Abstract
Childhood maltreatment is highly prevalent and serves as a risk factor for mental and physical disorders. Self-reported childhood maltreatment appears heritable, but the specific genetic influences on this phenotype are largely unknown. The aims of this study were to (1) identify genetic variation associated with self-reported childhood maltreatment, (2) estimate SNP-based heritability (h2snp), (3) assess predictive value of polygenic risk scores (PRS) for childhood maltreatment, and (4) quantify genetic overlap of childhood maltreatment with mental and physical health-related phenotypes, and condition the top hits from our analyses when such overlap is present. Genome-wide association analysis for childhood maltreatment was undertaken, using a discovery sample from the UK Biobank (UKBB) (n = 124,000) and a replication sample from the Psychiatric Genomics Consortium-posttraumatic stress disorder group (PGC-PTSD) (n = 26,290). h2snp for childhood maltreatment and genetic correlations with mental/physical health traits were calculated using linkage disequilibrium score regression. PRS was calculated using PRSice and mtCOJO was used to perform conditional analysis. Two genome-wide significant loci associated with childhood maltreatment (rs142346759, p = 4.35 × 10−8, FOXP1; rs10262462, p = 3.24 × 10−8, FOXP2) were identified in the discovery dataset but were not replicated in PGC-PTSD. h2snp for childhood maltreatment was ~6% and the PRS derived from the UKBB was significantly predictive of childhood maltreatment in PGC-PTSD (r2 = 0.0025; p = 1.8 × 10−15). The most significant genetic correlation of childhood maltreatment was with depressive symptoms (rg = 0.70, p = 4.65 × 10−40), although we show evidence that our top hits may be specific to childhood maltreatment. This is the first large-scale genetic study to identify specific variants associated with self-reported childhood maltreatment. Speculatively, FOXP genes might influence externalizing traits and so be relevant to childhood maltreatment. Alternatively, these variants may be associated with a greater likelihood of reporting maltreatment. A clearer understanding of the genetic relationships of childhood maltreatment, including particular abuse subtypes, with a range of phenotypes, may ultimately be useful in in developing targeted treatment and prevention strategies.
Subject terms: Medical genetics, Psychiatric disorders
Introduction
The lifetime prevalence of childhood physical, sexual, and emotional maltreatment ranges from 8% to 36%1. In addition to being highly prevalent, such childhood abuse is associated with the development of mental disorders, including depression2,3, and physical ill health, including non-communicable diseases4,5. Although these associations are now well established, estimates of effect size vary considerably across epidemiological studies, likely reflecting methodological challenges, including uncertainty about how best to assess childhood maltreatment6.
A twin-based study found that retrospective reports of childhood maltreatment has a heritability of 6%7. Although the idea that childhood maltreatment is heritable may seem counter-intuitive, work on behavior genetics has long documented the heritability of many exposures perceived as environmental. Such heritability is referred to as gene–environment correlation (rGE), and three potential rGE mechanisms to explain the heritability of childhood maltreatment may be posited. First, a “passive” rGE: parental genes affecting parental behavior may influence the childhood environment (e.g. aggressive parents may be more likely to physically punish their children8). Second, an “active” rGE: individuals with genetic variants associated with certain behavioral phenotypes may be more at risk of selecting or creating adverse situations (e.g. risk-taking is heritable and children who are high in risk-taking may be exposed to more trauma)9,10. Third, an “evocative” rGE: genetic variation may influence child behavior, which in turn is associated with responses to the child (e.g. genetic factors may influence infant “difficultness”, which in turn is associated with maternal hostile-reactive behavior that is correlated with child abuse11,12). The latter two rGEs are sometimes collectively referred to as non-passive correlations7.
While a number of key risk factors for childhood maltreatment, including child behavioral characteristics and parental mental health, have been investigated6, studies have seldom focused on associated genetic variation. The few genetic association studies of childhood maltreatment have only considered variants in candidate genes13 and have had insufficient power to detect the small polygenic effect sizes typically associated with behavioral phenotypes14. Also, there are no studies of the genetic overlap of childhood maltreatment with mental and physical health-related traits, using genome-wide single nucleotide polymorphism (SNP) data. Knowledge of specific genetic variation for childhood maltreatment, the heritability of this phenotype, the polygenic risk, and the genetic overlap with other traits may be useful in informing our understanding of the risk factors, the etiology, and the outcomes of childhood maltreatment. This, in turn, may have implications for the design of prevention and treatment programs for adverse health outcomes. For example, environmental exposures that play a causal role in impacting health outcomes are likely to mediate any observed associations between genetic variants and that health outcome (e.g. early loss of a parent may lead to depression, with such loss then mediating the association between heritability of early parental loss and depression). Thus, preventative strategies would focus on decreasing the risk conferred by the environmental exposure without needing to specifically consider the genetic influences on the health outcome9 (e.g. development of programs for children who have experienced early loss).
The PGC-PTSD has collaborated to obtain access to well-powered genetic studies of trauma and PTSD that have allowed a number of key genetic questions in this field to be investigated15–17, providing a unique opportunity to address knowledge gaps in the area of childhood maltreatment. This study aims to: (1) identify genetic variants associated with childhood maltreatment using a genome-wide association study (GWAS) design, (2) quantify the heritability of childhood maltreatment using SNP-based methods, (3) assess the predictive value of polygenic risk scores (PRS) for childhood maltreatment, and (4) assess the degree of genetic overlap of childhood maltreatment with mental and physical health-related phenotypes, and condition the top genome-wide hits from our analyses when such overlap is present.
Materials and methods
Participating studies
Nineteen studies, comprising subjects of European ancestry only, were used in this analysis. The discovery dataset consisted of 124,711 individuals with available childhood maltreatment data from the UK Biobank (UKBB)18, and the replication sample comprised 26,290 individuals—a subset of the PGC-PTSD Freeze 1.5 dataset (PGC1.5)17. The details of these studies, including the demographics and instruments used to assess maltreatment can be found in Supplementary Table 1. We have complied with relevant ethical regulations for work with human subjects. All subjects provided written informed consent and studies were approved by the relevant institutional review boards and the UCSD IRB (protocol #16097×).
Phenotype harmonization
For the childhood maltreatment phenotype, Childhood Trauma Questionnaire (CTQ) scores on physical, sexual, and emotional abuse19 were obtained from the participating studies. From this, an overall childhood maltreatment count score of 0–3 was constructed, based on a count of the three abuse categories listed above. An individual was considered to have endorsed a childhood abuse category if they scored in the moderate to extreme range for that particular category, per established cut-offs20 (Supplementary Table 2). If CTQ data were not available, the event assessment during childhood (occurring before 18 years of age) that was most validated for that particular study was obtained, providing a count of the total number of different categories of reported childhood events (e.g. physical, sexual, or severe emotional abuse) along with the range of possible scores for the measure. The reported maltreatment exposure from the UKBB dataset comprised a score of three items where participants were asked whether they were (i) “physically abused by family as a child”, (ii) “sexually molested as a child”, and whether they (iii) “felt hated by family member as a child”. The childhood maltreatment count score, whether it was generated from the CTQ or another instrument, was used as the main outcome measure in the association analysis. The range and mean of maltreatment count scores for each study can be seen in Supplementary Table 1.
Global ancestry determination, genotyping quality control, and imputation
Study participants from the PGC-PTSD were genotyped with a number of different arrays (Supplementary Table 1). Genotype data were quality controlled and processed using the standard PGC pipeline, Ricopili-MANC (https://sites.google.com/a/broadinstitute.org/ricopili/ and https://github.com/orgs/Nealelab/teams/ricopili) as part of the PGC-PTSD Freeze 2 data analysis17,21. This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative. A detailed outline of these methods can be found in ref. 17. Briefly, ancestry was determined with pre-QC genotypes using a SNPweights panel of 10,000 ancestry informative markers from a reference panel comprising 2911 subjects from 71 diverse populations and six continental groups (https://github.com/nievergeltlab/global_ancestry). Samples with estimated > 90% European ancestry were classified as European. Samples were excluded if they had call rates < 98%, deviated from the expected inbreeding coefficient (fhet < −0.2 or >0.2), or had a sex discrepancy between reported and genotypic sex (based on inbreeding coefficients calculated from SNPs on the X chromosome). Markers were excluded if they had call rates < 98%, >2% difference in missing genotypes between PTSD cases and controls, or were monomorphic. Markers with a Hardy–Weinberg equilibrium (HWE) p < 1 × 10−6 in controls were excluded from all subjects. Principal components (PCs) were calculated using the smartPCA algorithm in EIGENSTRAT22. Pre-phasing and phasing was performed using SHAPEIT2 v2.r83723. Imputation was performed with IMPUTE2 v2.2.224 using the 1000 Genomes (1000G) phase 3 data25 as the reference.
Details regarding the QC, imputation, and ancestry determination of the UKBB dataset can be found in ref. 26. Briefly, study participants were genotyped with two custom genotyping arrays (with ∼800,000 markers). A two-stage imputation was performed using the Haplotype Reference Consortium (HRC)27 and the UK10K28 as the reference panels. Variants in the UKBB dataset were filtered to include only those with a minor allele frequency (MAF) of > 1% and an INFO score of > 0.4. Related individuals (third degree and closer) and those with a genotyping call rate < 98% were excluded. Ancestry was determined by 4-means clustering on the first two PCs provided by the UKBB29. Additional principal component analysis was conducted on the European-only data subset using flashpca230.
Main GWAS
GWAS analysis was conducted separately for each study. Best-guess genotypes were tested for association to self-reported childhood maltreatment using an ordinal logistic regression model with age, sex, and the first five PCs included as covariates. Variants with a MAF < 0.5% and a genotyping rate < 98% were excluded, for all studies except the UKBB. These analyses were implemented in PLINK 1.931 using the plug-in Rserve. To ensure computational efficiency, linear regression models were run for 4 of the larger contributing studies (NSS1; NSS2; PPDS; and UKBB, N = 143,392 subjects)17. For the NSS1; NSS2; and PPDS studies, age, sex, and 5 PCs were included as covariates in the regression model. For the UKBB dataset, the regression analysis was implemented in BGenie v1.232 with age, sex, 6 PCs, batch, and site included as covariates. All tests performed were two-sided.
Meta-analysis
As both linear and ordinal logistic models were implemented in the GWASs, which resulted in different effect statistics, fixed effects meta-analysis was conducted across studies using p-values and direction of effect, weighted according to the effective sample size as the analysis scheme, in METAL (v. March 25 2011)33. Effective sample sizes (Neff) for ordinal logistic regressions were calculated as Neff = harmonic mean*n levels of childhood maltreatment, and for linear regressions as Neff = ((1−probability of having a zero score) × mean of nonzero data)34. Heterogeneity across datasets was tested using the Cochran’s Q-test for heterogeneity, also implemented in METAL. Only variants with an INFO score of >0.8 and a conservative MAF of >5% were included in the meta-analysis, except where otherwise indicated in the results. Forest plots were generated for genome-wide significant hits using the R package meta35.
Functional mapping and annotation
Genome-wide significant hits identified from the GWAS meta-analysis were annotated using the web-based tool FUnctional Mapping and Annotation (FUMA) v1.3.4c36. Default settings were used and annotations were based on the human genome assembly GRCh37 (hg19). The SNP2GENE module was used to identify genomic risk loci and these were mapped to protein-coding genes within a 10 kb window. An r2 of ≥ 0.6 was used to identify variants in LD with lead SNPs. The 1000G European Phase 3 was used as the reference dataset. Variants were functionally annotated using ANNOVAR, combined dependent depletion (CADD), RegulomeDB (RDB), and chromatin states (only tissues/cells from brains were included). The NHGRI-EBI GWAS catalog was used to determine any previous associations with the identified risk variants. The GTEx v7 brain tissue, RNAseq data from the CommonMind Consortium and the BRAINEAC database were used to perform eQTL mapping for significant SNP–gene pairs (FDR q < 0.05).
A gene-based analysis was performed within FUMA using MAGMA whereby SNPs were mapped to 18,989 protein-coding genes. Genome-wide significance was set at a Bonferroni-corrected threshold p < 2.63 × 10−6. In addition, gene-based test statistics were used to determine whether specific biological pathways are associated with childhood maltreatment. This was performed for 10,678 curated gene sets and GO terms obtained from MsigDB, using MAGMA. The significance threshold was set at a Bonferroni-corrected threshold of p = 4.68 × 10−6 (0.05/10,678).
Heritability estimation
Linkage disequilibrium score regression (LDSR) is a technique for quantifying polygenicity and confounding, such as population stratification, in GWAS summary statistics37. This is accomplished by evaluating the relationship between linkage disequilibrium (LD) scores (the average squared correlation of a SNP with all neighboring SNPs) and SNP test statistics. Using this approach, the LDSR intercept was used to estimate the proportion of inflation in test statistics due to polygenic signal (rather than inflation due to population stratification and cryptic relatedness), with the Eq. (1)—(LDSR intercept−1)/(mean observed chi-square−1)17. Using GWAS summary statistics, SNP-based heritability (h2snp) was calculated, which is one of the applications of LDSR.
Polygenic risk scoring
Using PRSice v2.1.3.beta38, PRS were calculated in target samples (PGC1.5) based on SNP effect sizes from childhood maltreatment GWAS in non-overlapping discovery/training samples (UKBB). Multiple p-value thresholds (PT) (0.001, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 1) were generated using the best guess genotype data of target samples. Variants with a MAF < 5% were excluded from the discovery dataset. As a default in PRSice, LD pruning was performed whereby variants were pruned if they were nearby (within 250 kb) and in LD (r2 > 0.1) with the leading variant (lowest p-value) in a given region. For this analysis, a rescaled childhood maltreatment phenotype was generated whereby the childhood maltreatment score for each individual was divided by the theoretical maximum score for a given study. Best-fit PRS (at PT = 0.0354) were used to predict childhood maltreatment status as a quantitative trait, adjusting for five PCs and dummy study indicator variables. As women in PGC1.5 experienced significantly more childhood maltreatment than men, we generated PRS in women and men separately. The proportion of variance explained by PRS was estimated as the difference in Nagelkerke’s R2 between the full model (which includes PRS plus covariates) and the null model (which only has the covariates). PRS prediction plots were based on quantiles of PRS, with effect sizes calculated in reference to the lowest quantile. p-values for PRS were derived from a likelihood ratio test comparing the two models. The significance threshold was set at a Bonferroni-corrected threshold of p = 0.006 (0.05/8).
Genetic correlation
Another application of LDSR is the measurement of genetic correlation, i.e. the degree and direction of shared genetic effects between different traits37,39. Cross-cohort genetic correlation (rg) was calculated using LDSR. The web-based interface for LDSR, LD Hub, was used to further calculate pairwise genetic correlations between childhood maltreatment and 247 non-UKBB traits of interest including psychiatric, anthropomorphic, smoking behavior, reproductive, aging, education, autoimmune, and cardio-metabolic categories.
Conditional analyses of childhood maltreatment top hits
To evaluate if the effects of top variants in the UKBB GWAS and meta-analysis were specific to childhood maltreatment, we conditioned childhood maltreatment on genetically correlated traits using the multi-trait conditional and joint analysis (mtCOJO)40 feature in GCTA41. Data for major depressive disorder (MDD)42 (from https://www.med.unc.edu/pgc/results-and-downloads) was used to minimize sample overlap with the UK Biobank data. The effect of the correlated trait on childhood maltreatment was estimated using a generalized summary-data based Mendelian randomization analysis of significant LD independent SNPs (r2 < 0.05, based on 1000G Phase 3 CEU samples). The threshold for significance was set at p < 5 × 10−6, due to having less than the required 10 significant independent SNPs at the program default of p < 5 × 10−8, for the correlated trait.
Results
GWAS and meta-analysis
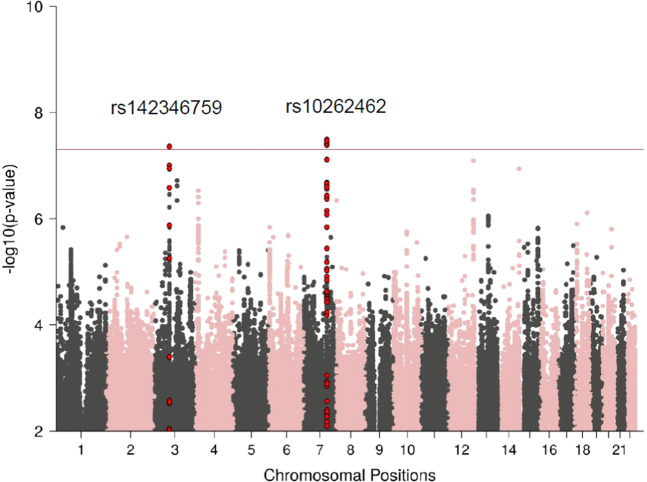
We report GWAS results from our discovery dataset (UKBB) (n = 124,711) and meta-analysis (n = 151,001). In our UKBB discovery dataset, we identified two genome-wide significant loci (p < 5 × 10−8) associated with childhood maltreatment (Table 1 and Fig. 1), rs142346759 (chr3, beta = 0.015, p = 4.35 × 10−8) and rs10262462 (chr7, beta = −0.016, p = 3.24 × 10−8). These variants remained significant in the meta-analysis (Table 1, Supplementary Figs. 1 and 2). Additional variants on chromosome 7 (rs1859100, beta = 0.015, p = 3.91 × 10−8) and chromosome 12 (rs917577, beta = 0.017, p = 2.64 × 10−8) (Supplementary Fig. 3), also achieved genome-wide significance in the meta-analysis. Running an ordinal regression on these hits in the UKBB led to similar results (data not shown). None of these hits were replicated in PGC1.5 (Table 1 and Supplementary Fig. 4).
Table 1.
Genome-wide significant hits in the UK Biobank, PGC-PTSD Freeze 1.5 (PGC 1.5), and meta-analyses.
| Variant | Chr | Position (bp) | Gene | A1 | A2 | A1 freq | Discovery: UK Biobank | Replication: Freeze 1.5 | Meta-analysis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Z-score | Beta | p-Value | n | Z-score | Betaa | P-value | I² (%) | P-het | n | Z-score | Betaa | p-Value | I² (%) | P-het | n | |||||||
| rs142346759 | 3 | 71,362,232 | FOXP1 | G | A | 0.039 | 5.476 | 0.015 | 4.35E−08 | 124,711 | 1.490 | 0.016 | 0.136 | 5 | 0.394 | 10,858b | 5.674 | 0.033 | 1.40E−08 | 0 | 0.491 | 135,569b |
| rs1859100 | 7 | 114,194,615 | FOXP2 | G | T | 0.597 | 5.517 | 0.016 | 3.45E−08 | 124,711 | 0.846 | 0.007 | 0.397 | 0 | 0.978 | 14,495 | 5.495 | 0.015 | 3.91E−08 | 0 | 0.973 | 139,206 |
| rs10262462 | 7 | 114,180,062 | FOXP2 | A | G | 0.403 | −5.528 | −0.016 | 3.24E−08 | 124,711 | −0.729 | −0.006 | 0.466 | 0 | 0.969 | 14,495 | −5.468 | −0.015 | 4.56E−08 | 0 | 0.958 | 139,206 |
| rs917577 | 12 | 126,548,817 | Intergenic | C | G | 0.265 | 5.367 | 0.015 | 8.02E−08 | 124,711 | 1.476 | 0.016 | 0.14 | 0 | 0.781 | 11,463 | 5.564 | 0.017 | 2.64E−08 | 0 | 0.831 | 136,174 |
aAn approximation was used to transform the Z-statistics from the effective sample-size-weighted meta-analysis (the output of the software METAL) into a beta value74. This was calculated on the scale of the MRS dataset
bAs this variant has a MAF of < 5% in the UKBB, only studies with a minor allele count of at least five alleles were included in the meta analysis
Fig. 1. Manhattan plot of UKBB GWAS for childhood maltreatment, showing the top variants.
The horizontal line represents genome-wide significance at p < 5 × 10−8.
Quantile–quantile (qq) plots indicate minimal inflation of p-values across studies (Supplementary Figs. 5–7). Using the LDSR intercept method, polygenic effects account for 93% and 94% of the observed inflation in test statistics for the UKBB dataset (intercept = 1.0096, SE = 0.0064) and meta-analysis (intercept = 1.0095, SE = 0.0077), respectively (Supplementary Figs. 5 and 7), consistent with minimal population stratification and cryptic relatedness.
Integration with functional genomic data
Using the web-based tool FUMA, the two UKBB GWAS hits were each annotated to two genes, FOXP1 and FOXP2 (Table 2). Gene-based analysis of the UKBB GWAS summary statistics further identified three gene-wide significant genes, KIF26B (p = 1.67 × 10−7), CNTNAP5 (p = 8.89 × 10−7), and EXOC2 (p = 2.04 × 10−6) from a total of 18,989 protein-coding genes. Gene-set analysis did not reveal any significant pathways associated with childhood maltreatment. Limited functionality of the two risk variants (rs142346759 and rs10262462) was observed (Table 2). One of the SNPs in LD for the risk variant on chromosome 3, rs142346759, obtained a CADD score of >12.37, indicating that this SNP may be deleterious. Six of the SNPs in LD with the risk variant on chromosome 7, rs10262462, had a CADD score of >12.37. No significant eQTLs were identified for either risk locus.
Table 2.
Functional mapping and annotation of UKBB GWAS and meta-analysis.
| Sample | GWAS hit lead variant | GWAS p-value | Position (hg19) | #SNPs in LD (r2 > 0.6) | Genomic coordinated risk locus (hg19) | Predicted genes in risk locus | SNPs in LD with CADD scores > 12.37 | SNPs in LD with RegulomeDB scores < 5 | Chromatin state analysis (Roadmap Epigenomics) in neuronal cell lines/tissuesa | eQTL (28 neuronal tissue/cell lines from CommonMind Consortium, BRAINEAC or GTEx v7) |
|---|---|---|---|---|---|---|---|---|---|---|
| UKBB GWAS | rs142346759 | 4.35E−08 | 3: 71362232 | 5 | chr3:71355932–71417539 | FOXP1 intronic | rs34936081 | rs13083684 (intronic) | Mainly quiecent | None |
| rs10262462 | 4.36E−08 | 7: 114180062 | 77 | chr7:114015707–114287116 | FOXP2 intronic | rs7785701,rs6466488,rs10249234,rs9332390,rs66823671,rs12533005 | rs2396753 (intronic), rs12536335 (intronic) | Mainly quiecent | None | |
| Meta-analysis | rs1859100 | 3.91E−08 | 7: 114194615 | 74 | chr7:114015707–114287116 | FOXP2 intronic | rs66823671,rs71149745,rs7785701,rs12533005,rs9332390,rs10249234, rs1476535, rs6466488 | rs2396753 (intronic), rs12536335 (intronic), rs9332390 (intronic) | Mainly quiecent | None |
| rs917577 | 2.64E−08 | 12:126548817 | 27 | chr12:126548816–126585545 | Intergenic | None | None | Mainly quiecent | None |
aIn neuronal cell lines/tissues E053, E054, E067, E068, E069, E070, E071, E072, E073, E074, E081, E082, E125
The chromosome 7 variant identified in the meta-analysis, rs1859100, also mapped to the gene FOXP2 and is located in the same genomic risk locus (chr7:114,015,707–114,287,116 base pairs) as rs10262462. The other hit observed in the meta-analysis, rs917577, was mapped to an intergenic region on chromosome 12. This variant obtained an RDB categorical score of 2B, indicating that it is likely to affect transcription factor binding. No eQTLs exist in the selected tissue types for this region (Table 2).
Heritability of reported childhood maltreatment
GWAS summary statistics were used to estimate the h2snp of childhood maltreatment with the tool LDSR (Table 3). The h2snp was estimated at 0.057 (p = 1.60 × 10−32) for the UKBB discovery dataset and 0.123 (p = 0.002) for PGC1.5. The h2snp for the meta-analysis was 0.057 (p = 4.48 × 10−46).
Table 3.
Heritability estimates based on LD-score regression (LDSR).
| Sample | n | h2snp | SE | Z | p-value |
|---|---|---|---|---|---|
| UKBB | 124,711 | 0.057 | 0.005 | 11.40 | 1.60E−32 |
| PGC1.5 | 26,290 | 0.123 | 0.040 | 3.08 | 2.00E−03 |
| Meta-analysis | 151,001 | 0.057 | 0.004 | 14.25 | 4.48E−46 |
Estimates are calculated for the UK biobank (UKBB), the PGC-PTSD Freeze 1.5 (PGC1.5), and meta-analysis
Polygenic risk scoring
We assessed the predictive value of PRS for childhood maltreatment, using our largest cohort, the UKBB, as a training sample. Our analyses showed a highly significant increase in effect size to develop childhood maltreatment across PRS quantiles in the PGC1.5 target sample, with a variance explained of r2 = 0.0025 (p = 1.8 × 10−15). Participants in the 5th quantile of genetic risk had significantly higher childhood maltreatment scores than subjects in the 1st quantile (beta = 0.042, p = 4.78 × 10−16; Supplementary Fig. 8). Since women reported significantly more childhood maltreatment than men (PGC1.5 mean childhood maltreatment: women = 0.32, men = 0.127, p < 1 × 10−80), PRS were also calculated separately for women and men. When stratified by sex, PRS had significantly higher explanatory power in women (r2 = 0.0053) relative to men (r2 = 0.0015) (p = 0.0002, Supplementary Fig. 8).
Genetic correlations of reported childhood maltreatment with other traits and disorders
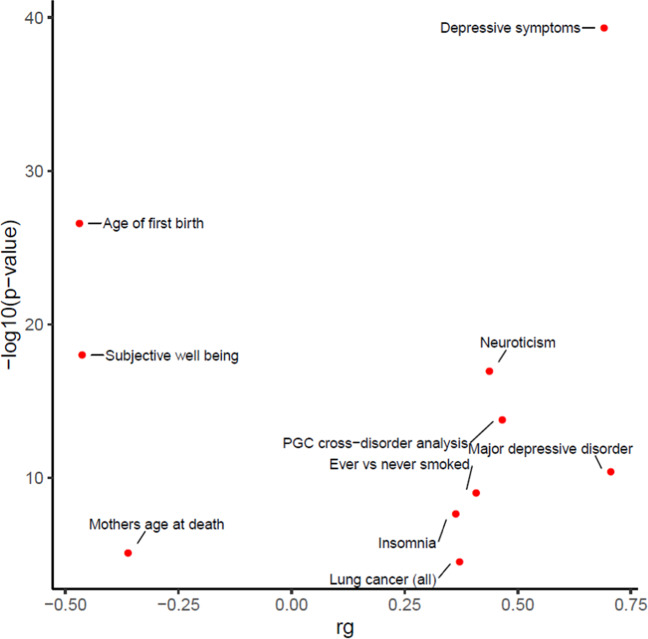
All pairwise genetic correlations are listed in Supplementary Table 3. The rg for childhood maltreatment between the UKBB and PGC1.5 datasets was 0.63 (p = 3.28 × 10−6). To determine whether there is significant genetic overlap between childhood maltreatment and other traits and disorders, pairwise genetic correlations were calculated using the web-based tool LD Hub. A total of 27 significant correlations (Bonferroni-corrected p-value threshold = 0.05/247 = 0.0002) were found between childhood maltreatment in the meta-analysis and 247 non-UKBB traits. The top 10 highest genetic correlations are plotted in Fig. 2 with depressive symptoms (rg = 0.70, p = 4.65 × 10−40) having the most significant correlation with childhood maltreatment. There were also positive genetic correlations with “MDD” (rg = 0.71, p = 4.13 × 10−11), “PGC cross-disorder analysis” (rg = 0.47, p = 1.62 × 10−14) and “neuroticism” (rg = 0.44, p = 1.14 × 10−17). Significant negative genetic correlations between childhood maltreatment and “age of first birth” (rg = −0.47, p = 2.61 × 10−27), “subjective well-being” (rg = −0.46, p = 1.00 × 10−18), and “mother’s age at death” (rg = −0.36, p = 7.42 × 10−6) were also observed.
Fig. 2.
Top ten genetic correlations between several groups of traits (from psychiatric, anthropomorphic, smoking behavior, reproductive, aging, education, autoimmune, and cardio-metabolic categories) and childhood maltreatment (meta-analysis).
Conditional analyses of childhood maltreatment top hits
As depressive symptoms (rg = 0.70, p = 4.65 × 10−40) and MDD (rg = 0.71, p = 4.13 × 10−11) were the most genetically correlated with childhood maltreatment, we conditioned the top hits from our meta-analysis for the effects of MDD using publicly available summary statistics for MDD42. We found that effect sizes for the four top hits for childhood maltreatment remained similar when adjusted for the effects of MDD (Supplementary Table 4). These findings indicate that the genetic variants identified here are specific to childhood maltreatment when tested in the context of MDD, the disorder genetically most significantly correlated with childhood maltreatment.
Discussion
The main findings of this study were that (1) variants located in the genes FOXP1 and FOXP2 and on chromosome 12 are significantly associated with childhood maltreatment, (2) the SNP-based estimate of childhood maltreatment is ~6%, (3) PRS of self-reported childhood maltreatment derived from a discovery cohort can significantly predict this phenotype in a target cohort, with 0.25% of variance explained, and (4) childhood maltreatment is significantly genetically correlated with “depressive symptoms” and “MDD”, “neuroticism”, “age of first birth”, and “subjective well-being”, despite showing evidence that our top hits may be specific to childhood maltreatment when conditioning on MDD.
Two genome-wide loci for childhood maltreatment identified in our discovery dataset were also significant in the meta-analysis: rs142346759 (chr3p13), an intronic variant in FOXP1 and rs10262462 (chr7q31.1) an intronic variant located in FOXP2. Both genes form part of the forkhead box superfamily of transcription factors which are widely expressed, and which play important roles during development and adulthood. FOXP1 and FOXP2 fall under the FOXP sub-family (also comprising FOXP3 and FOXP4) which has functions in oncogenic and tumor suppressive pathways43. FOXP2 contains highly conserved genomic sites, including an intronic region within this gene, located about 107 kb downstream from our risk variant44. FOXP1 and FOXP2 have ~60% homology at the amino acid level (https://www.ncbi.nlm.nih.gov/books/NBK7023/) and both proteins have been implicated in cognitive disorders, including expressive language impairment45. In the meta-analysis, we observed an additional genome-wide variant, located in an intergenic region on chromosome 12, but as this variant does not map to a particular gene, its possible biological mechanism is unclear.
Notably, variation within FOXP1 has been found to have associations with language impairment, internalizing symptoms, and externalizing symptoms46. FOXP2 has mainly been investigated in regards to speech and language development47, but has also been found to be associated with depression48 and attention deficit hyperactivity disorder (ADHD)49. Further, an intronic variant in the FOXP2 gene, rs727644, has been associated with risk-taking behavior50,51. While most work on childhood maltreatment has emphasized subsequent risk for mental and physical disorders, it is possible that externalizing behaviors increase risk for childhood trauma52, consistent with a non-passive rGE mechanism. Alternatively, phenotypes such as depression or neuroticism may increase the likelihood of individuals recalling childhood maltreatment53,54.
In this study we estimated SNP-based heritability for childhood maltreatment to be ~6%. A first possibility, in line with a link between FOXP variants and externalizing symptoms, is that genetic factors influence environmental factors indirectly through temperament and behavior9. A second possibility, consistent with the link of FOXP variants to internalizing symptoms and depression, is that genetic factors influence the recall of childhood maltreatment. In particular, retrospective assessment of childhood maltreatment may be limited by recall bias and the respondent’s subjective assessment of the event55,56. Indeed, a recent systematic review found very low concordance between prospective and retrospective measures of childhood maltreatment57 and those who retrospectively report childhood adversity were at greater risk for having psychopathology than those who prospectively reported childhood maltreatment58.
A twin-based study estimated the heritability of reported childhood maltreatment (comprising physical, and sexual maltreatment and neglect) to be 6%7, the same as our SNP-based estimate. As twin-based studies capture latent heritability across the entire genome, these heritability estimates are generally higher than SNP-based heritability estimates, which are limited to common variation and by the number of markers present and tagged on the genotyping array used15. However, in this twin study, when considering each maltreatment category separately, the heritability of childhood physical maltreatment, sexual maltreatment, and neglect was 28%, 0%, and 24%, respectively. This suggests that only physical abuse and neglect are heritable and that sexual abuse is not genetically influenced. It is notable that these twin data, then, do not support an rGE for some abuse types (i.e. sexual abuse).
We show that PRS derived from the UKBB was significantly predictive of childhood maltreatment in PGC1.5, explaining 0.25% of the variance for this exposure. Although the variance explained is relatively modest, we expect greater prediction accuracy with future larger sample sizes. When stratifying by sex, PRS had significantly higher explanatory power in women relative to men. This is expected as women had higher mean self-reported childhood maltreatment scores than men in PGC1.5.
The PRS results suggests a polygenic architecture for self-reported childhood maltreatment but does not indicate the mechanism by which genetic factors are able to influence this exposure. However, our finding of positive genetic correlations between childhood maltreatment, depressive symptoms, and MDD provides support for the hypothesis that genetic factors predisposing to reporting early life maltreatment overlap with those underlying depression. Genetic correlations between depression, stressful life events, and lifetime trauma have led to the hypothesis that genes increasing risk for the development of depression predispose individuals to entering into adverse environments59,60. Depressed individuals with and without trauma exposure differ in associated genetic variation, with trauma-exposed individuals having greater SNP-based heritability, supporting this hypothesis26,61. On the other hand, polygenic scores for MDD were associated with greater reporting of stressful life events in individuals with MDD62. Indeed, current mood can influence the recall of childhood experiences, and individuals with current depression are at an increased likelihood of reporting early life adversity63. Notably, although we show that childhood maltreatment is significantly genetically correlated with depression, results from our conditional GWAS analysis indicates that our top four hits are specific to self-reported childhood maltreatment, favoring a non-passive rGE mechanism for childhood maltreatment.
In addition to depression, we found significant positive genetic correlations between childhood maltreatment and “neuroticism” and “PGC cross-disorder analysis” (comprised of GWAS summary statistics of five psychiatric disorders: autism spectrum disorder, attention deficit-hyperactivity disorder, bipolar disorder, MDD, and schizophrenia). We observed negative genetic correlations of childhood maltreatment with “age of first birth” and “subjective well-being”. Associations between early life maltreatment and each of these phenotypes have previously been observed61,64–72. Further investigation is required to delineate the mechanisms that play a role in the relationship between childhood maltreatment and these outcomes.
Our study had a number of limitations that deserve emphasis. First, the genetic correlation between the UKBB and PGC1.5 datasets was only 0.63, indicating differences between the datasets, which possibly explains the non-replication of our top hit and of greater SNP heritability in PGC1.5. The UKBB dataset comprises healthy volunteers who are typically of a higher socioeconomic status and in better overall health than the general population of comparable age73, and the findings reported here may not be generalizable to the general population. However, it is also worth noting that the top hits were significant in the meta-analysis, where additional hits for childhood maltreatment were detected in an intergenic region on chromosome 12. Second, although many of the study sites included in the final meta-analysis utilized the well-validated CTQ, childhood maltreatment was measured in a diversity of ways across the different studies. Thus, our main phenotype was not homogenous and may reflect different aspects of childhood maltreatment in different contributing studies.
This is the first large-scale genetic study to identify specific variants associated with self-reported childhood maltreatment. Variation in FOXP genes and the polygenic architecture associated with childhood maltreatment may put individuals at greater risk for maltreatment. Alternatively, however, these variants may be associated with a greater likelihood of reporting maltreatment, given the high genetic correlation between childhood maltreatment and depression, and neuroticism. Using the available data, we are unable to indicate definitively which of these explanations is a better one, and it is possible that different mechanisms have more robust explanatory power in accounting for different abuse subtypes as well as different associated psychopathologies. A clearer understanding of the genetic relationships of childhood maltreatment, including particular abuse subtypes, with a range of different phenotypes, may ultimately be useful in developing targeted treatment and prevention strategies.
Disclaimer
The views, opinions, and/or findings contained in this report are those of the authors and should not be construed as official Department of the Army position, policy, or decision, unless so designated by other official documentation. Citations of commercial organizations or trade names in this report do not constitute an official Department of the Army endorsement or approval of the products or services of these organizations.
Supplementary information
Acknowledgements
This work was funded by Cohen Veterans Bioscience, the NIMH/U.S. Army Medical Research and Materiel Command Grant R01MH106595 to C.M.N., I.L., K.J.R. and K.C.K., One Mind, and supported by 5U01MH109539 to the Psychiatric Genomics Consortium. Statistical Analysis were carried out on the NL Genetic Cluster computer (URL) hosted by SURFsara. Genotyping of samples was supported in part through the Stanley Center for Psychiatric Genetics at the Broad Institute of MIT and Harvard. This research has been conducted using the UK biobank resource under application number 16577. This work would not have been possible without the contributions of the investigators who comprise the PGC-PTSD working group, and especially the more than 206,000 research participants worldwide who shared their life experiences and biological samples with PGC-PTSD investigators. Full acknowledgements for each study are in the Supplementary Note.
Data availability
The full meta-analysis summary statistics are available for download from the Psychiatric Genomics Consortium at https://www.med.unc.edu/pgc/results-and-downloads/. Access to individual-level data for available datasets may be requested through the PGC Data Access Portal at https://www.med.unc.edu/pgc/shared-methods/data-access-portal/. All other data that support the findings of this study are available from the corresponding author upon request.
Code availability
GWAS and associated scripts are available at https://github.com/nievergeltlab/ct_gwas/.
Conflict of interest
H.R.K. is a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative (ACTIVE), which in the last 3 years was supported by AbbVie, Alkermes, Amygdala Neurosciences, Arbor, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, and Pfizer. H.R.K. and J.G. are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. In the past 3 years, D.J.S. has received research grants and/or consultancy honoraria from Lundbeck and Sun. In the past 3 years, R.C.K. received support for his epidemiological studies from Sanofi Aventis; was a consultant for Johnson & Johnson Wellness and Prevention, Sage Pharmaceuticals, Shire, Takeda; and served on an advisory board for the Johnson & Johnson. Services Inc. Lake Nona Life Project. Kessler is a co-owner of DataStat, Inc., a market research firm that carries out healthcare research. M.B.S. has in the past 3 years been a consultant for Aptinyx, Bionomics, Dart Neuroscience, Janssen, Jazz Pharmaceuticals, Neurocrine Biosciences, Oxeia Biopharmaceuticals, and Pfizer. R.Y. is a co-inventor of the following patent application: “Genes associated with posttraumatic-stress disorder. European Patent# EP 2334816 B1.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-020-0706-0).
References
- 1.Child maltreatment. Global estimates. http://apps.who.int/gho/data/view.main.VIOLENCECHILDMALTREATMENTv (2016).
- 2.McLaughlin KA, et al. Childhood adversities and post-traumatic stress disorder: evidence for stress sensitisation in the World Mental Health Surveys. Br. J. Psychiatry. 2017;211:280–288. doi: 10.1192/bjp.bp.116.197640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copeland WE, et al. Association of childhood trauma exposure with adult psychiatric disorders and functional outcomes. JAMA Network Open. 2018;1:e184493–e184493. doi: 10.1001/jamanetworkopen.2018.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu A, McLaughlin KA, Misra S, Koenen KC. Childhood maltreatment and health impact: the examples of cardiovascular disease and type 2 diabetes mellitus in adults. Clin. Psychol. 2017;24:125–139. doi: 10.1111/cpsp.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suglia SF, et al. Childhood and adolescent adversity and cardiometabolic outcomes: a scientific statement from the American Heart Association. Circulation. 2018;137:e15–e28. doi: 10.1161/CIR.0000000000000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koenen, K. C., Roberts, A. L., Stone, D. M. & Dunn, E. C. in The Impact of Early Life Trauma on Health and Disease: The Hidden Epidemic, Vol. 1 (eds Lanius, R. A., Vermetten, E. & Pain, C.) Ch. 2 (Cambridge University Press, United Kingdom, 2010).
- 7.Schulz-Heik RJ, et al. Investigation of genetically mediated child effects on maltreatment. Behav. Genet. 2009;39:265–276. doi: 10.1007/s10519-009-9261-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knafo A, Jaffee SR. Gene–environment correlation in developmental psychopathology. Dev. Psychopathol. 2013;25:1–6. doi: 10.1017/S0954579412000855. [DOI] [PubMed] [Google Scholar]
- 9.Jaffee SR, Price TS. Gene–environment correlations: a review of the evidence and implications for prevention of mental illness. Mol. Psychiatry. 2007;12:432. doi: 10.1038/sj.mp.4001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutter M. Gene–environment interdependence. Dev. Sci. 2007;10:12–18. doi: 10.1111/j.1467-7687.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 11.Boivin M, et al. The genetic-environmental etiology of parents’ perceptions and self-assessed behaviours toward their 5-month-old infants in a large twin and singleton sample. J. Child Psychol. Psychiatry. 2005;46:612–630. doi: 10.1111/j.1469-7610.2004.00375.x. [DOI] [PubMed] [Google Scholar]
- 12.Berlin LJ, Dodge KA, Reznick JS. Examining pregnant women’s hostile attributions about infants as a predictor of offspring maltreatment. JAMA Pediatrics. 2013;167:549–553. doi: 10.1001/jamapediatrics.2013.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rehan W, et al. Gene–environment correlation between the dopamine transporter gene (DAT1) polymorphism and childhood experiences of abuse. J. Interpers. Violence. 2018;33:2059–2072. doi: 10.1177/0886260515622299. [DOI] [PubMed] [Google Scholar]
- 14.Munafo MR, Flint J. Dissecting the genetic architecture of human personality. Trends Cogn. Sci. 2011;15:395–400. doi: 10.1016/j.tics.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Duncan LE, et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol. Psychiatry. 2018;23:666–673. doi: 10.1038/mp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Merwe C, et al. Concordance of genetic variation that increases risk for anxiety disorders and posttraumatic stress disorders and that influences their underlying neurocircuitry. J. Affect. Disord. 2019;245:885–896. doi: 10.1016/j.jad.2018.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nievergelt CM, et al. International meta-analysis of PTSD genome-wide association studies identifies sex-and ancestry-specific genetic risk loci. Nat. Commun. 2019;10:1–16. doi: 10.1038/s41467-019-12576-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen NE, Sudlow C, Peakman T, Collins R. UK biobank data: come and get it. Sci. Transl. Med. 2014;6:224. doi: 10.1126/scitranslmed.3008601. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein DP, et al. Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abuse Negl. 2003;27:169–190. doi: 10.1016/s0145-2134(02)00541-0. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein, D. P. & Fink, L. Childhood Trauma Questionnaire: A Retrospective Self-report: Manual (The Psychological Corporation, Orlando, 1998).
- 21.Lam, M. et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics10.1093/bioinformatics/btz633 (2019). [DOI] [PMC free article] [PubMed]
- 22.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 24.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman, J. et al. Genome-wide gene-environment analyses of major depressive disorder and reported lifetime traumatic experiences in UK Biobank. Mol. Psychiatry10.1038/s41380-019-0546-6 (2020). [DOI] [PMC free article] [PubMed]
- 27.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter K, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren HR, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham G, Qiu Y, Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics. 2017;33:2776–2778. doi: 10.1093/bioinformatics/btx299. [DOI] [PubMed] [Google Scholar]
- 31.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou B, Shi J, Whittemore AS. Optimal methods for meta‐analysis of genome‐wide association studies. Genet. Epidemiol. 2011;35:581–591. doi: 10.1002/gepi.20603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarzer G. meta: an R package for meta-analysis. R News. 2007;7:40–45. [Google Scholar]
- 36.Watanabe K, Taskesen E, Van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulik-Sullivan BK, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Euesden J, Lewis CM, O’reilly PF. PRSice: polygenic risk score software. Bioinformatics. 2014;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Z, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripke S, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam EW-F, Brosens JJ, Gomes AR, Koo C-Y. Forkhead box proteins: tuning forks for transcriptional harmony. Nat. Rev. Cancer. 2013;13:482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 44.Atkinson EG, et al. No evidence for recent selection at FOXP2 among diverse human populations. Cell. 2018;174:1424–1435. e1415. doi: 10.1016/j.cell.2018.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bacon C, Rappold GA. The distinct and overlapping phenotypic spectra of FOXP1 and FOXP2 in cognitive disorders. Hum. Genet. 2012;131:1687–1698. doi: 10.1007/s00439-012-1193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siper PM, et al. Prospective investigation of FOXP1 syndrome. Mol. Autism. 2017;8:57. doi: 10.1186/s13229-017-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fisher SE, Scharff C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009;25:166–177. doi: 10.1016/j.tig.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Arnau-Soler A, et al. Genome-wide by environment interaction studies of depressive symptoms and psychosocial stress in UK Biobank and Generation Scotland. Transl. Psychiatry. 2019;9:14. doi: 10.1038/s41398-018-0360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demontis D, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clifton EA, et al. Genome-wide association study for risk taking propensity indicates shared pathways with body mass index. Commun. Biol. 2018;1:36. doi: 10.1038/s42003-018-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strawbridge RJ, et al. Genetics of self-reported risk-taking behaviour, trans-ethnic consistency and relevance to brain gene expression. Transl. Psychiatry. 2018;8:178. doi: 10.1038/s41398-018-0236-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui N, Liu J. Cognitive and behavioral risk factors for child physical abuse among Chinese children: a multiple-informant study. Child Adolesc. Psychiatry Ment. Health. 2016;10:36. doi: 10.1186/s13034-016-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernet CZ, Stein MB. Relationship of childhood maltreatment to the onset and course of major depression in adulthood. Depression Anxiety. 1999;9:169–174. [PubMed] [Google Scholar]
- 54.Stein MB, Jang KL, Taylor S, Vernon PA, Livesley WJ. Genetic and environmental influences on trauma exposure and posttraumatic stress disorder symptoms: a twin study. Am. J. Psychiatry. 2002;159:1675–1681. doi: 10.1176/appi.ajp.159.10.1675. [DOI] [PubMed] [Google Scholar]
- 55.Widom CS, Morris S. Accuracy of adult recollections of childhood victimization, part 2: childhood sexual abuse. Psychol. Assess. 1997;9:34–46. [Google Scholar]
- 56.Hardt J, Rutter M. Validity of adult retrospective reports of adverse childhood experiences: review of the evidence. J. Child Psychol. Psychiatry. 2004;45:260–273. doi: 10.1111/j.1469-7610.2004.00218.x. [DOI] [PubMed] [Google Scholar]
- 57.Baldwin JR, Reuben A, Newbury JB, Danese A. Agreement between prospective and retrospective measures of childhood maltreatment: a systematic review and meta-analysis. JAMA Psychiatry. 2019;76:584–593. doi: 10.1001/jamapsychiatry.2019.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newbury JB, et al. Measuring childhood maltreatment to predict early-adult psychopathology: comparison of prospective informant-reports and retrospective self-reports. J. Psychiatr. Res. 2018;96:57–64. doi: 10.1016/j.jpsychires.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kendler KS, Kessler RC, Neale MC, Heath AC, Eaves LJ. The prediction of major depression in women: toward an integrated etiologic model. Am. J. Psychiatry. 1993;150:1139–1139. doi: 10.1176/ajp.150.8.1139. [DOI] [PubMed] [Google Scholar]
- 60.Kendler KS, Karkowski-Shuman L. Stressful life events and genetic liability to major depression: genetic control of exposure to the environment? Psychol. Med. 1997;27:539–547. doi: 10.1017/s0033291797004716. [DOI] [PubMed] [Google Scholar]
- 61.Peterson RE, et al. Molecular genetic analysis subdivided by adversity exposure suggests etiologic heterogeneity in major depression. Am. J. Psychiatry. 2018;175:545–554. doi: 10.1176/appi.ajp.2017.17060621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mullins N, et al. Polygenic interactions with environmental adversity in the aetiology of major depressive disorder. Psychol. Med. 2016;46:759–770. doi: 10.1017/S0033291715002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Colman I, et al. Consistency in adult reporting of adverse childhood experiences. Psychol. Med. 2016;46:543–549. doi: 10.1017/S0033291715002032. [DOI] [PubMed] [Google Scholar]
- 64.Rosenman S, Rodgers B. Childhood adversity and adult personality. Aust. N. Z. J. Psychiatry. 2006;40:482–490. doi: 10.1080/j.1440-1614.2006.01826.x. [DOI] [PubMed] [Google Scholar]
- 65.Pesonen AK, et al. Reproductive traits following a parent–child separation trauma during childhood: a natural experiment during World War II. Am. J. Hum. Biol. 2008;20:345–351. doi: 10.1002/ajhb.20735. [DOI] [PubMed] [Google Scholar]
- 66.Matheson SL, Shepherd AM, Pinchbeck RM, Laurens KR, Carr VJ. Childhood adversity in schizophrenia: a systematic meta-analysis. Psychol. Med. 2013;43:225–238. doi: 10.1017/S0033291712000785. [DOI] [PubMed] [Google Scholar]
- 67.Palmier-Claus JE, Berry K, Bucci S, Mansell W, Varese F. Relationship between childhood adversity and bipolar affective disorder: systematic review and meta-analysis. Br. J. Psychiatry. 2016;209:454–459. [Google Scholar]
- 68.Kerns CM, Newschaffer CJ, Berkowitz S, Lee BK. Brief report: examining the association of autism and adverse childhood experiences in the National Survey of Children’s Health: the important role of income and co-occurring mental health conditions. J. Autism Dev. Disord. 2017;47:2275–2281. doi: 10.1007/s10803-017-3111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bjorkenstam E, Bjorkenstam C, Jablonska B, Kosidou K. Cumulative exposure to childhood adversity, and treated attention deficit/hyperactivity disorder: a cohort study of 543 650 adolescents and young adults in Sweden. Psychol. Med. 2018;48:498–507. doi: 10.1017/S0033291717001933. [DOI] [PubMed] [Google Scholar]
- 70.Corcoran M, McNulty M. Examining the role of attachment in the relationship between childhood adversity, psychological distress and subjective well-being. Child Abuse Negl. 2018;76:297–309. doi: 10.1016/j.chiabu.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 71.Mandelli L, Petrelli C, Serretti A. The role of specific early trauma in adult depression: a meta-analysis of published literature. Childhood trauma and adult depression. Eur. Psychiatry. 2015;30:665–680. doi: 10.1016/j.eurpsy.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 72.Li M, D’arcy C, Meng X. Maltreatment in childhood substantially increases the risk of adult depression and anxiety in prospective cohort studies: systematic review, meta-analysis, and proportional attributable fractions. Psychol. Med. 2016;46:717–730. doi: 10.1017/S0033291715002743. [DOI] [PubMed] [Google Scholar]
- 73.Fry A, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full meta-analysis summary statistics are available for download from the Psychiatric Genomics Consortium at https://www.med.unc.edu/pgc/results-and-downloads/. Access to individual-level data for available datasets may be requested through the PGC Data Access Portal at https://www.med.unc.edu/pgc/shared-methods/data-access-portal/. All other data that support the findings of this study are available from the corresponding author upon request.
GWAS and associated scripts are available at https://github.com/nievergeltlab/ct_gwas/.