal novel genetic determinants of CD47-SIRPα binding, we performed a fluorescence-activated cell sorting (FACS)-based haploid genetic screen, using an antibody against human CD47 (αhCD47-CC2C6) that binds to the SIRPα recognition site15. Analysis of gene-trap integration sites in cells with impaired αhCD47-CC2C6 binding revealed two strong hits, the CD47 gene itself, and the gene encoding glutaminyl-peptide cyclotransferase-like (QPCTL, isoQC) (Fig. 1a)(see Supplementary Table 1 for a full overview of screen results). QPCTL has been described as a Golgi-resident enzyme16 that, like its secreted family member QPCT, can catalyse the cyclisation of N-terminal glutamine and glutamic acid residues on target proteins into an N-terminal pyroglutamate-residue (pGlu)17,18. Pyroglutamate modification of proteins by QPCT has been implicated in Huntington’s Disease19 and Alzheimer’s Disease20–22. Notably, crystallographic analysis of the CD47 protein previously demonstrated the presence of an N-terminal pyroglutamate that, based on its hydrogen bonding, was considered to be involved in SIRPα binding23. However, this pyroglutamate was assumed to be formed spontaneously upon cleavage of the CD47 signal sequence24.
Fig. 1. Identification of QPCTL as a modulator of CD47-SIRPα binding.
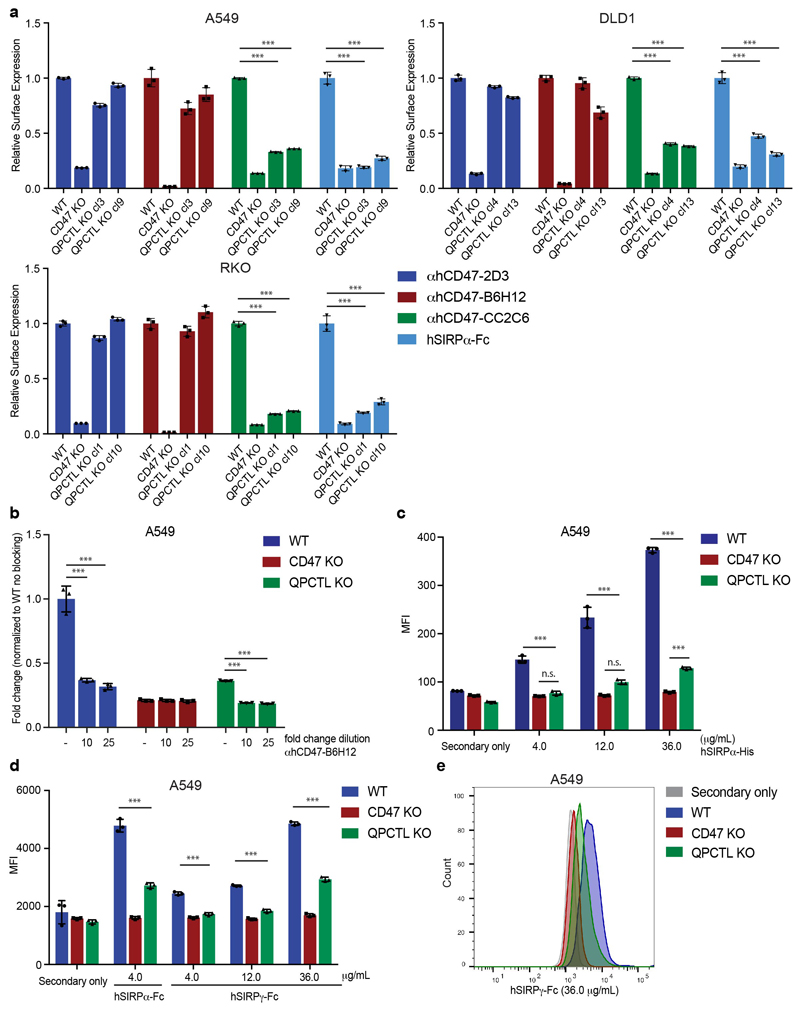
a, Flow-cytometry-based haploid genetic screen for modulators of CD47, as detected by anti-human CD47 antibody clone CC2C6 (αhCD47-CC2C6) binding. Dots represent individual genes; x axis indicates the number of disruptive insertions per gene; y axis shows the frequency of independent insertions in cells with high CD47 expression (CD47-CC2C6HIGH channel) over the frequency of insertions in cells with low CD47 expression (CD47-CC2C6LOW channel) for each gene. Light-blue and orange dots indicate genes with significant enrichment of insertions within the CD47-CC2C6HIGH and CD47-CC2C6LOW populations, respectively. Green dots represent CD47 and QPCTL. Significant enrichment of insertions (total amount of insertions n=3,254,889) was determined by two-sided Fisher’s Exact test with multiple comparison correction (FDR-corrected P < 0.05).
b, Flow cytometry plot of surface binding of anti-human CD47 antibody clone B6H12 (αhCD47-B6H12) and αhCD47-CC2C6 to HAP1 WT, CD47 KO and QPCTL KO (cl21) cells. Data are representative of two independent experiments with similar results (n=3 biological replicates per experiment).
c, Cell surface binding of anti-human CD47 antibody clone 2D3 (αhCD47-2D3), αhCD47-B6H12 and αhCD47-CC2C6 to HAP1 WT, CD47 KO or QPCTL KO cells, as determined by flow cytometry. Values indicate MFI relative to WT cells stained with the same reagent. Data represent n=3 biological replicates and mean ± s.d. of triplicates. ***P=0.0001 by one-way analysis of variance (ANOVA).
d, Cell surface binding of human SIRPα-Fc (hSIRPα-Fc) to HAP1 WT, CD47 KO or QPCTL KO cells (cl10 and cl21), as determined by flow cytometry. Values indicate MFI relative to WT cells. Data represent n=3 biological replicates and mean ± s.d. of triplicates. ***P=0.0001 by one-way ANOVA.
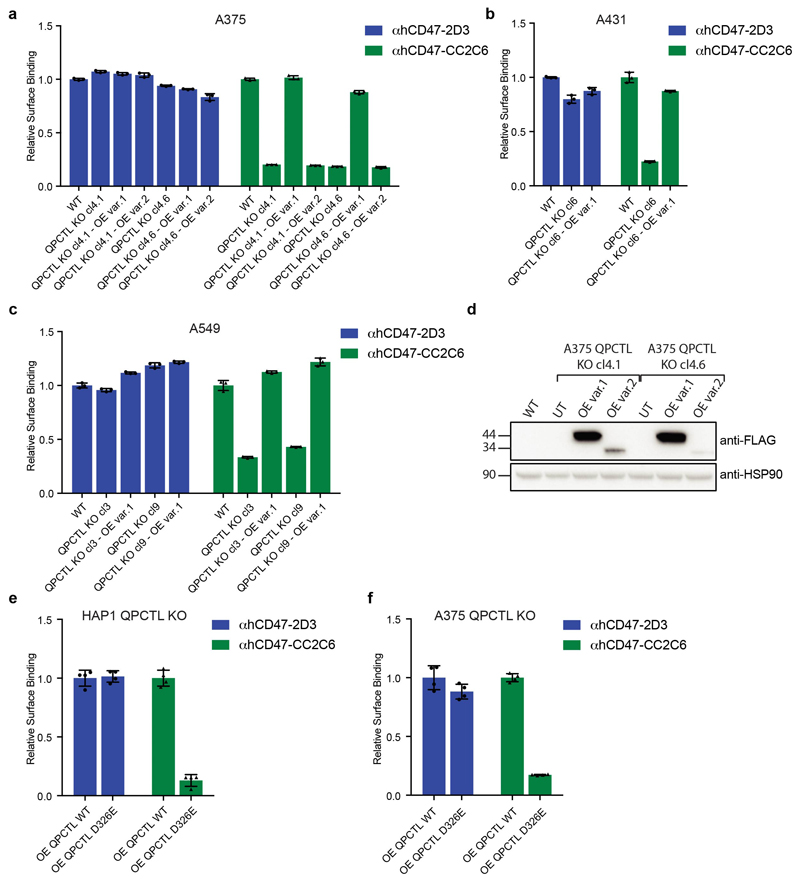
e, Cell surface binding of αhCD47-2D3, αhCD47-B6H12, αhCD47-CC2C6 and hSIRPα-Fc to WT, CD47 KO and QPCTL KO (cl4.1 and cl4.6) A375 melanoma cells and to WT, CD47 KO and QPCTL KO (cl6) A431 epidermoid carcinoma cells, as determined by flow cytometry. Values indicate MFI relative to WT cells stained with the same reagent. Data represent n=3 biological replicates and mean ± s.d. of triplicates. ***P≤0.0001 by one-way ANOVA (left graph) or unpaired two-sided t-test (right graph).
Data are representative of one (a), or at least two (b-e) independent experiments.
MI, mutation index; MFI, mean fluorescence intensity; WT, wild-type; KO, knock-out.
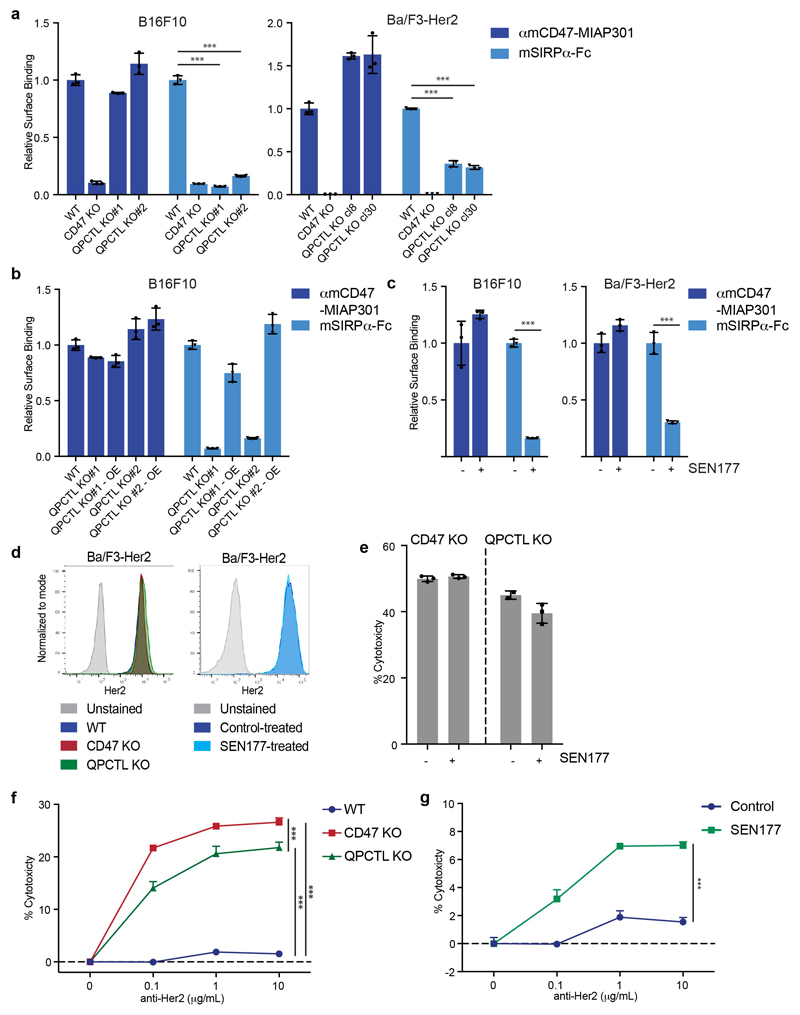
To determine how QPCTL influences the CD47 protein we generated CD47-deficient and QPCTL-deficient HAP1 cells. As expected, CD47 deficiency led to impaired binding of both recombinant human SIRPα (hSIRPα-Fc) and all anti-CD47 antibodies tested (αhCD47-CC2C6, αhCD47-2D3, αhCD47-B6H12). In contrast, QPCTL-knockout selectively affected binding of recombinant hSIRPα and αhCD47-CC2C6, while overall cell surface CD47 levels, as determined by binding of other anti-CD47 antibodies (αhCD47-2D3 and αhCD47-B6H12), remained unaltered (Fig. 1b, c and d). The role of QPCTL as a modifier of CD47 was not restricted to HAP1 but was likewise observed in malignant melanoma (A375), epidermoid carcinoma (A431), lung cancer (A549), colorectal cancer (DLD1) and rectal carcinoma (RKO) cancer cells (Fig. 1e and Extended Data Fig. 1a). Low-level residual binding of monomeric hSIRPα protein to QPCTL knockout cells was only observed at high concentrations, indicating that the binding affinity of SIRPα to CD47 is to a large extent dependent on QPCTL (Extended Data Fig. 1b, c). Binding of SIRPγ, a CD47 ligand without known signalling function that is primarily expressed on T cells25, was likewise affected by QPCTL deficiency (Extended Data Fig. 1d and e). Reconstitution experiments demonstrated that the CD47-modifying activity is encoded by QPCTL transcript variant 1 (Extended Data Fig. 2a-d), and introduction of a catalytically dead QPCTL variant (D326E, generated based on homology with QPCT)26 demonstrated that the enzymatic activity of QPCTL is essential for its role as a CD47 modifier (Extended Data Fig. 2e, f).
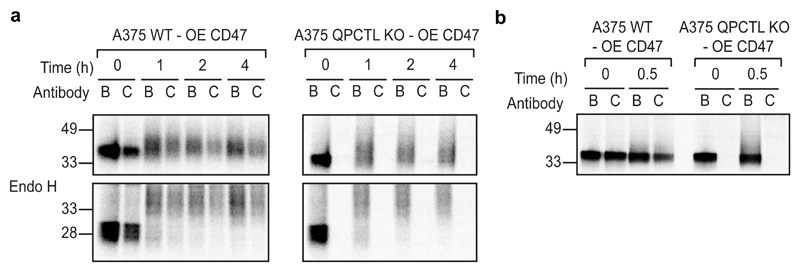
To assess where in the protein-life cycle CD47 is modified by QPCTL, the fate of CD47 molecules in wild-type and QPCTL-knockout melanoma cells transduced with an HA-tagged CD47 gene product was analysed by pulse-chase analysis. Comparison of immunoprecipitates obtained with αhCD47-CC2C6 and αhCD47-B6H12 revealed a selective loss of the CD47 conformation recognized by αCD47-CC2C6 in QPCTL deficient cells (Fig. 2a). Notably, at all time-points analysed no discernible levels of CD47 protein could be isolated by αhCD47-CC2C6, indicating that pyroglutamate formation on CD47 is strictly dependent on QPCTL, and does not involve an appreciable level of spontaneous conversion. QPCTL-mediated CD47 modification occurs very early in the protein life-cycle, as demonstrated by the presence of αhCD47-CC2C6-reactive CD47 molecules that are sensitive to deglycosylation by endoglycosidase H, indicating endoplasmic reticulum/early Golgi residence (Fig. 2a), and as demonstrated by the fact that a maximal level of pyroglutamate-modified CD47 is already reached after a 10’ minute labelling (Fig. 2b). To reveal possible additional regulators of formation of the SIRPα binding site with high sensitivity, we performed a 2-dimensional genetic screen (see Extended Data Fig. 3a) to distinguish genes that influence binding of both αhCD47-CC2C6 and αhCD47-B6H12 (i.e. general CD47 protein regulators), from genes that influence binding of αhCD47-CC2C6 but not αhCD47-B6H12 (i.e. selective regulators of the SIRPα binding site). This screen confirmed the role of QPCTL as a regulator of the the SIRPα binding site, but also identified HSPA13, encoding the Heat shock protein HSPA13, as a modifier of αhCD47-CC2C6, but not αhCD47-B6H12, binding (Extended Data Fig. 3b-e). HSPA13 has been described as an ER-resident protein27, consistent with a possible role of HSPA13 in regulating the efficiency of CD47 modification by QPCTL.
Fig. 2. Pyroglutamate formation occurs early in the CD47 protein life-cycle and is fully dependent on QPCTL.
a, SDS-PAGE analysis of αhCD47-B6H12 (B) or αhCD47-CC2C6 (C) immunoprecipitates from CD47-HA-overexpressing WT or QPCTL KO (cl4.1) A375 melanoma cells after a 0, 1, 2 or 4 hours (h) chase period following a 30’ 35S methionine/cysteine labelling.
b, SDS-PAGE analysis of αhCD47-B6H12 (B) or αhCD47-CC2C6 (C) immunoprecipitates from CD47-HA-overexpressing WT or QPCTL KO (cl4.1) A375 melanoma after a 0 or 30’ chase following a 10’ 35S methionine/cysteine labelling.
Data are representative of two (a, b) independent experiments with similar results. Blot images are cropped to show the relevant bands, and molecular mass markers are indicated (in kD). See Source Data for the uncropped western blots.
OE, over expression; B, αhCD47-B6H12; C, αhCD47-CC2C6.
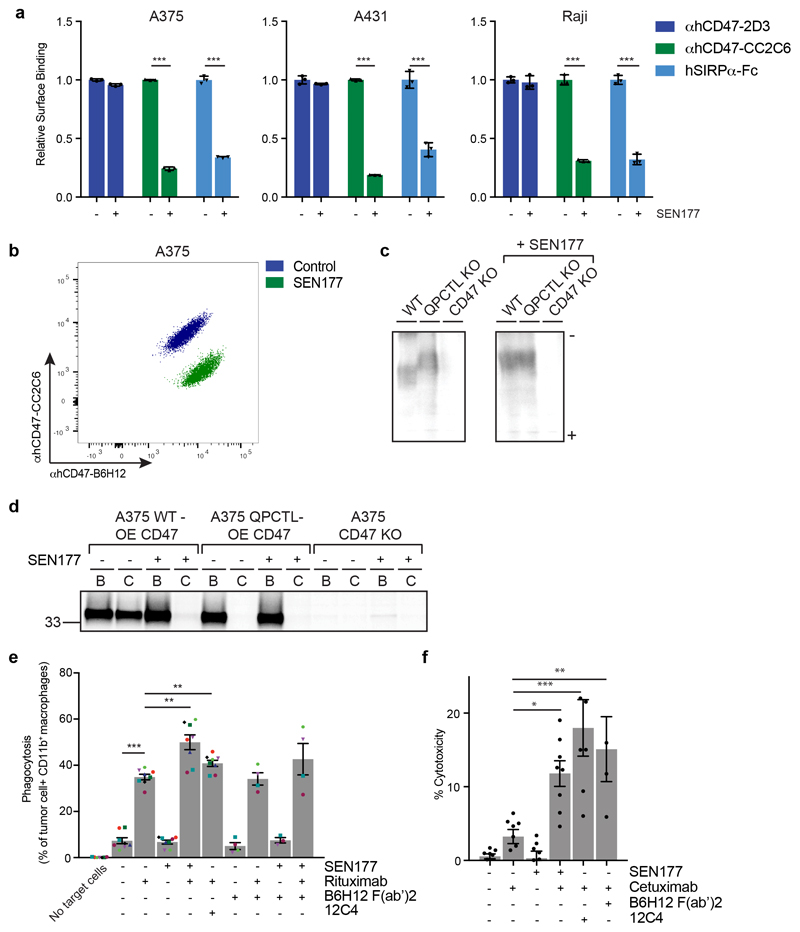
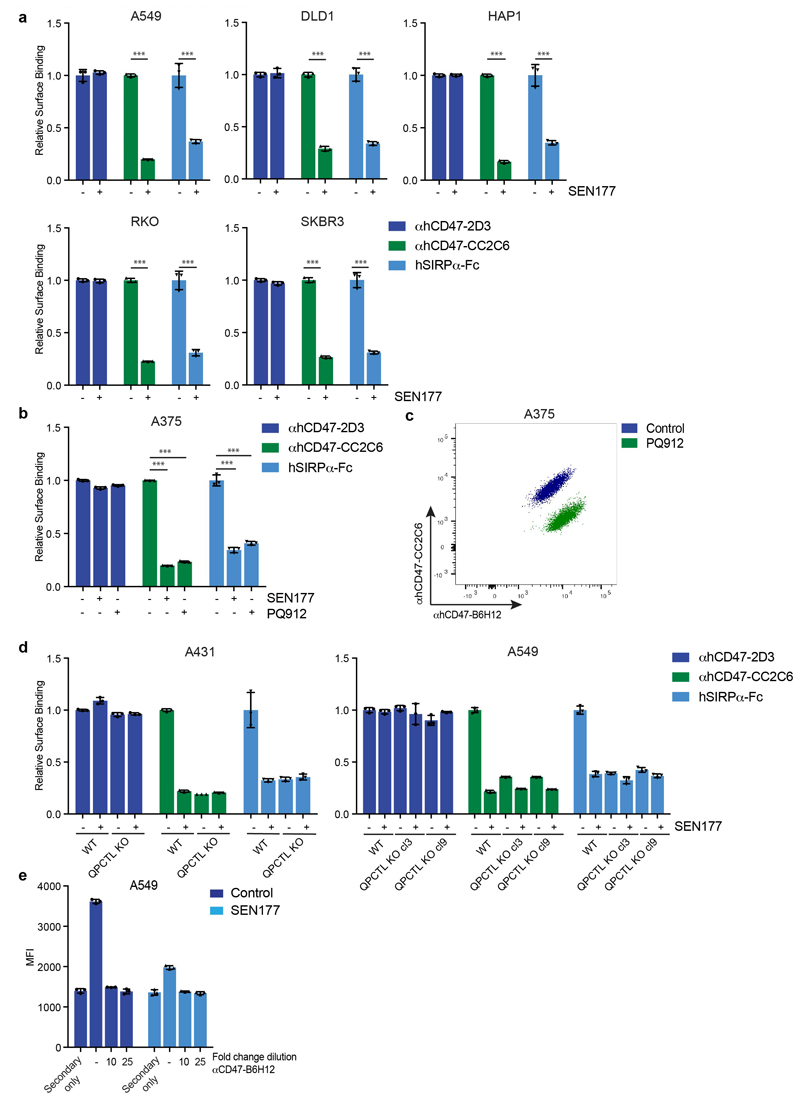
To prevent the generation of pathogenic pyroglutamate-modified proteins implicated in Huntington’s Disease and Alzheimer’s Disease by the QPCTL family member QPCT, glutaminyl cyclase inhibitors have been developed and are undergoing clinical evaluation19,28,29. To determine whether small molecule inhibition of QPCTL could be used to influence CD47 binding to SIRPα, we selected two QPCT inhibitors that inhibit QPCTL with high efficiency19,30. Treatment with the glutaminyl cyclase inhibitor SEN177 (IC50 of 0.013μM for QPCTL) reduced hSIRPα-Fc staining for 8 out of 8 cell lines tested (Fig. 3a and b, Extended Data Fig. 4a), and to the same extent as QPCTL gene deletion (Fig. 1c-e, Extended Data Fig. 1a), while CD47 surface levels remained unaffected. Treatment with the glutaminyl cyclase inhibitor PQ912 yielded similar results (Extended Data Fig. 4b and c).
Fig. 3. Synergy between blockade of CD47 pyroglutamate formation and tumor opsonization in tumor cell killing by macrophages and neutrophils.
a, Cell surface binding of αhCD47-2D3, αhCD47-CC2C6 and hSIRPα-Fc to control (DMSO)-treated (-) or SEN177-treated (+) melanoma (A375), epidermoid carcinoma (A431) and Burkitt’s lymphoma (Raji) cells, as determined by flow cytometry. Values indicate MFI relative to WT cells stained with the same reagent. Data represent n=3 biological replicates and mean ± s.d. of triplicates. ***P≤0.000373 by unpaired two-sided t-test.
b, Flow cytometry plot of surface binding of αhCD47-B6H12 and αhCD47-CC2C6 to control-treated or SEN177-treated melanoma (A375) cells. Data are representative of two independent experiments with similar results (n=3 biological replicates per experiment).
c, Isoelectric focusing analysis of αhCD47-B6H12 immunoprecipitates from CD47-HA-overexpressing WT, CD47-HA-overexpressing QPCTL KO, or CD47 KO melanoma (A375) cells left untreated (-) or treated with SEN177 (+). Data are representative of two independent experiments with similar results. Gel image is cropped to show the relevant bands. See Source Data for the uncropped gel.
d, SDS-PAGE analysis of αhCD47-B6H12 (B) or αhCD47-CC2C6 (C) immunoprecipitates from CD47-HA-overexpressing WT, QPCTL KO (cl4.1), or CD47 KO melanoma (A375) cells after a 30’ 35S methionine/cysteine labelling in the presence (+) or absence (-) of SEN177. Data are from one experiment. Blot image is cropped to show the relevant bands, and molecular mass markers are indicated (in kD). See Source Data for the uncropped western blot.
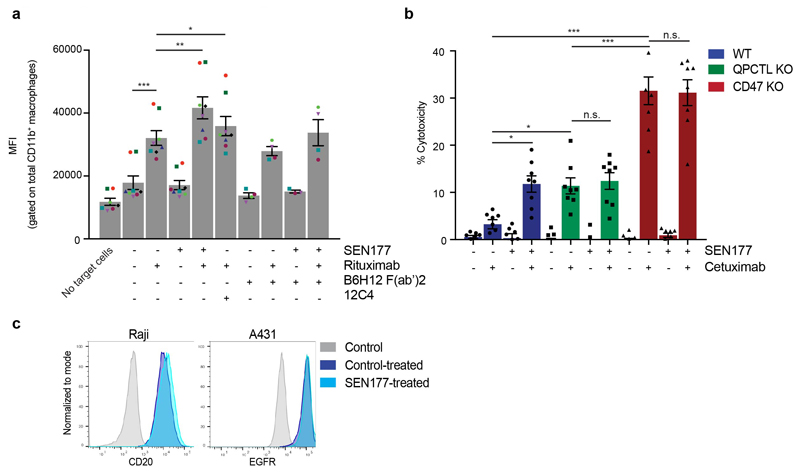
e, Phagocytosis of control-treated (DMSO) (-) or SEN177-treated (+) Burkitt’s lymphoma (Raji) cells in the presence or absence of the anti-human CD20 antibody rituximab, CD47-blocking F(ab’)2 fragment B6H12, or SIRPα blocking antibody 12C4 by human macrophages, as determined by ImageStream analysis. Symbols represent individual donors. Data represent mean ± s.d. of independent donors. ***P<0.0001; 0.0027≥**P≥0.0024 by one-way ANOVA.
f, Specific lysis of control-treated (-) or SEN177-treated (+) epidermoid carcinoma (A431) cells by human neutrophils in the presence or absence of the anti-human EGFR antibody cetuximab, CD47-blocking F(ab’)2 fragment B6H12, or SIRPα blocking antibody 12C4 in a 4 hour 51Cr-release assay. Data represent mean ± s.d. of independent donors. *P=0.0207; **P=0.0055; ***P<0.0001 by one-way ANOVA.
Data are representative of at least two independent experiments (a, b, e, f), evaluating effector cells from 4 donors (for B6H12(Fab’)2 conditions) or 8 donors (all other conditions) (e, f).
Upon cyclisation of an N-terminal residue to form a pyroglutamate, a leaving amino group is replaced by a hydroxyl group31, thereby altering the isoelectric point (pI) of the molecule. We used one-dimensional isoelectric focusing (1D-IEF) to visualise this alteration of pI of HA-tagged CD47 in wild-type and QPCTL-knockout A375 melanoma cells. CD47 from QPCTL-knockout cells was characterized by an increased pI as compared to CD47 from wild-type cells (Fig. 3c). Furthermore, treatment with SEN177 increased the pI of CD47 present in wild-type cells to that of QPCTL-knockout cells, but did not affect the pI of CD47 molecules isolated from QPCTL-knockout cells. As a second approach to assess to what extent CD47 modification can be influenced by small molecule inhibition, we performed Western blot analysis of immunoprecipitates of HA-tagged CD47 from untreated or SEN177-treated cells. In line with the IEF data, SEN177 resulted in a near-complete inhibition of pGlu-CD47 formation (Fig. 3d). Finally, SEN177-treatment of QPCTL-deficient lung cancer (A549) and epidermoid carcinoma (A431) cells did not further reduce SIRPα binding (Extended Data Fig. 4d and e). Together these data demonstrate 1). that QPCTL activity is critical for the formation of pGlu-modified CD47, 2). that glutaminyl cyclase inhibitors alter the CD47 protein by inhibiting QPCTL function, and 3). that the resulting block in pGlu-modified CD47 is near complete.
A hypothesis arising from the above data is that QPCTL deletion or inhibition could limit signalling through the CD47-SIRPα axis, thereby enhancing the capacity of macrophages and neutrophils to kill opsonized tumor cells by antibody-dependent cellular phagocytosis (ADCP) and antibody-dependent cellular cytotoxicity (ADCC). In vitro phagocytosis assays revealed that SEN177 treatment increased ADCP of anti-CD20 treated Burkitt’s lymphoma (Raji) cells by human macrophages to the same or higher extent than treatment with the combination of rituximab and the SIRPα-blocking agent 12C4, or the combination of rituximab and the CD47-blocking agent B6H12 F(ab’)27 (Fig. 3e, Extended Data Fig. 5 and Extended Data Fig. 6a). Likewise, both SEN177-treatment and knockout of QPCTL increased neutrophil-mediated ADCC of EGFR-expressing A431 epidermoid carcinoma cells in the presence of the EGFR-specific antibody cetuximab (Fig. 3f and Extended Data Fig. 6b and c). Thus, inhibition or knockout of QPCTL leads to increased killing of target cells by both human neutrophils and macrophages, indicating functional disruption of the CD47-SIRPα inhibitory pathway.
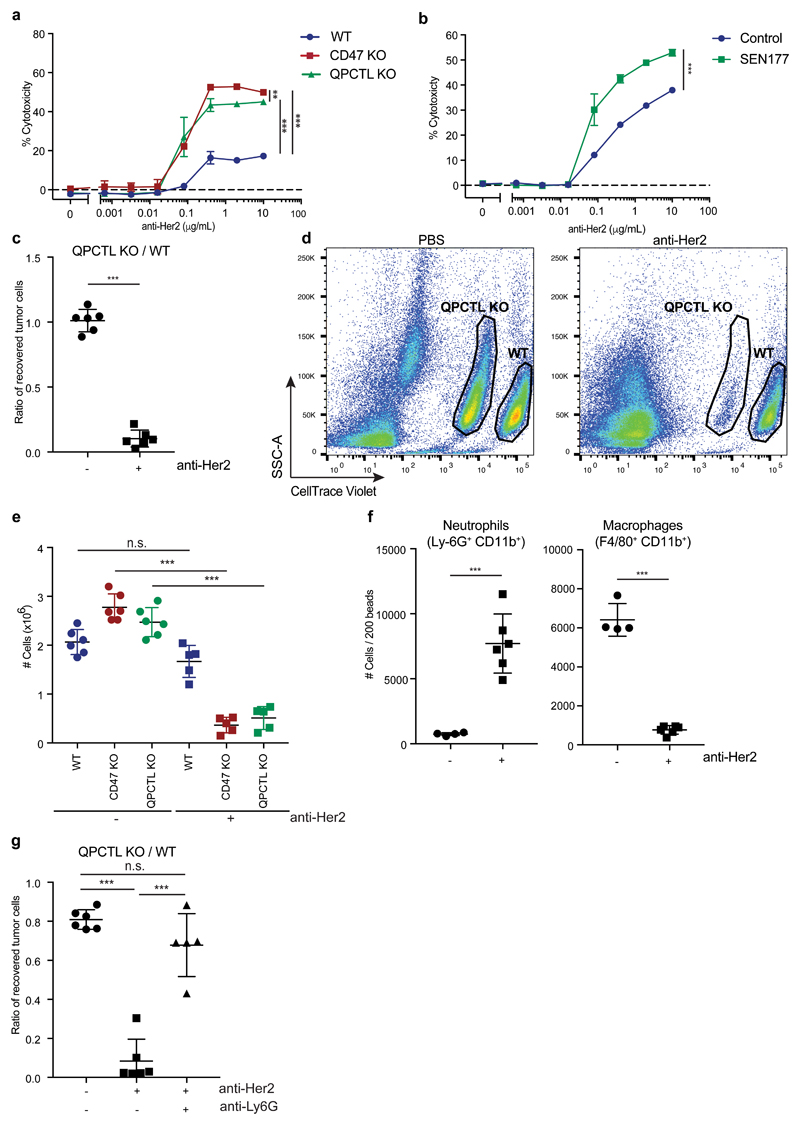
The role of QPCTL as a modifier of the CD47 protein is conserved in mice. Specifically, deletion of QPCTL in either B16F10 melanoma cells or Ba/F3 pro-B cells reduced binding of murine SIRPα (mSIRPα-Fc), and this could be restored by lentiviral overexpression of QPCTL (Extended Data Fig. 7a and b). Likewise, treatment with SEN177 led to reduced binding of SIRPα without altering total CD47 surface levels (Extended Data Fig. 7c). Next, we utilized Ba/F3 cells that express human Her2 to evaluate whether inhibition of pyroglutamate formation could increase killing of tumor cells by human neutrophils in the presence of anti-Her2 antibody. Similar to what was observed for human target cells, both QPCTL-knockout and SEN177 treatment synergized with anti-Her2 treatment to induce neutrophil-mediated lysis of tumor cells (Fig. 4a and b and Extended Data Fig. 7d). Killing efficiency of CD47- and QPCTL-deficient tumor cells by neutrophils was not further enhanced by SEN177-treatment, demonstrating that the functional effects of SEN177 are dependent on the CD47 pathway (Extended Data Fig. 7e). The same effects of QPCTL deletion and small molecule inhibition were observed when using murine immune cells isolated from whole blood as effector cells (Extended Data Fig. 7f and g).
Fig. 4. QPCTL deficiency and QPCTL inhibition enhance tumor cell control by tumor-specific antibodies.
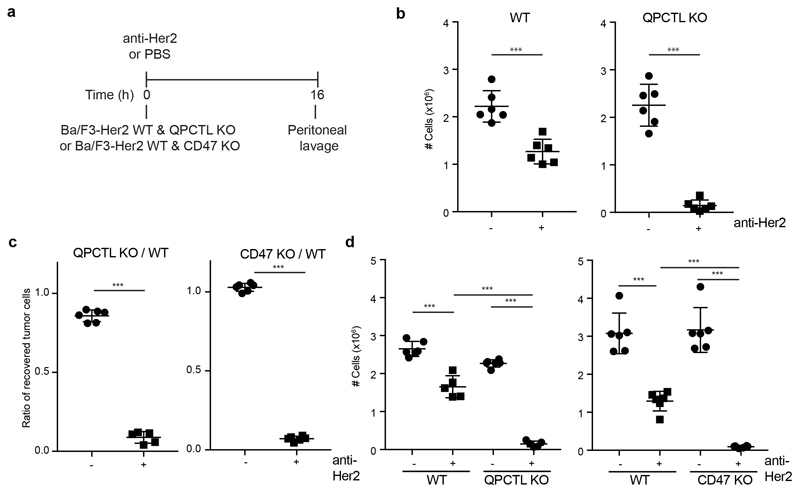
a, Specific lysis of WT, CD47 KO and QPCTL KO Her2-expressing murine pro-B cells (Ba/F3-Her2) by human neutrophils in the presence or absence of anti-Her2 (IgA1) in a 4 hour 51Cr-release assay. Data represent n=3 biological replicates and depict mean ± s.d. of one representative donor. ***P<0.0002; **P=0.087 by one-way ANOVA.
b, Specific lysis of control (DMSO)-treated (-) or SEN177-treated (+) Her2-expressing murine pro-B (Ba/F3-Her2) by human neutrophils in the presence or absence of anti-Her2 (IgA1) in a 4 hour 51Cr-release assay. Data represent n=3 biological replicates and depict mean ± s.d. of one representative donor. ***P=0.0001 by unpaired two-sided t-test.
c, In vivo killing of target cells in mice injected with a 1:1 mixture of WT and QPCTL KO Her2-expressing murine pro-B cells (Ba/F3-Her2) and treated with control (PBS) or anti-Her2 (IgA1) antibody. Data represent the ratio between QPCTL KO and WT Ba/F3-Her2 cells in mice treated with PBS (dots) or anti-Her2 (IgA1) (squares). n=6 animals per group. Data represent mean ± s.d. of independent mice. ***P=0.0001 by unpaired two-sided t-test.
d, Representative flow analysis plots of (c) of recovered WT and QPCTL KO tumor cells in mice treated with control (PBS) or anti-Her2 (IgA1). Data are representative of three independent experiments with similar results.
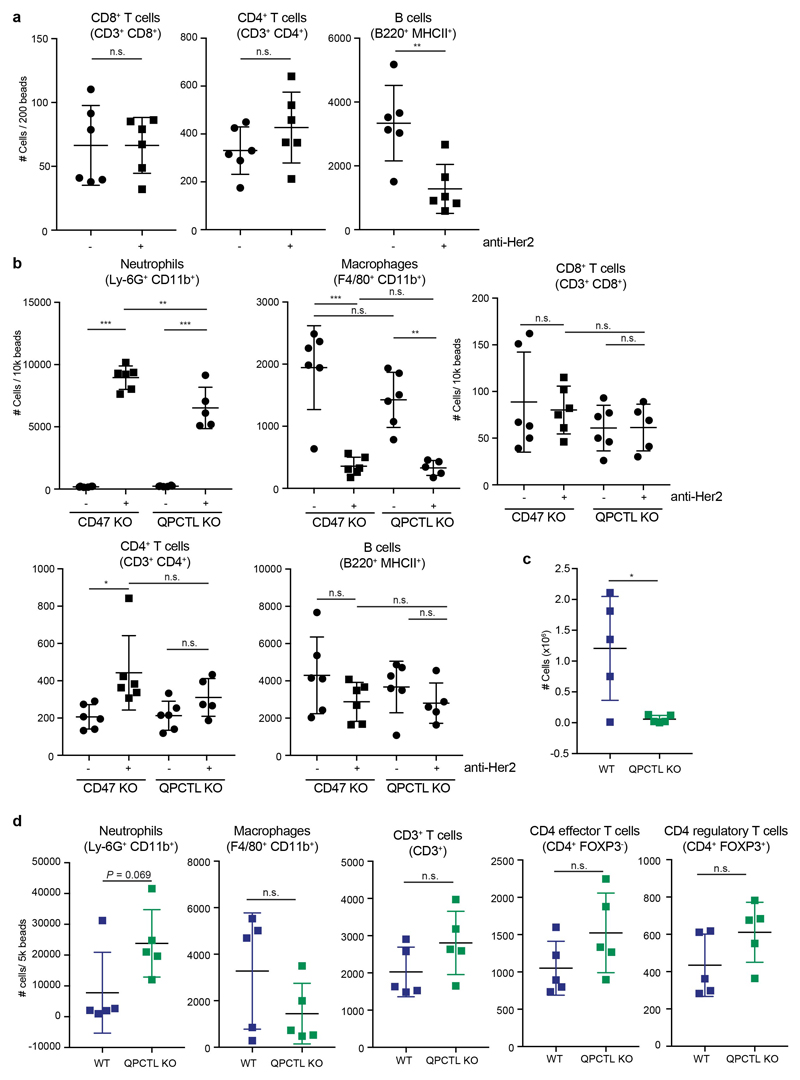
e, In vivo killing of target cells in mice injected with a 1:1:1 mixture of WT, CD47 KO and QPCTL KO Ba/F3-Her2 cells and treated with control (PBS) or anti-Her2 (IgA1) antibody. Data represent the number of cells in mice treated with PBS (dots) or anti-Her2 (IgA1) (squares). n=6 control-treated animals; n=5 anti-Her2-treated animals. Data represent mean ± s.d. of independent mice. ***P<0.0001 by one-way ANOVA; n.s., not significant.
f, Number of peritoneal neutrophils (Ly-6G+CD11b+) and macrophages (F4/80+CD11b+) present in recipients of a 1:1 mixture of WT and QPCTL KO Her2-expressing murine pro-B cells that were either treated with PBS (-) or with anti-Her2 (IgA1) (+) antibody, 16 hours after treatment. n=4 control-treated animals; n=6 anti-Her2-treated animals. Data represent mean ± s.d. of independent mice. ***P≤0.0003 by unpaired two-sided t-test.
g, In vivo killing of target cells in mice injected with a 1:1 mixture of WT and QPCTL KO Ba/F3-Her2 cells and treated with control (PBS), anti-Her2 (IgA1) antibody or anti-Her2 (IgA1) antibody plus neutrophil-depleting antibody anti-Ly6G. Data represent the ratio between QPCTL KO and WT Ba/F3-Her2 cells in mice treated with PBS (dots), anti-Her2 (IgA1) (squares) or anti-Her2 (IgA1) plus anti-Ly6G (pyramids). n=6 control-treated animals; n=6 anti-Her2-treated animals, n=5 anti-Her2- plus anti-Ly6G-treated animals. Data represent mean ± s.d. of independent mice. ***P<0.0001 by one-way ANOVA; n.s., not significant.
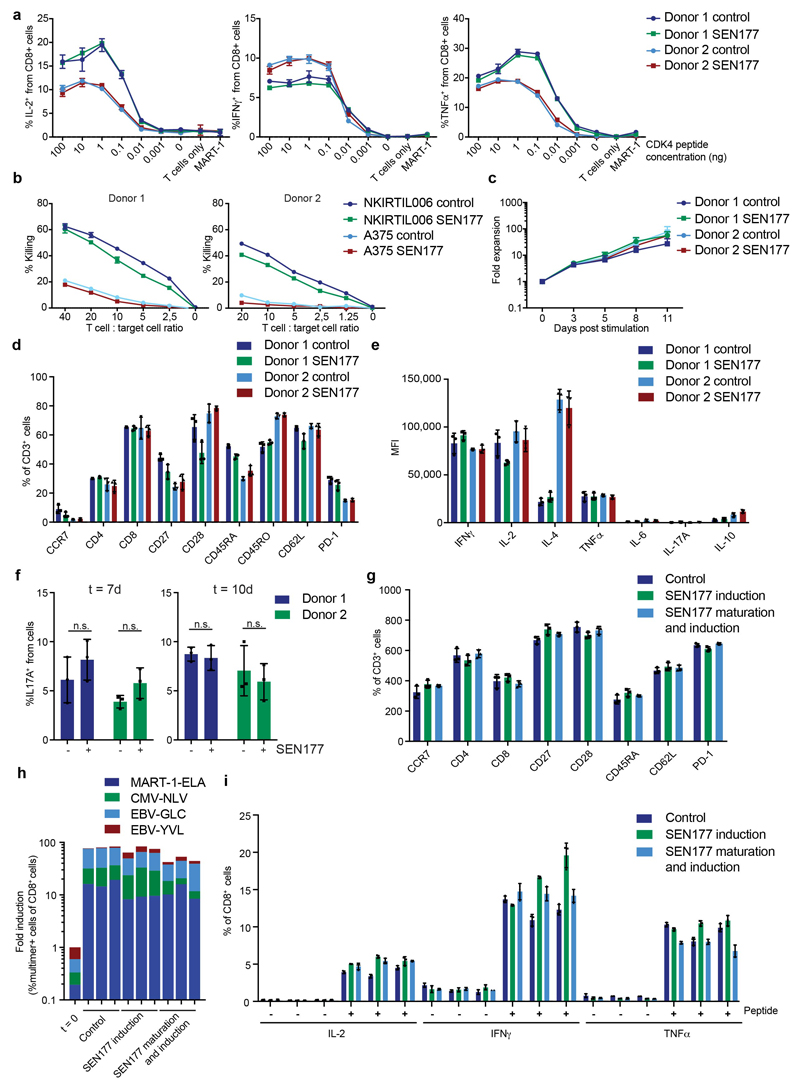
Having established QPCTL as a modifier of the mouse CD47 –SIRPα checkpoint, we next used Her2-expressing Ba/F3 cells in a short-term syngeneic peritoneal tumor model to assess the role of QPCTL in tumor cell killing in vivo32 (L.W. Treffers, T.T. Broeke, T. Rösner, J.H.M. Jansen, M. van Houdt, S. Kahl, K. Schornagel, P.J.J.H. Verkuijlen, K. Franke, T.W. Kuijpers, T.K. van den Berg, T.Valerius, J.H.W. Leusen, H.L. Matlung). Human FcαRI transgenic (Tg) BALB/c mice were injected with a 1:1 mixture of wild-type and QPCTL-deficient cells or, as a comparison, wild-type and CD47-deficient cells. Subsequently, mice were treated with anti-Her2 antibody or PBS, and after 16 hours the ratio of QPCTL-deficient versus wild-type cells was analysed (Extended Data Fig. 8a). In PBS-treated mice, the ratio of QPCTL deficient cells versus wild-type cells remained unaffected, indicating that QPCTL does not influence short-term tumor cell engraftment. In contrast, in mice treated with anti-Her2 a profound killing of QPCTL-deficient tumor cells over wild-type cells was observed (ratio WT: QPCTL deficient in anti-Her2 treated mice of 10 to 1) (Fig. 4c and d, Extended Data Fig. 8b). Of note, the enhanced tumor cell killing achieved by blockade of pyroglutamate formation was similar to that achieved by full genetic deficiency of the CD47 checkpoint (Extended Data Fig. 8c and d), also when CD47 and QPCTL deficient tumor cells were co-injected (Fig. 4e). The selective killing of QPCTL (and CD47)-deficient cells in anti-Her2-treated mice was accompanied by a large influx of neutrophils, suggesting a positive feedback loop that enhances anti-tumor immunity (Fig. 4f and Extended Data Fig. 9a and b). Similarly, injection of QPCTL-deficient cells in anti-Her2-treated mice led to a profound increase in neutrophils relative to recipients of wild-type cells, with only small changes in T cell populations (Extended Data Fig. 9c and d). Notably, the selective killing of QPCTL-deficient cells in anti-Her2-treated mice was largely abrogated by the pre-treatment with anti-Ly6G antibody (Fig. 4g), indicating that neutrophils are the dominant effector cells that eliminate QPCTL-deficient cells in this model. In solid tumor models, activity of CD47 blockade has been shown to depend on both innate immune cells and T cells33–36. It remains to be established whether in vivo QPCTL inhibition would alter intratumoral T cell activity. However, as shown by a series of in vitro assays, both the expansion, differentiation, cytokine production, and killing capacity of human T cells is compatible with small molecule inhibition of QPCTL (Extended Data Fig. 10).
We conclude that QPCTL is a critical modifier of the CD47 protein and, as such, a regulator of the CD47-SIRPα pathway. Prior research has provided strong evidence that targeting of the CD47 axis can promote tumor control by both innate and adaptive immune cells, and clinical development of CD47 antagonists is ongoing13,37. The poor PK of SEN177 in mice (a drop in serum concentration of about 1,000 fold in an 8 hour time period, data not shown), precludes in vivo experiments using this inhibitor, but it will be interesting to assess whether pharmacological manipulation of QPCTL activity with other existing30 or novel inhibitors can be utilized to promote tumor immunity. CD47 molecules in tumor cells in which pGlu formation is blocked do display a low remaining affinity for SIRPα, as shown by residual binding of monomeric SIRPα at high concentration. However, blockade of pGlu formation enhances anti-tumor activity of myeloid cells to a similar extent as antibody blockade of CD47 or SIRPα (Fig. 3e and f), and deletion of CD47 and QPCTL likewise leads to comparable tumor control in vivo (Fig. 4e), suggesting that this residual affinity does not have significant functional effects. Furthermore, QPCTL inhibitors should not be affected by the described antigen sink of anti-CD47 therapy due to CD47 expression on erythrocytes and other cells34. In addition, the fact that CD47 molecules that newly arrive at the cell surface upon QPCTL inhibition already lack the pGlu modification may potentially form an advantage relative to antagonistic antibodies that need to compete with SIRPα in the tumor microenvironment. Phase I/ 2a studies that evaluate QPCT/QPCTL inhibitors for the treatment of neurodegenerative diseases have not revealed any significant toxicities30,38, and the development of QPCTL inhibitors that may either be used in isolation or in combination with CD47-blocking antibodies will be a useful goal. Finally, from a biological perspective it is interesting to note that human and parasitic genomes encode pyroglutamate aminopeptidases that can remove N-terminal pyroglutamate residues from proteins39 and stimulation with LPS leads to increased expression of pyroglutamate aminopeptidase 140. Whether such aminopeptidases can act as external modifiers of the CD47–SIRPα signalling pathway remains to be established.
Materials and Methods
Haploid genetic flow cytometry-based screen
In order to identify genetic modulators of CD47, as measured by binding of the anti-human CD47-antibody CC2C6 (αhCD47-CC2C6) that recognizes the SIRPα binding site, mutagenized HAP1 cells were prepared using gene-trap retrovirus expressing blue fluorescent protein (BFP), as described previously 41. Briefly, 50 million HAP1 cells were seeded and transduced with virus from two combined harvests on three consecutive days in the presence of 8 μg/mL protamine sulfate (Sigma). The mutagenized cell library was then expanded to 30 T175 flasks at a confluence of approximately 80%. Subsequently, cells were dissociated with trypsin, washed once with PBS (Lonza) and stained with FITC-labelled αhCD47-CC2C6 (Biolegend) at a 1:80 dilution for 30 minutes in 20 mL PBS containing 0.5% w/v bovine serum albumin (BSA; Sigma) and 0.2% w/v sodium azide (Sigma) at 4°C, protected from light. Subsequently, cells were washed three times with PBS containing 10% FCS and stained with FITC-labelled polyclonal goat anti-mouse IgG: Poly4053 (Biolegend) at a 1:100 dilution for 30 minutes in PBS containing 0.5% w/v BSA and 0.2% w/v sodium azide at 4°C, protected from light. Following two washes with PBS containing 10% FCS and one wash with PBS, cells were fixed with BD Fix Buffer I (BD biosciences) for 10 minutes at 37°C. After two washes with PBS containing 10% FCS, cells were filtered through a 40 μm strainer (BD Falcon) before isolation of the two cell populations of interest (i.e. ‘CD47-CC2C6LOW’ and ‘CD47-CC2C6HIGH’) by fluorescence-activated cell sorting. Specifically, the first cell population, referred to as ‘CD47-CC2C6LOW’, constitutes the approximately 1-2% of cells with the lowest level of αCD47-CC2C6 binding. The second cell population, referred to as ‘CD47-CC2C6HIGH’, constitutes the approximately 1-2% of cells with the highest level of αCD47-CC2C6 binding. To reduce potential confounding effects of diploid cells that are heterozygous for alleles carrying gene-trap integrations, cell sorting was restricted to cells with a 1n DNA content, as determined by staining with propidium iodide (PI) (Life Technologies). Cell sorting was carried out on a Biorad S3 Cell sorter until approximately 10 million cells were collected for each population. Sorted cells were pelleted and genomic DNA was isolated using a DNA mini kit (Qiagen), according to manufacturer’s protocol. To assist de-crosslinking of genomic DNA, cell pellets were resuspended in PBS supplemented with Proteinase K (Qiagen) followed by overnight incubation at 56°C in lysis buffer AL (Qiagen) with agitation. Insertion sites present in both sorted cell populations were amplified and mapped to the human genome, as described previously41, using a Linear AMplification polymerase chain reaction (LAM-PCR) on the total yield of isolated genomic DNA. Samples were submitted for deep sequencing and gene-trap insertion sites were mapped and analyzed as follows: insertion sites were retrieved from trimmed reads (50b) that aligned unambiguously to Hg19 using Bowtie42 allowing one mismatch. Using intersectBed, aligned reads were mapped to non-overlapping Refseq gene-coordinates. Intragenic gene-trap insertions in sense orientation within its gene were considered disruptive and kept for further analysis. For each gene, the number of unique disruptive insertions was compared between the CD47-CC2C6LOW and CD47-CC2C6HIGH population. Genes that were significantly enriched for insertions in either of the two populations (two-sided Fisher’s Exact test with Benjamini-Hochberg multiple testing correction, P< 0.05) were considered as regulators of CD47-CC2C6 binding. To reflect the directionality of the effect on CD47 abundance, a mutational index (MI)-score was calculated as follows:
For those genes for which disruptive insertions were identified in only one of the two populations, 1 insertion was assigned to the other population to allow inclusion of these genes in visualization plots.
For the “4-way sort” genetic screen, cells were double-stained with APC-labelled αhCD47-antibody B6H12 (αhCD47-B6H12) (eBioscience) and FITC-labelled αhCD47-CC2C6 (BioLegend) at a 1:80 dilution in 30 mL PBS containing 10% FCS for 60 minutes at 4°C, protected from light. Cells were washed once with ice-cold PBS containing 10% FCS and once with ice-cold PBS. Cells were then fixed with BD Fix Buffer I for 10 minutes at 37°C, washed once with PBS containing 10% FCS, and stained for 5 minutes at room temperature in PBS containing 10% FCS with 1 μg/mL DAPI (Invitrogen) to visualize cells in G1 phase. Cells were washed once in PBS containing 10% FCS and filtered through a 40 μm strainer before sorting the populations of interest on a BD FACSAria II. Specifically, the 1-2% of cells exhibiting the highest (B6H12HIGH/CC2C6HIGH) or lowest (B6H12LOW/CC2C6LOW) signal for both antibodies, as well as the 1-2% of the cell population exhibiting the highest or lowest ratio in signal for the two antibodies (i.e. B6H12LOW/CC2C6HIGH; B6H12HIGH/CC2C6LOW), were sorted until 12 million cells per channel were collected. Samples were further processed as described above. The number of unique disruptive insertions for each gene was compared between the B6H12HIGH/CC2C6HIGH and B6H12LOW/CC2C6LOW or B6H12LOW/CC2C6HIGH and B6H12HIGH/CC2C6LOW channels, respectively.
Cell lines
HAP1 cells have been described previously43. A375, A431, A549, DLD1, JY, Raji, RKO and SKBR3 cells were purchased from American Type Culture Collection (ATCC). B16F10 cells were kindly provided by D. Peeper. Ba/F3 cells are bone-marrow-derived, immortalized cells which are IL-3-dependent, as described in44. NKIRTIL006 cell line was generated from a patient treated at the Netherlands Cancer Institute.
HAP1 cells were cultured in IMDM (ThermoFisher Scientific) supplemented with 10% Fetal Calf Serum (FCS, Sigma), 100 U/mL penicillin (Roche) and 100 μg/mL streptomycin (Roche) (penicillin/streptomycin) and L-glutamine. NKIRTIL006 cells were cultured in IMDM supplemented with 10% FCS and penicillin/streptomycin. A375, A549 and B16F10 were cultured in DMEM supplemented with 10% FCS and penicillin/streptomycin. A431, DLD1, Raji and JY cells were cultured in RPMI supplemented with 10% FCS and penicillin/streptomycin. Ba/F3-Her2 cells were cultured in RPMI supplemented with 10% FCS, penicillin/streptomycin, 0.2 ng/mL mouse IL-3 (Immunotools) and 5.0 μg/mL puromycin. SKBR3 cells were cultured in IMDM supplemented with 20% FCS and 100 U/mL penicillin/streptomycin. Cells were cultured at 37°C and 5% CO2.
Flow cytometry
The following antibodies and recombinant extracellular domains of SIRPα were used to analyse cell surface CD47 binding: anti-human CD47: CC2C6 (BioLegend), 2D3 (BioLegend), B6H12 (BioLegend); anti-mouse CD47: MIAP301 (BioLegend); recombinant human SIRP alpha/CD172a Fc Chimera Protein (R&D Systems); recombinant human SIRP alpha/CD172a His-Tag Protein (R&D Systems); recombinant human SIRP gamma/CD172g Fc Chimera Protein (R&D Systems); recombinant mouse SIRP alpha/CD172a Fc Chimera Protein (R&D Systems); goat anti-mouse IgG Fc: Poly4053 (BioLegend); anti-human IgG Fc: HP6017 (BioLegend),
Binding to cell surface CD47 was assessed by staining of cells with fluorchrome labelled antibodies directed against human CD47 (clones CC2C6, 2D3, B6H12) or mouse CD47 (clone MIAP301) at a dilution of 1:50 (or 1:80 in case of αCD47-CC2C6/αCD47-B6H12 double staining) in PBS containing 0.5% w/v BSA (Sigma) and 0.2% w/v sodium azide (Sigma) (“FACS buffer”) for 20 to 30 minutes, at 4°C, protected from light. SIRPα or SIRPγ binding to CD47 was assessed by incubating cells with recombinant human SIRPα (hSIRPα-Fc) (4 μg/mL), His-tagged recombinant human SIRPα (SIRPα-His) (4, 12 or 36 μg/mL), recombinant mouse SIRPα (mSIRPα-Fc) (2 μg/mL) or recombinant human SIRPγ (hSIRPγ-Fc) (4, 12 or 36 μg/mL) in FACS buffer for 30 minutes, at room temperature, protected from light. To assess the nature of residual binding of SIRPα to SEN177-treated and QPCTL KO cells, indicated cells were pre-incubated with αCD47-B6H12 (1:25 or 1:10 dilution) for 5 minutes before proceeding with SIRPα-Fc staining as described above. After one or two washes with FACS buffer to remove unbound antibody or recombinant protein, cells initially stained with recombinant proteins were immunostained with a fluorochrome labelled mouse antibody against human IgG (HP6017), mouse monoclonal antibody against Hexa histidine tag (J095G46), or a goat polyclonal antibody against mouse IgG (Poly4053), at a dilution of 1:100 or 1:200 for 30 minutes, at 4°C, protected from light. After indicated antibody stainings, cells were washed with FACS buffer to remove unbound antibody and DAPI or PI was added to allow dead cell exclusion.
The following antibodies were used to analyse antigen expression on target cells: anti-human CD20: 2H7 (BioLegend), Cetuximab (Merck Sorono), anti-human CD340 (erbB2/HER-2): 24D2 (BioLegend).
Surface expression of CD20 on control (DMSO)-treated or SEN177-treated Raji cells was assessed by staining cells with a fluorochrome labelled antibody directed against human CD20 (2H7, Biolegend) at a 1:100 dilution. Surface expression of EGFR on WT, CD47 KO, QPCTL KO, control (DMSO)-treated or SEN177-treated A431 cells was assessed by staining cells with Cetuximab (Merck Sorono) (1.0 μg/mL) at 4°C, protected from light. After one wash with FACS buffer to remove unbound antibody, cells were stained with a fluorchrome labelled mouse antibody against human IgG (HP6017) at a dilution of 1:100, or with with a fluorochrome labelled goat F(ab’)2 fragment against human IgG (polyclonal) at a l:1,000 dilution. Surface expression of Her2 on WT, CD47 KO, QPCTL KO, control (DMSO)-treated or SEN177-treated Ba/F3-Her2 cells was assessed by staining cells with fluorochrome labelled antibodies directed against human Her2 (24D2, Biolegend) at a 1:25 or 1:100 dilution for 20 minutes, at 4°C, protected from light. After indicated antibody stainings, cells were washed with FACS buffer to remove unbound antibody and DAPI or PI was added to allow dead cell exclusion.
The following antibodies were used to analyse surface expression and intracellular cytokine production of human T cells: anti-human CD8: SK1 (BioLegend); anti-human CD4: SK3 (eBioscience); anti-human IFNγ: B27 (BD Biosciences); anti-human TNFα: Mab11 (BD Biosciences) and anti-human IL-2: MQ1-17H12 (BD Biosciences). The following antibodies were used to analyse expression of phenotypic markers on T cells: anti-human CD4: SK3 (BD Biosciences); anti-human CD27: L128 (BD Biosciences); anti-human CD28: CD28.2 (BD Biosciences); anti-human CD45RA: MEM-56 (Invitrogen); anti-human CD45RO: UCHL1 (Invitrogen); anti-human CCR7: 150503 (BD Biosciences); anti-human PD-1: J105 (eBioscience); anti-human CD62L: SK11 (BD Biosciences). The following antibodies were used to analyse Th17 differentiation: anti-human CD4: SK3 (BD Biosciences); anti-human IL-4: MP4-25D2 (BioLegend); anti-human IL17A: N49-653 (BD Biosciences); anti-human IFNγ: B27 (BD Biosciences).
Cell surface expression of indicated markers was assessed by staining of T cells with fluorochrome-labelled antibodies in FACS buffer for 20-30 minutes, at 4°C, protected from light. In the case of subsequent intracellular cytokine analysis, cells were washed once with FACS buffer and fixed and permeabilized using Fixation/Permeabilization Solution Kit (BD Biosciences), according to the manufacturer’s protocol. Intracellular cytokine production was assessed by staining T cells with indicated fluorochrome-labelled antibodies in BD Perm/Wash™ buffer for 20-30 min, at 4°C, protected from light. After incubation, cells were washed twice with BD Perm/Wash™ buffer before resuspension in FACS buffer for analysis. Fixable Violet Dead Cell Stain kit 405 nm (Invitrogen) or LIVE/DEAD Fixable Near-IR Dead Cell Stain kit for 633 or 635 nm excitation (ThermoFisher Scientific) was used to allow for dead cell exclusion.
The following antibodies were used for in vivo immune infiltrate analysis: anti-mouse B220: RA3-6B2 (BioLegend); anti-mouse CD3ε: 145-2C11 (BioLegend); anti-mouse MHC class II: M5/114.15.2 (BioLegend); anti-human CD89: A59 (BioLegend); anti-mouse CD8α: 53-6.7 (BioLegend or eBioscience); anti-mouse Ly-6G: 1A8 (BioLegend); anti-mouse CD45: 30-F11 (BioLegend); anti-mouse CD4: RM4-5 or GK5.1 (BioLegend or eBioscience); anti-mouse FcγRIV: CD16.2, 9E9 (BioLegend); anti-mouse F4/80: BM8 or EMR1 (Biolegend); anti-mouse CD11b: m1/70 (BD Biosciences); anti-mouse Siglec-F: E50-2440 (BD Biosciences); anti-mouse I-A/I-E: M5 (BioLegend); anti-mouse FOXP3; 150D (BioLegend).
Ba/F3-Her2 and effector cells retrieved from the peritoneum of FcαRI transgenic BALB/c mice were analyzed after incubation with 5% normal mouse serum (nms) for 45 minutes at 4-7°C. To analyse receptor surface expression, cells were stained for 45-60 minutes at 4-7°C with indicated fluorochrome labelled antibodies. LIVE/DEAD Fixable Near-IR Dead Cell Stain kit for 633 or 635 nm excitation (ThermoFisher Scientific) was used to allow for dead cell exclusion. In the case of subsequent intracellular staining, cells were washed and fixed and permeabilized using eBioscience™ FoxP3/Transcription Factor Staining Buffer Set (Invitrogen), according to manufacturer’s protocol. Next, cells were incubated with 5% nms in PBS and intracellular expression was assessed by staining of cells with indicated fluorochrome-labelled antibodies in Permeabilization Buffer for 30-40 min, at room temperature, protected from light. Cells were washed with Permeabilization Buffer to remove unbound antibody and resuspended in FACS buffer for further analysis. FMO controls were used to identify marker-positive cell populations.
Measurements were performed on an LSRII, LSRFortessa or FACSCantoII (BD Biosciences). Data were analyzed using FACS Diva software (BD Biosciences) or FlowJo software.
Vector generation
The cDNA of the two human transcript variants of QPCTL, glutaminyl-peptide cyclotransferase-like, transcript 1 (RefSeq: NM_017659.3) and glutaminyl-peptide cyclotransferase-like, transcript 2 (RefSeq: NM_001163377.1), were ordered as codon optimized gBlock Gene Fragments (IDT DNA) encoding an N-terminal FLAG tag. QPCTL(1) consists of 7 exons, whereas QPCTL(2) consists of 6 exons, lacking exon 3 that encodes the amino acid sequence: FLEATLRSLTAGWHVELDPFTASTPLGPVDFGNVVATLDPRAARHLTLACHYDSKLFPPGSTPFVGATDSAVPCALLLELAQALDLELSRAKKQ. The cDNA of the mus musculus transcript variant of QPCTL, (RefSeq: NM_026111.3) (“mQPCTL”), was ordered as a codon optimized gBlock Gene Fragment. A codon optimized variant of the cDNA of CD47 transcript variant 2 (RefSeq: NM_198793.2), the most abundant transcript variant of CD47 containing the long 3’ untranslated region 45 was generated as a gBlock Gene Fragment that encodes a C-terminal HA-tag. The lentiviral pCDH-CMV-MCS-EF1-Puro vector encoding C-terminal His-tagged human QPCTL-WT (ENST00000012049.9) and the QPCTL (D326E) mutant were generated by cloning gBlock Gene Fragments digested with SpeI and EcoR1 into pCDH-CMV-MCS-EF1-Puro digested with NheI and EcoRI. All other constructs were cloned into the pCDH-CMV-MCS-EF1-Puro (CD510-B1) vector containing a puromycin selection cassette, or in the pCDH-CMV-MCS-mPGK-BSR vector (kindly provided by R. Agami) containing a blasticidin selection cassette using the restriction sites EcoRI and NotI. Constructs were verified by Sanger sequencing.
CRISPR/Cas9-mediated generation of CD47, QPCTL knockout cells
To generate CD47- and QPCTL-knockout HAP1 cells, cells were co-transfected with PX330 vector encoding sgRNA targeting the QPCTL or CD47 gene and a plasmid containing an expression cassette for a guide RNA targeting the zebrafish TIA gene (5’-GGTATGTCGGGAACCTCTCC-3’ (SEQ ID NO:5)) followed by a CMV promotor sequence driving expression of a blasticidin resistance gene flanked by two TIA target sites 46. Co-transfection of these plasmids occasionally results in the incorporation of the blasticidin resistance cassette at the site of the targeted genomic locus by non-homologous end joining, rendering cells resistant to blasticidin while also providing a genomic tag at the site of mutation. Four days after DNA transfection, culture medium was supplemented with 20 μg/mL blasticidin (Invivogen). Surviving colonies were clonally expanded and their mutations and/or genomic incorporation of the blasticidin resistance gene were verified by PCR and Sanger sequencing.
To generate CD47- and QPCTL-knockout A431, A375, A549, DLD1, RKO and SKBR3 cell lines, cells were transfected with pLentiCRISPR v.2 vector (Addgene 52961) encoding sgRNA targeting the QPCTL or CD47 gene. To generate HSPA13-knockout cells, HAP1 cells were transfected with pLentiCRISPR v.2 vector encoding sgRNA targeting the HSPA13 gene. One day after transfection, culture medium was supplemented with 2 μg/mL puromycin for two days. Single-cell clones were expanded to obtain clonal knockout populations.
To generate bulk CD47- and QPCTL-knockout B16F10 cells, cells were transfected with pLentiCRISPR v.2. vector encoding sgRNA targeting the murine QPCTL or CD47 gene. One day after transfection, culture medium was supplemented with 2 μg/mL puromycin for two days. Selected cells were expanded and sorted on the basis of αmCD47-MIAP301LOW mSIRPα-FcLOW (in case of CD47 knockout) and αmCD47-MIAP301HIGH mSIRPα-FcLOW (in case of QPCTL knockout) to obtain bulk knockout populations.
Her2-expressing Ba/F3 cells were generated by retroviral transduction with human HER2 (pMX-puro-Her2), and positive clones were selected using puromycin. To generate Ba/F3-Her2 CD47 and QPCTL-knockout cells, nucleofection was used to deliver pLentiCRISPR v.2. vectors encoding sgRNA targeting either the murine QPCTL or CD47 gene, together with a plasmid containing a Cas9, blasticidin resistance, and GFP expression cassette. One day after nucleofection, culture medium was supplemented with 2 μg/mL blasticidin for two days. Selected cells were expanded and sorted to obtain bulk knockout populations. Next, single cells were isolated and expanded to obtain clonal knockout populations.
Gene disruption was validated by sequence analysis of the relevant gene locus, by TIDE analysis 47 and, in case of CD47, by flow cytometry.
Generation and analysis of CD47 and QPCTL overexpressing cells
A375 wild-type and QPCTL-knockout cells (clone 4.1) were transduced with a lentiviral pCDH-Puro vector containing cDNA encoding CD47 plus a C-terminal HA-tag (CD47-HA) and that includes the 3’ long untranslated region of CD47. Two days after transduction, cells were selected with 2 μg/mL puromycin for two to three days.
A375 QPCTL-knockout cells (clone 4.1 and clone 4.6), A431 QPCTL-knockout cells (clone 6 and clone 11) and A549 QPCTL-knockout cells (clone 3 and clone 9) were transduced with a lentiviral pCDH-Puro vector containing cDNA encoding human QPCTL(1)-FLAG (and also QPCTL(2)-FLAG in case of A375), as described above. B16F10 QPCTL-knockout cells (bulk KO#1 and KO#2) and Ba/F3-Her2 QPCTL-knockout cells (clone 8 and clone 30) were transduced with a lentiviral vector containing cDNA encoding mouse QPCTL-FLAG, as described above. Two days after transduction, cells were selected with 2 μg/mL puromycin for two to three days.
SEN177 and PQ912 treatment
For flow cytometry analysis, cells were plated in triplicate in the appropriate medium containing 0.03% (v/v) DMSO (vehicle control), 10 μM SEN177 (Sigma Aldrich), or 10 μM PQ912 (Syncom). DMSO or inhibitor was refreshed every day and after four days, cells were analysed by flow cytometry.
Immunoprecipitation, SDS-PAGE, western blot analysis and isoelectric focusing
For SDS-PAGE and western blot analysis, cells were plated to obtain 70-90% confluency the next day. At the day of analysis, cells were washed with PBS and lysed with RIPA buffer (1% Triton, 0.1% SOC, 0.1% SDS,1 mM EDTA, 10 mM Tris pH 8.0, 140 mM NaCl) supplemented with protease inhibitor cocktail (Roche) and 1mM PMSF (Sigma). After a 30 minute incubation on ice, cell lysates were centrifuged at 13,300 g for 20 minutes at 4°C. Supernatants were subsequently processed and protein concentrations were measured using the Pierce BCA Protein Assay Kit, according to the manufacturer’s instructions (ThermoFisher Scientific). Equal amounts of protein supernatants were subsequently processed using a Novex NuPAGE Electrophoresis System (ThermoFisher Scientific) and Trans-Blot Turbo Transfer System (Bio-Rad), according to the manufacturers’ instructions. QPCTL(1)-FLAG or QPCTL(2)-FLAG expression was detected using anti-FLAG® M2 (Sigma) (1:1,000 dilution) and anti-mouse HRP (1:10,000 dilution).
For immunoprecipitation and pulse-chase analysis, A375 WT, QPCTL KO cells overexpressing CD47-HA, or CD47 KO cells were plated to obtain 70-90% confluency the next day and where indicated, treated with 10 μM SEN177 inhibitor for 16 hours. At the day of analysis, cells were starved in methionine- and cysteine-free medium for 1 hour at 37°C. Subsequently, cells were pulse-labelled with 0.75 mCi/600 μL [35S]Cys/[35S]Met (PerkinElmer) for the indicated time period. Cells were washed with PBS to remove residual [35S]Cys/[35S]Met and then cultured in regular medium with 1 mM ‘cold’ methionine and cysteine for the indicated time period. Cells were lysed and [35S]Cys/[35S] incorporation was measured via TCA precipitation of aliquots of lysates on 3 MM Whatman paper and counting in a Perkin Elmer LSC 2800 ultima gold scintillation counter. Next, for pre-clearing and IP purposes, purified mouse IgG1 kappa isotype control (Biolegend, 400102), anti-human CD47 antibody B6H12.2 (Novus, NBP2-31106), and purified anti-human CD47 antibody CC2C6 (Biolegend, 323102) was bound to protein-G-coated Dynabeads (ThermoFisher Scientific), according to the manufacturer’s instructions. Protein lysates were incubated with mouse IgG1 kappa isotype control/bead mixtures for 1 hour at 4°C to reduce unspecific binding. Next, pre-cleared protein supernatants were incubated with bead-bound anti-human CD47 B6H12.2 or anti-human CD47 CC2C6 at 4 °C overnight. Immunoprecipitates were either left untreated, treated with Endoglycosidase H (EndoH, New England Biolabs), or treated with N-glycosidase F (PNGase F, New England Biolabs), according to the manufacturer’s instructions. Next, imunoprecipitates were heated at 50°C for 10’ minutes in 2x Laemmli buffer, and analysed using a Novex NuPAGE Gel Electrophoresis System (ThermoFisher Scientific). Gels were treated with 1 M Na salicylate pH 5.6 before drying and then analysed on Fujifilm BAS-MP phosphor imager screens at 4 °C. Screens were analysed on a Typhoon FLA 9500 laser scanner.
1D-IEF was performed essentially as described48. Immunoprecipitates from indicated cell lines were prepared as described above and were eluted with 30 μL IEF buffer (9.0M urea, 2% Triton-X100, 2% (v/v) Ampholite pH 3-10, 5% beta-mercapto-ethanol), and samples were analysed on freshly prepared IEF gels (9.5M urea, 2% Triton-X100, 4.5% Acrylamide / 0.24% bis-Acrylamide, 4% Ampholite pH 5-7, 1% Ampholite pH 3-10, 0.4% Ampholite pH 6.5-9), which was run for 16 hours. Gels were fixed with 10% acetic acid, 45% methanol and next treated with 1 M Na salicylate pH 5.6 before drying. Gels were analysed on phosphor imager screens at 4°C or on Amersham Hyperfilm ECL (GE Healthcare) at -80°C. The phosphor imager screens were analysed on a Typhoon FLA 9500 laser scanner and the films were developed in a Kodak X-omat 2000 processor.
Generation of genetically labelled Raji Turquoise-LMNB1
The LMNB1 CDS without start codon was PCR amplified with BglII forward primer and MfeI reverse primer (Invitrogen), cloned into the BglII-EcoRI site of pE-Turquoise-C1 (Takara Bio), and sequence verified. From this, the AgeI-SalI digested Turquoise-LMNB1 fragment was cloned into pRRL PPT SFFV GFP prester SIN, in which GFP was removed by AgeI and partial SalI digestion. Lentiviral particles were produced by transient transfection of pRRL PPT SFFV prester SIN -Turquoise-LMNB1 and pMDLg/pRRE, RSVrev and pCMV-VSVg in 293T cells with TransIT-LT1 (Mirus Bio). Two or three days after transfection, supernatant of transfected cells was filtered through 0.45 μM filters and added to 5×105 Raji cells. Turquoise positive cells were selected by FACS sorting.
In vitro antibody-dependent cellular phagocytosis (ADCP)
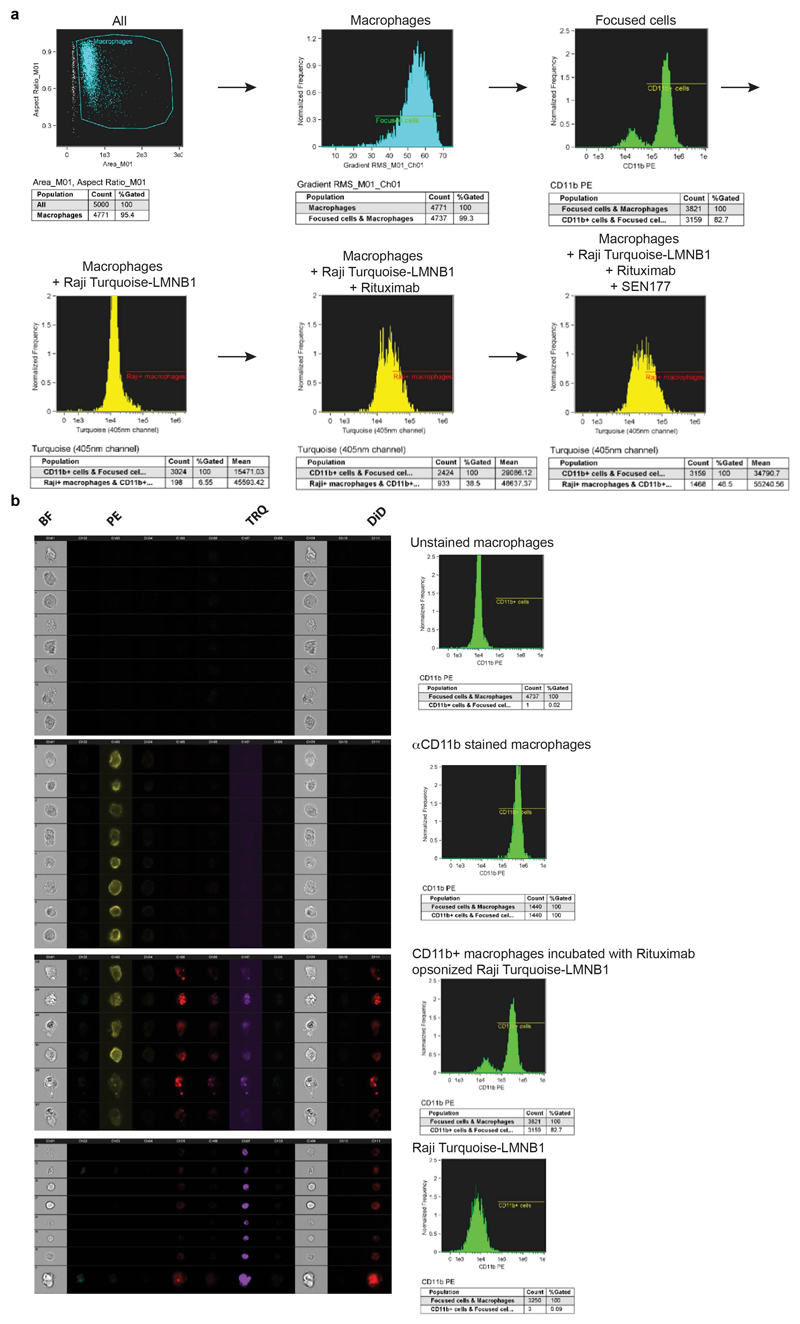
Human monocyte-derived macrophages served as effector cells and Raji B cell lymphoma with genetically labelled nuclei (Raji Turquoise-LMNB1, fluorescence detectable in 405nm channel) were used as target cells. Peripheral blood mononuclear cells (PBMC) were isolated from peripheral blood of healthy individuals collected by Sanquin Blood Supply (Amsterdam, The Netherlands).
Monocyte isolation was performed by gradient centrifugation on Percoll (Pharmacia), followed by negative selection of monocytes by magnetic-activated cell separation (Miltenyi Biotec B.V.). Freshly isolated monocytes were cultured for 7-8 days at 37°C and 5% CO2 in IMDM medium supplemented with 10% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine (Gibco Invitrogen), and 20 ng/mL human macrophage colony-stimulating factor (M-CSF, eBioscience) to allow differentiation into macrophages. One day before phagocytosis assays, 10 ng/mL of recombinant human IL-10 (PeproTech) was added. At the day of assay, culture medium was replaced by fresh IMDM medium containing 0.5% of FCS, where indicated in the presence of anti-SIRPα antibody 12C4 or F(ab)2’ fragment of anti-CD47 antibody B6H127. Raji Turquoise-LMNB1 cells were cultured 5 days prior to the experiment in the presence of either vehicle control (DMSO) or SEN177 (10 μM). Inhibition of SIRPα binding to CD47 on target cells by prior culture in the presence of SEN177 was monitored by flow cytometry at the end of each target cell culture. For ADCP, target cells were first labelled with the membrane dye DiIC18 å(5) solid (1,1'-dioctadecyl-3,3,3',3'-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (ThermoFisher Scientific,) for 30 minutes at 37°C, washed with PBS and subsequently incubated for 3 hours in the presence or absence of the opsonizing anti-CD20 antibody rituximab (2 μg/mL) and macrophages (E:T ratio of 1:2). Cells were washed with PBS and detached using citric saline solution (135 mM potassium chloride, 15 mM sodium citrate, Sigma-Aldrich). To discriminate between macrophages and Raji Turquoise-LMNB1 cells during analysis, cells were incubated with PE-labeled anti-human CD11b: M1/70 (eBioscience). Internalization of tumor cells by macrophages was determined by imaging flow cytometry (Image Stream, Amnis, EMD, Millipore, Seattle, WA, USA) using the following gating strategy: in focus macrophages were gated based on CD11b expression. Subsequently, the fraction of CD11b+ macrophages positive for Raji Turquoise-LMNB1 was analyzed based on fluorescence in the 405nm channel (analysis software IDEAS, Amnis Corporation). Extent of phagocytosis was evaluated both as the percentage of Raji Turquoise-LMNB1 positive CD11b+ macrophages, and as mean fluorescence intensity (MFI) of CD11b+ cells in the 405 nm channel.
In vitro antibody-dependent cellular cytotoxicity (ADCC)
For ADCC assays of 51Cr-labeled A431 target cells by human effector cells, neutrophils were isolated as described previously 49. After isolation, neutrophils were cultured for 30 minutes in HEPES buffered saline solution (containing 132 mM of NaCl, 6.0 mM of KCl, 1.0 mM of CaCl2 1.0 mM of MgSO4, 1.2 mM of potassium phosphate, 20 mM of HEPES. 5.5 mM of glucose, and 0.5% (wt/vol) human serum albumin, pH 7.4) in the presence of 10 ng/mL clinical grade recombinant GM-CSF (Peprotech) at a concentration of 5×106 cells/mL. Indicated target cells were labelled with 100 μCi 51Cr (Perkin-Elmer) for 90 minutes at 37 ºC. After 3 washes with PBS, CD47-SIRPα interactions were blocked by incubating A431 WT cells (1×106) with 20 μg/mL anti-CD47 F(ab’)2 antibody (clone B6H12), or incubating PMNs with the anti-SIRPα antibody 12C4 (10 μg/mL)7, where indicated. After 2 washes with PBS, 5×103 target cells were incubated in RPMI culture medium supplemented with 10% FCS for 4 hours at 37ºC and 5% CO2 in a 96-well U-bottom plate together with PMNs (E:T ratio of 50:1), in the presence of the appropriate antibodies. Following co-culture, supernatants were harvested and analyzed in a Wallac gamma counter. Percentage of cytotoxicity was calculated as [(experimental cpm- spontaneous cpm)/ (total cpm- spontaneous cpm)] x 100%. All experiments were performed in triplicate.
ADCC of 51Cr-labeled Ba/F3-Her2 target cells by human effector cells was described previously 50. Briefly, 1×106 target cells were labelled with 100 μCi (3.7MBq) 51Cr for 2 hours. After extensive washing, cell numbers were adjusted to 1×105/mL. The polymorphonuclear leukocyte (PMN) fraction from peripheral blood of healthy donors (UMC Utrecht, Utrecht) was isolated by Ficoll/Histopaque separation (GE Healthcare; Sigma-Aldrich). Effector cells and target cells were added to round-bottom microtiter plates (Corning Incorporated) (E:T ratio of 40:1), in the presence of the indicated concentration of IgA1 anti-Her2 antibody. After 4 hours incubation at 37°C, 51Cr release was measured. Percentage specific lysis was calculated using the following formula described above. The production and activity of IgA1 anti-Her2 antibody has been described previously51. For experiments with SEN177, 10 μM SEN177 or vehicle control (DMSO) was added three days before the assay, added freshly on the day of the assay, and kept present during the assay. All experiments were performed in triplicate.
ADCC of 51Cr-labeled Ba/F3-Her2 target cells by mouse effector cells was carried out as described52. In brief, to obtain mouse effector cells, blood was collected from pegylated granulocyte colony-stimulating factor (G-CSF)–stimulated human FcαRI transgenic Balb/c mice from the retro-orbital plexus into Li-heparin tubes. Erythrocytes were lysed by incubation in water for 30 minutes, and total leukocytes were resuspended in medium (half the volume of the original blood volume). 50 μL of total leukocytes, containing ~70% PMNs, were added per well.
In vivo killing assays
The peritoneal Ba/F3 tumor model in human FcαRI transgenic mice has been described previously32. Briefly, Ba/F3-Her2 and Ba/F3-Her2 CD47 KO or Ba/F3-Her2 QPCTL KO cells were labelled with 8-10 μM or 0.4-2 μM CT violet (Invitrogen, Thermofisher) respectively, for 15 minutes at room temperature. Subsequently, 1:1 mixtures of Ba/F3-Her2 and Ba/F3-Her2 CD47 KO cells, or Ba/F3-Her2 and Ba/F3-Her2 QPCTL KO cells were generated. For experiments that utilize 1:1:1 mixtures, Ba/F3-Her2 cells were labelled with 1 μM CFSE, Ba/F3-Her2 CD47 KO cells with 8 μM CT Violet, and Ba/F3-Her2 QPCTL KO cells with 0.4 μM CT violet. Subsequently, human FcαRI transgenic mice and littermate controls were injected intraperitoneally with 1×107 cells in 200 μL PBS, or 1.5×107 cells in 300 μL PBS for 1:1:1 mixtures, as described in32. For experiments that utilize homogenous cell populations, Ba/F3-Her2 cells and Ba/F3-Her2 QPCTL KO were labelled with 0.8 μM CT Violet. Directly after injection of tumor cells, mice received PBS or 100 μg IgA1 anti-Her2 by intraperitoneal injection. Sixteen hours after injection, mice were euthanized, the peritoneal cavity was washed with PBS containing 5 mM EDTA, and the absolute number of Ba/F3-Her2, Ba/F3-Her2 CD47 KO, and Ba/F3-Her2 QPCTL KO cells was determined by flow cytometry using TruCount tubes (BD Biosciences). Indicated effector cell types were measured in the peritoneum by staining with the indicated antibodies and quantification relative to a constant amount of Sulfate Latex Beads (ThermoFisher Scientific). For the depletion of neutrophils, 200 μg of anti-mouse Ly6G (1A8, BXCell) was injected i.p. into mice on day -2 and day 0 of the experiment.
Intracellular cytokine production by human T cells
Human peripheral blood T cells (Sanquin Blood Supply) were activated and transduced with a retroviral vector encoding the CDK4R24C-specific, essentially as described previously53.
Following transduction, T cells were cultured for 7 to 14 days in RPMI containing 5% Human Serum (Sigma), supplemented with 10 ng/mL IL-15 (Peprotech) and 50 U/mL IL-2 (Novartis), followed by a 4 day culture period in control (DMSO)- or SEN177-containing (10 μM) medium. Activity of SEN177 was confirmed by comparison of αhCD47-CC2C6/αCD47-B6H12 ratio in SEN177-treated cells and control (DMSO)-treated cells, as analyzed by flow cytometry. Expression of the CDK4R24C-specific TCR was confirmed by flow cytometry (data not shown). JY cells were pulsed with CDK4R24C peptide at indicated concentrations, or as a control with MART-126-35 A>L peptide, for 1 hour, at 37 °C. Next, 2 × 105 transduced T cells were incubated with 2 × 105 peptide-pulsed JY cells or non-pulsed JY cells in the presence of 1 μL/mL Brefeldin A (GolgiPlug, BD Biosciences) for 5 hours, at 37 °C and 5% CO2. Next, cells were washed and stained with specific antibodies as described above, and analysed for cytokine production, as determined by flow cytometry.
In vitro tumor cell killing by human T cells
Human HLA-A*02:01-positive melanoma lines that carry (NKIRTIL006) or lack (A375) a mutant CDK4 allele were plated in 96-well plates (Greiner Bio-One) and labelled with 100 μCi 51Cr (Amersham, Gent, Belgium) for 1 hour at 37 °C. Cells were washed with RMPI containing 10% FCS and penicillin/streptomycin to remove unincorporated 51Cr before addition of CDK4-specific TCR-transduced T cells at the indicated target: T cell ratio’s. Cells were incubated for 4 hours, at 37 °C in 200 μL medium. Next, plates were centrifuged for 1 minute at 246g and 50 μL medium was harvested, plated on LumaPlate-96 plates (Perkin Elmer), dried overnight and analyzed on a Top Counter (Perkin-Elmer). Specific cytotoxicity was calculated according to the formula described before.
In vitro proliferation, phenotypic analysis and cytokine secretion of human T cells
Peripheral blood lymphocytes from two donors were stimulated with anti-CD3/CD28 beads (ThermoFisher Scientific) at a 2:1 bead to T cell ratio. Cells were cultured for a two week period in RPMI with 5% HS, IL-2 (50 IU/mL), IL-15 (10 ng/mL) in the presence of SEN177 (10 μM) or control (DMSO). Every 2 to 3 days, cells were counted and re-plated at a density of 0.25 × 106 cells/mL. Surface expression of phenotypic markers was analysed on day 0, 5, 10 (data not shown) and 14 with the indicated antibodies. On day 14, T cells were re-stimulated with anti-CD3/CD28 beads (1:1 bead to T cell ratio), supernatants were collected 38 hours after stimulation, and levels of IL-2, IL-4, IL-6, IL-10, IFNγ, IL17A and TNFα were measured using the Cytometric Bead Array Human Th1/Th2/Th17 Cytokine Kit (BD Biosciences, San Jose, CA), according to the manufacturer’s protocol.
In vitro induction of antigen-specific T cells by autologous antigen-presenting cells
In vitro T cell induction was performed under three conditions; 1) T cell induction by matured autologous dendritic cells (DCs) in the presence of control (DMSO), 2) T cell induction by matured autologous dendritic cells, in which T cell induction was performed in the presence of 10 μM SEN177, or 3) T cell induction by matured autologous dendritic cells, in which both the maturation of autologous DCs and T cell induction was performed in the presence of 10 μM SEN177.
For the maturation of autologous DCs, monocytes from a healthy donor were isolated by CD14 selection using LS MACS columns (Miltenyi Biotec) and the purity of the obtained population was assessed by flow cytometry. Next, CD14+ cells were plated in CellGenix GMP Dendritic Cell Medium, supplemented with 400 U/mL IL-4 (CellGenix) and 800 U/mL GM-CSF (CellGenix) to generate immature DCs. After 5 days, immature DCs were loaded with he following long and short peptides: EBV BMLF-1 (RMQAIQNAGLCTLVAMLEETIFWLQ), CMV pp65 (WQAGILARNLVPMVATVQGQNLKYQ), EBV BRLF-1 (IACPIVMRYVLDHLIVVTDRFFIQA), MART-1 (HSYTTAEELAGIGILTVILGVLLLI), in which the sequences depicted in bold reflect previously identified CD8+ T cell epitopes. After 1 hour, “DC maturation mix” containing 400 U/mL IL-4, 800 U/mL GM-CSF, 10 ng/mL TNF-α (CellGenix) 10 ng/mL IL-1ß (CellGenix), 10 ng/mL IL-6 (CellGenix) and 0.5 μg/mL PGE1 (Pfizer) was added to DC medium, and DCs were cultured for 2 days.
For T cell inductions, mature peptide-loaded DCs were cultured with autologous hPBMCs in a 1:10 DC to hPBMC ratio in 20% AIM V Serum Free Medium (ThermoFisher Scientific) and 80% RPMI (Life Technologies), supplemented with 5% Human Serum, 1% GlutaMAX (ThermoFisher Scientific), penicillin/streptomycin, 5 ng/mL IL-7 (CellGenix) and 5 ng/mL IL-15 (CellGenix). Fresh IL-7, IL-15 (and control or SEN177)was added every 2-3 days, and cells were split 2 times a week.
After 11 days, the percentage of T cells specific for the indicated CMV, EBV and MART-1 epitopes was determined by combinatorial pMHC-multimer staining, as described previously54, and data were compared to values at baseline. Functionality of induced T cell populations was examined in a co-culture after 6 days. Induced T cell populations were cultured in the presence of autologous peptide-loaded DCs for 5 hours, and then analyzed for intracellular cytokine production, as determined by flow cytometry. After 12 days, surface expression of phenotypic markers was analysed by flow cytometry, as described previously.
In vitro differentiation of human T cells into Th17 cells
CD4+ T cells were isolated from human peripheral blood using the Dynabeads™ Untouched™ Human CD4 T Cells Kit (ThermoFisher Scientific), according to the manufacturer’s protocol. Purity of the obtained CD4+ T cell population was analysed by flow cytometry (Donor 1: 93%; Donor 2: 94%). Next, CD4+ T cells were cultured at a density of 0.5×106 cells/mL in RPMI medium containing 10% HS, anti-CD3/CD28 beads (2:1 bead to T cell ratio), 10 ng/mL IL-6 (R&D Systems), 10 ng/mL IL-1ß (R&D Systems), 10 ng/mL TGF-ß1 (R&D Systems), 10 ng/mL IL-23 (R&D Systems), 10 μg/mL anti-IL-4 (MP4-25D2, BD Biosciences), and 10 μg/mL anti-IFNγ (B27, BD Biosciences) (“Th17 differentiation medium”). Medium was refreshed every 2-3 days and cells were split at day 4 and 7. On day 7 and 10, cells were harvested and re-stimulated with 50 ng/mL Phorbol 12-myristate 13-acetate (PMA) (BD Biosciences) and 1 μg/mL ionomycin (BD Biosciences) in the presence of 1 μL/mL Brefeldin A for 5 hours, at 37 °C and 5% CO2. Next, cells were harvested and stained using the indicated antibodies, and the percentage of IL17A+ cells was quantified by flow cytometry.
Statistical analysis
Data are reported as mean ± s.d. as specified. Statistical significance was determined using Student’s t-test or one-way ANOVA as inidicated (GraphPad Prism version 7). All Student’s t-tests were two-sided under the assumption of equal variance between samples. All one-way ANOVA tests were corrected for multiple comparisons using statistical hypothesis testing. Differences were considered statistically significant if P<0.05. The n values used to calculate statistics, the type of replicates and the relevant significant P values have been noted in the figure legends.
Extended Data
Extended Figure 1. QPCTL deletion leads to reduced binding of recombinant SIRPα and SIRPγ.
a, Cell surface binding of αhCD47-2D3, αhCD47-B6H12, αhCD47-CC2C6 and hSIRPα-Fc to WT, CD47 and QPCTL KO lung cancer (A549) (upper left panel), colorectal cancer (DLD1) (upper right panel) and rectal carcinoma (RKO) (lower left panel) cells, as determined by flow cytometry. b, Cell surface binding of human SIRPα-Fc (hSIRPα-Fc) to lung cancer (A549) WT, CD47 KO and QPCTL KO cells alone or in combination with blocking antibody αhCD47-B6H12, as determined by flow cytometry. c, Cell surface binding of indicated concentrations of human SIRPα-His (hSIRPα-His) to WT, CD47 KO and QPCTL KO lung cancer (A549) cells, as determined by flow cytometry. d, Cell surface binding of indicated concentrations of hSIRPα-Fc and human SIRPγ-Fc (SIRPγ-Fc) to WT, CD47 KO and QPCTL KO lung cancer (A549) cells, as determined by flow cytometry. e, Representative plot (see d) of hSIRPγ-Fc binding (36.0 μg/mL) to WT, CD47 KO and QPCTL KO lung cancer (A549) cells.
Values in a-d indicate MFI relative to WT cells stained with the same reagent (a), MFI relative to WT cells without blocking antibody (b) or MFI values (c, d).
Data represent n=3 biological replicates (a-e) and mean ± s.d. of triplicates (a-d). ***P≤0.0001 by one-way ANOVA with multiple comparison correction (a-c) or unpaired two-sided t-test (d); n.s., not significant. Data are representative of at least two (a-e) independent experiments.
MFI, mean fluorescence intensity; WT, wild-type; KO, knock-out.
Extended Figure 2. QPCTL regulates binding of αhCD47-CC2C6.
a, Cell surface binding of αhCD47-2D3 and αhCD47-CC2C6 to WT, QPCTL KO and QPCTL KO cells reconstituted with FLAG-tagged cDNA of QPCTL isoform 1 (OE var.1) or QPCTL isoform 2 (OE var.2) melanoma (A375) (a) epidermoid carcinoma (A431) (b) and lung cancer (A549) cells (c), as determined by flow cytometry. d, Western blot analysis of WT, QPCTL KO and QPCTL KO melanoma (A375) cells reconstituted with FLAG-tagged cDNA of QPCTL isoform 1 (OE var.1) or QPCTL isoform 2 (OE var.2). Blot image has been cropped to show the relevant bands, and molecular mass markers are indicated (in kD). See Source Data for the uncropped western blot. e, Cell surface binding of αhCD47-CC2C6 and αhCD47-2D3 to HAP1 QPCTL KO cells reconstituted with QPCTL var.1 or a catalytically inactive QPCTL variant (QPCLT var.1 D326E), as determined by flow cytometry. f, Cell surface binding of αhCD47-CC2C6 and αhCD47-2D3 to QPCTL KO melanoma (A375) cells reconstituted with QPCTL var.1 or QPCTL var.1 (D326E), as determined by flow cytometry. Values in a-c, e, f indicate MFI relative to WT cells stained with the same reagent. Data represent n=3 biological replicates and mean ± s.d. of triplicates (a-c, e-f).
Data are representative of at least two (a-c, d, e-f) independent experiments.
OE, over-expression.
Extended Figure 3. Two-dimensional genetic screen to reveal selective modifiers of αhCD47-CC2C6 binding.
a, Schematic overview of FACS-based haploid genetic screen on αhCD47-BH612 (“B6H12”) and αhCD47-CC2C6 (“CC2C6”) double-stained cells, employing a 4-way sorting strategy to distinguish regulators that affect CD47 levels or that affect the αhCD47-CC2C6/SIRPα binding site of CD47. b, c, Results of the genetic screen for general CD47 modulators (b) and αhCD47-CC2C6/SIRPα binding site regulators (c). Dots represent individual genes. The relative mutation frequency (MI) in the B6H12HIGH/CC2C6HIGH versus B6H12LOW/CC2C6LOW (b) and B6H12LOW/CC2C6HIGH versus B6H12HIGH/CC2C6LOW cell population (c) is plotted against the total number of insertions mapped per gene. Significantly enriched genes (FDR-corrected P<0.05) are colored according to channel (B6H12HIGH/CC2CHIGH, green; B6H12LOW/CC2CLOW, dark blue; B6H12LOW/CC2CHIGH, light blue; B6H12HIGH/CC2C6LOW, orange) and selected regulators are labelled. n=3,253,240 (b) and n=3,209,992 insertions (c) were identified and data were analysed by two-sided Fisher’s Exact test with multiple comparison correction. d, Combined results for 4-way-sort screen. Dots represent individual genes. x-axis shows relative mutation frequency (MI) for the B6H12HIGH/CC2C6HIGH versus B6H12LOW/CC2C6LOW cell population, y-axis shows relative mutation frequency for the B6H12LOW/CC2C6HIGH versus B6H12HIGH/CC2C6LOW cell population. Selected regulators are highlighted. e, Ratio of αhCD47-CC2C6 and αhCD47-BH612 cell surface binding to HAP1 WT, CD47 KO, QPCTL KO and HSPA13 KO cells, as determined by flow cytometry. Values indicate ratio of MFI of αhCD47-CC2C6/αhCD47-BH612 for each cell population. Data represent n=3 biological replicates and mean ± s.d. of triplicates. ***P=0.0001 by one-way ANOVA with multiple comparison correction.
Data are representative of one (a-d) or two (e) independent experiments
MI, mutation index; WT, wild-type; KO, knockout; MFI, mean fluorescence intensity.
Extended Figure 4. Glutaminyl cyclase inhibition leads to reduced binding of recombinant hSIRPγ.
a, Cell surface binding of αhCD47-2D3, αhCD47-CC2C6 and hSIRPα-Fc to control (DMSO)-treated (-) or SEN177-treated (+) lung cancer (A549), colorectal (DLD1), HAP1, rectal carcinoma (RKO) and breast cancer (SKBR3) cells, as determined by flow cytometry. Data represent n=3 biological replicates and mean ± s.d. of triplicates. ***P≤0.000695 by unpaired two-sided t-test. b, Cell surface binding of αhCD47-2D3, αhCD47-CC2C6 and hSIRPα-Fc to control (DMSO)-treated (-), SEN177-treated, and PQ912-treated melanoma (A375) cells, as determined by flow cytometry. Data represent n=3 biological replicates and mean ± s.d. of triplicates. ***P=0.0001 by one-way ANOVA with multiple comparison correction. c, Flow cytometry plot of surface binding of αhCD47-B6H12 and αhCD47-CC2C6 to control-treated and PQ912-treated melanoma (A375) cells. Data are representative of two independent experiments with similar results (n=3 biological replicates per experiment). d, Cell surface binding of αhCD47-2D3, αhCD47-CC2C6 and hSIRPα-Fc to control (DMSO)-treated (-) and SEN177-treated (+) wild-type and QPCTL-knockout epidermoid carcinoma (A431) and lung cancer (A549) cells, as determined by flow cytometry. Data represent n=3 biological replicates and mean ± s.d. of triplicates. e, Cell surface binding of secondary antibody alone, hSIRPα-Fc (followed by secondary antibody) or hSIRPα-Fc in the presence of the CD47 blocking antibody αhCD47-B6H12 (followed by secondary antibody) to control (DMSO)-treated (-) or SEN177-treated lung cancer (A549) cells, as determined by flow cytometry. Data represent n=3 biological replicates and mean ± s.d. of triplicates. Values indicate MFI relative to WT cells stained with the same reagent (a, b, d) or MFI (e).
Data are representative of one (e) or at least two independent experiments (a-d)
Extended Figure 5. Analysis of phagocytosis.
a,b Representative images of gating strategy and phagocytosis (a) and examples of staining (b), as determined by ImageStream analysis. Data are representative of four independent experiments with similar results.
Extended Figure 6. Synergy between blockade of CD47 pyroglutamate formation and tumors opsonization in tumor cell killing by macrophages and neutrophils.
a, MFI of lamin B-Turquoise of the total CD11b+ macrophage population in samples incubated with control (DMSO)-treated (-) or SEN177-treated (+) Turquoise-expressing Burkitt’s lymphoma (Raji) cells in the presence or absence of the anti-human CD20 antibody Rituximab, CD47-blocking F(ab’)2 fragment B6H12, or SIRPα blocking antibody 12C4, as determined by ImageStream analysis. Symbols represent individual donors. Data represent mean ± s.d. of independent donors. ***P<0.0001; **P=0.0016; *P=0.0256 by one-way ANOVA with multiple comparison correction. b, Specific lysis of control (DMSO)-treated (-) or SEN177-treated (+) WT, QPCTL KO or CD47 KO epidermoid carcinoma (A431) cells by human neutrophils in the presence or absence of the anti-human EGFR antibody cetuximab in a 4 hour 51Cr-release assay. Data represent mean ± s.d. of independent donors. ***P<0.0001; 0.0325≥*P≥0.0207 by one-way ANOVA with multiple comparison correction. n.s.; not significant. c, Flow cytometry plot of cell surface binding of anti-human CD20 antibody to Burkitt’s lymphoma (Raji) cells (left panel) and anti-human EGFR antibody to epidermoid carcinoma (A431) cells (right panel) treated with control (DMSO) or SEN177 for 4 days.
Data are representative of one (c) or at least three independent experiments (a, b) representing 4 donors (for B6H12(Fab’)2 conditions), 8 donors (all other conditions) (a), and 8 donors (b).
Extended Figure 7. QPCTL deficiency and QPCTL inhibition enhances tumor-specific antibody-induced killing of mouse tumor cells by mouse effector cells.
a, Cell surface binding of anti-mouse CD47 antibody MIAP301 (αmCD47-MIAP301) and mouse SIRPα-Fc (mSIRPα-Fc) to WT, CD47 KO and QPCTL bulk KO (KO#1 and KO#2) murine melanoma (B16F10) cells, and WT, CD47 KO and QPCTL KO (cl8 and cl30) Her2-expressing mouse pro-B (Ba/F3-Her2) cells, as determined by flow cytometry. b, Cell surface binding of αmCD47-MIAP301 and mSIRPα-Fc to WT, QPCTL KO or QPCTL KO murine melanoma (B16F10) cells reconstituted with the murine QPCTL cDNA (OE), as determined by flow cytometry. c, Cell surface binding of αmCD47-MIAP301 and mSIRPα-Fc to control (DMSO)-treated (-) or SEN177-treated (+) murine melanoma (B16F10) or Her2-expressing murine pro-B (Ba/F3-Her2) cells, as determined by flow cytometry. (a-c) Data represent n=3 biological replicates and mean ± s.d. of triplicates. ***P≤0.0001 by one-way ANOVA with multiple comparison correction (a) or unpaired two-sided t-test (c). d, Flow cytometry plots of cell surface binding of anti-human Her2 antibody to WT, CD47 KO or QPCTL KO Ba/F3-Her2 cells (left), or control (DMSO)-treated or SEN177-treated Ba/F3-Her2 cells (right), as determined by flow cytometry. Data are representative of two independent experiments with similar results (n=3 biological replicates per experiment) (left graph) or one experiment with two biological replicates (right graph). e, Specific lysis of control (DMSO)-treated (-) and SEN177-treated (+) CD47 KO and QPCTL KO murine pro-B cells (Ba/F3-Her2) by human neutrophils in the presence of anti-Her2 (IgA1) in a 4 hour 51Cr-release assay. Data are representative of n=3 biological replicates and represent ± s.d. of triplicates of one representative donor. f, Specific lysis of WT, CD47 KO and QPCTL KO murine pro-B cells (Ba/F3-Her2) by murine immune cells isolated from whole blood in the presence or absence of anti-Her2 (IgA1) in a 4 hour 51Cr-release assay. Data are representative of n=3 biological replicates and represent mean ± s.d. of triplicates of one representative donor. ***P≤0.0007 by one-way ANOVA with multiple comparison correction. g, Specific lysis of control (DMSO)-treated (-) or SEN177-treated (+) murine pro-B cells (Ba/F3-Her2) by murine immune cells isolated from whole blood in the presence or absence of anti-Her2 (IgA1) in a 4 hour 51Cr-release assay. Data represent n=3 biological replicates and represent mean ± s.d. of triplicates of one representative donor. ***P<0.0001 by unpaired two-sided t-test. Values in a-c indicate MFI relative to WT cells stained with the same reagent. Data are representative of at least two independent experiments (a-g).
Extended Figure 8. QPCTL deficiency leads to enhanced tumor cell control by tumor specific antibodies.
a, Schematic representation of in vivo set-up. b, Absolute number (see Fig. 4c) of recovered tumor cells from mice injected with 1:1 mixtures of WT and QPCTL KO Ba/F3-Her2 cells that were then treated with control (PBS) (-) or anti-Her2 (IgA1) (+). n=6 control-treated animals; n=6 anti-Her2-treated animals. ***P≤0.0003 by unpaired two-sided t-test. c, Ratio of in vivo killing of target cells in mice injected with a 1:1 mixture of WT and CD47-KO cells, or a 1:1 mixture of WT and QPCTL KO Ba/F3-Her2 cells, and that were either treated with control (PBS) (-) or anti-Her2 (IgA1) antibody (+).n=6 control-treated animals (left graph); n=5 anti-Her2-treated animals (left graph); n=6 control-treated animals (right graph); n=6 anti-Her2-treated animals (right graph). ***P<0.0001 unpaired two-sided t-test. d, Absolute number (see Extended Data Fig. 8c) of recovered tumor cells in mice injected with a 1:1 mixture of WT and CD47 KO cells, or a 1:1 mixture of WT and QPCTL KO Ba/F3-Her2 cells, and that were either treated with control (PBS) (-) or anti-Her2 (IgA1) antibody (+). n=6 control-treated animals (left graph); n=5 anti-Her2-treated animals (left graph); n=6 control-treated animals (right graph); n=6 anti-Her2-treated animals (right graph). ***P<0.0001 by one-way ANOVA with multiple comparison correction; n.s., not significant. Dots represent mice treated with control (PBS), squares represent mice treated with anti-Her2 (IgA1) (b-d) and represent mean ± s.d. of individual mice (b-d). Data are representative of two independent experiments (b) or one experiment (c, d).
Extended Figure 9. QPCTL deficiency in combination with tumor specific antibodies leads to an enhanced neutrophil influx.
a, Absolute number (see Fig. 4c, d and f) of CD8+ T (CD3+ CD8+), CD4+ T (CD3+ CD4+) or B (B220+ MHCII+) cells present in mice that received a 1:1 mixture of WT and QPCTL KO Ba/F3-Her2 cells, and that were either control (PBS)-treated (-) or treated with anti-Her2 (IgA1) (+). n=6 control-treated animals; n=6 anti-Her2-treated animals. **P=0.0050. by unpaired two-sided t-test; n.s., not significant. b, Absolute number of peritoneal neutrophils (Ly-6G+/CD11b+), macrophages (F4/80+ CD11b+), CD8+ T (CD3+/CD8+), CD4+ T (CD3+/CD4+) and B (B220+/MHCII+) cells present in recipients of a 1:1 mixture of WT and QPCTL KO Ba/F3-Her2 cells that were control (PBS)-treated (-) or treated with anti-Her2 (IgA1) (+)(see Extended Data Fig. 8c). n=6 control-treated animals injected with mixture of WT/CD47 KO cells; n=6 anti-Her2-treated animals injected with mixture of WT/CD47 KO cells; n=6 control-treated animals injected with mixture of WT/QPCTL KO cells; n=5 anti-Her2-treated animals injected with mixture of WT/QPCTL KO cells. ***P<0.0001; 0.0015≤**P≤0.0022; *P=0.0226 by one-way ANOVA with multiple comparison correction; n.s., not significant. c, Absolute number of recovered tumor cells in recipients of WT (blue) or QPCTL KO (green) Ba/F3-Her2 cells that were treated with anti-Her2 (IgA1) antibody. n = 5 animals injected with WT cells. n = 5 animals injected with QPCTL KO cells. *P=0.0161 by unpaired two-sided t-test; n.s., not significant. d, Absolute number of peritoneal neutrophils (Ly-6G+/CD11b+), macrophages (F4/80+ CD11b+) and CD3+ T cells, CD4 effector (CD4+/FOXP3-) and CD4 regulatory (CD4+/FOXP3+) T cells present in recipients of WT (blue) or QPCTL KO (green) Ba/F3-Her2 cells that were treated with anti-Her2 (IgA1) (see Extended Data Fig. 7g). n = 5 animals injected with WT cells. n = 5 animals injected with QPCTL KO cells. n.s., not significant.
Dots represent mice treated with control (PBS), squares represent mice treated with anti-Her2 (IgA1) (a-d) and represent mean ± s.d. of individual mice (a-d). Data are representative of two independent experiments (a) or one experiment (b-d).
Extended Figure 10. Expansion, differentiation, cytokine production and killing capacity of human T cells are unaltered by glutaminyl cyclase inhibition.
To determine the effect of glutaminyl cyclase inhibition on different aspects of T cell function, in vitro T cell cytokine production and secretion, T cell phenotype, T cell killing capacity, and T cell induction by autologous dendritic cells (DCs) was assessed in the presence of control (DMSO) or SEN177. a and b, CDK4 TCR-transduced T cells (CDK4-specific T cells) from two donors were cultured for 4 days in the presence of control (DMSO) or SEN177 a, Intracellular cytokine production of control (DMSO)-treated or SEN177-treated CDK4-specific T cells upon co-culture with JY cells pulsed with the indicated concentrations of CDK4 or MART-1 peptide, as analysed by flow cytometry. Values indicate percentage of IL-2+ (left), IFNγ+ (middle) and TNFα+ (right) CD8+ cells of total CD8+cells. Data represent n=3 biological replicates and mean ± s.d. of triplicates. b, Specific lysis of CDK4+ (NKIRTIL006) and CDK4- (A375) tumor cells by control (DMSO)-treated or SEN177-treated CDK4-specific T cells, as determined in a 4 hour 51Cr-release assay. Data represent n=3 (Donor 1), n=2 (Donor 2 – NKIRTIL006 cocultures) or n=1 (donor 2 - A375 cocultures) biological replicates and mean ± s.d of triplicates (Donor 1). c-e, Peripheral Blood Lymphocytes (PBLs) from two donors were isolated, stimulated with anti-CD3/CD28 beads (2:1 bead to T cell ratio), and cultured for two weeks in the presence of control (DMSO) or SEN177. Data represent n=3 biological replicates and mean ± s.d. of triplicates. c, Fold expansion of control (DMSO)-treated or SEN177-treated CDK4-specific T cells over a period of 11 days. d, Surface expression of CCR7, CD4, CD8, CD27, CD28, CD45RA, CD45RO, CD62L and PD-1 on CD3+ T cells after 14 days of control (DMSO) treatment or SEN177 treatment, as determined by flow cytometry. Values indicate percentage of CD3+ cells positive for the indicated marker. e, IFNγ, IL-2, IL-4, TNFα, IL-6, IL-17A and IL-10 secretion of T cells after 14 days of control (DMSO) treatment or SEN177 treatment and subsequent re-stimulation with anti-CD3/CD28 beads, as determined by CBA bead array. Values indicate MFI. f, Differentiation of control (DMSO)-treated (-) or SEN177-treated (+) CD4+ T cells into Th17 cells after 7 days (t= 7d) or 10 days (t= 10d) of culture, as determined by flow cytometry. Values indicate the percentage of IL17A+ cells of total lymphocytes. Data represent n=3 biological replicates and mean ± s.d. of triplicates. n.s., not significant by unpaired two-sided t-test. h and i, T cells were induced by autologous dendritic cells (DCs) under three conditions; control (DMSO) was present during DC maturation and during T cell induction (“control”), control (DMSO) was present during DC maturation and SEN177 was present during T cell induction (“SEN177 induction)”, or SEN177 was present during both DC maturation and T cell induction (“SEN177 maturation and induction”). h, Surface expression of CCR7, CD4, CD8, CD27, CD28, CD45RA, CD62L and PD-1 on CD3+ T cells 12 days after start of induction by autologous DCs. Values reflect percentage of CD3+ cells positive for the indicated marker. g, Percentage of MHC multimer positive CD8+ T cells before induction (t = 0) and 11 days after start of induction by autologous DCs, as determined by flow cytometry. Values indicate the percentage of MHC multimer (HLA-A*02:01 multimers loaded with the MART-1-ELA, CMV-NLV, EBV-GLC or EBV-YVL epitopes) CD8+ cells of total CD8+ cells. i, Intracellular cytokine production of CD8+ T cells 6 days after start of induction and subsequent re-stimulation with unloaded DCs (-) or peptide-loaded DCs (+).Values indicate percentage of IL-2+ (left), IFNγ+ (middle) and TNFα+ (right) CD8+ cells of total CD8+ cells.
Data are representative of one (a-i) independent experiment.
Supplementary Material
Acknowledgments
We thank R. Mezzadra, M. Wellenstein, C. Sun and members of the Schumacher, Brummelkamp, Scheeren, Leusen, van den Berg and Haanen laboratories for discussions, and Olaf van Tellingen, the NKI-AVL Preclinical Intervention Unit and the NKI-AVL flow facility for technical support and input.
Funding
This work was supported by ERC AdG SENSIT (to T.N.S.), the Institute for Chemical Immunology (to T.N.S., J.N. and M.V.), ERC StG CHEMCHECK (to M.V.), KWF grant 10300 (to T.K.v.d.B.), KWF grant 11537 (to H.L.M.), a LUMC fellowship and the Errol McDowell Cancer Foundation (to F.A.S.), NWO Vici Grant 016.Vici.170.033, KWF grant NKI 2015-7609, the Cancer Genomics Center (CGC.nl), and the Ammodo KNAW Award 2015 for Biomedical Sciences (to T.R.B.), and KWF grant UU 2015-7650 (to J.H.W.L.).
Footnotes
Ethical Compliance
Animal experiments were in compliance with all relevant ethical regulations approved by the IVD committee (Utrecht, the Netherlands). Blood samples from healthy donors was collected after informed consent. The use of human blood samples was in compliance with all relevant ethical regulations approved by the Sanquin Ethics Advisory Council of Sanquin Blood Supply (Amsterdam, the Netherlands).
Reporting summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All sequencing datasets have been deposited in the NCBI Sequence Read Archive under accession number SRP144590. In addition, all processed screen results are accessible in an interactive database (https://phenosaurus.nki.nl/). All data presented in this manuscript are available from the corresponding authors upon reasonable request
Author contributions
M.E.W.L. conceived the project, designed and performed experiments, interpreted data and co-wrote the manuscript. M.R., A.F. an T.R.B. designed, performed and interpreted the haploid genetic screens. M.T. and J.N. designed, performed and interpreted biochemical data. J.H.M.J., A.M.B. and J.H.W.L. designed, performed and interpreted anti-Her2 in vitro and in vivo data, and J.H.W.L. co-wrote the manuscript. K.F., H.L.M. and T.K.v.d.B. designed, performed and interpreted in vitro data with human effector cells. S.v.d.S. supported and performed flow cytometry analyses. R.G.-E. and N.A.M.B. designed, performed and interpreted in vitro studies with human T cells. J.H.v.d.B. and J.B.A.G.H. supervised analyses of T cell reactivity. K.A.M. performed and interpreted experiments. M.V. designed experiments and provided reagents. F.A.S. and T.N.S. conceived the project, designed experiments, interpreted data and co-wrote the manuscript.
Competing interest
M.E.W.L., M.R., T.R.B., F.A.S., J.H.W.L. and T.N.S. are inventors on a patent application that covers manipulation of the CD47 – SIRPα axis via QPCTL.
T.N.S. is advisor for Adaptive Biotechnologies, AIMM Therapeutics, Allogene Therapeutics, Amgen, Merus, and Neon Therapeutics, is recipient of grant/research support from MSD, Bristol-Myers Squibb, and Merck KgaA, is stockholder in AIMM Therapeutics, Allogene Therapeutics, Merus, Neogene Therapeutics and Neon Therapeutics, and is venture partner at Third Rock Ventures. T.R.B. is cofounder and SAB member of Haplogen GmbH and cofounder and managing director of Scenic Biotech. J.H.W.L. is founder, advisor and shareholder of TigaTx. J.H.B. is recipient of grant/ research support from Bristol-Myers Squibb, Medimmune and Neon Therapeutics. K.F., H.L.M. and T.K.v.d.B are recipients of research support from Synthon Biopharmaceuticals BV. J.B.A.G.H. is advisor to Bristol-Myers Squibb, MSD, Novartis, Roche/Genentech, Pfizer, IPSEN, AZ/MedImmune, Bayer, Seattle Genetics, Immunocore, Gadeta, Neon Therapeutics, and Celsius Therapeutics, and is recipient of grant/ research support from Bristol-Myers Squibb, MSD, Novartis, Neon Therapeutics.
References
- 1.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Science Translational Medicine. 2016;8:328rv4–328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaiswal S, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majeti R, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiskopf K. Cancer immunotherapy targeting the CD47/SIRPα axis. Eur J Cancer. 2017;76:100–109. doi: 10.1016/j.ejca.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Willingham SB, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci USA. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matlung HL, Szilagyi K, Barclay NA, van den Berg TK. The CD47-SIRPα signaling axis as an innate immune checkpoint in cancer. Immunol Rev. 2017;276:145–164. doi: 10.1111/imr.12527. [DOI] [PubMed] [Google Scholar]
- 7.Zhao XW, et al. CD47-signal regulatory protein-α (SIRPα) interactions form a barrier for antibody-mediated tumor cell destruction. Proc Natl Acad Sci USA. 2011;108:18342–18347. doi: 10.1073/pnas.1106550108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao MP, et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Science Translational Medicine. 2010;2:63ra94–63ra94. doi: 10.1126/scitranslmed.3001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, et al. SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature. 2017;544:493–497. doi: 10.1038/nature22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao MP, et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 2010;142:699–713. doi: 10.1016/j.cell.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiskopf K, et al. Engineered SIRPα variants as immunotherapeutic adjuvants to anticancer antibodies. Science. 2013;341:88–91. doi: 10.1126/science.1238856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ring NG, et al. Anti-SIRPα antibody immunotherapy enhances neutrophil and macrophage antitumor activity. Proc Natl Acad Sci USA. 2017;114:E10578–E10585. doi: 10.1073/pnas.1710877114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Advani R, et al. CD47 Blockade by Hu5F9-G4 and Rituximab in Non-Hodgkin’s Lymphoma. N Engl J Med. 2018;379:1711–1721. doi: 10.1056/NEJMoa1807315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casey SC, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seiffert M, et al. Human signal-regulatory protein is expressed on normal, but not on subsets of leukemic myeloid cells and mediates cellular adhesion involving its counterreceptor CD47. Blood. 1999;94:3633–3643. [PubMed] [Google Scholar]
- 16.Cynis H, et al. Isolation of an isoenzyme of human glutaminyl cyclase: retention in the Golgi complex suggests involvement in the protein maturation machinery. J Mol Biol. 2008;379:966–980. doi: 10.1016/j.jmb.2008.03.078. [DOI] [PubMed] [Google Scholar]
- 17.Schilling S, et al. Identification of human glutaminyl cyclase as a metalloenzyme. Potent inhibition by imidazole derivatives and heterocyclic chelators. J Biol Chem. 2003;278:49773–49779. doi: 10.1074/jbc.M309077200. [DOI] [PubMed] [Google Scholar]
- 18.Stephan A, et al. Mammalian glutaminyl cyclases and their isoenzymes have identical enzymatic characteristics. FEBS J. 2009;276:6522–6536. doi: 10.1111/j.1742-4658.2009.07337.x. [DOI] [PubMed] [Google Scholar]
- 19.Jimenez-Sanchez M, et al. siRNA screen identifies QPCT as a druggable target for Huntington's disease. Nat Chem Biol. 2015;11:347–354. doi: 10.1038/nchembio.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saido TC, et al. Dominant and differential deposition of distinct β-amyloid peptide species, AβN3(pE), in senile plaques. Neuron. 1995;14:457–466. doi: 10.1016/0896-6273(95)90301-1. [DOI] [PubMed] [Google Scholar]
- 21.Tekirian TL, Yang AY, Glabe C, Geddes JW. Toxicity of Pyroglutaminated Amyloid β-Peptides 3(pE)-40 and -42 Is Similar to That of Aβ1-40 and -42. Journal of Neurochemistry. 2002;73:1584–1589. doi: 10.1046/j.1471-4159.1999.0731584.x. [DOI] [PubMed] [Google Scholar]
- 22.Russo C, et al. Pyroglutamate-modified amyloid β-peptides - AβN3(pE) - strongly affect cultured neuron and astrocyte survival. Journal of Neurochemistry. 2002;82:1480–1489. doi: 10.1046/j.1471-4159.2002.01107.x. [DOI] [PubMed] [Google Scholar]
- 23.Hatherley D, et al. Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47. Mol Cell. 2008;31:266–277. doi: 10.1016/j.molcel.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 24.Ho CCM, et al. ‘Velcro’ engineering of high affinity CD47 ectodomain as signal regulatory protein α (SIRPα) antagonists that enhance antibody-dependent cellular phagocytosis. J Biol Chem. 2015;290:12650–12663. doi: 10.1074/jbc.M115.648220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooke G, Holbrook JD, Brown MH, Barclay AN. Human lymphocytes interact directly with CD47 through a novel member of the signal regulatory protein (SIRP) family. J Immunol. 2004;173:2562–2570. doi: 10.4049/jimmunol.173.4.2562. [DOI] [PubMed] [Google Scholar]
- 26.Huang K-F, Wang Y-R, Chang E-C, Chou T-L, Wang AH-J. A conserved hydrogen-bond network in the catalytic centre of animal glutaminyl cyclases is critical for catalysis. Biochem J. 2008;411:181–190. doi: 10.1042/BJ20071073. [DOI] [PubMed] [Google Scholar]
- 27.Grizenkova J, et al. Overexpression of the Hspa13 (Stch) gene reduces prion disease incubation time in mice. Proc Natl Acad Sci USA. 2012;109:13722–13727. doi: 10.1073/pnas.1208917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schilling S, et al. Glutaminyl cyclase inhibition attenuates pyroglutamate Abeta and Alzheimer's disease-like pathology. Nat Med. 2008;14:1106–1111. doi: 10.1038/nm.1872. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann T, et al. Glutaminyl Cyclase Inhibitor PQ912 Improves Cognition in Mouse Models of Alzheimer's Disease-Studies on Relation to Effective Target Occupancy. J Pharmacol Exp Ther. 2017;362:119–130. doi: 10.1124/jpet.117.240614. [DOI] [PubMed] [Google Scholar]
- 30.Lues I, et al. A phase 1 study to evaluate the safety and pharmacokinetics of PQ912, a glutaminyl cyclase inhibitor, in healthy subjects. Alzheimer's & Dementia: Translational Research & Clinical Interventions. 2015;1:182–195. doi: 10.1016/j.trci.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang KF, Liu YL, Cheng WJ, Ko TP, Wang AHJ. Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation. Proceedings of the National Academy of Sciences. 2005;102:13117–13122. doi: 10.1073/pnas.0504184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boross P, et al. IgA EGFR antibodies mediate tumour killing in vivo. EMBO Mol Med. 2013;5:1213–1226. doi: 10.1002/emmm.201201929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, et al. CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat Med. 2015;21:1209–1215. doi: 10.1038/nm.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingram JR, et al. Localized CD47 blockade enhances immunotherapy for murine melanoma. Proc Natl Acad Sci USA. 2017;114:10184–10189. doi: 10.1073/pnas.1710776114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manguso RT, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sockolosky JT, et al. Durable antitumor responses to CD47 blockade require adaptive immune stimulation. Proc Natl Acad Sci USA. 2016;113:E2646–54. doi: 10.1073/pnas.1604268113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ansell S, et al. A Phase 1 Study of TTI-621, a Novel Immune Checkpoint Inhibitor Targeting CD47, in Patients with Relapsed or Refractory Hematologic Malignancies. Blood. 2016;128:1812. [Google Scholar]
- 38.Scheltens P, et al. Safety, tolerability and efficacy of the glutaminyl cyclase inhibitor PQ912 in Alzheimer's disease: results of a randomized, double-blind, placebo-controlled phase 2a study. Alzheimers Res Ther. 2018;10:107. doi: 10.1186/s13195-018-0431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morty RE, Bulau P, Pellé R, Wilk S, Abe K. Pyroglutamyl peptidase type I from Trypanosoma brucei: a new virulence factor from African trypanosomes that de-blocks regulatory peptides in the plasma of infected hosts. Biochem J. 2006;394:635–645. doi: 10.1042/BJ20051593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong Q, et al. A Ultrasensitive Near-Infrared Fluorescent Probe Reveals Pyroglutamate Aminopeptidase 1 Can Be a New Inflammatory Cytokine. Adv Sci (Weinh) 2018;5 doi: 10.1002/advs.201700664. 1700664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 42.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bracke M, Lammers JW, Coffer PJ, Koenderman L. Cytokine-induced inside-out activation of FcalphaR (CD89) is mediated by a single serine residue (S263) in the intracellular domain of the receptor. Blood. 2001;97:3478–3483. doi: 10.1182/blood.v97.11.3478. [DOI] [PubMed] [Google Scholar]
- 45.Berkovits BD, Mayr C. Alternative 3' UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lackner DH, et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun. 2015;6 doi: 10.1038/ncomms10237. 10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168–e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neefjes JJ, Breur-Vriesendorp BS, van Seventer GA, Iványi P, Ploegh HL. An improved biochemical method for the analysis of HLA-class I antigens. Definition of new HLA-class I subtypes. Hum Immunol. 1986;16:169–181. doi: 10.1016/0198-8859(86)90046-7. [DOI] [PubMed] [Google Scholar]
- 49.Kuijpers TW, et al. Membrane surface antigen expression on neutrophils: a reappraisal of the use of surface markers for neutrophil activation. Blood. 1991;78:1105–1111. [PubMed] [Google Scholar]
- 50.Brandsma AM, et al. Simultaneous Targeting of FcγRs and FcαRI Enhances Tumor Cell Killing. Cancer Immunol Res. 2015;3:1316–1324. doi: 10.1158/2326-6066.CIR-15-0099-T. [DOI] [PubMed] [Google Scholar]
- 51.Meyer S, et al. Improved in vivo anti-tumor effects of IgA-Her2 antibodies through half-life extension and serum exposure enhancement by FcRn targeting. MAbs. 2016;8:87–98. doi: 10.1080/19420862.2015.1106658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Haij S, et al. In vivo cytotoxicity of type I CD20 antibodies critically depends on Fc receptor ITAM signaling. Cancer Res. 2010;70:3209–3217. doi: 10.1158/0008-5472.CAN-09-4109. [DOI] [PubMed] [Google Scholar]
- 53.Gee MH, et al. Antigen Identification for Orphan T Cell Receptors Expressed on Tumor-Infiltrating Lymphocytes. Cell. 2018;172:549–563.e16. doi: 10.1016/j.cell.2017.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hadrup SR, et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nature Methods. 2009;6:520–526. doi: 10.1038/nmeth.1345. 2009 6:7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.