Abstract
Patients infected with the Dengue virus (DENV) often present with a massive generation of DENV-specific antibody-secreting cells (ASCs) in the blood. In some cases, these ASCs represent more than 50% of the circulating B cells, a higher magnitude than those induced by other infections, vaccinations, and plasma cell lymphomas. However, it remains unclear how the DENV infection elicits this colossal response. To address this issue, we utilised an in vitro strategy to induce human PBMCs of healthy individuals incubated with DENV particles (DENV4 TVP/360) to differentiate into ASCs. As controls, PBMCs were incubated with a mitogen cocktail or supernatants of uninfected C6/36 cells (mock). The ASC phenotype and function were increasingly detected in the DENV and mitogen-cultured PBMCs as compared to mock-treated cells. In contrast to the in vivo condition, secreted IgG derived from the PBMC-DENV culture was not DENV-specific. Lower ASC numbers were observed when inactivated viral particles or purified B cells were added to the cultures. The physical contact was essential between B cells and the remaining PBMCs for the DENV-mediated ASC response. Considering the evidence for the activation of the tryptophan metabolism detected in the serum of Dengue patients, we assessed its relevance in the DENV-mediated ASC differentiation. For this, tryptophan and its respective metabolites were quantified in the supernatants of cell cultures through mass spectrophotometry. Tryptophan depletion and kynurenine accumulation were found in the supernatants of PBMC-DENV cultures, which presented enhanced detection of indoleamine 2,3-dioxygenase 1 and 2 transcripts as compared to controls. In PBMC-DENV cultures, tryptophan and kynurenine levels strongly correlated to the respective ASC numbers, while the kynurenine levels were directly proportional to the secreted IgG titers. Contrastingly, PBMCs incubated with Zika or attenuated Yellow Fever viruses showed no correlation between their kynurenine concentrations and ASC numbers. Therefore, our data revealed the existence of distinct pathways for the DENV-mediated ASC differentiation and suggest the involvement of the tryptophan metabolism in this cellular process triggered by flavivirus infections.
Keywords: Dengue virus, B cell differentiation, antibody-secreting cells, tryptophan metabolism, flaviviruses
Introduction
Dengue is an acute infectious disease caused by Dengue virus (DENV) transmission through mosquito bites (Aedes aegypti or Aedes albopictus). This flavivirus-related disease has been widespread in tropical countries. Its global burden reaches nearly eight billion dollars/year due to the morbidity and mortality derived from almost 400 million infections. Moreover, 40% of the world population remains at risk for that viral infection (1). All four DENV serotypes (1–4) can similarly infect and elicit pathophysiology in humans. Regarding the clinical forms of the disease, DENV-infected individuals can be anything from asymptomatic to displaying mild or even severe symptoms that range from hemorrhage or shock syndrome to death (2).
This viral infection triggers robust innate and adaptive immune responses. About the innate responses, DENV particles stimulate the interferon (IFN) production (3, 4) upon the cell invasion and replication that occurs in multiple cell subsets, including monocytes as well as T and B lymphocytes (5). The augmented expression of IFN-stimulated genes ends up activating multiple cell signalling pathways in several cell subsets. Among them, the IFN-gamma interference is distinguished in the activity of some metabolic enzymes, such as the indoleamine 2,3-dioxygenase (IDO) (6). During the DENV infection, IDO promotes the tryptophan degradation, resulting in the accumulation of kynurenine and subsequent metabolites in the patient serum concomitantly with the symptomatology (7). Regarding the adaptive responses, T and B cells are activated upon DENV infection, leading to the further cell differentiation and secretion of DENV-specific neutralizing antibodies (nAbs). Despite the potent nAbs generated against the primary DENV serotype infection, some of them are quite effective against a different viral serotype from a secondary infection (8–10).
Intriguingly, the DENV infection triggers a substantial and transient antibody-secreting cell (ASC) response simultaneously to the symptomatology. Those DENV infection-elicited ASCs are highly specific to viral envelope epitopes and represent more than 50% of all circulating B cells on average. That percentage can still rise if a more severe DENV infection or a secondary viral exposure with a distinct serotype is considered (11, 12). Quantitatively, this DENV-specific ASC response presents a much higher magnitude than the counterparts induced by viral infections (Ebola, Andes, Influenza) (13–15), immunisations (attenuated Yellow Fever virus 17DD or inactivated Influenza vaccine) (11–16), and plasma cell lymphoma (17).
In order to elucidate the underlying requirements behind this massive DENV-induced ASC response seen in humans, we initially developed an in vitro assay based on the culture of PBMCs from healthy individuals. Then, we investigated the influence of certain parameters over the B cell ability to acquire the ASC phenotype and function: (a) viable vs. inactivated DENV particles; (b) PBMCs vs. purified CD19+ B cells; and (c) lack of cell-cell contact between purified CD19+ B cells and remaining PBMCs. Also, we evaluated whether those PBMC cultures with DENV or flaviviruses, such as Zika and Yellow Fever, had the tryptophan metabolism activated and whether they correlated with their respective ASC generation and IgG secretion.
Materials and Methods
Viruses, Blood Samples, and Cell Cultures
All flavivirus strains used in this study are part of the viral collection CVAM from the Laboratório de Virologia Molecular of the Instituto Carlos Chagas/Fiocruz-PR (ICC/Fiocruz-PR) (Brazil). Among them, we tested Dengue viruses [the lab-adapted DENV4 TVP/ 360 and clinical isolate DENV4 LRV13/422 (Genbank accession number KU513442 and KU513441, respectively)], Zika viruses [an African lineage (ZIKV-MR766) (18) and ZIKV-PE243, a Brazilian isolate from Asian lineage] (19), and the attenuated Yellow Fever virus used for vaccination (YFV 17DD) (20). All the DENV and ZIKV strains used in this study were previously expanded and isolated from C6/36 cells (A. albopictus mosquitoes) except for the attenuated yellow fever virus strain 17DD particles that were grown in Vero cell cultures.
Eight to ten millilitres of peripheral blood were collected from healthy adult individuals at the University of São Paulo and Instituto Carlos Chagas, Fiocruz/PR, Brazil. After the Ficoll gradient isolation, PBMCs were cultivated in different conditions to have their B cells differentiated into antibody-secreting cells (ASCs). For this, PBMCs were incubated at 37°C in a 5% CO2 incubator with individual flaviviruses with a multiplicity of infection (MOI) of 10 for 7 days (5). As controls, PBMCs were cultivated with the supernatant of uninfected C6/36 cells (Mock) or a mitogen cocktail [Pokeweed mitogen (PWM), Streptococcus aureus cowan (SAC), CpG ODN 2006 and β-mercaptoethanol] as previously described (21). To have their potential to differentiate into ASCs, CD19+ B cells were isolated from PBMCs through positive selection with magnetic beads (Miltenyi) before being stimulated. All experimental protocols and procedures were reviewed and approved by the Ethics Committee regulated by the Conselho Nacional de Ética em Pesquisa (Process No. 68875417.9.0000.0067).
Flow Cytometry
The cell staining for the ASC phenotype (CD20neg CD27hi CD38hi) was performed on the seventh day of culture with harvested cells using the described antibodies (Supplementary Table 1). Cell staining was performed for 30 min at 4°C in the dark. After two washing steps, the cell acquisition was done in a BD FACSCanto™ II from the PDTIS flow cytometry facility (ICC/Fiocruz-PR). The details of these analyses were evaluated with the FlowJo software.
B Cell ELISPOT
The enumeration of IgG-secreting cells was performed on day 7 of those cultures as previously described (22). A heatmap was designed based on Z-scores. Due to the wide range of spot enumeration per sample, those values were transformed into a normalised basic unit of comparison called the Z-score. Positive values indicated that the data were above average, and negative values were below average.
Mass Spectrometry
The supernatants of cell cultures from day 7 were gathered (nearly 300 μL) and analysed for the presence of tryptophan-derived metabolites. For this, they were initially incubated with 700 μL of a 0.1% acetic acid solution containing methanol:acetone (1:1). Then, 10 μL of a standard mix solution (MLT-D4 e Trp-D5) was added before homogenisation with a vortex for 1 min. The homogenised solution was stored for 30 min at −20°C for protein precipitation. A 14,000 × g centrifugation was applied to the tubes for 10 min at 4°C, and the supernatants were transferred to new tubes under the nitrogen gas atmosphere for drying. Samples were reconstituted in 100 μL of a solution containing water:methanol (9:1), and the injection volume corresponded to 2 μL applied at a flow rate of 200 μL/min into a LC-MS/MS [LC 1250 Bin Pump VL and 1260 HiP ALS autosampler coupled to a triple quadrupole 6460 mass spectrometer (Agilent Technologies, CA, USA)]. The dependence linearity of the concentration response was verified by regression analysis. The electrospray ion source (ESI) was operated in the positive mode. MS data were acquired by multiple reaction monitoring (MRM) mode and evaluated through the MassHunter Quantitative Analysis software (Agilent Technologies). A Luna C18 [150 mm × 2 mm, 3 μm] reversed-phase column (Phenomenex, CA, USA) was used for the LC separation. Chromatography was performed through a gradient elution composed by water, 0.5% formic acid and 2 mM ammonium formate in an mix of acetonitrile:water (9:1) (adjusted with 0.5% formic acid). Data normalisation for each metabolite were performed by subtracting the sample values from those obtained with culture medium kept under the same conditions without cells.
cDNA Synthesis and Quantitative Real Time PCR
The gene expression analyses were measured through quantitative real time PCR (qRT-PCR) with a 7500 FAST Real Time System thermocycler™ (Thermo Fisher Scientific) as described below. Briefly, 2 μg of total RNA of those cell cultures were used as templates for the cDNA synthesis. For this, we used the SuperScript Vilo Master Mix™ (Life Technologies) and ultra-pure water as recommended by the manufacturer. The reaction was run in a thermocycler with adjusted settings: 65°C for 5 min, 50°C for 120 min, and 95°C for 5 min. cDNA samples were stored in a −20°C freezer until the use.
Different genes potentially related to the ASC differentiation were targeted (SYK, IL-10, SRC, TNFS13B, IDO1, and IDO2) with those cDNAs. Those targets were amplified with the SYBR™ Green Master Mix (Thermo Fisher Scientific) and the following primers (Supplementary Table 2). The PCR settings were 50°C for 2 min, 95°C for 10 min, 45 cycles of 95°C for 15 s, and 60°C for 1 min. The relative gene expression (ΔCt) was calculated by the difference between the cycle threshold (Ct) of the target gene and the Ct of the reference gene. The fold change data were obtained through the formula 2∧-ΔΔCt = (ΔCt of treated group – ΔCt of the control group) (23), using the Mock conditions as the control group.
The presence of intracellular viral particles was also estimated from the cell cultures through qRT-PCR. DENV-specific primers (Supplementary Table 2) were used to amplify those cDNAs with the SYBR™ Green Master Mix. For this, the PCR settings were 50°C for 2 min, 95°C for 10 min, 45 cycles of 95°C for 15 s, and 67°C for 1 min.
ELISA
Plasma samples derived from the healthy individuals were assessed for the presence of anti-DENV1-4 IgG antibodies through ELISA assays against NS1 and EDIII recombinant proteins and viral particles. Briefly, ELISA plates were coated with a pool of recombinant proteins (DENV1-4 rNS1 or EDIII - 550 ng/well) or viral particles (DENV1-4 - 200 ng/well) in carbonate buffer (pH 9.6) overnight at 4°C. After four washing steps with a 0.5% Tween 20-PBS solution, the plate wells were blocked for 2 h at 37°C with a 3% skimmed milk solution containing 0.5% fetal bovine serum (FBS) in a 1X PBS–Tween 0.05% solution. Serum samples were further diluted 1:100 and added to the plates in a serial dilution of 2. After a washing step with a 1X PBS–Tween 0.05% solution, a goat anti-IgG human secondary antibody was added to the plates in a 1:4,000 dilution, which incubation lasts 2 h. Revelation of the assay was performed with a specific buffer (citrate phosphate buffer (pH 5.8), o-Phenylenediamine and hydrogen peroxide) for 15 min in the dark upon another washing step. The revelation was stopped with 2N sulphuric acid (H2SO4), and the absorbance was measured at 492 nm in an ELISA plate reader (Epoch).
Flavivirus Titration
Viral titers of DENV4 TVP and ZIKV PE243 were determined in the supernatants of cell cultures on day 7. Briefly, 100 μL of frozen supernatants were thawed and serially diluted 6-fold in Leibovitz's medium 15 (L-15) without FBS and supplemented with 0.26% of tryptose and 25 μg/mL gentamicin. Each dilution was inoculated into C6/36 cells (105 cells/well in 24-well-plates) in duplicate in a well-containing 105 cells and the culture was kept at 28°C for 90 min for the virus adsorption. Then, the inoculum was removed out from the plate and 0.5 mL of CMC overlay medium (L-15 supplemented with 5% SFB, 0.26% tryptose, 25 μg/mL gentamicin, 1.6% carboxymethylcellulose) was added in each plate well. Plates were sealed and incubated for 7 days at 28°C. Then, cells were washed thrice with 1X PBS and fixed with 3% paraformaldehyde solution upon incubation for 20 min. After three washing steps with PBS 1X, cells were permeabilised with 0.5% Triton X-100 solution for 4 min. Cells were washed thrice again with 1X PBS and stained with the anti-envelope protein of flaviviruses 4G2 antibody (ATCC® HB-112™) and diluted 1:200 at 37°C for 60 min. Plates were washed with 1X PBS thrice before their incubation with the anti-mouse alkaline phosphatase (AP) secondary antibody (Promega, Madison, WI, USA) and diluted 1:7,500 at 37°C for 60 min. After the last washing step with 1X PBS, focus forming units (FFUs) were detected by adding a solution of NBT/BCIP (Promega, Madison, WI, USA) as a substrate. Foci were counted and expressed as FFU C6/36/ml.
Statistical Analyses
These analyses were performed with the software GraphPad Prism for Windows, version 6.0 (GraphPad Software, Inc., La Jolla, CA, EUA). The parametric quantitative variables were compared through a T-test or One-way ANOVA with multiple comparisons through the Bonferroni test. The non-parametric quantitative variables were compared through the Mann-Whitney or by Kruskal-Wallis tests with multiple comparisons through the Dunn's test. A p-value (p < 0.05) represented a significant difference among two groups.
Results
DENV Particles Promote the Acquisition of the ASC Phenotype and Function by Human PBMCs in vitro
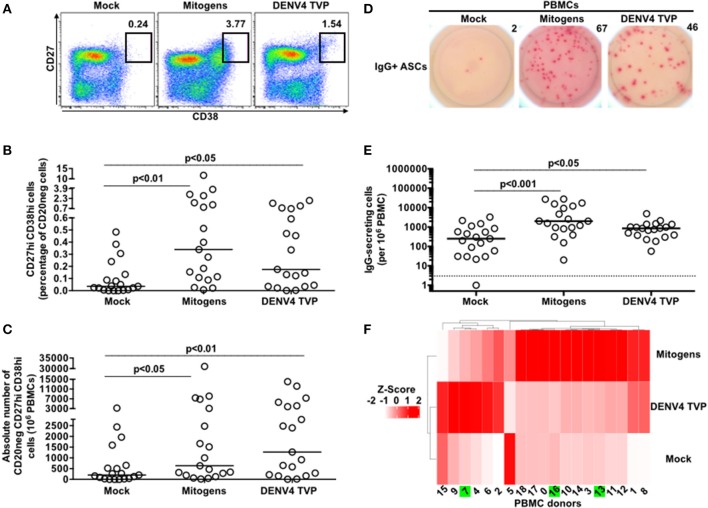
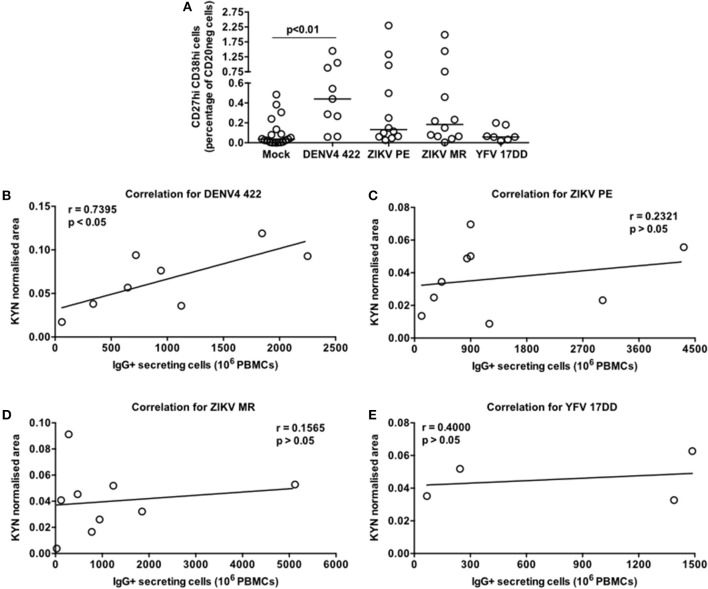
To estimate the ability of DENV particles to elicit the massive B cell differentiation into ASCs, as seen with human infections, we quantified the ASC phenotype and function in human PBMCs cultivated with the DENV4 TVP strain (TVP/360), a laboratory-adapted reference strain (24). For this, PBMCs derived from 19 healthy individuals were incubated for 7 days with DENV4 TVP particles at MOI of 10. As controls, PBMCs were cultivated with the supernatant of uninfected C6/36 cells (Mock) or a cocktail of Mitogens (CpG ODN, Pokeweed mitogen, and Staphylococcus aureus cowan). For the ASC phenotype analysis, we assessed the CD20neg CD27hi CD38hi cell phenotype in the cultures through FACS (Supplementary Figure 1 and Supplementary Table 1). A significant increase in the frequency of cells with the ASC phenotype was detected upon cell culture either with DENV4 TVP (p < 0.05; Median and St Dev – 0.175 ± 0.558%) or Mitogens (p < 0.01; Median and St Dev – 0.339 ± 2.731%) in comparison to the mock (Median and St Dev – 0.036 ± 0.144%) (Figures 1A,B). In terms of absolute number of ASCs detected per 106 PBMCs, both DENV4 TVP (p < 0.01; Median and St Dev – 1,272 ± 4,099) or Mitogen cultures (p < 0.05; Median and St Dev – 635 ± 7,170) presented an increased ASC number in comparison to the mock-treated cells (Median and St Dev – 200 ± 948) (Figure 1C). Subsequently, we evaluated whether the incubation with DENV4 TVP prompted the ASC function (i.e., antibody secretion) in those cells through ELISPOT. A significant augment in the frequency of IgG-secreting cells was enumerated with either the DENV4 TVP (p < 0.05; Median and St Dev – 853 ± 1,069%) or Mitogens (p < 0.001; Median and St Dev – 2,100 ± 9,014) in relation to the mock-treated culture (Median and St Dev – 250 ± 866) (Figures 1D,E). Similar data were also obtained when IgM-secreting ASCs were counted (Supplementary Figure 2). Regarding the IgG secretion, PBMCs cultured with DENV particles were previously shown to present IgG in the supernatants at day 12 of the culture (25). Our data showed that total IgG could be found already at day 7 of the culture (Supplementary Figure 3A). However, all the detected IgG seem to be unspecific to DENV particles (Supplementary Figures 3B,C).
Figure 1.
Dengue virus promotes the B cell differentiation into antibody-secreting cells (ASCs) in vitro disregarding pre-existing DENV immunity. (A) Representative FACS data displaying the percentage of cells with the ASC phenotype at day 7 of culture. (B,C) Percentage and absolute number of ASCs quantified through FACS for different PBMC cultures derived from 19 healthy donors. (D) Representative data obtained from 3.3 × 104 PBMCs harvested at day 7 of culture and added per well of ELISPOT plates to enumerate IgG-secreting cells in each culture condition. (E) Magnitude of IgG-secreting cells detected per 106 PBMCs through ELISPOT. (F) Heatmap based on Z scores derived from ELISPOT data for each culture condition. Green boxes represent the PBMC donors that presented pre-existing immunity to DENV.
Since two different stimuli (DENV4 TVP and Mitogens) were capable of promoting the ASC phenotype and function, we asked whether such activation could be indiscriminately elicited by themselves. For this, we approached the ELISPOT numbers for IgG-secreting cells that represented not the real cell frequency but the variations among the different stimuli for each sample (Z-score). Interestingly, a group of six samples (#2, 4, 6, 7, 9, and 15) clearly presented a trend to respond better to DENV4 TVP rather than to Mitogens. In contrast, the 13 remaining samples displayed the opposite behaviour (Figure 1F). To verify whether the DENV-responsive samples in the ELISPOT assay were derived from subjects previously infected with DENV, ELISAs were performed to quantify plasma IgG specific to DENV1-4 NS1 and EDIII recombinant proteins and viable DENV particles. For this, plasma samples derived from 13 out of 19 individuals evaluated in this study were used. Although some samples displayed higher optical densities (OD) than the negative control sample against the three different DENV targets, none of them presented OD values as high as the positive control sample did (Supplementary Figures 4A–C). Thus, these data rule out the participation of DENV-specific memory B cells driving the ASC differentiation in this in vitro model.
Inactivated DENV Particles Do Not Activate the ASC Differentiation by Human PBMCs
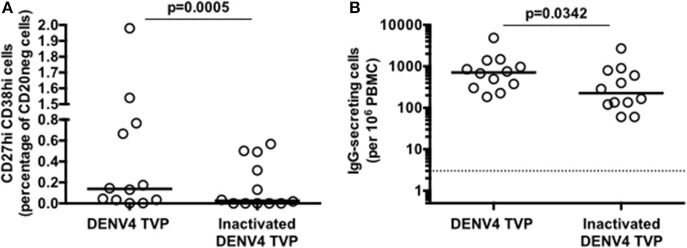
To address whether the DENV particle integrity had a role in the B cell differentiation in vitro, we cultivated PBMCs with either whole or inactivated DENV4 TVP (UV-treated) particles for 7 days. PBMCs lost the ability to induce ASCs either phenotypically upon the incubation with inactivated viral particles [p < 0.0005 – Whole (Median and St Dev – 0.14 ± 0.66) vs. Inactivated particles (Median and St Dev – 0.03 ± 0.23)] and functionally [p < 0.0342 – Whole (Median and St Dev – 712 ± 1,274) vs. Inactivated particles (Median and St Dev – 225 ± 742)] (Figures 2A,B, respectively). The ASC generation provided by this model of cell culture can therefore be observed only with whole DENV particles.
Figure 2.
Inactivated Dengue virus particles do not have the same ability to trigger the B cell differentiation into antibody-secreting cells (ASCs) as whole particles do. (A) Percentage of ASCs quantified through FACS and (B) magnitude of IgG-secreting cells detected through ELISPOT at day 7 of PBMC cultures derived from 12 healthy donors.
Viable DENV Particles Are Unable to Trigger the ASC Function in Purified CD19+ B Cell Culture
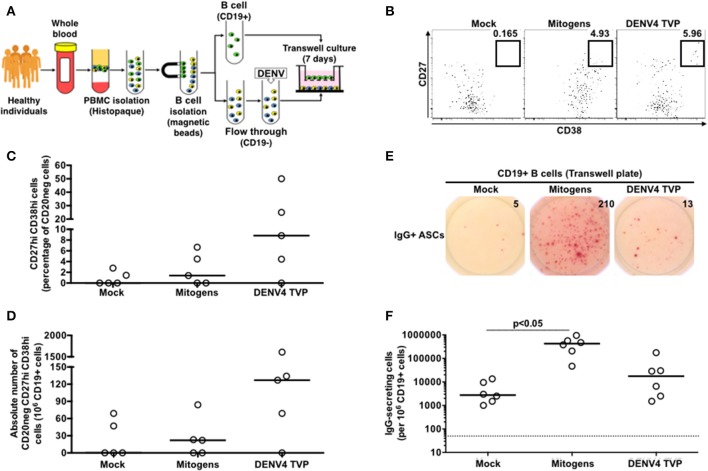
Since only viable DENV4 TVP particles could stimulate the acquisition of the ASC phenotype and function in human PBMCs in vitro, can these viral particles reproduce the same cell differentiation in purified B cells? CD19+ B cells were isolated from the same tested PBMCs with anti-human CD19-specific magnetic beads and cultivated with viable viral particles for the enumeration of IgG-secreting cells at day 7. Meanwhile, Mitogens induced a significant CD19+ B cell differentiation into ASCs in comparison to Mock-treated cells [p < 0.05; Mitogens (Median and St Dev – 34,560 ± 23,174) vs. Mock (Median and St Dev – 550 ± 431)]. DENV4 TVP particles showed no significant activation of this process [p > 0.05; DENV4 TVP (Median and St Dev – 1,125 ± 710)] (Figures 3A,B). Hence, we suggest that the ASC differentiation is dependent on either physical cell contact or the secretion of soluble B cell stimulator by the remaining PBMCs in PBMCs cultivated with DENV4 TVP.
Figure 3.
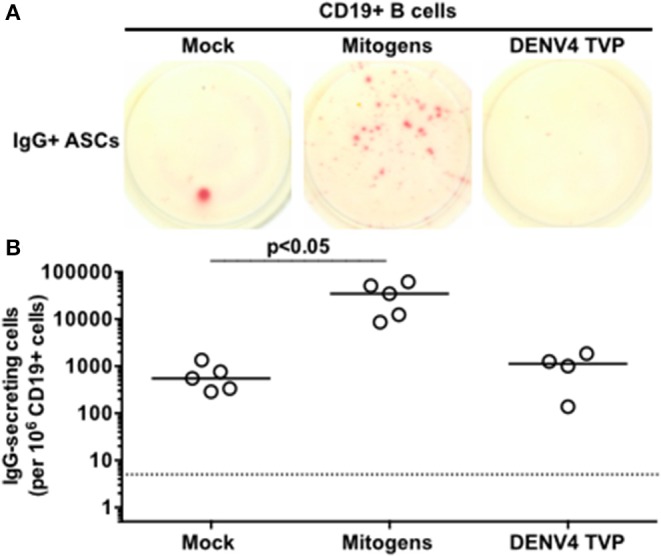
Dengue virus does not trigger the ASC differentiation when incubated with isolated B cells. CD19+ B cells were isolated from PBMCs of six donors and five of them were tested in this assay, except for DENV4 TVP incubation (n = 4 samples). (A) Representative data obtained from 6.6 × 104 CD19+ B cells harvested at day 7 of culture and added per well of ELISPOT plates to enumerate IgG-secreting cells in each culture condition. (B) Magnitude of IgG-secreting cells detected per 106 CD19+ B cells through ELISPOT.
DENV-Mediated ASC Differentiation Is Dependent on the B Cell-Remaining PBMC Contact
To investigate whether the cell physical contact was critical for the DENV4 TVP-mediated ASC differentiation detected in vitro, we cultivated purified CD19+ B cells with the remaining PBMCs infected with DENV4 TVP (in a 1:10 ratio respectively) in a transwell plate. CD19+ B cells were harvested on day 7 of culture and probed for the ASC phenotype and function. Although DENV4 TVP particles elicited higher ASC responses in the culture, measured by the cell percentage (Median and St Dev – 8.82 ± 20.4%) and absolute numbers of CD20neg CD27hi CD38hi cells (Median and St Dev – 127 ± 684), those numbers did not differ from the mock condition (ASC %: Median and St Dev – 0 ± 1.25%; ASC absolute number: Median and St Dev – 0 ± 33). Moreover, Mitogens were not able to significantly increase the ASC frequency (ASC %: Median and St Dev – 1.4 ± 3.0%; ASC absolute number: Median and St Dev – 22 ± 34) (Figures 4A–D). Furthermore, whereas DENV4 TVP generated similar IgG+ ASC numbers (Median and St Dev – 17.5 × 103 ± 67.2 × 103) in comparison to mock (2.75 × 103 ± 5.11 × 103), Mitogens elicited a massive IgG+ ASC differentiation in the transwell culture (p < 0.05 – Median and St Dev – 425 × 103 ± 311 × 103) (Figures 4E,F). To fully differentiate into ASCs in our model, CD19+ B cells must have the physical contact with the DENV4 TVP exposed-remaining PBMCs.
Figure 4.
DENV-mediated ASC differentiation is dependent on the physical contact between CD19+ B cells and the remaining PBMCs. (A) CD19+ B cells were isolated from PBMCs (n = 5) and cultured without physical contact with the flow through (CD19– cells) (in a 1:10 ratio, respectively) in transwell plates. CD19+ B cells were harvested at day 7 of culture and had the ASC phenotype (B–D) and function (E,F) assessed. (F) A total of 2.2 × 104 CD19+ cells were added per well, except for the Mitogen culture (2.2 × 103 CD19+ cells).
Tryptophan Metabolism Distinguishes the ASC Differentiation Mediated by DENV and Mitogens in vitro
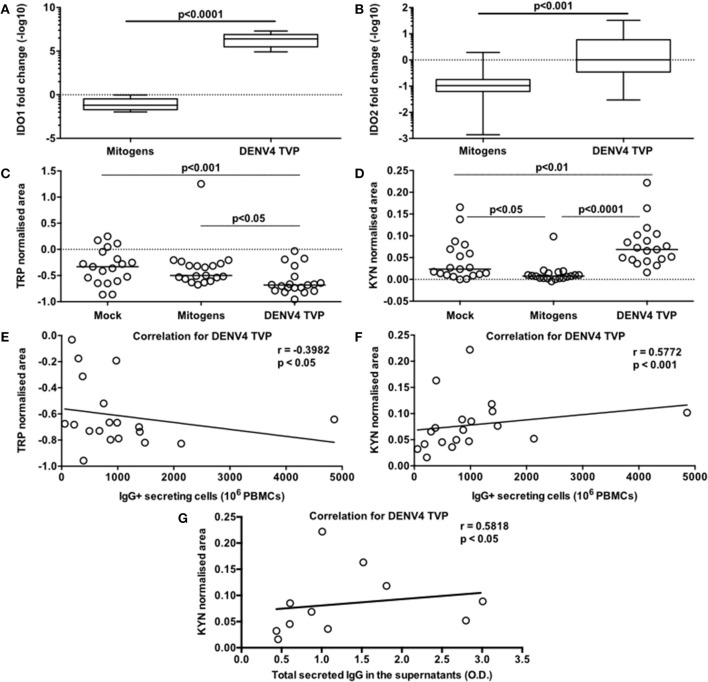
DENV4 TVP and Mitogens clearly induced phenotypical and functional ASC differentiation when incubated with PBMCs. Is this cellular process regulated by the same metabolic pathway for both stimulators? To answer this question, we initially investigated the transcriptome of those PBMC cultures through qRT-PCR. As Dengue patients seem to present with lower tryptophan and higher kynurenine concentrations in the serum (7), we decided to include IDO1 and IDO2 genes in the analysis. Surprisingly, we observed increased amounts for both IDO transcripts in the PBMC-DENV4 TVP culture (p < 0.0001 – IDO1 – Median and St Dev – 2.5 × 106 ± 6.6 × 106; p < 0.001 – IDO2 – Median and St Dev – 1.01 ± 13.07) in comparison to the Mitogen counterpart (IDO1 – Median and St Dev – 0.065 ± 0.296; IDO2 – Median and St Dev – 0.105 ± 0.668) (Figures 5A,B, respectively). Other targets associated to the cellular signalling (SYK and SRC) and the B cell differentiation into ASCs (TNFS13B and IL-10) also had their expression evaluated. For all of them (TNFS13B, IL-10, SYK, and SRC), the DENV4 TVP-stimulated PBMCs displayed increased transcript amounts with regard to Mitogen-exposed cells (Supplementary Figures 5A–D). Although either DENV4 TVP and Mitogens were capable of fully differentiating PBMC-containing CD19+ B cells into ASCs, our qRT-PCR data suggest that each stimulator could elicit different metabolic and signalling pathways during this in vitro process.
Figure 5.
Activation of the tryptophan metabolism is strongly associated with the DENV-mediated ASC differentiation detected in PBMCs in vitro. Different PBMC cultures derived from 19 healthy donors had their cells and supernatants harvested at day 7 of the culture. To verify the participation of the tryptophan metabolism in this cellular process, the following parameters were measured: (A,B) gene expression levels of IDO1 and IDO2 respectively in the cells; (C) Tryptophan (Trp) consumption and (D) Kynurenine (Kyn) accumulation in the supernatants; (E) correlation between the Trp consumption and ASC numbers (counted through ELISPOT); correlations between the Kyn accumulation and ASC numbers (F) or the total amount of secreted IgG in the supernatants (G). (A,B) Data are represented as boxes and whiskers (10–90% percentile). Dashed line represents the gene expression within the Mock culture condition. (C,D) Dashed lines represent the standard amount of the metabolite found in the culture medium kept under the same conditions, but without cells.
Considering the data paucity regarding the metabolism influence on the B cell response and higher concentrations of serum kynurenine found in Dengue patients (7), we also assessed the participation of the tryptophan metabolism in our model. For this, we measured the normalised areas occupied by the tryptophan and kynurenine peaks obtained through mass spectrometry in the culture supernatants at day 7. The PBMCs cultured with DENV4 TVP showed the highest tryptophan consumption (Median and St Dev: 0.682 ± 0.252) in comparison to the Mock (p < 0.001 – Median and St Dev: −0.333 ± 0.332) or Mitogen cultures (p < 0.05 – Median and St Dev: −0.499 ± 0.419) (Figure 5C). Moreover, the supernatants from the PBMC-DENV4 TVP culture had the highest kynurenine concentration (Median and St Dev – 0.07 ± 0.05) in relation to the Mock (p < 0.01 – Median and St Dev – 0.024 ± 0.047) or Mitogen counterparts (p < 0.0001 – Median and St Dev – 0.008 ± 0.022) (Figure 5D). Notably, the tryptophan degradation and kynurenine accumulation detected in the DENV4 TVP-treated cultures presented correlations with the magnitude of the IgG+ ASCs enumerated at day 7 (respectively r = −0.3982; p < 0.05; r = 0.5772; p < 0.001) (Figures 5E,F) and the titers of secreted IgG (r = 0.5818; p < 0.05) (Figure 5G). Other metabolites downstream of this metabolic pathway (5-hydroxyindoleacetic, anthranilic, kynurenic, quinolinic, and nicotinic acids) were also detected in higher amounts for DENV4 TVP in comparison to Mitogen cultures (Supplementary Figures 6A–E).
Activation of the Tryptophan Metabolism Portrays the Flavivirus-Mediated ASC Response in vitro
Since the DENV-induced ASC differentiation described herein was performed with a laboratory adapted reference viral strain we sought to determine whether the exposure to a clinical DENV isolate and other flaviviruses could elicit similar responses. Then, we selected the DENV4 LRV13/422 (24) as well as two Zika virus strains (ZIKV PE243 and ZIKV MR766) and the Yellow fever virus vaccine strain (YFV 17DD) for further analysis. We detected a significant augment of CD20neg CD27hi CD38hi cells only in the PBMC-DENV4 LRV13/422 culture in comparison to mock (p < 0.01 – DENV4 LRV13/422 – Median and St Dev – 0.44 ± 0.476; Mock – Median and St Dev – 0.036 ± 0.144). The remaining flaviviruses showed similar ASC percentages to mock (Figure 6A), whereas DENV infections have been associated with a massive ASC generation in vivo (11, 12) as well as ZIKV infection in humans (26, 27) or macaques (28), and the vaccination with YFV 17DD does not activate the same phenomenon (11). Considering that a direct correlation between kynurenine levels in the supernatants and the number of functional IgG+ ASC or secreted IgG titers were found for DENV4 TVP (Figures 5F,G), we evaluated whether other flavivirus would display it as well. Among all flaviviruses tested in this in vitro model, only the DENV4 LRV13/422 presented a similar correlation (r = 0.7395; p < 0.05) (Figures 6B–E). Therefore, the direct correlation between the kynurenine levels and functional IgG+ ASCs was only pointed out for DENV particles, which have been strongly linked to the ASC responses seen in human infections.
Figure 6.
Contrasting pattern of the tryptophan metabolism induced in PBMC cultures stimulated by a DENV4 clinical isolate (422) or flaviviruses that elicit low ASC responses in vivo (ZIKV MR766, ZIKV PE243, and attenuated Yellow Fever 17DD). Different PBMC cultures derived from healthy donors had cells and supernatants obtained at day 7 of the cultures. (A) ASC percentage quantified through FACS for different flavivirus-stimulated PBMC cultures. (B–E) Correlation between the Kyn levels and ASC numbers (counted through ELISPOT) derived from each cell culture.
Discussion
Although it is well-established that DENV infection causes a massive and transient DENV-specific ASC response during the disease symptomatology in patients (11, 12), the mechanisms related to this cellular process are still poorly understood. In vivo, the major DENV targets are monocytic cells (29). Among them, CD14+ CD16+ monocytes were described as capable to stimulate the B cell differentiation into ASCs in vitro (30). B cells are also susceptible to the DENV invasion (25), and infected cells have been found in the blood of Dengue patients (3). If B cells need to be infected to initiate that enormous ASC response, it has not been elucidated yet.
Correa and colleagues sought to clarify the DENV infection-mediated ASC differentiation through an in vitro strategy in which PBMCs from healthy donors were incubated with DENV particles. Only indirect suppositions of cellular responses could be taken from that study, though, as all measurements were based on the antibody secretion detected from culture supernatants. Whenever “cellular data” were analysed in that regard, purified B cells were selected for the incubation with DENV particles (25). However, our data showed that the functional ASC differentiation does not occur under that B-cell culture setting (Figure 3). On the other hand, purified B cells could clearly differentiate into ASCs upon incubation with mitogens (Figure 3) as previously demonstrated (21). It is known that the B cell differentiation into ASCs can be facilitated by the activation of receptors related to the innate immunity, such as Toll-like receptors (TLR) 3, 4, 7/8, and 9 (31). If viable DENV particles do not contain sufficient amounts of those TLR ligands in their composition, it has to be defined. Whereas the DENV NS1 protein was described to activate immune cells through TLR4 (32, 33), the DENV NS4 seems to be related to autophagy (34), which also has been associated with the ASC differentiation (35). More specifically, the DENV NS4 seems to interact with TAX1BP1 (36), restricting the Blimp1 expression (37) that is critical for the B cell differentiation into ASCs. Of note, TAXBP1 KO mice present lower numbers of germinal centers in lymphoid organs as well as titers of antigen-specific antibodies (37).
Herein, we investigated the cellular basis of the DENV-mediated ASC generation using the same in vitro model previously described (25). We demonstrated an increased cell frequency with the ASC phenotype and function upon the PBMC culture with DENV4 TVP particles (Figure 1). Although the ASC numbers and percentage can rise up in Dengue patients if a secondary or a more severe DENV infection is considered (11, 12), our in vitro cellular differentiation model has shown to be independent of pre-existing DENV immunity (Supplementary Figure 4). To date, the literature is ambiguous regarding whether this DENV-mediated ASC differentiation is or is not related to DENV-specific memory B cells (10, 38, 39). Furthermore, cell cultures prepared with inactivated DENV particles did not allow the phenotypical and functional ASC differentiation (Figure 2) as reported in vitro and in vivo (25, 40, 41). In order to be differentiated into ASCs, our data also pointed out that B cells must have physical contact with the remaining PBMCs in the presence of DENV particles (Figure 4). In this context, a co-culture between B cells and CD14+ dermal monocytic cells presented higher frequency of ASCs and augmented antibody secretion in vitro. Whereas, the addition of TLR ligands to the co-cultures enhanced the B cell ability to differentiate into ASCs, the lack of physical contact among those cells diminished the ASC frequency and antibody production. In another co-culture experiment between B cells and CD14+ CD16+ monocytes, the ASC differentiation was dependent on the expression of BAFF, APRIL, and IL-10 (30).
Since viral loads are fundamental in the ASC differentiation process for infections, such as SIV (Silveira et al., unpublished data) and HIV (42), we wondered the DENV influence in our model. Neither viable DENV particles in the supernatants, nor viral transcripts were detected in the cells at day 7 of culture (data not shown). Since DENV-infected B cells have been found up to day 5 of cultures (5), earlier sample collection would potentially aid to define the role of DENV loads in our model. Recently, a model of human DENV infection presented a direct correlation between viremia and the magnitude of ASC response (29). However, other technologies would need to be applied to define the source of the DENV load for this B cell differentiation: monocytes or lymphocytes. Currently, DENV-infected cells are identified based on invasive methods, such as cell permeabilisation or nucleic acid isolation. These strategies inhibit the cell tracking to answer questions like “Can a DENV-infected B cell differentiate into an ASC? Are the B-cell viral loads associated to its ability to further differentiate into an ASC?” The use of an infective fluorochrome-labeled DENV clone may overcome those issues.
Multiple metabolic pathways have been associated to the ASC differentiation, such as glycolysis, oxidative phosphorylation, glucose uptake, inositol phosphate, and sterol metabolisms (43, 44). Furthermore, the DENV infection can stimulate the activation of the tryptophan metabolism. An elevated kynurenine concentration, the major product resultant from the tryptophan consumption, has been detected in the serum of DENV- and HIV-infected patients (7, 45, 46). This metabolite has been associated with a tolerizating immune response, especially for T cells (47–49). It does not seem to be the case for DENV, HIV, and SIV infections since they enhance the immunological status of patients (50–54). Another common characteristic of these viral infections is the induction of hypergammaglobulinemia (55–59), a hallmark of B cell activity that augments the ASC frequencies (29, 60, 61). The major difference between these scenarios is the target choice. Whereas a potent IgG+ ASC response is driven to the viral envelope during Dengue (11), the number of HIV-specific IgG+ ASCs display a low percentage of the total counterpart (60).
B cell responses have had their tryptophan metabolism evaluated in different models. The Zostavax booster vaccination elicited the expression of tryptophan metabolism-related genes, which were closely connected to genes linked to the ASC response (7). Among the relevant genes derived from this metabolic pathway are IDO1 or IDO2 (61). In addition, immunised IDO1 KO mice displayed greater humoral responses than controls with T-cell-independent antigens due to their higher capacity to proliferate and secrete antibodies (62). In a murine model of Citrobacter rodentium-induced colitis, IDO-deficient mice also displayed higher titers of IgA and IgG than controls (63). Genes with expressions tightly bound to the IDO activity, such as eukaryotic initiation factor 2 alpha (eIF2alpha) and kinase general control non-derepressible-2 (GCN2), can enhance the BLIMP1 (or PRDM1) expression in a tryptophan-depleted condition. On the other hand, the BLIMP1 protein binds to the IDO promoter, repressing its expression (64). The tryptophan degradation also modifies the expression of SLC7A5 (65) and WARS during this process in cancer cells (66) and ASCs (67). Regarding cancer cells, the prognostic of diffuse large B-cell lymphoma, mainly composed by ASCs, and its outcome for a patients have been strongly connected to the IDO expression and the presence of determined serum kynurenine concentrations (68–71). The clinical progression of multiple myeloma, also characterised by transformed long-lived ASCs, has also been connected to the IDO activity (72). In our model, while no tryptophan degradation was identified in the mitogen-driven ASC differentiation, a strong activation of this amino acid metabolism seemed to occur during the DENV-mediated ASC generation in vitro. Either a lab-adapted or a clinical DENV isolate stimulated the tryptophan consumption, accumulating kynurenine in the supernatants of the PBMC-DENV cultures. Overall, an increased expression of IDO1 and IDO2 transcripts was noticed in those cell cultures. More importantly, the tryptophan and kynurenine concentrations were directly correlated to the magnitude of an DENV-specific ASC response in vitro (Figure 5). Downstream metabolites were also detected in higher amounts in the PBMC-DENV cultures than in controls (Supplementary Figure 4). In corroboration with these data, we also observed increased amounts of transcripts related to the ASC survival or differentiation, such as TNFS13B and IL-10 (Supplementary Figure 5) as previously described (30, 73–75). Thus, it is suggested that the tryptophan degradation would be the cause and not consequence of the DENV-mediated ASC differentiation in vitro, as it seems to occur earlier than the BLIMP1 expression (64). The role of this metabolic pathway in the massive ASC response detected in Dengue patients is still to be ascertained.
Other flaviviruses present lower potential to induce robust ASC responses in vivo, as the DENV does. Among them, we tested two ZIKV strains (26–28) and the attenuated YFV 17DD (11) in our model. Their ASC data obtained in vitro reinforced the same scenario observed in vivo (Figure 6A). In terms of magnitude, ZIKV or YFV 17DD-treated PBMC cultures had lower median kynurenine concentrations than those treated with DENV particles. Moreover, the cultures with ZIKV or YFV 17DD showed no correlation between their kynurenine concentrations and the respective ASC magnitudes (Figures 6B–D). It is likely that relevant factors for the ASC differentiation, such as BAFF, APRIL, or IL-10 were reduced in those flavivirus-PBMC cultures in comparison to the DENV counterpart. Thus, our data suggests that the activation of the tryptophan metabolism as a biomarker of the DENV-mediated ASC responses in vivo.
Conclusions
The in vitro model of B cell differentiation into ASCs has provided relevant evidence on how this phenomenon operates. Indeed, the presence of mitogens or DENV particles in the PBMC cultures did elicit the ASC differentiation. Regarding the viral participation in this cell differentiation process, it requires viable particles rather than inactivated forms in order to stimulate the acquisition of the ASC phenotype and function. Moreover, those PBMC-containing B cells requested the cell–cell contact with the remaining PBMCs to fully differentiate into ASCs in the presence of DENV particles. In contrast to the mitogen-mediated ASC differentiation, we demonstrated that both laboratory-adapted and clinically isolated DENV particles can activate the tryptophan metabolism in PBMC cultures in a magnitude directly correlated with the generated ASC numbers. Flaviviruses known to stimulate inferior ASC responses in vivo, such as Zika and the attenuated Yellow Fever viruses, presented no correlation between their abilities to stimulate tryptophan degradation and their PBMC-derived ASC differentiation in vitro. Other conditions capable of triggering this amino acid degradation pathway, such as HIV and SIV infection, B cell lymphoma, and multiple myeloma, show similarities with the DENV-mediated B cell differentiation. Thus, the activation status of the tryptophan metabolism may affect the B cell fate during an infectious disease by stimulating or not the ASC differentiation. Furthermore, such information may help researchers to understand why specific adjuvants stimulate stronger humoral responses than others in vaccine formulations. Eventually, strategies based on the effects of the tryptophan metabolism may be applied to improve B cell responses against other pathogens or diseases.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
All blood samples evaluated in this study were voluntarily donated by healthy adults from the University of São Paulo and Instituto Carlos Chagas/Fiocruz-PR (ICC/Fiocruz-PR) (Brazil). All patients provided written informed consent prior to the study enrollment. Experimental protocols and procedures were reviewed and approved by the Institutional Ethics Committee (CAEE 68875417.9.0000.0067) regulated by the Conselho Nacional de Ética em Pesquisa.
Author Contributions
VB conducted the execution of all experiments, except the ELISA, and contributed to the statistical analyses, review, and interpretation of data sets. AHDC contributed to the experiment execution, review, and interpretation of data sets. MB and MS generated proteomics data. PG-D provided bioinformatics support. SP conducted the ELISA. LF provided the ELISA kit and contributed to the experiment execution, review, and interpretation of data sets. HN provided bioinformatics data and support, and contributed to review and interpretation of data sets. AC contributed to the generation of proteomic data, review, and interpretation of data sets. PW provided viral strains and contributed to the experiment execution, generation of samples, review, and interpretation of data sets. ES conceptualised and directed the project, provided the statistical analyses, interpreted the data sets, and prepared the manuscript with input from all authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Juliano Bordignon and Dr. Claudia N. Duarte dos Santos Laboratório de Virologia Molecular of the Instituto Carlos Chagas/Fiocruz-PR (ICC/Fiocruz-PR) (Brazil) for their contribution to the experiment execution, discussions, and critical reading of this manuscript. We also thank the Program for Technological Development in Tools for Health-PDTIS FIOCRUZ for the use of Flow Cytometry Facility at the Instituto Carlos Chagas/Fiocruz-PR (Brazil).
Footnotes
Funding. This study was supported by CNPq (Grant No. 420616/2016-0), Fapesp (Grant No. 2017/11931-6), and Programa de Apoio aos Novos Docentes (University of São Paulo). VB was a Capes and CNPq fellow during different periods of her graduation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.00020/full#supplementary-material
References
- 1.World Health Organization (2019). Fact sheet: Dengue and Severe Dengue. Available online at: https://www.who.int/en/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed August 31, 2019).
- 2.Khetarpal N, Khanna I. Dengue fever: causes, complications, and vaccine strategies. J Immunol Res. (2016) 2016:6803098. 10.1155/2016/6803098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurane I, Ennis FA. Production of interferon alpha by dengue virus-infected human monocytes. J Gen Virol. (1988) 69:445–9. 10.1099/0022-1317-69-2-445 [DOI] [PubMed] [Google Scholar]
- 4.Kurane I, Meager A, Ennis FA. Induction of interferon alpha and gamma from human lymphocytes by dengue virus-infected cells. J Gen Virol. (1986) 67:1653–61. 10.1099/0022-1317-67-8-1653 [DOI] [PubMed] [Google Scholar]
- 5.Silveira GF, Wowk PF, Cataneo AHD, dos Santos PF, Delgobo M, Stimamiglio MA, et al. Human T lymphocytes are permissive for dengue virus replication. J Virol. (2018) 92:e02181-17. 10.1128/JVI.02181-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor MW, Feng G. Relationship between interferon-γ, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. (1991) 5:2516–22. 10.1096/fasebj.5.11.1907934 [DOI] [PubMed] [Google Scholar]
- 7.Becerra A, Warke RV, Xhaja K, Evans B, Evans J, Martin K, et al. Increased activity of indoleamine 2,3-dioxygenase in serum from acutely infected dengue patients linked to gamma interferon antiviral function. J Gen Virol. (2009) 90:810–7. 10.1099/vir.0.004416-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai W-Y, Lai C-Y, Wu Y-C, Lin H-E, Edwards C, Jumnainsong A, et al. High-avidity and potently neutralizing cross-reactive human monoclonal antibodies derived from secondary dengue virus infection. J Virol. (2013) 87:12562–75. 10.1128/JVI.00871-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. (2015) 16:170–7. 10.1038/ni.3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Priyamvada L, Cho A, Onlamoon N, Zheng N-Y, Huang M, Kovalenkov Y, et al. B cell responses during secondary dengue virus infection are dominated by highly cross- reactive, memory-derived plasmablasts. J Virol. (2016) 90:5574–85. 10.1128/JVI.03203-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrammert J, Onlamoon N, Akondy RS, Perng GC, Polsrila K, Chandele A, et al. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J Virol. (2012) 86:2911–8. 10.1128/JVI.06075-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Bates TM, Cordeiro MT, Nascimento EJM, Smith AP, Soares de Melo KM, McBurney SP, et al. Association between magnitude of the virus-specific plasmablast response and disease severity in dengue patients. J Immunol. (2013) 190:80–7. 10.4049/jimmunol.1103350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McElroy AK, Akondy RS, Davis CW, Ellebedy AH, Mehta AK, Kraft CS, et al. Human Ebola virus infection results in substantial immune activation. Proc Natl Acad Sci USA. (2015) 112:4719–24. 10.1073/pnas.1502619112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellebedy AH, Jackson KJL, Kissick HT, Nakaya HI, Davis CW, Roskin KM, et al. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat Immunol. (2016) 17:1226–34. 10.1038/ni.3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García M, Iglesias A, Landoni VI, Bellomo C, Bruno A, Córdoba MT, et al. Massive plasmablast response elicited in the acute phase of hantavirus pulmonary syndrome. Immunology. (2017) 151:122–35. 10.1111/imm.12713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. (2008) 453:667–71. 10.1038/nature06890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gawoski JM, Ooi WW. Dengue fever mimicking plasma cell leukemia. Arch Pathol Lab Med. (2003) 127:1026–7. [DOI] [PubMed] [Google Scholar]
- 18.Dick GWA. Zika virus. I Isolations and serological specificity. Trans R Soc Trop Med Hyg. (1952) 46:509–20. 10.1016/0035-9203(52)90042-4 [DOI] [PubMed] [Google Scholar]
- 19.Donald CL, Brennan B, Cumberworth SL, Rezelj VV, Clark JJ, Cordeiro MT, et al. Full genome sequence and sfRNA interferon antagonist activity of Zika virus from Recife, Brazil. PLoS Negl Trop Dis. (2016) 10:e0005048. 10.1371/journal.pntd.0005048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchevsky RS, da Luz Leal M, Homma A, Coutinho ESF, Camacho LAB, Jabor AV, et al. Molecular and phenotypic analysis of a working seed lot of yellow fever virus 17DD vaccine strain produced from the secondary seed lot 102/84 with an additional passage in chicken embryos. Biologicals. (2006) 34:191–7. 10.1016/j.biologicals.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 21.Crotty S, Aubert RD, Glidewell J, Ahmed R. Tracking human antigen-specific memory B cells: a sensitive and generalized ELISPOT system. J Immunol Methods. (2004) 286:111–22. 10.1016/j.jim.2003.12.015 [DOI] [PubMed] [Google Scholar]
- 22.Smith K, Garman L, Wrammert J, Zheng N-Y, Capra JD, Ahmed R, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. (2009) 4:372–84. 10.1038/nprot.2009.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenneth JL, Schmittgenb T. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. (2001) 25:402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 24.Kuczera D, Bavia L, Mosimann ALP, Koishi AC, Mazzarotto GACA, Aoki MN, et al. Isolation of dengue virus serotype 4 genotype II from a patient with high viral load and a mixed Th1/Th17 inflammatory cytokine profile in South Brazil. Virol J. (2016) 13:93. 10.1186/s12985-016-0548-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Correa ARV, Berbel ACER, Papa MP, De Morais ATS, Peçanha LMT, De Arruda LB. Dengue virus directly stimulates polyclonal B cell activation. PLoS ONE. (2015) 10:e0143391. 10.1371/journal.pone.0143391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers TF, Goodwin EC, Briney B, Sok D, Beutler N, Strubel A, et al. Zika virus activates de novo and cross-reactive memory B cell responses in dengue-experienced donors. Sci Immunol. (2017) 2:eaan6809. 10.1126/sciimmunol.aan6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricciardi MJ, Magnani DM, Grifoni A, Kwon YC, Gutman MJ, Grubaugh ND, et al. Ontogeny of the B- and T-cell response in a primary Zika virus infection of a dengue-naïve individual during the 2016 outbreak in Miami, FL. PLoS Negl Trop Dis. (2017) 11:e0006000. 10.1371/journal.pntd.0006000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silveira ELV, Rogers KA, Gumber S, Amancha P, Xiao P, Woollard SM, et al. Immune cell dynamics in rhesus macaques infected with a brazilian strain of Zika virus. J Immunol. (2017) 199:1003–11. 10.4049/jimmunol.1700256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nivarthi UK, Tu HA, Delacruz MJ, Swanstrom J, Patel B, Durbin AP, et al. Longitudinal analysis of acute and convalescent B cell responses in a human primary dengue serotype 2 infection model. EBioMedicine. (2019) 41:465–78. 10.1016/j.ebiom.2019.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwissa M, Nakaya HI, Onlamoon N, Wrammert J, Villinger F, Perng GC, et al. Dengue virus infection induces expansion of a CD14+CD16+monocyte population that stimulates plasmablast differentiation. Cell Host Microbe. (2014) 16:115–27. 10.1016/j.chom.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pone EJ, Lou Z, Lam T, Greenberg ML, Wang R, Xu Z, et al. B cell TLR1/2, TLR4, TLR7 and TLR9 interact in induction of class switch DNA recombination: modulation by BCR and CD40, and relevance to T-independent antibody responses. Autoimmunity. (2015) 48:1–12. 10.3109/08916934.2014.993027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modhiran N, Watterson D, Muller DA, Panetta AK, Sester DP, Liu L, et al. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci Transl Med. (2015) 7:304ra142 10.1126/scitranslmed.aaa3863 [DOI] [PubMed] [Google Scholar]
- 33.Modhiran N, Watterson D, Blumenthal A, Baxter AG, Young PR, Stacey KJ. Dengue virus NS1 protein activates immune cells via TLR4 but not TLR2 or TLR6. Immunol Cell Biol. (2017) 95:491–5. 10.1038/icb.2017.5 [DOI] [PubMed] [Google Scholar]
- 34.McLean JE, Wudzinska A, Datan E, Quaglino D, Zakeri Z. Flavivirus NS4A-induced autophagy protects cells against death and enhances virus replication. J Biol Chem. (2011) 286:22147–59. 10.1074/jbc.M110.192500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold J, Murera D, Arbogast F, Fauny JD, Muller S, Gros F. Autophagy is dispensable for B-cell development but essential for humoral autoimmune responses. Cell Death Differ. (2016) 23:853–64. 10.1038/cdd.2015.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khadka S, Vangeloff AD, Zhang C, Siddavatam P, Heaton NS, Wang L, et al. A physical interaction network of dengue virus and human proteins. Mol Cell Proteomics. (2011) 10:M111.012187. 10.1074/mcp.M111.012187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsushita N, Suzuki M, Ikebe E, Nagashima S, Inatome R, Asano K, et al. Regulation of B cell differentiation by the ubiquitinbinding protein TAX1BP1. Sci Rep. (2016) 6:31266. 10.1038/srep31266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Appanna R, Srinivasan KG, Xu MH, Toh YX, Velumani S, Carbajo D, et al. Plasmablasts during acute dengue infection represent a small subset of a broader virus-specific memory B cell pool. EBioMedicine. (2016) 12:178–88. 10.1016/j.ebiom.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tas JMJ, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, et al. Visualizing antibody affinity maturation in germinal centers. Science. (2016) 351:1048–54. 10.1126/science.aad3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rizvi N, Chaturvedi UC, Mathur A. Obligatory role of macrophages in dengue virus antigen presentation to B lymphocytes. Immunology. (1989) 67:38–43. [PMC free article] [PubMed] [Google Scholar]
- 41.Yam-Puc JC, García-Cordero J, Calderón-Amador J, Donis-Maturano L, Cedillo-Barrón L, Flores-Romo L. Germinal center reaction following cutaneous dengue virus infection in immune-competent mice. Front Immunol. (2015) 6:188. 10.3389/fimmu.2015.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cepok S, von Geldern G, Nolting T, Grummel V, Srivastava R, Zhou D, et al. Viral load determines the B-cell response in the cerebrospinal fluid during human immunodeficiency virus infection. Ann Neurol. (2007) 62:458–67. 10.1002/ana.21195 [DOI] [PubMed] [Google Scholar]
- 43.Price MJ, Patterson DG, Scharer CD, Boss JM. Progressive upregulation of oxidative metabolism facilitates plasmablast differentiation to a T-independent antigen. Cell Rep. (2018) 23:3152–9. 10.1016/j.celrep.2018.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S, Sullivan NL, Rouphael N, Yu T, Banton S, Maddur MS, et al. Metabolic phenotypes of response to vaccination in humans. Cell. (2017) 169:862–77.e17. 10.1016/j.cell.2017.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werner ER, Fuchs D, Hausen A, Jaeger H, Reibnegger G, Werner-Felmayer G, et al. Tryptophan degradation in patients infected by human immunodeficiency virus. Biol Chem Hoppe Seyler. (1988) 369:337–40. 10.1515/bchm3.1988.369.1.337 [DOI] [PubMed] [Google Scholar]
- 46.Huengsberg M, Winer JB, Gompels M, Round R, Ross J, Shahmanesh M. Serum kynurenine-to-tryptophan ratio increases with progressive disease in HIV-infected patients. Clin Chem. (1998) 44:858–62. [PubMed] [Google Scholar]
- 47.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med. (1999) 189:1363–72. 10.1084/jem.189.9.1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hwu P, Du MX, Lapointe R, Do M, Taylor MW, Young HA. Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J Immunol. (2000) 164:3596–9. 10.4049/jimmunol.164.7.3596 [DOI] [PubMed] [Google Scholar]
- 49.Grohmann U, Orabona C, Fallarino F, Vacca C, Calcinaro F, Falorni A, et al. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat Immunol. (2002) 3:1097–101. 10.1038/ni846 [DOI] [PubMed] [Google Scholar]
- 50.Kurane I, Innis BL, Nimmannitya S, Nisalak A, Meager A, Janus J, et al. Activation of T lymphocytes in dengue virus infections. High levels of soluble interleukin 2 receptor, soluble CD4, soluble CD8, interleukin 2, and interferon-gamma in sera of children with dengue. J Clin Invest. (1991) 88:1473–80. 10.1172/JCI115457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haltaufderhyde K, Srikiatkhachorn A, Green S, Macareo L, Park S, Kalayanarooj S, et al. Activation of peripheral T follicular helper cells during acute dengue virus infection. J Infect Dis. (2018) 218:1675–85. 10.1093/infdis/jiy360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Utay NS, Hunt PW. Role of immune activation in progression to AIDS. Curr Opin HIV AIDS. (2016) 11:131–7. 10.1097/COH.0000000000000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spudich SS. Immune activation in the central nervous system throughout the course of HIV infection. Curr Opin HIV AIDS. (2016) 11:226–33. 10.1097/COH.0000000000000243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. Natural SIV hosts: showing AIDS the door. Science. (2012) 335:1188–93. 10.1126/science.1217550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oleske J, Minnefor A, Cooper R, Thomas K, Dela Cruz A, Ahdieh H, et al. Immune deficiency syndrome in children. JAMA J Am Med Assoc. (1983) 249:2345–9. 10.1001/jama.1983.03330410031024 [DOI] [PubMed] [Google Scholar]
- 56.Chess Q, Daniels J, North E, Macris NT. Serum immunoglobulin elevations in the acquired immunodeficiency syndrome (AIDS): IgG, IgA, IgM, and IgD. Diagn Immunol. (1984) 2:148–53. [PubMed] [Google Scholar]
- 57.Castella A, Croxson TS, Mildvan D, Witt DH, Zalusky R. The bone marrow in AIDS. A histologic, hematologic, and microbiologic study. Am J Clin Pathol. (1985) 84:425–32. 10.1093/ajcp/84.4.425 [DOI] [PubMed] [Google Scholar]
- 58.Mizuma H, Zolla-Pazner S, Litwin S, el-Sadr W, Sharpe S, Zehr B, et al. Serum IgD elevation is an early marker of B cell activation during infection with the human immunodeficiency viruses. Clin Exp Immunol. (1987) 68:5–14. [PMC free article] [PubMed] [Google Scholar]
- 59.Lane HC, Masur H, Edgar LC, Whalen G, Rook AH, Fauci AS. Abnormalities of B-cell activation and immunoregulation in patients with the acquired immunodeficiency syndrome. N Engl J Med. (1983) 309:453–8. 10.1056/NEJM198308253090803 [DOI] [PubMed] [Google Scholar]
- 60.Buckner CM, Moir S, Ho J, Wang W, Posada JG, Kardava L, et al. Characterization of plasmablasts in the blood of HIV-infected viremic individuals: evidence for nonspecific immune activation. J Virol. (2013) 87:5800–11. 10.1128/JVI.00094-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Badawy AAB. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int J Tryptophan Res. (2017) 10:1178646917691938. 10.1177/1178646917691938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shinde R, Chaudhary K, Shimoda M, Pacholczyk G, McGaha T. Indoleamine 2,3-dioxygenase inhibits B cell immune response to T independent antigens (IRM8P.710). J Immunol. (2014) 192(Suppl. 1) 127.11. 10.4049/jimmunol.1402854. [DOI] [Google Scholar]
- 63.Harrington L, Srikanth CV, Antony R, Rhee SJ, Mellor AL, Hai NS, et al. Deficiency of indoleamine 2,3-dioxygenase enhances commensal-induced antibody responses and protects against Citrobacter rodentium-induced colitis. Infect Immun. (2008) 76:3045–53. 10.1128/IAI.00193-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barnes NA, Stephenson SJ, Tooze RM, Doody GM. Amino acid deprivation links BLIMP-1 to the immunomodulatory enzyme indoleamine 2,3-dioxygenase. J Immunol. (2009) 183:5768–77. 10.4049/jimmunol.0803480 [DOI] [PubMed] [Google Scholar]
- 65.Sinclair LV, Neyens D, Ramsay G, Taylor PM, Cantrell DA. Single cell analysis of kynurenine and system L amino acid transport in T cells. Nat Commun. (2018) 9:1981. 10.1038/s41467-018-04366-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adam I, Dewi DL, Mooiweer J, Sadik A, Mohapatra SR, Berdel B, et al. Upregulation of tryptophanyl-tRNA synthethase adapts human cancer cells to nutritional stress caused by tryptophan degradation. Oncoimmunology. (2018) 7:e1486353. 10.1080/2162402X.2018.1486353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Human Protein Atlas Blood Atlas: WARS - Monaco Scaled Dataset. Protein Atlas Version 19 (2019). Available online at: https://www.proteinatlas.org/ENSG00000140105-WARS/blood (accessed September 18, 2019).
- 68.Ninomiya S, Tsurumi H, Hara T, Goto N, Yoshikawa T, Kitagawa J, et al. Serum concentration of L-kynurenine can determine clinical outcome of patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood. (2008) 112:2812 10.1182/blood.V112.11.2812.2812 [DOI] [Google Scholar]
- 69.Finger E, Wong J, Gray J, Rock D, Loberg R, Fitzpatrick D, et al. Utilization of metabolomics to identify biomarkers in hematological malignancies: role of IDO and the tryptophan pathway. Blood. (2017) 130(Suppl. 1):5100 10.1182/blood.V130.Suppl_1.5100.5100 [DOI] [Google Scholar]
- 70.Yoshikawa T, Hara T, Tsurumi H, Goto N, Hoshi M, Kitagawa J, et al. Serum concentration of L-kynurenine predicts the clinical outcome of patients with diffuse large B-cell lymphoma treated with R-CHOP. Eur J Haematol. (2010) [DOI] [PubMed] [Google Scholar]
- 71.Fornecker LM, Muller L, Bertrand F, Paul N, Pichot A, Herbrecht R, et al. Multi-omics dataset to decipher the complexity of drug resistance in diffuse large B-cell lymphoma. Sci Rep. (2019) 9:895. 10.1038/s41598-018-37273-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romano A, Parrinello NL, Todoerti K, La Cava P, Puglisi F, Giallongo C, et al. Tryptophan deprivation promotes an adaptive response and contributes to bioenergetics in multiple myeloma. Blood. (2018) 132:4511 10.1182/blood-2018-99-116447 [DOI] [Google Scholar]
- 73.Heine G, Drozdenko G, Grün JR, Chang H-D, Radbruch A, Worm M. Autocrine IL-10 promotes human B-cell differentiation into IgM- or IgG-secreting plasmablasts. Eur J Immunol. (2014) 44:1615–21. 10.1002/eji.201343822 [DOI] [PubMed] [Google Scholar]
- 74.Avery DT, Kalled SL, Ellyard JI, Ambrose C, Bixler SA, Thien M, et al. BAFF selectively enhances the survival of plasmablasts generated from human memory B cells. J Clin Invest. (2003) 112:286–97. 10.1172/JCI18025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Gallastegui N, Rosenblatt JD. Regulatory B cells in anti-tumor immunity. Int Immunol. (2015) 27:521–30. 10.1093/intimm/dxv034 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.