Abstract
Mitochondria are indispensable organelles that perform critical cellular functions, including energy metabolism, neurotransmission, and synaptic maintenance. Mitochondrial dysfunction and impairment in the organellar homeostasis are key hallmarks implicated in the progression of neurodegenerative disorders. The members of DJ-1/ThiJ/PfpI family are highly conserved, and loss of DJ-1 (PARK7) function in humans results in the impairment of mitochondrial homeostasis, which is one of the key cellular etiology implicated in the progression of Parkinson's Disease. However, the underlying molecular mechanism involved in mitochondrial maintenance and other cellular processes by DJ-1 paralogs is poorly understood. By utilizing genetic approaches from S. cerevisiae, we uncovered intricate mechanisms associated with the mitochondrial phenotypic variations regulated by DJ-1 paralogs. The deletion of DJ-1 paralogs led to respiratory incompetence and the accumulation of enhanced functional mitochondrial mass. The lack of DJ-1 paralogs also displayed enriched mitochondrial interconnectivity due to upregulation in the fusion-mediating proteins, facilitated by the elevation in the basal cellular ROS and oxidized glutathione levels. Intriguingly, these mitochondrial phenotypes variations cause cell size abnormalities, partially suppressed by reestablishing redox balance and upregulation of fission protein levels. Besides, in the absence of DJ-1 paralogs, cells exhibited a significant delay in the cell-cycle progression in the G2/M phase, attributed to mitochondrial hyperfusion and partial DNA damage. Additionally, the aberrations in mitochondrial dynamics and cell-cycle induce cell death mediated by apoptosis. Taken together, our findings first-time provide evidence to show how DJ-1 family members regulate mitochondrial homeostasis and other intricate cellular processes, including cell cycle and apoptosis.
Keywords: Parkinson disease, DJ-1, Mitochondria, Hsp31, Apoptosis
Highlights
-
•
Lack of DJ-1 members causes respiratory incompetence and elevated basal ROS levels.
-
•
Enhanced ROS and GSSG levels promote increased mitochondrial mass and hyperfusion.
-
•
Mitochondrial hyperfusion together with ROS-induced DNA damage cause G2/M arrest.
-
•
Impairment in cell cycle progression triggers apoptotic cell death in yeast.
1. Introduction
The DJ-1/ThiJ/PfpI superfamily is an evolutionarily conserved multifunctional group of proteins widely distributed across the species [1]. In bacteria, DJ-1 ortholog referred to as Hsp31, required for acid-stress adaptation, and functions as novel protease and glyoxalase [2,3]. In Saccharomyces cerevisiae, four orthologs identified collectively named as Hsp31 (Hsp31, Hsp32, Hsp33, Hsp34) mini-family members belong to DJ-1 superfamily (Table 1). Among these paralogs, Hsp31is found to be the closest relative to the bacterial Hsp31 [4,5]. Yeast Hsp31 functions as a robust methylglyoxalase critical for the maintenance of cellular glutathione (GSH) and redox homeostasis [6]. Besides, Hsp31 paralogs play an important role upstream of the TORC1 signaling pathway, essential for diauxic-shift and cell survival in the stationary phase [7,8]. The regulation of the TORC1 function through Hsp31 members is mediated by a negative regulator, Sfp1. Moreover, the lack of Hsp31 members showed decreased proteasome activity resulting in the accumulation of aberrant proteins and eventually causing cell death [9]. Recent evidence also suggests that yeast Hsp31 displays acid-stress resistance [10], modulates the protein aggregation and pathogenesis in models of Parkinson's Disease (PD), Alzheimer's (AD), and Huntington diseases (Table 1) [11,12].
Table 1.
Summary of the Function(s) of Sc Hsp31 paralogs and human DJ-1.
| Function(s) | Hsp31 | Hsp32 | Hsp33 | Hsp34 | hDJ-1 | References |
|---|---|---|---|---|---|---|
| Sequence Homology within the paralogs and hDJ-1 |
70% (16–35%) |
99.5% (16–35%) |
99.5% (16–35%) |
99.5% (16–35%) |
_ | [4,5] |
| Catalytic site residues, (Cys, His, Glu) |
Yes | Yes | Yes | Yes | Yes | [4,13,14] |
| Mutations linked to early onset of familial PD |
_ | _ | _ | _ | Yes | [15] |
| Role in protein quality control | Yes | Yes | Yes | Yes | Yes | [16,17] |
| Methylglyoxalase Activity | Yes | ND | ND | ND | Yes | [6,18] |
| Role in Oxidative Stress | Yes | Yes | Yes | Yes | Yes | [6,7,19] |
| Regulation of 20S proteasome complex |
ND | ND | ND | ND | Yes | [20] |
| Role in cell survival during Diauxic shift |
Yes | Yes | Yes | Yes | ND | [7] |
| Role in Autophagy | Yes | Yes | Yes | Yes | Yes | [7,21] |
| Localization of the protein¥ | Cytosolic | Cytosolic | Cytosolic | Cytosolic | Cytosolic/ Nuclei |
[6,22] |
| Mitochondrial Translocation* | Yes# | ND | ND | ND | Yes | [6,23] |
ND: Not Determined, ¥: Without Stress, *: During Oxidative stress, #: >50% of the total protein.
Human DJ-1 encoded by PARK7, a recessive novel gene, is responsible for the early onset of a familial form of PD (Table 1) [15]. Human DJ-1 is a multitasking protein involved in several important functions, including redox-based sensing, transcription regulation, and RNA binding [17,24,25]. Knockdown of DJ-1 by RNA-interference rendered cells more susceptible to oxidative injury [26]. Recent findings suggest that human DJ-1 functions as novel glyoxalase and deglycase, required for the repair of glycated forms of protein and DNA [27,28]. Besides, DJ-1 also inhibits protein aggregation by regulating chaperone-mediated autophagy, thereby aiding amelioration of PD-progression [29,30]. Also, DJ-1 has been shown to be a regulator of ubiquitin-independent 20S proteasomal degradation pathway, thus signifying its critical role in the maintenance of protein homeostasis, a key hallmark of several neuronal disorders, including PD (Table 1) [20].
Apart from the role in proteostasis, early evidence suggests a positive correlation between DJ-1 cellular function and mitochondrial phenotypic variations [31]. In mammalian cells, loss of function of DJ-1 induced mitochondrial depolarization, fragmentation, and was associated with the accumulation of dysfunctional mitochondria in primary neurons leading to reduced cell viability [32]. DJ-1 deficient cells exhibited aberrant mitochondrial morphology in the mouse brain, neuron, and lymphoblast cell lines [33]. Importantly, DJ-1 homologs in human and yeast are known to localize to mitochondria in response to oxidative stress to protect organellar functions [6,23]. Furthermore, DJ-1 has been shown to interact with the mitochondrial electron transport chain (ETC) complex-I to regulate its activity [34]. Moreover, overexpression of human DJ-1 functionally complemented Hsp31 and suppressed mitochondrial superoxide levels in yeast, thus highlighting the functional conservation across species (Table 1) [6]. This growing evidence suggests the possible existence of functional crosstalk between DJ-1 paralogs and the mitochondrial homeostasis. However, the direct involvement of DJ-1 paralogs in the regulation of mitochondrial life cycle and the underlying mechanism is poorly understood.
The current study highlights a key lacuna in the field by addressing how DJ-1 paralogs directly regulate mitochondrial health by altering the redox status of the cell. Loss of DJ-1 paralogs attenuates the organellar life cycle by elevating the fusion protein levels leading to enhanced mitochondrial interconnectivity. These mitochondrial phenotypic variations manifest as a delay in the cell cycle progression, cell size abnormalities, and induction of apoptosis. The mitochondrial phenotypes are partially rescued by restoring the redox balance and upregulating the fission protein. In conclusion, our findings provide direct evidence in revealing a constitutive function of DJ-1 homologs in the maintenance of mitochondrial quality control and cell viability.
2. Results
2.1. Loss of DJ-1 homologs in S. cerevisiae enhances respiratory incompetency
Knockdown of DJ-1 using RNA interference leads to mitochondrial dysfunction in neurons [31]. In the line of this evidence, a DJ-1 ortholog in yeast (Hsp31) showed an exclusive functional specificity in the maintenance of mitochondrial integrity under oxidative stress conditions [6]. However, the critical function of DJ-1 proteins in the maintenance of mitochondrial health under normal physiological conditions is still elusive. DJ-1 homologs are highly conserved across phylogeny including yeast, which possesses 4 paralogs. By utilizing the ease of yeast genetic manipulation and traceability, we uncovered the functional significance of DJ-1 members in the maintenance of mitochondrial health under normal physiological conditions.
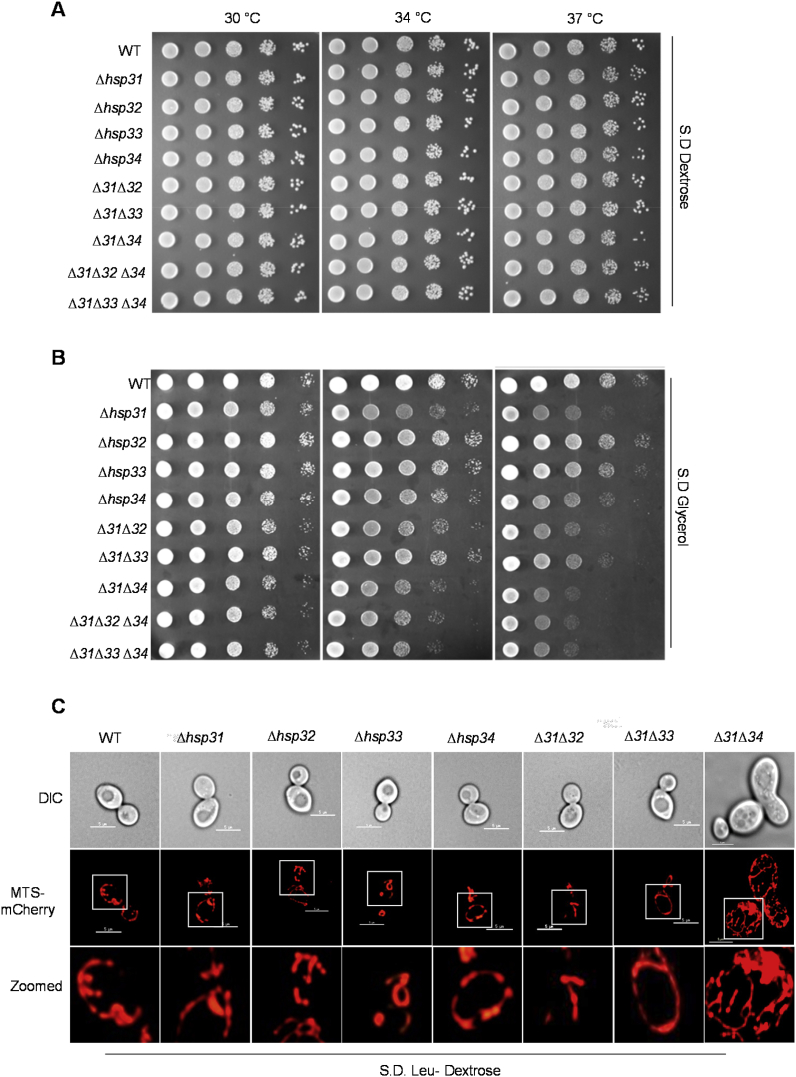
To understand the role of DJ-1 members in mitochondrial health, the growth phenotypes were examined for the individual deletion of Hsp31 paralogs in different growth media. The deletion of Hsp31 members showed no obvious growth defects in the complete (S.D. Dextrose) medium (Fig. 1A). At the same time, single deletion strains showed a variable degree of growth sensitivity in the non-fermentable carbon source (S.D. Glycerol) (Fig. 1B). Among them, Δhsp31 (Δ31) showed maximum growth impairment at 34 °C and 37 °C, followed by Δhsp34 (Δ34), with a moderate growth sensitivity. On the other hand, Δhsp32 (Δ32) or Δhsp33 (Δ33) did not exhibit a significant growth defect as compared to wild type (WT). To test the additive functional overlap, a double deletion (Δ31Δ34) strain constructed, which showed robust synergistic growth sensitivity in S.D. Glycerol as compared to Δ31Δ32 and Δ31Δ33 strains (Fig. 1B). Furthermore, additional deletions in combinations such as Δ31Δ32Δ34 and Δ31Δ33Δ34 did not enhance the sensitivity significantly, suggesting that Hsp34 plays a crucial role in sensitizing Hsp31's cellular function (Fig. 1B).
Fig. 1.
Deletion of Hsp31 paralogs exhibited variable growth phenotypes. (A, B) Growth phenotype analysis. Cells grown till mid-log phase were harvested and spotted on YP rich medium containing different carbon sources, Dextrose, A and Glycerol, B. Images were captured after 38 h and 50 h of incubation in case of dextrose and glycerol, respectively. (C) Assessment of mitochondrial integrity. Yeast strains expressing MTS-mCherry grown in YP-Dextrose containing media till mid-log phase were subjected to microscopic analysis to visualize mitochondria. Scale bar (5 μm). Images were zoomed up to ~2X and represented (bottom panels).
The respiration incompetence in yeast is very well correlated to organellar functions, hence, tested for mitochondrial integrity in the deletion strains. To analyze, mitochondria was decorated with MTS-mCherry and visualized under the microscope at 100 x resolution. As indicated in Fig. 1C, all the single deletion strains displayed comparable mitochondrial network structures. However, in the combination of deletions, only Δ31Δ34 strain showed increased mitochondrial reticularity and volume in dextrose media as compared to other strains (Fig. 1C). These experimental findings provide early evidence for cooperative functional sensitization of Hsp31 by Hsp34 paralog in the maintenance of mitochondrial health.
2.2. DJ-1 homologs regulate mitochondrial mass and turnover
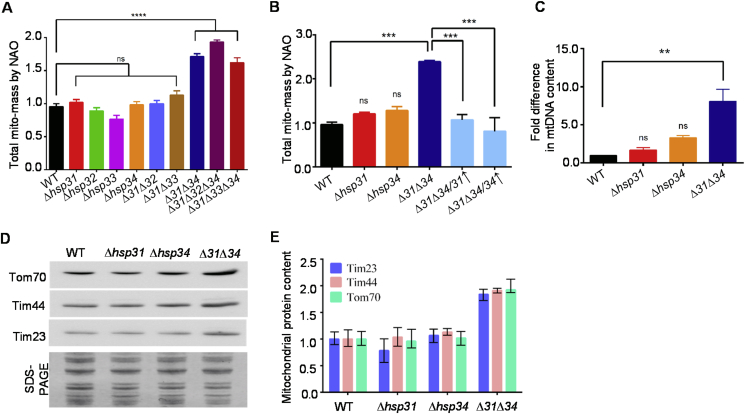
Based on our preliminary microscopic analysis it is evident that the deletion of DJ-1 paralogs leads to alterations in the mitochondrial content. To evaluate quantitatively, Nonyl-Acridine Orange (NAO) was utilized, which preferentially stains cardiolipin of the inner membrane of mitochondria [35]. Upon flow cytometry analysis, the deletion of individual paralogs did not show a significant difference in the mitochondrial mass. However, the yeast deletion strains (Δ31Δ34, Δ31Δ32Δ34, Δ31Δ33Δ34) exhibited a significant enhancement in the total mitochondrial mass, consistent with microscopic observations (Fig. 2A). To further ascertain that specificity of mitochondrial mass increment is due to the Hsp31/34, the paralogs were overexpressed in Δ31Δ34 strain, which significantly restored the mitochondrial mass comparable to WT (Fig. 2B). This proportional increment in the mass was further supported by an enhancement in the total mitochondrial DNA (mtDNA) measured by qPCR and protein levels by western analysis in Δ31Δ34 strain (Fig. 2C, 2D & 2E).
Fig. 2.
DJ-1 paralogs regulate mitochondrial turnover.(A, B) Assessment of mitochondrial mass. Yeast cells were grown till mid-log phase and treated with cardiolipin stain, NAO, and subjected to flow cytometry. Median fluorescence values obtained from FACS analysis were plotted as a bar graph. (C) qPCR analysis. Relative mitochondrial DNA content was measured by quantifying mitochondrial gene COX2 using qPCR. (D, E) Quantitation of mitochondrial protein content. Total lysates prepared from respective yeast strains were analyzed by immunoblotting using mitochondrial-protein specific antibodies. The Coomassie staining of mitochondrial proteins served as equal loading controls (D). Mitochondrial-specific proteins quantitated by densitometry analysis using Multi gauge software and the relative values levels were represented in a bar graph (E).
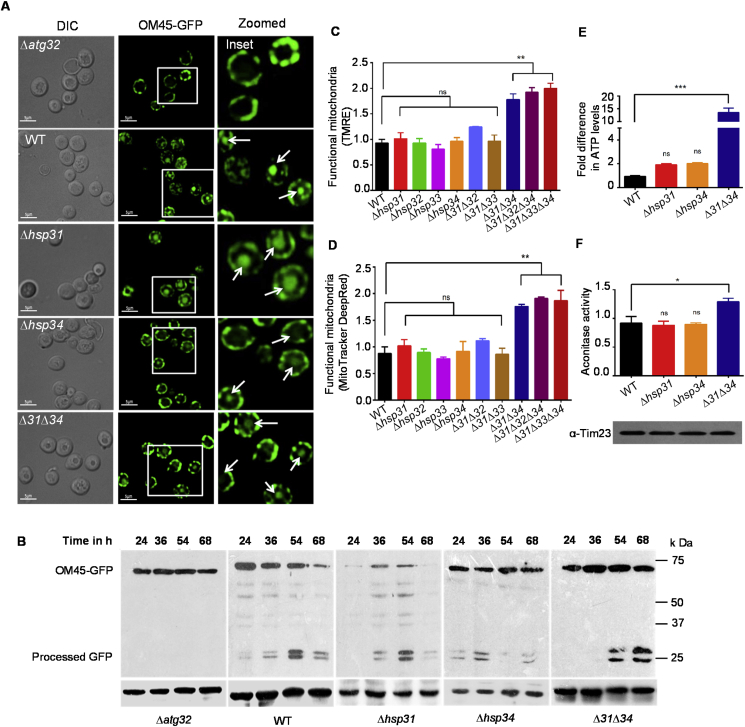
To rule out the possibility that accumulation of mitochondrial mass is not due to the impairment in the mitophagy process, the organelle turnover was monitored in Δ31Δ34 strain. The mitophagy in Δ31Δ34 cells was measured by tagging the mitochondrial outer membrane protein (OM45) with GFP followed by microscopic and western analysis [36]. In a positive control, most of the GFP fluorescence found restricted to the mitochondrial compartment in the deletion of mitophagy receptor (Δatg32) due to impairment in the mitophagy process (Fig. 3A) [36]. On the other hand, WT cells showed the occurrence of mitophagy as the GFP fluorescence equally partitioned between the vacuole and mitochondrial compartment. Likewise, both Δhsp31 and Δhsp34 single deletion strains showed a comparable distribution pattern of GFP fluorescence in the vacuolar and mitochondrial compartment. However, an insignificant reduction in the mitophagy was observed in Δ31Δ34 cells as compared to WT (Fig. 3B). This slight reduction in the mitophagy is perhaps due to impaired mitochondrial fragmentation in Δ31Δ34 cells, which is a prerequisite to induce mitophagy [37,38]. These observations were further supported by measuring the rate of GFP-processing by immuno-detection, wherein, unlike single deletion strains, Δ31Δ34 showed a minor reduction in mitophagy, especially in the early time intervals of 24 and 36 h (Fig. 3B).
Fig. 3.
Role of Hsp31/34 paralogs in mitophagy.(A) Mitophagy induction in WT, Δhsp31, Δhsp34, and Δ31Δ34. Cells expressing Om45-GFP were grown till mid-log phase in YP-glycerol medium for 40 h, subsequently the GFP localization was analyzed by fluorescence microscopy. Δatg32 strain was used as a positive control for mitophagy deficiency. Vacuolar localization of Om45-GFP, a marker of mitophagy indicated with white arrows. Scale bars (5 μm). Images were zoomed up to ~2X and represented (rightmost panels). (B) Mitophagy induction by western analysis. Total lysates were separated using SDS-PAGE and immunodecorated with anti-GFP and anti-YDJ1 antibodies. (C, D) Measurement of functional mitochondrial mass by flow cytometry. Yeast cells were grown till mid-log phase and treated with 10 μM TMRE (C), 10 μM Mito Tracker Deep Red (D) followed by flow cytometry analysis. (E) ATP measurement. Mitochondrial ATP levels was measured in purified mitochondrial fractions isolated from indicated yeast strains. (F) Enzyme assay. Aconitase activity was measured in mitochondrial fractions from indicated yeast strains. Error bars represent the standard deviation in median values from 3 biological replicates. Asterisks indicate the p-value, *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To test the functionality, mitochondria from different strains were subjected to Tetramethylrhodamine Ethyl Ester (TMRE) and MitoTracker Deep-Red dyes staining, which are sensitive to membrane polarity (ψm) [39], an indicator of functionality. Interestingly, flow cytometry analysis with both dyes highlights that Δ31Δ34 cells show a healthy membrane potential, suggesting the accumulation of functional mitochondria. Convincingly, similar to NAO-staining, double deletion mutant showed enhanced functional mitochondrial mass as compared to controls (Fig. 3C and 3D). These data further supported by the enhancement in the mitochondrial ATP levels in Δ31Δ34 cells, which is an indicator of functional mass (Fig. 3E). As additional evidence, the activity of a mitochondrial enzyme aconitase was measured. In corroboration, the isolated mitochondrial fractions from Δ31Δ34 cells showed enhanced aconitase activity as compared to control strains (Fig. 3F). In summary, our findings indicate that the DJ-1 paralogs, specifically Hsp31/34 play a crucial role in the maintenance of appropriate mitochondrial volume and functionality in yeast.
2.3. Deletion of DJ-1 paralogs induce mitochondrial hyperfusion
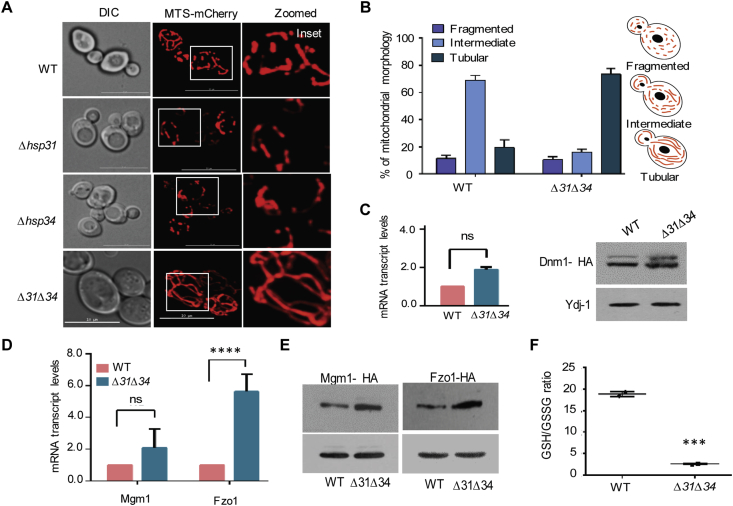
To understand the mitochondrial dynamics, the morphological changes in Δ31Δ34 strain was assessed microscopically by decorating with MTS-mCherry. The single deletion strains (Δ31 or Δ34) did not show any significant changes in the mitochondrial morphology as compared to WT, which showed intermediate tubular and lesser hyperfused structures (Fig. 4A, B). On the contrary, Δ31Δ34 cells exhibited extensive hyperfused mitochondrial network structures as compared to other control strains (Fig. 4A, compare panels 1-3 with 4). An increased mitochondrial fusion could be due to a stress-responsive adaptive mechanism to spare mitochondria from autophagic degradation [37]. Moreover, Hsp31 paralogs in yeast were shown to be critical in the autophagic process during diauxic shift in the growth phase [7]. To test whether mitochondrial hyperfusion is an adaptive response to the diauxic shift, the morphology was assessed as a function of time during the growth phase. As indicated in Fig. S1A, Δ31Δ34 cells showed accumulation of hyperfused mitochondrial network as early as 8 h of growth. Also, Δ31Δ34 cells showed a robust reticulate hyperfused mitochondrial network within 12 h of growth as compared to 24 h at 30 °C (Fig. S1A, panels 4 & 6). At the same growth conditions, WT showed intermediate and fragmented structures (Fig. S1A, panels 3 & 5), thereby highlighting that the observed hyperfused mitochondria in Δ31Δ34 cells are not due to the diauxic shift in the stationary phase. At the same time, it is not unreasonable to believe that such morphological changes could be triggered by internal stress due to a combined loss of Hsp31/34 paralogs.
Fig. 4.
Loss of DJ-1 paralogs alters mitochondrial integrity.(A) Yeast strains (WT, Δhsp31, Δhsp34, and Δ31Δ34 cells) harboring MTS-mCherry construct grown till mid-log phase and subjected to microscopy for the visualization of mitochondria. (B) Qualitative assessment of mitochondrial morphology. The mitochondrial morphology from WT and Δ31Δ34 cells containing MTS-mCherry analyzed using ImageJ 1.0. (n ≥ 150). Error bars in the graphs represent the std. dev. in the percentage of population. (C-E) Evaluation of levels of mitochondrial dynamic proteins. Measurement of the mRNA transcript level of mitochondrial fission-fusion genes by q-PCR analysis and relative steady-state levels of proteins by western blotting Dnm1 (C), mRNA transcript levels of Mgm1 and Fzo1 (D), protein levels of Mgm1 and Fzo1 (E). Cytosolic Ydj-1 used as an internal loading control. (F) GSH/GSSG ratio. Whole-cell lysates from WT and Δ31Δ34 cells subjected to the assessment of cellular reduced/oxidized (GSH/GSSG) glutathione ratio. Error bars in the graphs represent the std. dev. in the mean values from 3 biological replicates. Asterisks indicate the p-value significance, *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
To delineate the mechanism of mitochondrial hyperfusion in Δ31Δ34 cells, mRNA transcript, and steady-state levels of mitochondrial fission protein, Dnm1 was measured. The q-PCR and western analysis did not reveal any significant changes albeit minor (1.4-fold increment) in the expression of Dnm1 transcript as well as protein levels (Fig. 4C). Also, the recruitment to the outer mitochondrial membrane was relatively unaltered as analyzed by GFP tagging of Dnm1 in Δ31Δ34 cells (Fig. S1B). In contrast, significantly elevated levels of mitochondrial fusion mediating proteins, Fzo1, and Mgm1was observed. The level of Fzo1 was enhanced by 5–6 folds (transcript and protein levels) in Δ31Δ34 cells (Fig. 4D&E). Relatively, a mild increment in the Mgm1 levels (2–3 folds) was observed in Δ31Δ34 cells (Fig. 4D and 4E). The oxidized glutathione (GSSG) shown to favor the mitochondrial hyperfusion by modulating Mfn2, a homolog of Fzo1 in mammalian cells [40]. Since Hsp31 homologs are involved in the redox regulation, the relative GSSG levels assessed by measuring the GSH/GSSG ratio in Δ31Δ34 cells. In agreement with the mammalian system, a significant enhancement in the GSSG was observed in Δ31Δ34 cells [40] (Fig. 4F). In conclusion, our findings highlight that Hsp31/34 paralogs efficiently regulate mitochondrial dynamics by altering the fusion protein levels mediated perhaps through changes in the cellular glutathione content.
2.4. DJ-1 homologs regulate mitochondrial biogenesis by altering basal redox levels
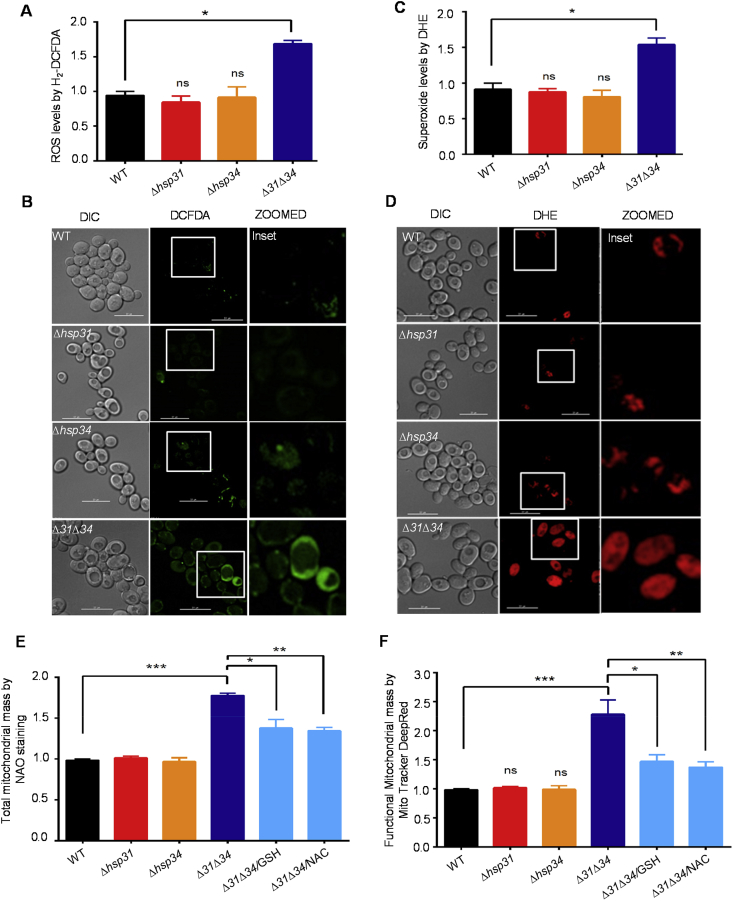
Growing evidence suggests that redox species (ROS) act as signaling molecules and regulate mitochondrial biogenesis [41]. Previously, our results provided evidence to highlight that Hsp31 provides robust cytoprotection against exogenous peroxide as well as methylglyoxal stress in yeast. Besides these observations, Δ31Δ34 cells also exhibited a reduction in the GSH levels, which is an indicator of oxidative stress [6]. Therefore, to evaluate the effect of redox species, the basal intrinsic ROS levels were measured in different media conditions. The yeast cells were grown in liquid media treated with ROS-reactive dye, H2-DCFDA and fluorescence were quantitated by flow cytometric measurements. The basal intrinsic ROS levels significantly elevated in Δ31Δ34 cells grown in dextrose media compared to control strains (Fig. 5A). This increment in the basal ROS levels essentially does not manifest any growth defects (Fig. 1A). These observations was further supported by the microscopic analysis were Δ31Δ34 cells displayed significant enhancement in the green fluorescence, an indicator of elevated ROS levels (Fig. 5B). The data further validated by using another superoxide reactive dye Dihydroethidium (DHE), which also showed a similar pattern of enhanced fluorescence in the Δ31Δ34 cells (Fig. 5C & 5D).
Fig. 5.
DJ-1 paralogs regulate basal redox status of the cell.(A, B) Measurements of overall cellular ROS levels. An equivalent amount of cells from WT, Δhsp31, Δhsp34, and Δ31Δ34 strains were harvested and treated with H2-DCFDA and subsequently analyzed using flow cytometry. Median fluorescence values were obtained from FACS analysis plotted and represented as a bar graph (A). Similarly, H2-DCFDA treated strains were analyzed for cellular ROS levels by microscopic analysis, left panels (DIC image), middle panels (DCFDA fluorescence), and right panels (merge) (B). (C, D) Measurement of cytosolic superoxide levels. The yeast strains (WT, Δhsp31, Δhsp34, and Δ31Δ34) were grown till mid-log phase and treated with 10 μM oxidation-sensitive dye, DHE, and subjected to flow cytometry (C) and microscopic analysis (D). Scale bar (10 μm). All the panels imaged at identical exposure conditions. (E, F) Effect of ROS scavengers on mitochondrial mass. Yeast strains were grown overnight in YPD (Δ31Δ34 grown in YPD in the presence of either GSH or NAC) media and treated with 10 μM NAO-cardiolipin stain (E), and MitoTracker Deep-Red (F) followed by flow cytometry analysis. Error bars represent the standard deviation in the median values from 3 biological replicates. Asterisks indicate the p-value, *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The increment in the basal redox levels may be the initial trigger for the mitochondrial remodeling, which is an adaptive response to cell survival. Under this circumstance, the mitochondrial phenotypes should be reversed upon the depletion of ROS levels. To test this hypothesis, Δ31Δ34 cells were grown in the presence of ROS-scavenging agents, GSH and N-Acetyl Cysteine (NAC). Interestingly, there was a significant restoration in the total mitochondrial mass upon NAO staining in Δ31Δ34 cells comparable to WT (Fig. 5E). A similar restoration in the functional mitochondrial mass was observed in the presence of GSH and NAC treatment using Mitotracker DeepRed staining (Fig. 5F). To further ascertain the redox-homeostatic function of the Hsp31 paralogs is critical in maintaining the mitochondrial health, methylglyoxalase-deficient mutant (Hsp31C138A), which exhibits elevated ROS levels was overexpressed in Δ31Δ34 cells. Intriguingly, the mutant failed to restore the functional mitochondrial mass and morphology as compared to WT Hsp31 overexpression, suggesting that glyoxalase activity is critical, in vivo (Figs. S2A and S2B). In conclusion, our results provide compelling evidence to demonstrate that the elevation in the basal ROS triggers the mitochondrial remodeling in Δ31Δ34 cells without compromising the growth, thus corroborating with the earlier findings.
2.5. Loss of DJ-1 members exhibits a delay in cell-cycle progression
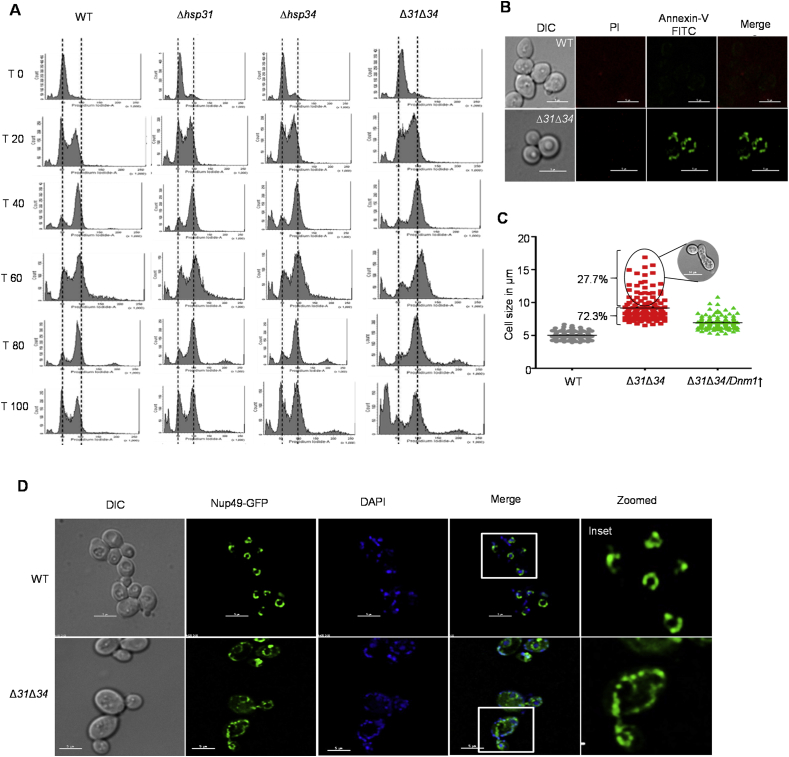
The changes in the mitochondrial dynamics is tightly connected to the cell-cycle progression [42]. To test, the yeast cells were arrested in G1-phase using α-factor and released synchronously in dextrose media, and samples subjected to FACS analysis [43]. Unlike WT, the Δ31Δ34 cells exhibited a significant delay in G2/M progression (Fig. 6A, 60 min). This delay was even evident in the second round of the cell cycle (Fig. 6A, 100 min). At the same time, Δ31 and Δ34 strains did not display any delay in the cell cycle progression (Fig. 6A). In addition to that, a sizable population of Δ31Δ34 cells exhibited aneuploidy condition (>2c) (Figs. S3A and S3B). In cell cycle progression, the G2/M transition is also closely connected to the mitochondrial fission process. Since there is a delay in G2/M progression, we hypothesized that it may due to impairment in the mitochondrial fragmentation before segregation to the daughter cells. To test, yeast cells were treated with nocodazole drug, which arrests the cells in G2/M-phase, and monitored for the changes in the mitochondrial morphology. A distinct fragmentation of mitochondria observed in WT, which is essential to trigger mitotic division to segregate mitochondria to the daughter cells (Fig. S3C, top panel). On the other hand, impaired mitochondrial fragmentation was observed in Δ31Δ34 cells with retention of reticular tubular network structures. Furthermore, these mitochondrial phenotypes rescued upon overexpression of fission protein, Dnm1 (Fig. S3C, middle & last panels).
Fig. 6.
Loss of DJ-1 paralogs promotes delayed cell-cycle progression and apoptosis.(A) Cell-cycle analysis. Yeast strains grown in liquid media were synchronized in G1 using α-factor. Released cells were collected at different time intervals and subjected to flow cytometry after PI staining. (B) Apoptotic assay. Cells grown till mid-log were stained with Annexin-V/PI followed by microscopic analysis for apoptotic induction. (C) Assessment of cell morphology and size. The yeast strains (WT, Δ31Δ34, and Δ31Δ34/Dnm1↑ cells) were microscopically examined to measure the individual cell size using Image J. The values plotted using Graph Pad Prism 5.0. (WT, n = 126; Δ31Δ34, n = 158; Δ31Δ34/Dnm1↑, n = 116). (D) Nuclear morphology analysis. The yeast cells (WT and Δ31Δ34) expressing Nup49-GFP were grown till mid-log phase and analyzed microscopically to determine nuclear morphology.
Since the overexpression of Dnm1 induces mitochondrial fission in the G2/M phase, we asked whether this leads to restoration in the cell cycle progression in Δ31Δ34 cells. A significant restoration in the cell cycle progression was observed in Δ31Δ34 cells, however, not similar to WT (Fig. S4A). This raises an intriguing question of what other possibilities lead to delay in cell cycle progression. Owing to the function of Hsp31 paralogs in the maintenance of redox homeostasis, elevated ROS levels can induce cell cycle arrest due to DNA damage. Therefore, we tested for the occurrence of DNA damage in Δ31Δ34 cells. Interestingly, upon spot analysis in the media containing DNA damaging agents such as Mehtylmethane sulfonate (MMS) and hydroxyurea (HU), Δ31Δ34 cells showed significant growth impairment (Fig. S4B). To investigate whether the delay in G2/M progression may be a consequence of activation of a checkpoint pathway resulting from enhanced spontaneous DNA damage, we assessed the phosphorylation status of checkpoint proteins, Rad9, and Rad53. As compared to WT, we did not observe a significant change in the phosphorylated forms of Rad9 and Rad53 in Δ31Δ34 cells (Fig. S4C). This suggests that lack of Hsp31/34 paralogs induces partial DNA damage leading to a delay in cell cycle progression. In summary, our findings highlight that lack of Hsp31/34 paralogs causes a significant delay in the cell cycle progression due to mitochondrial hyperfusion and elevated DNA damage.
2.6. Deletion of DJ-1 homologs leads to apoptosis in yeast
Perturbations in the cell cycle progression are often associated with the manifestation of apoptosis [44]. The induction of apoptosis was monitored by Annexin-V/Propidium Iodide (PI) staining. Upon microscopic analysis, Δ31Δ34 cells showed positive staining towards Annexin-V, suggesting induction of apoptosis (Fig. 6B). Besides, Δ31Δ34 cells showed an elongated aberrant cell morphological feature, which is a hallmark of apoptotic induction [45] (Fig. S5A). The average size of Δ31Δ34 cells measured to be around ~ 8–9 μm against WT of 5–6 μm in the growth phase. A significant population of Δ31Δ34 cells (20–30 %) showed enhancement in the cell size ranging from 11 to 17 μm at 12 and 24 h of growth, respectively (Figs. 6C and S5B). Similarly, chromatin fragmentation or deformations in nuclei structures was monitored by GFP-tagging for the nuclear protein, Nup49. A substantial nuclear membrane fragmentation is observed upon microscopic analysis as Nup49-GFP was found to be distributed throughout cytosol in Δ31Δ34 cells, unlike WT (Fig. 6D). A similar nuclear fragmentation was also observed in Δ31Δ34 cells upon DAPI staining highlighting the induction of apoptosis (Fig. S5C). In conclusion, our findings indicate that the loss of Hsp31/34 paralogs induces apoptotic mediated cell death in S. cerevisiae.
3. Discussion
Maintenance of mitochondrial health is vital for cellular physiology. Quality surveillance of mitochondria is mainly achieved by mitophagy, a process intimately linked to its dynamics. In humans, mitophagy is promoted by PINK1, a mitochondria-targeted kinase, and Parkin, a cytosolic ubiquitin ligase [46]. Loss of function mutations in PINK1 and Parkin is attributed to the accumulation of dysfunctional mitochondria, one of the leading causes of neurodegeneration. Hence, uncovering multiple mechanisms that regulate organelle quality control processes are of paramount importance for cell survival.
In humans, DJ-1 functions in parallel with the PINK1/Parkin pathway and loss of its activity results in mitochondrial fragmentation under oxidative stress conditions [31]. Our findings highlight that loss of DJ-1 paralogs (Hsp31/34) elicits alterations in the mitochondrial biogenesis and dynamics, which are critical in the regulation of key physiological processes like cell cycle, apoptosis and organelle quality [47]. The increment in the functional mitochondrial volume is primarily induced due to enhancement in the basal ROS levels, which is critically maintained by DJ-1 paralogs. The elevation in the basal ROS levels is likely a consequence of methylglyoxal mediated toxicity since DJ-1 paralogs are involved in the suppression of harmful effects of MGO. This was further evidenced by the fact that the glyoxalase deficient mutant failed to restore the mitochondrial integrity upon overexpression. At the cellular level, the upregulation of mitochondria is observed as an adaptive compensatory response mechanism to maintain its functions by buffering the harmful effects elicited by the ROS surge. A similar ROS-linked mitochondrial upregulation was reported previously via activation of PGC-1α in humans as well as under caloric restriction conditions [48]. Such organellar adaptation mechanisms are reversible by treatment with ROS scavengers like GSH or NAC, which significantly restores the dynamic mitochondrial equilibrium in the absence of Hsp31/34 paralogs. This underscores the significance of DJ-1 paralogs in the maintenance of organellar homeostasis by regulating the redox status of the cell.
Mitochondrial functions and plasticity are stringently regulated at multiple levels in response to metabolic alterations under stress conditions. For instance, during autophagy, mitochondria forms elongated tubular structures to escape from the degradation process, which is induced by the impairment of Drp1 localization onto the outer membrane [37]. Under stress conditions, Opa1 promotes mitochondrial interconnectivity, facilitated by mitochondrial scaffolding protein, SLP-2 [49]. In a similar line, DJ-1 paralogs regulate mitochondrial phenotypic variations by altering levels of fusion-mediated proteins, Fzo1 and Mgm1. The interconnectivity of organelle is dependent on the levels of oxidized glutathione (GSSG), which is critically maintained by DJ-1 paralogs. Elevated GSSG levels are known to induce mitochondrial fusion in mammalian cells by promoting trans-dimerization of the outer membrane fusion protein, Mfn1 [40]. Our data support this idea that perhaps DJ-1 paralogs mediate inter-organelle interactions by altering GSSG levels in response to basal ROS levels (Fig. 7). The upside of mitochondrial hyperfusion could be a short-term benefit to cells as a pro-survival response against intrinsic stress in the absence of DJ-1 paralogs. This is well-correlated with increased mitochondrial ATP levels in Δ31Δ34 cells, generally associated with mitochondrial fusion to confer the resistance against stress [49].
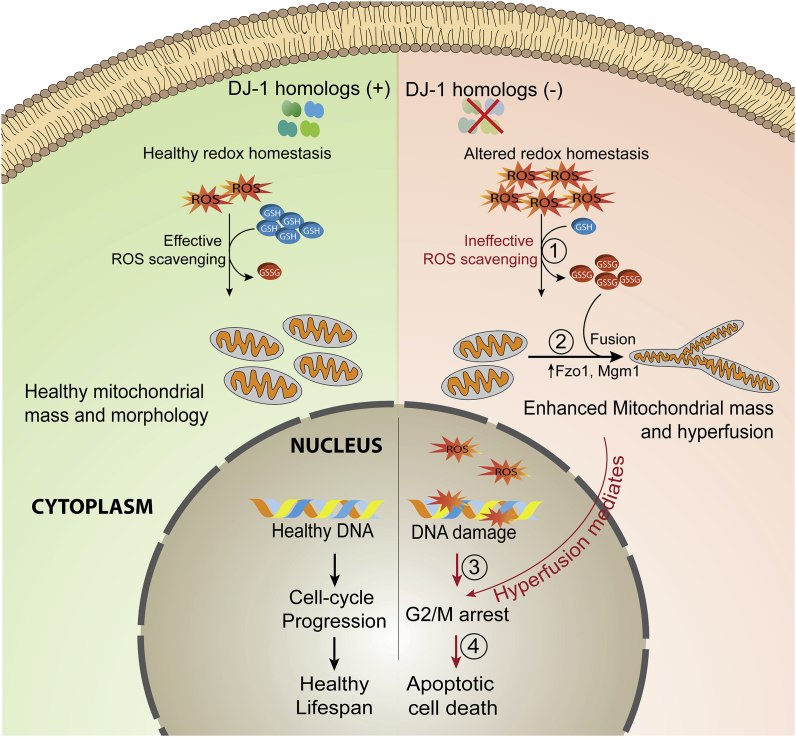
Fig. 7.
Mechanistic highlights of DJ-1 paralogs role in the maintenance of mitochondrial health. Loss of DJ-1 paralogs results in altered redox homeostasis leading to elevation of cellular GSSG levels (1). The enhancement in the basal ROS also upregulates levels of mitochondrial fusion proteins, Fzo1, and Mgm1 promoting robust mitochondrial interconnectivity (2). Furthermore, the enhanced basal ROS level induces partial nuclear DNA damage, and together with mitochondrial hyperfusion delays cell cycle progression (3). DJ-1 paralog mediated mitochondrial phenotypic variation manifest into alterations in cell size, reduced chronological life span followed with apoptotic mediated cell death (4).
Mitochondrial fission-fusion processes are well-coordinated with cell cycle progression ensuring accurate spatial organelle segregation during cell division [42]. Loss of Drp1 function promotes mitochondrial hyperfusion resulting in cell cycle arrest at the G2/M phase and exhibits a high degree of aneuploidy [50]. The downside of persistent mitochondrial interconnectivity observed in the absence of DJ-1 paralogs leads to G2/M accumulation accompanied by aneuploidy [50]. Unlike WT, Δ31Δ34 cells treated with nocodazole failed to undergo mitochondrial fragmentation, which was restored partially upon overexpression of Dnm1 fission-protein. Besides the mitochondrial phenotypic variations, a partial accumulation of DNA damage also contributed to the delayed cell cycle progression in Δ31Δ34 cells without activating the checkpoint pathway. A ROS-mediated abnormal cell cycle progression is often associated with the induction of apoptosis as revealed by gross cell morphological changes, and nuclear disintegration in the absence of DJ-1 paralogs (Fig. 7).
In summary, we uncover the functional diversity of eukaryotic DJ-1 paralogs in the maintenance of mitochondrial homeostasis by regulating the basal redox status of the cell. As mitochondrial respiration is susceptible to several intrinsic and extrinsic factors, it is imperative to maintain a threshold functional volume in high energy-consuming post-mitotic cells, including neurons and cardiomyocytes. Mitochondrial dysfunction and phenotypic variations critically influence the manifestation of cellular etiologies associated with familial PD-linked mutations in DJ-1. Therefore, our current findings highlight a significant lacuna in our understanding of the underlying mechanisms of how DJ-1 paralogs are involved in the maintenance of mitochondrial health. Based on our results, we hypothesize that loss of function of mammalian DJ-1 orthologs may be responsible for an adaptive mitochondrial stress-response, which could eventually trigger the cell death. Moreover, DJ-1 could serve as an interesting therapeutic enzyme target to restore the mitochondrial equilibrium by modulating its methylglyoxalase activity which is linked to several neurodegenerative diseases, including PD, AD that is associated with impaired mitochondrial homeostasis.
4. Materials and methods
Generation of yeast strains and plasmid construction. The single knockouts of Δhsp31, Δhsp32, Δhsp33, Δhsp34 and double knockout of Δhsp31Δhsp34 generated in the haploid yeast strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) as mentioned in the earlier study (Supplemental Table S1) [6]. For the generation of Δhsp31Δhsp32 and Δhsp31Δhsp33, we amplified hphNT1 cassette using suitable primers P1/P2 (Supplemental Table S2) and transferred into Δhsp31 background strain. Similarly, for the generation of Δhsp31Δhsp32Δhsp34 and Δhsp31Δhsp33Δhsp34, the cassette was generated by using primers P1/P2 and transferred into Δhsp31Δhsp34 double knockout strain. The positive colonies screened using a similar method described earlier [6].
For mitophagy analysis, mitochondrial Om45 was tagged by PCR-based gene integration of DNA fragment of GFP at 3′ end of OM45 via homologous recombination using suitable primers P3/P4, as explained earlier [51]. Mitochondrial dynamics related proteins such as Fzo1, Mgm1, and Dnm1 were tagged at the C-terminus with heme agglutinin (HA) using hphNT1 cassette as described earlier [51]. For Dnm1 localization studies, the C-terminus of the corresponding ORF was fused with GFP in the genomic DNA using hphNT1 as a selectable marker. For the assessment of nuclear morphology, Nup49, a nuclear membrane protein was tagged with GFP at the C- terminus, using hphNT1 as a selection marker.
For complementation assays, Hsp31 and Hsp34 ORFs were amplified using P5/P6 and P7/P8 primers respectively and cloned under TEF promoters in pRS415 centromeric vector using BamHI and SalI restriction enzymes. For visualization of mitochondria, MTS-mCherry constructs were generated using a similar method as described earlier [6] and expressed in respective yeast strains.
Media and growth conditions. The yeast strains were grown in standard YP-Dextrose media. For mitochondrial phenotype analysis, cells were grown in S.D. Glycerol (2 %) media. For organelle turnover analysis, YP-lactic acid (2 %) media used to induce mitophagy. To test the effects of ROS scavengers on the mitochondrial content, GSH or NAC was added directly to YP-Dextrose media. For cell cycle analysis, strains were grown in S.D. complete media and then released in YP-Dextrose.
Estimation of total and functional mitochondrial mass. The total mitochondrial mass was estimated using a cardiolipin specific dye, NAO (Nonyl-Acridine Orange). The cells were grown in a respective liquid medium to mid-log phase (A600 ~ 0.6–0.8), harvested, and resuspended in 1X PBS (Phosphate Buffered Saline) along with 10 μM NAO dye. For the assessment of functional mitochondrial mass, cells were resuspended with either 10 μM TMRE (Tetramethylrhodamine ethyl ester) or 0.5 μM Mitotracker DeepRed at 30 °C for 20 min. Subsequently, the cells were washed with 1X PBS once and analyzed on flow cytometry (BD FACS Verse). The median values obtained for each strain represented as a bar graph using GraphPad Prism 6.0.
Assessment of aconitase activity and mitochondrial ATP. Mitochondria were isolated from respective yeast strains using earlier published protocols [52]. Aconitase assay performed as per previous protocols [53]. Mitochondrial lysate (50 μg) prepared in lysis buffer (50 mM Tris-Cl, pH 8.0, 50 mM NaCl) containing 1.0 % Deoxycholic acid at 4 °C for 10 min. The reaction was initiated by adding aconitase buffer containing sodium citrate dihydrate. The absorbance kinetics measured at 235 nm for 3 min using a spectrophotometer.
Mitochondrial ATP content was measured using the Mitochondrial ToxGlo™ Assay kit (Promega). Briefly, 10 μg of mitochondria resuspended in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, pH 7.2) was subjected to repeated cycles of freeze-thaw to ensure complete mitochondrial lysis. Subsequently, ATP-detection reagent was added to each well and mixed for 5 min. The luminescence measured using TECAN-infinite pro-microplate reader. The final readouts are plotted and represented as a bar graph using GraphPad Prism 6.0.
Microscopic analysis. Yeast strains were grown till mid-log phase and treated with either cytosolic ROS specific dye, 100μM H2-DCF-DA, (2′,7′-Dichlorodihydrofluorescein Diacetate) or cytosolic superoxides specific dye, 10μM DHE (Dihydroethidium) (Life Technologies) for 30 min at 30 °C followed by microscopic analysis. For mitochondrial morphological studies, cells transformed with the MTS-mCherry construct were grown at 30 °C till the mid-log phase (A600 ~ 0.6–0.8). An equivalent number of cells were collected by centrifugation and resuspended in 1X PBS. To check mitochondrial morphology at the G2/M phase, cells arrested with Nocodazole (Sigma) at a final concentration of 15 μg/ml. The Imaging was performed by mounting yeast cells on to 2.0 % agarose pads. All the images captured using a Cool SNAP HQ2 camera on the Delta Vision Elite microscope using a 100 x objective lens.
Flow cytometry analysis. For ROS measurement, yeast strains were grown in liquid YPD medium at 30 °C till mid-log phase. An equivalent number of cells were harvested and resuspended in either H2-DCF-DA or DHE (Life Technologies) and incubated at 30 °C for 20 min, followed by flow cytometry analysis (BD FACS Verse). The respective median values are taken and plotted as a bar graph. For cell cycle progression analysis, cultured cells at A600 ~ 0.1–0.2 synchronized using alpha-factor (Sigma) at a final concentration of 50 ng/ml. The released yeast cells were collected at different time intervals and fixed in 70 % ethanol overnight at 4 °C. The cells washed with 1X PBS, mildly sonicated (26 % amplitude four to five pulses), and treated with 0.25 mg/ml RNase A (US Biologicals) at 37 °C overnight. Propidium Iodide (Life Technologies) added to a final concentration of 16 μg/ml, and FACS analysis performed using BD Verse. The data were analyzed using BD FACS Diva software.
Apoptotic assay by FITC Annexin V staining. Apoptosis was measured using the FITC AnnexinV apoptosis detection kit (BD Pharmingen). Briefly, cells grown till mid-log phase (A600 ~ 0.6–0.8) were collected and washed in sorbitol buffer (1.2 M sorbitol, 35 mM potassium phosphate, 0.5 mM MgCl2, pH 6.8), digested with 15 U/ml zymolyase (US Biologicals) in sorbitol buffer for 2 h at 28 °C. The yeast cells were harvested, washed in annexin binding buffer, and resuspended along with 2 μl of FITC Annexin V and 2 μl of Propidium Iodide. The samples were incubated at room temperature for 20 min and then mounted on agarose pads for imaging or subjected for FACS analysis.
Statistical analysis. All statistical analyses performed using GraphPad Prism 6.0 software. Error bars represent the standard deviation derived from three biological replicates. For significance analysis, one-way ANOVA with Tukey's multiple comparison test and Dunnet's multiple comparisons test to compare between columns and against WT used respectively. Asterisks used in the figures represent the following significance values: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001 and ****, p ≤ 0.0001.
Miscellaneous. Total RNA was extracted from yeast cells using Tri-reagent (Sigma). qPCR performed using the corresponding cDNA as a template and suitable primers listed in Supplemental Table S2. The Ct values obtained from the raw data represented in the form of a bar graph. Yeast cell lysates prepared using 10 % Trichloroacetic acid precipitation to perform the western analysis.
Funding information
This work was supported by DST-SERB Grant File No. CRG/2018/001988 and Department of Biotechnology (DBT-IISC Partnership Program Phase-II (No. BT/PR27952/IN/22/212/2018), and DST-FIST Programme-Phase III (No. SR/FST/LSII-045/2016(G), DST-Swarnajayanthi Fellowship (DST/SJF/LS-01/2011–2012) (to P.D.S.).
Authors contribution
P.D.S and K.B designed the study, analyzed the data, and wrote the paper. K.B performed the experiments. V.V, S.S, and S.N helped in conducting mitophagy and DNA damage related assays. V.V and G.S conducted experiments related to Fig. S2. D.P conducted experiments related to qPCR. V.V helped in discussions and editing the manuscript. K.B, V.V, and S.S prepared the figures. All authors reviewed the results and approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they do not have any potential conflicts of interest pertinent to this article to disclose.
Acknowledgement
Yeast Tim44-specific antibody is a kind gift from Prof. Elizabeth A. Craig, University of Wisconsin-Madison, USA. We thank the Flow Cytometry facility of the Indian Institute of Science. We thank Dr. Lakshmi Mahendrawada for valuable discussions in conducting cell cycle experiments. K.B acknowledges the Council of Scientific and Industrial Research (CSIR-India) for providing senior research fellowship. S.N and D.P acknowledge postdoctoral fellowships from SERB and UGC-Dr. DS Kothari, respectively.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101451.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Fig. S1. Loss of Hsp31 paralogs induces mitochondrial hyperfusion. (A) Assessment of mitochondrial morphology as a function of time. WT and Δ31Δ34 cells expressing MTS-mCherry were grown till indicated time-points (8 h, 12 h, and 24 h) in S. D. Leu-medium and subjected for microscopic analysis. Scale bar (10 μm). (B) Evaluation of Dnm1 localization. WT and Δ31Δ34 cells expressing pRS415-MTS-mCherry and Dnm1-GFP were grown till mid-log phase followed by microscopic analysis using a 100 x objective lens. Images were zoomed up to ~2X level and represented (rightmost panels). Scale bar (10 μm).
Fig. S2. Methylglyoxalase-deficient Hsp31 mutant fails to rescue the mitochondrial phenotypes. (A) Analysis of mitochondrial morphology. WT, Δ31Δ34, Δ31Δ34/Hsp31WT, and Δ31Δ34/Hsp31C138A mutant cells expressing MTS-mCherry were grown till the mid-log phase and subjected for microscopic analysis. Scale bar (10 μm). (B) Evaluation of mitochondrial mass. Indicated yeast strains were grown till the mid-log phase and stained with TMRE dye. Cells were analyzed by flow cytometry for the functional mitochondrial mass. Error bars represent the standard deviation in the median values from 3 biological replicates. Asterisks indicate the p-value, *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Fig. S3. Hsp31/34 paralogs regulate cell cycle and aneuploidy. (A, B) Assessment of aneuploidy. The yeast cells stained with Propidium iodide analyzed for the population having DNA >2C marked as aneuploidy condition (A). The gated cell population (>2C) quantified and represented in the bar graph (B). (C) Assessment of mitochondrial morphology at the G2/M phase. The yeast strains (WT, Δ31Δ34, Δ31Δ34/Dnm1↑) expressing MTS-mCherry arrested at the G2/M phase using nocodazole treatment followed by fluorescence microscopic analysis to monitor mitochondrial morphology. Images were zoomed up to ~2X level and represented (rightmost panels). Scale bar (10 μm). Error bars represent the standard deviation in mean fluorescence intensity from 3 biological replicates. Asterisks indicate the p-value, *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Fig. S4. Mitochondrial hyperfusion and DNA damage delays cell cycle progression. (A) Cell-cycle analysis by flow cytometry. The G1 synchronized cells from strains (WT, Δ31Δ34, and Δ31Δ34/Dnm1↑) released and collected at indicated time-intervals stained with propidium iodide (16 ng/mL) followed by flow cytometric measurements. The G2/M cell cycle delay in Δ31Δ34 is indicated with arrows at 60 min and 80 min time interval. (B) DNA damage analysis. The yeast strains (WT and Δ31Δ34) were grown till the mid-log phase, serially diluted and spotted on YP-Dextrose medium containing DNA damaging agents (0.05% MMS, 200 mM HU). Images captured after 36 h of incubation at 28 °C. (C) Assessment of DNA damage checkpoint activation. Total lysates prepared from WT, Δ31Δ34 cells expressing Rad9-HA, and Rad53-HA were analyzed by western blotting using HA-specific antibody. Phosphorylated forms of Rad9 and Rad53 marked with an arrow. WT cells treated with a DNA damaging agent (MMS) was used as a positive control to obtain phosphorylated forms. The non-specific is band highlighted with an asterisk.
Fig. S5. The deletion of Hsp31/34 paralogs displays apoptotic features. (A, B) Evaluation of cell size and morphology. The yeast strains (WT, Δhsp31, Δhsp34, and Δ31Δ34) were grown until the mid-log phase, and the cell morphology analyzed by microscopy (A). Similarly, yeast strains (WT, Δ31Δ34, Δ31Δ34/Dnm1↑) were grown till 24 h, and cell size was measured. The values are represented as a scattered plot (WT, n = 143; Δ31Δ34, n = 150; Δ31Δ34/Dnm1↑, n = 149) (B). (C) Analysis of nuclear morphology by DAPI staining. The yeast strains (WT and Δ31Δ34), grown until the mid-log phase was stained with DAPI and analyzed by microscopy.
References
- 1.Lucas J.I., Marin I. A new evolutionary paradigm for the Parkinson disease gene DJ-1. Mol. Biol. Evol. 2007;24(2):551–561. doi: 10.1093/molbev/msl186. PubMed PMID: 17138626. [DOI] [PubMed] [Google Scholar]
- 2.Subedi K.P., Choi D., Kim I., Min B., Park C. Hsp31 of Escherichia coli K-12 is glyoxalase III. Mol. Microbiol. 2011;81(4):926–936. doi: 10.1111/j.1365-2958.2011.07736.x. PubMed PMID: 21696459. [DOI] [PubMed] [Google Scholar]
- 3.Mujacic M., Baneyx F. Chaperone Hsp31 contributes to acid resistance in stationary-phase Escherichia coli. Appl. Environ. Microbiol. 2007;73(3):1014–1018. doi: 10.1128/AEM.02429-06. PubMed PMID: 17158627; PubMed Central PMCID: PMC1800746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson M.A., St Amour C.V., Collins J.L., Ringe D., Petsko G.A. The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: a member of the DJ-1/ThiJ/PfpI superfamily. Proc. Natl. Acad. Sci. U.S.A. 2004;101(6):1531–1536. doi: 10.1073/pnas.0308089100. PubMed PMID: 14745011; PubMed Central PMCID: PMC341769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Y., Ringe D., Wilson M.A., Ondrechen M.J. Identification of functional subclasses in the DJ-1 superfamily proteins. PLoS Comput. Biol. 2007;3(1) doi: 10.1371/journal.pcbi.0030010. PubMed PMID: 17257049. [DOI] [PubMed] [Google Scholar]
- 6.Bankapalli K., Saladi S., Awadia S.S., Goswami A.V., Samaddar M., D'Silva P. Robust glyoxalase activity of Hsp31, a ThiJ/DJ-1/PfpI family member protein, is critical for oxidative stress resistance in Saccharomyces cerevisiae. J. Biol. Chem. 2015;290(44):26491–26507. doi: 10.1074/jbc.M115.673624. PubMed PMID: 26370081; PubMed Central PMCID: PMC4646309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller-Fleming L., Antas P., Pais T.F., Smalley J.L., Giorgini F., Outeiro T.F. Yeast DJ-1 superfamily members are required for diauxic-shift reprogramming and cell survival in stationary phase. Proc. Natl. Acad. Sci. U.S.A. 2014;111(19):7012–7017. doi: 10.1073/pnas.1319221111. PubMed PMID: 24706893; PubMed Central PMCID: PMC4024926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson M.A. Metabolic role for yeast DJ-1 superfamily proteins. Proc. Natl. Acad. Sci. U.S.A. 2014;111(19):6858–6859. doi: 10.1073/pnas.1405511111. PubMed PMID: 24785296; PubMed Central PMCID: PMC4024854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padilla C.A., Barcena J.A., Lopez-Grueso M.J., Requejo-Aguilar R. The regulation of TORC1 pathway by the yeast chaperones Hsp31 is mediated by SFP1 and affects proteasomal activity. Biochim. Biophys. Acta Gen. Subj. 2019;1863(3):534–546. doi: 10.1016/j.bbagen.2018.12.011. PubMed PMID: 30578832. [DOI] [PubMed] [Google Scholar]
- 10.Natkanska U., Skoneczna A., Sienko M., Skoneczny M. The budding yeast orthologue of Parkinson's disease-associated DJ-1 is a multi-stress response protein protecting cells against toxic glycolytic products. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864(1):39–50. doi: 10.1016/j.bbamcr.2016.10.016. PubMed PMID: 27984092. [DOI] [PubMed] [Google Scholar]
- 11.Aslam K., Tsai C.J., Hazbun T.R. The small heat shock protein Hsp31 cooperates with Hsp104 to modulate Sup35 prion aggregation. Prion. 2016;10(6):444–465. doi: 10.1080/19336896.2016.1234574. PubMed PMID: 27690738; PubMed Central PMCID: PMC5161301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai C.J., Aslam K., Drendel H.M., Asiago J.M., Goode K.M., Paul L.N. Hsp31 is a stress response chaperone that intervenes in the protein misfolding process. J. Biol. Chem. 2015;290(41):24816–24834. doi: 10.1074/jbc.M115.678367. PubMed PMID: 26306045; PubMed Central PMCID: PMC4598993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo P.C., Zhou Y.Y., Ma X.X., Li W.F. Structure of Hsp33/YOR391Cp from the yeast Saccharomyces cerevisiae. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2010;66(Pt 12):1557–1561. doi: 10.1107/S1744309110039965. PubMed PMID: 21139195; PubMed Central PMCID: PMC2998354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao X., Tong L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson's disease. J. Biol. Chem. 2003;278(33):31372–31379. doi: 10.1074/jbc.M304221200. PubMed PMID: 12761214. [DOI] [PubMed] [Google Scholar]
- 15.Bonifati V., Rizzu P., Squitieri F., Krieger E., Vanacore N., van Swieten J.C. DJ-1(PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol. Sci. : Off. J. Ital. Neurol. Soc. Ital. Soc. Clin. Neurophysiol. 2003;24(3):159–160. doi: 10.1007/s10072-003-0108-0. PubMed PMID: 14598065. [DOI] [PubMed] [Google Scholar]
- 16.Amm I., Norell D., Wolf D.H. Absence of the yeast Hsp31 chaperones of the DJ-1 superfamily perturbs cytoplasmic protein quality control in late growth phase. PloS One. 2015;10(10) doi: 10.1371/journal.pone.0140363. PubMed PMID: 26466368; PubMed Central PMCID: PMC4605529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shendelman S., Jonason A., Martinat C., Leete T., Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004;2(11):e362. doi: 10.1371/journal.pbio.0020362. PubMed PMID: 15502874; PubMed Central PMCID: PMC521177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J.Y., Song J., Kwon K., Jang S., Kim C., Baek K. Human DJ-1 and its homologs are novel glyoxalases. Hum. Mol. Genet. 2012;21(14):3215–3225. doi: 10.1093/hmg/dds155. PubMed PMID: 22523093. [DOI] [PubMed] [Google Scholar]
- 19.Mitsumoto A., Nakagawa Y. DJ-1 is an indicator for endogenous reactive oxygen species elicited by endotoxin. Free Radic. Res. 2001;35(6):885–893. doi: 10.1080/10715760100301381. PubMed PMID: 11811539. [DOI] [PubMed] [Google Scholar]
- 20.Moscovitz O., Ben-Nissan G., Fainer I., Pollack D., Mizrachi L., Sharon M. The Parkinson's-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 2015;6:6609. doi: 10.1038/ncomms7609. PubMed PMID: 25833141. [DOI] [PubMed] [Google Scholar]
- 21.Thomas K.J., McCoy M.K., Blackinton J., Beilina A., van der Brug M., Sandebring A. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 2011;20(1):40–50. doi: 10.1093/hmg/ddq430. PubMed PMID: 20940149; PubMed Central PMCID: PMC3000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagakubo D., Taira T., Kitaura H., Ikeda M., Tamai K., Iguchi-Ariga S.M. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 1997;231(2):509–513. doi: 10.1006/bbrc.1997.6132. PubMed PMID: 9070310. [DOI] [PubMed] [Google Scholar]
- 23.Canet-Aviles R.M., Wilson M.A., Miller D.W., Ahmad R., McLendon C., Bandyopadhyay S. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. U.S.A. 2004;101(24):9103–9108. doi: 10.1073/pnas.0402959101. PubMed PMID: 15181200; PubMed Central PMCID: PMC428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J., Zhong N., Wang H., Elias J.E., Kim C.Y., Woldman I. The Parkinson's disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum. Mol. Genet. 2005;14(9):1231–1241. doi: 10.1093/hmg/ddi134. PubMed PMID: 15790595. [DOI] [PubMed] [Google Scholar]
- 25.van der Brug M.P., Blackinton J., Chandran J., Hao L.Y., Lal A., Mazan-Mamczarz K. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc. Natl. Acad. Sci. U.S.A. 2008;105(29):10244–10249. doi: 10.1073/pnas.0708518105. PubMed PMID: 18626009; PubMed Central PMCID: PMC2481328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y., Gehrke S., Haque M.E., Imai Y., Kosek J., Yang L. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc. Natl. Acad. Sci. U.S.A. 2005;102(38):13670–13675. doi: 10.1073/pnas.0504610102. PubMed PMID: 16155123; PubMed Central PMCID: PMC1224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mihoub M., Abdallah J., Richarme G. Protein repair from glycation by glyoxals by the DJ-1 family maillard deglycases. Adv. Exp. Med. Biol. 2017;1037:133–147. doi: 10.1007/978-981-10-6583-5_9. PubMed PMID: 29147907. [DOI] [PubMed] [Google Scholar]
- 28.Richarme G., Liu C., Mihoub M., Abdallah J., Leger T., Joly N. Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science. 2017;357(6347):208–211. doi: 10.1126/science.aag1095. PubMed PMID: 28596309. [DOI] [PubMed] [Google Scholar]
- 29.Xu C.Y., Kang W.Y., Chen Y.M., Jiang T.F., Zhang J., Zhang L.N. DJ-1 inhibits alpha-synuclein aggregation by regulating chaperone-mediated autophagy. Front. Aging Neurosci. 2017;9:308. doi: 10.3389/fnagi.2017.00308. PubMed PMID: 29021755; PubMed Central PMCID: PMC5623690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W., Zhu M., Wilson M.A., Petsko G.A., Fink A.L. The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J. Mol. Biol. 2006;356(4):1036–1048. doi: 10.1016/j.jmb.2005.12.030. PubMed PMID: 16403519. [DOI] [PubMed] [Google Scholar]
- 31.Thomas K.J., McCoy M.K., Blackinton J., Beilina A., van der Brug M., Sandebring A. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 2010;20(1):40–50. doi: 10.1093/hmg/ddq430. Epub 2010/10/14. doi: ddq430 [pii]10.1093/hmg/ddq430. PubMed PMID: 20940149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Petrie T.G., Liu Y., Liu J., Fujioka H., Zhu X. Parkinson's disease-associated DJ-1 mutations impair mitochondrial dynamics and cause mitochondrial dysfunction. J. Neurochem. 2012;121(5):830–839. doi: 10.1111/j.1471-4159.2012.07734.x. PubMed PMID: 22428580; PubMed Central PMCID: PMC3740560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao L.Y., Giasson B.I., Bonini N.M. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc. Natl. Acad. Sci. U.S.A. 2010;107(21):9747–9752. doi: 10.1073/pnas.0911175107. PubMed PMID: 20457924; PubMed Central PMCID: PMC2906840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi T., Ishimori C., Takahashi-Niki K., Taira T., Kim Y.C., Maita H. DJ-1 binds to mitochondrial complex I and maintains its activity. Biochem. Biophys. Res. Commun. 2009;390(3):667–672. doi: 10.1016/j.bbrc.2009.10.025. PubMed PMID: 19822128. [DOI] [PubMed] [Google Scholar]
- 35.Goswami A.V., Samaddar M., Sinha D., Purushotham J., D'Silva P. Enhanced J-protein interaction and compromised protein stability of mtHsp70 variants lead to mitochondrial dysfunction in Parkinson's disease. Hum. Mol. Genet. 2012;21(15):3317–3332. doi: 10.1093/hmg/dds162. PubMed PMID: 22544056; PubMed Central PMCID: PMC3392108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanki T., Wang K., Cao Y., Baba M., Klionsky D.J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17(1):98–109. doi: 10.1016/j.devcel.2009.06.014. PubMed PMID: 19619495; PubMed Central PMCID: PMC2746076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomes L.C., Di Benedetto G., Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13(5):589–598. doi: 10.1038/ncb2220. PubMed PMID: 21478857; PubMed Central PMCID: PMC3088644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan D.C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. PubMed PMID: 22934639. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J., Yi J., Fu R., Liu E., Siddique T., Rios E. Hyperactive intracellular calcium signaling associated with localized mitochondrial defects in skeletal muscle of an animal model of amyotrophic lateral sclerosis. J. Biol. Chem. 2010;285(1):705–712. doi: 10.1074/jbc.M109.041319. PubMed PMID: 19889637; PubMed Central PMCID: PMC2804218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattie S., Riemer J., Wideman J.G., McBride H.M. A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J. Cell Biol. 2017;217(2):507–515. doi: 10.1083/jcb.201611194. Epub 2017/12/08. doi: jcb.201611194 [pii]10.1083/jcb.201611194. PubMed PMID: 29212658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chevtzoff C., Yoboue E.D., Galinier A., Casteilla L., Daignan-Fornier B., Rigoulet M. Reactive oxygen species-mediated regulation of mitochondrial biogenesis in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2009;285(3):1733–1742. doi: 10.1074/jbc.M109.019570. Epub 2009/11/10. doi: M109.019570 [pii], PubMed PMID: 19897478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mishra P., Chan D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 2014;15(10):634–646. doi: 10.1038/nrm3877. PubMed PMID: 25237825; PubMed Central PMCID: PMC4250044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao H., Thorner J. Yeast mating pheromone alpha factor inhibits adenylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 1980;77(4):1898–1902. doi: 10.1073/pnas.77.4.1898. PubMed PMID: 6246513; PubMed Central PMCID: PMC348616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Endo K., Mizuguchi M., Harata A., Itoh G., Tanaka K. Nocodazole induces mitotic cell death with apoptotic-like features in Saccharomyces cerevisiae. FEBS Lett. 2010;584(11):2387–2392. doi: 10.1016/j.febslet.2010.04.029. PubMed PMID: 20399776. [DOI] [PubMed] [Google Scholar]
- 45.Madeo F., Frohlich E., Frohlich K.U. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 1997;139(3):729–734. doi: 10.1083/jcb.139.3.729. PubMed PMID: 9348289; PubMed Central PMCID: PMC2141703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. PubMed PMID: 19029340; PubMed Central PMCID: PMC2592826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otera H., Mihara K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J. Biochem. 2011;149(3):241–251. doi: 10.1093/jb/mvr002. PubMed PMID: 21233142. [DOI] [PubMed] [Google Scholar]
- 48.Wenz T. Regulation of mitochondrial biogenesis and PGC-1alpha under cellular stress. Mitochondrion. 2013;13(2):134–142. doi: 10.1016/j.mito.2013.01.006. PubMed PMID: 23347985. [DOI] [PubMed] [Google Scholar]
- 49.Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28(11):1589–1600. doi: 10.1038/emboj.2009.89. PubMed PMID: 19360003; PubMed Central PMCID: PMC2693158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian W., Choi S., Gibson G.A., Watkins S.C., Bakkenist C.J., Van Houten B. Mitochondrial hyperfusion induced by loss of the fission protein Drp1 causes ATM-dependent G2/M arrest and aneuploidy through DNA replication stress. J. Cell Sci. 2012;125(Pt 23):5745–5757. doi: 10.1242/jcs.109769. PubMed PMID: 23015593; PubMed Central PMCID: PMC4074216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21(11):947–962. doi: 10.1002/yea.1142. PubMed PMID: 15334558. [DOI] [PubMed] [Google Scholar]
- 52.Gambill B.D., Voos W., Kang P.J., Miao B., Langer T., Craig E.A. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol. 1993;123(1):109–117. doi: 10.1083/jcb.123.1.109. PubMed PMID: 8408191; PubMed Central PMCID: PMC2119813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saha P.P., Srivastava S., Kumar S.K.P., Sinha D., D'Silva P. Mapping key residues of ISD11 critical for NFS1-ISD11 subcomplex stability: implications IN the development OF mitochondrial disorder, COXPD19. J. Biol. Chem. 2015;290(43):25876–25890. doi: 10.1074/jbc.M115.678508. PubMed PMID: 26342079; PubMed Central PMCID: PMC4646244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.