Abstract
With a five-year survival rate of 9%, pancreatic ductal adenocarcinoma (PDAC) is the deadliest of all cancers. The rapid mortality makes PDAC difficult to research, and inspires a resolve to create reliable, tractable cellular models for preclinical cancer research. Organoids are increasingly used to model PDAC as they maintain the differentiation status, molecular and genomic signatures of the original tumour. In this paper, we present novel methodologies and experimental approaches to develop PDAC organoids from PDX tumours, and the simultaneous development of matched primary cell lines. Moreover, we also present a method of recapitulating primary cell line cultures to organoids (CLOs). We highlight the usefulness of CLOs as PDAC organoid models, as they maintain similar transcriptomic signatures as their matched patient-derived organoids and patient derived xenografts (PDX)s. These models provide a manageable, expandable in vitro resource for downstream applications such as high throughput screening, functional genomics, and tumour microenvironment studies.
Subject terms: Cancer models, Cancer models
Introduction
With a rapid progression and fatal outcome, pancreatic cancer is one of the deadliest of all cancers. Long-term survivors are limited to those with resected early stage tumours; however, overall survival rate is a dismal 9%, with a median survival time of 7–11 months1,2. As pancreatic cancer is notoriously asymptomatic at an early stage, 80% of patients are diagnosed after the cancer has metastasised, making them ineligible for resection, which is the only curative treatment. The number of cases of pancreatic cancer has been steadily rising since 20043. It is currently the fourth most common cause of cancer death in the US, and by 2030 it is estimated that it will surpass breast and colorectal cancer to become the second most common cause of death by cancer4. The majority of pancreatic cancers are in the exocrine pancreas (95%) known as pancreatic ductal adenocarcinoma (PDAC), and 5% are in the endocrine pancreas5. The leading epidemiological factors include smoking, obesity, type II diabetes mellitus and acute pancreatitis, which account for approximately 25% of PDAC cases6–9.
A limitation in the understanding of the disease progression and development of effective treatments in PDAC may be due the lack of in vitro patient tumour representative models. Established 2D cell lines are the most widely used model for the development and testing of chemotherapeutics for over 50 years10. The ability of cell lines to grow indefinitely makes them a low-cost, repeatable model, easy to manipulate and are an important base for discovery and proof-of-concept studies. Their importance in cancer research is indisputable, however, their use as a robust clinical model is questionable11. During passaging, cell lines undergo genetic modifications, such as copy number variation and point mutations12. Cell lines also have a high level of homogeneity, which does not represent the heterogenetic nature of PDAC tumours, and not all cancer subtypes are well represented as they are usually developed from aggressive and metastatic tumours, therefore tumour progression cannot be studied13. Deer et al.14 highlighted the genetic diversity and differences in adhesion, migration and tumorgenicity between the different PDAC cell lines in a detailed review of phenotypes and genotypes of PDAC cell lines. Primary tumour cultures derived from patient tumour or biopsy and grown in vitro in 2D tend to be heterogeneous, and are a better representation of the tumour of origin tissue as the cells are at an early passage number15. These models allow for the development of personalised cancer therapy as demonstrated through functional screening of chemotherapeutic drugs, modelling individual tumour response16. However, although an important tool in cancer research, there are limitations associated with primary cell culture, including the difficulty of obtaining tumour samples. Primary cell cultures are often difficult to establish as the initial sample may lack tumour cells; the outgrowth of stromal cells such as fibroblasts may overrun the culture, and the cells may only grow for a finite number of passages15,16.
Three-dimensional (3D) in vitro models can address the limitations of growing cancer in 2D. Unlike 2D cultures, 3D cell cultures are not grown attached to plastic, so they adopt a more physiologically relevant morphology and signalling pathways similar to in vivo conditions17. Like solid tumours, 3D cell cultures are exposed to complex physiological and heterogeneous environments, resulting in varied exposure to oxygen, nutrients and stresses. This allows for the study of cell-to-cell interaction, drug penetration, response and resistance18,19. The 3D culture model contains proliferating, quiescent, hypoxic and necrotic cells, whereas in 2D cell models, all the cells are in the same growth phase20. Organoids are 3D cultures derived from organ specific stem cells, allowing for the self-organisation of cells to resemble structures from within an organ or tumour21. These 3D system models the physiology, shape, dynamics and cell make-up of the cancer and produces a relevant and highly adaptable model system22. Organoids can be derived from embryonic stem cells, induced pluripotent stem cells, and both tumour and normal organ restricted adult stem cells (aSCs)21. Organoids can be maintained in a 3D matrix which supports cell growth, such as Matrigel or hydrogels which are laminin rich, mimicking the pancreatic microenvironment in vivo23. Organoid culture requires special media containing a cocktail of growth factors that mimic the microenvironments of organ stem cell niches24. Unlike established and primary cell lines, organoids display cell heterogeneity after several passages and organoids which have been developed from the pancreas can be expanded, managed and cryopreserved and PDAC organoids can proliferate indefinitely22. Boj et al.25 developed a method for the establishment of both normal and neoplastic organoids from human and murine pancreatic tissues. Orthotopic transplant of both the PDAC and normal organoids resulted in full tumour and ductal development respectively. These methods are useful for the further development of PDAC research, including the study of the tumour microenvironment, personalised medicine, furthering the understanding of the genetics and for testing novel therapeutics in PDAC.
Patient derived xenografts (PDX) are widely used animal models to study cancer. The patient tumour is grown subcutaneously or orthotopically in an immunodeficient mouse, and sub-cultured into new mice when the tumour reaches a sufficient size. PDX models allow for tumours to retain their cell-to-cell interactions, and orthotopic implantation allows for the tumour to grow in the same micro-environment as the original tumour26.
There are several advantages to using PDX tumours for the study of PDAC. PDX tumours are established from small amount of patient tumour; the tumours retain the original tumour heterogeneity, which can be lost in established cell lines. The PDX tumours also preserve the original tumours’ genetics and histological characteristics during passaging. These models are used as an unlimited resource for in vivo and ex vivo drug testing, which is invaluable in PDAC research. However, disadvantages to the use of PDX models include: expensive to develop, time consuming, dependent on the use of animals, require ethics approval and are subject to strict regulations27. PDX models also have a slow take rate, and can take months to develop tumours. Moreover, as the tumour is sub-cultured, the tumour-associated stroma is replaced by murine stromal cells, such as blood vessels and fibroblasts28. Finally, SCID-mice which are used for PDX biobanks lack immune systems, and this limits the testing of drugs such as immune modulators, which are increasingly being used in cancer treatments.
PDX models and 3D cancer organoid cultures are important preclinical in vitro live material models. However, limitations for both include considerable amount of time required and cost to expand cultures in vitro, which makes it difficult to scale-up and perform downstream high‐throughput analysis. Here, we report methodologies for the establishment of PDX-tumour organoids, isogenetically matched 2D primary cell line and recapitulated primary cell line into organoids (CLOs). We show that organoids and CLOs maintain the same molecular and transcriptomic signatures compared to the 2D primary cell line, indicating the robustness and usefulness of CLOs in preclinical studies of pancreatic cancer.
Results
Development of PDX organoids, isogenetically matched primary cell lines and 3D cell line organoids (CLOs) as novel models to study PDAC
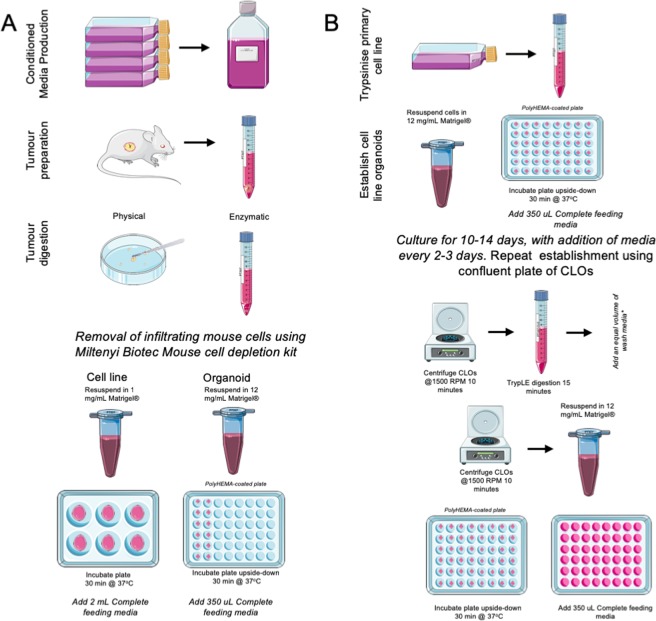
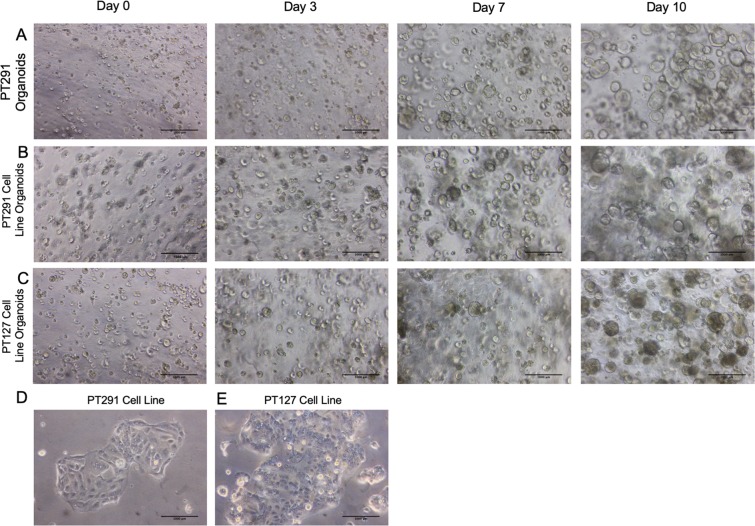
Recent progress with preclinical models to study PDAC has advanced our research into this deadly disease. We have built on the scientific advancements in PDX28–30 and cancer organoid models21,25,31,32 to bridge the in vivo/in vitro gap in PDAC research. Therefore, we developed a protocol to established organoids from PDX tumour with modified approaches from Boj et al. (Fig. 1A)25. Primary cell lines were simultaneously generated by maintaining organoid outgrowth cells in lower concentration extracellular matrix (ECM) gel derived from Engelbreth-Holm-Swarm murine sarcoma on adherent wells, and allowing the cells to attach and proliferate. This method allows for the rapid development of primary cell lines, which can be scaled up and manipulated. The 2D primary cells are cultured for two passages then recapitulated as 3D cultures (Fig. 1B) and embedded in high concentration ECM, which provides a scaffold for PDAC organoid growth. This novel method leads to the rapid development of cell line organoids (CLOs), which are morphologically similar to the matched organoids within only 3–7 days (Fig. 2A–C).
Figure 1.
Schematic diagram protocol for generating PDAC organoids, isogenetically matched 2D primary cell lines and cell line organoids (CLOs) from a primary cell line. (A) The steps involved in the simultaneous production of organoids and isogenetically matched 2D primary cell lines from a single PDX tumour sample, (B) Steps involved in the establishment of CLOs from a 2D primary cell line. This figure was created using Servier Medical Art templates, which have been modified. These images are licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com.
Figure 2.
Brightfield images at Day 0, Day 3, Day 7, Day 10 of (A) PT291 organoids (B) PT291 CLO (C) PT127 CLO (D) PT291 2D primary cell line and (E) PT127 2D primary cell line. Scale bars 1000 µm.
Confirmation of primary cell lines and organoid culture derived from human PDAC PDX tumour cells
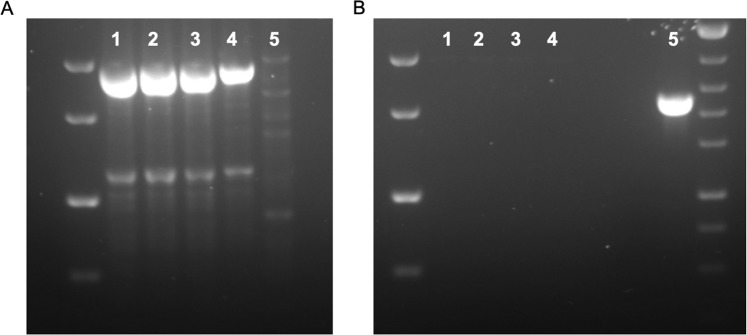
Limitations of the use of PDX tumours in cancer research is infiltrating mouse stroma cells into the PDX tumour. A mouse cell depletion kit was used for the removal of the infiltrating mouse cells. A two step, nested PCR method was used to confirm the absence of mouse cells in the primary cell lines and organoids33. Universal primers amplified a region common in both the human and mouse mitochondrial genomes (Fig. 3A). Mouse specific primers were used to identify if mouse-contaminating cells were present in the samples. Figure 3B confirms that no contaminating mouse mitochondrial DNA is present in the PDX derived organoids, 2D primary cell lines and CLOs.
Figure 3.
Electrophoresis gels of samples (1) PT291 primary cell line (2) PT127 primary cell line (3) PT291 organoids (4) BxPC3 (negative control) (5) Murine cell line C2C12 (positive control) for (A) Products of first PCR and (B) Nested PCR using mouse-specific primers.
Confocal microscopy reveals CLO models representative of derived organoid
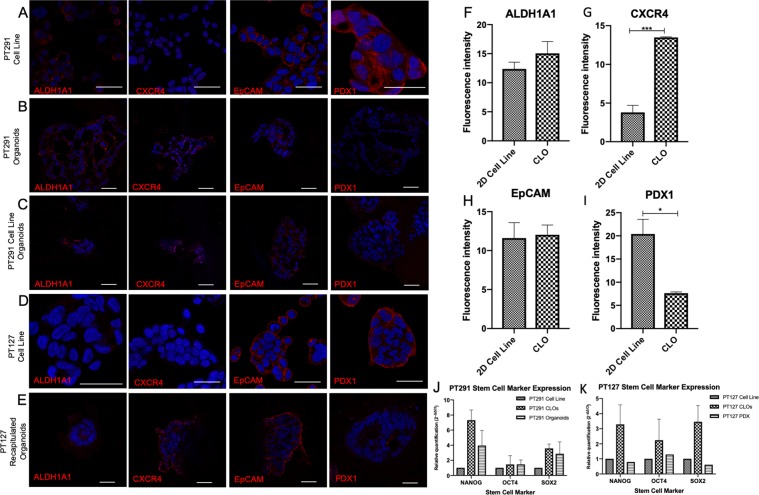
Confocal immunofluorescence was performed in order to determine whether the expression profile of key PDAC tumour initiating markers was maintained in organoids and CLOs. Organoid cultures and derived CLOs were enriched for ALDH1A1 and CXCR4 populations and displayed decreased expression of PDX1 compared to 2D primary cell lines; whereas a similar expression profile for EpCAM was observed in all samples (Fig. 4A–E). To further test for the presence of stem cell markers responsible for self-renewal and propagation in the organoids and CLOs, quantitate PCR (qPCR) was performed. Expression of specific markers OCT4, NANOG and SOX2 was increased in PT291 organoids and CLOs compared to the 2D primary cell lines (Fig. 4J), while PT127 PDX tumour had a similar expression profile to the PT127 CLOs compared to its 2D primary cell line (Fig. 4K).
Figure 4.
Confocal images of immunofluorescence staining images of ALDH1A1, CXCR4, EpCAM, and PDX1 (red) scale bars 40 μm in (A) PT291 2D primary cell line (B) PT291 organoids (C) PT291 CLO (D) PT127 2D primary cell line and (E) PT127 CLO. DAPI counterstain (blue). Images are representative of three independent experiments. (F–I) Analysis comparing the fluorescence intensity between PT291 CLOs and 2D cell lines in (F) ALDH1A1 (G) CXCR5 (H) EpCAM and (I) PDX1. (J,K) Relative quantification of stem cell markers NANOG, OCT4 and SOX2 assessed using qRT-PCR in (J) PT291 PDX derived cell line, organoids and CLOs, and (K) PT127 PDX derived cell line, CLOs and PDX tumour sample. Expression is shown relative to the expression in matched cell line. 18 S rRNA used as endogenous control n = 3.
RNA-seq transcriptomic analysis identifies novel signature defining recapitulated CLO
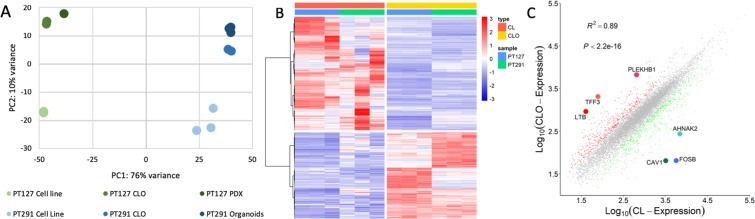
RNA-seq was performed to identify transcriptome signatures of genes dysregulated among PDX organoids/PDX tumour and corresponding CLOs compared to 2D primary cell lines. Principal component analysis (PCA) revealed PT291 organoids and CLOs clustered together; while PT127 PDX tumour and CLOs clustered together, separate from their corresponding isogenetically matched 2D primary cell line (Fig. 5A). Hierarchical clustering was performed of all differentially expressed genes in the grouped CLOs and 2D primary cell lines (PT291 and PT127) (Fig. 5B).
Figure 5.
(A) Principal component analysis was performed to compare the expression profiles of PT127 Cell line, PT127 CLO, PT127 PDX, PT291 cell line, PT291 CLO and PT291 organoids (B) Clustering of all differentially expressed genes (fold change >1, p-adj = 0.001) in CLOs and cell lines (PT127 and PT291) (C) Scatterplot – grouped PT291 and PT127 CLO vs CL, genes with the biggest log fold-change highlighted. FOSB (log fold change −6.496, p-adj 6.65E-50), CAV1 (log fold change −5.7652 p-adj 4.89E-75), AHNAK2 (log fold change −4.7934931, p-adj 9.25E-20), TFF3 (log fold change 4.8678, p-adj 5.86E-32), PLEKHB1 (log fold change 3.803, p-adj 2.59E-13), LTB (log fold change 3.633, p-adj 2.32E-11).
In Fig. 5C, a scatterplot reveals the most differentially expressed genes between grouped PT291 and PT127 CLOs compared to grouped PT291 and PT127 cell line. We identified FOSB (log fold change −6.496, p-adj 6.65E-50), CAV1 (log fold change −5.7652 p-adj 4.89E-75) and AHNAK2 (log fold change −4.7934931, p-adj 9.25E-20) as the three top down regulated genes in the grouped CLOs compared to the grouped 2D primary cell lines. TFF3 (log fold change 4.8678, p-adj 5.86E-32), PLEKHB1 (log fold change 3.803, p-adj 2.59E-13) and LTB (log fold change 3.633, p-adj 2.32E-11) were the top up regulated genes.
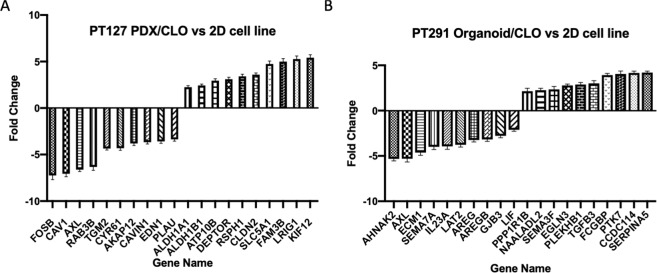
To further identify a specific transcriptomic signature common among the PDX tumour/organoid and CLOs we assessed the transcriptomic signature of the PT127 PDX-derived tumour and CLO compared to the 2D primary cell line, and the PT291 organoid and CLO versus the 2D primary cell line. We observed 144 genes up regulated and 130 genes down regulated (log2 fold change ≥2, p-adj 0.001) in both PT127 PDX tumour/CLO compared to the isogenetically matched cell line in 2D (Supplementary Tables 6 and 7). In PT291, 62 genes were up-regulated and 105 genes down regulated in the organoids and CLOs compared to the isogenetically matched 2D cell line (Supplementary Tables 8 and 9). The 10-most significantly (p-adj ≤ 7.15E-42) dysregulated genes in PT127 PDX and CLO compared to the 2D cell line are shown in Fig. 6A; KIF12 had the highest log2 fold change (log2 fold change 5.422, p-adj = 1.62E-62) and FOSB (log2 fold change −7.241, p-adj 1.82E-55) had the lowest fold change. The 10 most significant (p-adj ≤ 5.79E-14) up and down-regulated genes in PT291 organoids and CLOs compared to the isogenetically matched 2D cell line are outlined in Fig. 6B. SERPINA5 (log2 fold change 4.212, p-adj = 2.17E-14) had the highest log2 fold change, and AHNAK2 (log2 fold change −5.316, p-adj = 1.28E-110) displayed the lowest fold change.
Figure 6.
(A) The 10 most significant (p-adj ≤ 7.15E-42) up and down regulated genes in PT127 CLO/PDX compared to the matched 2D cell line (B) The 10 most significant genes in PT291 CLO/organoid vs 2D cell line.
Experimental Design
The method for the establishment of PDAC PDX organoids and isogenetically matched cell lines was modified from Boj et al.25. Cultures were prepared as outlined below: a mouse cell depletion kit for the removal of the infiltrating mouse cells in the PDX samples, with a PCR based method to confirm the removal of the mouse cells33. Wnt3a, Rspondin3 and Noggin conditioned media was made using cell line L-WRN (ATCC, CRL-3276), and was assessed for the levels for protein using the TOPFlash dual luciferase assay34.
Patient demographics and PDX information
PDAC primary tumour tissue specimens were obtained from Saint Vincent’s University Hospital Ethics and Medical Research Committee, Dublin, Ireland between 2013 and 2017. All patients underwent pancreatic adenocarcinoma resection. All donors agreed by written informed consent to donate tissue and for study participation. The specimen donor’s personal information was confidential and protected except the date of birth. All samples and methods used in this study were approved by Saint Vincent’s University Hospital Ethics and Medical Research Committee and conducted in accordance with the relevant guidelines and regulations in compliance with the Saint Vincent’s University Hospital Ethics and Medical Research Committee. PDAC PDX tissues were generated by subcutaneous seeding in severe combined immunodeficiency disease (SCID) mice. Primary patient samples were confirmed as PDAC by a pathologist (N.S.) in Saint Vincent’s University Hospital, and PDX samples were also confirmed by pathology examination to maintain the human tumour content and morphology of the original tumour.
All animal work has received ethical approval from the DCU Research Ethics Committee (DCUREC/2012/202) and was licensed by Department of Health (B100–4501) all methods were performed in accordance with the relevant guidelines and regulations in compliance with the DCU Research Ethics Committee and the Department of Health.
PDX tumour development
After initial macroscopic pathological confirmation, material remaining after diagnostic sampling was cold transferred in RPMI media containing 1% Pen Step, 1% fungazone to DCU.
The tumour was cut into implant sized pieces (<2 mm) and rinsed with fresh serum-free RPMI media following transport. Severe combined immunodeficiency (SCID), CB17/lcr-Prkdcscid/lcrCrl mice (Charles River, UK) were implanted subcutaneously with fresh patient tumour material. Under anaesthesia (isoflourane, O2 carrier gas) a small incision was made in the skin of the left flank of the animal. The tumour piece was placed in the pocket under the skin and the wound sealed with a single staple. The animals were monitored post-surgery, and staple removal was within 10 days. Animals were monitored weekly for body weight and tumour development. Mice were monitored for tumour development for up to 1 year post implantation. Animal welfare monitoring criteria included tumour volume, tumour axis, body weight and condition, where maximum tumour volume is <2000 mm3, and maximum tumour axis is <20 mm. A decrease in body weight of >10% resulted in increased monitoring, with body weight decrease of 20% resulting in humane euthanasia. Tumour measurements were by calliper measurement and tumour volume was calculated as (HxDxW)/2, where H indicated height, D indicated depth and W indicated width. At experiment end, the animals were humanely euthanized and the tumour was harvested. PDX samples were confirmed by pathology examination to maintain the human tumour content and morphology of the original tumour.
PDX tumour preparation
The media for organoid establishment and culture is represented in Supplementary Tables 1–4. Firstly, a 21 mL solution of 20:1 PBS to PB Buffer (Miltenyi Biotec, 130-091-376), and a 2% PenStrep in PBS solution was prepared. The PDAC PDX tumour sample was received on ice, in basal media. Using a Pasteur pipette, all media was removed and 10 mL of the PBS-PenStrep solution was added to the sample, and the tube was shaken for 20 seconds. The tumour sample was allowed to settle, and the PBS was aspirated. The process was repeated with 10 mL and 15 mL washes. In a petri-dish, the tumour was chopped finely using a scalpel and forceps. The sample was placed in a 50 mL Falcon tube, and 10 mL digestion buffer (Supplementary Table 3) was added. The tube was placed on a rocking table at room temperature for 15 minutes. The tumour sample was removed to a laminar flow hood. The digested tumour sample was put through a MACS SmartStrainers (70 µm) (Miltenyi Biotec, 130-098-462) using a cell scraper into a fresh 50 mL falcon tube. The cell strainer was rinsed with 20 mL wash media (Supplementary Table 2). The cells were centrifuged at 1500 RPM for 7 minutes at room temperature (RT).
Mouse cell depletion kit
Following centrifugation, the media was aspirated, and the pellet was resuspended in 5 mL PB buffer, making a single cell suspension. Using Trypan blue (Sigma, T8154O), the cells were counted and a cell suspension of 2 × 106 cells was centrifuged at 1500 RPM for 10 minutes at RT, and resuspended in 80 μL PB buffer, and 20 μL of the mouse cell depletion kit antibody cocktail (Miltenyi Biotec, 130-104-694). This suspension was incubated at 4 °C for 15 minutes. Following incubation, 400 μL of PB buffer was added to the cell suspension and mixed thoroughly. An LS column (Miltenyi Biotec, 130-042-401) was placed in the MidiMacs Separator (Miltenyi Biotec, 130-042-302) and rinsed with 3 mL PB buffer. The cell suspension was pipetted into the column, and the flow-through was collected in a 15 mL tube. The column was washed twice with 1 mL of PB buffer, and the flow-through was collected in the same tube. The cells were centrifuged at 1000 RPM for 5 minutes at 4 °C. All media was removed, using a P200 to avoid disrupting the pellet.
Organoid establishment
The cells were resuspended in 50 μL ECM (Sigma, E1270, Lot 087M177V 8–12 mg/mL) per 1.5 × 105 cells. 20 μL of organoid:12 mg/ml ECM solution was pipetted into poly-HEMA coated 48-well plate. The plate was placed in an incubator upside down at 37 °C for 20 minutes for ECM to polymerise. 350 μL of the Complete Human Feeding Media (CHFM) (Supplementary Table 4) with RhoK inhibitor was added to each well of the 48 well plate. Two days after establishing the tumour sample, cells were fed using CHFM with RhoK inhibitor, then subsequently fed using CHFM without RhoK inhibitor (Supplementary Table 4).
Establishment of isogenetically matched 2D primary cell line from organoids
Protocol was followed as outlined above in “mouse cell depletion kit”. On day zero, 1.5 × 105 cells were mixed in 100 μL of ECM diluted to 1 mg/mL and 50 μL was plated per well in a non-poly-HEMA coated 24 well plate. Cells were fed 500 μL of the CHFM with RhoK inhibitor (Supplementary Table 4). Cells were cultured as above for a further 10 days, or until the cells began to adhere to the bottom of the plate, feeding with CHFM. To passage cells, 1 mL TrypLE (Gibco, 12605010) was added to each well until the cells detached, and 1 mL of DMEM High-Glucose Glutamax (Gibco, 10566–016) with 10% FBS was added to stop trypsinisation. Cells were grown in a 6-well plate, then up scaled to T25 cm3 flask. Once in the T25 cm3 flask, the media was changed from CHFM to 50:50 GlutaMax DMEM and L-WRN Conditioned Media with 10% FBS and 1% PenStrep.
Passaging organoids
A fresh poly-HEMA coated 48 well plate was preheated to 37 °C. The media from each well was removed to a 30 mL tube, and the well was washed with 300 μL ice cold PBS, which was placed in the tube, and centrifuged at 1500 RPM for 10 minutes at 4 °C. Pellets were washed with 500 μL of ice cold PBS to removed remaining ECM, and centrifuged at 1500 RPM for 10 minutes at 4 °C. 3 mL TrypLE (Gibco, 12605010) was added to the pellet and placed at 37 °C for 15 minutes. The pellet disrupted using “hard pipetting” (using a P1000 pipette, the organoids were pipetted quickly and vigorously). DMEM with 10% serum was added to stop trypsinisation. The organoids established as previously outlined above, and fed with RhoK inhibitor supplemented in CHFM for two days after trypsinisation, and then subsequently fed using CHFM without RhoK inhibitor.
CLO establishment
Using early passage 2D primary cell lines, which had been established from organoids, cells were trypsinised, and a cell suspension of 5 × 104 cells per well to be seeded was made, and centrifuged at 1500 RPM for 10 minutes at 4 °C. Cells were resuspended in 20 μL ECM per well, and plated and fed as outlined above. Two weeks after establishment, cells were passaged as organoids, as outlined above.
Species confirmation PCR
DNA was extracted from organoids and primary cell lines using DNeasy Blood & Tissue Kit (Qiagen, 69504). PCRs were performed using MyTaq™ PCR Kit. The first PCR is performed using a universal primer pair complementary to conserved sequences in cytochrome B and 16S rRNA genes.
100 ng DNA, 10 pmol Primers (Table 1), 0.5 µl MyTaq HS Red Mix, 2 × 12.5 µL Water (ddH2O) up to 25 µl. Amplification was carried out in G-Storm, at 94 °C for 5 minutes, 59 °C for 5 minutes, followed by 35 cycles of 72 °C for 2.5 minutes, 94 °C for 30 seconds, 59 °C for 45 seconds, and followed by a final annealing step of 72 °C for 10 minutes. Samples were stored at 4 °C.
Table 1.
Primers for PCR.
| Forward 5′−3′ | Reverse 5′−3′ | |
|---|---|---|
| Universal | THGTHSAATGAATCTGAGGVGGVT | CGATGTTGGATCAGGACATC |
| Mouse | GCACTGAAAATGCTTAGATGGATAATTG | CCTCTCATAAACGGATGTCTAG |
The amplified product from the first PCR was diluted 1:10 with ddH2O. 1 μL PCR product, 10 pmol Primers (Table 1), 0.5 µl MyTaq HS Red Mix, 2 × 12.5 µl Water (ddH2O) up to 25 µL. Amplification was carried out in G-Storm, 94 °C for 5 minutes, 60 °C for 5 minutes, followed by 30 cycles of 72 °C for 1.5 minutes, 94 °C for 30 seconds, 60 °C for 30 seconds, followed by a final annealing step of 72 °C for 10 minutes. Samples were stored at 4 °C. 5 μL of second PCR product was run on a 2% agarose (Sigma), stained with SafeView (NBS Biologicals, NBS-SV1), visualized under UV light, and imaged. A 100 bp DNA Ladder (ThermoFisher, 15628019) was applied as a size marker.
Brightfield imaging
Brightfield imaging of PT127 CLOs and PT291 CLOs and matched organoids was performed on days 0, 3, 7, and 10 to determine the morphological changes of the cells. Pictures were taken using a Canon EOS 550D camera (Canon GmbH, Krefeld, Germany) through a Leica DM IL LED inverted microscope (Leica Microsystems, Wetzlar, Germany). Scale bars were added using ImageJ.
Immunofluorescence
Cell lines
A cell suspension of 5 × 104 in 500 μL was seeded in a glass bottomed 8-well plate (Ibidi, 80827), and grown overnight. Media was removed from the cells, and washed three times in a PBS 0.1% Tween 20 (Sigma, P1379) and 2% BSA (Sigma, A9418) solution. Cells were fixed in ice-cold methanol for 5 minutes at −20 °C, and washed in the PBS-T-BSA solution.
Organoids and CLOs
Four wells of organoids at day 7 were combined and centrifuged at 1500 RPM for 10 minutes in a 1.5 mL Eppendorf tube. All media was removed from the cell pellet, and iPGell (Funakoshi, PG20–1) was used according to the manufacturer’s protocol. Gel plugs organoids were placed in optimal cutting temperature (OCT) embedding matrix gel (Tissue-Tek, KMA-0100–00A) in a disposable histology mold (Lecia, 3803025) and placed at −80 °C overnight. Organoids were cut into 10 μm sections using a cryostat (Leica, CM 1900) which was cooled to −25 °C prior to use. OCT embedded organoids were mounted onto the freezing block using OCT, and faced off by cutting 60 μm sections until the organoids were exposed. The organoids were cut into 10 μm sections and placed onto SuperFrost Plus slides (ThermoFisher, 4591PLUS4), and stored at 4 °C until further use.
Cells were blocked for 1 hour 30 minutes in 10% Goat Serum (Gibco, 16210064), the primary antibody (Supplementary Table 5) was made in the PBS-T-BSA solution, and was added to the cells overnight at 4 °C. The cells were washed three times in the PBS-T-BSA solution, and the secondary antibody was made up in the PBS-T-BSA solution and added to the cells. The cells were incubated on a rocker for an hour at room temperature in the dark. The secondary antibody was removed, and the cells were washed three times in the PBS-T-BSA solution, DAPI (1:2500) was added to the cells for 3 minutes, and the cells were washed three times in PBS-T-BSA solution. The 24 well coverslips were mounted using ProLong Gold Antifade Mountant (Invitrogen, P36930), and allowed to dry for 24 hours. The immunofluorescence was observed using a Leica TCS SP8 STED super-resolution microscope equipped with a CCD camera and 100 × oil immersion objective. DAPI was excited with a 405 nm picoquant laser unit and emission captured between 387 and 474 nm. Alexa Fluor 488 was excited at 499 nm with emission captured between 490 and 566 nm. Images were acquired whereby combinations of excitation and emission wavelengths for specific dyes were applied sequentially.
Quantitative reverse transcription PCR (qRT-PCR)
A High Capacity cDNA Reverse Transcription Kit (Applied Biosystems 4368814) was used to synthesis cDNA from RNA. Master mix was prepared as described in the protocol. A G-Storm Thermocycler (Model GS1, Somerton Biotechnology Centre) was used to perform cDNA synthesis at 25 °C for 10 minutes for annealing, 37 °C for 2 hours for cDNA synthesis, 85 °C for 5 minutes for enzyme inactivation, and storage at 4 °C. Following synthesis, cDNA was stored at −20 °C. TaqMan qRT-PCR was used to assess NANOG, OCT4 and SOX2 expression in the PDX, organoid, CLO, and cell line panels. TaqMan Gene expression assays (Thermo Scientific, NANOG Hs02387400; POU5F1 Hs04260367 and SOX2 Hs01053049), TaqMan Fast Advanced Master Mix (Applied Biosystems, 4444556), RT-PCR Grade Water (Thermo Scientific, AM9935) and cDNA template were prepared as described in the assay protocol using 20 ng of cDNA, in MicroAmp Fast Optical 96 well reaction plates (Applied Biosystems, 4346907). The plate was sealed using Adhesive PCR Plate Seals (Thermo Scientific, AB0558) and centrifuged briefly. Using an Applied Biosystems 7900 Real-Time PCR System qRT-PCR was performed using 50 °C for 2 minutes, 95 °C for 20 seconds, and 40 cycles of 95 °C for 1 second and 60 °C for 20 seconds.
RNA-sequencing
Organoids and CLOs were grown as described for 14 days. TRI-reagent® (Sigma) was used to lyse cells, and RNA isolation was performed using Direct-zol RNA Miniprep Plus Kit (Zymo Research, R2072). RNA samples were quality controlled with an RNA Screen Tape (Agilent) and quantified with Qubit RNA HS Assay kit. Sequencing libraries were prepared with 200 ng of total RNA for all samples. The Nugen Universal Plus mRNA protocol was followed as recommended by the manufacturer. Libraries were amplified by 15 cycles of PCR. Libraries were sequenced in one NextSeq550 run with the NextSeq500/550 High Output Kit v2.5 (300 cycles), sequencing was done with Paired End 150 nt reads and 8 nt indexes. Libraries were loaded at 1.4 pM, 5% PhiX control was used.
RNA analysis
Raw sequencing files were merged for each sample to generate a single fastq file per sample. Cutadapt was used to remove adapter sequences. Trimmed reads were aligned against the Homo sapiens Ch37 genome (hg19) with hisat2 using default parameters. The resulting sam file was piped into samtools for bam conversion and sorting. The resulting featureCounts object can easily be manipulated to generate a counts matrix for DESeq2. This counts generation is written to counts.tsv for further analysis. Up/down regulated genes: differential expression genes (p < 0.001 and log2 fold-change ≧ 2) are detected using DESeq235 after filtering out reads count less than 10. Compare CLOs with CLs; compare PT127 PDX, CLO with CL; compare PT291 organoids, CLO with CL. The correlation of paired groups - for each sample, reads count is normalised to TPM. Median is applied if there is more than one sample in a group. Linear regression was performed to identify the relationship of paired groups: PT291 ORG and PT127 PDX; PT127 PDX and PT127 CLO; PT291 ORG and PT291 CLO, (p < 0.05). Raw and processed RNA-seq files are available at GEO (accession number GSE144737).
Discussion
In this study, we present a method for the establishment of organoids, and isogenetically matched 2D primary cell lines from PDAC PDX tumour samples and recapitulation of the cell lines to CLOs, which we show are morphologically, molecularly and transcriptomicly similar to their PDX derived tumour organoids. This method can be utilised for all cancer types, allowing laboratories to utilise PDX samples already available for the development of organoids and primary cell lines. While PDX tumours remain stable during passaging, tumour-associated stroma is replaced with murine stromal cells36, with up to 90% of the cells in a PDX tumour being murine stroma28. When establishing primary cell lines from PDX samples, these murine fibroblasts can often quickly out-grow and outnumber the PDAC cancer cells in vitro, resulting in a completely unrepresentative model for PDAC37. In order to overcome this issue, we used mouse cell depletion kit, which results in the removal of infiltrating mouse cells from the tumour sample33. Utilisation of the mouse cell depletion kit to develop organoids and 2D primary cell lines from PDX tumours ensures the starting material is pure tumour tissue.
While organoids are a more representative model of PDAC ex vivo, they are expensive to culture, require time-consuming maintenance and can be difficult to scale up in short periods. In order to overcome these issues, we highlight the establishment of cell line organoids (CLOs). The CLOs are developed from 2D primary cell lines within two passages, grown under the same conditions as PDX organoids. These CLOs were imaged, and found to be morphologically similar to the matched organoids. To validate CLOs as representative organoid models, we molecularly profiled the expression of tumour initiating markers in our cells. Expression of ALDH1A1 and CXCR4 was low in both the PT291 and PT127 2D primary cell lines, but consistently expressed in organoid and CLOs. ALDH activity selectively defines an enhanced tumour-initiating cells38. ALDH1A1 is involved in retinoic acid metabolism, and has a role in proliferation and differentiation39. The expression of the transmembrane glycoprotein, EpCAM was retained in all models, and is known to be highly expressed in PDAC and established cell lines40,41.
Interestingly, PDX1 expression was low in CLOs compared to 2D primary cell lines. PDX1 is a transcription factor which is essential for pancreatic development and differentiation of all pancreatic cell lineages42. The over expression of PDX1 in PDAC highlighted its role as an oncogene43. However, PDX1 expression may be linked to the changing stages during cancer development and progression. PDX1 loss or downregulation is associated with a more aggressive phenotype, and loss of PDX1 observed in subset of cells undergoing EMT44,45. Furthermore, upregulation of key stem cell markers, transcription factors NANOG, OCT4 and SOX2 in CLOs compared to 2D primary cultures indicates a pluripotent stem cell state and suppression of lineage-specific genes in these models46.
RNA-seq analysis highlighted by PCA that the PT291 organoids and CLOs clustered together as did the PT127 PDX-tumour and CLOs compared to their respective 2D primary cell lines. Additionally, hierarchical clustering comparing the dysregulated genes between PT291 and PT127 CLOs versus the 2D primary cell lines, revealed a clear signature pattern as shown by heat map (Fig. 5B). The three top up and down regulated genes in the CLOs vs primary cell lines were TFF3, PLEKHB1, LTB and FOSB, CAV1, AHNAK2, respectively. Interestingly, FOSB is a proto-oncogene, and CAV1 expression has been linked with mammary tumorigenesis and increased lung metastasis47,48. AHNAK2 has also previously been found to be upregulated in PDAC49. The up regulated signature also contains increased expression of genes associated with cilia (KIF12, RSPH1 and CCDC114)50–52. Cilia have been found to play an important role in modulating signalling cascades including the WNT signalling pathway, and are essential in pancreatic tissue organisation53,54. The upregulation of stem cell markers ALDH1A1, ALDH1B1 and LRIG1 may indicate restored embryonic programs and dedifferentiation pattern among our CLOs39,55,56. This dedifferentiation theory is supported by a previous study which has shown that when implanted orthotopically, organoids recapitulate tumour progression from precursor PanIn lesions to metastatic carcinoma25. Interestingly, genes down regulated in PDX tumour/organoids and CLOs include genes which have increased expression in cancers, including potential PDAC biomarkers (CYR61, PLAU, FOSB, ECM1, LIF, GJB3 and CAV1)48,49,57–62. There is also an increased expression of tumour suppressor genes LRIG1 and SEMA3F55,63. This gene signature also highlights the “phenotypic switch” of the 2D the primary cell lines as the cells invade through ECM, attach and proliferate, resulting in increased motility, invasion and proliferation of cells by up-regulation of oncogenic pathways.
A limitation of our study was the use of non-metastatic surgical samples which were implanted as subcutaneous PDX tumours which did not metastasise in the mouse. In order to establish CLOs as an invasive disease model for PDAC, future work will include the establishment of preclinical models from biopsies of metastatic sites. Further limitations of our study and others studying PDAC organoids25,46,64,65 include the lack of a relevant microenvironment integrating the immune and stromal components, as tumour derived organoids exclusive express ductal cells, but not of other pancreatic cell lines46. Tsai et al.66 constructed complex organotypic models containing tumour, stromal and immune components of the tumour microenvironment. This study showed spheroids derived from established PDAC cell line, Panc-1, a mouse xenograft cell line, and the A549 lung cancer cell line are phenotypically distinct from primary organoids as they formed homogenous, non-lumen forming spheres. However, the cell line models used by Tsai et al.66 are not primary cultures, and were not compared to their isogenetically matched organoid or PDX tumour, therefore are not directly comparable and tumour to tumour bias may exist.
Conclusions
As thirty years of cancer research have done little to improve patient outcomes, additional disease modelling strategies are required to complement existing techniques and advance PDAC research. Here, we demonstrate that CLOs established from 2D primary cell lines are a physiologically relevant in vitro organoid-similar model to study cancer. CLOs have the molecular and transcriptomic phenotype resembling organoids, however offer more flexibility, expandability and utility for downstream translational research and for the development of personalised treatment of PDAC patients.
Supplementary information
Acknowledgements
Confocal imaging was carried out at the Nano Research Facility in Dublin City University which was funded under the Programme for Research in Third Level Institutions (PRTLI) Cycle 5. The PRTLI is co-funded through the European Regional Development Fund (ERDF), part of the European Union Structural Funds Programme 2011–2015. This research was supported by a research grant from Science Foundation Ireland (SFI) under the Starting Investigator Research Grant (SIRG) Programme. Grant number 15/SIRG/3482 (N.W). Authors also acknowledge CellFit (COST Action CA16119) and PRECODE (European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement Nº 861196). S.R.N was a recipient of an EACR Travel Fellowship. We would also like to thank Charlotte Andrieu for her bioinformatic assistance.
Author contributions
The project was conceptualized by N.W. N.W. and J.C. oversaw the project. S.R.N. developed the methodology and performed the establishment, characterisation and growth of organoids, CLOs and 2D matched primary cell lines. C.Z. performed genomic evaluation of RNA-seq data. F.O.N. and S.R. performed mouse xenograft experiments. N.S. performed pathologic evaluation. AM.L. and Y.L. provided intellectual input. S.R.N. and N.W. wrote the manuscript text. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-59368-7.
References
- 1.Chiorean EG, Coveler AL. Pancreatic cancer: optimizing treatment options, new, and emerging targeted therapies. Drug. Des. Devel. Ther. 2015;9:3529–3545. doi: 10.2147/DDDT.S60328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rawla Prashanth, Sunkara Tagore, Gaduputi Vinaya. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World Journal of Oncology. 2019;10(1):10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yancik R, Ries LA. Cancer in older persons. Magnitude of the problem–how do we apply what we know? Cancer. 1994;74:1995–2003. doi: 10.1002/1097-0142(19941001)74:7+<1995::AID-CNCR2820741702>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 5.Becker AE, Hernandez YG, Frucht H, Lucas AL. Pancreatic ductal adenocarcinoma: Risk factors, screening, and early detection. World J. Gastroenterol. 2014;20:11182–11198. doi: 10.3748/wjg.v20.i32.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch SM, et al. Cigarette smoking and pancreatic cancer: a pooled analysis from the pancreatic cancer cohort consortium. Am. J. Epidemiol. 2009;170:403–413. doi: 10.1093/aje/kwp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stolzenberg-Solomon RZ, Amundadottir LT. Epidemiology and inherited predisposition for sporadic pancreatic adenocarcinoma. Hematol. Oncol. Clin. North Am. 2015;29:619–640. doi: 10.1016/j.hoc.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arslan AA, et al. Anthropometric measures, body mass index, and pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan) Arch. Intern. Med. 2010;170:791–802. doi: 10.1001/archinternmed.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elena JW, et al. Diabetes and risk of pancreatic cancer: a pooled analysis from the pancreatic cancer cohort consortium. Cancer Causes Control. 2013;24:13–25. doi: 10.1007/s10552-012-0078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucey, B. P., Nelson-Rees, W. A. & Hutchins, G. M. Henrietta Lacks, HeLa cells, and cell culture contamination. Archives of Pathology and Laboratory Medicine, 10.1043/1543-2165-133.9.1463 (2009). [DOI] [PubMed]
- 11.Gillet J.-P., Varma S., Gottesman M. M. The Clinical Relevance of Cancer Cell Lines. JNCI Journal of the National Cancer Institute. 2013;105(7):452–458. doi: 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson-Rees W. A., Owens R. B., Arnstein P., Kniazeff A. J. Source, alterations, characteristics and use of a new dog cell line (Cf2Th) In Vitro. 1976;12(10):665–669. doi: 10.1007/BF02797468. [DOI] [PubMed] [Google Scholar]
- 13.Goodspeed A., Heiser L. M., Gray J. W., Costello J. C. Tumor-Derived Cell Lines as Molecular Models of Cancer Pharmacogenomics. Molecular Cancer Research. 2015;14(1):3–13. doi: 10.1158/1541-7786.MCR-15-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deer EL, et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–35. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zieba J, et al. Sensitivity of neoplastic cells to senescence unveiled under standard cell culture conditions. Anticancer Res. 2015;35:2759–68. [PubMed] [Google Scholar]
- 16.Kodack DP, et al. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Rep. 2017;21:3298–3309. doi: 10.1016/j.celrep.2017.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia (New York, N.Y.) 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoma CR, Zimmermann M, Agarkova I, Kelm JM, Krek W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Advanced Drug Delivery Reviews. 2014;69–70:29–41. doi: 10.1016/j.addr.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Zanoni M, et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016;6:19103. doi: 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JB. Three-dimensional tissue culture models in cancer biology. Semin. Cancer Biol. 2005;15:365–377. doi: 10.1016/j.semcancer.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Lancaster M. A., Knoblich J. A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125–1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 22.Clevers Hans. Modeling Development and Disease with Organoids. Cell. 2016;165(7):1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Fang-Xu, Harrison Leonard C. Laminin-1 and epidermal growth factor family members co-stimulate fetal pancreas cell proliferation and colony formation. Differentiation. 2005;73(1):45–49. doi: 10.1111/j.1432-0436.2005.07301002.x. [DOI] [PubMed] [Google Scholar]
- 24.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–21. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boj Sylvia F., Hwang Chang-Il, Baker Lindsey A., Chio Iok In Christine, Engle Dannielle D., Corbo Vincenzo, Jager Myrthe, Ponz-Sarvise Mariano, Tiriac Hervé, Spector Mona S., Gracanin Ana, Oni Tobiloba, Yu Kenneth H., van Boxtel Ruben, Huch Meritxell, Rivera Keith D., Wilson John P., Feigin Michael E., Öhlund Daniel, Handly-Santana Abram, Ardito-Abraham Christine M., Ludwig Michael, Elyada Ela, Alagesan Brinda, Biffi Giulia, Yordanov Georgi N., Delcuze Bethany, Creighton Brianna, Wright Kevin, Park Youngkyu, Morsink Folkert H.M., Molenaar I. Quintus, Borel Rinkes Inne H., Cuppen Edwin, Hao Yuan, Jin Ying, Nijman Isaac J., Iacobuzio-Donahue Christine, Leach Steven D., Pappin Darryl J., Hammell Molly, Klimstra David S., Basturk Olca, Hruban Ralph H., Offerhaus George Johan, Vries Robert G.J., Clevers Hans, Tuveson David A. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell. 2015;160(1-2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung Jaeyun, Seol Hyang Sook, Chang Suhwan. The Generation and Application of Patient-Derived Xenograft Model for Cancer Research. Cancer Research and Treatment. 2018;50(1):1–10. doi: 10.4143/crt.2017.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Festing Simon, Wilkinson Robin. The ethics of animal research. EMBO reports. 2007;8(6):526–530. doi: 10.1038/sj.embor.7400993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneeberger Valentina E., Allaj Viola, Gardner Eric E., Poirier J. T., Rudin Charles M. Quantitation of Murine Stroma and Selective Purification of the Human Tumor Component of Patient-Derived Xenografts for Genomic Analysis. PLOS ONE. 2016;11(9):e0160587. doi: 10.1371/journal.pone.0160587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eirew P, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jimeno A., Rubio-Viqueira B., Rajeshkumar N.V., Chan A., Solomon A., Hidalgo M. A Fine-Needle Aspirate-Based Vulnerability Assay Identifies Polo-Like Kinase 1 as a Mediator of Gemcitabine Resistance in Pancreatic Cancer. Molecular Cancer Therapeutics. 2010;9(2):311–318. doi: 10.1158/1535-7163.MCT-09-0693. [DOI] [PubMed] [Google Scholar]
- 31.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 32.Tiriac H, et al. Successful creation of pancreatic cancer organoids by means of EUS-guided fine-needle biopsy sampling for personalized cancer treatment. Gastrointest. Endosc. 2018;87:1474–1480. doi: 10.1016/j.gie.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono Kazumi, Satoh Motonobu, Yoshida Touho, Ozawa Yutaka, Kohara Arihiro, Takeuchi Masao, Mizusawa Hiroshi, Sawada Hidekazu. Species identification of animal cells by nested PCR targeted to mitochondrial DNA. In Vitro Cellular & Developmental Biology - Animal. 2007;43(5-6):168–175. doi: 10.1007/s11626-007-9033-5. [DOI] [PubMed] [Google Scholar]
- 34.Veeman Michael T., Slusarski Diane C., Kaykas Ajamete, Louie Sarah Hallagan, Moon Randall T. Zebrafish Prickle, a Modulator of Noncanonical Wnt/Fz Signaling, Regulates Gastrulation Movements. Current Biology. 2003;13(8):680–685. doi: 10.1016/S0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 35.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 10.1186/s13059-014-0550-8. (2014) [DOI] [PMC free article] [PubMed]
- 36.Hidalgo Manuel, Amant Frederic, Biankin Andrew V., Budinská Eva, Byrne Annette T., Caldas Carlos, Clarke Robert B., de Jong Steven, Jonkers Jos, Mælandsmo Gunhild Mari, Roman-Roman Sergio, Seoane Joan, Trusolino Livio, Villanueva Alberto. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discovery. 2014;4(9):998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Agosto Sabrina, Andreani Silvia, Scarpa Aldo, Corbo Vincenzo. Preclinical Modelling of PDA: Is Organoid the New Black? International Journal of Molecular Sciences. 2019;20(11):2766. doi: 10.3390/ijms20112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim MP, et al. ALDH Activity Selectively Defines an Enhanced Tumor-Initiating Cell Population Relative to CD133 Expression in Human Pancreatic Adenocarcinoma. PLoS One. 2011;6:e20636. doi: 10.1371/journal.pone.0020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomita, H., Tanaka, K., Tanaka, T. & Hara, A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget, 10.18632/oncotarget.6920. (2016) [DOI] [PMC free article] [PubMed]
- 40.Skoda Jan, Hermanova Marketa, Loja Tomas, Nemec Pavel, Neradil Jakub, Karasek Petr, Veselska Renata. Co-Expression of Cancer Stem Cell Markers Corresponds to a Pro-Tumorigenic Expression Profile in Pancreatic Adenocarcinoma. PLOS ONE. 2016;11(7):e0159255. doi: 10.1371/journal.pone.0159255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng, Y. et al. Cytoplasmic EpCAM over-expression is associated with favorable clinical outcomes in pancreatic cancer patients with hepatitis B virus negative infection. Int. J. Clin. Exp. Med. (2015). [PMC free article] [PubMed]
- 42.Dassaye Reshmi, Naidoo Strini, Cerf Marlon E. Transcription factor regulation of pancreatic organogenesis, differentiation and maturation. Islets. 2015;8(1):13–34. doi: 10.1080/19382014.2015.1075687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Shihe, Ballian Nikiforos, Belaguli Narasimhaswamy S., Patel Sanjeet, Li Min, Templeton Nancy Smyth, Gingras Marie-Claude, Gibbs Richard, Fisher William, Brunicardi F. Charles. PDX-1 Acts as a Potential Molecular Target for Treatment of Human Pancreatic Cancer. Pancreas. 2008;37(2):210–220. doi: 10.1097/MPA.0b013e31816a4a33. [DOI] [PubMed] [Google Scholar]
- 44.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 45.Roy Nilotpal, Takeuchi Kenneth K., Ruggeri Jeanine M., Bailey Peter, Chang David, Li Joey, Leonhardt Laura, Puri Sapna, Hoffman Megan T., Gao Shan, Halbrook Christopher J., Song Yan, Ljungman Mats, Malik Shivani, Wright Christopher V.E., Dawson David W., Biankin Andrew V., Hebrok Matthias, Crawford Howard C. PDX1 dynamically regulates pancreatic ductal adenocarcinoma initiation and maintenance. Genes & Development. 2016;30(24):2669–2683. doi: 10.1101/gad.291021.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hohwieler Meike, Müller Martin, Frappart Pierre-Olivier, Heller Sandra. Pancreatic Progenitors and Organoids as a Prerequisite to Model Pancreatic Diseases and Cancer. Stem Cells International. 2019;2019:1–11. doi: 10.1155/2019/9301382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams Terence M., Medina Freddy, Badano Ines, Hazan Rachel B., Hutchinson John, Muller William J., Chopra Neeru G., Scherer Philipp E., Pestell Richard G., Lisanti Michael P. Caveolin-1 Gene Disruption Promotes Mammary Tumorigenesis and Dramatically Enhances Lung Metastasisin Vivo. Journal of Biological Chemistry. 2004;279(49):51630–51646. doi: 10.1074/jbc.M409214200. [DOI] [PubMed] [Google Scholar]
- 48.Cervantes-Madrid Diana Lizeth, Nagi Sabah, Asting Gustafsson Annika. FosB transcription factor regulates COX-2 expression in colorectal cancer cells without affecting PGE2 expression. Oncology Letters. 2017;13(3):1411–1416. doi: 10.3892/ol.2017.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu, D., Wang, J., Shi, X., Yue, B. & Hao, J. AHNAK2 is a potential prognostic biomarker in patients with PDAC. Oncotarget, 10.18632/oncotarget.15990 (2017). [DOI] [PMC free article] [PubMed]
- 50.Mrug Michal, Zhou Juling, Yang Chaozhe, Aronow Bruce J., Cui Xiangqin, Schoeb Trenton R., Siegal Gene P., Yoder Bradley K, Guay-Woodford Lisa M. Genetic and Informatic Analyses Implicate Kif12 as a Candidate Gene within the Mpkd2 Locus That Modulates Renal Cystic Disease Severity in the Cys1cpk Mouse. PLOS ONE. 2015;10(8):e0135678. doi: 10.1371/journal.pone.0135678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cano David A., Sekine Shigeki, Hebrok Matthias. Primary Cilia Deletion in Pancreatic Epithelial Cells Results in Cyst Formation and Pancreatitis. Gastroenterology. 2006;131(6):1856–1869. doi: 10.1053/j.gastro.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 52.Li Ping, He Yani, Cai Guangyan, Xiao Fei, Yang Jie, Li Qinggang, Chen Xiangmei. CCDC114 is mutated in patient with a complex phenotype combining primary ciliary dyskinesia, sensorineural deafness, and renal disease. Journal of Human Genetics. 2018;64(1):39–48. doi: 10.1038/s10038-018-0514-z. [DOI] [PubMed] [Google Scholar]
- 53.Goetz Sarah C., Anderson Kathryn V. The primary cilium: a signalling centre during vertebrate development. Nature Reviews Genetics. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.diIorio Philip, Rittenhouse Ann R., Bortell Rita, Jurczyk Agata. Role of cilia in normal pancreas function and in diseased states. Birth Defects Research Part C: Embryo Today: Reviews. 2014;102(2):126–138. doi: 10.1002/bdrc.21064. [DOI] [PubMed] [Google Scholar]
- 55.Powell Anne E., Wang Yang, Li Yina, Poulin Emily J., Means Anna L., Washington Mary K., Higginbotham James N., Juchheim Alwin, Prasad Nripesh, Levy Shawn E., Guo Yan, Shyr Yu, Aronow Bruce J., Haigis Kevin M., Franklin Jeffrey L., Coffey Robert J. The Pan-ErbB Negative Regulator Lrig1 Is an Intestinal Stem Cell Marker that Functions as a Tumor Suppressor. Cell. 2012;149(1):146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ioannou Marilia, Serafimidis Ioannis, Arnes Luis, Sussel Lori, Singh Surendra, Vasiliou Vasilis, Gavalas Anthony. ALDH1B1 is a potential stem/progenitor marker for multiple pancreas progenitor pools. Developmental Biology. 2013;374(1):153–163. doi: 10.1016/j.ydbio.2012.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chatterjee, M. et al. Caveolin-1 is associated with tumor progression and confers a multi-modality resistance phenotype in pancreatic cancer. Sci. Rep., 10.1038/srep10867. (2015) [DOI] [PMC free article] [PubMed]
- 58.Tsai Miaw-Sheue, Bogart Daphne F, Castañeda Jessica M, Li Patricia, Lupu Ruth. Cyr61 promotes breast tumorigenesis and cancer progression. Oncogene. 2002;21(53):8178–8185. doi: 10.1038/sj.onc.1205682. [DOI] [PubMed] [Google Scholar]
- 59.Harris N L E, Vennin C, Conway J R W, Vine K L, Pinese M, Cowley M J, Shearer R F, Lucas M C, Herrmann D, Allam A H, Pajic M, Morton J P, Biankin A V, Ranson M, Timpson P, Saunders D N. SerpinB2 regulates stromal remodelling and local invasion in pancreatic cancer. Oncogene. 2017;36(30):4288–4298. doi: 10.1038/onc.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Luping, Yu Jiyao, Ni Jian, Xu Xue-Ming, Wang Jianjin, Ning Haoyong, Pei Xu-Fang, Chen Jinguo, Yang Shanmin, Underhill Charles B., Liu Lei, Liekens Joeri, Merregaert Joseph, Zhang Lurong. Extracellular matrix protein 1 (ECM1) is over-expressed in malignant epithelial tumors. Cancer Letters. 2003;200(1):57–67. doi: 10.1016/S0304-3835(03)00350-1. [DOI] [PubMed] [Google Scholar]
- 61.Saito Tomoaki, Kasamatsu Atsushi, Ogawara Katsunori, Miyamoto Isao, Saito Kengo, Iyoda Manabu, Suzuki Takane, Endo-Sakamoto Yosuke, Shiiba Masashi, Tanzawa Hideki, Uzawa Katsuhiro. Semaphorin7A Promotion of Tumoral Growth and Metastasis in Human Oral Cancer by Regulation of G1 Cell Cycle and Matrix Metalloproteases: Possible Contribution to Tumoral Angiogenesis. PLOS ONE. 2015;10(9):e0137923. doi: 10.1371/journal.pone.0137923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bressy Christian, Lac Sophie, Nigri Jérémy, Leca Julie, Roques Julie, Lavaut Marie-Nöelle, Secq Véronique, Guillaumond Fabienne, Bui Thi-Thien, Pietrasz Daniel, Granjeaud Samuel, Bachet Jean-Baptiste, Ouaissi Mehdi, Iovanna Juan, Vasseur Sophie, Tomasini Richard. LIF Drives Neural Remodeling in Pancreatic Cancer and Offers a New Candidate Biomarker. Cancer Research. 2017;78(4):909–921. doi: 10.1158/0008-5472.CAN-15-2790. [DOI] [PubMed] [Google Scholar]
- 63.Roche, J. et al. Distinct 3p21.3 deletions in lung cancer and identification of a new human semaphorin. Oncogene (1996). [PubMed]
- 64.Hou S, et al. Advanced Development of Primary Pancreatic Organoid Tumor Models for High-Throughput Phenotypic Drug Screening. SLAS Discov. Adv. life Sci. R D. 2018;23:574–584. doi: 10.1177/2472555218766842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cristobal A, Van Den Toorn HWP, Van De Wetering M, Clevers H, Heck AJR. Personalized Proteome Profiles of Healthy and Tumor Human Colon Organoids Reveal Both Individual Diversity and Basic Features of Colorectal Cancer. CellReports. 2017;18:263–274. doi: 10.1016/j.celrep.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 66.Tsai, S. et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models, 10.1186/s12885-018-4238-4 (2018). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.