Abstract
Homologous recombination repair (HRR) pathway deficiency (HRD) is involved in the tumorigenesis and progression of high-grade serous ovarian carcinoma (HGSOC) as well as in the sensitivity to platinum chemotherapy drugs. In this study, we obtained data from The Cancer Genome Atlas (TCGA) on HGSOC and identified scores for the loss of heterozygosity, telomeric allelic imbalance, and large-scale state transitions, and calculated the HRD score. We then investigated the relationships among the score, genetic/epigenetic alterations in HRR-related genes, and the clinical data. We found that BRCA1/2 mutations were enriched in the group with HRD scores ≥63. Compared with the groups with scores ≤62, this group had a good prognosis; we thus considered HRD scores ≥63 to be the best cutoff point for identifying HRD cases in HGSOC. Classification of HGSOC cases by the HRD status revealed a better prognosis for HRD cases caused by genetic alterations (genetic HRD) than those caused by epigenetic changes and those caused by undetermined reasons (p = 0.0002). Among cases without macroscopic residual tumors after primary debulking surgery, 11 of 12 genetic HRD cases survived after the median observation period of 6.6 years, showing remarkably high survival rates (p = 0.0059). In conclusion, HGSOC can be classified into subtypes with different prognoses according to HRD status. This classification could be useful for personalized HGSOC treatment.
Subject terms: Cancer genomics, Ovarian cancer
Introduction
Ovarian cancer has the worst prognosis of all gynecologic malignancies1. Specifically, high-grade serous ovarian cancer (HGSOC), which is the most common type of ovarian cancer, is mostly advanced stage III–IV disease at the time of diagnosis2. The primary treatment approach for this cancer is a combination of debulking surgery and a platinum-chemotherapy. However, the majority of ovarian cancer patients experience tumor recurrence; they develop chemotherapy resistance that ultimately becomes lethal3.
HGSOC is characterized by chromosomal instability due to homologous recombination repair (HRR) pathway deficiency (HRD)4. Germline BRCA1/2 mutations, somatic BRCA1/2 mutations, and BRCA gene promotor methylations are well-known causes of HRD, but other genetic abnormalities of the HRR pathway could also cause HRD4,5, although no consensus has been reached. The presence of HRD results in irreparable DNA damage from platinum-containing drugs, which leads to cell death. Moreover, an underlying HRD in tumor cells makes the cells sensitive to PARP inhibitors. PARP inhibitors bind to and trap PARP1 and PARP2 on DNA at the sites of single-strand breaks, which results in the generation of double-strand breaks. In cancer cells with HRD, double-strand DNA breaks are repaired by error-prone pathways (i.e. nonhomologous end joining), ultimately leading to cell death6.
HRD causes characteristic genomic scar signatures, namely, the loss of heterozygosity (LOH)7, telomeric allelic imbalance (TAI)8, and large-scale state transitions (LST)9. The HRD score is the sum of these scar signature scores10. The HRD score correlates with sensitivity to niraparib, which is a PARP inhibitor11.
In The Cancer Genome Atlas (TCGA) project, about half of HGSOC cases are reported to have HRD due to an HRR pathway abnormality4. However, the relationship between the HRD score and HRR pathway gene abnormalities other than those in BRCA1/2 has not been thoroughly investigated. We thus investigated the connection between HRD status and HRR pathway gene abnormalities in HGSOC data in the TCGA, and we show that HGSOC can be classified according to HRD status into subtypes with different prognoses.
Results
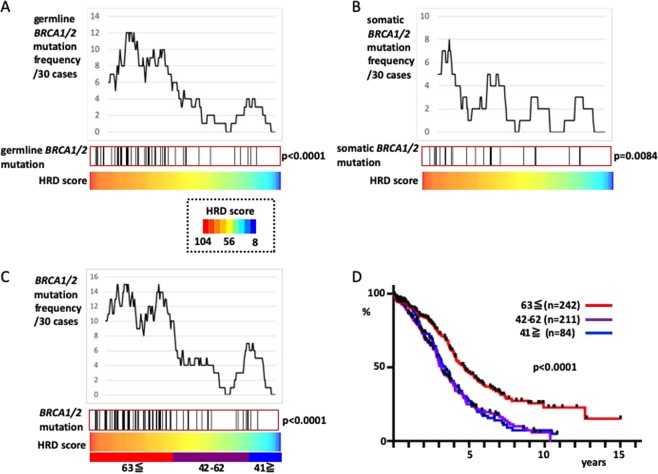
First, we defined HRD cases by investigating the distribution of cases that harbored germline BRCA1/2 or somatic BRCA1/2 mutations. Compared with the cases without germline BRCA1/2 mutations, those cases with germline BRCA1/2 mutations had higher scores for LOH7 (p = 0.0018), TAI8 (p < 0.0001), and LST9 (p < 0.0001). Cases with somatic BRCA1/2 mutations scored higher for LOH compared with those without somatic BRCA1/2 mutations (p = 0.0018; p = 0.28 and 0.06 for TAI and LST scores, respectively). HRD scores (TAI + LST + LOH) were higher in those cases with germline BRCA1/2 mutations or somatic BRCA1/2 mutations, compared with those without (Fig. 1A,B; p = 0.0001 and 0.0084 for germline BRCA1/2 mutations and somatic BRCA1/2 mutations, respectively). When germline BRCA1/2 mutations and somatic BRCA1/2 mutations were analyzed together as BRCA mutations, it was found that BRCA mutation cases had high HRD scores (p < 0.0001, Fig. 1C).
Figure 1.
Definition of HRD cases according to HRD scores. The 296 cases for which both exome sequence and SNP array data were available were sorted by HRD score. (A) Distribution of cases with germline BRCA1/2 mutations. The upper graph shows the frequency of germline BRCA1/2 mutation cases by sliding the windows of 30 cases. The black bars in the central box represent germline BRCA1/2 mutations cases. (B) Distribution of cases with somatic BRCA1/2 mutations. (C) germline BRCA1/2 mutations and somatic BRCA1/2 mutations are presented together as BRCA1/2 mutations; these cases are concentrated in HRD scores ≥63. The cutoff HRD score ≥42, which is generally used, is also shown. D) Comparison of overall survival rate by HRD scores; 537 cases with known prognosis and SNP array data were divided according to HRD scores ≥63, 42–62, and ≥41.
Until now, the proposed cutoff value for the HRD score has been ≥4210. In the report where this cutoff was proposed, the ovarian cancer and breast cancer datasets were analyzed together. Therefore, we also analyzed the breast cancer TCGA dataset12, and found that generally, breast cancer had lower HRD scores than HGSOC (Supplementary Fig. 1A, p < 0.0001), but higher HRD scores were still associated with BRCA mutation cases (p < 0.0001, Supplementary Fig. 1B). Combined analysis of ovarian and breast cancer datasets from 1257 cases showed that the distribution of BRCA mutation cases was bimodal. Indeed, there also seemed to be a cutoff at HRD scores ≥42 (Supplementary Fig. 1C). However, in the analysis of ovarian cancer alone, BRCA mutation cases were more likely to have HRD scores ≥63 (Fig. 1C). BRCA mutations represented 38% of the cases with HRD scores ≥63 (49/128), 10% with HRD scores between 42 and 62 (12/118), and 10% with HRD scores 41 and below (5/50); no enrichment of BRCA mutation cases with HRD scores from 42 to 62 was observed compared with those with scores ≤41. An analysis of the prognosis revealed overlap in the survival curves of patients with HRD scores of 42–62 (n = 211) and ≤41 (n = 84), but the survival rate of patients with HRD scores ≥63 (n = 242) was clearly better than that of the other two groups (Fig. 1D, p < 0.0001). Thus, we designated an HRD score ≥63 as the cutoff.
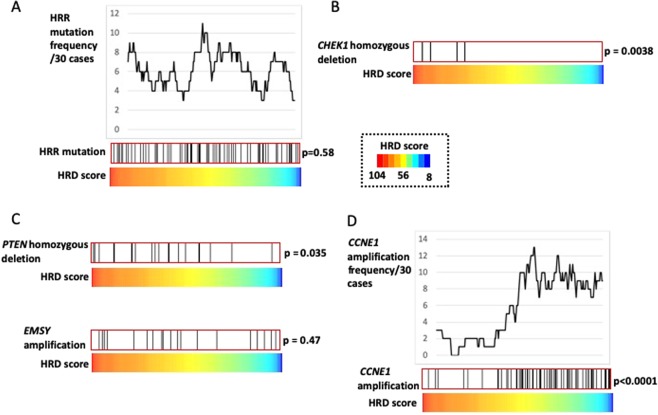
Subsequently, we investigated the connection between the HRD score and overall HRR pathway gene mutations other than BRCA mutations but found none (Fig. 2A, p = 0.58). Moreover, no difference was observed in survival rates between patients with and without these HRR mutations (p = 0.78, data not shown). However, individual analyses of these HRR mutations revealed that the CHEK1 homozygous deletions was associated with high HRD scores (Fig. 2B, Supplementary Fig. 2, p = 0.0038). The PTEN homozygous deletions and EMSY amplifications are candidate mutations for causing HRD5. Of these, PTEN homozygous deletions were positively correlated with HRD scores (p = 0.035), whereas EMSY amplifications were not (Fig. 2C, Supplementary Fig. 2). CCNE1 amplifications, a genetic abnormality that characterizes HR-proficient HGSOC5, were clearly associated with low HRD scores (Fig. 2D; p < 0.0001).
Figure 2.
Relationship between HRD scores and mutations in HRR-related genes other than BRCA1/2. Dividing 296 cases according to HRD scores shows the distribution of cases with mutations. (A) Distribution of cases with HRR pathway gene mutations other than those in BRCA1/2, including homozygous deletions. (B) CHEK1 homozygous deletions had high HRD scores among the HRR pathway gene mutations other than those in BRCA1/2. (C) PTEN homozygous deletions and EMSY amplifications are two gene mutations that may cause HRD. (D) Relationship between CCNE1 amplifications and HRD score; CCNE1 amplification cases were enriched in low HRD scores.
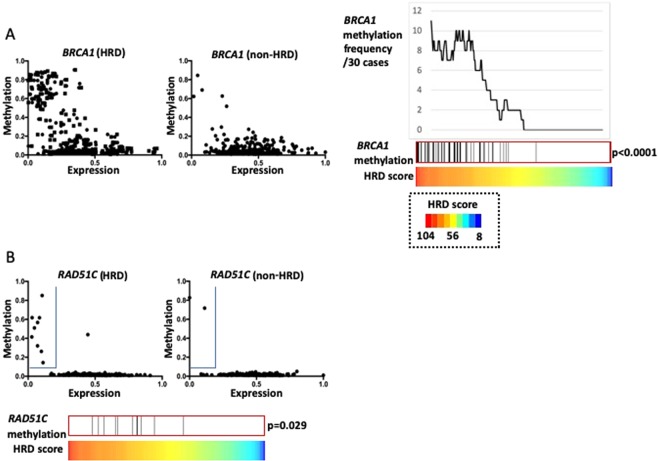
Next, we analyzed DNA methylation in the promoter region of HRR-related genes. First, we investigated BRCA1 methylation. Of the nine BRCA1 methylation probes, the β value of cg04658354 was most negatively correlated with the BRCA1 mRNA expression level (r = −0.49). When BRCA1 mRNA expression and cg04658354 β values were plotted, markedly more methylated cases with a high β value and low mRNA expression were observed in the HRD group than in the non-HRD group (Fig. 3A). The 56 BRCA1 methylated cases described in the TCGA paper4 revealed higher HRD scores than non-methylated cases (Fig. 3A, p < 0.0001). Similarly, RAD51C methylation probe cg14837411 showed strong negative correlation with the mRNA expressions (r = −0.40). Cases with methylation, which were defined by a β value > 0.1 and a normalized expression <0.2, were concentrated in the high HRD cases (p = 0.029, Fig. 3B). Among the other HRR pathway-related genes, the only one with a strong negative correlation between the methylation probe β value and gene expression was PTEN cg21573601 (r = −0.40). However, the maximum β value of this probe was low (0.12), and it is thus unclear whether this gene was actually methylated. Candidate PTEN methylation cases selected using a cutoff β value > 0.03 and a normalized expression <0.4 had high HRD scores (p = 0.0022, Supplementary Fig. 3).
Figure 3.
Relationship between HRD status and DNA promoter methylation of HRR-related genes. (A) BRCA1 methylation. Left: HRD cases; Center: non-HRD cases. X-axis: Average values of two BRCA1 mRNA expression probes normalized to 0–1. Y-axis: β value of cg04658354. Right: Distribution of BRCA1 methylated cases. (B) RAD51C methylation. X-axis: Average values of two RAD51C mRNA expression probes normalized to 0–1. Y-axis: β value of cg14837411. RAD51C methylation cases are surrounded by a blue line in the upper-left region. The frequency of RAD51C methylation differs greatly between HRD cases (left) and non-HRD cases (right).
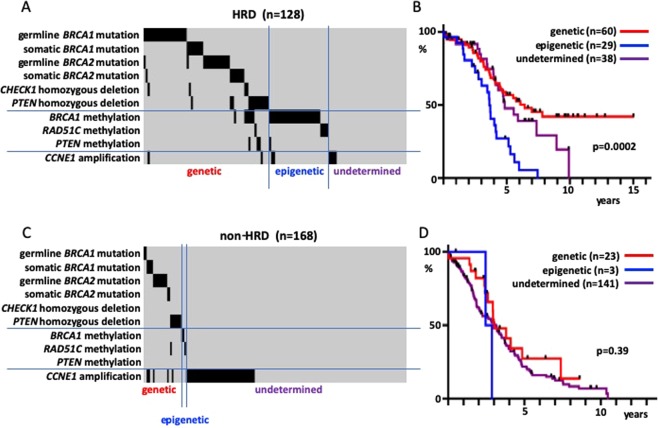
Considering the above findings, the HRD cases for which both SNP array and exome sequence data were available (n = 128) were classified according to the underlying molecular mechanism. Those cases that had one of the six factors that could induce HRD genetically (germline BRCA1 mutations, somatic BRCA1 mutations, germline BRCA2 mutations, somatic BRCA2 mutations, CHEK1 homozygous deletions, and PTEN homozygous deletions) were classified as genetic HRD. Other cases that had BRCA1 methylations or RAD51C methylations were classified as epigenetic HRD, while the remainder were classified as undetermined HRD. There was no case with PTEN candidate methylation alone (Fig. 4A). The survival rates of these three groups (comprising 127 patients for which the overall survival (OS) data were available) revealed a good prognosis for genetic HRD and a poor prognosis for epigenetic HRD (Fig. 4B, p = 0.0002). No difference was found in the prognosis between genetic HRD cases with BRCA1/2 mutations and those with CHEK1 homozygous deletions or PTEN homozygous deletions. Since there were only a few RAD51C methylation cases among the epigenetic HRD cases, it was difficult to compare their prognoses with those of the BRCA1 methylation cases (Supplementary Fig. 4A). A similar classification of the non-HRD cases (n = 168; n = 167 for those with OS information; Fig. 4C) revealed no difference in survival rates (Fig. 4D, p = 0.39). Additionally, a survival rate comparison between HRD and non-HRD cases with the aforementioned six factors causing genetic HRD revealed a better prognosis for HRD cases (Supplementary Fig. 4B, p = 0.019).
Figure 4.
Relationship between the molecular mechanism of HRD and prognosis; sorting of cases by mechanism into the genetic mutation (genetic), DNA methylation (epigenetic), and unknown mechanism (undetermined) groups. (A) Classification of HRD cases. (B) Survival rate of HRD cases. (C) Classification of non-HRD cases. (D) Survival rate of non-HRD cases.
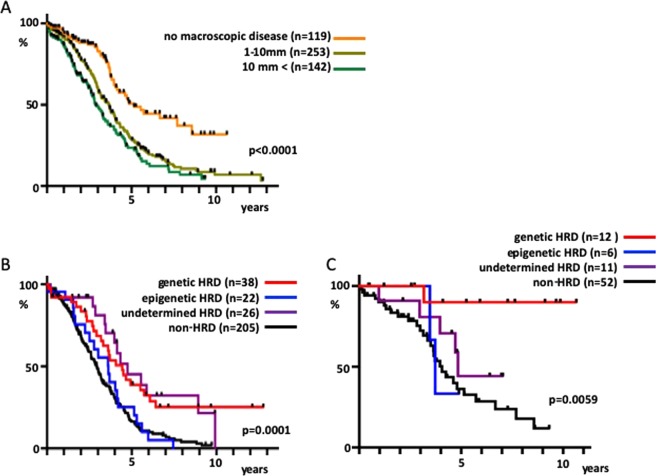
Finally, in the analysis, we included data on residual tumor at the time of surgery. The 514 cases were sorted by tumor size if data on residual tumor at the time of surgery were available, and a poor long-term prognosis for those groups with residual tumors ≥1 mm was revealed regardless of size (Fig. 5A, p < 0.0001). Among the cases with residual tumor ≥1 mm, approximately 25% of genetic HRD and undetermined HRD patients had a long-term survival >7 years, whereas non-HRD and epigenetic HRD patients had low long-term survival rates (Fig. 5B, p = 0.0001). Intriguingly, among patients with no macroscopic diseases, genetic HRD patients had a very high survival rate, and 11 of 12 patients survived after a median observation period of 6.6 years (Fig. 5C, p = 0.0059).
Figure 5.
Relationship among residual tumor at the time of surgery, HRD status, and prognosis. (A) The 514 cases were sorted by size of the residual tumor if data on residual tumor at the time of surgery were available (no macroscopic disease, 1–10 mm and >10 mm) and their survival rates were compared. (B) Cases with residual tumor ≥1 mm and available SNP array data were divided into the HRD and non-HRD groups. HRD cases with exome sequencing data were further sorted by molecular mechanism into genetic HRD, epigenetic HRD, and undetermined HRD to compare the survival rate. (C) Similarly, cases with no macroscopic disease were classified by the HRD status for survival rate comparison.
Discussion
HGSOC data from the TCGA became publicly available in 2011 and have been analyzed in numerous reports since then. However, in this study, we still found some important observations from these data. Our contributions are as follows: (1) establishing a cutoff value to identify HRD from the analysis of HGSOC data alone (2); a comprehensive analysis of HRR pathway genes (3); proposal of a classification method for HRD cases according to cause; and (4) integration of residual tumor data.
A cutoff HRD score ≥42 was previously proposed in a study that combined ovarian and breast cancer SNP array data from the TCGA and two other datasets from custom panels10. After running a similar analysis that combined the breast cancer and HGSOC TCGA datasets, BRCA1/2 mutations seemed to be enriched at HRD scores ≥42 (Supplementary Fig. 1C). However, a comparison of the HGSOC and breast cancer data revealed a large gap in HRD scores (Supplementary Fig. 1A). In another study, a pan-cancer analysis using the TCGA data revealed that the TAI, LST, and LOH scores of HGSOC, with a median of 24, 20, and 15, respectively, were higher than those of cancers derived from other organs; the corresponding median scores for breast cancer were 12, 8, and 8, respectively, for TAI, LST, and LOH, which were approximately half those of HGSOC13. Another study calculated the HRD scores of formalin-fixed paraffin-embedded samples using an OncoScan SNP array and found that HGSOC had far higher HRD scores than ovarian clear cell carcinoma, as over 80% of cases scored ≥4214. HRD scores thus vary greatly between organ of origin and tissue type. As the aim of our study was to classify HGSOC by HRD status, we based our analyses on HGSOC data only and adopted an HRD score ≥63 as the cutoff. As a result, a little less than half of HGSOC patients were classified in the HRD group (Fig. 1C). In other studies, authors performed SNP arrays with a custom panel, classified a little less than half of their samples as HRD with a cutoff HRD score ≥42, and found a high frequency of BRCA mutations15,16. The ratio of HRD is consistent with findings from our study even though the cutoff point differs. In order to clarify why such a difference occurred, it would be beneficial if the raw SNP array data that were used to generate the HRD scores for research purposes would be open to the public.
The question of whether HRR pathway gene mutations other than BRCA1/2 actually cause HRD has not been thoroughly investigated. Our TCGA data analyses indicated that these overall mutations are unrelated to the HRD score (Fig. 2A) and prognosis and that they are not involved in HRD or platinum sensitivity. Among these overall genes, only those cases with CHEK1 homozygous deletions displayed high HRD scores (Fig. 2B). CHEK1 is required for commencing HRR in cases of DNA double-strand breaks17, which means that CHEK1 homozygous deletions are likely to cause HRD. Moreover, PTEN homozygous deletions correlated with high HRD scores (Fig. 2C). Therefore, we suggest that the only genetic changes that are involved in HRD in HGSOC besides the germline/somatic mutations in BRCA1/2 are CHEK1 and PTEN homozygous deletions. In support of our results, the HRD-associated LOH score is high in ovarian cancer with PTEN homozygous deletions, but ovarian cancer cases with ATM, ATR, FANCA, FANCD2, FANCM, and PALB2 mutations do not have high LOH scores7.
It is well known that HRD is caused by promoter methylation of BRCA15; indeed, HRD scores were high in cases in which BRCA1 was methylated (Fig. 3A). The TCGA database only lists 168 genes including BRCA1 because of the strict standards for β value and mRNA expression for the selection of methylated genes4. In addition, tumors with methylated RAD51C also transform to HRD5 and have high LOH scores7. We focused on the negative correlation between the RAD51C β value and gene expression and found, as in previous reports, that RAD51C is often methylated in HRD cases (Fig. 3B).
We classified the HRD cases and found that epigenetic HRD cases had a poor prognosis, just like non-HRD cases (Fig. 4B). Previous studies have reported the correlation between BRCA1 and RAD51C methylation and a good prognosis in ovarian cancer18,19. However, many recent studies do not support these findings20–23. In some cases, methylated BRCA1 in tumors was found to be demethylated after chemotherapy24,25. The presence of BRCA1 methylation before treatment was associated with post relapse platinum resistance in a study of triple-negative breast cancer26. These results suggest that demethylation of methylated BRCA1 occurs more readily than genetic changes such as the reversion mutation in BRCA1/2 mutation cases24,27. Our findings for the elevated HRD scores in the tumors with BRCA1 or RAD51C methylations (Fig. 3) indicate that there is a poor prognostic subtype in the high-HRD score cases and warrants HRD-positive cases should be classified into epigenetic and non-epigenetic cases when conducting clinical trials.
Compared with genetic HRD cases, non-HRD cases had a poorer prognosis despite the presence of HRD-inducing genetic changes (Supplementary Fig. 3B). Presumably, tumors that managed to regain DNA repair functions through some mechanism became dominant in such cases, even though the DNA repair defects is initially important to carcinogenesis. In HGSOC with BRCA abnormalities, tumor infiltration by lymphocytes was augmented28,29, which was likely due to tumor immunity triggered by neo-antigen production as a result of DNA repair disorders. In the course of tumor progression, some tumors that regain DNA repair function may become dominant as the tumors interact with immune cells.
Finally, we performed analyses that integrated residual tumors after surgery. The HGSOC TCGA dataset comprises patients who underwent primary debulking surgery. We found that over 90% of genetic HRD patients who had no residual tumor are long-term survivors (Fig. 5C). Consistent with our data, stage III HGSOC cases who survived more than ten years were associated with germline or somatic BRCA1/2 mutations and no residual disease after primary debulking surgeries30. This finding indicates that genetic HRD patients may be those who should undergo the most extensive surgery in order to extend their survival. Presumably, it is possible that tumor cells which harbor genetic alterations like reversion mutations24,27 could be minimized in genetic HRD cases by complete resection. Whether or not a similarly high survival rate could be obtained in genetic HRD patients with complete surgery by interval debulking surgery following neoadjuvant chemotherapy remains to be elucidated. Several recent studies have evaluated interval debulking surgery31,32; genomic analyses of such clinical study samples would provide the answer to this question.
In conclusion, we classified the HGSOC cases into genetic HRD, epigenetic HRD, undetermined HRD, and non-HRD, each of which had a different prognosis. We found that in the genetic HRD fraction, a high survival rate could be increased by complete resection. Our data could be the foundational basis for future clinical studies in HGSOC.
Materials and Methods
Datasets
Raw data from the Affymetrix Genome-Wide Human SNP Array 6.0 and the HT Human Genome U133 Array (Affymetrix) as well as clinical data on HGSOC4 and breast invasive carcinoma12 were obtained from the GDC Data Portal (https://portal.gdc.cancer.gov). In addition, processed data from Infinium HumanMethylation27 BeadChip (Illumina) was obtained from the GDAC Firehose (http://gdac.broadinstitute.org).
Data processing
SNP array data of cases for which tumor samples could be matched with a normal sample were extracted. For both tumor and normal sample data, the log R ratio and B-allele frequency of each probe was calculated using Affymetrix Power Tools and PennCNV33, and segmented copy number data were generated using allele-specific copy number analysis of tumors34. Gene expression data were normalized by the Robust Multi-array Average method using the “affy” package of R.
HRD score analysis
Genomic scar signatures were analyzed using an R program, as previously described13, and TAI, LST, and LOH scores were calculated. Briefly, TAI was defined as the number of regions of allelic imbalance that extended to the telomeres after deciding upon the major copy number state to be used to adjust biases in samples with an uneven chromosome count. LOH was defined as the number of chromosomal LOH regions longer than 15 Mb. LST was defined as the number of break points between regions longer than 10 Mb after filtering out regions shorter than 3 Mb. The HRD score was calculated as the sum of the TAI, LST, and LOH scores. The calculated scores are shown in Supplementary Table 1 (ovarian cancer) and in Supplementary Table 2 (breast cancer).
Homologous recombination repair (HRR)-related gene analysis
According to the ARIEL3 study and other reports35,36, BRCA1, BRCA2, ATM, ATR, BARD1, BLM, BRIP1, CDK12, CHEK1, CHEK2, FANCA, FANCC, FANCD2, FANCE, FANCF, FANCI, FANCL, FANCM, MRE11, NBN, PALB2, RAD50, RAD51, RAD51B, RAD51C, RAD51D, RAD52, RAD54L, and RPA1 were identified as HRR pathway genes. Cases of germline mutations or somatic mutations including homozygous deletions were identified. Moreover, cases of EMSY amplification and PTEN homozygous deletions, which may cause HRD, and the CCNE1 amplifications5, which are associated with HR proficiency, were identified. These cases were identified using the cBioPortal for Cancer Genomics (http://www.cbioportal.org)37,38.
The correlation analysis of HRR pathway genes was performed using promotor methylation DNA microarray data and mRNA expression microarray data. When multiple mRNA expression probes existed, average of these probes was calculated for each case and then normalized from 0 to 1. Subsequently, we selected methylation probes which β values showed strong negative correlation (r < −0.30) with the mRNA expressions. If multiple methylation probes were selected for one gene, the most negatively correlated probe was used.
Heatmap
A heatmap of the HRD score was created using R 3.2.2 and the MATLAB RGB color specifier. Other heatmaps were drawn using Java TreeView version 1.1.6r4.
Statistical analysis
We determined the presence of genetic abnormalities, and using the Mann–Whitney U test, we compared the TAI, LST, LOH, and HRD scores between tumors with genetic abnormalities and those without them. We also compared the HRD scores between HGSOC and breast cancer using the Mann-Whitney U test. Correlation analyses were performed using the Pearson correlation coefficient. Survival function was estimated using a Kaplan-Meier analysis, and survival curves were compared between groups using the log-rank test. All statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, San Diego) with an α of 0.05.
Supplementary information
Supplementary Figure 1, 2, 3, 4, Figure Legends, Table 1, 2.
Acknowledgements
Noriomi Matsumura reports that this work was supported by JSPS KAKENHI Grant Number 18H02947.
Author contributions
H.T. designed the study and prepared the manuscript draft. N.M. and H.N. contributed in analyzing the data as well as in drafting the manuscript. S.T. and M.M. collected and interpreted the data and conducted a comprehensive literature review related to this project. All authors thoroughly reviewed the manuscript. All authors read and approved the final version of this manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-59671-3.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA. Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Seidman JD, et al. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int. J. Gynecol. Pathol. 2004;23:41–44. doi: 10.1097/01.pgp.0000101080.35393.16. [DOI] [PubMed] [Google Scholar]
- 3.Miller DS, et al. Phase II evaluation of pemetrexed in the treatment of recurrent or persistent platinum-resistant ovarian or primary peritoneal carcinoma: a study of the Gynecologic Oncology Group. J. Clin. Oncol. 2009;27:2686–2691. doi: 10.1200/JCO.2008.19.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ledermann JA, Pujade-Lauraine E. Olaparib as maintenance treatment for patients with platinum-sensitive relapsed ovarian cancer. Ther. Adv. Med. Oncol. 2019;11:1758835919849753. doi: 10.1177/1758835919849753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abkevich V, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkbak NJ, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popova T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 10.Telli ML, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin. Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirza MR, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marquard AM, et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark. Res. 2015;3:9. doi: 10.1186/s40364-015-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao A, et al. Genomic scar signatures associated with homologous recombination deficiency predict adverse clinical outcomes in patients with ovarian clear cell carcinoma. J. Mol. Med. (Berl). 2018;96:527–536. doi: 10.1007/s00109-018-1643-8. [DOI] [PubMed] [Google Scholar]
- 15.Stronach EA, et al. Biomarker Assessment of HR Deficiency, Tumor BRCA1/2 Mutations, and CCNE1 Copy Number in Ovarian Cancer: Associations with Clinical Outcome Following Platinum Monotherapy. Mol. Cancer Res. 2018;16:1103–1111. doi: 10.1158/1541-7786.MCR-18-0034. [DOI] [PubMed] [Google Scholar]
- 16.Patel JN, et al. Characterisation of homologous recombination deficiency in paired primary and recurrent high-grade serous ovarian cancer. Br. J. Cancer. 2018;119:1060–1066. doi: 10.1038/s41416-018-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 18.Ignatov T, et al. BRCA1 promoter methylation is a marker of better response to platinum-taxane-based therapy in sporadic epithelial ovarian cancer. J. Cancer Res. Clin. Oncol. 2014;140:1457–1463. doi: 10.1007/s00432-014-1704-5. [DOI] [PubMed] [Google Scholar]
- 19.Bai X, et al. BRCA1 promoter hypermethylation in sporadic epithelial ovarian carcinoma: Association with low expression of BRCA1, improved survival and co-expression of DNA methyltransferases. Oncol. Lett. 2014;7:1088–1096. doi: 10.3892/ol.2014.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruscito I, et al. BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients–a study of the tumour Bank ovarian cancer (TOC) and ovarian cancer diagnosis consortium (OVCAD) Eur. J. Cancer. 2014;50:2090–2098. doi: 10.1016/j.ejca.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Sun T, et al. Genetic Versus Epigenetic BRCA1 Silencing Pathways: Clinical Effects in Primary Ovarian Cancer Patients: A Study of the Tumor Bank Ovarian Cancer Consortium. Int. J. Gynecol. Cancer. 2017;27:1658–1665. doi: 10.1097/IGC.0000000000001071. [DOI] [PubMed] [Google Scholar]
- 22.Bernards SS, et al. Clinical characteristics and outcomes of patients with BRCA1 or RAD51C methylated versus mutated ovarian carcinoma. Gynecol. Oncol. 2018;148:281–285. doi: 10.1016/j.ygyno.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Zhu X, Zhao L, Lang J. The BRCA1 Methylation and PD-L1 Expression in Sporadic Ovarian Cancer. Int. J. Gynecol. Cancer. 2018;28:1514–1519. doi: 10.1097/IGC.0000000000001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patch AM, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 25.Prieske K, et al. Loss of BRCA1 promotor hypermethylation in recurrent high-grade ovarian cancer. Oncotarget. 2017;8:83063–83074. doi: 10.18632/oncotarget.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tutt A, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat. Med. 2018;24:628–637. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakai W, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAlpine JN, et al. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod. Pathol. 2012;25:740–750. doi: 10.1038/modpathol.2011.211. [DOI] [PubMed] [Google Scholar]
- 29.George J, et al. Nonequivalent gene expression and copy number alterations in high-grade serous ovarian cancers with BRCA1 and BRCA2 mutations. Clin. Cancer Res. 2013;19:3474–3484. doi: 10.1158/1078-0432.CCR-13-0066. [DOI] [PubMed] [Google Scholar]
- 30.Yang SYC, et al. Landscape of genomic alterations in high-grade serous ovarian cancer from exceptional long- and short-term survivors. Genome Med. 2018;10:81. doi: 10.1186/s13073-018-0590-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vergote I, et al. Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N. Engl. J. Med. 2010;363:943–953. doi: 10.1056/NEJMoa0908806. [DOI] [PubMed] [Google Scholar]
- 32.Kehoe S, et al. Primary chemotherapy versus primary surgery for newly diagnosed advanced ovarian cancer (CHORUS): an open-label, randomised, controlled, non-inferiority trial. Lancet. 2015;386:249–257. doi: 10.1016/S0140-6736(14)62223-6. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coleman RL, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bajrami I, et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014;74:287–297. doi: 10.1158/0008-5472.CAN-13-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1, 2, 3, 4, Figure Legends, Table 1, 2.