In our recent Communications Biology article, we reported on the biophysical mechanism of resistance for polymyxin antibiotics in bacterial membranes. The emergence of plasmid-borne colistin resistance poses a threat to our last line of defense against many pathogens. Here, we outline the current understanding of mcr-1-mediated polymyxin resistance, and propose future directions for membrane-targeting antibiotic research.
Subject terms: Microbiology, Membrane biophysics
Adree Khondker and Maikel Rheinstadter discuss how bacteria escape being killed by polymyxin antibiotics. Touching on their recent Communications Biology paper, they elaborate on the mechanism by which the bacterial membrane becomes resistant and on future directions to take in order to understand this phenomenon.
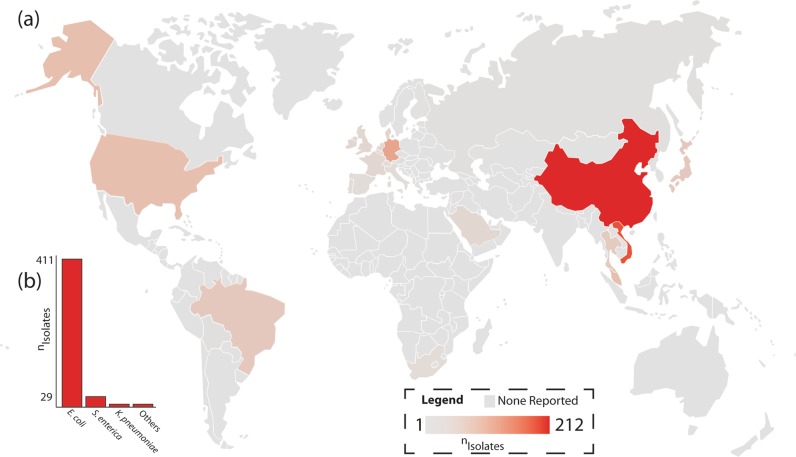
Polymyxin antibiotics, such as colistin, have important roles in both medicine and agriculture. However, the use of polymyxins for the latter endangered its use in the former. The first transmissible plasmid-bearing resistance to colistin was reported by Liu and colleagues in 2016, and isolates of Escherichia coli harboring this resistance have been found in livestock across southeastern Asia1. Expression of the resistance gene, mcr-1, causes modification of lipid A in the bacterial outer surface, resulting in reduced affinity for polymyxins. Worryingly, the gene can be readily passed between different bacterial strains making widespread polymyxin resistance inevitable2. Wang and colleagues reported on the global distribution of mcr-1 and showed the resistance gene is currently present across five continents, as shown in Fig. 1 (ref. 3).
Fig. 1. Geographic distribution of polymyxin resistance.
a Global incidence of mcr-1-harboring isolates per country and b by bacterial strain, data used from ref. 3.
Polymyxin antibiotics kill bacteria by damaging their cell membranes, but now bacteria have figured out how to inhibit this process. Biophysical tools, such as diffraction and molecular dynamics computer simulations, have provided important insights into these mechanisms4,5. In order to prolong the lifespan of current polymyxins and develop new ones, it is critical to gain a detailed understanding of the biophysics of polymyxin–bacteria interactions.
How do polymyxins damage bacterial membranes?
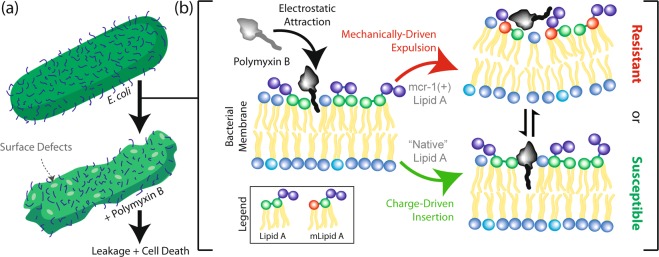
The earliest high-resolution studies showed that polymyxins can kill bacteria by puncturing holes into their outer surface, and causing leakage of internal contents6–8. Polymyxin B and colistin are both cationic antimicrobial peptides that are attracted to the net anionic bacterial outer membrane, resulting in an electrostatic attraction between drug and target. The polymyxin creates local curvature in the membrane, while the membrane itself repels the insertion process. At a critical point, the hydrophobic tail of the polymyxin can insert into the bacterial membrane and create a lipid defect by separating lipids away from one another. When fully inserted into the membrane core, the polymyxins are highly mobile and start forming aggregates in the membrane core, leading to the increased water intake and structural instabilities. The accumulation of those defects in the membrane eventually results in the permeation of water across the bilayer, dysfunction of membrane proteins, the formation of membrane pores at high concentrations, and subsequent membrane collapse. This mechanism has been supported experimentally by our work and others9–12, and is pictured in Fig. 2. The higher the net negative bacterial membrane charge, the more susceptible bacteria are to the formation of these defects.
Fig. 2. Mechanism of resistance for polymyxin antibiotics conferred by mcr-1.
a General mode of polymyxin activity, and b biophysical mechanism of action for polymyxin-induced membrane damage in general membranes, as described in Khondker et al.11. mLipid A is modified lipid A.
Indeed, increasing polymyxin concentration is proportional to membrane damage and bacterial cell death; however, there are inconsistencies at higher concentrations with regards to damage. Currently the pore formation models attribute these to aggregation effects, or polymyxin pores in the form of a barrel stave. At high concentrations of polymyxin, the polymyxin molecules will form aggregates on top of the bacterial surface that can create large physical defects via the carpet model of insertion5,9,13. One of our current goals is to sensitively measure concentration-dependent effects of polymyxin on membrane damage using a lipid-based biosensor that detects passivating currents through a membrane layer in polymyxin-enriched environments. These data will be important to determine threshold concentrations that may be necessary for bactericidal effects.
The nonspecific nature of polymyxin interactions with membranes occurs in the absence of biochemical binding to specific targets. This can also lead to unwanted side effects through the damage of renal epithelial cells, giving rise to nephrotoxicity. We have previously reported that membrane cholesterol, for instance, is crucial for the suppression of polymyxin-induced damage in kidney membrane mimics9. Cells in the renal papillary ducts, which do not contain stiffening membrane cholesterol, seem especially susceptible to polymyxins. Whereas cholesterol did not significantly prevent polymyxin insertion into the membrane, it prevented membrane collapse and subsequent cell death by stabilizing the membrane structure.
Combining colistin with numerous antibiotics have shown synergistic effects, and this offers a promising approach to overcome bacterial resistance. Specifically, clarithromycin in combination with polymyxin showed efficacy in mouse models to eliminate mcr-1 infection and improve survival14. The two biophysical processes to explain this phenomenon likely involve either a direct drug–drug interaction between polymyxin and other antibiotics or that the damage done to the bacterial membranes increases permeability to antibiotics with intracellular targets.
How are resistant membranes different?
The two balancing forces that determine whether polymyxins can insert into membranes and create damage are electrostatic attraction and the “elastic” resistance of the membrane against penetration. If the charge difference is larger than the repulsive forces, the polymyxin antibiotic will eventually be able to penetrate and create membrane damage. Liu and colleagues showed that in mcr-1-expressing bacteria, a negatively charged phosphate on each lipid A in the bacterial outer membrane is replaced by a small neutral ethanolamine moiety in highly virulent pathogens15. The loss of a negatively charged group in the bacterial membrane reduces the affinity of the cationic polymyxin. Moreover, addition of the ethanolamine to lipids across the bacterial surface increases the volume of the membrane core and intermolecular attraction between adjacent lipids, ultimately increasing membrane stability and resistance to mechanical compression and membrane collapse. Paracini et al. showed that polymyxin B activity is dependent on a gel to liquid crystalline phase transition in complex membrane models, and modifications to the structure of lipid A plays a determining role in the phase of the bacterial outer membrane16. Altogether, mcr-1 expression was found to affect the global physical properties of bacterial membranes, making resistant bacteria less attracted to the polymyxin, and less susceptible to polymyxin insertion (Fig. 2).
Notably, polymyxin resistance can also be triggered by two-component signal transduction systems, such as PmrAB and PhoPQ, in response to environmental conditions, such as irregular local cation concentrations17–19. In Acinetobacter baumannii, polymyxin resistance can be conferred by complete loss of negatively charged lipopolysaccharide on the membrane surface20. The physical principles behind the resistance is similar; the loss of the charge and increase in membrane rigidity will independently or synergistically confer resistance to polymyxin antibiotics.
We are currently also focusing our attention on the similarities between polymyxins and other membrane-damaging antibiotics. The underlying mechanisms of these antibiotics are likely also concentration dependent, with a regime where defects are created, and another where pores form in the bacteria cell walls. A unified model may explain the contrasting results with regards to different mechanisms of polymyxin resistance.
Concluding remarks
Polymyxin antibiotics have provided a critical option for clinicians in treating complex multidrug-resistant infections. With advances in biophysical imaging techniques and increasing computational power, it has become possible to observe each subsequent step from polymyxin binding to membrane damage, while measuring the physical effects on the structure of the bacterial outer membrane. With a better understanding of strain-specific resistance, novel lipopeptides based on the polymyxins may be developed, which also overcome toxicity concerns. Fortunately, we are seeing these derivatives in preclinical studies, and increased attention from the biophysical community on developing antimicrobial peptides as a whole21,22. Li, Nation, and Kaye have recently edited the book “Polymyxin Antibiotics: From Laboratory Bench to Bedside”, which provides a comprehensive understanding on the current state of polymyxin antibiotics23.
With regards to the urgency of polymyxin resistance, it is important to consider what can be done on a short term. Indeed, one must restrict polymyxin use in agriculture, or we will soon find ourselves without our last-line defense against multidrug-resistant infections.
Acknowledgements
Warmest thanks to our collaborators on the original Communications Biology article. The authors were funded by the Canadian Institutes of Health Research (CIHR), Natural Sciences and Engineering Research Council of Canada (NSERC), the Canadian Foundation for Innovation (CFI), and the Province of Ontario and McMaster University.
Author contributions
A.K. and M.C.R. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liu YY, et al. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Paterson DL, Harris PN. Colistin resistance: a major breach in our last line of defence. Lancet Infect. Dis. 2016;16:132–133. doi: 10.1016/S1473-3099(15)00463-6. [DOI] [PubMed] [Google Scholar]
- 3.Wang R, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018;9:1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Huang HW. Observation of a membrane fusion intermediate structure. Science. 2002;297:1877–1879. doi: 10.1126/science.1074354. [DOI] [PubMed] [Google Scholar]
- 5.Berglund NA, et al. Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: a molecular dynamics study. PLoS Computational Biol. 2015;11:e1004180. doi: 10.1371/journal.pcbi.1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 7.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiological Rev. 1985;49:1. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teuber M, Bader J. Action of polymyxin B on bacterial membranes. Arch. Microbiol. 1976;109:51–58. doi: 10.1007/BF00425112. [DOI] [PubMed] [Google Scholar]
- 9.Khondker A, et al. Membrane cholesterol reduces polymyxin B nephrotoxicity in renal membrane analogs. Biophysical J. 2017;113:2016–2028. doi: 10.1016/j.bpj.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu L, Wan M, Zhang S, Gao L, Fang W. Polymyxin B loosens lipopolysaccharide bilayer but stiffens phospholipid bilayer. Biophysical J. 2019;118:138–150. doi: 10.1016/j.bpj.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khondker A, et al. Membrane charge and lipid packing determine polymyxin-induced membrane damage. Commun. Biol. 2019;2:67. doi: 10.1038/s42003-019-0297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupuy FG, et al. Selective interaction of Colistin with lipid model membranes. Biophysical J. 2018;114:919–928. doi: 10.1016/j.bpj.2017.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta. 1999;1462:55–70. doi: 10.1016/S0005-2736(99)00200-X. [DOI] [PubMed] [Google Scholar]
- 14.MacNair CR, et al. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 2018;9:458. doi: 10.1038/s41467-018-02875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu YY, et al. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrobial Agents Chemother. 2017;61:e00580–17.. doi: 10.1128/AAC.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paracini N, Clifton LA, Skoda MW, Lakey JH. Liquid crystalline bacterial outer membranes are critical for antibiotic susceptibility. Proc. Natl Acad. Sci. USA. 2018;115:E7587–E7594. doi: 10.1073/pnas.1803975115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trimble MJ, Mlynárčik P, Kolář M, Hancock RE. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016;6:a025288. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arroyo LA, et al. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrobial Agents Chemother. 2011;55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schurek KN, et al. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrobial Agents Chemother. 2009;53:4345–4351. doi: 10.1128/AAC.01267-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moffatt JH, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrobial Agents Chemother. 2010;54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaara M. New polymyxin derivatives that display improved efficacy in animal infection models as compared to polymyxin B and colistin. Medicinal Res. Rev. 2018;38:1661–1673. doi: 10.1002/med.21494. [DOI] [PubMed] [Google Scholar]
- 22.Velkov T, Roberts KD. in. Cham: (Springer; 2019. pp. 343–362. [Google Scholar]
- 23.Li, J., Nation, R. L., & Kaye, K. S. Polymyxin Antibiotics: from Laboratory Bench to Bedside (Springer Nature Switzerland AG, 2019).