Abstract
Treatment of community urinary tract infections (UTIs) caused by extended-spectrum β lactamase (ESBL)- producing Escherichia coli (E. coli) is more expensive than treating ESBL-negative opposites. Evaluation of the prevalence of ESBL-production among urinary E. coli isolates is crucial due to its great impact on the choice of proper antimicrobials. Accordingly, the aim of this work was to detect and characterize ESBL-producing E. coli isolated from outpatients with signs of UTIs in Upper Egypt. Urinary E. coli isolates were identified by 16S rRNA and their ESBL-production was confirmed by Modified Double Disc Synergy Test (MDDST) and ESBL- CHROMagar media. Isolates were then subjected to Polymerase Chain Reaction (PCR) for new Clermont phylogrouping, ESBL genes detection and CTX-M typing. The study enrolled 583 patients with clinically diagnosed UTIs. Uropathogens were found in 400 urine samples (68.6%) out of which 134 E. coli isolates were identified. Among the examined uropathogenic E. coli (UPEC), 80 (59.7%) were recognized as ESBL-producers. Greater than half of the ESBL-producers were multi-drug resistant (MDR) (62%). All of them were susceptible to meropenem. Most of the E. coli isolates were distributed in 4 phylogenetic groups: B2 = 42 (52.5%), F = 17 (21.25%) and Clade I or II = 10 (12.5%). The predominant gene types were TEM 60 (75%) and CTX-M gene 45 (56.25%). The CTX-M-1 group was the most prevalent (62.2%), including the CTX-M-15 enzyme, followed by the CTX-M-2 group, CTX-M-8 group and CTX-M-9 group. In conclusion, the results present alarming evidence of a serious spread of ESBL genes in Egypt, especially the epidemiological CTX-M 15, with the potential for the dissemination of MDR UPEC strains in the community.
Subject terms: Microbiology, Medical research
Introduction
Extended-spectrum β-lactamases (ESBLs) are plasmid-mediated β-lactamases recognized for their ability to hydrolyze 3rd- and 4th-generation cephalosporins (oxyimino-cephalosporin) and monobactams but not cephamycin or carbapenems. Additionally, these enzymes are repressed by β-lactamase inhibitors as clavulanic acid and tazobactam. Typically, an ESBL evolved from a narrow spectrum parent, for example from blaTEM-1, blaTEM-2, or blaSHV1. Recently, a new class of ESBL genes called blaCTX-M have appeared and posed a great burden on the health environment2. The worldwide dissemination of blaCTX-M producing E. coli has been increasing, and are now known to be the main ESBL genes3. The first analysis and alignment of the amino acid sequences of the CTX-M variants categorized these enzymes into five clusters: CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-254. Although different informs about CTX-M β-lactamases have been available, updated data about dispersion of CTX-M producing isolates, molecular epidemiology, protein plasticity, evolution and origin of the bla genes, influence of antibiotic use, and patients risk factors are still lacking in some regions5.
The spread of ESBLs in Enterobacteriaceae has become an ever-increasing problem6, with a global rise of ESBL-producing Enterobacteriaceae7. One of the most frequently found Enterobacteriaceae harboring ESBL genes is Escherichia coli (E. coli), where multi-drug resistance (MRD) due to ESBL production is rapidly becoming a threat to the community8. Spreading rates of nosocomial ESBL producing E. coli are markedly variable, with flat trends in Europe ~15%, and increasing trends in North America from 7.8% in 2010 to 18.3% in 20149. In fact, propagation of MDR and ESBL-producing E. coli strains reduces the treatment preferences. It is also mandatory to be informed with the predominant resistant pattern of any region, which could assist in proper antimicrobial therapy10.
A recent study done by our group11, showed a disturbingly high level of ESBL producers in the urine of asymptomatic pregnant women. Thus, we aimed at evaluating their incidence in patients suffering from UTIs in the community. To the best of our knowledge, there are no previous epidemiological data regarding the phylogenetic grouping of ESBL-producing E. coli causing UTIs in Egypt. Accordingly, a phenotypic and a genotypic evaluation of ESBL- producing E. coli was carried out, followed by phylogenetic grouping of the obtained isolates.
Results
Isolation of E coli from urine samples
Out of 583 urine samples obtained from patients suffering from UTIs, 400 isolates were confirmed as positive cultures. 134 of these cultures were confirmed to be E. coli (33.5%) (Figure S1).
Phenotypic detection for esbl production
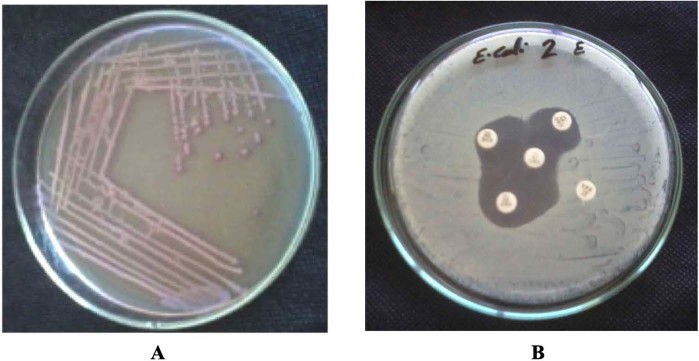
Out of 134 E. coli isolates, 80 (∼60%) were ESBL producers by CHROMagar ESBL screening and 75 (56%) using Modified Double Disc Synergy Test (MDDST) (Fig. 1). There was no statistical significance between CHOROMagar and MDDST regarding detection of ESBL-production (P > 0.5; X² = 0.245). Different patterns of synergism of 3rd generation cephalosporin and 4th generation cephalosporin with Amoxicillin Clavulanate using MDDST were observed on testing 80 CHROMAgar ESBL producing E. coli isolates (Table 1).
Figure 1.
(A) E. coli on chromogenic agar. (B) Detection of ESBL production tested by the Modified Double Disc Synergy Test (MDDST). The test was done by using a disc of amoxicillin-clavulanate (20/10 μg) along with four cephalosporins; cefotaxime, ceftriaxone, cefpodoxime and cefepime.
Table 1.
MDDST for 80 possible ESBL producing E. coli isolates:.
| Pattern of synergism | Number | Percentage (%) | Cause of resistance |
|---|---|---|---|
| No synergism of 3GC and 4GC with AMC | 5 | 6.25% | Non-ESBLs |
| Synergism of both 3GC and 4GC with AMC | 75 | 93.75% | ESBLs |
| Synergism of 3GC only with AMC | 0 | 0% | ESBLs |
Antibiotic susceptibility pattern of ESBL producing E. coli isolates
The overall antimicrobial resistance in ESBL producing E. coli isolates is summarized in Table 2. The rates of antibiotic resistant E. coli were 100% for penicillin group (ampicillin (AMP), piperacillin (PRL), ofloxacin (OFX)), cephems (cefotaxime (CTX), ceftazidime (CAZ), Cefpodoxime (CPD), cefepime (FEP)) and monobactams (aztreonam (ATM)), 82.5% for ampicillin/sulbactam (AMC), 76.25 for sulphamethoxazole-trimethoprim (SXT), 28.75% for Chloramphenicol (C), 23.75% for gentamicin (CN), 13.75% for ciprofloxacin (CIP), and 0% for meropenem (MEM). Most of isolates (>80%) were susceptible to ciprofloxacin, while all the isolates were susceptible to meropenem. About 62% of the isolates were MDR with resistance to one antimicrobial agent in at least 3 different groups.
Table 2.
Antibiotic susceptibility pattern of ESBL producing E. coli.
| Sensitivity of isolates | Antimicrobial | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPD | ATM | CIP | AMP | CAZ | AMC | MEM | CN | CTX | OFX | C | PRL | SXT | FEP | |
| R | 95% | 71.25% | 13.75% | 90% | 75% | 82.5% | 0% | 23.75% | 97.5% | 17.5% | 28.75% | 47.5% | 76.25% | 50% |
| I | 3.75% | 11.25% | 0% | 2.5% | 11.25% | 5% | 0% | 2.5% | 0% | 0% | 6.25% | 32.5% | 5% | 20% |
| S | 1.25% | 17.5% | 86.25% | 7.5% | 13.75% | 12.5% | 100% | 73.75% | 2.5% | 82.5% | 65% | 20% | 18.75% | 30% |
R = resistant, I = Intermediate, S = sensitive.
Phylo-Grouping profile of isolates
Phylogenetic grouping was done according to modified Clermont method12. Forty-two isolates (52.5%) were classified into B2 group, 17 isolates (21.25%) in Group F, 10 isolates (12.5%) in Clade I or II, while 11 isolates (13.75%) did not belong to any group (Fig. 2) (Table 3).
Figure 2.
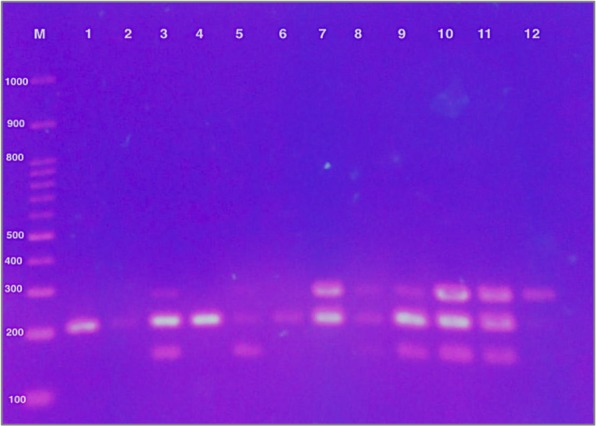
Quadruplex PCR profiles of Clermont phylo-typing method of E. coli isolates. Lane M contained 100 bp marker; Lane 3, 7, 8, 9,10 and 11, group B2; Lane 12, group F; Lane 1,2, 4 and 6, Clade I or II; Lane 5, unknown.
Table 3.
Phylogenetic analysis of 80 E. coli isolates (Positive for ESBL genes) Causing urinary tract infection derived from various hospitals in Egypt.
| Phylogenetic groups | Genes Combination Profile | Number of isolates | (%) of isolates |
|---|---|---|---|
| A | (arpA+, chuA-, yjaA-, TspE4.C2-) | 0 | 0% |
| B1 | (arpA+, chuA-, yjaA-, TspE4.C2+) | 0 | 0% |
| F | (arpA-, chuA+, yjaA-, TspE4.C2-) | 17 | 21.25% |
| B2 | (arpA-, chuA+, yjaA+, TspE4.C2-) | 10 | B2 = 42 (52.5%) |
| B2 | (arpA-, chuA+, yjaA+,TspE4.C2+) | 32 | |
| B2 | (arpA-, chuA+, yjaA-,TspE4.C2+) | 0 | |
| A or C | (arpA+, chuA-, yjaA+,TspE4.C2-) | 0 | 0 |
| D or E | (arpA+, chuA+, yjaA-,TspE4.C2-) | 0 | 0 |
| D or E | (arpA+, chuA+, yjaA-,TspE4.C2+) | 0 | 0 |
| E or clade I | (arpA+, chuA+, yjaA+, TspE4.C2-) | 0 | 0 |
| Clade I or II | (arpA-, chuA-, yjaA+, TspE4.C2-) | 10 | 12.5% |
| Clade III, IV or V | (arpA-, chuA+(476)C, yjaA-, TspE4.C2-) | 0 | 0 |
| Unknown | (arpA-, chuA-, yjaA-,TspE4.C2+) | 4 | Unknown = 11 (13.75%) |
| Unknown | (arpA-, chuA-, yjaA+,TspE4.C2+) | 3 | |
| Unknown | (arpA+, chuA-, yjaA+, TspE4.C2+) | 0 | |
| Unknown | (arpA+, chuA+, yjaA+, TspE4.C2+) | 0 | |
| Unknown | (arpA-, chuA-, yjaA-, TspE4.C2-) | 4 |
Molecular detection of ESBL genes
The predominant ESBL gene in this study was blaTEM, which was found in 60 isolates (75%) (Figure S2). The blaCTX-M gene was found in 45 isolates (56.25%), whereas blaSHV was found in only 15 isolates (18.75%) (Figure S3 & S4). Most of the isolates (66.25%) showed coexistences of more than one gene, with (28.75%) of ESBL-producing E. coli harboring blaTEM, blaSHV, and blaCTX−M. Coexistences of two genes was also observed where blaTEM and blaCTX−M were detected in (21.25%) of the isolates, blaTEM and blaSHV in (6.25%) of the isolates, while blaSHV and blaCTX−M were found in (10%) of the isolates. About 25% of the isolates harbored blaTEM alone, and 13.75% had blaCTX-M alone. None of the isolates had blaSHV alone. Distribution of tested ESBL genes among different groups is found in Table 4.
Table 4.
Frequency of the different ESBL types among ESBL producing E. coli isolates of the different phylogenic types.
| ESBL-gene | blaTEM | blaSHV | blaCTX-M | blaCTX-M Sub-types | ||||
|---|---|---|---|---|---|---|---|---|
| CTX-M-1 (n = 22) | CTX-M-2 (n = 12) | CTX-M-8 (n = 10) | CTX-M-9 (n = 1) | CTX-M-25 (n = 0) | ||||
| Phylo-group | ||||||||
| B2 | 30 | 9 | 37 | 17 | 12 | 7 | 1 | 0 |
| F | 6 | 1 | 2 | 2 | 0 | 0 | 0 | 0 |
| Clade I/II | 15 | 3 | 1 | 1 | 0 | 0 | 0 | 0 |
| unknown | 9 | 2 | 5 | 2 | 0 | 3 | 0 | 0 |
| Total of ESBL isolates | 60 | 15 | 45 | 22 | 12 | 10 | 1 | 0 |
Characterization of CTX-M-producing E. coli clinical isolates
Out of the 45 blaCTX-M isolates, Group 1 enzymes were found in 22 isolates (48.9%) (Figure S5 & S6), while CTX-M 15 enzymes were found in 11 isolates (24.4%) (Figure S7). Group 2 enzymes were produced by 12 (26.7%) isolates. The rest of the isolates included: 10 isolates that produced group 8 CTX-M enzymes and one isolate that produced group 9 enzyme. On the other hand, 3 E. coli isolates harbored both group 1 and group 8 enzymes. There were no producers of group 25 CTX-M enzymes detected in this study.
Discussion
Investigating the prevalence of antimicrobial resistance rates is of great importance in both creating strategies for empirical treatment and in evaluating the existing guidelines. The frequencies and types of infections caused by ESBL-producing Enterobacteriaceae have increased dramatically in the past few decades with disparity between different institutions and countries. Since the beginning of the new millennium, E. coli has become the most commonly isolated ESBL-producing bacteria worldwide with CTX-M ESBLs being the most frequently isolated types13. This upsurge in ESBL-producing E. coli adds a great burden to the treatment of community-onset UTIs as such isolates are frequently multidrug-resistant, with increased chance of treatment failure14–16. Since there is no comprehensive surveillance of community-acquired UTIs caused by E. coli in Egypt, this study was aimed at evaluating the prevalence and the mechanisms underlying their ESBL production. To the best of our knowledge, this is the first article from Upper Egypt to report the characteristics of ESBL-producing E. coli from community-onset UTIs.
Out of a total of 134 clinical isolates of E. coli, 80 isolates (59.7%) were ESBL positive. This high frequency of ESBLs is comparable to those found in Egypt by Al-Agamy et al. and Abdel-Moaty et al.17, where 52% of the detected isolates were ESBL producers, however, their study did not demonstrate whether the isolates were community or hospital-acquired. On the other-hand, our reported incidence is much higher than that reported earlier in Egypt by Fam et al., with a lower prevalence of 17% among community-acquired UTIs18, suggesting an increasing trend in the incidence of ESBLs-producing E. coli in Egypt. Compared to countries in the same region, our studies were also higher than that found in Lybia (6.7%)19 and Emirates (39%)20. However, comparable results (67%) were obtained by Zorgani et al. (in a different larger-scale study done in Lybia), who reported a high incidence of ESBL-producers among hospital isolates21.
Within different ESBLs, CTX-M enzymes are the most predominant in different epidemiological settings, which have outnumbered other ESBL enzymes such as TEM and SHV22,23, with more than 172 CTX-M variants reported to date. Al-Agamy et al. reported the first detection of CTX-M β-lactamase production by urinary nosocomial E. coli isolates in Egypt. They found a high incidence of ESBLs-producing isolates in a single hospital (60.9%)24. In this study the predominant gene types were blaTEM in 60 isolates (75%) and bla CTX-M in 45 isolates (56.25%), while blaSHV was found in only 15 isolates (18.75%). The relatively higher frequency of blaCTX-M among our community isolates is concurring with the fact that CTX-M ESBLs originate from environmental bacteria unlike TEM- or SHV-ESBLs25. These findings are in agreement with previous studies done in Egypt where blaCTX-M was prevailing24,26.
Interestingly, blaTEM was detected in most isolates although it is commonly found in hospital strains, this could probably be due to previous contact with hospitals.
Among blaCTX-M, blaCTX-M-15 was the most prevalent (37.8%) in our study, which concurs with various reports demonstrating the extensive worldwide dissemination of blaCTX-M-15 mediated by clonally related E. coli strains27.
Regarding the susceptibility profile of ESBLs-producing isolates; all of the isolates were resistant to cephems, which is concurring with previous studies28,29.
Our detected resistance rates with ampicillin/sulbactam (AMC) (82.5%) and sulphamethoxazole-trimethoprim (SXT) (76.25%) were higher than that previously reported26,30,31 but in agreement with Abdel-Moaty et al.17.
On the other hand, about 13% of the isolates were resistant to ciprofloxacin using CLSI disk breakpoints32, which is much lower than previously reported in Egypt by Abdel-Moaty et al.17 or in the Middle East region19,33. This decreased resistance rate could be due to a better understanding and a wiser use of fluoroquinolones in UTI cases.
In addition, all the isolates were susceptible to meropenem, which is in consensus with a previous study done in the same region on asymptomatic urinary carriers of ESBL-producer strains11, who reported that all ESBL producers were sensitive to imipenem (100%).
Alteration in the phylogenetic types are important in identifying novel groups of emerging bacteria that are better recognized as a result of this analysis. Phylogenetic grouping in this study was done according to a modified Clermont method12, which was done for the first time on the Egyptian isolates. This new method used modified primers for chuA, yjaA and TspE4.C2, which eradicated some primer mismatches. The most imperative benefit of this method was its power to distinguish strains belonging to phylogroups C, E, F and clade I. In this study most of the isolates were in group B2 (52.5%), followed by group F (21.25%), Clade I or II (12.5%), while 13.75% were of unknown type. The high frequency of phylogenetic group of B2 (52, 5%) is comparable to previous studies34–36, where the B2 subgroup was the most common group especially among CTX-M15 strains, as well as phylogroup F, which is closely similar to phylogroup B237.
The presence of Clade I or II isolates (cryptic lineages which are phenotypically similar to E. coli but genetically divergent38,39) require further investigation as this is the first report for the presence of such clades among extra intestinal isolates in Egypt. Interestingly, cryptic isolates found in this study harbor a variety of ESBLs: TEM, SHV and CTX-M, suggesting a threatening horizontal gene transfer in our community. Environmental spread of ESBL-producing E. coli could be attributed to the release of wastewater into rivers40,41, where mobile genetic elements are allowed to transfer ESBL-production from environmental bacteria to human and animals42.
In conclusion, our data underscores the importance of continuous surveillance of antimicrobial resistance in community E. coli isolates and shows the alarming increases of ESBL-production among such strains. Public health efforts should focus on the correct use of antibiotics to limit their dissemination and further investigation of molecular epidemiology of ESBLs in various clinical samples would be promising to obtain a better database for ESBL-producing E. coli in Egypt.
Methods
Study design
This cross-sectional study was conducted to assess the prevalence and antimicrobial resistance pattern of ESBL-producing E. coli isolates from patients with suspected community-onset UTIs during the period of August 2016 to February 2018 from Minia General Hospital, Kidney Hospital, Suzan Mubarak Hospital and Liver Virus Unit in Minia Governorate (located in Upper Egypt). Community-onset infections were defined as infections that have an onset in less than 48 hours of hospital admission or that present in the outpatients’43. Study recruits in this work were patients ≥18 years with symptoms of suspected urinary tract infections, attending the outpatient’s clinics or admitted to the inpatient’s wards (within 48 hours of admission). Written informed consent was obtained from the patients prior to data collection. The methods were carried out in accordance with the relevant guidelines and regulations. All experimental protocols were approved by the Ethics Committee of the Faculty of Science, Beni-Suef University. Patients with history of antibiotics intake within the last 2 weeks were excluded.
Sample size
Before the study, the number of required patients was determined after a power calculation according to data obtained in a previous study carried in Assiut, Egypt44. In that study the frequency of ESBL-producing E. coli was about 6.8%. A sample size of 80 patients in the group was determined to provide 80% power and 5% type I error with precision of 5.5% using the following equation:
Z1−α/2 = Is standard normal variate (at 5% type 1 error (P < 0.05) it is 1.96.
P = Expected proportion in population. d = Absolute error or precision.
Isolation of E. coli from urine samples
A total of 583 midstream urine samples were collected by giving a sterile, dry, wide necked plastic container to every patient and were transported to the laboratory for processing within 2–4 h of collection. Positive cultures were identified by detection of at least 105 CFU/ml45, after inoculation of a 10 μl (0.01 ml) of the urine sample into MacConkey agar (Oxoid, UK) and its incubation at 37 °C for 24 hours. E. coli identification was further done by inoculation on Eosin Methylene Blue (EMB) Agar plate and aerobic incubation at 37 °C for 24 hours. Gram staining of suspected colonies was performed (Gram negative bacilli). Citrate Utilization test was used to differentiate E. coli from other lactose-fermenters (Only negative with E. coli). E. coli strains were further confirmed by complete 16S rRNA detection by uniplex PCR.
Phenotypic detection of ESBL production
All E. coli isolates were initially screened for ESBL production by inoculation on CHROMagar ESBL (CHROMagar, F-75006, Paris, France). All E. coli isolates, which showed dark pink to reddish color colonies on CHROMagar ESBL media were selected for further confirmation by the Modified Double Disc Synergy Test (MDDST)46. Briefly, isolates were inoculated on a plate containing a disc of amoxicillin-clavulanate (20/10 μg) at the center along with three 3rd generation cephalosporins; cefotaxime, ceftriaxone, cefpodoxime and a 4th generation cephalosporin; cefepime placed at 15 mm and 20 mm, respectively, centre to centre to that of the amoxicillin-clavulanate disc47. Any increase in the zone towards the disc of amoxicillin-clavulanate was considered as positive for ESBL production.
Antibiotic susceptibility pattern of E. coli isolates
Antimicrobial susceptibility testing for phenotypically confirmed ESBL-isolates was determined by the disk diffusion method with reference to the standards of the Clinical and Laboratory Standards Institute32. The quality of antibiotic sensitivity was confirmed by using E. coli ATCC 25922 as a reference strain. Testing of antimicrobial susceptibilities of the following antibiotics was carried out: ampicillin (AMP 10 μg), piperacillin (PRL 100 μg), ofloxacin (OFX 5 μg), cefotaxime (CTX 30 μg), ceftazidime (CAZ 30 μg), cefpodoxime (CPD 10 μg), cefepime (FEP 30 μg), aztreonam (ATM), ampicillin/sulbactam (AMC 10/10 μg), sulphamethoxazole-trimethoprim (SXT 1.25/23.75 μg), chloramphenicol (C), gentamicin (CN 10 μg), ciprofloxacin (CIP 5 μg) and meropenem (MEM 10 μg) (Oxoid, UK). Multidrug resistance (MDR) was identified as having resistance to three or more classes of antibiotics.
DNA extraction
Crude genomic bacterial DNA from all isolates with positive ESBL-screening results was extracted and purified using DNA extraction kits (Thermo Scientific, Gene JET Genomic, DNA Purification Kit, USA), according to manufacturer’s instructions.
Phylogenetic grouping by quadruplex PCR
A quadruplex polymerase chain reaction (PCR)12 modified from the original triplex PCR method by Clermont and colleagues48 was used to group the E. coli isolates phylogenetically. This method was used to allocate the ESBL-producing E. coli isolates based on the presence or absence of 4 genes (arpA (400 bp), chuA (288 bp), yjaA (211 bp), TspE4.C2 (152 bp)) and allocating E. coli isolates into 1 of 8 phylo-groups (A, B1, B2, C, D, E, F and cryptic clade).
Multiplex PCR was carried out in a 25 μL reaction mixture, including 12.5 μl of MyTaq Red Mix (Bioline, USA Inc.), 1 μL (10 μM) each primer, 2.5 μL (nuclease free water), and 2 μL template DNA. Amplification was carried out as follows: initial denaturation at 94 C° (4 min), 30 cycles of denaturation at 94 °C (5 sec), annealing at 59 °C (10 sec) and elongation at 72 °C (10 sec), followed by final extension at 72 °C (5 min). The PCR products were analyzed electrophoretically by running the PCR product through 1% (w/v) agarose in Tris-borate-EDTA (TBE) at 90 V for 35 min and visualization under UV transillumination.
PCR amplification for 16s rrna, uniplex PCR for blatem, and duplex PCR for blashv and blactx-m amplification
Quality control of the DNA extraction was carried out by testing all extracted isolates for 16S rRNA by uniplex PCR49. This was followed by PCR detection of ESBL genes (blaTEM, blaSHV, blaCTX-M genes) and subgrouping for blaCTX-M genes into the 5 major groups (CTX-M-1(including CTX-M-15), CTX-M-2, CTX-M-8, CTX-M-9 and CTX-M-25), using primer pairs listed in Table 5 and amplification conditions as described previously49–52. The amplification reactions were carried out in 25 μl volumes containing 12.5 μl of MyTaq Red Mix (Bioline, USA Inc.), 1 μl of each primer (10 pmol/μl) and 2 μl of 100 ng/μl chromosomal DNA.
Table 5.
Primers used in this work.
| Gene | Primer (5′-3′) | size (bp) | ref. |
|---|---|---|---|
| 16s rRNA |
F-AGT TTG ATC MTG GCT CAG R-GGA CTA CHA GGG TAT CTA AT |
797 | 49 |
| blaTEM |
F-ATG AGT ATT CAA CAT TTC CG R-CCA ATG CTT AAT CAG TGA GG |
858 | 50 |
| blaSHV |
F-ATG CGT TAT ATT CGC CTG TG R-AGC GTT GCC AGT GCT CGA TC |
862 | 50 |
| Universal blaCTX-M |
F-5′-SCS ATG TGC AGY ACC AGT AA R-5′-CCG CRA TAT GRT TGG TGG TG |
554 | 51,52 |
| blaCTX-M-1 |
F-5′-AAA AAT CAC TGC GCC AGT TC R-5′-AGC TTA TTC ATC GCC ACG TT |
415 | 51,52 |
| blaCTX-M-2 |
F-5′-CGA CGC TAC CCC TGC TAT T R-5′-CCA GCG TCA GAT TTT TCA GG |
552 | 51,52 |
| blaCTX-M-8 |
F- 5′-TCG CGT TAA GCG GAT GATGC R-5′-AAC CCA CGA TGT GGG TAG C |
666 | 51,52 |
| blaCTX-M-9 |
F-5′-CAA AGA GAG TGC AAC GGA R-TG 5′-ATT GGA GGT TCA TCA CC |
205 | 51,52 |
| blaCTX-M-25 |
F-5′-GCA CGA TGA CAT TCG GG R-5′-AAC CCA CGA TGT GGG TAG C |
327 | 51,52 |
| blaCTX-M-15 |
F-5′-ATAAAACCGGCAGCGGTG R-5′-GAATTTGACGATCGGGG |
500 | 51,52 |
| chuA |
F-5′- GACGAACCAACGGTCAGGAT R-5′- TGCCGCCAGTACCAAAGACA |
279 | 12 |
| yjaA |
F-5′- TGAAGTGTCAGGAGACGCTG R-5′- ATGGAGAATGCGTTCCTCAAC |
211 | 12 |
| TspE4C2 |
F-5′- GAGTAATGTCGGGGCATTCA R-5′- CGCGCCAACAAAGTATTACG |
152 | 12 |
| arpA |
5′-AACGCTATTCGCCAGCTTGC-3′ 5′-TCTCCCCATACCGTACGCTA-3′ |
400 | 12 |
Statistical analysis
Analyses of data were performed by SPSS software (version 23) and proportions were compared using the Chi-square test (χ2 test) to determine the significant differences in resistance. Differences were considered significant at P < 0.05. Graphics were performed using Excel 2010. For description statistics, data are presented as mean standard deviation for continuous variables, as well as frequency and percentage for categorical variables.
Data availability
Supplementary files are added.
Supplementary information
Author contributions
N.A.H. and A.S.K. conceived and designed the study. N.A.H., E.M.F. carried out the experiments and collected the data. N.A.H., E.M.F. and A.S.K. wrote the manuscript. N.A.H., A.S.K., A.M.H. and M.A.F. critically revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-59772-z.
References
- 1.Rupp ME, Fey PD. Extended spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Drugs. 2003;63:353–365. doi: 10.2165/00003495-200363040-00002. [DOI] [PubMed] [Google Scholar]
- 2.Livermore DM, et al. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 2007;59:165–174. doi: 10.1093/jac/dkl483. [DOI] [PubMed] [Google Scholar]
- 3.Pitout JD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 2008;8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naseer U, Sundsfjord A. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug. Resist. 2011;17:83–97. doi: 10.1089/mdr.2010.0132. [DOI] [PubMed] [Google Scholar]
- 6.Copăcianu B, Tuchiluş C, Poiata A, Iancu L. Research regarding extended-spectrum beta-lactamases produced by enterobacteria strains. Rev. medico-chirurgicala a Societatii de. Medici si Naturalisti din. Iasi. 2010;114:896–899. [PubMed] [Google Scholar]
- 7.Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017;72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 8.Calbo E, et al. Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2006;57:780–783. doi: 10.1093/jac/dkl035. [DOI] [PubMed] [Google Scholar]
- 9.Lob SH, Biedenbach DJ, Badal RE, Kazmierczak KM, Sahm DF. Antimicrobial resistance and resistance mechanisms of Enterobacteriaceae in ICU and non-ICU wards in Europe and North America: SMART 2011–2013. J. Glob. Antimicrob. Resist. 2015;3:190–197. doi: 10.1016/j.jgar.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Jena J, Sahoo RK, Debata NK, Subudhi E. Prevalence of TEM, SHV, and CTX-M genes of extended-spectrum β-lactamase-producing Escherichia coli strains isolated from urinary tract infections in adults. Biotech. 2017;7:244. doi: 10.1007/s13205-017-0879-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Youssef MM, Rizk HA, Hassuna NA. Phenotypic and Genotypic Characterization of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae in Asymptomatic Bacteriuria in Pregnancy. Microb. Drug. Resist. 2019;25:731–738. doi: 10.1089/mdr.2018.0088. [DOI] [PubMed] [Google Scholar]
- 12.Clermont O, Christenson JK, Denamur E, Gordon DM. The C lermont E scherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environm Microbiol. Rep. 2013;5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 13.Chong Y, Shimoda S, Shimono N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Gen. Evol. 2018;61:185–188. doi: 10.1016/j.meegid.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Organization., W. H. Antimicrobial resistance e global report on surveillance, </bitstream/10665/112642/1/9789241564748_eng.pdf?uaZ1 (2014).
- 15.Esteve-Palau E, et al. Clinical and economic impact of urinary tract infections caused by ESBL-producing Escherichia coli requiring hospitalization: a matched cohort study. J. Infect. 2015;71:667–674. doi: 10.1016/j.jinf.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Lee, D. S., Lee, S.-J. & Choe, H.-S. Community-Acquired Urinary Tract Infection by Escherichia coli in the Era of Antibiotic Resistance. BioMed Res Inter. 2018 (2018). [DOI] [PMC free article] [PubMed]
- 17.Abdel-Moaty MM, Mohamed WS, Abdel-All SM, El-Hendawy HH. Prevalence and molecular epidemiology of extended spectrum β-lactamase producing Escherichia coli from hospital and community settings in Egypt. JAPS. 2016;6:042–047. doi: 10.7324/JAPS.2016.600107. [DOI] [Google Scholar]
- 18.Fam N, et al. CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb. Drug. Resist. 2011;17:67–73. doi: 10.1089/mdr.2010.0063. [DOI] [PubMed] [Google Scholar]
- 19.Abujnah AA, et al. Multidrug resistance and extended-spectrum β-lactamases genes among Escherichia coli from patients with urinary tract infections in Northwestern Libya. Libyan J. Med. 2015;10:26412–26412. doi: 10.3402/ljm.v10.26412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Zarouni M, Senok A, Rashid F, Al-Jesmi SM, Panigrahi D. Prevalence and Antimicrobial Susceptibility Pattern of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in the United Arab Emirates. MPP. 2008;17:32–36. doi: 10.1159/000109587. [DOI] [PubMed] [Google Scholar]
- 21.Zorgani A, Almagatef A, Sufya N, Bashein A, Tubbal A. Detection of CTX-M-15 among uropathogenic Escherichia coli isolated from five major hospitals in Tripoli, Libya. OMJ. 2017;32:322. doi: 10.5001/omj.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D’Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 2013;303:305. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Bush, K. Past and Present Perspectives on beta-Lactamases. Antimicrob Agents Chemother. 62, 10.1128/AAC.01076-18 (2018). [DOI] [PMC free article] [PubMed]
- 24.Al-Agamy MHM, Ashour MSE-D, Wiegand I. First description of CTX-M β-lactamase-producing clinical Escherichia coli isolates from Egypt. Inter. J. Antimicrob. Agents. 2006;27:545–548. doi: 10.1016/j.ijantimicag.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Chong Y, Ito Y, Kamimura T. Genetic evolution and clinical impact in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Gen. Evol. 2011;11:1499–1504. doi: 10.1016/j.meegid.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Fam NS, El-Damarawy M. CTX-M15 extended-spectrum-β-lactamase detected from intensive care unit of an Egyptian medical research institute. Res. J. Med. Sci. 2008;3:84–91. [Google Scholar]
- 27.Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: origin and diffusion. Front. Microbiol. 2012;3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melzer M, Petersen I. Mortality following bacteraemic infection caused by extended spectrum beta-lactamase (ESBL) producing E. coli compared to non-ESBL producing E. coli. J. Infect. 2007;55:254–259. doi: 10.1016/j.jinf.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Alfola, M. M. H. R., Kamel, Z., Nada, M. G. E. D., Rashed, L. A. & El-Awady, B. A. Phenotypic and Genotypic Characterization of ESBL-Producing Escherichia coli and Klebsiella pneumonia isolates from Patient’s Urine specimens. IAJAA. 6 (2017).
- 30.Algan O, et al. Holocene coastal change in the ancient harbor of Yenikapı–İstanbul and its impact on cultural history. Quat. Res. 2011;76:3. doi: 10.1016/j.yqres.2011.04.002. [DOI] [Google Scholar]
- 31.Meier S, Weber R, Zbinden R, Ruef C, Hasse B. Extended-spectrum β-lactamase-producing Gram-negative pathogens in community-acquired urinary tract infections: an increasing challenge for antimicrobial therapy. Infect. 2011;39:333–34. doi: 10.1007/s15010-011-0132-6. [DOI] [PubMed] [Google Scholar]
- 32.Institute, C. L. a. S. In Performance standards for antimicrobial disk susceptibility tests. M02 standard, Clinical Laboratory and Standards Institute (Wayne, 2018).
- 33.Mnif B, et al. Molecular epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli in Tunisia and characterization of their virulence factors and plasmid addiction systems. BMC Microbiol. 2013;13:147. doi: 10.1186/1471-2180-13-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iranpour, D. et al. Phylogenetic groups of Escherichia coli strains from patients with urinary tract infection in Iran based on the new Clermont phylotyping method. BioMed Res Inter. 2015 (2015). [DOI] [PMC free article] [PubMed]
- 35.Dureja C, Mahajan S, Raychaudhuri S. Phylogenetic distribution and prevalence of genes encoding class I integrons and CTX-M-15 extended-spectrum β-lactamases in Escherichia coli isolates from healthy humans in Chandigarh, India. PLoS One. 2014;9:e112551. doi: 10.1371/journal.pone.0112551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonçalves LF, et al. Multidrug resistance dissemination by extended-spectrum β-lactamase-producing Escherichia coli causing community-acquired urinary tract infection in the Central-Western Region, Brazil. J. Glob. Antimicrob. Resist. 2016;6:1–4. doi: 10.1016/j.jgar.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Jaureguy F, et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Gen. 2008;9:560. doi: 10.1186/1471-2164-9-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walk ST, et al. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 2009;75:6534–6544. doi: 10.1128/AEM.01262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walk, S. The “Cryptic” Escherichia, 10.1128/EcoSal Plus.ESP-0002–2015 (2015). [DOI] [PubMed]
- 40.Bréchet C, et al. Wastewater treatment plants release large amounts of extended-spectrum β-lactamase–producing Escherichia coli into the environment. Clin. Infect. Dis. 2014;58:1658–1665. doi: 10.1093/cid/ciu190. [DOI] [PubMed] [Google Scholar]
- 41.Hawkey PJ. Multidrug-resistant Gram-negative bacteria: a product of globalization. J. Hosp. Infect. 2015;89:241–247. doi: 10.1016/j.jhin.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Pehrsson EC, et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman ND, et al. Health care–associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann. Int. Med. 2002;137:791–797. doi: 10.7326/0003-4819-137-10-200211190-00007. [DOI] [PubMed] [Google Scholar]
- 44.Salah M, Azab M, Halaby H, Hanora A. Mutations in β-lactamases detected in multidrug resistant gram negative bacteria isolated from community acquired urinary tract infections in Assiut, Egypt. Afr. J. Microbiol. Res. 2016;10:1938–1943. doi: 10.5897/AJMR2016.8150. [DOI] [Google Scholar]
- 45.CDC/HICPAC. Urinary Tract Infection (UTI) Event for Long-Term Care Facilities. (2009).
- 46.Kaur J, Chopra S, Sheevani GM. Modified double disc synergy test to detect ESBL production in urinary isolates of Escherichia coli and Klebsiella pneumoniae. JCDR. 2013;7:229. doi: 10.7860/JCDR/2013/4619.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Env. Microbiol. 2000;4555-4558:66. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibreel, T. M. Molecular Epidemiology of Uropathogenic Escherichia coli in North West England and characterisation of the ST131 clone in the region, University of Manchester (2011).
- 50.Tofteland S, et al. Effects of phenotype and genotype on methods for detection of extended-spectrum-β-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J. Clin. Microbiol. 2007;45:199–205. doi: 10.1128/JCM.01319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2005;57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 52.Malato S, Fernández-Ibáñez P, Maldonado MI, Blanco J, Gernjak W. Decontamination and disinfection of water by solar photocatalysis: recent overview and trends. Catal. Today. 2009;147:1–59. doi: 10.1016/j.cattod.2009.06.018. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary files are added.