Abstract
Exosomes perform important functions for intercellular communication through extracellular signaling pathways, leading to the regulation of important biological processes, including cell proliferation, but also systemic dysfunctions such as preeclampsia (PE). However, the inhibitory effects of mesenchymal stem cell (MSCs)-derived exosomes in PE remain largely unknown. Thus, we assessed the possibility that exosomes could transport long non-coding RNA H19 and the correlation between H19 and the apoptosis of trophoblast cells. The expression of microRNA let-7b and forkhead box protein O1 (FOXO1) was characterized in placental tissues of PE patients. Gain- and loss-of-function experiments were performed to examine the roles of FOXO1 and let-7b in trophoblast cells. Interactions between let-7b and H19 as well as between let-7b and FOXO1 were confirmed by a dual-luciferase reporter assay, RNA pull-down, and RNA immunoprecipitation. HTR-8/SVneo cells were co-cultured with exosomes derived from MSCs overexpressing H19, followed by invasion, migration, and apoptosis assessments of trophoblast cells. We found that let-7b was highly expressed and FOXO1 was poorly expressed in placental tissues of PE patients. Furthermore, H19 acts as a competitive endogenous RNA against let-7b, and let-7b directly targeted FOXO1. Moreover, H19 could be transferred to trophoblast cells via MSC-secreted exosomes. MSC-derived exosomes overexpressing H19 decreased let-7b, increased FOXO1, and activated the protein kinase B (AKT) signaling pathway, thus increasing invasion and migration and inhibiting apoptosis of trophoblast cells. These results suggest that MSC-derived exosomes overexpressing H19 may be a novel direction for therapeutic strategies against PE.
Keywords: preeclampsia, long non-coding RNA H19, let-7b, FOXO1, mesenchymal stem cells, exosomes, AKT signaling pathway
Introduction
As a gestational hypertensive syndrome, preeclampsia (PE), is the major cause of fetal and maternal morbidity and mortality worldwide and it also leads to serious maternal complications, such as eclampsia, stroke, and organ failure.1,2 PE affects approximately 76,000 pregnant women and leads to 500,000 infant deaths worldwide every year.3 Multiple risk factors are related to the occurrence and progression of PE, which include nulliparity, diabetes, hypertension, previous PE, high maternal weight, twin pregnancy, and environmental and vascular-mediated factors.4, 5, 6 Some immunological factors are also known to increase the risks of PE, but the etiology of PE remains largely unknown and no available cure exists with the exception of pregnancy termination.7,8 Interestingly, abnormal proliferation and function of mesenchymal stem cells (MSCs) have been shown to localize at the maternal-fetal interface in PE patients.9 In addition, MSCs have been implicated in capillary formation and trophoblast cell invasion in PE.10 Exosomes secreted by MSCs have been reported to have a therapeutic potential that curbs cell apoptosis and induces angiogenesis in placental tissues.11 Exosomes are small intraluminal vesicles secreted from various cells, which can deliver intracellular contents, such as noncoding RNAs, cellular proteins, and messenger RNA (mRNA).12 The expression of some specific exosomal long noncoding RNAs (lncRNAs) has been reported to be associated with patients’ clinicopathological characteristics and can work as potential biomarkers.13
Furthermore, inhibition of let-7b is linked to the induction of forkhead box O1 (FOXO1) phosphorylation in hepatocarcinogenesis.14 Shallow trophoblast invasion and impaired vascular remodeling of spiral arteries have been associated with early-onset PE.15 As a transcription factor, FOXO1 upregulation was found to promote adhesion and migration of trophoblast cells, thus inhibiting cell motility in PE.16 Autophagy plays a critical role in the induction of cell death and thus the survival capacity of villous trophoblasts; alterations in autophagic protein expression are associated with pathological pregnancies such as PE.17 Upregulation of FOXO1 is known to induce autophagy through activation of the protein kinase B (AKT)1 signaling pathway.18 In addition, downregulation of CXC chemokine receptor 2 was associated with induction of PE through inhibition of trophoblast invasion by decreasing matrix metalloproteinase 2 and 9 via the AKT signaling pathway.19 An increased level of villous trophoblast apoptosis in pregnancies was induced by PE.20 It has been reported that inhibition of H19 could reduce trophoblast cell migration and invasion through repressing type III transforming growth factor β (TGF-β) receptor in placentae.21 Furthermore, disruption of imprinting orders in insulin-like growth factor 2/H19 by hormone-regulated placenta was associated with induction of PE.22 Moreover, recent reports show that H19 can act as a molecular “sponge” to sequester and regulate microRNA (miRNA) let-7 in muscle cells.23
Based on the aforementioned findings, we hypothesized that H19 delivered by exosomes from MSCs could enhance trophoblast cell invasion, migration, and suppression of apoptosis in PE. Thus, in the present study, we explored the role of H19 and its downstream effects in the progression of PE.
Results
FOXO1 Is Poorly Expressed in Placental Tissues in PE Patients
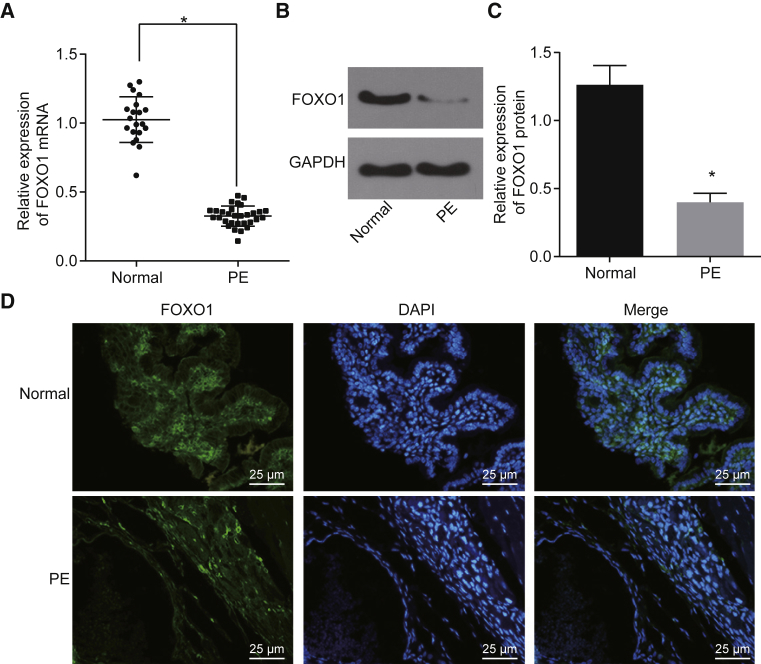
Clinical data from 30 pregnancies with PE and 20 healthy pregnancies were analyzed (Table 1). The patient samples were matched so that there was no significant difference in the maternal age, maternal body mass index (BMI), gestational age, fetal weight, and placental weight between the two groups. The blood pressure (BP) and urinary protein level of patients with PE were substantially greater when compared with those of healthy pregnant women (p < 0.05). To investigate whether the expression of the FOXO1 gene was altered in patients with PE, quantitative reverse transcription polymerase chain reaction (qRT-PCR) and western blot analysis were performed. Low expression of FOXO1 was found in placental tissues of PE patients relative to that in healthy pregnant women (p < 0.05; Figures 1A–1C). Furthermore, immunofluorescence staining showed that the expression of FOXO1 was decreased in placental tissue of PE patients compared to that of healthy pregnant women (Figure 1D). These data suggest that FOXO1 is downregulated in placental tissues of PE patients.
Table 1.
Maternal and Fetal Baseline Demographic Data of the PE Patients and Normal Pregnant Women
| Clinical Indicators | PE (n = 30) | Normal Pregnant Women (n = 20) |
|---|---|---|
| Maternal age (years) | 30.97 ± 2.92 | 31.45 ± 3.93 |
| BMI (kg/m2) | 25.60 ± 2.19 | 22.43 ± 2.31 |
| Gestational age (weeks) | 38.77 ± 2.64 | 39.20 ± 1.32 |
| Fetal weight (g) | 3,214.17 ± 513.44 | 3,374.50 ± 261.19 |
| Placental weight (g) | 512.73 ± 81.85 | 548.80 ± 65.66 |
| BP (mm Hg) | 144/12 | 112/72 |
| Urinary protein | ++ | − |
PE, preeclampsia; BMI, body mass index; BP, blood pressure.
Figure 1.
Downregulation of FOXO1 Is Found in Placental Tissues of PE Patients
(A–D) The expression of FOXO1 in placental tissues of PE patients and healthy pregnant women was measured by qRT-PCR and western blot analysis (A–C) and was detected by immunofluorescence staining (D) (original magnification, ×400). *p < 0.05 compared with healthy pregnant women. The data are expressed as mean ± standard deviation. Comparisons between two groups were conducted by means of an unpaired t test (normal, n = 20; PE, n = 30) PE, preeclampsia.
Overexpression of FOXO1 Promotes Migration and Invasion and Inhibits Apoptosis of Trophoblast Cells
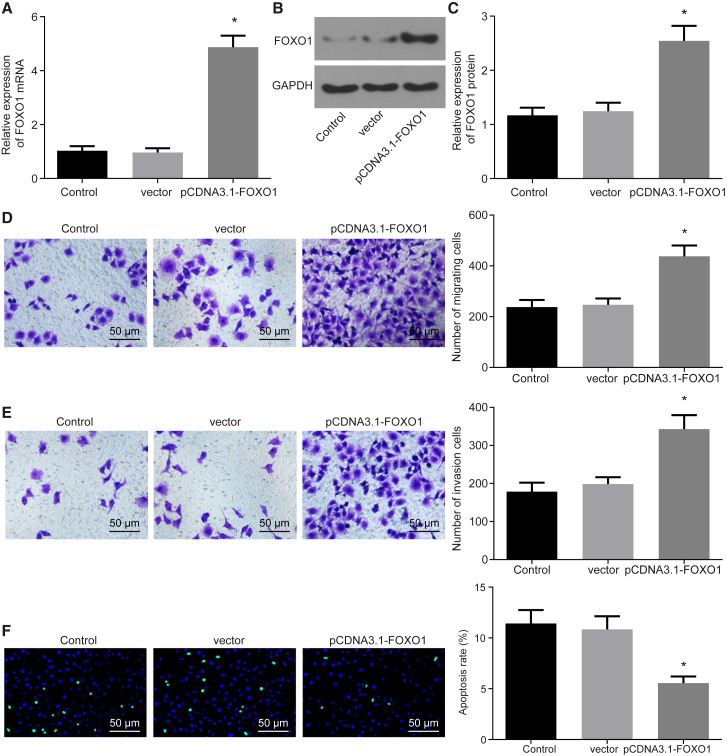
Next, we set out to investigate the effects of FOXO1 in trophoblast cells. qRT-PCR and western blot analysis were performed to detect the expression of FOXO1 in HTR-8/SVneo cells transfected with pCDNA3.1-FOXO1; a Transwell assay was set to test the migration and invasion of trophoblast cells, and terminal deoxynucleotidyltransferase (TdT)-mediated deoxyuridine triphosphate (dUTP) nick end labeling (TUNEL) staining was performed to detect cell apoptosis. Cells transduced with pCDNA3.1-FOXO1 increased the FOXO1 expression in HTR-8/SVneo cells, as expected (Figures 2A–2C), which in turn induced HTR-8/SVneo cell migration and invasion and reduced cell apoptosis (p < 0.05; Figures 2D–2F).
Figure 2.
Overexpression of FOXO1 Enhances HTR-8/SVneo Cell Migration and Invasion and Represses Apoptosis
(A) FOXO1 expression in HTR-8/SVneo cells measured by qRT-PCR. (B) Band diagram of FOXO1 expression in HTR-8/SVneo cells determined by Western blot analysis. (C) Histogram of FOXO1 expression in HTR-8/SVneo cells determined by Western blot analysis. (D) Cell migration of HTR-8/SVneo cells detected by Transwell assay (×100). (E) Cell invasion of HTR-8/SVneo cells detected by Transwell assay (×100). (F) The apoptosis of HTR-8/SVneo cells was detected by TUNEL staining (original magnification, ×200). *p < 0.05 compared with the control group (untransfected cells). The data are expressed as mean ± standard deviation. Comparisons among multiple groups were analyzed by one-way analysis of variance with Tukey’s post hoc test. The experiment was repeated three times.
Downregulation of let-7b Induces Cell Migration and Invasion while Suppressing Apoptosis by Upregulating FOXO1 in Trophoblast Cells
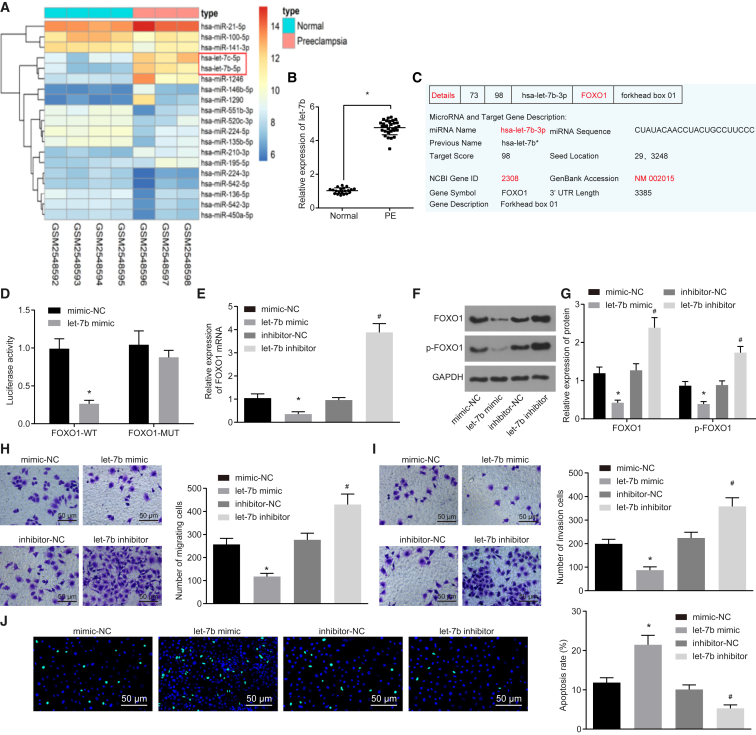
We then analyzed the expression of let-7b and the relationship between let-7b and FOXO1 in PE patients. let-7b was found to be highly expressed in PE patients after analysis of PE-related microarray data in GEO: GSE96985 (Figure 3A). qRT-PCR results also confirmed that the let-7b expression was higher in the placental tissues of patients with PE than that of placental tissues in healthy pregnant women (p < 0.05; Figure 3B). Using online analysis software, we uncovered predicted binding sites between FOXO1 and let-7b based on their gene sequences (Figure 3C). The molecular interaction between FOXO1 and let-7b was further verified by a dual-luciferase reporter gene assay. Compared with the negative control (NC) group, the luciferase activity of FOXO1-wild-type (WT) was reduced by a let-7b mimic (p < 0.05), while mutation of the binding sites abolished the repressive effect of let-7b (p > 0.05; Figure 3D). let-7b overexpression or knockdown in HTR-8/SVneo cells further verified the strong negative correlation with FOXO1 (p < 0.05; Figures 3E–3G). At the same time, cell migration, invasion, and apoptosis were detected by a Transwell assay and TUNEL staining (Figure 3H–3J). Cells treated with a let-7b inhibitor induced cell migration and invasion and reduced cell apoptosis, while cells transfected with a let-7b mimic decreased cell migration and invasion and increased cell apoptosis. Taken together, downregulation of let-7b can enhance cell migration and invasion, at the same time suppressing cell apoptosis, by negatively regulating FOXO1.
Figure 3.
Downregulation of let-7b Induces Cell Migration and Invasion and Inhibits Cell Apoptosis by Upregulating FOXO1
HTR-8/SVneo cells were transfected with inhibitor-NC, let-7b inhibitor, mimic-NC, and let-7b mimic vectors. (A) Analysis of PE-related dataset GEO: GSE96985. (B) The let-7b expression in the placental tissues of PE patients was performed by qRT-PCR. (C) The let-7b and FOXO1 binding site was predicted online. (D) Relationship between FOXO1 and let-7b was verified by detecting the luciferase activity of FOXO1-WT and FOXO1-Mut in HTR-8/SVneo cells. (E) FOXO1 mRNA expression determined by qRT-PCR. (F) Band diagram of FOXO1 and p-FOXO1 protein expressions determined by Western blot analysis. (G) Statistical chart of FOXO1 and p-FOXO1 protein expression determined by Western blot analysis. (H and I) The migration and invasion of HTR-8/SVneo cells were measured by Transwell assay. (J) The apoptosis of HTR-8/SVneo cells was detected by using TUNEL staining (original magnification, ×200). *p < 0.05 compared with the normal (normal placenta) or mimic-NC groups (cells transfected with mimic-NC); #p < 0.05 compared with the inhibitor-NC group (cells transfected with inhibitor-NC). The data are expressed as mean ± standard deviation. Comparisons between two groups were conducted by means of an unpaired t test. The experiment was repeated three times.
H19 Competitively Binds to let-7b
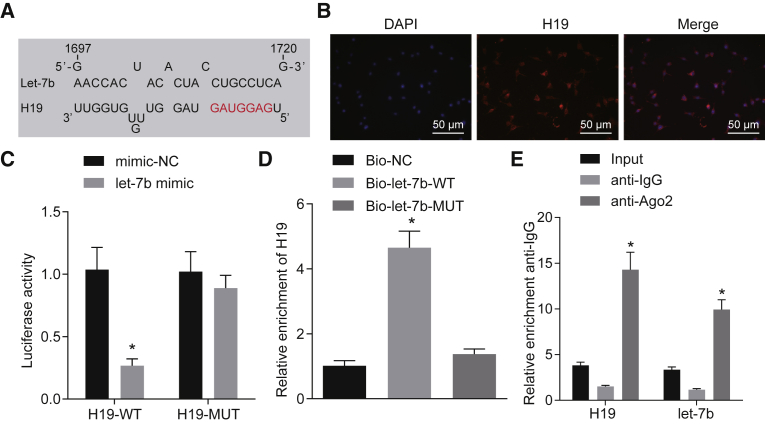
According to bioinformatics analysis, a binding interaction between lncRNA H19 and let-7b was predicted (Figure 4A). A fluorescence in situ hybridization (FISH) experiment substantiated that H19 was mainly located in the cytoplasm (Figure 4B) and, furthermore, the molecular interaction between H19 and let-7b was verified by a dual-luciferase reporter gene assay. The let-7b mimic significantly reduced luciferase activity of H19-WT (p < 0.05), while the let-7b mimic had no significant effect on the luciferase activity of H19-mutant (Mut) (Figure 4C). To further test the relationship between let-7b and H19, RNA immunoprecipitation (RIP) and RNA pull-down assays were carried out. Results showed that bio-let-7b-WT was able to pull down H19 RNA (p < 0.05), while the corresponding bio-let-7b-Mut had no effect on H19 expression (Figure 4D). According to the RIP results, the enrichment of Argonaute 2 (Ago2) antibody in H19 and let-7b RNA augmented notably (p < 0.05; Figure 4E). Taken together, these results strongly suggest that H19 can bind to let-7b.
Figure 4.
H19 Binds to let-7b
(A) The binding sequences of H19 and let-7b were predicted by bioinformatics analysis. (B) The subcellular localization of H19 was verified by using RNA-FISH (original magnification, ×200). (C) The relationship between H19 and let-7b was verified by the dual-luciferase reporter gene assay. *p < 0.05 compared with the mimic-NC group. (D) Cells were delivered with mimic-NC and let-7b mimic vectors. The relationship between let-7b and H19 was tested by RNA pull-down. *p < 0.05 compared with the bio-NC group. (E) H19 and let-7b bound to Ago2 protein were detected by RIP. *p < 0.05 compared with the vector group. The data are expressed as mean ± standard deviation. Comparisons between two groups were conducted by means of an unpaired t test. Comparisons among multiple groups were analyzed by one-way analysis of variance with Tukey’s post hoc test. The experiment was repeated three times.
MSCs Are Successfully Cultured and Exosomes Are Successfully Extracted
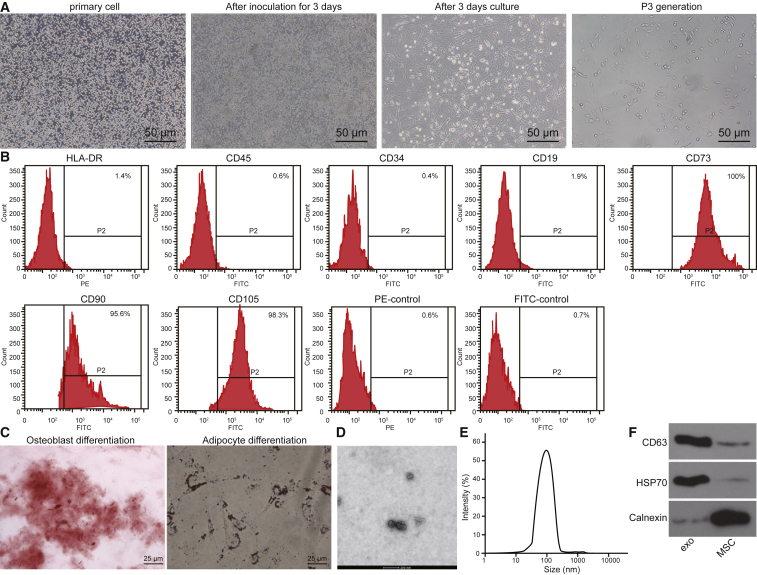
MSCs were allowed to adhere to cell culture plates for 3 days after inoculation. Cells had a typical spindle shape and grew in a spiral-shaped or cluster formation with clear nuclei, which were typical cellular properties of MSCs (Figure 5A). Flow cytometry was used to analyze the surface markers of MSCs. The expression percentages of CD73, CD90, CD105, CD34, CD45, CD19, and human leucocyte antigen-G (HLA-DR) were 100%, 95.6%, 98.3%, 0.4%, 0.6%, 1.9%, and 1.4%, respectively (Figure 5B). Next, we identified the osteogenic and adipogenic differentiation ability of the MSC cells. After 21 days of osteogenic induction culture, the cells overlapped in layers and formed calcified nodules containing a small amount of mineral deposits, which indicated that they had the potential to differentiate into osteoblasts (Figure 5C). After 25 days of adipogenic induction culture, lipid deposition occurred in the cells, and the lipid droplets gradually became larger or beaded, indicating that cells had a potential to also differentiate into adipocytes (Figure 5C).
Figure 5.
MSCs Were Successfully Cultured and Identified, and Exosomes Were Successfully Extracted from MSCs
(A) Morphological observation of MSCs was performed (original magnification, ×200). (B) The surface antigen expression of MSCs was detected by flow cytometry. (C) Osteogenesis and adipogenic induction cultures of MSCs were conducted (original magnification, ×400). (D) Identification of exosomes was performed by TEM. (E) The diameter of exosome particles was measured by dynamic light scattering. (F) The expression of CD63 and HSP70 in exosomes was further detected by western blot analysis. *p < 0.05 compared with the MSCs. The data are expressed as mean ± standard deviation. Comparisons between two groups were conducted by means of an unpaired t test. The experiment was repeated three times.
Finally, transmission electron microscopy (TEM) of exosomes extracted from MSCs revealed the presence of spherical structures 30–120 nm in diameter (Figure 5D). Dynamic light scattering measurements verified the vesicle size distribution (Figure 5E). The expression of exosome surface markers CD63 and heat shock protein 70 (HSP70) as well as the negative marker calnexin was detected by western blot analysis. The exosome was positive for CD63 and HSP70 and negative for calnexin (Figure 5F). Overall, we were able to successfully culture MSCs and extract the exosomes they produce.
H19 from MSC-Derived Exosomes Inhibits let-7b and Induces FOXO1 Expression in Human Trophoblast Cells
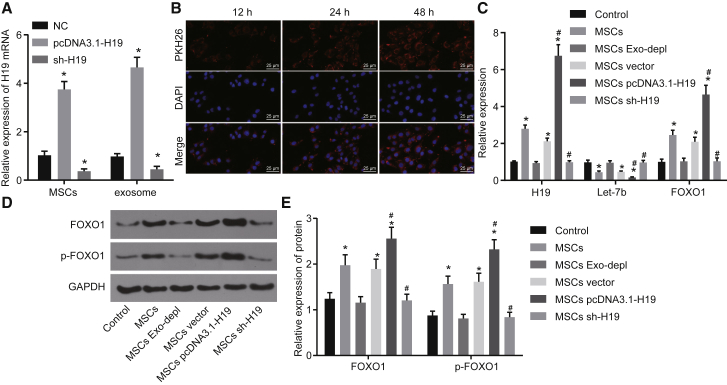
To investigate whether exosomes released by MSCs could transfer H19 to HTR-8/SVneo cells, the expression of H19 in MSCs and exosomes derived from MSCs was detected by qRT-PCR. H19 was expressed at a high level in H19-overexpressing MSCs and exosomes derived from H19-overexpressing MSCs (p < 0.05; Figure 6A). Then, exosomes traced by PKH26 were co-cultured with HTR-8/SVneo cells for 12, 24, and 48 h, and the exosomes that were taken up by HTR-8/SVneo cells were observed under a confocal fluorescence microscope. Our results showed that as the co-culture time was prolonged, more and more HTR-8/SVneo cells became positive for green fluorescence (Figure 6B). After co-culture for 48 h, the uptake of PKH26-labeled exosomes by HTR-8/SVneo cells was evident, suggesting that exosomes can be transferred from MSCs to HTR-8/SVneo cells.
Figure 6.
H19 Overexpression in MSCs Downregulates let-7b and Upregulates FOXO1 in HTR-8/SVneo Cells
(A) The expression of H19 in MSCs and in the exosomes derived from MSCs was detected by qRT-PCR. (B) Uptake of exosomes in HTR-8/SVneo cells was observed by confocal fluorescence microscopy (original magnification, ×400). (C–E) HTR-8/SVneo cells were co-cultured with MSCs or MSC-derived exosomes treated with GW4869, MSC-derived exosomes overexpressing H19, or MSCs transfected with sh-H19. (C) Expression of H19, let-7b, FOXO1, and p-FOXO1 in HTR-8/SVneo cells measured by qRT-PCR. (D) Band diagram of FOXO1 and p-FOXO1 protein expression determined by Western blot analysis. (E) Histogram of FOXO1 and p-FOXO1 protein expression determined by Western blot analysis. *p < 0.05 compared with the NC (untransfected MSCs) or control group (HTR-8/SVneo cells); #p < 0.05 compared with the MSCs. The data are expressed as mean ± standard deviation. Comparisons among multiple groups were analyzed by one-way analysis of variance with Tukey’s post hoc test. The experiment was repeated three times.
To determine whether H19 transferred in vitro can effectively regulate let-7b and FOXO1 expression in HTR-8/SVneo cells, qRT-PCR and western blot analysis were performed to detect the expression of let-7b, FOXO1, and phosphorylated FOXO1 in HTR-8/SVneo cells co-cultured with MSCs. The results showed no significant changes in the expression of H19, let-7b, FOXO1, and phosphorylated FOXO1 in HTR-8/SVneo cells co-cultured with MSCs and treated with GW4869. In contrast, the expression of let-7b in HTR-8/SVneo cells co-cultured with MSCs overexpressing H19 was significantly decreased, while H19, FOXO1, and phosphorylated FOXO1 expression was profoundly increased, which were opposite to the changes caused by silencing of H19 in MSCs (p < 0.05; Figures 6C–6E). Taken together, our results suggest that H19 transferred in vitro can effectively inhibit let-7b expression and in turn upregulate expression of FOXO1 and phosphorylated FOXO1 in HTR-8/SVneo cells.
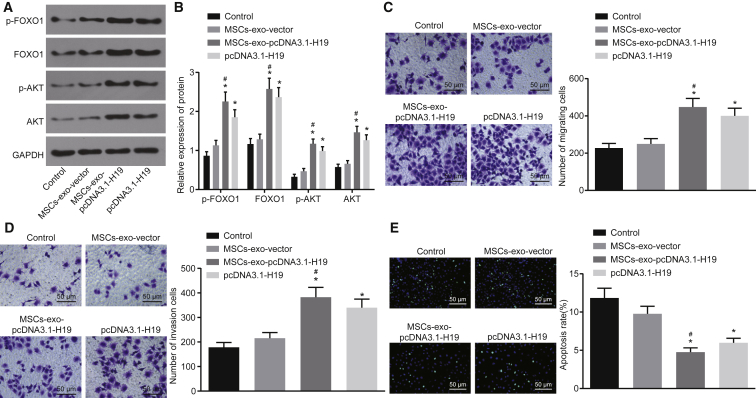
MSC-Derived Exosomal H19 Potentiates Cell Migration and Invasion and Reduces Apoptosis of Trophoblast Cells
In order to identify the role of the H19/let-7b/FOXO1 axis in migration, invasion, and apoptosis of trophoblast cells and whether this acts through the protein kinase B (AKT) signaling pathway, we investigated these processes using qRT-PCR, western blot analysis, Transwell assay, and TUNEL staining, respectively. When HTR-8/SVneo cells were co-cultured with exosomes from MSCs overexpressing H19, increased expression of FOXO1, phosphorylated FOXO1, and AKT induced cell migration and invasion, but significantly decreased expression of let-7b and reduced cell apoptosis (p < 0.05; Figure 7A–7E). Taken together, overexpression of H19 in MSCs and release via exosomes inhibited the expression of let-7b, increased FOXO1 expression, activated the AKT signaling pathway, and led to enhanced cell migration and invasion and lower apoptosis rates of trophoblast cells.
Figure 7.
MSCs Overexpressing H19 via Exosomes Promote Migration and Invasion and Suppress Apoptosis of Trophoblast Cells
HTR-8/SVneo cells were transfected with pcDNA3.1-H19 or co-cultured with exosomes derived from MSCs overexpressing H19. (A and B) The expression of FOXO1 and AKT, as well as the extents of FOXO1 and AKT phosphorylation, was detected by western blot analysis. (C) Cell migration of HTR-8/SVneo cells measured by Transwell assay (×100). (D) Cell invasion of HTR-8/SVneo cells measured by Transwell assay (×100). (E) Cell apoptosis of HTR-8/SVneo cells measured by TUNEL assay (×100). *p < 0.05 compared with the control group (HTR-8/SVneo cells transfected with pcDNA3.1-NC); #p < 0.05 compared with the MSC-exo-vector group. The data are expressed as mean ± standard deviation. Comparisons among multiple groups were analyzed by one-way analysis of variance with Tukey’s post hoc test. The experiment was repeated three times.
Discussion
PE is linked to decreased invasion of fetal trophoblast cells and injured remodeling of maternal spiral arteries, which results in poor uterine placental perfusion.24,25 miRNAs were found to interact with lncRNAs and regulate each other’s biological processes; in particular, the sponge-like function of lncRNAs leads to complex miRNA interactions.26 Our study explored the role of MSC-secreted exosomal H19 as a competitive endogenous RNA (ceRNA) for let-7b with downstream effects on the regulation of FOXO1 and AKT signaling in trophoblast cells. We found that restoration of MSC-secreted exosomal H19 and silencing of let-7b promoted trophoblast cell migration and invasion and suppressed apoptosis by increasing FOXO1 expression and activating the AKT signaling pathway.
We found that let-7b is highly expressed, whereas FOXO1 expression is downregulated, in trophoblast cells of PE patients compared to healthy controls. Our results are consistent with a study from Noack et al.27 showing that let-7b is induced in patients suffering from severe PE. Furthermore, overexpression of let-7 in endothelial cells from PE notably correlated with the expression of genes related to endothelial dysfunction and pathophysiology development, suggesting a critical role of let-7 in PE.28 In addition, low expression of FOXO1 was found in villous trophoblasts, while overexpression of FOXO1 contributed to abnormal trophoblast differentiation and inhibition of apoptosis in mild and severe PE.29 More importantly, an increase of FOXO1 was reported to promote adhesion and migration of trophoblast cells, thus preventing the progression of PE.16 Similarly, in this study, induction of FOXO1 was suggested to promote the migration and invasion of trophoblast cells and inhibit their apoptosis. We also showed that FOXO1 was negatively regulated by let-7b, indicating that let-7b-associated functions in PE could be targeted through FOXO1 regulation. A previous study showed that FOXO3 is a gene target of let-7c, and that expression of let-7b is inhibited by FOXO3 in breast cancer.30 Therefore, overexpression of FOXO1 and inhibition of let-7b may hold potential as a therapeutic target in PE. Recently, it was found that lncRNAs can act as ceRNAs to mediate gene expression involved in disease progression.31 Accumulating evidence reveals that overexpression of H19 inhibits let-7 and could work as a ceRNA in different cancers.32,33 Herein, we report that H19 acts as a ceRNA of let-7b and induces FOXO1 expression in trophoblast cells of PE patients, consistent with the previous reports. Furthermore, our study showed that H19 could be delivered to the trophoblast cells by MSC-secreted exosomes. A recent study suggested that MSCs might exert their therapeutic function via exosomes.11 Exosomes work as biological cargo vessels, which lately have been established as an important mediator of intercellular communication through transfer of mRNAs, miRNAs, and noncoding RNAs.34 A previous study found that the expressions of certain exosomal lncRNAs are linked to patients’ clinicopathological characteristics and have the potential to work as biomarkers.13 It has been shown that miRNAs transferred by MSC-derived exosomes can inhibit human umbilical vein endothelial cell capillary formation and trophoblast invasion, thus affecting the pathogenesis of PE.35 In this study, we carried out a series of experiments to see whether exosomal H19 secreted from MSCs could be transferred into trophoblast cells, thereby modulating the progression of PE. These findings suggested that MSC-released exosomal H19 can play a role in trophoblast dysfunction, influencing trophoblast invasion and migration, hence leading to the development of PE.
Finally, we demonstrated that upregulation of H19 transferred by MSC-derived exosomes enhanced migration and invasion while inhibiting apoptosis of trophoblast cells in PE by downregulating let-7b, upregulating FOXO1, and activating the AKT signaling pathway. Upregulation of H19 induced migration and invasion of extravillous trophoblast cells in placentae with fetal growth restriction.21 Furthermore, it has been revealed that overexpression of H19 enhanced invasion of trophoblast cells by activating the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathways.36 Inhibition of let-7b and let-7g was found to activate the AKT signaling pathway in gastric tumorigenesis.37 Moreover, Szydłowski et al.38 showed that FOXO1, as a critical effector of the AKT signaling pathway, could continuously affect cell cycle and apoptosis via continuous steps in B cell differentiation. Another study indicated that activation of the AKT signaling pathway is associated with induced trophoblast proliferation, migration, and invasion, as well as hormone secretion in PE.39 In addition, inhibition of the transforming acidic coiled-coil protein 3 suppressed the migration and invasion of HTR-8/SVneo cells through suppression of the PI3K/AKT signaling pathway, indicating that activation of the AKT signaling pathway could induce cell migration and invasion in PE.40
Taken together, the present study revealed that exosomes carrying H19 could be effectively internalized by trophoblast cells. In addition, we provide evidence that exosomes carrying H19 work as a ceRNA against let-7 to upregulate FOXO1 and induce the activation of the AKT signaling pathway in trophoblast cells. Exosomes carrying H19 could potentially promote migration and invasion by binding to let-7, while inhibiting the apoptosis of trophoblast cells in patients suffering from PE. Further studies are needed to improve the purity of MSC-derived exosomes and to elucidate the function of the H19/let-7/FOXO1/AKT axis in an animal model for PE.
Materials and Methods
Ethics Statement
This study was conducted under the approval of the Ethics Committee of The Second Hospital of Jilin University. All participating patients signed informed consent documentation. All experiments were conducted in strict accordance with the Declaration of Helsinki.
Study Subjects
Thirty samples of placenta from PE patients in The Second Hospital of Jilin University from December 2017 to May 2018 were collected, and 20 healthy pregnant women in the same period of pregnancy served as normal controls. Shortly after delivery from the uterus (within 30 min), placental villous tissue samples (0.5 × 0.5 cm) were harvested from the maternal side of the placenta at a depth of 5 mm underneath the outermost layer of the placenta, adjacent to the insertion of the umbilical cord. The samples were rinsed in sterile phosphate-buffered saline (PBS) to remove excessive blood and were immediately frozen in liquid nitrogen and stored at −80°C until further analysis.41 Enrolled pregnant women had matched gestational weeks, age, height, and weight, regular menstruation, and normal pregnancy. All pregnancies with confounding factors, including smoking and drinking, were excluded from this study. Pregnancies with chronic hypertension, renal disease, cancers, or pregnancy complications such as gestational diabetes were also excluded. Enrolled pregnant women were in their first pregnancy with a singleton. The gestational weeks were carefully checked according to the last menstruation and the early pregnancy ultrasound. The PE diagnostic criteria were based on previous studies.42 Mild PE was defined as BP ≥ 140/90 mm Hg and urinary protein (+) after 20 weeks of gestation. Severe PE was defined as BP ≥ 160/110 mm Hg and urinary protein (++) after 20 weeks of gestation.
RNA Isolation and Quantification
Total RNA from placenta tissues or trophoblast cells was extracted according to instructions of the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The concentration and quality of total RNA were determined by NanoDrop 2000 (NanoDrop Technologies, Wilmington, DE, USA). The RNA was reversely transcribed into cDNA according to the instructions of the PrimeScript RT reagent kit with a genomic DNA (gDNA) eraser (RRO37A, Takara, Dalian, Liaoning, China). Reverse transcription was performed using the TaqMan miRNA reverse transcription kit (4366596, Thermo Fisher Scientific, Waltham, MA, USA), and let-7b expression was measured by a TaqMan miRNA assay according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, qRT-PCR experiments were performed on an ABI 7500 quantitative real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) using the SYBR Premix Ex Taq (Tli RNaseH Plus) kit (RR820A, Takara Bio, Shiga, Japan). The quantification of target genes was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), while that of miRNAs was normalized by U6. All primers were supplied by Shanghai Gene Pharma (Shanghai, China) and are listed in Table 2. The fold changes were calculated by means of relative quantification (2−ΔΔCt method).
Table 2.
Primer Sequences for qRT-PCR
| Gene | Primer Sequence |
|---|---|
| H19 | F: 5′-TACAACCACTGCACTACCTG-3′ |
| R: 5′-TGGAATGCTTGAAGGCTGCT-3′ | |
| let-7b | F: 5′-CTATACAACCTACTGCCTTCCC-3′ |
| FOXO1 | R: 5′-GCTGTCAACGATACGCTACCTA-3′ |
| F: 5′-CAGCCCTGGATCACAGTTTT-3′ | |
| R: 5′-CATCCCCTTCTCCAAGATCA-3′ | |
| U6 | F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | |
| GAPDH | F: 5′-ATGGAGAAGGCTGGGGCTC-3′ |
| R: 5′-AAGTTGTCATGGATGACCTTG-3′ |
FOXO1, forkhead box O1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; F, forward; R, reverse.
Western Blot Analysis
In strict accordance with the manufacturer’s instructions, high-efficiency radioimmunoprecipitation assay (RIPA) lysis buffer (C0481, Sigma-Aldrich, St. Louis, MO, USA) was used to extract total protein from placenta tissues or trophoblast cells at 4°C for 15 min. Total protein was quantified using a bicinchoninic acid protein reagent kit (23227, Thermo Fisher Scientific, Waltham, MA, USA). Lysates were centrifuged at 4°C at 15,000 rpm for 15 min. Protein extracts were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. After blockade of the membrane with 5% bovine serum albumin at room temperature for 1 h, target proteins were probed with primary antibodies FOXO1 (1:2,000, 18592-1-AP, Proteintech Group, Chicago, IL, USA), phosphorylated (p-)AKT (1:1,000, ab38449, Abcam, Cambridge, UK), AKT (1:500, ab8805, Abcam, Cambridge, UK), and GAPDH (1:5,000, ab8245, Abcam, Cambridge, UK) at 4°C overnight. Subsequently, horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG; 1:20,000, Abcam, Cambridge, UK) was added for 1.5 h at room temperature. GAPDH served as an internal reference.43
Immunofluorescence Assay
After blocking with 5% goat serum for 1 h at room temperature, the sections were incubated with the primary antibodies rabbit anti-mouse antibody against α-smooth muscle actin (α-SMA; 1:50, ab223068, Abcam, Cambridge, UK), collagen I (1:500, ab34710, Abcam, Cambridge, UK), and FOXO1 polyclonal antibody (1:100, 18592-1-AP, Proteintech Group, Chicago, IL, USA) overnight at 4°C. Then, sections were incubated with fluorescein-labeled goat anti-rabbit antibody against IgG (1:1,000, A21235, Alexa Fluor 488, Thermo Fisher Scientific, Waltham, MA, USA) for 1 h in the dark at room temperature. Sections were stained for 30 min in the dark at room temperature with 4′,6-diamidino-2-phenylindole (DAPI). Imaging was carried out at 525 nm and 470 nm using confocal laser scanning microscopy (Olympus, Tokyo, Japan).44
Isolation and Identification of MSCs
Human MSCs were isolated as previously described.45 In more detail, bone marrow aspirates (20 mL) were collected during orthopedic surgery with a bone marrow biopsy needle inserted through the cortical bone. The collected aspirates were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Biowest, Nuaillé, France) and 100 U/mL penicillin and streptomycin (Gibco, Life Technologies, Grand Island, NY, USA) in a 37°C and 5% CO2 incubator. When cell confluence reached 80%–90%, the cells were passaged and divided into bottles. The cells were collected for use at the third passage.
MSCs at passage 3 were resuspended in PBS after being detached by trypsin (Gibco by Life Technologies, Grand Island, NY, USA), and the cell concentration was adjusted to 1 × 106 cells/mL. A total of 200 μL of cell suspension was dispensed into Eppendorf tubes and probed with 5 μL of monoclonal antibody, fluorescein isothiocyanate-conjugated antibodies against CD19 (ab24936), CD34 (ab18227), CD45 (ab27287), CD73 (ab239246), CD90 (ab226), CD105 (ab53318), polyethylene glycol/Cy5-conjugated antibody against HLA-DR (ab95830), or with appropriate isotype control IgG (all from Abcam, Cambridge, UK) at 4°C for 15 min in the dark. Excess antibodies were removed by centrifugation at 1,000 rpm for 5 min with 2 mL of PBS. Then, MSCs were resuspended in 400 μL of PBS containing 0.5% paraformaldehyde and were fully mixed. Surface markers of MSCs were detected by flow cytometry.
To evaluate the differentiation potential of MSCs, osteogenic or adipogenic differentiation was induced using osteogenic (MUBMX-90021) or adipogenic (MUBMX-90031) media according to the manufacturer’s instructions. The osteogenic or adipogenic differentiation was identified by alizarin red staining (A5533-25G, Sigma-Aldrich, St. Louis, MO, USA) and oil red O staining (O1391-250 ML, Sigma-Aldrich, St. Louis, MO, USA), respectively.
Extraction and Identification of Exosomes from MSCs
Exosomes were removed from the serum by ultracentrifugation at 100,000 × g overnight at 4°C. MSCs at passage 3 were cultured overnight in DMEM. When cells reached 80%–90% confluence, the supernatant was collected after 24 h of culture in medium free of exosomes. The collected supernatant was centrifuged at 500 × g for 15 min and at 2,000 × g for 20 min at 4°C to remove cell debris and apoptotic bodies, and the obtained supernatant was centrifuged at 100,000 × g for 1 h at 4°C. The pellet was then suspended in serum-free DMEM containing 25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.4) and submitted to a second ultracentrifugation in the same conditions. The pellet was stored at −80°C until further use.46
TEM was used to identify exosomes. A total of 30 μL of exosomes was added to the formvar-coated copper grid, allowed to stand for 1 min, and the liquid was blotted from the side with filter paper. Then, exosomes were added with 30 μL of phosphotungstic acid (pH 6.8), and the mixture was counterstained for 5 min at room temperature. Next, exosomes were heated under an incandescent lamp and imaged with TEM.
Western blot analysis was used to characterize the isolated exosomes according to methods described previously.47 We next incubated the samples overnight with antibodies against CD63 (1:1,000, ab134041, Abcam, Cambridge, UK), HSP70 (1:1,000, ab134041, Abcam, Cambridge, UK), and the endoplasmic reticulum marker calnexin (1:1,000, ab112995, Abcam, Cambridge, UK). After a rinse in 0.15 M NaCl buffer, the samples were negatively stained, as already described. To evaluate particle size of exosomes, dynamic light scattering measurements were performed using a Zetasizer Nano ZS90 instrument (Malvern Instruments, Malvern, UK).
Transfection of Trophoblast Cells with DNA Plasmid
Human trophoblast HTR-8/SVneo cells (CRL-3271, American Type Culture Collection, Manassas, VA, USA) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% FBS and a mixture of penicillin-streptomycin (100 U/mL, 1:1) in a 5% CO2 incubator at 37°C. Cell detachment was performed with 0.25% trypsin, and HTR-8/SVneo cells were passaged at a ratio of 1:3. Experiments were carried out in HTR-8/SVneo cells seeded on six-well plates at a density of 4 × 105 cells/well that were 80%–90% confluent and then transfected with pCDNA3.1-FOXO1, let-7b mimic, and let-7b inhibitor plasmids as well as their corresponding controls using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). The cells were incubated for 48 h in the transfection mixture, after which the medium was replaced with RPMI 1640 medium containing 10% FBS (Santa Cruz Biotechnology, Santa Cruz, CA, USA). All plasmids were purchased from Shanghai Gene Pharma (Shanghai, China). Each experiment was repeated three times.
Twenty-four hours before transfection, cells were plated at a density of 2 × 105 cells/well in antibiotic-free DMEM. When MSCs reached 80%–90% confluence, the transfection was performed according to the instructions of Mirus Bio TransIT-LT1 transfection reagent (MIR5404, Mirus Bio, Madison, WI, USA), and DMEM was renewed at 6 h after transfection. MSCs were introduced with vector, pcDNA3.1-H19, and sh-H19.
Co-culture of MSCs, Exosomes, and Human Trophoblast Cells
To inhibit the exosome release from MSCs, MSCs were treated with the exosome inhibitor GW4869. MSCs were plated in a six-well plate at a density of 1 × 106 cells/well. When cells reached 80%–90% confluence, MSCs were incubated in culture media containing 0.005% dimethyl sulfoxide or GW4869 (D1692-5MG, Sigma-Aldrich, St. Louis, MO, USA) for 24 h.
The basolateral chamber of a 24-well Transwell chamber was covered with MSCs (1 × 104 cells/well), while the apical chamber was covered with HTR-8/SVneo cells. The insert aperture between the apical and basolateral chambers was 0.4 um. After co-culture for 24 h, HTR-8/SVneo cells were isolated. Then, the expression of let-7b and FOXO1 was detected by qRT-PCR and western blot analysis. The exosomes extracted from MSCs were labeled with PKH26 (red, MINI26-1KT; Sigma-Aldrich, St. Louis, MO, USA). MSCs were delivered with pcDNA3.1-NC plasmids or pcDNA3.1-H19 plasmids before co-culture. The fluorescently labeled and MSC-derived exosomes were co-cultured with HTR-8/SVneo culture medium with 50%–60% confluence in a 24-well plate for 48 h. Cells were observed under an inverted fluorescence microscope. The expression of FOXO1, AKT, H19, let-7b, and p-AKT was detected by qRT-PCR and western blot analysis.
Transwell Assay for Cell Migration and Invasion
Cell migration and invasion assays were performed using Transwell inserts (8-μm pore size polycarbonate membrane; Corning Life Sciences, Corning, NY, USA). A total of 600 mL of RPMI 1640 medium with 20% FBS was added into the basolateral chamber and equilibrated at 37°C for 1 h. The trophoblast cells (1 × 106 cells/well) after 48 h of transfection resuspended in FBS-free RPMI 1640 medium were placed into the apical chamber. The cells were allowed to migrate or invade for 24 h at 37°C with 5% CO2. The cells that failed to migrate or invade on the inner surface of the Transwell were carefully removed using a cotton swab. The cells that had penetrated to the bottom side of the membrane were then fixed with 5% glutaraldehyde and stained with 0.1% crystal violet for 5 min at 4°C. Then, the cells in five high-power fields were observed under an inverted fluorescence microscope (TE2000, Nikon Imaging (China) Sales, Beijing, China).
TUNEL Staining
TUNEL reaction mixture was prepared according to a TUNEL cell apoptosis detection kit (green fluorescence, C1088, Beyotime Institute Biotech, Shanghai, China). The treatment group was added with 50 μL of TdT + 450 μL of fluorescein-labeled dUTP solution, while the NC group was treated with only 50 μL of fluorescein-labeled dUTP solution. Then, cells were incubated at 37°C for 60 min in the dark. After that, the cells were sealed with an anti-fluorescence quenching liquid and observed under a fluorescence microscope with excitation wavelength of 450 nm and emission wavelength of 550 nm (green fluorescence).
Dual-Luciferase Reporter Gene Assay
The 3′ untranslated region (3′ UTR) dual-luciferase reporter vector of FOXO1 and the mutant plasmids with mutated let-7b binding site were separately constructed, named PGLO-FOXO1-WT and PGLO-FOXO1 mutant (Mut). H19-WT and let-7b binding site mutation-derived plasmids were separately constructed: PGLO-H19-WT and PGLO-H19-Mut. Then, 293T cells were co-transfected with the above reporters, let-7b-mimic, and NC plasmids. After transfection for 24 h, the cells were lysed and centrifuged at 12,000 rpm for 1 min. Luciferase activity was measured by using a dual-luciferase reporter gene system (Dual-Luciferase reporter assay system, E1910, Promega, Madison, WI, USA). The rate of firefly luciferase activity to Renilla luciferase activity served as the relative luciferase activity.
RNA FISH
Bioinformatics tools (http://lncatlas.crg.eu/) were used to obtain the subcellular localization information of H19. The subcellular localization of H19 in trophoblast cells was identified by FISH according to the instructions of the Ribo lncRNA FISH Probe Mix (Red) (RiboBio, Guangzhou, Guangdong, China). In brief, the cover glass was placed in a 24-well plate, and the cells were seeded at 6 × 104 cells/well. The cover glass was fixed with 1 mL of 4% paraformaldehyde. After being treated with protease K (2 μg/mL), glycine, and acetylation reagents, the cells were pre-hybridized with 250 μL of pre-hybridization solution at 42°C for 1 h and hybridized with 250 μL of hybridization solution containing 300 ng/mL probe at 42°C overnight. The cells were treated with PBS/Tween 20-diluted DAPI (1:800) to stain the nucleus for 5 min. Then, the cells were sealed with anti-fluorescence quencher. After that, the cells were observed and imaged under a fluorescence microscope (Olympus, Tokyo, Japan) with five different fields.
RNA Pull-Down
The HTR-8/SVneo cells were lysed (Ambion, Austin, TX, USA). Then, the lysate was incubated with 3 μg of biotinylated let-7b probe for 2 h at room temperature. After that, the cell lysate was incubated with pre-coated M-280 streptavidin magnetic beads (S3762, Sigma-Aldrich, St. Louis, MO, USA) for 4 h at 4°C. The bound RNA was further purified by TRIzol, and then H19 enrichment was detected by qRT-PCR.
RIP
The binding of H19 and Ago2 protein was verified and conducted according to the instructions of the RIP kit (Millipore, Bedford, MA, USA). Human trophoblast cells were placed in an ice bath for 5 min with RIPA lysis buffer (P0013B, Beyotime Biotechnology, Shanghai, China). Part of the cell extract was used as input, and the other part was incubated with antibodies and magnetic beads. The magnetic beads-antibody complexes were resuspended in 900 μL of RIP wash buffer. Samples were set on a magnetic base to collect the magnetic bead-protein complexes. RNAs in the samples and input were separately extracted after detachment by protease K for the subsequent quantitative real-time PCR detection. The antibodies for RIP included Ago2 (1:50, ab32381, Abcam, Cambridge, UK), and IgG (1:100, ab109489, Abcam, Cambridge, UK) as the NC.
Statistical Analysis
All data were processed by SPSS 21.0 statistical software (IBM, Armonk, NY, USA). Normal distribution and variance homogeneity tests were performed. Data were presented as mean ± standard deviation when it conformed to the normal distribution. When the normal distribution and homogeneity of the variance were violated, the interquartile range was applied. Comparisons between two groups were conducted by means of an unpaired t test, and the skewed distribution data were tested using a nonparametric Wilcoxon signed rank test. Comparisons among multiple groups were assessed by one-way analysis of variance with Tukey’s post hoc test. Statistical significance was indicated when with p < 0.05.
Author Contributions
Y.C., H.D., M.W., W.Z., S.G., N.L., Y.L., Y.T., Y.W., and F.W. designed the study. Y.C. collated the data, carried out data analyses, and produced the initial draft of the manuscript. H.D., M.W., W.Z., S.G., N.L., Y.L., Y.T., Y.W., and F.W. contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We would like to acknowledge the helpful comments on this paper received from our colleagues. This study was supported by the Department of Science and Technology of Jilin Province (20170204045SF).
References
- 1.Bibbins-Domingo K., Grossman D.C., Curry S.J., Barry M.J., Davidson K.W., Doubeni C.A., Epling J.W., Jr., Kemper A.R., Krist A.H., Kurth A.E., US Preventive Services Task Force Screening for preeclampsia: US Preventive Services Task Force recommendation statement. JAMA. 2017;317:1661–1667. doi: 10.1001/jama.2017.3439. [DOI] [PubMed] [Google Scholar]
- 2.Ho L., van Dijk M., Chye S.T.J., Messerschmidt D.M., Chng S.C., Ong S., Yi L.K., Boussata S., Goh G.H., Afink G.B. ELABELA deficiency promotes preeclampsia and cardiovascular malformations in mice. Science. 2017;357:707–713. doi: 10.1126/science.aam6607. [DOI] [PubMed] [Google Scholar]
- 3.Turanov A.A., Lo A., Hassler M.R., Makris A., Ashar-Patel A., Alterman J.F., Coles A.H., Haraszti R.A., Roux L., Godinho B.M.D.C. RNAi modulation of placental sFLT1 for the treatment of preeclampsia. Nat. Biotechnol. 2018;36:1164–1175. doi: 10.1038/nbt.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farzaneh F., Tavakolikia Z., Soleimanzadeh Mousavi S.H. Assessment of occurrence of preeclampsia and some clinical and demographic risk factors in Zahedan city in 2017. Clin. Exp. Hypertens. 2019;41:583–588. doi: 10.1080/10641963.2018.1523919. [DOI] [PubMed] [Google Scholar]
- 5.Liao Y., Liu X.H., Tan J., He G.L., Yang H.M., Chen M. [Development of a predictive model for adverse outcomes of preeclampsia] Sichuan Da Xue Xue Bao Yi Xue Ban. 2018;49:797–802. [PubMed] [Google Scholar]
- 6.Paré E., Parry S., McElrath T.F., Pucci D., Newton A., Lim K.H. Clinical risk factors for preeclampsia in the 21st century. Obstet. Gynecol. 2014;124:763–770. doi: 10.1097/AOG.0000000000000451. [DOI] [PubMed] [Google Scholar]
- 7.Jääskeläinen T., Heinonen S., Kajantie E., Kere J., Kivinen K., Pouta A., Laivuori H., FINNPEC Study Group Cohort profile: the Finnish Genetics of Pre-eclampsia Consortium (FINNPEC) BMJ Open. 2016;6:e013148. doi: 10.1136/bmjopen-2016-013148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunnarsson R., Åkerström B., Hansson S.R., Gram M. Recombinant alpha-1-microglobulin: a potential treatment for preeclampsia. Drug Discov. Today. 2017;22:736–743. doi: 10.1016/j.drudis.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Li X., Song Y., Liu F., Liu D., Miao H., Ren J., Xu J., Ding L., Hu Y., Wang Z. Long non-coding RNA MALAT1 promotes proliferation, angiogenesis, and immunosuppressive properties of mesenchymal stem cells by inducing VEGF and IDO. J. Cell. Biochem. 2017;118:2780–2791. doi: 10.1002/jcb.25927. [DOI] [PubMed] [Google Scholar]
- 10.Ji L., Zhang L., Li Y., Guo L., Cao N., Bai Z., Song Y., Xu Z., Zhang J., Liu C., Ma X. miR-136 contributes to pre-eclampsia through its effects on apoptosis and angiogenesis of mesenchymal stem cells. Placenta. 2017;50:102–109. doi: 10.1016/j.placenta.2017.01.102. [DOI] [PubMed] [Google Scholar]
- 11.Xiong Z.H., Wei J., Lu M.Q., Jin M.Y., Geng H.L. Protective effect of human umbilical cord mesenchymal stem cell exosomes on preserving the morphology and angiogenesis of placenta in rats with preeclampsia. Biomed. Pharmacother. 2018;105:1240–1247. doi: 10.1016/j.biopha.2018.06.032. [DOI] [PubMed] [Google Scholar]
- 12.Patel N.A., Moss L.D., Lee J.Y., Tajiri N., Acosta S., Hudson C., Parag S., Cooper D.R., Borlongan C.V., Bickford P.C. Long noncoding RNA MALAT1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J. Neuroinflammation. 2018;15:204. doi: 10.1186/s12974-018-1240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y.N., Zhang R., Du J.W., Yuan H.H., Li Y.J., Wei X.L., Du X.X., Jiang S.L., Han Y. Predictive role of UCA1-containing exosomes in cetuximab-resistant colorectal cancer. Cancer Cell Int. 2018;18:164. doi: 10.1186/s12935-018-0660-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin K., Su K.K., Li T., Zhu X.Q., Wang Q., Ge R.S., Pan Z.F., Wu B.W., Ge L.J., Zhang Y.H. Hepatic premalignant alterations triggered by human nephrotoxin aristolochic acid i in canines. Cancer Prev. Res. (Phila.) 2016;9:324–334. doi: 10.1158/1940-6207.CAPR-15-0339. [DOI] [PubMed] [Google Scholar]
- 15.Saito S., Nakashima A. A review of the mechanism for poor placentation in early-onset preeclampsia: the role of autophagy in trophoblast invasion and vascular remodeling. J. Reprod. Immunol. 2014;101–102:80–88. doi: 10.1016/j.jri.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Chen C.P., Chen C.Y., Wu Y.H., Chen C.Y. Oxidative stress reduces trophoblast FOXO1 and integrin β3 expression that inhibits cell motility. Free Radic. Biol. Med. 2018;124:189–198. doi: 10.1016/j.freeradbiomed.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Hutabarat M., Wibowo N., Huppertz B. The trophoblast survival capacity in preeclampsia. PLoS ONE. 2017;12:e0186909. doi: 10.1371/journal.pone.0186909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J., Liao W., Yang J., Ma K., Li X., Wang Y., Wang D., Wang L., Zhang Y., Yin Y. FOXO3 induces FOXO1-dependent autophagy by activating the AKT1 signaling pathway. Autophagy. 2012;8:1712–1723. doi: 10.4161/auto.21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu D., Hong H., Huang X., Huang L., He Z., Fang Q., Luo Y. CXCR2 is decreased in preeclamptic placentas and promotes human trophoblast invasion through the Akt signaling pathway. Placenta. 2016;43:17–25. doi: 10.1016/j.placenta.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Cao C., Li J., Li J., Liu L., Cheng X., Jia R. Long non-coding RNA uc.187 is upregulated in preeclampsia and modulates proliferation, apoptosis, and invasion of HTR-8/SVneo trophoblast cells. J. Cell. Biochem. 2017;118:1462–1470. doi: 10.1002/jcb.25805. [DOI] [PubMed] [Google Scholar]
- 21.Zuckerwise L., Li J., Lu L., Men Y., Geng T., Buhimschi C.S., Buhimschi I.A., Bukowski R., Guller S., Paidas M., Huang Y. H19 long noncoding RNA alters trophoblast cell migration and invasion by regulating TβR3 in placentae with fetal growth restriction. Oncotarget. 2016;7:38398–38407. doi: 10.18632/oncotarget.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song X., Luo X., Gao Q., Wang Y., Gao Q., Long W. Dysregulation of lncRNAs in placenta and pathogenesis of preeclampsia. Curr. Drug Targets. 2017;18:1165–1170. doi: 10.2174/1389450118666170404160000. [DOI] [PubMed] [Google Scholar]
- 23.Gao Y., Wu F., Zhou J., Yan L., Jurczak M.J., Lee H.Y., Yang L., Mueller M., Zhou X.B., Dandolo L. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014;42:13799–13811. doi: 10.1093/nar/gku1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zadora J., Singh M., Herse F., Przybyl L., Haase N., Golic M., Yung H.W., Huppertz B., Cartwright J.E., Whitley G. Disturbed placental imprinting in preeclampsia leads to altered expression of DLX5, a human-specific early trophoblast marker. Circulation. 2017;136:1824–1839. doi: 10.1161/CIRCULATIONAHA.117.028110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant I., Cartwright J.E., Lumicisi B., Wallace A.E., Whitley G.S. Caffeine inhibits EGF-stimulated trophoblast cell motility through the inhibition of mTORC2 and Akt. Endocrinology. 2012;153:4502–4510. doi: 10.1210/en.2011-1930. [DOI] [PubMed] [Google Scholar]
- 26.Paraskevopoulou M.D., Vlachos I.S., Karagkouni D., Georgakilas G., Kanellos I., Vergoulis T., Zagganas K., Tsanakas P., Floros E., Dalamagas T., Hatzigeorgiou A.G. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noack F., Ribbat-Idel J., Thorns C., Chiriac A., Axt-Fliedner R., Diedrich K., Feller A.C. miRNA expression profiling in formalin-fixed and paraffin-embedded placental tissue samples from pregnancies with severe preeclampsia. J. Perinat. Med. 2011;39:267–271. doi: 10.1515/jpm.2011.012. [DOI] [PubMed] [Google Scholar]
- 28.Caldeira-Dias M., Luizon M.R., Deffune E., Tanus-Santos J.E., Freire P.P., Carvalho R.F., Bettiol H., Cardoso V.C., Antonio Barbieri M., Cavalli R.C., Sandrim V.C. Preeclamptic plasma stimulates the expression of miRNAs, leading to a decrease in endothelin-1 production in endothelial cells. Pregnancy Hypertens. 2018;12:75–81. doi: 10.1016/j.preghy.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Sheridan R., Belludi C., Khoury J., Stanek J., Handwerger S. FOXO1 expression in villous trophoblast of preeclampsia and fetal growth restriction placentas. Histol. Histopathol. 2015;30:213–222. doi: 10.14670/hh-30.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopkins B.L., Nadler M., Skoko J.J., Bertomeu T., Pelosi A., Shafaei P.M., Levine K., Schempf A., Pennarun B., Yang B. A peroxidase peroxiredoxin 1-specific redox regulation of the novel FOXO3 microRNA target let-7. Antioxid. Redox Signal. 2018;28:62–77. doi: 10.1089/ars.2016.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han C.L., Ge M., Liu Y.P., Zhao X.M., Wang K.L., Chen N., Hu W., Zhang J.G., Li L., Meng F.G. Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell Death Dis. 2018;9:617. doi: 10.1038/s41419-018-0496-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W.T., Ye H., Wei P.P., Han B.W., He B., Chen Z.H., Chen Y.Q. lncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J. Hematol. Oncol. 2016;9:117. doi: 10.1186/s13045-016-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris D.A., Patel S.H., Gucek M., Hendrix A., Westbroek W., Taraska J.W. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS ONE. 2015;10:e0117495. doi: 10.1371/journal.pone.0117495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motawi T.M.K., Sabry D., Maurice N.W., Rizk S.M. Role of mesenchymal stem cells exosomes derived microRNAs; miR-136, miR-494 and miR-495 in pre-eclampsia diagnosis and evaluation. Arch. Biochem. Biophys. 2018;659:13–21. doi: 10.1016/j.abb.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 36.Xu J., Xia Y., Zhang H., Guo H., Feng K., Zhang C. Overexpression of long non-coding RNA H19 promotes invasion and autophagy via the PI3K/AKT/mTOR pathways in trophoblast cells. Biomed. Pharmacother. 2018;101:691–697. doi: 10.1016/j.biopha.2018.02.134. [DOI] [PubMed] [Google Scholar]
- 37.Kang W., Tong J.H., Lung R.W., Dong Y., Yang W., Pan Y., Lau K.M., Yu J., Cheng A.S., To K.F. let-7b/g silencing activates AKT signaling to promote gastric carcinogenesis. J. Transl. Med. 2014;12:281. doi: 10.1186/s12967-014-0281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szydłowski M., Jabłońska E., Juszczyński P. FOXO1 transcription factor: a critical effector of the PI3K-AKT axis in B-cell development. Int. Rev. Immunol. 2014;33:146–157. doi: 10.3109/08830185.2014.885022. [DOI] [PubMed] [Google Scholar]
- 39.Li M., Cheng W., Luo J., Hu X., Nie T., Lai H., Zheng X., Li F., Li H. Loss of selenocysteine insertion sequence binding protein 2 suppresses the proliferation, migration/invasion and hormone secretion of human trophoblast cells via the PI3K/Akt and ERK signaling pathway. Placenta. 2017;55:81–89. doi: 10.1016/j.placenta.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Zhu X., Cao Q., Li X., Wang Z. Knockdown of TACC3 inhibits trophoblast cell migration and invasion through the PI3K/Akt signaling pathway. Mol. Med. Rep. 2016;14:3437–3442. doi: 10.3892/mmr.2016.5659. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y., Ma Y. Promoter methylation status of WNT2 in placenta from patients with preeclampsia. Med. Sci. Monit. 2017;23:5294–5301. doi: 10.12659/MSM.903602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lain K.Y., Roberts J.M. Contemporary concepts of the pathogenesis and management of preeclampsia. JAMA. 2002;287:3183–3186. doi: 10.1001/jama.287.24.3183. [DOI] [PubMed] [Google Scholar]
- 43.Rojo A.I., Medina-Campos O.N., Rada P., Zúñiga-Toalá A., López-Gazcón A., Espada S., Pedraza-Chaverri J., Cuadrado A. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: role of glycogen synthase kinase-3. Free Radic. Biol. Med. 2012;52:473–487. doi: 10.1016/j.freeradbiomed.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Huang Y., Fan X., Tao R., Song Q., Wang L., Zhang H., Kong H., Huang J. Effect of miR-182 on hepatic fibrosis induced by Schistosomiasis japonica by targeting FOXO1 through PI3K/AKT signaling pathway. J. Cell. Physiol. 2018;233:6693–6704. doi: 10.1002/jcp.26469. [DOI] [PubMed] [Google Scholar]
- 45.Nakamizo A., Marini F., Amano T., Khan A., Studeny M., Gumin J., Chen J., Hentschel S., Vecil G., Dembinski J. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res. 2005;65:3307–3318. doi: 10.1158/0008-5472.CAN-04-1874. [DOI] [PubMed] [Google Scholar]
- 46.Potian J.A., Aviv H., Ponzio N.M., Harrison J.S., Rameshwar P. Veto-like activity of mesenchymal stem cells: functional discrimination between cellular responses to alloantigens and recall antigens. J. Immunol. 2003;171:3426–3434. doi: 10.4049/jimmunol.171.7.3426. [DOI] [PubMed] [Google Scholar]
- 47.Zhang G., Zou X., Miao S., Chen J., Du T., Zhong L., Ju G., Liu G., Zhu Y. The anti-oxidative role of micro-vesicles derived from human Wharton-Jelly mesenchymal stromal cells through NOX2/gp91(phox) suppression in alleviating renal ischemia-reperfusion injury in rats. PLoS ONE. 2014;9:e92129. doi: 10.1371/journal.pone.0092129. [DOI] [PMC free article] [PubMed] [Google Scholar]