Abstract
Trichosporonaceae incorporates six genera of physiologically and ecologically diverse fungi including both human pathogenic taxa as well as yeasts of biotechnological interest, especially those oleagenic taxa that accumulate large amounts of single cell oils (SCOs). Here, we have undertaken comparative genomic analysis of thirty-three members of the family with a view to gain insight into the molecular determinants underlying their lifestyles and niche specializations. Phylogenomic analysis revealed potential misidentification of three strains which could impact subsequent analyses. Evaluation of the predicted proteins coding sequences showed that the free-living members of the family harbour greater numbers of carbohydrate active enzymes (CAZYmes), metallo- and serine peptidases compared to their host-associated counterparts. Phylogenies of selected lipid biosynthetic enzymes encoded in the genomes of the studied strains revealed disparate evolutionary histories for some proteins inconsistent with the core genome phylogeny. However, the documented oleagenic members distinctly cluster based on the constitution of the upstream regulatory regions of genes encoding acetyl-CoA carboxylase (ACC), ATP-citrate synthase (ACS) and isocitrate dehydrogenase [NADP] (ICDH), which are among the major proteins in the lipid biosynthetic pathway of these yeasts, suggesting a possible pattern in the regulation of these genes.
Subject terms: Molecular evolution, Taxonomy, Genomics
Introduction
The basidiomycetous fungal family Trichosporonaceae belongs to the order Trichosporonales, the class Tremellomycetes, and subphylum Agaricomycotina and incorporates morphologically and physiologically diverse, aromatic compound-assimilating yeasts1. Recently the taxonomy of this family was revised to include six genera, namely Apiotrichum, Cutaneotrichosporon, Effuseotrichosporon, Haglerozyma, Trichosporon (type genus) and Vanrija. This revision was based on phylogenetic analysis of seven markers, namely LSU (D1/D2 domains) and SSU rRNA, the Internal Transcribed Spacer (ITS) and the protein coding genes RPB1, RPB2, TEF1 and CYTB and a combination of morphological, biochemical and physiological characteristics1,2. Members of the Trichosporonaceae show a global distribution and have been recovered from a wide range of environments. Cutaneotrichosporon spp. are most frequently associated with a human host, and may represent opportunistic human pathogens. Trichosporon spp. form part of the natural microflora on human and animal skin and result in a non-serious mycosis of hair termed white piedra3. However, they have also been implicated in trichosporonosis, a collection of opportunistic infections caused by a number of species, including Trichosporon asahii, T. asteroides and T. ovoides4. By contrast Apiotrichum and Vanrija spp. are generally free-living and have been isolated from water bodies, food sources and rotten wood (Table 1).
Table 1.
Genome features of thirty-three Trichosporonaceae and two outgroup species included in the analysis.
| Strains | Accession | BioProject | Isolation source/locality | Sequencing technology | Assembly Size (bp) | # Scaffolds | N50 (bp) | % G + C | # Genes | # tRNA | # Proteins | Unique Proteins | Prots atleast 1 orthologue | Comp* | Dup¥ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Apiotrichum brassicae JCM 1599T | BCJI00000000 | PRJDB3695 | Cabbage purchased in a market/Tokyo, Japan | HiSeq. 2500 | 23,647,732 | 16 | 2,410,059 | 56.47 | 8,334 | 422 | 7,912 | 1,204 | 6,708 | 91.3 | 0.2 |
| Apiotrichum domesticum JCM 9580T | BCFW00000000 | PRJDB3573 | Rotten wooden sideboard (pneumonitis patient), Japan | HiSeq. 2500 | 24,510,922 | 28 | 3,306,260 | 58.52 | 8,536 | 379 | 8,157 | 582 | 7,575 | 91.1 | 0.6 |
| Apiotrichum gamsii JCM 9941T | BCJN00000000 | PRJDB3703 | Moist humus around roots/Colombia | HiSeq. 2500 | 24,609,388 | 29 | 3,284,570 | 61.14 | 8,608 | 385 | 8,223 | 1,628 | 6,595 | 96.3 | 1.2 |
| Apiotrichum gracile JCM 10018 | BCJO00000000 | PRJDB3704 | Sour milk/Germany | HiSeq. 2500 | 24,114,851 | 17 | 4,554,691 | 59.23 | 9,556 | 918 | 8,638 | 1,677 | 6,961 | 90.2 | 0.5 |
| Apiotrichum laibachii JCM 2947T | BCKV00000000 | PRJDB3730 | Soil | HiSeq. 2500 | 30,616,633 | 26 | 2,819,049 | 59.63 | 11,708 | 1,530 | 10,178 | 1,886 | 8,292 | 92.8 | 0.5 |
| Apiotrichum montevideense JCM 9937 | BCFV00000000 | PRJDB3572 | Water purification tank/Montevideo, Uruguay | HiSeq. 2500 | 24,872,216 | 61 | 1,972,470 | 58.17 | 8,550 | 375 | 8,175 | 599 | 7,576 | 91.9 | 0.4 |
| Apiotrichum porosum DSM 27194 | RSCE00000000 | PRJNA506377 | Grassland /Germany | PacBio | 25,479,456 | 32 | 1,376,709 | 59.15 | 9,729 | 576 | 9,153 | 555 | 8,598 | 96.7 | 0.9 |
| Apiotrichum porosum JCM 1458T | BCJG00000000 | PRJDB3693 | Exudate of Taxus baccata/Hamburg, Germany | HiSeq. 2500 | 25,989,348 | 37 | 3,276,928 | 59.05 | 9,816 | 546 | 9,270 | 662 | 8,608 | 97.2 | 0.9 |
| Apiotrichum veenhuisii JCM 10691T | BCKJ00000000 | PRJDB3717 | Buffalo dung/Minturno, Campania, Italy | HiSeq. 2500 | 31,617,680 | 35 | 2,551,142 | 59.59 | 10,127 | 1,019 | 9,108 | 1,778 | 7,330 | 92.2 | 0.7 |
| Cutaneotrichosporon cutaneum ACCC 20271 | LTAL00000000 | PRJNA313001 | Hangzhou oil refinery/China | Illumina MiSeq | 30,717,177 | 21 | 5,629,136 | 57.13 | 9,586 | 878 | 8,708 | 1,386 | 7,322 | 94.1 | 0.4 |
| Trichosporon akiyoshidainum HP2023 | PQXP00000000 | PRJNA428315 | Ryzosphere/Las Yungas rainforest, Argentina | Illumina MiSeq | 30,042,643 | 1067 | 53,521 | 60.69 | 11,492 | 1,292 | 10,200 | 1,746 | 8,454 | 90.5 | 4.3 |
| Cutaneotrichosporon arboriformis JCM 14201T | BEDW00000000 | PRJDB5900 | Urine of chronic renal failure patient/Hokkaido, Japan | Illumina MiSeq | 19,894,493 | 28 | 1,599,305 | 60.59 | 7,697 | 143 | 7,554 | 1,357 | 6,197 | 88.7 | 0.5 |
| Cutaneotrichosporon curvatus JCM 1532T | BCJH00000000 | PRJDB3694 | Sputum of tubercular patient/The Netherlands | HiSeq. 2500 | 18,637,344 | 75 | 571,590 | 57.89 | 7,137 | 151 | 6,986 | 293 | 6,693 | 88.4 | 0.3 |
| Cutaneotrichosporon curvatus SBUG-Y 855 | LDEP00000000 | PRJNA281029 | Sputum of tubercular patient/Netherlands | Illumina GAIIx | 16,443,618 | 354 | 62,631 | 59.41 | 6,628 | 151 | 6,477 | 246 | 6,231 | 80.8 | 0.6 |
| Cutaneotrichosporon cutaneum B3 | LRUG00000000 | PRJNA310294 | Soil/Nanchang, Jiangxi, China | Illumina HiSeq | 38,696,417 | 592 | 116,762 | 60.30 | 14,851 | 448 | 14,403 | 1,004 | 13,399 | 94.1 | 55.7 |
| Cutaneotrichosporon cutaneum JCM 1462 | BCKU00000000 | PRJDB3729 | Probably human clinical specimen | HiSeq. 2500 | 23,155,501 | 98 | 744,446 | 61.97 | 9,635 | 399 | 9,236 | 1,756 | 7,480 | 88.3 | 0.4 |
| Cutaneotrichosporon cyanovorans JCM 31833 | BEDZ00000000 | PRJDB5903 | Cyanide contaminated soil /Sasolburg, South Africa | Illumina MiSeq | 19,941,766 | 90 | 582,111 | 58.01 | 7,086 | 150 | 6,936 | 993 | 5,943 | 87.5 | 0.2 |
| Cutaneotrichosporon daszewskae JCM 11166 | BEDX00000000 | PRJDB5901 | Skin/Germany | Illumina MiSeq | 17,225,847 | 12 | 2,186,752 | 60.98 | 8,162 | 199 | 7,963 | 1,697 | 6,266 | 85.2 | 0.5 |
| Cutaneotrichosporon dermatis JCM 11170 | BCKR00000000 | PRJDB3725 | Human infected skin/Tübingen, Germany | HiSeq. 2500 | 23,337,637 | 37 | 2,690,106 | 60.05 | 8,945 | 297 | 8,648 | 511 | 8,137 | 92.1 | 0.3 |
| Cutaneotrichosporon mucoides JCM 9939T | BCJT00000000 | PRJDB3710 | Meningitis case/Belgium | HiSeq. 2500 | 40,783,511 | 84 | 1,668,497 | 60.14 | 15,434 | 507 | 14,927 | 1,016 | 13,911 | 94.4 | 60.8 |
| Cutaneotrichosporon oleaginosum ATCC 20509T | MATS00000000 | PRJNA327102 | Dairy plant | PacBio | 19,908,169 | 16 | 2,509,747 | 60.59 | 8,302 | 266 | 8,036 | 169 | 7,867 | 89.6 | 0.4 |
| Cutaneotrichosporon oleaginosum IBC0246 | JZUH00000000 | PRJNA239490 | - | Illumina | 19,835,558 | 180 | 216,041 | 60.74 | 8,267 | 267 | 8,000 | 197 | 7,803 | 89.1 | 0.5 |
| Trichosporon asahii JCM 2466T | BCLT00000000 | PRJDB3696 | Human, Japan | HiSeq. 2500 | 24,687,929 | 36 | 2,256,092 | 59.45 | 9,439 | 502 | 8,937 | 210 | 8,727 | 94.4 | 1.1 |
| Trichosporon asahii var. asahii CBS 2479T | ALBS00000000 | PRJNA164647 | Trichosporia cutis proriasiformis progressiva, Japan | 454; Illumina | 24,540,311 | 78 | 1,660,894 | 59.01 | 9,364 | 497 | 8,867 | 287 | 8,580 | 93.2 | 0.5 |
| Trichosporon asahii var. asahii CBS 8904 | AMBO00000000 | PRJNA172216 | Maize cobs§ Illinois Peoria USA | 454; Illumina | 25,299,608 | 194 | 3,223,897 | 58.91 | 9,816 | 518 | 9,298 | 810 | 8,488 | 93.7 | 0.7 |
| Trichosporon coremiiforme JCM 2938T | JXYL00000000 | PRJDB3697 | Head lesion caused by beesting/Turrialba, Costa Rica | Illumina HiSeq | 42,353,277 | 190 | 1,468,092 | 59.60 | 15,969 | 908 | 15,061 | 2,104 | 12,957 | 95.3 | 69.5 |
| Trichosporon faecale JCM 2941T | JXYK00000000 | PRJDB3698 | Human feces | Illumina HiSeq | 24,653,913 | 32 | 3,676,950 | 60.21 | 9,494 | 471 | 9,023 | 1,233 | 7,790 | 93.6 | 0.7 |
| Trichosporon inkin JCM 9195 | JXYM00000000 | PRJDB3701 | Human | Illumina HiSeq | 20,339,538 | 18 | 2,739,924 | 62.69 | 7,700 | 318 | 7,382 | 654 | 6,728 | 89.3 | 0.2 |
| Trichosporon ovoides JCM 9940 | JXYN00000000 | PRJDB3702 | White piedra infected scalp | Illumina HiSeq | 40,322,879 | 116 | 2,824,449 | 60.22 | 15,163 | 584 | 14,579 | 2,470 | 12,109 | 96.4 | 70 |
| Pascua guehoae JCM 10690 | BCJX00000000 | PRJDB3714 | Meadow soil/Renswoude, the Netherlands | HiSeq. 2500 | 33,698,914 | 35 | 2,162,681 | 59.17 | 11,097 | 1,032 | 10,065 | 3,277 | 6,788 | 91.9 | 1.0 |
| Prillingera fragicola JCM 1530T | BEDY00000000 | PRJDB5902 | Strawberry /Market in Akihabara, Tokyo, Japan | Illumina MiSeq | 20,263,689 | 21 | 1,928,959 | 58.21 | 8,282 | 297 | 7,985 | 1,620 | 6,365 | 94 | 0.7 |
| Vanrija humicola JCM 1457T | BCJF00000000 | PRJDB3692 | Soil | HiSeq. 2500 | 22,653,840 | 10 | 3,082,120 | 62.74 | 9,255 | 279 | 8,976 | 495 | 8,481 | 93.7 | 0.4 |
| Vanrija humicola UJ1 | BFAH00000000 | PRJDB6593 | Soil | Illumina HiSeq. 2500 | 22,628,423 | 46 | 1,340,400 | 62.76 | 9,270 | 296 | 8,974 | 483 | 8,491 | 93.5 | 0.4 |
| Takashimella koratensis JCM 12878T | BCKT00000000 | PRJDB3728 | Leaf of Lagerstroemia calyculata/Thailand | HiSeq. 2500 | 25,142,362 | 31 | 1,454,648 | 54.94 | 9,382 | 54 | 9,328 | 3,020 | 6,308 | 95.1 | 0.2 |
| Takashimella tepidaria JCM 11965T | BCKS00000000 | PRJDB3726 | Stream water/Japan | HiSeq. 2500 | 22,370,450 | 44 | 1,186,041 | 44.66 | 6,989 | 59 | 6,930 | 1,035 | 5,895 | 90 | 0.2 |
*Completeness and ¥duplication determined using BUSCO based on basidiomycota_odb9. §Human pathogenic (superficial and systemic).
While the Trichosporonaceae include several opportunistic human pathogens, there has also been increased interest in these taxa for a broad range of biotechnological applications. Most pertinently, members of the Trichosporonaceae are known to produce and accumulate large amounts of single cell oil (SCO) relative to their dry biomass5–11, with up to 70% w/dwbiomass (weight/dry weight of biomass) accumulated by Cutaneotrichosporon oleaginosus12. Furthermore, they are amenable to large-scale fermentations as they are not as sensitive as other oleaginous yeasts to fermentation inhibitors including furanes and phenolic compounds8. These factors make members of the Trichosporonales suitable candidates in a wide range of biotechnological applications such as the production of oleo-chemicals and biofuels13,14.
The rapid development of genome sequencing technologies and bioinformatics has been pivotal in shaping our understanding of fungal genetics. Since the publication of the first fungal genome, Saccharomyces cerevisiae, in 199615, fungal genomics has experienced rapid development. As of June 2019, 5,269 fungal genome assemblies have been deposited in the NCBI database16. With the increasing availability of fungal genomes, recent works have harnessed the information contained in the genomes to develop more robust taxonomic frameworks for several fungal taxa. For instance, Takashima et al.17–19 have pioneered and variously reported a genome-based characterisation and phylogenetic analysis of the order Trichosporonales using 24 haploid and 3 natural hybrid genomes. Furthermore genome sequencing provides access to the full complement of proteins encoded on a fungal genome, which can serve as resource for modelling functional capacities of the fungal strains and to further their use as biological resources in a wide range of biotechnological applications20.
In the current study, we have employed comparative genomic strategies to study thirty-three members of the family Trichosporonaceae. Phylogenomic analysis identified three mis-classified taxa within this family, while genes coding for enzymes involved in oleagenicity and their regulatory regions show evolutionary patterns distinct from the genome scale phylogeny. Furthermore, the genome comparisons highlighted a range of genetic determinants underlying the distinct lifestyles and niche specialisations of the different taxa within this family.
Results and Discussion
Genomic characteristics of the Trichosporonaceae
The genomes of thirty-three taxa belonging to the genera Apiotrichum (nine strains), Cutaneotrichosporon (twelve strains), Pascua guehoae, Prillingera fragicola, Trichosporon (eight strains) and Vanrija (two strains) were incorporated in the analyses. Twenty-nine of the strains have haploid genome structures, while three strains, namely C. mucoides JCM 9939T, T. ovoides JCM 9940 and T. coremiiforme JCM 2938T, have been shown to comprise hybrid genomes18. In this study, genome duplication and phylogenetic analyses revealed one additional strain, C. cutaneum B3 to comprise of a hybrid genome. Two strains of Takashimella (belonging to the closely related family Tetragoniomycetaceae were included as outgroups. A survey of the origin of the Trichosporonaceae strains shows a wide geographic distribution of the organisms with isolates obtained from food, decomposing wood, human body, soils, water bodies, among others (Table 1). The two outgroup strains have originated from two distinct sources; leaf of plant and stream water. However, majority of members of the genus Trichosporon and Cutaneotrichosporon species, for which the genome sequences are available, are either associated with human or animal skin while genomes of isolates from insect1,21 are not available. This may reflect preference for the sequencing of clinically important strains. The phylogenomic analyses of thirty-three members of the family Trichosporonaceae, including Apiotrichum porosum DSM 27194 and one putative hybrid genome strain, C. cutaneum B3 are presented here. The estimated genome sizes of the thirty-three Trichosporonaceae strains ranged between 16.4 and 42.4 Mb with an average G + C content range of 56.5–62.8%. The N50, which is the contig/scaffold size for which at least 50% of the assembly is contained in equal or larger contigs/scaffolds, ranged between 53.5Kb in T. akiyoshidainum HP2023 and 5.6 Mb in C. cutaneum ACCC 20271, indicating wide variety in assembly quality. However, previous studies have shown that large N50 values may arise because of erroneous concatenation of contigs, thereby limiting the value of this metric in evaluating assembly quality22. The largest genome sizes (average of 40.5 Mb) are observed for the four hybrid genomes incorporated in the analysis. Among the haploid genomes, the largest genome sizes belong to the yeast strains that are predominantly isolated from various soil types. Prediction of protein encoding gene models revealed that the genomes of these fungi encode between 6,477 (C. curvatus SBUG-Y 855) and 15,061 (T. coremiiforme JCM 2938T) proteins. Evaluation of the predicted protein models using the BUSCO23 basidiomycota_odb9, which includes 1335 single copy genes/proteins, revealed that the genome completeness of the yeast strains included in this study ranged between 80.8 and 97.2% (Table 1). Additionally, BUSCO23 analysis revealed extensive protein duplication ranging between 55.7 to 70% in the four hybrid genomes that harbour the largest genome sizes. In contrast, the two outgroup species have genome sizes of 22.4 and 25.1 Mb and G + C content of 44.66 and 54.94% for Takashimella tepidaria JCM 11965T and T. koratensis JCM 12878T, respectively.
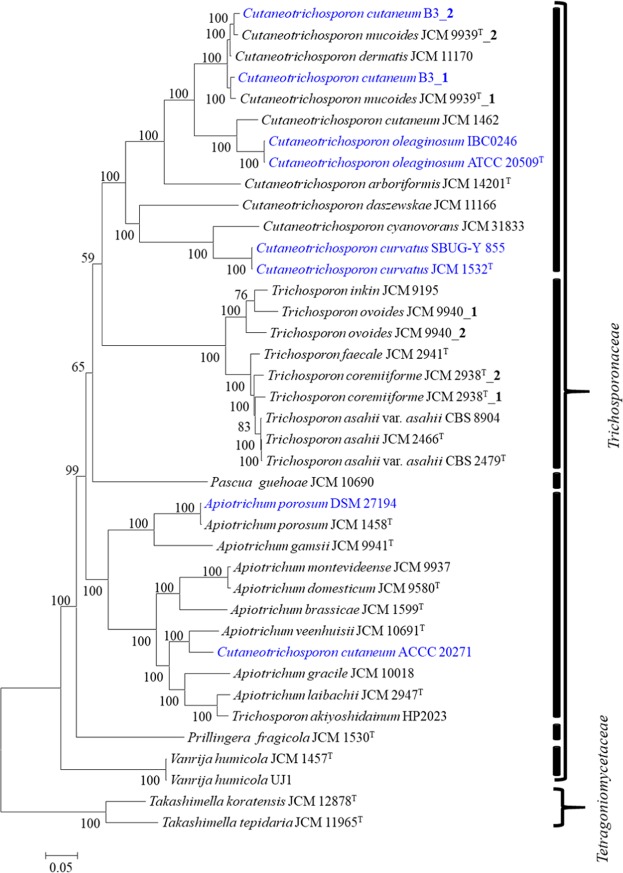
Genome-wide phylogenetic analysis reveals several misclassifications in the Trichosporonaceae
Orthologous proteins conserved among all compared taxa were identified using Proteinortho524. A total of 1,351 proteins are common to all the studied strains, including the outgroups. However, to put the hybrid genomes into phylogenomic perspective, 405 orthologous proteins present solely in single copies among the haploid genomes and only in duplicate copies in the hybrid genomes were used to reconstruct the phylogeny of the Trichosporonaceae. The trimmed concatenated protein alignment comprised 223,082 amino acids in length. The resultant maximum likelihood phylogeny (Fig. 1) shows the clustering of the Trichosporonaceae into six distinct clades. Eleven of the twelve Cuteaneosporotrichon, seven of the eight Trichosporon and all nine Apiotrichum strains incorporated in the study fall into three separate clades congruent with the distinct Trichosporonaceae genera that they represent1,2.
Figure 1.
Phylogenomic analysis of members of the family Trichosporonaceae. The maximum likelihood (ML) tree was inferred from the concatenated protein alignment (223,082 amino acids) of 405 proteins present in single copies among the haploid genomes and only in duplicate copies in the hybrid genomes. The phylogeny was generated using IQ-TREE version 1.6.7 based on the LG + F + R10 model. The ML was generated with confidence values based on 1,000 bootstrap replicates. The documented oil accumulating members of the family are indicated in blue fonts. The labels ‘_1’ and ‘_2’ indicate the two sets of single copy orthologs (SCOs) in the hybrid genomes, where the letter shows higher amino acid similarity to the closest haploid genome.
While three clear genus clades can be observed in the single copy orthologues phylogeny (SCOP), two taxa, namely C. cutaneum ACCC 20271 and T. akiyoshidainum HP2023 are clearly delineated within the Apiotrichum clade in the SCOP, and should thus be reassigned to the latter genus. As has previously been observed through separation of subgenomes18,25, the duplicate orthologue copies (here referred to as ‘strain number’_1 and _2) in the three described hybrid genomes form distinct branches but are still retained within their genus clades (Fig. 1). When considering the fourth putative hybrid genome identified in this study, C. cutaneum B3, B3_1 clusters with C. mucoides JCM 9939T_1, while B3_2 also clusters with C. mucoides JCM 9939T_2, suggesting that the two strains are likely to have shared similar evolutionary history including episodes of hybridization. In addition to evidence from gene duplication (55.7%) determined using BUSCO23 basidiomycota_odb9, BLASTP analyses showed that C. dermatis JCM 11170 shares 92.41% and 97.92% amino acid similarity among the 405 single copy orthologues (SCO) with those of C. cutaneum B3_1 and B3_2, respectively. In additon, the 405 SCO sets of B3_1 and B3_2 shared on average 92.76% amino acid similarity, further proving support for the distinct origin of the duplicated single copy orthologue sets.
Differences in the proteolytic and carbohydrate metabolic enzyme complements of the Trichosporonaceae may influence their lifestyles
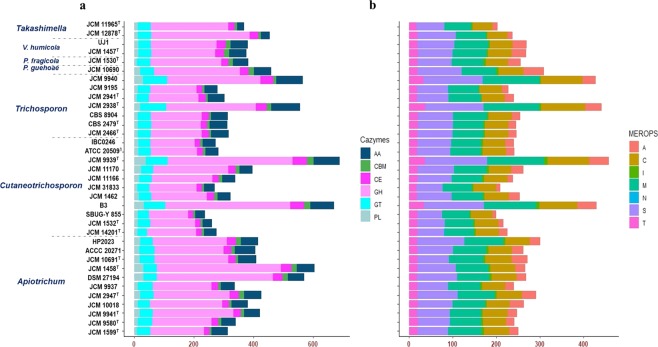
To further enhance our understanding of various functional and adaptational capacities of the studied strains, proteins annotated as Carbohydrate-Active enZYmes (CAZYmes) and proteolytic enzymes (MEROPS) were identified and compared (Fig. 2a). The presence of these proteins can provide an indication of the ranges of possible carbohydrate and protein substrates utilised by an organism. CAZYmes represent a broad scope of proteins associated with the assembly, modification and degradation of various types of carbohydrates26 and are curated in the Carbohydrate-Active EnZYmes database (http://www.cazy.org). The Cutaneotrichosporon strains displaying hybrid genomes showed the highest numbers of CAZYmes; 671 in Cutaneotrichosporon cutaneum B3 and 689 in Cutaneotrichosporon mucoides JCM 9939T8 (Supplementary Fig. 1) Aside from these hybrid genome taxa, the genomes of the two Apiotrichum porosum strains encode the highest numbers of CAZYmes (570 & 604 proteins) with ~68%, of these belonging to the class of glycoside hydrolases (GH). Similarly, GHs form the largest proportion of the CAZYmes in all studied strains. Considering the average CAZYme numbers within each genus, the Apiotrichum species also harbour the most CAZYmes (average 421), followed by Vanrija (379), Trichosporon (378) and Cutaneotrichosporon (365). However, the single available genome of Pascua guehoae also encodes 460 CAZYmes. Within the genera, Trichosporon has the highest average number of CAZYmes linked to auxillary activities (AA) and glycosyltransferases (GTs) encompassing 65 and 55, respectively and Vanrija harbours the highest average number of carbohydrate-binding modules (CBM) and carbohydrate esterases (CE) with 17 and 30, respectively while the highest mean number of glycoside hydrolases, 261 and polysaccharide lyases (PL), 19 was recorded in Apiotrichum and Cutaneotrichosporon, respectively. Abundance of CAZYmes has been linked to the various fungal adaptations with saprophytic fungi harbouring larger numbers of these enzymes compared to their parasitic counterparts27. This feature may readily be inferred from the current comparison, where on the average the free-living fungi of the genera Apiotrichum and Vanrija harbour greater numbers of CAZYmes than the predominantly host-associated Trichosporon and Cutaneotrichosporon taxa. Furthermore, the abundance of GHs and CEs28 in Apiotrichum and Vanrija, respectively may reflect their capacity to breakdown and utilise wide range of substrates. These taxa are frequently isolated from soil and other environments where they degrade and subsist on various forms of complex substrates29.
Figure 2.
Comparison of number of proteins associated with (a) CAZymes and (b) MEROPS among thirty-three strains of Trichosporonaceae. CAZymes; AA: auxillary activities, CBM: carbohydrate-binding modules, CE: carbohydrate esterases, GH: glycoside hydrolases and GT: glycosyltransferases. MEROPS; A: aspartic peptidases, C: cysteine peptidases, M: metallo-peptidases, N: asparagine peptide lyases, S: serine peptidases, T: threonine peptidases, and I: protease inhibitors.
Proteolytic enzymes are proteins that hydrolyse peptide bonds and are widely distributed across all domains of life with estimates showing that they comprise ~2% of all proteins encoded on the genomes of organisms across all domains of life30. These enzymes form an important component of the biomass degradation capacities of both fungi and bacteria31 and their distribution is reflective of the lifestyle of the organisms. For instance, comparison of pathogenic and non-pathogenic Pseudogymnoascus strains revealed a marked underrepresentation of proteases in the former relative to the latter organisms32. To predict these enzymes, proteins of the organisms included in this study were searched against the manually curated enzymes in the MEROPS database33. All seven classes of MEROPS, namely aspartic peptidases (A), cysteine peptidases (C), metallo-peptidases (M), asparagine peptide lyases (N), serine peptidases (S), threonine peptidases (T), and protease inhibitors (I) are represented in the genomes of the thirty-three Trichosporonaceae, comprising approximately 3% of the proteins of the organisms (Fig. 2b). As observed with the CAZYmes, the hybrid genomes in the genera Trichosporon and Cutaneotrichosporon harbour the most abundant peptidases, ranging between 428 and 458 proteins. Omitting the hybrid genomes, the highest average number of the MEROPS was observed among the Vanrija and Apiotrichum species, with 264 and 270 proteins, respectively. The three most abundant MEROPS belong to the class S (56–143 proteins), M (63–130 proteins) and C (49–101 proteins) across the different genera. However, asparagine peptide lyase (N), which is the only member of the MEROPS that is not a peptidase34, appears to be restricted to only five of the strains; Apiotrichum domesticum JCM 9580T (1 protein), Apiotrichum laibachii JCM 2947T (1 protein), Cutaneotrichosporon arboriformis JCM 14201T (2 proteins), Trichosporon faecale JCM 2941T (1 protein) and Trichosporon inkin JCM 9195 (1 protein). Serine and metallo-peptidases are widely distributed in fungi and may reflect the capacity of these organisms to use proteinaceous substrates35,36. However, serine peptidases contents have been shown to be determined by both proteome size and lifestyle of fungi. Parasitic fungi, often associated with reduced genomes/proteomes and those involved in symbiosis have been shown to harbour less serine proteases37. The predominance of serine peptidases S (average 81 and 82 proteins, respectively) and metallo-peptidases (average of 77 and 78 proteins, respectively) among the mainly soil inhabiting Vanrija and Apiotrichum spp. reflect their versatility in sequestering a wide range of complex substrates in their environment. Cysteine peptidase were reported as pivotal in sustaining parasitic lifestyles38. Among the Trichosporonaceae, the upper range of the cysteine peptidases are seen among the predominantly host-associated Trichosporon (an average 66 proteins) and Cutaneotrichosporon (on average 60 proteins) strains, while Apiotrichum spp. and Vanrija strains only had on average 56 and 53 of these proteins encoded on their genomes, respectively.
Phylogeny of oleagenic proteins and promoter regions of their genes highlights the complex evolution of lipid biosynthetic pathway
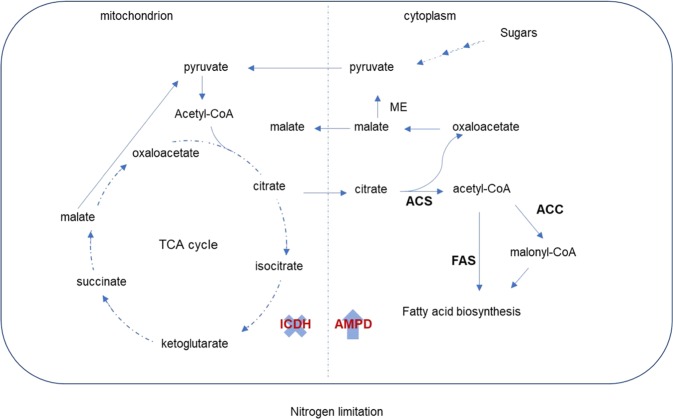
The biochemical production and accumulation of single cell oil in fungi has received extensive interest because these organisms could serve as eco-friendly sources of lipids and other important biochemicals with a wide range of biotechnological applications7,39. To provide additional insights into the genomic basis of oil accumulation among the compared strains, six proteins involved in the biochemical pathway (Fig. 3) central to lipid production and accumulation were analysed. These were acetyl-CoA carboxylase (ACC), AMP deaminase (AMPD), ATP-citrate synthase (ACS), fatty acid synthase subunits alpha and beta (FASI & II) and isocitrate dehydrogenase [NADP] (ICDH). Understanding the structure of the regulatory elements of the genes that code for these proteins may be pivotal in deciphering approaches for enhanced oil production. For instance, an increase in lipid accumulation was achieved through the overexpression of ACC under various promoter systems40,41. As such, the transcription factor binding domains (TFBDs) 600 bp upstream of these genes were analysed.
Figure 3.
Illustration of the initiation of the biochemical oil production in yeasts showing the steps within the pathway catalysed by the studied enzymes under nitrogen limitation. ACC, acetyl-CoA carboxylase, AMPD, AMP deaminase, ACS, ATP-citrate synthase, FASI & II, fatty acid synthase subunits alpha and beta and ICDH, isocitrate dehydrogenase [NADP]. × and ↑ indicates the inhibition of ICDH and increased activity of AMPD under nitrogen limitation. Modified from7
Evaluation of the proteomes of the yeasts included in this study reveals that orthologues of the selected proteins occur in all of the strains studied, with the exception of T. asahii var. asahii CBS 8904 and T. akiyoshidainum HP2023 in which orthologues of ACC are absent and C. curvatus SBUG-Y 855, which does encode an orthologue of AMP on its genome. The hybrid genomes of C. cutaneum B3, T. coremiiforme JCM 2938, T. ovoides JCM 9940 and C. mucoides JCM 9939 harbour two copies of FASI &II, ACC and ACS. However, only JCM 2938 retains the duplicate copy of ICDH, while AMPD is present in two copies in B3 and JCM 2938. Given the essential nature of these proteins, it is likely that the absence of some of the orthologues is associated with the level of genome completeness rather than the lack of the affected function.
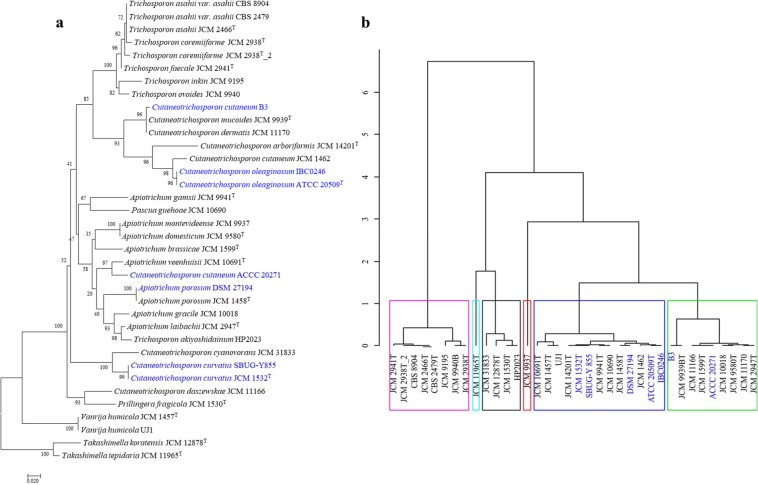
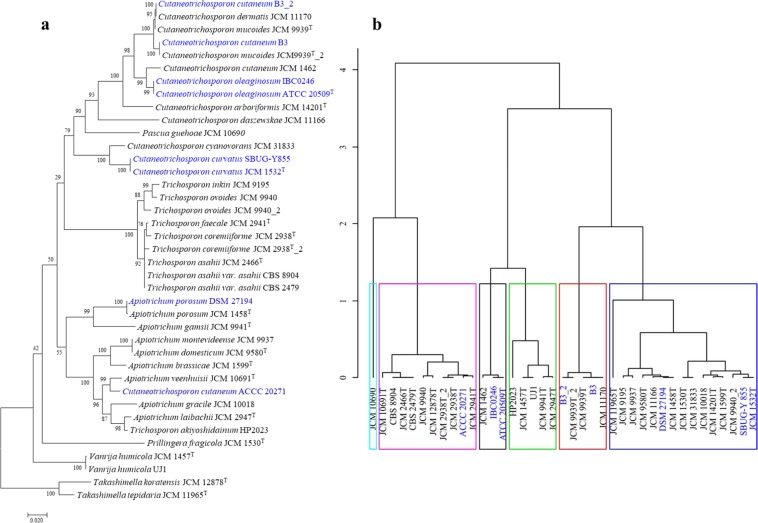
Oil production in yeasts has been linked to nutrient limitation, where the organisms channel carbon flux to lipid instead of energy production5,7. Two enzymes directly associated with this function are AMPD and ICDH with the former shown to enhance the depletion of AMP and consequently playing a role in the inhibition of ICDH42. Comparison of a phylogeny on the basis on the AMPD amino acid sequences (Supplementary Fig. 2,a) showed that, apart from the placement of V. humicola, this tree shows a similar topology and clustering as the SCOP. Clustering of the strains based on the distribution and abundance of TFBDs upstream of the AMPD gene (Supplementary Fig. 2,b) shows distinct grouping of the organisms suggesting disparate evolution of this regulatory region. In the ICDH tree (Fig. 4a), only the Trichosporon species showed a coherent grouping while members of the genus Cutaneotrichosporon, including the known oleaginous strain C. curvatus show incongruent branching pattern relative to the SCOP, indicating distinct evolutionary history of the ICDH gene. Comparison of the TFBDs of the ICDH gene revealed that these fungi form six distinct clusters (Fig. 4b) with the documented oleaginous strains A. porosum, C. curvatus and C. oleaginosum, clustering together thereby suggesting a possible similarity in the regulation of the ICDH gene among these strains. Two other reported oil accumulating yeast, namely C. cutaneum B3 and C. cutaneum ACCC 20271 are also closely clustered with the rest of the oleaginous strains. Discussion on the affiliation of the two strains has been presented above. The predicted TFBDs of ICDH include binding motifs for Gis1; Gat1p, Gln3p, Gzf3p; and Gln3p all of which have been implicated in the regulation of gene expression under nutrients starvation, including amino acids and nitrogen limitations43–45.
Figure 4.
Evolutionary analyses of the ICDH protein and the upstream region of its gene among thirty-three strains of Trichosporonaceae. (a) ML tree of ICDH (380 amino acids long trimmed alignment) generated using IQ-TREE version 1.6.7 with confidence values based on 1,000 bootstrap replicates. (b) Distribution of predicted transcription factor binding sites 600 nucleotide bases upstream of the transcription initiation site of ICDH gene clustered using hierarchical clustering on principal components (HCPC) in R. The documented oil accumulating members of the family are indicated in blue fonts in the phylogeny and with blue arrows in the HCPC.
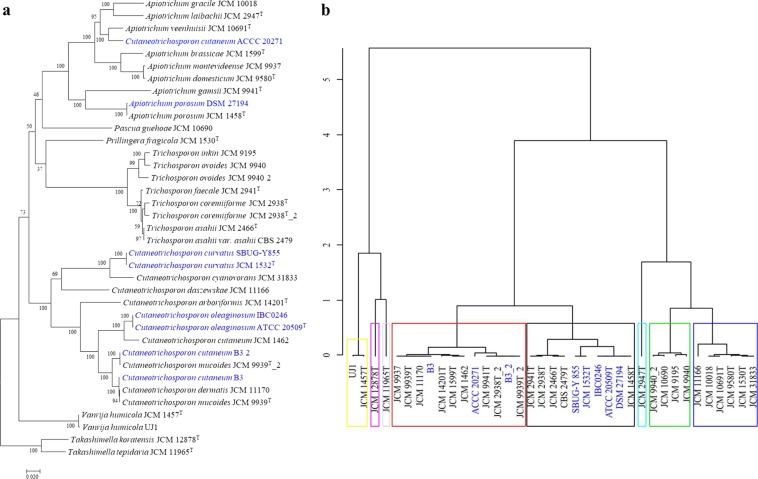
Suppression of ICDH, which is considered as a feature specific to oleaginous yeasts5 results in the accumulation of citrate in the mitochondrion. The citrate is then transferred into the cytoplasm where ACS catalyses its conversion into to acetyl-CoA and oxaloacetate. Evaluation of the ACS phylogeny (Fig. 5a) showed similar branching pattern with the SCOP. However, P. guehoae is placed within the well supported Cutaneotrichosporon clade. However, based on the TFBDs of ACS, the strains group into six distinct clusters (Fig. 5b) with two of the known oleaginous strains, A. porosum and C. curvatus, clustering together. In addition to the Gis1p, Msn2p, Msn4p, Rph1p, YER130C binding domains, which are known to regulate gene expression under nutrients limitation and stress45, the regulatory region of ACS includes the Adr1p TFBD. Adr1p is a carbon source-responsive transcription factor involved in the regulation of genes associated with ethanol, glycerol, and fatty acid utilization and peroxisome biogenesis46–48. As reflected in the characteristic clustering of A. porosum and C. curvatus, each of the strains carries two putative binding sites for Adr1p compared to C. oleaginosum which harbours four such TFBDS.
Figure 5.
Evolutionary analyses of the ACS protein and the upstream region of its gene among thirty-three strains of Trichosporonaceae. (a) ML tree of ACS (1,097 amino acids long trimmed alignment) generated using IQ-TREE version 1.6.7 with confidence values based on 1,000 bootstrap replicates. (b) Distribution of predicted transcription factor binding sites 600 nucleotide bases upstream of the transcription initiation site of ACS gene clustered using hierarchical clustering on principal components (HCPC) in R. The documented oil accumulating members of the family are indicated in blue fonts in the phylogeny and with blue arrows in the HCPC.
One of the products of the cleavage of citrate, acetyl-CoA, is either directly channelled to fatty acids synthesis via the FAS complex (catalysed by FASI &II) or converted into malonyl-CoA, which is subsequently directed to fatty acid synthesis. The latter reaction is catalysed by ACC. Incongruent with the SCOP, the ACC of Apiotrichum and Trichosporon species as well as those of Pascua guehoae and Prillingera fragicola appear to share similar evolutionary history clustering distinctly from the Cutaneotrichosporon species (Fig. 6a). The TFBDs of the ACC gene grouped the studied strains into eight distinct clusters (Fig. 6b). Based on this grouping, the five documented oleaginous yeasts assemble in two close clades. In addition to previously discussed putative sites for transcription factors regulating genes under nutrients limitation, adaptation to stress and utilisation of ethanol, glycerol, and fatty acid, the TFBDs of the ACC gene include a putative binding site for the zinc cluster protein Gsm1p and the basic helix-loop-helix transcription factor Pho4p. Gsm1p has been predicted to regulate energy metabolism49,50 while Pho4p was shown to be activated in response to phosphate limitation and controls genes of the phosphatase regulon and an inorganic phosphate (Pi) transport system in Saccharomyces cerevisiae51,52. Pi limitation has been used as an alternative means of inducing oil accumulation in oleaginous yeast53. The phylogeny generated based on FAS subunits (Supplementary Fig. 3c,e) revealed a clustering similar to that observed in the SCOP with exception of the placements of P. guehoae and P. fragicola in both trees and the distinct grouping of C. curvatus and C. cyanovorans in FASII (Supplementary Fig. 2,f). This may indicate a disparate evolution of the FASII genes in the latter strains. In terms of the TFBDs, the oleaginous strains group in separate clusters for both FASI & II (Supplementary Fig. 2d,f), indicating a more complex evolution of these genomic regions. However, the TFBDs of both genes include Gis1p, Msn2p, Msn4p, Rph1p, YER130C binding sites which are involved in gene regulation under nutrient starvation45 while the FASI regulatory region harbours Adr1p46–48 and Gsm1p49,50 binding domains and that of FASII includes Pho4p49,50 TFBDs. On the overall, the prediction of the TFBDs could serve as a preliminary approach for the genomic exploration and identification of potential oleaginous yeast.
Figure 6.
Evolutionary analyses of the ACC protein and the upstream region of its gene among thirty-three strains of Trichosporonaceae. (a) ML tree of ACC (2094 amino acids long trimmed alignment) generated using IQ-TREE version 1.6.7 with confidence values based on 1,000 bootstrap replicates. (b) Distribution of predicted transcription factor binding sites 600 nucleotide bases upstream of the transcription initiation site of ACC gene clustered using hierarchical clustering on principal components (HCPC) in R. The documented oil accumulating members of the family are indicated in blue fonts in the phylogeny and with blue arrows in the HCPC.
Clustering of the fungal isolates based on the regulatory regions of genes encoding the enzymes that determine oil production pathway may be useful in selecting strains with similar pattern of putative regulatory mechanisms for subsequent characterisation. Considering the TFBDs clustering pattern of ICDH and ACC, seven strains namely, A. porosum JCM 1458T, A. gamsii JCM 9941T, A. brassicae JCM 1599T, A. laibachii JCM 2947T, C. arboriformis JCM 14201T, C. mucoides JCM 9939T and C. dermatis JCM 11170 are closely grouped with the oil accumulating isolates in the two clusters. Whereas the Cutaneotrichosporon species may not be excellent candidates because of their association with human host, the Apiotrichum species, all of which are free-living and isolated from various environments (Table 1) could potentially be oleagenic. A. porosum JCM 1458T and A. gamsii JCM 9941T, are the closest relatives of the oleagenic A. porosum DSM 27194.
Conclusion
Here, we have analysed the genomes of thirty-three members of the Trichosporonaceae, including five yeast, A. porosum, C. curvatus, C. oleaginosum, C. cutaneum B3 and C. cutaneum ACCC 20271 for which data regarding substantial lipid accumulation are available. Analysis of the whole genome phylogeny based on single copy orthologs shows that certain strains incorporated in the genera Trichosporon and Cutaneotrichosporon belong to the genus Apiotrichum. This highlights the need for the use of appropriate genomic evaluation schemes in the course of genome deposition in various databases. Comparison of the proteomes of these strains suggests functional diversification consistent with the various lifestyles and isolation sources of the studied organisms. For instance, abundance of the various CAZYmes and MEROPS signified the potential capacity of the yeast to degrade a wide variety of biomass, with distinct enzyme sets linked to these capacities in free-living and host-associated taxa within the Trichosporonaceae. The evaluation of selected genes coding for proteins involved in lipid biosynthesis and their corresponding transcription factor binding domains suggests a complex evolution with some level of conservation for the TFBDs of ACC, ACS and ICDH among the well-studied oil accumulating members of the family Trichosporonaceae. This indicates a possible similarity in terms of the regulation of the genes encoding these enzymes among the clustered strains. Further work should focus on investigating the specific binding potentials of the predicted TFBDs and their potential roles in oil production and accumulation in oleaginous yeast. Taken together, this information could be harnessed towards the selection of strains with potential functional capabilities that could be explored for the generation of environment friendly bioproducts, including single cell oils, biopharmaceuticals, and various raw materials in the food industry.
Methods
Genome sequences, gene predictions and annotation
Thirty-five genomes, comprising those of thirty-three members of the family Trichosporonaceae and two from the family Tetragoniomycetaceae (outgroup strains) were incorporated in this study (Table 1). Genome annotation was accomplished using the Funannotate pipeline (v. 1.5.0–8f86f8c)54. In brief, small duplicate contigs (clean) were removed, size sorted and renamed (sort) and repeat contains were masked using RepeatMasker v4.0.7 prior to gene prediction and annotation. Gene models were predicted using Augustus v3.2.3, GeneMark-ES v4.35, Evidence modeler v1.1.1 and tRNAscan-SE v1.3.1. For all gene prediction the Augustus training set for ‘cryptococcus’ was used. The predicted proteins were functionally annotated using Interproscan v.5.30–69.0, eggNOG-mapper v1.0.3.3-g3e22728, PFAM v.31.0, UniProtKB 2018_07, MEROPS v12.0, CAZYme (dbCAN v6.0), phobius v1.01 and SignalP v4.1. The completeness of the studied genomes was determined using BUSCO v3.0.3.
Phylogenomic analysis
Single copy orthologues conserved among the predicted protein sequences of the thirty-three Trichosporonaceae and two outgroup strains were identified using Proteinortho524 using all default parameters except percent amino acid identity which was set at 40%. To restrict the phylogeny to single copy orthologs (SCOs), the analysis included only proteins occurring in single copies among the haploid genomes and strictly in two copies for the hybrid genomes. The subgenome SCOs complement for each hybrid genome was determined by BLASTP comparison of the duplicate SCOs with the corresponding SCOs of the closest relative non-hybrid genomes18,25. The orthologous proteins were aligned using T-coffee v11.00.8cbe48655,56. The resultant alignment was concatenated and trimmed using Gblocks v0.9b57,58 with -b5 = h. The trimmed alignment was used to construct a Maximum likelihood (ML) tree using IQ-TREE version 1.6.759 based on the LG + F + R10 model (predicted using IQ-TREE) and 1,000 bootstrap replicates.
Evolutionary analysis of oleagenic proteins and promoter regions of their genes
Orthologs of selected proteins that play a major role in the biochemical pathways of lipid production in yeasts were selected among the Trichosporonaceae and Tetragoniomycetaceae based on BLASTP (percent identify cutoff value of 40%) using Proteinortho524. Individual orthologous proteins were aligned using T-coffee v11.00.8cbe48655,56 and manually inspected to ensure accuracy of the alignments. The alignments were trimmed using Gblocks v0.9b57,58 and Maximum likelihood (ML) trees were generated using IQ-TREE version 1.6.759 with 1,000 bootstrap replicates. Bedtools v2.27.160 was employed to extract the regulatory regions of the genes encoding these proteins comprising 600 nucleotide bases upstream of the transcription initiation site. Each set of the regulatory regions was scanned for putative transcription factor binding domains (TFBDs) using the tools in YEASTRACT61, a database that curates the transcription factors (TF) and their target regulatory binding sites in Saccharomyces cerevisiae. The variation in the distribution of the TFBDs among the studied strains was used to group them using hierarchical clustering on principal components (HCPC) computed in R.
Supplementary information
Acknowledgements
H.A. acknowledges funding from Alexander von Humboldt Foundation. Bioeconomy International BMBF (grant #031B0452) supported O.G. We acknowledge support by Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of Karlsruhe Institute of Technology.
Author contributions
H.A. and K.O. conceived and designed the study. H.A., K.O., O.G., A.N. and P.D. analysed the data and wrote the manuscript. All the authors have reviewed and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Habibu Aliyu, Email: habibu.aliyu@partner.kit.edu.
Katrin Ochsenreither, Email: katrin.ochsenreither@kit.edu.
Supplementary information
is available for this paper at 10.1038/s41598-020-59672-2.
References
- 1.Liu XZ, et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in Mycology. 2015;81:85–147. doi: 10.1016/j.simyco.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu XZ, et al. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Studies in Mycology. 2015;81:1–26. doi: 10.1016/j.simyco.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gueho E, De Hoog G, Smith MT. Neotypification of the genusTrichosporon. Antonie Van Leeuwenhoek. 1992;61:285–288. doi: 10.1007/BF00713937. [DOI] [PubMed] [Google Scholar]
- 4.Sugita T, Nishikawa A, Shinoda T. Rapid detection of species of the opportunistic yeast Trichosporon by PCR. J. Clin. Microbiol. 1998;36:1458–1460. doi: 10.1128/JCM.36.5.1458-1460.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adrio JL. Oleaginous yeasts: Promising platforms for the production of oleochemicals and biofuels. Biotechnol. Bioeng. 2017;114:1915–1920. doi: 10.1002/bit.26337. [DOI] [PubMed] [Google Scholar]
- 6.Kourist R, et al. Genomics and Transcriptomics Analyses of the Oil-Accumulating Basidiomycete Yeast Trichosporon oleaginosus: Insights into Substrate Utilization and Alternative Evolutionary Trajectories of Fungal Mating Systems. mBio. 2015;6:e00918–00915. doi: 10.1128/mBio.00918-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratledge C, Wynn JP. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002;51:1–52. doi: 10.1016/S0065-2164(02)51000-5. [DOI] [PubMed] [Google Scholar]
- 8.Bracharz F, Beukhout T, Mehlmer N, Brück T. Opportunities and challenges in the development of Cutaneotrichosporon oleaginosus ATCC 20509 as a new cell factory for custom tailored microbial oils. Microbial Cell Factories. 2017;16:178. doi: 10.1186/s12934-017-0791-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papanikolaou S, Aggelis G. Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production. European Journal of Lipid Science and Technology. 2011;113:1031–1051. doi: 10.1002/ejlt.201100014. [DOI] [Google Scholar]
- 10.Gorte O, Aliyu H, Neumann A, Ochsenreither K. Draft Genome Sequence of the Oleaginous Yeast Apiotrichum porosum (syn. Trichosporon porosum) DSM 27194. Journal of genomics. 2019;7:11. doi: 10.7150/jgen.32210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulze I, et al. Characterization of newly isolated oleaginous yeasts - Cryptococcus podzolicus, Trichosporon porosum and Pichia segobiensis. AMB Express. 2014;4:24. doi: 10.1186/s13568-014-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun MK, et al. Catalytic decomposition of the oleaginous yeast Cutaneotrichosporon oleaginosus and subsequent biocatalytic conversion of liberated free fatty acids. ACS Sustainable Chemistry & Engineering. 2019;7:6531–6540. doi: 10.1021/acssuschemeng.8b04795. [DOI] [Google Scholar]
- 13.Madani M, Enshaeieh M, Abdoli A. Single cell oil and its application for biodiesel production. Process Saf. Environ. Prot. 2017;111:747–756. doi: 10.1016/j.psep.2017.08.027. [DOI] [Google Scholar]
- 14.Ochsenreither K, Glück C, Stressler T, Fischer L, Syldatk C. Production strategies and applications of microbial single cell oils. Frontiers in microbiology. 2016;7:1539. doi: 10.3389/fmicb.2016.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goffeau A, et al. Life with 6000 Genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 16.Jenuth J. The NCBI. Publicly available tools and resources on the Web. Methods Mol. Biol. 2000;132:301–312. doi: 10.1385/1-59259-192-2:301. [DOI] [PubMed] [Google Scholar]
- 17.Takashima M, et al. Recognition and delineation of yeast genera based on genomic data: Lessons from Trichosporonales. Fungal Genet. Biol. 2019;130:31–42. doi: 10.1016/j.fgb.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Takashima M, et al. A Trichosporonales genome tree based on 27 haploid and three evolutionarily conserved ‘natural’ hybrid genomes. Yeast. 2018;35:99–111. doi: 10.1002/yea.3284. [DOI] [PubMed] [Google Scholar]
- 19.Takashima M, et al. Selection of Orthologous Genes for Construction of a Highly Resolved Phylogenetic Tree and Clarification of the Phylogeny of Trichosporonales Species. PLOS ONE. 2015;10:e0131217. doi: 10.1371/journal.pone.0131217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grigoriev IV, et al. Fueling the future with fungal genomics. Mycology. 2011;2:192–209. doi: 10.1080/21501203.2011.584577. [DOI] [Google Scholar]
- 21.Fuentefria AM, et al. Trichosporon insectorum sp. nov., a new anamorphic basidiomycetous killer yeast. Mycol. Res. 2008;112:93–99. doi: 10.1016/j.mycres.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Lischer HEL, Shimizu KK. Reference-guided de novo assembly approach improves genome reconstruction for related species. BMC Bioinformatics. 2017;18:474. doi: 10.1186/s12859-017-1911-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterhouse RM, et al. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lechner M, et al. Proteinortho: Detection of (Co-) orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sriswasdi S, et al. Global deceleration of gene evolution following recent genome hybridizations in fungi. Genome Res. 2016;26:1081–1090. doi: 10.1101/gr.205948.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Z, Liu H, Wang C, Xu J-R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14:274–274. doi: 10.1186/1471-2164-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park Y-J, Jeong Y-U, Kong W-S. Genome Sequencing and Carbohydrate-Active Enzyme (CAZyme) Repertoire of the White Rot Fungus Flammulina elastica. International journal of molecular sciences. 2018;19:2379. doi: 10.3390/ijms19082379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rytioja J, et al. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiology and molecular biology reviews: MMBR. 2014;78:614–649. doi: 10.1128/MMBR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neurath H. Proteolytic enzymes, past and future. Proceedings of the National Academy of Sciences. 1999;96:10962–10963. doi: 10.1073/pnas.96.20.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Da Silva RR. Bacterial and fungal proteolytic enzymes: production, catalysis and potential applications. Appl. Biochem. Biotechnol. 2017;183:1–19. doi: 10.1007/s12010-017-2427-2. [DOI] [PubMed] [Google Scholar]
- 32.Palmer JM, Drees KP, Foster JT, Lindner DL. Extreme sensitivity to ultraviolet light in the fungal pathogen causing white-nose syndrome of bats. Nature Communications. 2018;9:35. doi: 10.1038/s41467-017-02441-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawlings ND, et al. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2017;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlings ND, Barrett AJ, Bateman A. Asparagine Peptide Lyases A seventh catalytic type of proteolytic enzymes. J. Biol. Chem. 2011;286:38321–38328. doi: 10.1074/jbc.M111.260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Silva RR. Commentary: Fungal lifestyle reflected in serine protease repertoire. Frontiers in microbiology. 2018;9:467–467. doi: 10.3389/fmicb.2018.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva RRd, Cabral TPdF, Rodrigues A, Hamilton C. Production and partial characterization of serine and metallo peptidases secreted by Aspergillus fumigatus Fresenius in submerged and solid state fermentatio. Braz. J. Microbiol. 2013;44:235–243. doi: 10.1590/S1517-83822013000100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muszewska A, et al. Fungal lifestyle reflected in serine protease repertoire. Scientific Reports. 2017;7:9147. doi: 10.1038/s41598-017-09644-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atkinson HJ, Babbitt PC, Sajid M. The global cysteine peptidase landscape in parasites. Trends Parasitol. 2009;25:573–581. doi: 10.1016/j.pt.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellou S, et al. Microbial oils as food additives: recent approaches for improving microbial oil production and its polyunsaturated fatty acid content. Curr. Opin. Biotechnol. 2016;37:24–35. doi: 10.1016/j.copbio.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Gomma AE, Lee S-K, Sun SM, Yang SH, Chung G. Improvement in Oil Production by Increasing Malonyl-CoA and Glycerol-3-Phosphate Pools in Scenedesmus quadricauda. Indian J. Microbiol. 2015;55:447–455. doi: 10.1007/s12088-015-0546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Xu R, Wang R, Haque ME, Liu A. Overexpression of ACC gene from oleaginous yeast Lipomyces starkeyi enhanced the lipid accumulation in Saccharomyces cerevisiae with increased levels of glycerol 3-phosphate substrates. Biosci. Biotechnol. Biochem. 2016;80:1214–1222. doi: 10.1080/09168451.2015.1136883. [DOI] [PubMed] [Google Scholar]
- 42.Wynn JP, Hamid AA, Li Y, Ratledge C. Biochemical events leading to the diversion of carbon into storage lipids in the oleaginous fungi Mucor circinelloides and Mortierella alpina. Microbiology. 2001;147:2857–2864. doi: 10.1099/00221287-147-10-2857. [DOI] [PubMed] [Google Scholar]
- 43.Pedruzzi I, Bürckert N, Egger P, De Virgilio C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. The EMBO journal. 2000;19:2569–2579. doi: 10.1093/emboj/19.11.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coffman JA, Rai R, Cunningham T, Svetlov V, Cooper TG. Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen-catabolic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:847–858. doi: 10.1128/mcb.16.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orzechowski Westholm J, et al. Gis1 and Rph1 Regulate Glycerol and Acetate Metabolism in Glucose Depleted Yeast Cells. PLOS ONE. 2012;7:e31577. doi: 10.1371/journal.pone.0031577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gurvitz A, et al. Saccharomyces cerevisiae Adr1p Governs Fatty Acid β-Oxidation and Peroxisome Proliferation by RegulatingPOX1 and PEX11. J. Biol. Chem. 2001;276:31825–31830. doi: 10.1074/jbc.M105989200. [DOI] [PubMed] [Google Scholar]
- 47.Gurvitz A. A novel circuit overrides Adr1p control during expression of Saccharomyces cerevisiae 2-trans-enoyl-ACP reductase Etr1p of mitochondrial type 2 fatty acid synthase. FEMS Microbiol. Lett. 2009;297:255–260. doi: 10.1111/j.1574-6968.2009.01688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young ET, et al. Characterization of a p53-related Activation Domain in Adr1p That Is Sufficient for ADR1-dependent Gene Expression. J. Biol. Chem. 1998;273:32080–32087. doi: 10.1074/jbc.273.48.32080. [DOI] [PubMed] [Google Scholar]
- 49.Todd RB, Andrianopoulos A. Evolution of a Fungal Regulatory Gene Family: The Zn(II)2Cys6 Binuclear Cluster DNA Binding Motif. Fungal Genet. Biol. 1997;21:388–405. doi: 10.1006/fgbi.1997.0993. [DOI] [PubMed] [Google Scholar]
- 50.Deng Y, et al. Computationally analyzing the possible biological function of YJL103C-an ORF potentially involved in the regulation of energy process in yeast. Int. J. Mol. Med. 2005;15:123–127. [PubMed] [Google Scholar]
- 51.Zhou X, O’Shea EK. Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol. Cell. 2011;42:826–836. doi: 10.1016/j.molcel.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogawa N, Oshima Y. Functional domains of a positive regulatory protein, PHO4, for transcriptional control of the phosphatase regulon in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:2224–2236. doi: 10.1128/mcb.10.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, et al. Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnology for Biofuels. 2018;11:148. doi: 10.1186/s13068-018-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Funannotate: pipeline for genome annotation (2016).
- 55.Magis, C. et al. In Multiple Sequence AlignmentMethods 117–129 (Springer, 2014).
- 56.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 57.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 58.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt HA, Minh BQ, von Haeseler A, Nguyen L-T. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teixeira MC, et al. YEASTRACT: an upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res. 2017;46:D348–D353. doi: 10.1093/nar/gkx842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.