Abstract
Multi-drug resistant in Mycobacterium tuberculosis (M.tb) is considered as major bottleneck in the treatment and cure of tuberculosis (TB). Several anti-tubercular drugs fail in its efficacy due to drug-resistant M.tb developed mechanism for resistance. So, research across globe has been carried out to develop effective anti-TB drugs to improve the treatment of these strains. Traditional drug development methods have been proved unsuccessful as it fails to develop a broad-spectrum drug due to lack of structure based approach. Several studies have been conducted in this regard and identified several drug target sites that influence drug-resistant M.tb strains. In this study, the attempt was to study the interaction between the protein Arabinosyltransferase C with the two existing drugs (Ethambutol and Isoniazid) and five modified molecules derived from Ethambutol by calculating their binding affinity and mode of binding through molecular docking study using AutoDock 4. From the comparison study of the existing drug (EMB and INH) and the five proposed modified molecules (Emb1, Emb2, Emb3, Emb4 and Emb5), it is analysed that Emb1 and Emb3 with binding affinities -5.77 kcal/mol and -5.13 kcal/mol respectively can be considered as potential inhibitors of Arabinosyltransferase C in Mycobacterium tuberculosis which is responsible for cell wall synthesis. The facts provided may be further verified experimentally for future drug discovery process to make a stand against tuberculosis and contribute an advance research for worthy antimycobacterial strategies.
Keywords: Pharmaceutical chemistry, Binding affinity, Multi-drug resistant, Molecular docking, Anti-TB drugs, Tuberculosis
Pharmaceutical chemistry; Binding affinity; Multi-drug resistant; Molecular docking; Anti-TB drugs; Tuberculosis.
1. Introduction
Tuberculosis (TB) is one of the world's deadliest and infectious diseases caused by Mycobacterium tuberculosis (MTB) which infects around one fourth of the world's population. According to WHO, in 2016 around 10.4 million people were affected with TB and is a major cause of death of HIV positive people [1]. Multidrug resistant (MDR) bacteria are encountered to be one of the most predominant public health issues which creates havoc worldwide [2]. Among 6,00,000 cases of TB, 4,90,900 are multidrug resistant TB (MDR-TB) [1]. EMB (Ethambutol) and INH (Isoniazid) are anti mycobacterial drugs used massively for treatment of tuberculosis out of that the isoniazid (INH) is found to be resistant in recent days [3]. In the United States latest assess indicates that EMB resistance occurs in 2.3 % and 3.8 % of patients with new and recurrent tuberculosis, respectively [4]. A total list of 63 surveys of resistance to antituberculosis drugs conducted between 1985 and 1994 revealed that the rate of gained EMB resistance was above 13.7% in few countries [5]. Arabinosyltransferase C is a transferase enzyme which acts upon arabinose that occurs in furanose form which is involved in polymerisation of arabinogalactan that is an important and essential part of mycobacterium cell wall. EMB is used in fusion with isoniazid, pyrazinamide and rifampin(rifampicin) which hinder the biosynthesis of cell wall inhibiting the synthesis of arabinogalactan and lipoarabinaomannan(LAM) and act via three arabinosyl transferase i.eEmbA(Rv3794), EmbB(Rv3795) and EmbC(Rv3793). EmbA, EmbB and EmbC belongs to glycosyltransferase superfamily C(GT-C) [6]. EmbA and EmbB are involved in biosynthesis of arabinogalactan [7] whereas EmbC are associated with LAM synthesis [8]. LAM is a major component in many outlooks of inter relationship between Mycobacterium species and host cells [9, 10, 11]. EmbC is considered as the high quality target for ethambutol [12] as there is strong association between EmbC activity and size of LAM species produced from over expression of embC mutant of M.smegmatis with embCMtb. These Emb enzymes of M. tuberculosis exhibit a typical architecture of 13 transmembrane helices and have synchronicity with a hydrophilic C-terminal domain consist of residues from 719–1094 within the full-length enzyme [13, 14]. Structure-based drug discovery, computational approach furnishes a precious substitute to the expensive and tedious process of random screening whereas, ligand based computational virtual screening methods along with other methods are found to be important part that being used for de novo characterisation, identification of potential inhibitors and drug repurposing. The current study has been undertaken to evaluate the binding affinity of existing drugs Isoniazid and different functional derivatives of Ethambutol against Arabinosyltransferase C of Mycobacterium tuberculosis using molecular docking [15].
2. Materials and methods
The current investigation involved retrieval of the 3D structure of target enzyme and ligands from Protein Data Bank(https://www.rcsb.org/) and PubChem database(https://pubchem.ncbi.nlm.nih.gov/) respectively. The proposed modified structures of the ligand were designed in Argus lab(http://www.arguslab.com). Molecular docking experiments were performed by AutoDock 4 [16] whereas docking complexes were visualized by Discovery Studio 3.0 (http://www.3dsbiovia.com) and PyMol tool(https://pymol.org/).
2.1. Retrieval of target enzyme structure
Probable Arabinosyltransferase C (embc) enzyme was retrieved from the UniProt protein sequence database (http://www.uniprot.org) for collecting annotated sequence information. The secondary structure of C terminal extracellular domain of embc protein was analysed using PredictProtein (https://www.predictprotein.org/) tool in order to find out the composition of helices, sheets and coils that are helpful to understand the pattern of hydrogen bonds between the amine hydrogen and carboxyl oxygen atoms. Three dimensional crystal structure of the C terminal extracellular domain of Mycobacterium tuberculosis embc was retrieved from the Protein Data Bank (PDB) database (http://www.rcsb.org/pdb/home/home.do). Structural details pertaining to enzyme were retrieved from PDBsum server (http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html) which includes topology of the secondary structure, sheets, Beta alpha beta units, Beta hairpins, Beta bulges, strands, Helices, Helix-helix interactions, Beta turns, gammas turn and disulphide bridge. Prior to docking the water molecules, unwanted hetero atoms and other ligand compounds were removed through Pymol and energy minimisation was performed using ModRefiner algorithm to obtain more suitable and stable conformations of embc [17].
2.2. Ligand retrieval and modifications
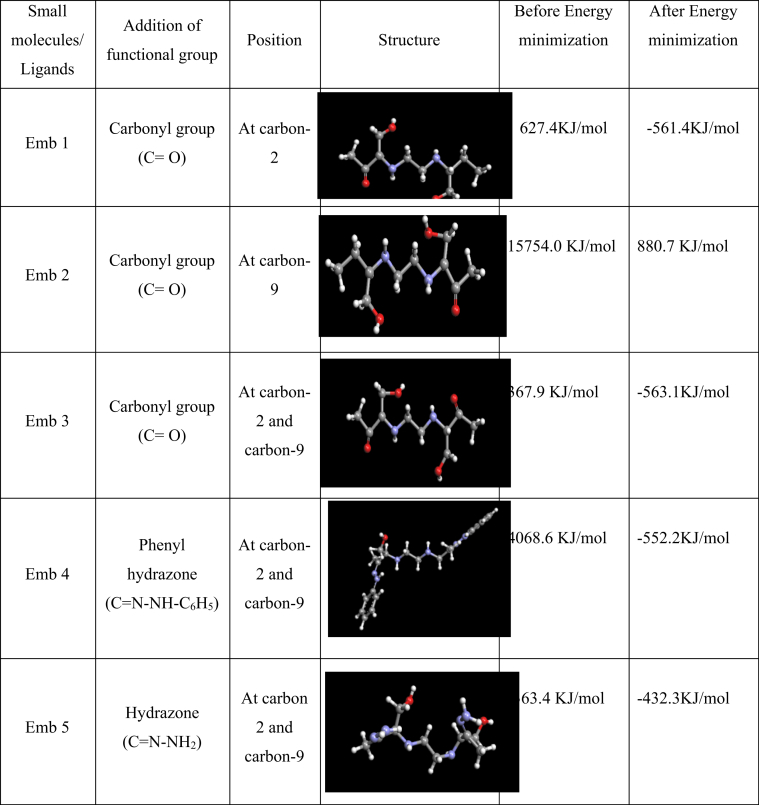
The structures of Ethambutol (CID: 14052) and Isoniazid (CID: 3767) were downloaded from the PubChem database (http://www.ncbi.nlm.nih.gov/pcc) in SDF format. The specified 3D structure of the ligands in SDF format were converted into PDB format using Pymol. The downloaded structures of Ethambutol were again modified using Argus lab (http://www.arguslab.com/arguslab.com/ArgusLab.html) through rational approach. The first three modified structures Emb1, Emb2 and Emb3 were modified at carbon-2, carbon-9 and both carbon-2 and 9 respectively by adding polar carbonyl groups. The fourth structure was modified at carbon 2 and 9 by adding Phenylhydrazone(C=N–NH–C6H5) and the fifth structure was modified at carbon 2 and 9 by adding Hydrazone (C=N–NH2). Then the energy minimisation of five modified ligand structures of Ethambutol were performed in UCSF Chimera [18] using Amber force field to make the molecule stable with less steric strain. Then these five modified structures of ethambutol along with the native ethambutol and isoniazid were used for molecular docking calculations.
2.3. Prediction of drug binding cavities
The specific Ethambutol and Isoniazid drug binding cavities in embc protein of Mycobacterium tuberculosis are not well characterised. The amino acid residues responsible for cavity formation in embc protein were identified through CASTp (Computed Atlas of Surface Topography of Proteins) (http://sts.bioe.uic.edu/castp/) web server. Moreover, the selection of binding pocket with largest volume and area allows free flexible rotation of ligands after binding.
2.4. Protein and ligand preparation
2.4.1. Protein
The energy minimised crystal structure of C-terminal domain of Arabinosyltransferase C (PDB ID: 3PTY) was selected for the molecular docking studies using AutoDock Tools 1.5.6. Then Kollaman charges were added to the protein molecule followed by addition of polar hydrogen atoms and saved the charged protein molecule in .pdbqt format.
2.4.2. Ligand
The native structures of Ethambutol and Isoniazid (standard) along with other five modified structures of Ethambutol (test ligand) were subjected to molecular docking study for prediction of possible binding pose with the target protein molecule(embc). The ligand preparation consists of series of steps that generates variation and optimisation of the structure and saved it as .pdbqt.
2.5. Molecular docking studies
The two native and five modified ligand structures were docked in the potential binding sites of embc protein by using AutoDock 4 package [16]. The AutoDock software performs the prediction of bound conformation based on free binding energies, which was calculated on the basis of the empirical force field and the Lamarackian Genetic Algorithm [16]. The AutoGrid module was used to create a grid box of dimension(50 × 66 × 74) A0 along with the direction with a spacing of 0.375 A0. The binding affinity between the ligand and target receptor was pursued by considering the hydrogen bond (intermolecular) interaction which was observed between the amino acid residues available on the target side with the functional group of the small molecules.
3. Results and discussion
3.1. Analysis of target enzyme molecular structure
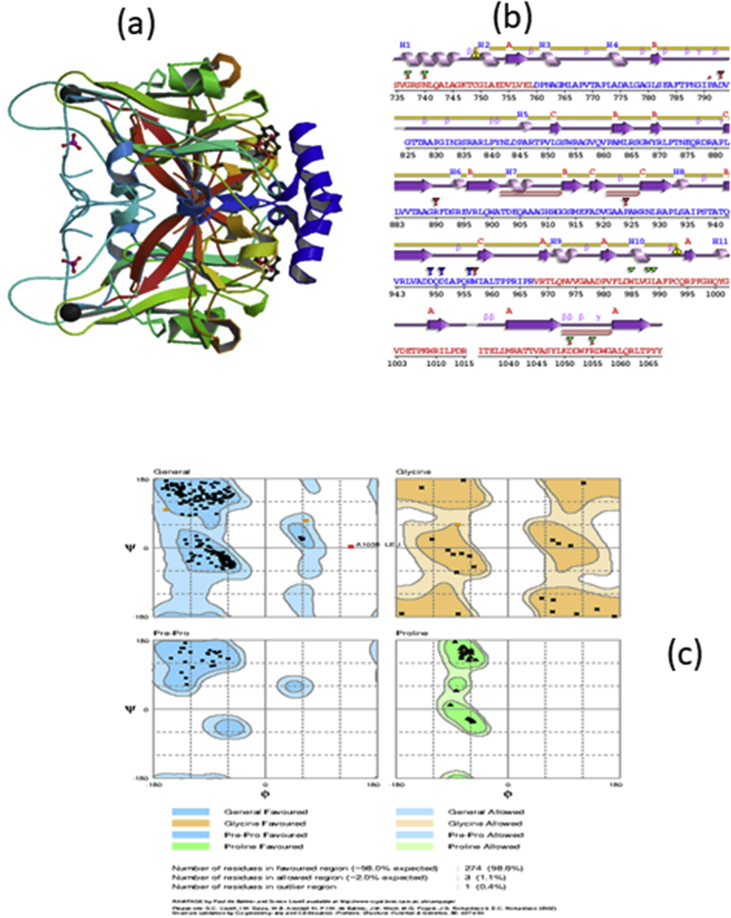
The Probable Arabinosyltransferase C protein(UniProt ID: P9WNL5) consists of 3 domain Arabinos_trans(PF04602), Arabino_trans_N(PF17689) and Arabino_trans_C(PF14896). Three dimensional crystal structure of the C terminal extracellular domain of Mycobacterium tuberculosis embc (PDB ID- 3PTY) with resolution 2.00 Ao was retrieved from the Protein Data Bank (PDB) database shown in Fig. 1. Secondary structure analysis revealed that the protein molecule is dominated by beta turns (22), strands(18) followed by alpha helices(11) (Table 1.). The three dimensional structure consists of chain-A (284 amino acids), two groups of ligands [AFO(Octyl alpha-D-Arabinofuranoside) & PO4(Phosphate ion)], one metal ion(Ca ion) and water molecules(113). The active sites of the protein embc were obtained through CASTp server. The positions are ASN740, ALA743, LEU744, LYS747, LEU751, ASP754, LEU1049, ASP1051, ASP1052 and ARG1055.
Fig. 1.
Structural information of Arabinosyl transferase C (a)3D Structure of Arabinosyl tansferase c (b) Secondary structure of 3pty mapped obtained using PDBsum (c) Ramachandran plot.
Table 1.
Details of structural elements of Arabinosyltransferase C.
| Secondary structural elements | Numbers |
|---|---|
| Sheets | 3 |
| Beta alpha beta units | 1 |
| Beta hairpins | 3 |
| Beta bulges | 3 |
| Strands | 18 |
| Helices | 11 |
| Helix-helix interactions | 2 |
| Beta turns | 22 |
| Gamma turns | 3 |
| Disulphide bridge | 1 |
3.2. Determination of the potential ligand structure
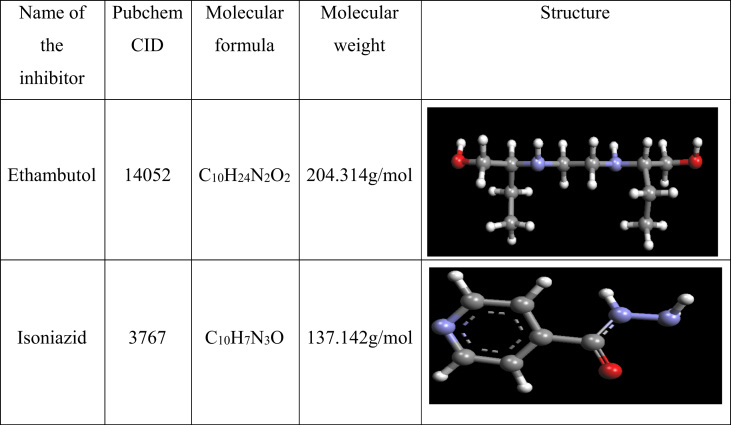
The native conformation of Ethambutol (C10H24N2O2; MW: 204.314 g/mol) with PubChem CID:14052 and Isoniazid(C10H7N3O; MW:137.14 g/mol) with PubChem CID: 3767 used as an antibiotic with bacteriostatic, antimicrobial and antitubercular properties (Fig. 2). The five modified Ethambutol structures (addition of functional groups such as carbonyl, phenyl hydrazone, hydrazone groups at different carbon positions) were used to find out their potential binding with the target protein(embc) (Fig. 3).
Fig. 2.
Structural information about Arabinosyltransferase inhibitors.
Fig. 3.
List of five modified Ethambutol structures.
3.3. Docking-based interaction studies
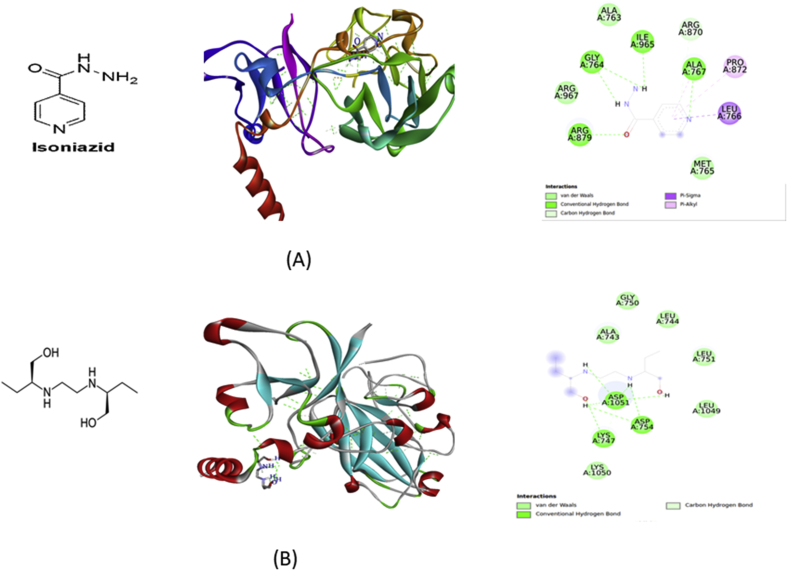
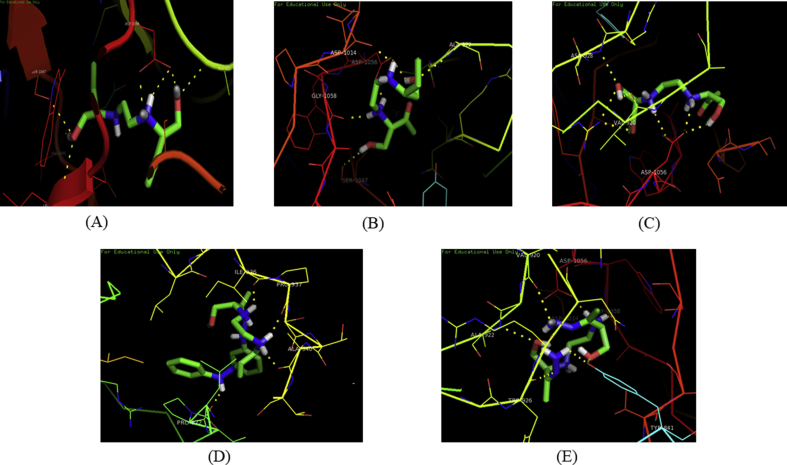
The molecular docking study was performed by using the enzyme Arabinosyltransferase C to identify the binding affinity and interaction of ligands taken with different essential amino acid residues on the C-terminal domain of EmbC. A specific ligand is stabilized energetically at a particular binding site of the protein structure by weak intermolecular interactions such as H-bonds or even with other noncovalent interactions such as ionic interactions, hydrophobic interactions, van der Waals forces which play a vital role in protein ligand interaction [19, 20]. Based on this background, docking was carried out against the receptor where the two standard ligands i.e Ethambutol formed eight conventional hydrogen bonds with binding energy -4.52 kcal/mol and Isoniazid formed five conventional hydrogen bonds,one carbon-hydrogen bonds, pi-sigma bond with LEU766 and pi-alkyl bond with PRO872 with binding energy -5.24 kcal/mol(Table 2) shown in Fig. 4. The modifications occurred in different position with addition of polar functional groups and further subjected to energy minimization are illustrated in Fig. 2. The five modified test ligands derived from Ethambutol i.e Emb1 forms hydrogen bond with binding energy -5.77 kcal/mol with crucial amino acid residues ALA922, ASP1056, SER1047 and VAL1045. Another molecule which was modified by adding carbonyl group at carbon-9 i.e Emb2 forms hydrogen bond with binding energy -4.06 kcal/mol with amino acid residues ALA922, ASP1056, SER1047, GLY1058 and ASP1014.The third molecule was modified at carbon-2 and carbon-9 i.e Emb3 forms hydrogen bonds with binding energy -5.14 kcal/mol with amino acid residues ASP1056, VAL920 and ASN928.The fourth molecule was modified at carbon 2 and 9 by adding phenyl hydrazone (C=N–NH–C6H5) i.e Emb4 with binding energy -4.79 kcal/mol with ILE936, PRO937, ALA940 and PRO872. At last the fifth molecule was modified at carbon 2 and 9 by hydrazone(C=N–NH2) i.e Emb5 forms hydrogen bonds with binding energy -2.22 kcal/mol with amino acid residues ASP1056, ASP1014, GLY1058, TYR841, VAL920, ALA922 and TYR926(Table 2) and shown in Fig. 5. Among these five modified molecules taken into consideration, Emb1 and Emb3 formed with binding energy -5.77 kcal/mol and -5.13 kcal/mol respectively can be predicted as energetically important for ligand binding forming hydrogen bonds which can be considered as the best ligand for inhibition of ArabinosyltransferaseC.
Table 2.
Docking calculation depicting interactions of target enzyme with native and modified ligand structures.
| Sl. no. | Modified molecule | Protein name | Amino acids | Types | Binding energies(kcal/mol) |
|---|---|---|---|---|---|
| 1 | Isoniazid | Arabinosyltransferase C | Ala 767, Arg 879, Gly764, Ile965, Arg 870 | Hydrogen bond | -5.24 |
| 2 | Ethambutol | Arabinosyltransferase C | Lys747, Asp1051, Asp754 | Hydrogen bond | -4.52 |
| 3 | Emb1 | Arabinosyltransferase C | Ala922, Asp1056, Ser1047, Val1045 | Hydrogen bond | -5.77 |
| 4 | Emb2 | Arabinosyltransferase C | Ala922, Asp1056, Ser1047, Gly1058, Asp 1014 | Hydrogen bond | -4.06 |
| 5 | Emb3 | Arabinosyltransferase C | Asp1056,Val920,Asn 928 | Hydrogen bond | -5.14 |
| 6 | Emb4 | Arabinosyltransferase C | Ile936,Pro937,Ala940,Pro872 | Hydrogen bond | -4.79 |
| 7 | Emb5 | Arabinosyltransferase C | Asp1056,Val920,Asp1014 Ala922,Gly1058,Trp926,Tyr841 | Hydrogen bond | -2.22 |
Fig. 4.
Interaction of Isoniazid (A) and Ethambutol (B) with Arabinosyltransferase C.
Fig. 5.
Interactions of the modified molecules Emb1(A), Emb2(B), Emb3(C), Emb4(D) and Emb5(E) with Arabinosyltransferase C.
4. Conclusion
This study involves the design of novel inhibitor molecules against the EmbC protein which involves in lipoarabinaomannan(LAM) symthesis in Mycobacterium tuberculosis. It revealed the binding modes of Ethambutol, Isoniazid and the five modified molecules derived from Ethambutol with the C-terminal domain of the Arabinosyltransferase (EmbC) of Mycobacterium tuberculosis. Isoniazid binds with Ala767, Arg879, and Gly767and Ile965 with binding affinity -5.24 kcal/mol whereas Ethambutol forms hydrogen bond with crucial amino acid residues such as Lys747, Asp1051 and Asp754 with binding affinity -4.52 kcal/mol. The five modified ethambutol ligands designed computationally also computed for their binding affinity with the target protein. Emb1 and Emb3 have shown more binding affinity i.e -5.77 kcal/mol and -5.13 kcal/mol respectively than that of original Ethambutol compound (-4.52 kcal/mol). With reference to the computed results, it has been observed that Emb1 shown better binding affinity than that of existing and modified ligands which may be considered as a potential drug target and can be encouraged for their anti-mycobacterial activity. The findings of this study may be further validated experimentally for future drug discovery process for combating tuberculosis. From these above observations, it can be stated that the proposed modified small molecules can provide a new area of research for novel anti-mycobacterial strategies.
Declarations
Author contribution statement
Nisha Das: Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Pradip Kumar Jena: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Sukanta Kumar Pradhan: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.World Health Organization . World Health Organization; 2016. World Malaria Report 2015. [Google Scholar]
- 2.van Duin D., Paterson D.L. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect. Dis. Clin. 2016;30:377–390. doi: 10.1016/j.idc.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marrakchi H., Lanéelle G., Quémard A. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology. 2000;146:289–296. doi: 10.1099/00221287-146-2-289. [DOI] [PubMed] [Google Scholar]
- 4.Bloch A.B., Cauthen G.M., Onorato I.M., Dansbury K.G., Kelly G.D., Driver C.R., Snider D.E. Nationwide survey of drug-resistant tuberculosis in the United States. Jama. 1994;271:665–671. [PubMed] [Google Scholar]
- 5.Cohn D.L., Bustreo F., Raviglione M.C. Drug-resistant tuberculosis: review of the worldwide situation and the WHO/IUATLD global surveillance project. Clin. Infect. Dis. 1997;24:S121–S130. doi: 10.1093/clinids/24.supplement_1.s121. [DOI] [PubMed] [Google Scholar]
- 6.Liu J., Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escuyer V.E., Lety M.A., Torrelles J.B., Khoo K.H., Tang Jyh-Bing, Rithner C.D., Frehel C., McNeil M.R., Brennan P.J., Chatterjee D. The role of the embA, B gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatisarabinogalactan. J. Biol. Chem. 2001;276:854–862. doi: 10.1074/jbc.M102272200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang N., Torrelles J.B., McNeil M.R., Escuyer V.E., Khoo K.H., Brennan P.J., Chatterjee D. The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol. Microbiol. 2003;50:69–76. doi: 10.1046/j.1365-2958.2003.03681.x. [DOI] [PubMed] [Google Scholar]
- 9.Chan J., Fan X.D., Hunter S.W., Brennan P.J., Bloom B.R. Lipoarabinomannan, a possible virulence factor involved in persistence of Mycobacterium tuberculosis within macrophages. Infect. Immun. 1991;59:1755–1761. doi: 10.1128/iai.59.5.1755-1761.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stokes R.W., Speert D.P. Lipoarabinomannan inhibits nonopsonic binding of Mycobacterium tuberculosis to murine macrophages. J. Immunol. 1995;155:1361–1369. [PubMed] [Google Scholar]
- 11.Underhill D.M., Ozinsky A., Smith K.D., Aderem A. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc. Natl. Acad. Sci. 1999;96:459–463. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goude R., Amin A.G., Chatterjee D., Parish T. The arabinosyltransferase EmbC is inhibited by ethambutol in mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2009;53:4138–4146. doi: 10.1128/AAC.00162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg S., Starbuck J., Torrelles J.B., Vissa V.D., Crick D.C., Chatterjee D., Brennan P.J. Roles of conserved proline and glycosyltransferase motifs of EmbC in biosynthesis of lipoarabinomannan. J. Biol. Chem. 2005;280:5651–5663. doi: 10.1074/jbc.M411418200. [DOI] [PubMed] [Google Scholar]
- 14.Seidel M., Alderwick L.J., Sahm H., Besra G.S., Eggeling L. Topology and mutational analysis of the single Emb arabinofuranosyltransferase of Corynebacterium glutamicum as a model of Emb proteins of Mycobacterium tuberculosis. Glycobiology. 2006;17:210–219. doi: 10.1093/glycob/cwl066. [DOI] [PubMed] [Google Scholar]
- 15.Kitchen D.B., Decornez H., Furr J.R., Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 2004;3:935. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 16.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 17.Yang J., Zhang Y. Protein structure and function prediction using I-TASSER. Curr. Protoc. Bioinform. 2015;52:5–8. doi: 10.1002/0471250953.bi0508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 19.Patil R., Das S., Stanley A., Yadav L., Sudhakar A., Varma A.K. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H., Huang D. Hydrogen bonding penalty upon ligand binding. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019923. [DOI] [PMC free article] [PubMed] [Google Scholar]