Abstract
During the course of survey, an incidence of 7.14–90% of apple mosaic disease (AMD) was recorded in apple orchards in Jammu and Kashmir among various commercially grown cultivars. The maximum incidence of mosaic disease was observed in cultivar Golden Delicious. In addition to mosaic, symptoms of chlorosis, necrosis and ring spots were also observed. In the present study association of Apple necrotic mosaic virus (ApNMV) was confirmed by RT-PCR and sequencing of whole coat protein gene in samples tested negative for Apple mosaic virus (ApMV) in DAS-ELISA. Out of 18 samples tested in RT-PCR, ten were found positive for ApNMV. Out of ten ApNMV positive samples, amplicon of 680 bp of samples representing five cultivars were sequenced and sequence analysis showed 89–91% sequence identity with ApNMV. The phylogenetic analysis grouped Indian isolates into two sub-clusters under one major cluster (ApNMV group). The sub-cluster-I, included ApNMV isolates from cultivars, Oregon Spur, Red Delicious and Fuji Aztec along with Chinese and Korean isolates. Sub-cluster-II included ApNMV isolates associated with Golden Delicious and Royal Delicious. The comparison of coat protein gene-based sequence identity matrix showed maximum and minimum similarity of 89–99% with ApNMV isolates from China. It also showed maximum identity with PNRSV (61.6%) and ApMV (52.8%) under subgroup 3 of genus Illarvirus. Our study indicates that the ApNMV is commonly associated with AMD in India and may be a major cause of the mosaic disease in apple cultivars. To the best of our knowledge, this is the first report of the association of ApNMV with apple mosaic disease from India.
Keywords: Apple, Mosaic, Illarvirus, Sequencing, Coat protein
Introduction
Apple (Malus domestica Borkh.) is commercially most important, remunerative and widely grown crop in temperate regions of the world. Fruits are valued for their attractiveness, taste, nutritional quality and as a source of earning foreign exchange (Ferree and Warrington 2003). In India, it is mainly cultivated in North Western Himalayan region with Jammu and Kashmir as the leading apple-growing state contributing 65% of the apple production followed by Himachal Pradesh and Uttarakhand (Muneer et al. 2017). Many biotic factors such as pathogens and pests are responsible for losses both quantitatively and qualitatively. Viral diseases are one of the important limiting factors affecting apple productivity. Viruses pass to the successive generations through propagating material and cause decline in health of trees (Sajad et al. 2019). There are at least 12 viruses and virus like pathogens, which affect apple trees (Nisar 2013) and majority of these viruses are latent in nature, producing no visible symptoms (Sajad et al. 2018). Among viral diseases, apple mosaic disease (AMD) is a severe threat to the development of the apple industry (Li et al. 2002). It has been shown to decrease the net photosynthetic rate of infected leaves from 2.93–45.83% (Chai et al. 2017) and decrease fruit yield by 30–50% (Cembali et al. 2003; Tombisana et al. 2009). The mosaic infected apple leaves show bright pale-yellow chlorotic spots and mosaic patterns (Petrzik and Lenz 2011). Trees with high disease severity lack vigor (Nemeth 1986); symptoms do not appear on branches or fruits (Grimova et al. 2016). Association of Apple necrotic mosaic virus (ApNMV) (Noda et al. 2017), Prunus necrotic ring spot virus (PNRSV) and Cucumber mosaic virus (CMV) has been associated with mosaic disease in addition to Apple mosaic virus (ApMV) (Hu et al. 2016a,b). Association of ApNMV with the mosaic disease of apple was shown recently in China (Noda et al. 2017), Korea (Cho et al. 2017) and crabapple in China (Hu et al. 2019). The ApNMV belongs to the genus Ilarvirus, family Bromoviridae. The genome consists of single-stranded positive-sense RNA molecules (RNA1, RNA2, and RNA3) and an encapsidated subgenomic RNA4. RNA1 and RNA2 each encode single replication-associated proteins. RNA3 encodes a movement protein (MP) and coat protein (CP), whereas RNA4 is a subgenomic RNA that functions as an mRNA for CP (Xing et al. 2018). In this study, the association of Apple necrotic mosaic virus with mosaic disease of apple was studied to know its occurrence in different varieties cultivated in Jammu and Kashmir. To the best of our knowledge, this is the first attempt to identify the association of Apple necrotic mosaic virus with apple mosaic disease in apple-growing regions of India.
Materials and methods
Incidence, collection of symptomatic and asymptomatic samples
The survey was conducted at different locations in five apple-growing districts of Jammu and Kashmir (Table 1). Plants were randomly selected for calculating the incidence of mosaic disease depending upon the cultivar present in orchard. Total twenty leaf samples showing mosaic and necrosis along with asymptomatic leaves were collected from cultivars, viz., Red Delicious, Golden Delicious, Red Gold, Royal Delicious, Fuji Aztec, Oregon Spur and Gala Mast. Leaf samples were collected randomly from different parts of the same tree during the months of April and August in the year 2018 and 2019 and used for virus detection. The collected samples were used either fresh or stored at − 80 °C for further analysis.
Table 1.
Mosaic affected leaf samples collection sites, symptoms, disease incidence and presence and absence of ApMV and ApNMV on different apple varieties
| Site of collection | Longitude/latitude | Cultivar | Symptoms | Incidence of mosaic (%) on cultivars | Presence of ApMV (DAS-ELISA)a | Presence of ApMV (RT-PCR | Presence of ApNMV (RT-PCR |
|---|---|---|---|---|---|---|---|
| Shopian | 33.7594° N/ 74.8039° E | Red Delicious | Mosaic and necrosis | 16.6 | − | − | − |
| Anantnag | 33.7050° N/ 75.2479° E | Mosaic | + | + | − | ||
| Pulwama | 33.9819° N/ 75.0144° E | Mosaic and necrosis | − | − | + | ||
| Srinagar | 34.1255° N/ 74.9443° E | Mosaic and necrosis | − | − | + | ||
| Budgam | 33.9349° N/ 74.6400° E | Mosaic and necrosis | − | − | + | ||
| Shopian | 33.7594° N/ 74.8039° E | Golden Delicious | Mosaic | 90 | + | + | − |
| Anantnag | 33.7411° N/ 75.1297° E | Mosaic and necrosis | − | − | + | ||
| Pulwama | 33.8202° N/ 74.8158° E | Mosaic | − | − | − | ||
| Srinagar | 34.1255° N/ 74.9443° E | Mosaic and necrosis | − | − | + | ||
| Budgam | 33.9451° N/ 74.7966° E | Mosaic and necrosis | − | − | + | ||
| Pulwama | 33.9819° N/ 75.0144° E | Royal Delicious | Mosaic and necrosis | 13.3 | − | − | + |
| Shopian | 33.7651° N/ 74.8441° E | Mosaic and necrosis | − | − | + | ||
| Srinagar | 33.9843° N/ 74.7991° E | Fuji Aztec | Mosaic | 7.60 | − | − | + |
| Pulwama | 33.8600° N/ 74.8289° E | Red Gold | Mosaic and necrosis | 13.25 | − | − | − |
| Pulwama | 33.8651° N/ 74.8756° E | Gala Mast | Mosaic and ring spots | 7.14 | − | − | − |
| Budgam | 33.9643° N/ 74.5082° E | Mosaic and ring spots | − | − | − | ||
| Srinagar | 34.1255° N/ 74.9443° E | Oregon spur | Mosaic and necrosis | 15.0 | − | − | + |
| Srinagar | 34.1255° N/ 74.9443° E | Mosaic and necrosis | − | − | − |
( +) positive for virus, (−) negative for virus, aO.D values for positive − 1.18 and above, negative − 0.38 and less
Double antibody sandwich-enzyme linked immunosorbent assay (DAS-ELISA)
The samples were tested serologically for Apple mosaic virus using DAS ELISA kit (BIOREBA Switzerland). The results were assessed by measuring the absorbance at 405 nm wavelength using ELISA reader (Thermo Scientific). The samples were considered infected when the ELISA readings were twice the mean absorbance reading of negative control.
Total RNA isolation and c-DNA synthesis
For detection of ApNMV and ApMV, total RNA was extracted from both symptomatic and asymptomatic leaves using QIAGEN RNeasy® Plant minikit (Qiagen Germany) as per manufactures instructions. The total RNA was quantified using Nanodrop (Thermo Scientific). The first strand of c-DNA was synthesized using M-MuLV reverse transcriptase c-DNA synthesis kit (NEB, New England Biolabs). The reaction mixture was spinned and incubated at 25 °C for 5 min, 42 °C for 59 min, and 65 °C for 5 min. Later cDNA was used for PCR amplification and the remaining quantity was stored at − 20 °C for further use.
Reverse transcriptase-PCR (RT-PCR)
The RT-PCR was carried out in 20 μl volume containing 12.5 μl of nuclease-free water, 2 μl of cDNA, 2 μl of 10 × standard taq buffer, 0.5 μl of 25 mM MgCl2, 1 μl of 2 mM dNTP mix, 0.4 units of Taq polymerase (NEB Applied Science, New England Biolabs), 1 μl each of (10 μM) specific forward and reverse primers of CP for ApNMV and ApMV with primer sequence F(5'-CTTGCGTGCAATCGATATGG-3'), R(5'-TCATCTCAACCTAGACATCC-3') (Noda et al. 2017) and F(5'-CTCAAGCGAACCCGAATAAGGGTAAGAA-3'), R(5'-TCGTCGATAAGTAGAACATTCGTCGGTATTGTC-3') (Grimova et al. 2016), respectively. The reaction was performed in a thermo cycler (Eppendorf Mastercycler gradient, USA) using same program for both the viruses, except the annealing temperatures. The program was set up for 35 cycle with denaturation at 94 °C for 30 s, annealing at 46 °C (ApNMV) and 53 °C (ApMV) for 40 s followed by extension at 72 °C for 30 s along with a final elongation step at 72 °C for 10 min. The PCR products were electrophoresed in 1% agarose gel in 0.5X TAE buffer (pH 8.0) at 80 V. For estimating the amplicon size, 1 kb DNA molecular ladder was used (NEB Applied Science, New England Biolabs) and electrophoresis was done for 1 h. The fragments were observed under UV lamp in gel-documentation (Bio Rad, Gel Doc XR system 170 – 8170).
Cloning and sequence analysis
The amplified DNA on the electrophoresis gel was eluted using mini elution gel extraction kit (Promega Wizard® SV Gel and PCR Clean-Up System, USA) as per manufactures instructions. The purified specific RT-PCR products were cloned in pGEM-T cloning vector (Promega, USA). The recombinant plasmid was transformed to competent DH5 α bacterial cell for the multiplication of the insert(s). The positive clones were confirmed through blue-white selection, colony PCR and restriction digestion. Positive clones of both the viruses were sequenced using two reactions in both directions from FIRST BASE laboratories Selangor, Malaysia. Sequences were assembled to generate the consensus sequence for phylogeny and sequence identity matrix using BioEdit Sequence Alignment Editor Version 7.0.9.0. The edited nucleotide sequence and amino acid sequences were confirmed through basic local alignment search tool (BLAST) analysis (https://www.ncbi.nlm.nih.gov/). The dendrogram was constructed using the MEGA7 (Molecular Evolutionary Genomics Analysis Version 7) software (Kumar et al. 2014). The ApMV and ApNMV sequences were submitted to GenBank.
Results
Disease incidence and symptomatology
During the course of the survey, the mosaic disease was recorded in the orchards in all varieties at all the locations surveyed. Among the seven varieties surveyed, Golden Delicious was found to be highly susceptible with 90% disease incidence followed by Red Delicious (16.6%), Oregon Spur (15.0%), Royal Delicious (13.3%), Red Gold (13.25%), Fuji Aztec (7.60%) and Gala Mast (7.14%). During the survey, symptoms such as chlorosis, mosaic, necrotic spots and ring spots were observed on different cultivars of apple (Fig. 1). Mosaic symptoms ranged from small pale-yellow spots scattered across an entire leaf or part of a leaf along with necrosis on some cultivars to large contiguous, chlorotic spots covering an entire leaf. The mosaic incidence on different varieties along with symptoms observed during survey is presented in Table 1.
Fig. 1.
Symptoms of mosaic (a) and mosaic along with necrosis (b) and mosaic with ring spots (c) on leaves of apple trees
DAS-ELISA
The sample leaves with pale-yellow mosaic patterns were believed to be infected with ApMV, but serologically the presence of ApMV was revealed two samples, one each from cultivars Golden Delicious and Red Delicious. The rest of the samples from all tested varieties were negative for ApMV in DAS-ELISA (Table 1).
PCR amplification, sequencing and BLAST analysis
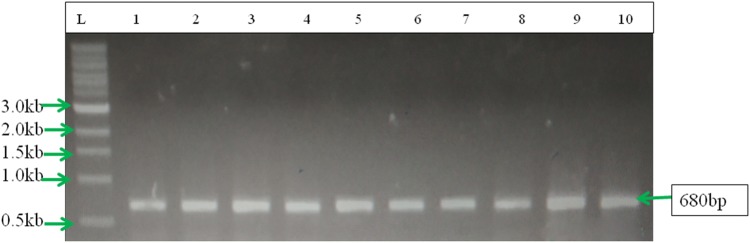
The ApMV negative samples of different apple varieties showing mosaic and other symptoms such as necrosis, ring spots, amplification of 650 bp was amplified from 10 samples of different cultivars (Fig. 2). Among the varities, out of 5 samples from Red Delicious and Golden Delicious, three were found positive for ApNMV, both the samples from varieties Royal Delicious and Oregon Spur and single sample from cultivar Fuji Aztec were found positive for ApNMV. The samples from varieties Gala Mast and Red Gold were found negative for both ApNMV and ApMV. The amplification of 550 bp for ApMV was observed in only two samples from varieties Red Delicious and Golden Delicious and no amplification was observed for ApNMV in both the samples. The presence and absence of ApMV and ApNMV on different apple cultivars is shown in Table 1. Five samples from five varieties, Red Delicious, Golden Delicious, Royal Delicious, Fuji Aztec and Oregon Spur infected with ApNMV were cloned and sequenced. The clones were confirmed through restriction digestion using EcoR1 enzyme and 650 bp for ApNMV were released after 3 h restriction digestion along with 3 kb vector (data not shown). The BLASTn result of all five sequences showed 89–91% nucleotide sequence identity and 90–100% query cover with the CP gene of the previously published ApNMV isolates (MG924901 and MG924900). Similarly, the sequences of ApMV showed 98% sequence similarity with published sequences of ApMV in GenBank (KY971019). The full coat protein gene sequences of all five isolates were deposited in GenBank and accession numbers (MN529261, MN529262, MN529263, MN627355 and MN627356) were received for all the submitted sequences.
Fig. 2.
PCR product (690 bp) amplified from CP region of Apple necrotic mosaic virus from different varieties of apple, L-1 kb ladder, 1: Red Delicious-3, 2: Red Delicious-4, 4: Golden Delicious-2, 5: Golden Delicious-4, 6: Royal Delicious-1, 7: Royal Delicious-2, 8: Fuji Aztec-1, 9: Oregon Spur-1, 10: Oregon Spur-2
Phylogenetic analysis
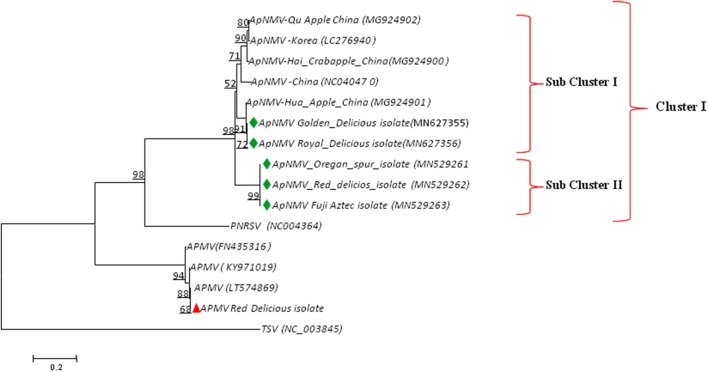
To understand the taxonomic position of ApNMV, phylogenetic analysis was carried out for our isolates along with three Chinese and one Korean ApNMV isolates using the neighbor-joining method. The phylogenetic analysis grouped the ApNMV isolates into one major cluster, which was further divided into sub-cluster-I and sub-cluster-II. The sub-cluster-I, included ApNMV isolates from cultivars, Golden Delicious and Royal Delicious along with Chinese and Korean isolates. Interestingly, sub-cluster-II included ApNMV isolates from Oregon Spur, Red Delicious and Fuji Aztec. In the present study the ApMV isolate from Golden Delicious variety in the present study formed separate cluster in phylogeny (Fig. 3). The Tobacco streak virus (TSV) was taken as out group in phylogenetic analysis, which formed separate clade in phylogenetic tree. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches.
Fig. 3.
Phylogenetic relationship of Apple necrotic mosaic virus using coat protein gene nucleotide sequence alignment through neighbor joining method at 1000 replications for each bootstrap value using the MEGA 7.0, symbols green and red in phylogeny represents our ApNMV and ApMV isolates. Tobacco streak virus (TSV) represents out-group in same genus
Relationship among ApNMV isolates and with other Illarviruses based on sequence identity matrix
Since little information is available on the homology between ApNMV and other illarviruses, sequence identity matrix was performed to confirm the sequence diversity at coat protein level. Comparison of CP gene sequence identity matrix between our ApNMV isolates with submitted isolates of ApNMV from China and Korea revealed maximum and minimum similarity of 89% (ApNMV-OS) and 99% (ApNMV-GD) with ApNMV-Hua China and ApNMV-Crabapple China isolates, respectively (Table 2 and Fig. 4). Two isolates (ApNMV-OS and ApNMV-GD) representing two subclusters in major cluster of phylogeny selected for comparison of CP gene sequence identity matrix with other Illarviruses revealed maximum identity of 61.6% and 59.9% with PNRSV, 52.8% and 52.5% with ApMV, respectively. The similarity with all other Illarviruses was below 35% (Table 3 and Fig. 5).
Table 2.
Percentage sequence similarity matrix of identified Apple necrotic mosaic virus from different varieties with submitted isolates from China and Korea
| Isolate/isolate | ApNMV-OS | ApNMV-RD | ApNMV-FA | ApNMV-GD | ApNMV-RLD | ApNMV-1 | ApNMV-2 | ApNMV-3 | ApNMV-4 | ApNMV-5 |
|---|---|---|---|---|---|---|---|---|---|---|
| ApNMV-OS | 100 | |||||||||
| ApNMV-RD | 98 | 100 | ||||||||
| ApNMV-FA | 100 | 98.1 | 100 | |||||||
| ApNMV-GD | 89.8 | 88.7 | 89.5 | 100 | ||||||
| ApNMV-RLD | 89.8 | 88.7 | 89.5 | 100 | 100 | |||||
| ApNMV-1 | 89.8 | 89.0 | 89.8 | 99.3 | 99.3 | 100 | ||||
| ApNMV-2 | 88.6 | 87.8 | 88.6 | 94.6 | 94.6 | 95.0 | 100 | |||
| ApNMV-3 | 88.3 | 87.8 | 88.3 | 93.4 | 93.4 | 93.7 | 97.7 | 100 | ||
| ApNMV-4 | 87.7 | 87.1 | 87.7 | 92.5 | 92.5 | 92.8 | 93.7 | 94.2 | 100 | |
| ApNMV-5 | 86.7 | 85.0 | 86.7 | 90.7 | 90.7 | 91.3 | 95.0 | 96.2 | 91.3 | 100 |
ApNMV-1 Apple necrotic mosaic virus Hua China (MG924901), ApNMV-2 Apple necrotic mosaic virus Hai_Crabapple_China (MG924900), ApNMV-3 Apple necrotic mosaic virus Qu_Apple China (MG924902), ApNMV-4 Apple necrotic mosaic virus China (NC040470), ApNMV-5 Apple necrotic mosaic virus Korea (LC276940), ApNMV-GD Apple necrotic mosaic virus Golden Delicious isolate, ApNMV-OS Apple necrotic mosaic virus Oregon spur isolate, ApNMV-RD Apple necrotic mosaic virus Red Delicious isolate, ApNMV-FA Apple necrotic mosaic virus Fuji Aztec isolate, ApNMV-RLD Apple necrotic mosaic virus Royal Delicious isolate
Fig. 4.
Two dimensional colour coded graphical representation of sequence identities based on coat protein between Indian ApNMV isolates from different varieties of apple with other ApNMV isolates from China and Korea. A scale with percent color code is presented
Table 3.
Percentage sequence similarity matrix of Apple necrotic mosaic virus with other illarviruses
| Illarviruses | ApNMV-GD | ApNMV-OS | TSV | TuAMV | ApMV | PDV | APLPV | PNRSV |
|---|---|---|---|---|---|---|---|---|
| ApNMV-GD | 100 | |||||||
| ApNMV-OS | 88.70 | 100.00 | ||||||
| TSV | 28.10 | 28.60 | 100.00 | |||||
| TuAMV | 32.40 | 31.50 | 30.60 | 100.00 | ||||
| ApMV | 52.50 | 52.80 | 28.10 | 33.80 | 100.00 | |||
| PDV | 32.80 | 32.70 | 25.30 | 32.80 | 33.40 | 100.00 | ||
| APLPV | 36.90 | 36.50 | 31.00 | 38.00 | 37.70 | 37.60 | 100.00 | |
| PNRSV | 59.90 | 61.60 | 32.40 | 32.00 | 57.10 | 33.50 | 37.40 | 100 |
ApNMV GD Apple necrotic mosaic virus Golden Delicious isolate, ApNMV OS Apple necrotic mosaic virus Oregon spur isolate, TSV Tobacco streak virus (NC003845), TuAMV Tulare apple mosaic virus (NC003835), ApMV Apple mosaic virus (NC003480), PDV Prune dwarf virus (NC008038), APLPV American plum line pattern virus (EF494413), PNRSV Prunus necrotic ringspot virus (NC004364)
Fig. 5.
Two dimensional colour coded graphical representation of sequence identities based on coat protein of Indian ApNMV isolates with other Illarviruses. A scale with percent color code is presented
Discussion
Apple mosaic is widely distributed viral disease in apple-growing regions throughout the world (Petrzik and Lenz 2011) leading to the major adverse effects to the yield and quality of apple fruits (Li et al. 2002). Presence of mosaic disease in different cultivars constitutes a potential threat to apple cultivation in India. The mosaic symptoms along with necrosis associated with mosaic disease has been reported from different countries including India (Moury et al. 2000; Helguera et al. 2002; Noda et al. 2017). During our survey, both types of symptoms, mosaic as well necrosis was observed on various cultivars of apple. The cultivar “Golden Delicious” was found most susceptible to mosaic disease, due to the presence of high disease incidence. The higher susceptibility of Golden Delicious cultivar to mosaic disease was also reported by several workers (Cembali et al. 2003; Padder et al. 2011; Katwal et al. 2016).
In India, until now, the causal agent of apple mosaic disease has been shown to be ApMV only, using either ELISA or RT-PCR (Lakshmi et al. 2011; Verma et al. 2014; Padder et al. 2011). However, in the present study 90% of symptomatic samples tested negative in ELISA/RT-PCR for ApMV. Our attempt to associate the ApNMV with apple trees negative for ApMV was successfully demonstrated and 60% of samples were ApNMV positive out of all samples processed by RT-PCR. Some of the samples from different varieties especially Gala mast and Red Gold were tested negative for both viruses, which may be because of low virus titer or due to the presence of various inhibitory compounds in the sap or high sensitivity of the virus to temperature fluctuation. However, it is not easy to precisely determine the causes of these divergences in virus load in plant tissues, specifically whether they are caused by true changes in virus concentrations or the reliability of detection methods influenced by inhibitors (Caglayan et al. 2006). There was significant sequence variability (10%) among the ApNMV isolates from various cultivars of apple. Similar results have been shown by (Grimova et al. 2013) using phylogenetic analysis of ApMV isolates from many different geographical locations and plant hosts. Sequence analysis of the complete CP sequences of ApNMV isolates showed that isolates are closely related to each other in their genomes (RNA3), demonstrating a common source of virus. The sequence comparison of complete nucleotide sequences of CP gene of our isolates showed maximum similarity with ApNMV followed by PNRSV and ApMV, hence was placed in the same subgroup 3 of the genus Ilarvirus as ApMV and PNRSV.
Earlier (Noda et al. 2017) reported a novel Ilarvirus (ApNMV) closely related to PNRSV and ApMV using next-generation sequencing analysis from mosaic diseased apple trees in Japan and China. Cho also reported the presence of ApNMV from apple trees infected with mosaic disease in Korea (Cho et al. 2017). Phylogenetic analysis based on CP gene placed ApNMV in the subgroup 3 of the genus Ilarvirus same as ApMV and PNRSV belongs, but shows clear distinction from both the viruses. The present ApNMV isolates formed two sub clusters, where sub-cluster-I, included ApNMV isolates from cultivars; Oregon Spur, Red Delicious and Fuji Aztec along with Chinese and Korean isolates. Interestingly, sub-cluster-II included only ApNMV isolates from Golden Delicious and Red Delicious, showed distinctiveness of these two isolates from others. According to the International Committee on Taxonomy of Viruses (ICTV) (https://ictv.global/report), species demarcation criteria of species in terms of sequence similarity in the genus Ilarvirus is still not defined and little information is available on the homology between ApNMV with other Illarviruses. The whole genome of ApNMV has been characterized and confirmed, that the genomic components are consistent with viruses in the genus Illarvirus subgroup 3 (Hu et al. 2019; Cho et al. 2017). From our results, it is evident that the ApNMV is also associated with AMD in India and is more common than ApMV. However, given the strong correlation between the ApNMV distributions among various cultivars, it is likely that ApNMV is a major cause of the mosaic symptoms in Indian apple cultivars.
The present study also revealed that not only ApNMV, but also ApMV was found to be associated with mosaic disease in few samples. Apart from ApNMV, the apple trees were also infected with Apple stem pitting virus (Foveavirus) and Apple stem grooving virus (Capillovirus), (data not shown), both the viruses being latent in nature (Sajad et al. 2018). Symptoms induced by ApMV or ApNMV are generally indistinguishable and therefore accurate diagnostics of viral infection in early stages of orchard establishment is indispensable, in view of viral dissemination through rootstock and bud wood. The occurrence of ApNMV obtained from different cultivars during the present study would be helpful in developing diagnostics for indexing of elite varieties, host range in woody trees like pome, stone fruits and production of clean and healthy planting material.
Acknowledgements
The authors are thankful to Head, Division of Plant Pathology, ICAR-IARI, New Delhi for providing financial assistance and facilities for carrying out the study.
Author contributions
SUN: Conducted survey, RT-PCR experiments, cloning and analyses of sequences of PCR products, wrote the manuscript. VKB: Conceptualized, planned experiments and wrote the manuscript, MKY: Conducted DAS-ELISA, Phylogenetic analysis, Sequence identity matrix. GPR: Conceptualized and wrote the manuscript and planned the experiments.
Compliance with ethical standards
Conflict of interest
Authors declare no conflict of interest.
Informed consent
No individual participant was included in the study.
Research involving in human and animal participants
This manuscript does not contain any experiments with human participants or animals performed by any of the authors.
References
- Caglayan K, Serce CU, Gazel M, Jelkmann W. Detection of four apple viruses by ELISA and RT-PCR assays in Turkey. Turkish J Agric For. 2006;30:241–246. [Google Scholar]
- Cembali TR, Folwell J, Wand-Schneider P, Eastwell KC, Howell WE. Economic implications of a virus prevention program in deciduous tree fruits in the US. Crop Prot. 2003;22:1149–1156. doi: 10.1016/S0261-2194(03)00156-X. [DOI] [Google Scholar]
- Chai GZ, Song LQ, Jiang ZW, Zhang XY, Zhang S, Liu MY, Tang Y, Sun YX, Zhao LL. The effect of apple mosaic on photosynthesis of different varieties of apple. Fruits. 2017;3:8–9. [Google Scholar]
- Cho IS, Chung BN, Yoon JY. First report of apple necrotic mosaic virus infecting apple trees in Korea. J Plant Pathol. 2017;99(3):799–818. [Google Scholar]
- Ferree DC, Warrington IJ, editors. Apples: botany, production and uses. Wallington, Oxford, UK: CABI; 2003. pp. 1–14. [Google Scholar]
- Grimova L, Winkowska L, Ryšánek P, Svoboda P, Petrzik K. Reflects the coat protein variability of apple mosaic virus host preference? Virus Genes. 2013;47:119–125. doi: 10.1007/s11262-013-0925-z. [DOI] [PubMed] [Google Scholar]
- Grimova L, Lucie W, Michal K, Pavel R. Apple mosaic virus. Phytopathol Mediterr. 2016;55(1):1–19. [Google Scholar]
- Helguera PR, Docampo DM, Nome F, Ducasse DA. Enhanced detection of Prune dwarf virus in peach leaves by immunocapture-reverse transcription-polymerase chain reaction with nested polymerase chain reaction (IC-RT-PCR nested PCR) J Phytopathol. 2002;150:94–96. doi: 10.1046/j.1439-0434.2002.00696.x. [DOI] [Google Scholar]
- Hu GJ, Dong YF, Zhang ZP, Fan XD, Ren F, Li ZN, Zhou J. First report of Prunus necrotic ringspot virus infection of apple in China. Plant Dis. 2016;100:1955. doi: 10.1094/PDIS-01-16-0079-PDN. [DOI] [Google Scholar]
- Hu Y, Shi HW, Jing CC, Li K, Sun XC, Zhou CY, Qing L. First report of Cucumber mosaic virus infecting apple in China. J Plant Pathol. 2016;98:181. [Google Scholar]
- Hu GJ, Dong YF, Zhang ZP, Fan XD, Ren F. Molecular characterization of Apple necrotic mosaic virus identified in crabapple (Malus spp.) tree of China. J Integr Agric. 2019;18:698–701. doi: 10.1016/S2095-3119(18)62116-1. [DOI] [Google Scholar]
- Katwal VS, Handa A, Thakur PD, Tomar M. Prevalence and serological detection of apple viruses in Himachal Pradesh. Plant Pathol J. 2016;15(2):40–48. doi: 10.3923/ppj.2016.40.48. [DOI] [Google Scholar]
- Kumar S, Lakhmir S, Raja R, Aijaz AZ, Vipin H. Simultaneous detection of major pome fruit viruses and a viroid. Indian J Microbiol. 2014;54(2):203–210. doi: 10.1007/s12088-013-0431-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmi V, Hallan V, Ram RV, Ahmed N, Zaidi AA, Varma A. Diversity of Apple mosaic virus isolates in India based on coat protein and movement protein genes. Indian J Virol. 2011;22:44–49. doi: 10.1007/s13337-011-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DH, Zhao HY, Hu ZQ, Hu XS, Zhang YH. Studies on the damage, loss of production and control to apple mosaic disease. J Northwest Sci-Tech Univ Agric For. 2002;30:77–80. [Google Scholar]
- Moury B, Cardin L, Onesto JP, Candresse T, Poupet A. Enzyme-linked immunosorbent assay testing of shoots grown in vitro and the use of immunocapture-reverse transcription polymerase chain reaction improve the detection of Prunus necrotic ringspot virus in rose. Phytopathol. 2000;90:522–528. doi: 10.1094/PHYTO.2000.90.5.522. [DOI] [PubMed] [Google Scholar]
- Muneer AS, Bhat KM, Mir JI, Mir MA, Nabi SU, Bhat MA, Hilal A, Wajida S, Shafia Z, Sumaira J, Waseem HR. Phenotypic and molecular screening for disease resistance of apple cultivars and selections against apple scab (Venturia inaequalis) Int J Chem Stud. 2017;5(4):1107–1111. [Google Scholar]
- Nemeth MV. Virus, mycoplasma, and rickettsia diseases of fruit trees. Hingham, MA, USA: Kluwer Academic; 1986. [Google Scholar]
- Nisar AD. Apple stem grooving virus-a review article. Int J Mod Plant Anim Sci. 2013;1:28–42. [Google Scholar]
- Noda H, Yamagishi N, Yaegashi H, Xing F, Xie J, Li S, Zhou T, Ito T, Yoshikawa N. Apple necrotic mosaic virus, a novel ilarvirus from mosaic-diseased apple trees in Japan and China. J Gen Plant Pathol. 2017;83:83–90. doi: 10.1007/s10327-017-0695-x. [DOI] [Google Scholar]
- Padder BA, Shah MD, Mushtaq A, Aflaq H, Sofi TA, Ahanger FA, Sahar S. Status of apple mosaic virus in Kashmir valley. Appl Biol Res. 2011;13(2):117–120. [Google Scholar]
- Petrzik K, Lenz O. Apple mosaic virus in pome fruits. In: Hadidi A, Barba M, Candresse T, Jelkmann W, editors. Virus and virus like diseases of pome and stone fruits. St. Paul, MN: APS Press; 2011. pp. 25–28. [Google Scholar]
- Sajad UN, Javid IM, Om CS, Desh BS, Shafia Z, Muneer AS, Kamran LM, AK Optimization of tissue and time for rapid serological and molecular detection of Apple stem pitting virus and Apple stem grooving virus in apple. Phytoparasitica. 2018;46(5):705–713. doi: 10.1007/s12600-018-0701-7. [DOI] [Google Scholar]
- Sajad UN, Manoj Y, Nida Y, Wasim HR, Kavi S, Saurabh D, Damini J. Apple mosaic disease: potential threat to apple productivity. EC Agric. 2019;5(10):614–618. [Google Scholar]
- Tombisana T, Tanuja R, Vipin H, Raja R, Zaidi AA. Molecular characterization of the Indian strain of Apple mosaic virus isolated from apple (Malus domestica) Phytoparasitica. 2009;37:375–379. doi: 10.1007/s12600-009-0041-8. [DOI] [Google Scholar]
- Verma RK, Ahmed N, Mir JI, Verma MK, Srivastava KK, Focktoo SZ, Rashid R, Shafi W. Detection of apple mosaic and chlorotic leaf spot viruses by DAS-ELISA from farmers orchards of Kashmir valley. Indian J Hort. 2014;71(4):567–570. [Google Scholar]
- Xing F, Robe BL, Zhang Z, Wang H, Li S. Genomic analysis, sequence diversity, and occurrence of Apple necrotic mosaic virus, a novel Ilarvirus associated with mosaic disease of apple trees in China. Plant Dis. 2018;102(9):1841–1847. doi: 10.1094/PDIS-10-17-1580-RE. [DOI] [PubMed] [Google Scholar]