Abstract
Based on C (wild) to T (mutant) transition at amino acid position 1432 bp of lpa1-1 gene, two dominant markers each specific to wild type (LPA1) and mutant (lpa1-1) allele were developed and validated across seven F2 populations. Joint segregation of these markers behaved in co-dominant fashion, clearly distinguishing heterozygote from two other homozygote genotypes. Full length sequence alignment between wild type (LPA2) and mutant (lpa2-1) allele revealed one transition mutation (A to G) and a co-dominant CAPS marker was developed which differentiated all three types of segregants across seven F2 populations. Across populations, segregants with lpa1-1/lpa1-1 (1.77 mg/g) and lpa2-1/lpa2-1 (1.85 mg/g) possessed significantly lower phytic acid compared to LPA1/LPA1 (2.58 mg/g) and LPA2/LPA2 (2.53 mg/g). Inorganic phosphorus was however higher in recessive homozygotes (lpa1-1/lpa1-1: 0.77 mg/g, lpa2-1/lpa2-1: 0.53 mg/g) than the dominant homozygotes (LPA1/LPA1: 0.33 mg/g, LPA2/LPA2: 0.19 mg/g). Overall, homozygous segregants of lpa1-1 and lpa2-1 showed 31% and 27% reduction of phytic acid, respectively. Analysis of phytate and inorganic phosphorous in the maize kernel in these segregating populations confirmed co-segregation of trait and markers specific to lpa1-1 and lpa2-1. This is the first report of the development of breeder-friendly gene-based markers for lpa1-1 and lpa2-1; and it holds great significance for maize biofortification.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-2113-x) contains supplementary material, which is available to authorized users.
Keywords: Maize, Bioavailability, Low phytate, Iron, Zinc, Markers
Introduction
Maize is the third most important food grain crop in India, next to rice and wheat (Gupta et al. 2019). It is being cultivated in 9.6 million ha of area with an annual production of 27.14 million metric tonnes in the country (AICRP-Maize Progress Report 2018). Considering the growing significance of maize as food and animal feed; enhancement of micronutrients in grain assumes great importance in the scenario of micronutrient deficiency which affects two billion people globally (De Steur et al. 2015; Hossain et al. 2019). Among micronutrients, deficiency of iron (Fe) and zinc (Zn) poses most serious health constraints worldwide (Bouis 2018). In developing countries, major portion of the population depends on plant-based foods particularly cereals, where lack of required levels of Fe and Zn cause severe metabolic disorders (Yadava et al. 2017). Breeding efforts were made to develop crop varieties with high kernel Fe and Zn, although success could not be achieved due to its polygenic nature and high genotype × environment interactions (Gupta et al. 2015a).
Bioavailability is the degree to which food nutrients are available for absorption and utilization in the body. One of the major anti-nutritional factors (e.g. phytic acid in maize kernel) plays a key role in reducing the bioavailability of kernel Fe and Zn to both humans and animals (Adams et al. 2000). Therefore, reducing the phytic acid content in crops like maize would be an important strategy in genetic bio-fortification and significantly improve the bioavailable kernel Fe and Zn content (Gupta et al. 2015b). Moreover, monogastric animals including humans, poultry and swine cannot digest phytic acid in their gut, so the phytate is expelled directly to the environment along with excreta posing a serious concern where the continuous expulsion of high phosphorous load causes pollution in the nearby water bodies (Jorquera et al. 2008). These low phytic acid (lpa) mutants are available in many crops, and maize is the first crop in which lpa mutations were isolated (Raboy et al. 2000). These lpa mutations hamper various steps in the phytic acid biosynthesis pathway thereby reducing the levels of phytic acid in the grain (Raboy et al. 2000). These mutants produce seeds that have normal levels of total phosphorous but greatly reduced levels of phytic acid phosphorous. These mutations, therefore, do not affect the ability of a plant to uptake phosphorous and its transportation to a developing seed; instead, block the ability of a seed to synthesize phosphorous into phytic acid (Pilu et al. 2003; Raboy 2009). Several lpa mutants have been isolated in maize viz. lpa1, lpa2, lpa3 and lpa241. Of these, lpa1-1 mutation causes up to 55–65% reduction of phytic acid in maize grain; and is due to a mutation in trans-membrane transporter protein (ZmMRP4) i.e., C to T transition which led to alanine to valine amino acid change (Shi et al. 2007). The lpa2-1 mutation causes 50% reduction in phytic acid and is due to mutation in inositol phosphate kinase (ZMIPK) enzyme (Raboy et al. 2000) and this lpa2-1 mutation may be due to a genomic sequence rearrangement in the ZmIpk gene (Shi et al. 2003). The maize lpa3 gene is located near adh1 locus on chromosome 1S, and it encodes myo-inositol kinase (MIK) which is an enzyme and causes reduction of phytic acid up to 50% (Shi et al. 2005). The lpa241 mutation has shown 90% reduction in phytic acid and tenfold increase in the grain-free phosphate, however, there is a 30% reduction in germination in mutant lines compared to the wild types (Pilu et al. 2003). Therefore, the use of mutations such as lpa241 has been limited by severe negative effects on seed viability, seed germination and plant growth, resulting in various levels of yield penalty (Pilu et al. 2003).
Currently, low phytic acid mutations are available in the temperate genetic background and it is very important to transfer them in the locally adapted maize lines. Non-availability of widely adapted low phytic acid maize inbreds poses serious limitation in breeding programme. Further, phenotypic selection for kernel phytic acid is destructive and can be done only after harvest, accounting for increase in cost, time and resources, hence these objectives can be achieved with the aid of marker-assisted selection (MAS).
Molecular markers are very useful tool for recognizing genomic regions responsible for the control of traits of interest and marker-assisted selection (Singh and Singh 2015). The use of molecular markers to select for the trait of interest not only saves time but also huge resources involved in the breeding programme (Muthusamy et al. 2014; Hossain et al. 2018; Sarika et al. 2018; Zunjare et al. 2018). Gene-based markers have polymorphic sites present within the gene, and it nullifies the chance of recombination between marker and the gene, which often lead to false positives, hence selection become more precise (Das et al. 2019). So far only few attempts have been made to develop markers for lpa gene(s). Naidoo et al. (2012) crossed two inbred parental lines viz. CM32 (temperate LPA line) and P16 (tropical wild type line) to produce F1 heterozygotes, designed SNP primers for lpa1-1 and used high resolution melt (HRM) analysis to successfully distinguish among the homozygous dominant (wild type), homozygous recessive (mutant) and heterozygous genotypes. Sureshkumar et al. (2014) reported a linked SSR marker umc2230 for lpa2-2gene. However, linked markers pose problems during the MAS process, as there is always a possibility of recombination between gene and the marker loci, thereby leading to false positives during the selection process. Therefore, the development of breeder friendly gene-based marker(s) for the lpa genes would be of immense significance in breeding for low phytate maize. The present study addressed this issue by developing gene-based markers for both lpa1-1 and lpa2-1genes and validated them in a set of seven segregating populations each, for its use in MAS.
Methods
Plant materials
A diverse panel of seven high phytic acid (wild type) genotypes viz.HKI161PV, HKI163PV, HKI193-1PV, HKI193-2PV, HKI323Q, HKI1105Q andHKI1128Q were selected for the study (Table 1). Four of these lines, HKI161-PV, HKI163-PV,HKI193-1PV and HKI193-2PV are QPM inbreds enriched with provitamin A, whileHKI323Q, HKI1105Q and HKI1128Q are QPM inbreds rich in lysine and tryptophan developed through marker-assisted selection (MAS) at ICAR-IndianAgricultural Research Institute (IARI), New Delhi. Three low phytic acid (lpa) mutants viz. A619 lpa1-1 (EC860912), A632 lpa 1-1 (EC860913) and A619 lpa2-1(EC860914) obtained from Dr. Victor Raboy, United States Department of Agriculture (USDA), Idaho, United States were also used in the study. Crosses were attempted between the normal (high phytate) lines and lpa mutants (lpa1-1 and lpa2-1) (Table S1).
Table 1.
Details of the wild type and mutant maize inbreds used in the study
| S. no. | Genotype | Pedigree | Source institution |
|---|---|---|---|
| Wild type | |||
| 1 | HKI161PV | (HKI161///HP704-23)-⊗-⊗-⊗-⊗ | ICAR-IARI, New Delhi |
| 2 | HKI163PV | (HKI163///HP704-22)-⊗-⊗-⊗-⊗ | ICAR-IARI, New Delhi |
| 3 | HKI193-1PV | (HKI193-1///HP704-23)-⊗-⊗-⊗-⊗ | ICAR-IARI, New Delhi |
| 4 | HKI193-2PV | (HKI193-2///HP704-22)-⊗-⊗-⊗-⊗ | ICAR-IARI, New Delhi |
| 5 | HKI323Q | (HKI323///HKI161)-⊗-⊗-⊗-⊗-⊗-⊗-⊗-⊗-⊗ | ICAR-IARI, New Delhi |
| 6 | HKI1105Q | (HKI1105///CML161)-⊗-⊗-⊗-⊗-⊗-⊗-⊗-⊗-⊗ | ICAR-IARI, New Delhi |
| 7 | HKI1128Q | (HKI1128///HKI193-1)-⊗-⊗-⊗-⊗-⊗-⊗-⊗-⊗ | ICAR-IARI, New Delhi |
| Mutant | |||
| 1 | A619 lpa1-1 | EC860912 | USDA-ARS, Idaho, USA |
| 2 | A632 lpa1-1 | EC860913 | USDA-ARS, Idaho, USA |
| 3 | A619 lpa2-1 | EC860914 | USDA-ARS, Idaho, USA |
⊗ Number of selfed generation
Isolation and quantification of DNA
Genomic DNA was isolated from leaves of young seedlings of each plant using standard CTAB procedure (Murray and Thompson 1980) optimized at Maize Genetics Unit, Division of Genetics ICAR-Indian Agricultural Research Institute (IARI), New Delhi. The DNA was dissolved in Tris-EDTAbuffer (10 mMTris: 1 mM EDTA) and quantified using a UV–Spectrophotometer (BenchTop Labsystems, US). The quality of DNA was checked using 0.8% agarose gel electrophoresis, followed by dilution with Tris–EDTA buffer to the concentration of 20 ng/μl, the final concentration required for PCR reaction.
Designing of primers to amplify the target genes
The primers for lpa1-1 and lpa2-1 has been designed using Primer3online (v.0.4.0) software (Rozen and Skaletsky 2000). The method of designing of primers for each of the genes has been described below.
- lpa1-1
The sequence information of lpa1-1 was available for both the mutant and the wild type in the public domain (Shi et al. 2007). Based on the available sequence data the primers were designed for the polymorphism between the mutant and the wild type (C to T transition) using Primer3 online software (v.0.4.0) (Table 2).
- lpa2-1
Sequence information of lpa2-1 mutant allele was not available in the public domain. Hence, overlapping primers from the sequence of lpa2-1 of B73 genome were designed (Accession No. NM001112431.2) to cover the full-length gene of 2.26 kb, seven overlapping primers each amplifying around 400–500 base pairs were designed (Table S2). Using the designed overlapping primer(s), the lpa2-1 gene was amplified in both the mutant and wild types. PCR products were custom sequenced in both the directions using forward and reverse primers with two replications (MacrogenInc., South Korea). The consensus sequence for each amplicon in the selected genotypes was generated using both forward and reverse sequence chromatograms and aligned with BioEdit programme (Hall 2011). The aligned sequences were used to analyze the presence of nucleotide polymorphisms specific to the mutant and the wild type. Specific primers were designed for the polymorphism that could differentiate the mutant and wild type.
Table 2.
Details of the primers designed for lpa1-1 gene
| S. no. | Name | Sequence (5–′3′) | No. of bases | Position of mismatch (from 3′ end) | Remarks |
|---|---|---|---|---|---|
| 1 | WTSM-F1 | TCGATGAGGCGACCGC | 16 | 3rd base | Non-specific |
| 2 | WTSM-F2 | TACTCGATGAGGCGACAGC | 19 | Nil | Highly specific |
| 3 | MSM-F1 | TTGGTACTCGATGAGGCGAAAGT | 23 | 4th base | Highly specific |
| 4 | MSM-F2 | GGTACTCGATGAGGCGACCGT | 21 | 3rd base | Non-specific |
| 5 | M-R1 | CATGGCAGATACGGGCTATT | 20 | Nil | Highly specific |
| 6 | M-R2 | CAATAACGGTGGGAATACGG | 20 | Nil | Non-specific |
WTSM wild type specific marker, MSM mutant specific marker, M marker, F forward primer, R reverse primer
PCR amplification of the and markers in the segregating populations
lpa1-1
The PCR was carried out in 20 μl reaction mixture containing 0.25 μM each primer (forward and reverse), 100 ng genomic DNA as template and 10 μl of master mix (GeneDirex Inc., Taiwan). The amplification was carried out with initial denaturation of 94 °C for 5 min, 35 cycles of denaturation at 94 °C for 45 s, annealing at 61 °C for 45 s, extension at 72 °C for 45 s and final extension step was carried out at 72 °C for 5 min. The above conditions were used for amplification using mutant specific primer; and the wild type-specific primers with annealing temperature at 61.5 °C.
lpa2-1
The PCR was carried out in 20 μl reaction mixture containing 0. 25 μM each primer (forward and reverse), 1000 ng genomic DNA as template and 10 μl of master mix GeneDirex Inc., Taiwan). The amplification was carried out with initial denaturation of 94 °C for 5 min, 35 cycles of denaturation at 94 °C for 45 s, annealing at 60 °C for 45 s, extension at 72 °C for 45 s and final extension step was carried out at 72 °C for 5 min.
The PCR product was digested using HindIII (New England BioLabs, USA) restriction enzyme as follows: 8 μl of PCR product, 2.5 μl of 1X digestion buffer (supplied with enzyme), 0.05 U of enzyme and made up to 25 μl with nuclease-free water. The mixture was then incubated at 37 °C for 2 h.
Resolution of digested PCR products and scoring of marker profiles
The final PCR amplified products for the markers were resolved through horizontal electrophoresis system at 120 V for 3 to 4 h using 1.0X TBE buffer. 2.0% agarose (SeaKemR LE Agarose, USA) gels stained with ethidium bromide (10 mg/ml) were used. A100 bp DNA ladder (G-Biosciences, USA) were loaded at bothends of gel and images were recorded using a gel documentation system (Alpha Innotech, USA), followed by the scoring of marker profiles. The amplicons were scored as alleles for the loci. The alleles were scored manually and allele sizes (base pairs) were determined comparing with 100 bp DNA ladder which was run parallel with the genotypes.
Quantification of phytic acid and inorganic phosphorous
Determination of phytic acid and inorganic phosphorous in the seeds of the mutant and wild type was carried out as per the method described by Lorenz et al. (2007) with minor modifications. 100 mg of ground kernels from each individual genotype was weighed and placed in 2 ml micro-centrifuge tube with 2 ml of 0.65 M HCl. The tubes were then shaken for overnight at room temperature at 120 rpm and then centrifuged at 12,000 rpm for 5 min. A total of 500 μl of the extract of the respective genotype was transferred to a clean 2 ml micro-centrifuge tube for estimation of phytic acid and to a 15 ml tube for estimation of inorganic phosphorus. Equal volumes of the phytic acid and inorganic phosphorous quantitative standards were used. Phytic acid dodecasodium salt from corn (Sigma) and KH2PO4 (HiMedia) were used as phytate and Pi standards, respectively. The Pi reagent was made immediately before use and it consisted of two parts of distilled H2O, one part each of 0.02 M ammonium molybdate, 0.57 M ascorbic acid and 3 M sulphuric acid. For the estimation of inorganic phosphorus, 1 ml of Pi reagent and 1 ml of distilled H2O were added to each tube. Once, the blue colour develops after 15 to 20 min of incubation at room temperature, the optical density (OD 820) was measured at 820 nm (Figure S1). For measurement of phytate, 1.25 ml of Wade reagent was added to each tube and allowed to react for 15 min at room temperature and after the development of pink colour the optical density at 490 nm (OD 490) was measured (Figure S1). Wade reagent consisted of 0.3 g 5-sulfosalycyclic acid, 0.03 g FeCl3.6H2O and 80 ml distilled H2O and could be stored in a refrigerator for 1 month. The above solution was refrigerated overnight and adjusted to a pH of 3.05 with NaOH the following day. After pH adjustment, dist. H2O was added to a final volume of 100 ml. Phytate was converted to phytate P by dividing phytate by 3.55 (Raboy and Dickinson, 1984).
Statistical analysis
Chi-square (χ2) test was performed using the standard procedure for testing the goodness of fit of the observed segregation pattern of the lpa1-1 and lpa2-1 genes in the respective F2 generation. ‘t’-test to differentiate the allelic class means was performed using Microsoft Excel (2010).
Results and discussion
Targeting the anti-nutritional factors that reduce the bioavailability of the micronutrients present in the diet of humans is a viable approach of genetic improvement of the targeted micronutrients due to profound effect of one or few genes (Gupta et al. 2015a). Low phytic acid is one of the anti-nutritional factors that chelates with positively charged mineral elements such as Fe and Zn and makes them unavailable to human metabolism (Zhou and Erdman 1995). Low phytate mutants were reported in maize and their role in reducing the phytic acid in maize grain is well established (Raboy et al. 2000).
lpa1-1
The lpa1-1 mutation is an EMS induced recessive mutation developed by Raboy et al. (2000) and it has been mapped to chromosome 1S. Shi et al. (2007) sequenced the full-length gene and reported that the lpa1-1 mutation is due to C to T transition in the ZmMRP4 gene which governs the membrane transporter protein. Though the sequence information for lpa1-1 was available, to the best of our knowledge, no report is available on the development of breeder friendly PCR based markers for its use in MAS. The sequence information (GenBank Accession Number: EF586878) was used to develop gene-based markers for lpa1-1. Allele-specific primers were designed using the wild type and the mutant sequence. While designing the primers, efforts were also made to have a mismatch in the penultimate bases (3–4 base from 3′end) to have higher specificity for primer binding (Liu et al. 2012). Based on the Tm value of each primer, the PCR conditions were optimised using gradient PCR. With the standardised cycle conditions, these primers were amplified in a set of mutant and wild type genotypes. From the different primer combinations (Table 2), the primer combination WTSM-F2 and M-R1 and MSM-F1 and M-R1 selected as wild type-specific marker (Fig. 1) and mutant specific marker (Fig. 2), respectively, based on their high specificity in amplification. Both of these markers work in dominant fashion. Genotyping of the seven segregating populations with the WTSM and showed that the marker segregation was as per the Mendelian fashion (3:1) (AA + Aa: aa) (Table 3). Similarly, the segregation pattern of the MSM also fit to the expected Mendelian segregation of 3:1 (aa + Aa: AA) across the seven populations (Table 4). Co-dominant markers are always preferred over dominant markers as they easily differentiate the homozygotes from the heterozygotes. However, in the present study only dominant markers (WTSM and MSM) for lpa1-1 has been developed. Hence, to differentiate the homozygous and the heterozygous genotypes, we analysed the joint-segregation of WTSM and MSM in all the seven segregating populations to differentiate heterozygote (Lpa1/lpa1)) from two different homozygotes (Lpa1/Lpa1 and lpa1/lpa1) (Fig. 3). Joint segregation of both WTSM and MSM has also revealed that the segregation is as per the expected Mendelian pattern across all the seven populations (Table 5).
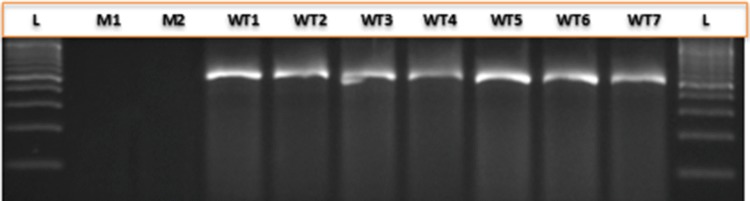
Fig. 1.
Segregation pattern of wild type specific marker (WTSM-LPA1) among the mutant and wild type genotypes. M1: A619 lpa1-1, M2 A632 lpa1-1, WT1 HKI161PV, WT2 HKI163PV, WT3 HKI193-1PV, WT4 HKI193-2PV, WT5 HKI323Q, WT6 HKI1105Q, WT7, HKI1128Q, L 100 bp DNA ladder
Fig. 2.
Segregation pattern of mutant specific marker (MSM-lpa1-1) among the mutant and wild type genotypes. M1: A619 lpa1-1, M2 A632 lpa1-1, WT1 HKI161PV, WT2 HKI163PV, WT3 HKI193-1PV, WT4 HKI193-2PV, WT5 HKI323Q, WT6 HKI1105Q, WT7, HKI1128Q, L 100 bp DNA ladder
Table 3.
Segregation pattern of wild type specific marker (WTSM-LPA1) for lpa1-1
| S. no. | Cross | Total no. of F2s genotyped | No. of genotypes with genotypic class | χ2 | P value | |
|---|---|---|---|---|---|---|
| Presence (A1A1/A1a1) | Absence (a1a1) | |||||
| 1 | HKI161PV × A619 lpa1-1 | 108 | 87 | 21 | 1.778ns | 0.182 |
| 2 | HKI163PV × A632 lpa1-1 | 95 | 75 | 20 | 0.790ns | 0.374 |
| 3 | HKI193-1PV × A619 lpa1-1 | 102 | 79 | 23 | 0.327ns | 0.568 |
| 4 | HKI193-2PV × A619 lpa1-1 | 104 | 82 | 22 | 0.821ns | 0.365 |
| 5 | HKI323Q × A632 lpa1-1 | 99 | 75 | 24 | 0.030ns | 0.030 |
| 6 | HKI1105Q × A632 lpa1-1 | 110 | 86 | 24 | 0.594ns | 0.441 |
| 7 | HKI1128Q × A619 lpa1-1 | 104 | 80 | 24 | 0.205ns | 0.441 |
A1A1 LPA1/LPA1, A1a1 LPA1/lpa1, a1a1 lpa1/lpa1, ns not significant
Table 4.
Segregation pattern of mutant specific marker (MSM-lpa1-1) for lpa1-1
| S. no. | Cross | Total no. of F2s genotyped | No. of genotypes with genotypic class | χ2 | P value | |
|---|---|---|---|---|---|---|
| Presence (a1a1/A1a1) | Absence (A1A1) | |||||
| 1 | HKI161PV × A619 lpa1-1 | 108 | 79 | 29 | 0.198ns | 0.657 |
| 2 | HKI163PV × A632 lpa1-1 | 95 | 67 | 28 | 1.014ns | 0.314 |
| 3 | HKI193-1PV × A619 lpa1-1 | 102 | 73 | 29 | 0.641ns | 0.424 |
| 4 | HKI193-2PV × A619 lpa1-1 | 104 | 76 | 28 | 0.205ns | 0.651 |
| 5 | HKI323Q × A632 lpa1-1 | 99 | 71 | 28 | 0.569ns | 0.451 |
| 6 | HKI1105Q × A632 lpa1-1 | 110 | 79 | 31 | 0.594ns | 0.441 |
| 7 | HKI1128Q × A619 lpa1-1 | 104 | 76 | 28 | 0.205ns | 0.651 |
A1A1 LPA1/LPA1, A1a1 LPA1/lpa1-1, a1a1 lpa1-1/lpa1-1, ns not significant
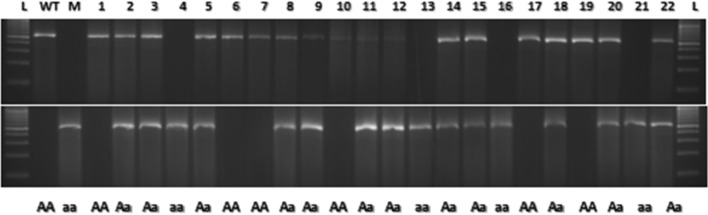
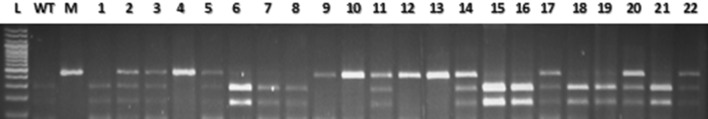
Fig. 3.
Joint segregation pattern of WTSM-LPA1 and MSM-lpa1-1 in HKI161PV × A619 lpa1-1. WT HKI161PV, M A619 lpa1-1, 1–22 F2 individuals, L 100 bp DNA ladder
Table 5.
Joint segregation pattern of WTSM-LPA1 and MSM-lpa1-1 marker for lpa1-1 used in the study
| S. no. | Cross | Total no. of F2s genotyped | No. of genotypes with genotypic class | χ2 | P value | ||
|---|---|---|---|---|---|---|---|
| A1A1 | A1a1 | a1a1 | |||||
| 1 | HKI161PV × A619 lpa1-1 | 108 | 29 | 58 | 21 | 1.778ns | 0.411 |
| 2 | HKI163PV × A632 lpa1-1 | 95 | 28 | 47 | 20 | 1.357ns | 0.507 |
| 3 | HKI193-1PV × A619 lpa1-1 | 102 | 29 | 50 | 23 | 1.357ns | 0.507 |
| 4 | HKI193-2PV × A619 lpa1-1 | 104 | 28 | 54 | 22 | 0.846ns | 0.655 |
| 5 | HKI323Q × A632 lpa1-1 | 99 | 28 | 47 | 24 | 0.576ns | 0.749 |
| 6 | HKI1105Q × A632 lpa1-1 | 110 | 31 | 55 | 24 | 0.890ns | 0.640 |
| 7 | HKI1128Q × A619 lpa1-1 | 104 | 28 | 52 | 24 | 0.308ns | 0.857 |
A1A1 LPA1/LPA1, A1a1a LPA1/lpa1-1, a1a1 lpa1-1/lpa1-1, ns not significant
Parents and individual segregants in all the F2 populations were phenotyped for kernel phytic acid phosphorus (PAP) and inorganic phosphorus (iP) content (Tables 6 and 7). The effect of lpa1-1 allele is prominent that the recessive homozygotes showed significantly lesser PAP and higher iP as compared to the dominant homozygotes and the heterozygotes. This scenario was repeatably observed across all the seven populations. Raboy (2009) has reported that use of lpa1-1 mutation leads to the reduction in PAP followed by corresponding increase in the iP. In this study, there was no significant difference observed between the dominant homozygotes and the heterozygotes, indicating the complete dominance of the trait. Raboy et al. (2000) reported that recessive allele of lpa1-1confered low phytic acid in the maize grain. Across the seven populations used in this study, ~ 25 to 36% (mean 31.2%) reduction in PAP was observed in the recessive homozygotes (lpa1/lpa1), when compared to the dominant homozygotes (LPA1/LPA1). Though there is a reduction in the PAP in the recessive lpa1-1 genotypes, the extent of reduction is relatively low as compared to the earlier reports (Raboy et al. 2000, Raboy 2001, 2002; Shi et al. 2007; Naidoo et al. 2012). About, 61 to 335% (mean 164.8%) of iP has increased in the lpa1/lpa1 genotypes as compared to LPA1/LPA1 genotypes. This has clearly shown the increase in iP content while the reduction in the phytic acid by the recessive lpa1-1 allele. The percentage of PAP to total phosphorus has also reduced to 70% in the homozygous recessive genotypes compared to the dominant homozygotes (87%) (Table 7).
Table 6.
Kernel phytic acid and inorganic phosphorus among wild type and mutant parents
| S. no. | Name of the genotype | Genotype | PAP (mg/g) | iP (mg/g) | TP (mg/g) | PAP/TP (%) |
|---|---|---|---|---|---|---|
| Wild type | ||||||
| 1 | HKI161PV | A1A1/A2A2 | 2.58 | 0.31 | 2.89 | 89.27 |
| 2 | HKI163PV | A1A1/A2A2 | 2.49 | 0.26 | 2.75 | 90.55 |
| 3 | HKI193-1PV | A1A1/A2A2 | 2.40 | 0.31 | 2.71 | 88.56 |
| 4 | HKI193-2PV | A1A1/A2A2 | 2.63 | 0.21 | 2.84 | 92.61 |
| 5 | HKI323Q | A1A1/A2A2 | 2.69 | 0.43 | 3.12 | 86.22 |
| 6 | HKI1105Q | A1A1/A2A2 | 2.71 | 0.25 | 2.96 | 91.55 |
| 7 | HKI1128Q | A1A1/A2A2 | 2.53 | 0.28 | 2.81 | 90.04 |
| Mutant | ||||||
| 1 | A619 lpa1-1 | a1a1/A2A2 | 1.79 | 0.85 | 2.64 | 67.80 |
| 2 | A632 lpa1-1 | a1a1/A2A2 | 1.81 | 0.91 | 2.72 | 66.54 |
| 3 | A619 lpa2-1 | a2a2/A1A1 | 1.83 | 0.62 | 2.45 | 74.69 |
PAP phytic acid phosphorous, iP inorganic phosphorous, TP total phosphorous, A1A1 LPA1/LPA1, A2A2 LPA2/LPA2, a1a1 lpa1-1/1lpa1-1, a2a2 lpa2-1/1lpa2-1
Table 7.
Kernel phytic acid and inorganic phosphorus in the different genotypic classes of each lpa1-1 F2 population
| S. no | Cross | Genotype | PAP (mg/g) | iP (mg/g) | TP (mg/g) | PAP/TP (%) | % Reduction in PAP (a1a1vs A1A1) | % Increase in iP (a1a1vs A1A1) |
|---|---|---|---|---|---|---|---|---|
| 1 | HKI161PV × A619 lpa1-1 | A1A1 | 2.53 | 0.29 | 2.82 | 89.7 | 32.0 | 151.7 |
| a1a1 | 1.72* | 0.73* | 2.45 | 70.5 | ||||
| A1a1 | 2.42 | 0.27 | 2.69 | 88.0 | ||||
| 2 | HKI163PV × A632 lpa1-1 | A1A1 | 2.61 | 0.46 | 3.07 | 82.4 | 34.5 | 104.3 |
| a1a1 | 1.71* | 0.94 | 2.65* | 64.7 | ||||
| A1a1 | 2.55 | 0.42 | 2.97 | 82.4 | ||||
| 3 | HKI193-1PV × A619 lpa1-1 | A1A1 | 2.43 | 0.29 | 2.72 | 87.3 | 24.7 | 100.0 |
| a1a1 | 1.83* | 0.58* | 2.41 | 75.9 | ||||
| A1a1 | 2.32 | 0.28 | 2.60 | 87.3 | ||||
| 4 | HKI193-2PV × A619 lpa1-1 | A1A1 | 2.73 | 0.17 | 2.90 | 92.5 | 32.6 | 335.3 |
| a1a1 | 1.84* | 0.74* | 2.58 | 71.4 | ||||
| A1a1 | 2.63 | 0.18 | 2.81 | 92.1 | ||||
| 5 | HKI323Q × A632 lpa1-1 | A1A1 | 2.66 | 0.49 | 3.15 | 81.2 | 29.7 | 61.2 |
| a1a1 | 1.87* | 0.79* | 2.66 | 70.8 | ||||
| A1a1 | 2.53 | 0.47 | 3.00 | 81.5 | ||||
| 6 | HKI1105Q × A619 lpa1-1 | A1A1 | 2.68 | 0.21 | 2.89 | 90.6 | 36.2 | 309.5 |
| a1a1 | 1.71* | 0.86* | 2.57 | 67.2 | ||||
| A1a1 | 2.59 | 0.24 | 2.83 | 89.4 | ||||
| 7 | HKI1128Q × A619 lpa1-1 | A1A1 | 2.41 | 0.38 | 2.79 | 86.3 | 29.0 | 91.2 |
| a1a1 | 1.71* | 0.73* | 2.44 | 70.0 | ||||
| A1a1 | 2.39 | 0.47 | 2.86 | 83.5 | ||||
| 8 | Average across populations | A1A1 | 2.58 | 0.33 | 2.91 | 87.1 | 31.2 | 164.8 |
| a1a1 | 1.77* | 0.77* | 2.54 | 70.1 | ||||
| A1a1 | 2.49 | 0.33 | 2.82 | 86.3 |
PAP phytic acid phosphorous, iP inorganic phosphorous, TP total phosphorous, A1A1 LPA1/1LPA1-1, A1a1 LPA1/1lpa1-1, a1a1 lpa1-1/lpa1-1
*Different from A1A1 and a1a1 at 5% level of significance
lpa2-1
Shi et al. (2007) reported that the lpa2-1 mutation is due to genomic sequence rearrangement in the ZmITPK gene, which governs the synthesis of inositol-4-phosphate from inositol-3-phosphate. The exact causative polymorphism and the mutant gene sequence has not been reported,. Unlike lpa1-1, the lpa2-1 mutant sequence could not be obtained from the public domain. To identify the causative polymorphism between the wild type and the mutant allele, we sequenced both the lpa2-1 and LPA2-1 alleles. From that information, a set of eight overlapping primers were designed using the sequence of the wild type available from the public domain (Accession No. NM001112431.2) to cover the full-length gene i.e., LPA2-1 allele of 2263 bp (Table S2). The consensus nucleotide sequence of lpa2-1 gene of the mutant line, A619 lpa2-1 (GenBank accession number-MN917647) and the wild type genotypes viz. HKI161PV (GenBank accession number-MN917648), HKI163PV (GenBank accession number-MN917649), HKI193-1PV (GenBank accession number-MN917650), HKI193-2PV (GenBank accession number-MN917651), HKI323Q (GenBank accession number-MN917652), HKI1105Q (GenBank accession number-MN917653) and HKI1128Q (GenBank accession number-MN917654) were generated and subsequently submitted to NCBI to have the accession number.
Sequence alignment between the mutant and seven wild type allele along with the B73 genome sequence identified an A to G transition between the wild type and mutant (Figure S2) at 90 bp position from the transcription initiation site. The nucleotide polymorphism does not change amino acid and codon with both nucleotide codes for glutamine. Though Shi et al. (2003), has reported genomic sequence rearrangement in the gene that alters the phytic acid concentration, we could identify a SNP in the gene that clearly differentiated the mutant lpa2-1 allele and wild type allele. A set of primers which amplifies the region involving the SNP that differentiated the mutant lpa2-1 allele and wild type allele were designed (Table S3), that could specifically amplify the polymorphic region (Fig. 4). The sequence was analysed and the presence of restriction site within the polymorphism (A to G), for HindIII restriction enzyme was identified. HindIII is a hexacutter restriction enzyme and the target site is AAGCTT. Restriction digestion of the mutant PCR product using HindIII generated an intact amplicon of 459 bp due to the absence of restriction site. On the other hand, wild type genotypes yielded two fragments of size 169 bp and 290 bp, owing to the presence of the restriction site (Fig. 5). Use of restriction enzyme has exactly differentiated the mutant and the wild type allele, resulting in a Cleaved Amplified Polymorphic Sequence (CAPS) as marker to select for lpa2-1 allele. Development and use of CAPS markers for the target gene has also been reported for disease resistance and nutritional quality in maize and other crops (Udoh et al. 2017). Based on the polymorphism obtained among the mutant and wild type, the CAPS marker developed in the study was selected for further genotyping the segregating populations (Fig. 6). Genotyping of the seven segregating populations with the CAPS marker revealed the marker segregation as per the Mendelian fashion (1: 2: 1) (AA: Aa: aa) (Table 8).
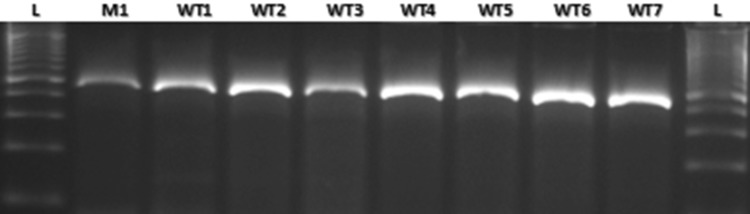
Fig. 4.
Amplification among the wild type and mutant allele using the primers designed for polymorphic region of lpa2-1. M A619 lpa2-1, WT1 HKI161PV, WT2 HKI163PV, WT3 HKI193-1PV, WT4 HKI193-2PV, WT5 HKI323Q, WT6 HKI1105Q, WT7 HKI1128Q, L 100 bp ladder
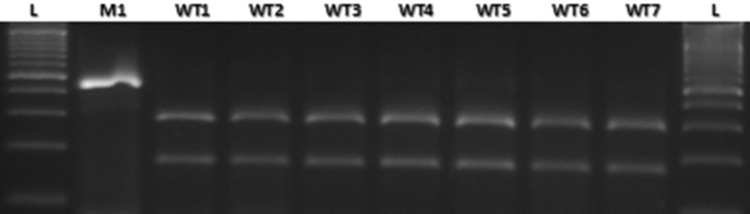
Fig. 5.
Amplification among the wild type and mutant allele using the CAPS marker. M A619 lpa2-1, WT1 HKI161PV, WT2 HKI163PV, WT3 HKI193-1PV, WT4 HKI193-2PV, WT5 HKI323Q, WT6 HKI1105Q, WT7 HKI1128Q, L 100 bp ladder
Fig. 6.
Segregation pattern of CAPS marker for lpa2-1 in HKI1128Q × A619lpa2-1. WT HKI1128Q, M A619 lpa2-1, 1–22 F2 individuals, L 100 bp DNA ladder
Table 8.
Segregation pattern of lpa2-1 gene using the CAPS marker developed in the study
| S. no. | Cross | Total no. of F2s genotyped | No. of genotypes with genotypic class | χ2 | P value | ||
|---|---|---|---|---|---|---|---|
| A2A2 | A2a2 | a2a2 | |||||
| 1 | HKI161PV × A619 lpa2-1 | 105 | 26 | 58 | 21 | 1.629ns | 0.443 |
| 2 | HKI163PV × A619 lpa2-1 | 98 | 28 | 51 | 19 | 1.816ns | 0.403 |
| 3 | HKI193-1PV × A619 lpa2-1 | 104 | 30 | 53 | 21 | 1.596ns | 0.450 |
| 4 | HKI193-2PV × A619 lpa2-1 | 107 | 24 | 54 | 29 | 0.477ns | 0.788 |
| 5 | HKI323Q × A619 lpa2-1 | 98 | 25 | 54 | 19 | 1.755ns | 0.416 |
| 6 | HKI1105Q × A619 lpa2-1 | 109 | 27 | 57 | 25 | 0.303ns | 0.860 |
| 7 | HKI1128Q × A619 lpa2-1 | 97 | 24 | 52 | 21 | 0.691 ns | 0.708 |
A2A2 LPA2/LPA2, A2a2 LPA2/lpa2-1, a2a2 lpa2-1/lpa2-1, ns not significant
Individual segregants in all the F2 populations were phenotyped for kernel phytic acid and inorganic phosphorus content. The same trend observed in lpa1-1 allele also observed in lpa2-1 in the case of inverse relationship between phytic acid and the inorganic phosphorous. Across the seven populations, ~ 25 to 30% (mean 26.9%) reduction in phytic acid was observed in the recessive homozygotes (lpa2/lpa2), compared to the dominant homozygotes (LPA2/LPA2). Shi et al. (2003) also reported a similar level of 28–30% reduction in phytic acid in the lpa2 mutants of maize. Though the reduction in the phytic acid was observed in the recessive lpa2-1 genotypes developed in the present study, the extent of reduction is relatively low compared to the earlier reports (Raboy et al. 2000, Raboy 2009). About, 96 to 258% (mean 179.8%) of inorganic phosphorus has increased in the lpa2/lpa2 genotypes as compared to LPA2/LPA2 genotypes. This clearly showed the increase in inorganic phosphorus content with the reduction in the phytic acid by recessive lpa2-1 allele. The percentage of phytic acid to total phosphorus was also reduced to 78% in the homozygous recessive genotypes compared to the dominant homozygotes (93%) (Table 9).
Table 9.
Kernel phytic and inorganic phosphorus in the different genotypic classes of each F2 population
| S. no. | Cross | Genotype | PAP (mg/g) | iP (mg/g) | TP (mg/g) | PAP/TP (%) | Reduction in PAP (%) | Increase in iP (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | HKI161PV × A619 lpa2-1 | A2A2 | 2.46 | 0.24 | 2.70 | 91.0 | 27.4 | 225.1 |
| a2a2 | 1.78* | 0.79* | 2.57 | 69.4 | ||||
| A2a2 | 2.43 | 0.32 | 2.75 | 88.4 | ||||
| 2 | HKI163PV × A619 lpa2-1 | A2A2 | 2.41 | 0.19 | 2.60 | 92.7 | 25.9 | 257.9 |
| a2a2 | 1.78* | 0.68* | 2.46 | 72.4 | ||||
| A2a2 | 2.34 | 0.20 | 2.54 | 92.1 | ||||
| 3 | HKI193-1PV × A619 lpa2-1 | A2A2 | 2.42 | 0.23 | 2.65 | 91.5 | 25.3 | 112.4 |
| a2a2 | 1.81* | 0.48* | 2.29 | 79.0 | ||||
| A2a2 | 2.33 | 0.23 | 2.55 | 91.1 | ||||
| 4 | HKI193-2PV × A619 lpa2-1 | A2A2 | 2.53 | 0.17 | 2.70 | 93.9 | 24.5 | 180.3 |
| a2a2 | 1.91* | 0.46* | 2.37 | 80.4 | ||||
| A2a2 | 2.45 | 0.18 | 2.63 | 93.2 | ||||
| 5 | HKI323Q × A619 lpa2-1 | A2A2 | 2.48 | 0.13 | 2.61 | 95.0 | 26.2 | 221.7 |
| a2a2 | 1.83* | 0.42* | 2.25 | 81.3 | ||||
| A2a2 | 2.43 | 0.23 | 2.66 | 91.3 | ||||
| 6 | HKI1105Q × A619 lpa2-1 | A2A2 | 2.68 | 0.18 | 2.87 | 93.6 | 29.6 | 165.3 |
| a2a2 | 1.89* | 0.49* | 2.38 | 79.5 | ||||
| A2a2 | 2.60 | 0.18 | 2.78 | 93.7 | ||||
| 7 | HKI1128Q × A619 lpa2-1 | A2A2 | 2.69 | 0.20 | 2.89 | 93.1 | 29.2 | 95.7 |
| a2a2 | 1.91* | 0.39* | 2.29 | 83.1 | ||||
| A2a2 | 2.69 | 0.21 | 2.90 | 92.9 | ||||
| 8 | Average across populations | A2A2 | 2.53 | 0.19 | 2.72 | 93.0 | 26.9 | 179.8 |
| a2a2 | 1.85* | 0.53* | 2.38 | 77.9 | ||||
| A2a2 | 2.47 | 0.22 | 2.69 | 91.8 |
PAP phytic acid phosphorus, iP inorganic phosphorus, TP total phosphorus, A2A2 LPA2/LPA2, A2a2 LPA2/lpa2-1, a2a2 lpa2-1/lpa2-1
*Different from A2A2 and a2a2 at 5% level of significance
The CAPS marker for lpa2-1 was able to differentiate the three possible genotypic classes (AA: Aa: aa). Their Mendelian segregation pattern and the phenotypic effects of the lpa2-1 allele in reducing the phytic acid with increase in inorganic phosphorus across all the seven segregating populations shows the robustness, reliability and feasibility of the marker developed in the study. Thus, the CAPS marker developed for lpa2-1 would offer tremendous assistance to breed for low phytate maize through MAS.
Impact of low phytic acid on the bioavailability of iron and zinc
Maize inbreds possessing 40 ppm of kernel Fe is available (Pandey et al. 2015; Prasanna et al. 2011) but inbreds with target level of 60 ppm is quite uncommon (Gupta et al. 2015b). Similarly, inbreds with ~ 30 ppm of kernel Zn is available (Pandey et al. 2015; Prasanna et al. 2011), but inbreds with the target level of 37 ppm to meet recommended daily allowance (RDA) is also not normally available (Gupta et al. 2015b). The polygenic nature of the trait, high influence of G × E and minor effects QTLs limit the development of inbreds with target level for Fe and Zn (Gupta et al. 2015a). However, considering the bioavailability of 5% for Fe and 25% for Zn, only 3 ppm and 9.25 ppm of Fe and Zn. respectively is available in human gut. On the other hand, the low phytic acid maize developed in the present study offers potential scope to meet the RDA due to its profound effects on reduction of phytic acids (Raboy et al. 2000; Raboy 2009). The average reduction of phytic acid observed in the present study was 30%, which will lead to increase in bioavailability of the Fe by 1.5% (30% of 5%) and Zn by 7.5% (30% of 25%). Therefore, use of the low phytate maize genotypes developed in the study will offer higher bioavailability of kernel Fe (6.5%) and Zn (32.5%). With the increased bioavailability of Fe, 2.6 ppm of kernel Fe (6.5% of 40 ppm) could be achieved even with the average level available in the germplasm. To achieve the target of 3 ppm, breeding for 45 ppm alone would be sufficient which can easily be achieved through breeding approaches. Most importantly, the target of Zn can easily be achieved by use of the low phytate maize genotypes developed in the study. With the average of 30 ppm and increased bioavailability of 32.5%, around 9.75 ppm of Zn (32.5% of 30 ppm) would be available in the diet, which is similar to the target level. Thus the low phytate maize genotypes developed in the study can be used for the development of low phytate maize hybrids which would offer potential scope on bio-fortification of kernel Fe and Zn in maize.
Conclusion
Maize serves as an important source of energy as food but possesses high concentration of phytic acid. Thus, reduction of the phytic acid in maize through genetic manipulation holds immense promise for enhancing the bioavailable Fe and Zn. This study successfully developed and validated gene-based markers in seven segregating populations each for lpa1-1 and lpa2-1 genes which offers tremendous benefit and assistance in accelerated development of low phytate maize genotypes. The low phytate maize genotypes developed across the populations in the present study would serve as a rich genetic resource in the future breeding programmes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
First author sincerely acknowledges Indian Council of Agricultural Research (ICAR) for providing Junior Research Fellowship (JRF) to pursue his M.Sc. programme. Sincere thanks are due to Dr. Victor Raboy, USDA for providing the lpa mutants used in the study.
Author contributions
Conduct of the experiments: KPA; Design of experiments: VM and FH; Generation of segregating populations: RUZ and VM; Field evaluation: VB, GC, SKJ and VM; Genotyping and sequence analysis: KPA, RC and SD; Biochemical analysis: KPA, SV and DKY; Statistical analysis: FH and VM; Drafting and editing of manuscript: KPA, VM and FH.
Compliance with ethical standards
Conflict of interest
Author declares that no conflict of interest exits.
Contributor Information
Krishnan P. Abhijith, Email: abhijithkpgen@gmail.com
Vignesh Muthusamy, Email: pmvignesh@yahoo.co.in, Email: vignesh@iari.res.in.
Rashmi Chhabra, Email: reshu0428@rediffmail.com.
Sweta Dosad, Email: swetadosad@gmail.com.
Vinay Bhatt, Email: vinaybhatt024@gmail.com.
Gulab Chand, Email: gulab.biotech@yahoo.com.
Sunil K. Jaiswal, Email: jaiswal1982@gmail.com
Rajkumar U. Zunjare, Email: raj_gpb@yahoo.com, Email: rajkumaruz@iari.res.in
Sujata Vasudev, Email: sujatavasudev@gmail.com, Email: sujatavasudev@iari.res.in.
Devendra K. Yadava, Email: dkygenet@gmail.com, Email: dkyadav@iari.res.in
Firoz Hossain, Email: fh_gpb@yahoo.com, Email: fhossain@iari.res.in.
References
- Adams C, Raboy V, Krebs N, Westcott J, Lei S, Hambidge M. The effect of low-phytic acid corn mutants on zinc absorption. FASEB J. 2000;14:510. [Google Scholar]
- AICRP-Maize progress Report (2018) ICAR-Indian Institute of Maize Research, PAU Campus, Ludhiana, pp 1–10
- Bouis H. Reducing mineral and vitamin deficiencies through biofortification: progress under HarvestPlus. Hidden Hunger. 2018;118:112–122. doi: 10.1159/000484342. [DOI] [PubMed] [Google Scholar]
- Das AK, Jaiswal SK, Muthusamy V, Zunjare RU, Chauhan HS, Chand G, Saha S, Hossain F. Molecular diversity and genetic variability of kernel tocopherols among maize inbreds possessing favourable haplotypes of γ-tocopherol methyl transferase (ZmVTE4) J Plant BiochemBiotechnol. 2019 doi: 10.1007/s13562-018-0470-x. [DOI] [Google Scholar]
- De Steur H, Blancquaert D, Strobbe S, Lambert W, Gellynck X, Van Der Straeten D. Status and market potential of transgenic biofortified crops. Nat Biotechnol. 2015;33:25–29. doi: 10.1038/nbt.3110. [DOI] [PubMed] [Google Scholar]
- Gupta HS, Hossain F, Muthusamy V. Biofortification of maize: an Indian perspective. Indian J Genet Plant Breed. 2015;75:1–22. doi: 10.5958/0975-6906.2015.00001.2. [DOI] [Google Scholar]
- Gupta HS, Hossain F, Nepolean T, Vignesh M, Mallikarjuna MG. Understanding genetic and molecular bases of Fe and Zn accumulation towards development of micronutrient-enriched maize. In: Rakshit A, editor. Nutrient use efficiency: from basics to advances. New York: Springer; 2015. pp. 255–282. [Google Scholar]
- Gupta HS, Hossain F, Muthusamy V, Zunjare RU. Marker-assisted breeding for enrichment of provitamin A in maize. In: Qureshi AMI, editor. Quality breeding in field crops. Basel: Springer Basel AG; 2019. pp. 139–157. [Google Scholar]
- Hall T. BioEdit: an important software for molecular biology. GERF Bull Biosci. 2011;2:60–61. [Google Scholar]
- Hossain F, Muthusamy V, Pandey N, Vishwakarma AK, Baveja A, Zunjare RU, Thirunavukkarasu N, Saha S, Manjaiah KM, Prasanna BM, Gupta HS. Marker-assisted introgression of opaque2 allele for rapid conversion of elite hybrids into quality protein maize. J Genet. 2018;97:287–298. doi: 10.1007/s12041-018-0914-z. [DOI] [PubMed] [Google Scholar]
- Hossain F, Sarika K, Muthusamy V, Zunjare RU, Gupta HS. Quality protein maize for nutritional security. In: Qureshi AMI, editor. Quality breeding in field crops. Basel: Springer Basel AG; 2019. [Google Scholar]
- Jorquera M, Martinez OZ, Maruyama F, Marschner P, de la Luz MM. Current and future biotechnological applications of bacterial phytases and phytase-producing bacteria. Microbes Environ. 2008;23:182–191. doi: 10.1264/jsme2.23.182. [DOI] [PubMed] [Google Scholar]
- Liu J, Huang S, Sun M, Liu S, Liu Y, Wang W, Hua W. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods. 2012;8:34. doi: 10.1186/1746-4811-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz AJ, Scott MP, Lamkey KR. Quantitative determination of phytate and inorganic phosphorus for maize breeding. Crop Sci. 2007;47:600–604. doi: 10.2135/cropsci2006.03.0177. [DOI] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy V, Hossain F, Thirunavukkarasu N, Choudhary M, Saha S, Bhat JS, Prasanna BM, Gupta HS. Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE. 2014;9:e113583. doi: 10.1371/journal.pone.0113583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo R, Watson GMF, Derera J, Tongoona P, Laing MD. Marker-assisted selection for low phytic acid (lpa1-1) with single nucleotide polymorphism marker and amplified fragment length polymorphisms for background selection in a maize backcross breeding programme. Mol Breed. 2012;30:1207–1217. doi: 10.1007/s11032-012-9709-8. [DOI] [Google Scholar]
- Pandey N, Hossain F, Kumar K, Vishwakarma AK, Muthusamy V, Manjaiah KM, Agrawal PK, Guleria SK, Reddy SS, Thirunavukkarasu N, Gupta HS. Microsatellite marker-based genetic diversity among quality protein maize (QPM) inbreds differing for kernel iron and zinc. Mol Plant Breed. 2015;6:1–10. doi: 10.5376/mpb.2015.06.0003. [DOI] [Google Scholar]
- Pilu R, Panzeri D, Gavazzi G, Rasmussen SK, Consonni G, Nielsen E. Phenotypic, genetic and molecular characterization of a maize low phytic acid mutant (lpa241) Theor Appl Genet. 2003;107:980–987. doi: 10.1007/s00122-003-1316-y. [DOI] [PubMed] [Google Scholar]
- Prasanna BM, Mazumdar S, Chakraborti M, Hossain F, Manjaiah KM, Agrawal PK, Gupta HS. Genetic variability and genotype × environment interactions for kernel iron and zinc concentrations in maize (Zea mays) genotypes. Indian J Agric Sci. 2011;81:704–711. [Google Scholar]
- Raboy V. Genetics and breeding of seed phosphorus and phytic acid. J Plant Physiol. 2001;158:489–497. doi: 10.1078/0176-1617-00361. [DOI] [Google Scholar]
- Raboy V. Progress in breeding low phytate crops. J Nutr. 2002;132:503–505. doi: 10.1093/jn/132.3.503S. [DOI] [PubMed] [Google Scholar]
- Raboy V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009;177:281–296. doi: 10.1016/j.plantsci.2009.06.012. [DOI] [Google Scholar]
- Raboy V, Dickinson DB. Effect of phosphorus and zinc nutrition on soybean seed phytic acid and zinc. Plant Physiol. 1984;75:1094–1098. doi: 10.1104/pp.75.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V, Gerbasi PF, Young KA, Stoneberg SD, Pickett SG, Bauman AT, Murthy PPN, Sheridan WF, Ertl DS. Origin and seed phenotype of maize low phytic acid 1–1 and low phytic acid 2–1. Plant Physiol. 2000;124:355–368. doi: 10.1104/pp.124.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Bioinformatics methods and protocols. Totowa, NJ: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sarika K, Hossain F, Muthusamy V, Zunjare RU, Baveja A, Goswami R, Gupta HS. Marker-assisted pyramiding of opaque2 and novel opaque16 genes for further enrichment of lysine and tryptophan in sub-tropical maize. Plant Sci. 2018;272:142–152. doi: 10.1016/j.plantsci.2018.04.014. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Wu Y, Hazebroek J, Meeley RB, Ertl DS. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 2003;131:507–515. doi: 10.1104/pp.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang H, Hazebroek J, Ertl DS, Harp T. The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 2005;42:708–719. doi: 10.1111/j.1365-313X.2005.02412.x. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Schellin K, Li B, Faller M, Stoop JM, Glassman K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat Biotechnol. 2007;25:930–937. doi: 10.1038/nbt1322. [DOI] [PubMed] [Google Scholar]
- Singh BD, Singh AK. Marker-assisted plant breeding: principles and practices. New Delhi, India: Springer; 2015. [Google Scholar]
- Sureshkumar S, Tamilkumar P, Senthil N, Nagarajan P, Thangavelu AU, Raveendran M, Vellaikumar S, Ganesan KN, Balagopal R, Vijayalakshmi G, Shobana V. Marker assisted selection of low phytic acid trait in maize (Zea mays L.) Hereditas. 2014;151:20–27. doi: 10.1111/j.1601-5223.2013.00030.x. [DOI] [PubMed] [Google Scholar]
- Udoh L, Gedil M, Parkes EY, Kulakow P, Adesoye A, Nwuba C, Rabbi IY. Candidate gene sequencing and validation of SNP markers linked to carotenoid content in cassava (Manihot esculenta Crantz) Mol Breed. 2017;37:1–12. doi: 10.1007/s11032-017-0718-5. [DOI] [Google Scholar]
- Yadava DK, Choudhury PR, Hossain F, Kumar D. Biofortified varieties: sustainable way to alleviate malnutrition. New Delhi: Indian Council of Agricultural Research; 2017. pp. 1–32. [Google Scholar]
- Zhou JR, Erdman JW. Phytic acid in health and disease. Food Sci Nutr. 1995;35:495–508. doi: 10.1080/10408399509527712. [DOI] [PubMed] [Google Scholar]
- Zunjare RU, Hossain F, Muthusamy V, Baveja A, Chauhan HS, Bhat JS, Thirunavukkarasu N, Saha S, Gupta HS. Development of biofortified maize hybrids through marker-assisted stacking of β-carotene hydroxylase, lycopene-ε-cyclase and opaque2 genes. Front Plant Sci. 2018;9:178. doi: 10.3389/fpls.2018.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.