Abstract
The white leg Litopenaeus vannamei shrimp is of importance to the eastern Pacific fisheries and aquaculture industry but suffer from diseases such as the recently emerged early mortality syndrome. Many bacterial pathogens have been identified but the L. vannamei microbiota is still poorly known. Using a next-generation sequencing (NGS) approach, this work evaluated the impact of the inclusion in the diet of mannan oligosaccharide, (MOS, 0.5% w/w), over the L. vannamei microbiota and production behavior of L. vannamei under intensive cultivation in Ecuador. The MOS supplementation lasted for 60 days, after which the shrimp in the ponds were harvested, and the production data were collected. MOS improved productivity outcomes by increasing shrimp survival by 30%. NGS revealed quantitative differences in the shrimp microbiota between MOS and control conditions. In the treatment with inclusion of dietary MOS, the predominant phylum was Actinobacteria (28%); while the control group was dominated by the phylum Proteobacteria (30%). MOS has also been linked to an increased prevalence of Lactococcus- and Verrucomicrobiaceae-like bacteria. Furthermore, under the treatment of MOS, the prevalence of potential opportunistic pathogens, like Vibrio, Aeromonas, Bergeyella and Shewanella, was negligible. This may be attributable to MOS blocking the adhesion of pathogens to the surfaces of the host tissues. Together, these findings point to the fact that the performance (survival) improvements of the dietary MOS may be linked to the impact on the microbiota, since bacterial lines with pathogenic potential towards shrimps were excluded in the gut.
Subject terms: Microbiome, Biological sciences
Introduction
The “white leg” Litopenaeus vannamei shrimp is originally from waters of the equatorial Pacific east coast. Commercial cultivation began in Ecuador in 1968 in the province El Oro where this study was performed1. The shrimp industry has also been significant in other regions, such as the Middle East, Southeast Asia, the Indian Subcontinent and China. The production of the shrimp industry in Latin American countries such as Brazil, Honduras, Mexico and Ecuador, is relevant within the aquaculture economic context in the region2. Despite this apparent success in terms of production expansion, global shrimp production continues to suffer significant losses due to the effects of a wide range of diseases3. In recent years, Acute Hepatopancreatic Necrosis Syndrome (AHPNS), has impacted L. vannamei production areas worldwide, leading to major economic losses and significantly affecting production4. This disease is caused by Vibrio parahaemolyticus, which colonizes the digestive tract and releases toxins that affect the liver and pancreas5.
Traditionally, fight diseases strategies in aquatic cultures have been based on the use of antibiotics and chemotherapeutics; currently, however, the biosafety directives of markets and the environmental legislation of producing countries have limited this approach. The employment of antibiotics for the control of the mass outbreaks of bacterial diseases in the cultivation of shrimp has raised the possibility of the emergence of bacterial strains resistant to antibiotics due to the selective pressure exerted by antibiotic residues in environment (e.g., bottoms, sediments) and by the unintentional exposure of cultivated or wild shrimp to antibiotics6–9. Currently, it is widely accepted that prevention is more advisable than treatment; thus, strategies have been developed to modify the intestinal microbiota with the goals of promoting colonization of beneficial bacteria and of preventing the colonization of potentially pathogenic bacteria9,10.
Considering the intestinal microbiota as the microbial ecosystem colonizing the digestive tract, which serves as the primary interaction surface between the external environment and the internal environment of any organism, numerous investigations in humans and other vertebrates have led to the comprehension of the essential importance of microbiota to health and welfare. In fish cultivation, evidence has been found regarding the contributions of microbiota to various aspects of the production of enzymes to improve nutrient availability and the competitive exclusion of potential pathogenic bacteria10–16. Other studies have provided evidence about the of microbiota´s function in host nutrition, specific tissues proliferation and immune mechanism regulation17–20. For example, bacterial strains derived from intestinal microbiota such as Pediococcus acidilactici21–23 and Bacillus subtilis24–26 have been tested in various aquaculture species and shown beneficial effects Thes.e advances have fostered a growing interest in microbiota modulation and their consequences on the productivity performance of animals under cultivation9,10,27. These supplements may be (i) probiotics, defined by Merrifield et al.28 as any microorganism supplied through the feed or culture water that benefits the host; (ii) prebiotics, defined by Bindels et al.29 as an indigestible compound which, via its metabolic breakdown by microorganisms in the intestine, acts on the profile and functionality of the intestinal microbiota, giving it a positive action on the host as an improvement of food growth and efficiency; or iii) symbiotic, which are defined by Gibson & Roberfroid (1995)30 as nutritional supplements containing a prebiotic and a probiotic that beneficially influence the host by acting on the survival and implantation of live microorganisms as dietary supplements in the gastrointestinal tract.
Mannan Oligosaccharides (MOS) are glucides obtained from the Saccharomyces cerevisiae yeast cell31,32. The use of MOS to block pathogen colonization derives from the conception that certain polysaccharides could be used to block the mechanism of recognition and adhesion of potential pathogens to molecules on the surfaces of host tissues (competition for attachment sites). This action would reduce the adhesion of the pathogens to the digestive tract, leaving them to be excreted in the feces. This may lead to the improvement of the integrity and performance of the intestinal epithelial barrier16.
There are no previous publications using deep sequencing (next generation sequencing, NGS) to address effects of MOS in L. vannamei microbiota. Several investigations have reported the effects of MOS on crustacean culture performance including parameters such as growth rates, survival, hemocyte proliferation, polyphenoloxidase activity, and changes in gastrointestinal (GI) morphology. In terms of microbiological analyses, only quantitative variation of aerobic bacteria has been described33–43. However, these previous investigations were conducted under laboratory conditions and involved only cultivable bacteria. Therefore, these observations have not been validated at a commercial farming scale, limiting the application of those prebiotics.
For cultivated crustaceans such as L. vannamei, information on the functionality and structuring of microbiota is very limited. Latterly, advances in NGS have allowed develop research on crustaceans microbiota, as recently addressed in some studies44–48. These investigations have been based on modulating the L. vannamei microbiota through nutritional challenges in laboratory conditions. However, information is lacking regarding the microbiota-host relationship, including evidence of cause and effect relationships on the physiology of the host and consequences for productivity. Considering this deficiency, our research was aimed to use NGS to assess the effects of the dietary inclusión of a prebiotic (MOS) on both: production parameters in intensive cultivation conditions and composition of the intestinal microbiobiota of L. vannamei in Ecuador.
Results
Productivity response indexes to the inclusion of MOS
The shrimp population in all the ponds (MOS and control) was completely removed because the entire biomass was harvested for the market. This action was coordinated with the new moon phase because this was the period during which the largest proportion of the shrimp was in Stage C (anecdysis) of the molting cycle49. The influence of the lunar cycle on L. vannamei commercial culture, has long been recognized in Ecuador from the observation of molting patterns and activity cycles in aquaculture production ponds, and its follow-up is financially relevant due to the fact that soft shrimp cause high post-harvest handling losses50. Therefore, because ecdysis corresponds to the renewal of the exoskeleton during the molting cycle, for practical reasons, the harvest should be performed during anecdysis51. Cultivation outcomes (Fig. 1) revealed better performance in ponds whose populations received feed supplemented with MOS, based on the total harvested biomass (B) and the performance by cultivation area (R). The comparative analysis in Table 1 reflects significant increases in B and R associated with the treatment with 0.5% MOS and highlights a 34% increase in survival (S). Meanwhile, the productivity response, expressed in terms of average weight (W), food conversion (CV) and weight gain (WG), exhibited no significant dissimilarities (p > 0.05).
Figure 1.

Productivity outcomes. Graphic representation of the average harvested biomass (Kg) and yield (Kg/Ha) of L. vannamei in ponds supplemented with mannan oligosaccharides (MOS) and ponds with a commercial diet (Control).
Table 1.
Indexes used for evaluation of the productivity outcome. (W; CV; WG; B; R), Welch’s t-test74. (S), Contingency tables using Pearson’s X2 test75. Source of data: Industrial biomass payment report.
| Productive Performance Indicators | 0.5% MOS | Control | P value |
|---|---|---|---|
| Average weight W (g) | 15.6 ± 0.6 | 15.5 ± 0.7 | 0.978 |
| Food Conversion CV | 1.2 ± 0.1 | 1.63 ± 0.02 | 0.081 |
| Weight Gain WG (g/d) | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.861 |
| Average Biomass B (kg) | 4830 ± 189 | 2565 ± 201 | 0.028* |
| Yield R (kg/Ha) | 9660 ± 378 | 5131 ± 402 | 0.028* |
| Survival Rate S (%) | 70.3 ± 6.6 | 37.5 ± 4.7 | 2.2E-16** |
16S rRNA gene sequencing
Sequencing of the 16S amplicons with Ion Torrent technology provided a total of 1,303,492 sequences, having an average-quality ratio by sequence (Phred Score) of 29, corresponding to a base call accuracy of 99.9% (Supplementary Table 1) plus a probability of error of 0.00126. The data of the sequences were deposited in the Sequence Read Archive of the National Center for Biotechnological Information (SRA, NCBI) in the framework of the BioProject (PRJNA352369). After dereplication and removal of chimeras, singletons and Archaea sequences, were a global of 470,639 sequences of high quality (Table 2), that were classified to a total of 2065 operational taxonomic units (OTUs), 97% of which were classified by sequence identity. Bacteria were the focus of this study, and archaeal sequences were detected unintentionally due to mismatches in the primers used. The rarefaction curves for OTUs observed and the Chao1 alpha diversity index approached the saturation phase, indicating good sample coverage, while the asymptotic distribution of the curves implies that they are comparable (Fig. 2).
Table 2.
Numbers of sequences per sample.
| Pond | Condition | Sequences/sample |
|---|---|---|
| P2 | MOS | 40,814 |
| P2 | MOS | 42,611 |
| P2 | MOS | 46,966 |
| P6 | MOS | 30,625 |
| P6 | MOS | 36,558 |
| P6 | MOS | 42,725 |
| P10 | Control | 56,553 |
| P10 | Control | 70,436 |
| P10 | Control | 103,351 |
| Total | 470,639 |
Figure 2.
Rarefaction curves. (a) Alpha diversity index Chao1: Species richness estimators based upon the number of rare OTUs found in a sample. (b) OTUs observed: Richness is expressed as the number of observed OTUs. Curves were averaged by ■ Control, ■ 0.5% MOS for a sampling depth of 29,000 sequences. Errors bars represent standard error.
Alpha diversity
The alpha diversity of the microbiota was slightly higher in shrimps that were fed with the control diet compared to shrimps that were fed with the MOS diet 0.5% (Fig. 2). However, this difference was not significant (Table 3).
Table 3.
Comparison of the alpha diversity in the microbiota of L. vannamei supplemented with MOS (nonparametric t-test) for a sampling depth of 29,000 sequences.
| Process | Control mean | MOS mean | t stat | P-value |
|---|---|---|---|---|
| Chao1 | 830 | 795 | 0.491 | 0.639 |
| OTUs Observed | 671 | 636 | 0.866 | 0.398 |
Beta diversity
The non-directional variation of the beta diversity in a community is a measurement of the change of the community structure as a response to environmental or experimental factors52. Beta diversity linked to L. vannamei bacterial communities in the two conditions, control and 0.5% MOS dietary supplementation was investigated through PCoA (Fig. 3). The first 2 categories cover a total of 54.26% of the variation (first component, 29.3%; second component, 24.9%). The results of the similarity analysis test (ANOSIM), with R = 0.6 ≈ 1 (p = 0.016), showed that there are significant variations among the microbiota of the animals treated with MOS and those of the control animals. The multivariate analysis of the taxa that support this diversity is described in the following sections.
Figure 3.

Principal component analysis (PCoA) normalized distribution plot. Blue dots correspond to microbiota of shrimp supplemented with MOS, and red dots correspond to microbiota of shrimp fed the commercial diet as described in Material and Methods.
Comparison of the intestinal microbiota
The prevailing sequences linked to MOS treatment at the Phylum level were mostly Actinobacteria (28%), Proteobacteria (20%), Verrucomicrobia (13%), Chloroflexi (7%) and Firmicutes (6%), whereas the microbiota in the control group were dominated by Proteobacteria (30%), Bacterioidetes (22%), Actinobacteria (11%), Chloroflexi (10%) and Firmicutes (5%) (Fig. 4). In total, 39 phyla were detected.
Figure 4.
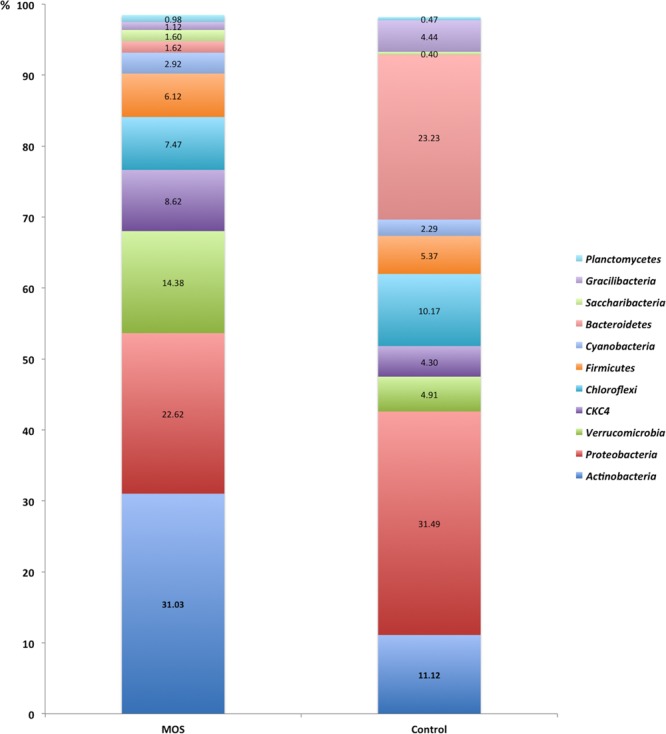
Comparison of relative abundance at the phylum level. Shrimp microbiota composition (relative to OTU composition) at the phylum level. Comparison between Control and MOS shrimp microbiota including the 11 phyla showing the highest abundance. Taxonomic summary of the observed relative abundance of abundant phyla across all samples divided by culture condition.
Identification by taxonomic analysis allocated a total of 847 genera. The most abundant genera under MOS treatment were uncultured Xanthomonadales (9%), uncultured CKC4 (9%), uncultured Chthoniobacterales LD29 (8%), uncultured Propionibacteriaceae (6%) and uncultured Caldilineaceae (5%). In contrast, the control shrimp microbiota was composed by Bergeyella (10%), uncultured Caldilineaceae (9%), Shewanella (7%) and Microvirga (6%) (Fig. 5).
Figure 5.
Comparison of the relative abundance of the most abundant genera between Control and MOS shrimp microbiota. Microbiota composition (relative to OTUs composition) at genus level, between Control and MOS supplemented microbiota. Taxonomic summary of observed relative abundance of abundant genera across all samples divided by culture condition.
Identifying OTUs that differed significantly between treatments
The analysis of the differential abundance considered only those OTUs with more than 100 sequences. This analysis revealed a total of 4 OTUs that were present at significantly higher levels in the MOS treatment than in the Control (p < 0.05), as follows: OTUs 62 and 43, uncultured Verrucomicrobiaceae; OTU 14, Lactococcus; and OTU 86, Chlamydia. OTUs with significantly lower presence in the microbiota of shrimp fed MOS than in control shrimp were as follows: OTU 7, Shewanella sp.; and OTU 1, Bergeyella (Table 4).
Table 4.
OTUs with differential abundance levels observed in the analysis of differential abundance based on MetagenomeSeq’s fitZIG algorithm86 for OTUs with more than 100 counts.
| OTU | MOS 0.5% | Control | FisherP | FisherAdjP | AdjPvalues | Taxonomy |
|---|---|---|---|---|---|---|
| OTU_14 | 6,945 | 380 | 1 | 1 | 0.046 | Lactococcus |
| OTU_43 | 1,990 | 12 | 0.083 | 1 | 0.046 | Uncultured Verrucomicrobiaceae |
| OTU_62 | 1214 | 4 | 0.083 | 1 | 0.046 | Uncultured Verrucomicrobiaceae |
| OTU_86 | 611 | 3 | 0.083 | 1 | 0.046 | Chlamydia psittaci 6BC |
| OTU_1 | 28 | 23,154 | 1 | 1 | 0.046 | Uncultured Bergeyella |
| OTU_7 | 9 | 15,376 | 0.5 | 1 | 0.046 | Shewanella sp. MOLA 59 |
Linear discriminant analysis (LDA) effect size (LEfSe)
Using LEfSe, microbiota were compared between the Control and MOS treatments in order to identify taxa consistently associated with each condition (MOS or Control) that consistently explain their differences based on the effect size. The results of this analysis are shown in Figs. 6 and 7 and reflect the fact that MOS treatment significantly promoted 27 taxa (LDA > 2.5). Among these groups are highlighted taxa for the phylum Actinobacteria, including the orders Micrococcales, Corynebacteriales and Frankiales and the genus Actinomadura. In contrast, only 12 taxa were associated with the control group, and these taxa mainly belonged to the phylum Proteobacteria. The taxa associated with potential crustacean pathogens, such as the order Aeromonadales, the family Aeromonadaceae, the species Aeromonas caviae and the order Vibrionales, family Vibrionaceae and genus Vibrio, stood out distinctly.
Figure 6.
Circular cladogram information of results from LEfSe, reporting OTUs identified with significant differences p < 0.05; LDA > 2.5. Cladogram illustrating the phylogenetic relationship amongst the significantly differentiating bacterial taxa. The dots in the center present the OTUs at phylum level, whereas the outer circle of dots present the OTUs at genus level. The colors of the dots indicate the treatment (MOS or Control diet) in which the respective OTUs are most abundant. The explanation of the colors is given in the upper left corner. Yellow color indicates OTUs that showed similar abundance in both treatments. Phyla, classes, orders, families and genera that were significantly different between treatments are named along the right side of the figure.
Figure 7.

Graphical summary of LEfSe results. Comparison of LDA effect size of the significantly differentiating bacterial taxa. The histogram shows the LDA scores computed for significantly abundant taxa between Control and MOS shrimp microbiota. The histogram represents the most significantly abundant taxa between MOS and Control shrimp microbiota. The explanation of the colors is given in the upper left corner.
Discussion
To the best of our knowledge, this article presents the first description of L. vannamei microbiota´s diversity and taxa composition, under MOS dietary treatment in a commercial aquaculture facility. There are no previous NGS-based reports addressing the effect of MOS on the microbiota of L. vannamei. Productivity parameters have been reported previously, but those reports have been limited to laboratory conditions at the experimental level. In one study with 1080 shrimp distributed in 36 1-m3 tanks, Zhang et al.40 reported a 66% increase in weight gain (WG; P < 0.05) in the group treated with 0.2% MOS. In another study with 270 shrimp in 18 0.128-m3 aquariums, Genc & Ebeoglu 201341 reported a 17% increase in survival (S) after increasing MOS to 0.4%, but these results relied on the salinity of the environment (38‰). Our results under intensive commercial cultivation conditions, with 1,780,000 juvenile L. vannamei in 4 0.5-ha ponds, 5‰ salinity and 0.5% MOS, contrast with these laboratory conditions because, unlike the report by Zhang et al.40, we found no significant differences in growth parameters between the animals supplemented with MOS and the control treatment. Our increase in harvested biomass (B) in the pond that received the 0.5% MOS treatment was a response to the significant increase in S (>30%). The observed effect matches results from S Genc & Ebeoglu, 201341, but our results obtained at 5‰ contrasted with those in the previous study, which reported a positive response only to salinities greater than 38‰.
The promotion of changes in the composition of the intestinal microbiota is a key component of the case for using MOS as prebiotics, but this aspect has also been one of the least developed. The composition of the microbiota of shrimp under control conditions is consistent with previous research on this species, agreeing that Proteobacteria is the dominant phylum, with relative abundance levels between 28 and 70%44–48,53–57. Other phyla showing high abundance under control conditions were Bacteroidetes (22%) and Actinobacteria (11%), whose values matched those in some of the previous studies; however, overall, those studies showed variable relative abundance levels.
The effect of the inclusion of MOS on the microbiota of shrimp is reflected in the relative abundance at the phylum level, with the predominance of Actinobacteria over Proteobacteria and without any previous background about the displacement of the phylum Proteobacteria by the phylum Actinobacteria in association with a dietary intervention in this species. However, using NGS, it has been reported for L. vannamei that dietary supplementation with the supernatant from the bacterial strain HC-246 or with cornstarch as a source of carbohydrates45 leads to changes associated with increases in Actinobacteria and Firmicutes, respectively, without affecting the position of Proteobacteria as the dominant phylum.
Previous studies of the effect of the inclusion of MOS on microbiota are based on measurements of cultivable bacteria counts, and most of those studies are focused on fish. There is no published work on the modulation of microbiota in L. vannamei by dietary MOS. However, reports in other crustaceans have described reductions in the counts of Vibrio36,37,42. This reduction in colonization of the digestive tract by potential pathogens would lead to an improvement in the integrity and functionality of the epithelial intestinal barrier16. Consequently, the absence of potential pathogens in MOS-treated shrimp promoted the presence of uncultured Verrucomicrobiaceae and Lactococcus. Competition between Aeromonas and Lactococcus for binding sites has been previously demonstrated in vitro58. Furthermore, genomes of Lactococcus spp. have revealed the presence of adhesins associated with their permanence in the gut59. Considering the impact of AHPNS on the production of L. vannamei worldwide4, our results showing the reduction of the genus Vibrio to negligible levels in a commercial culture facility are of particular relevance; previous studies only involved laboratory-scale assays36,37,42. This work allows us to infer that MOS could constitute an important tool for preventing AHPNS, which remains one of the most interesting open fields for exploration based on our results.
The change in the intestinal microbiota is reflected in the improved survival and better productivity outcomes and is consistent with the concept that Actinobacteria produce beneficial secondary metabolites for the host, such as antimicrobial factors and growth promoters4,60,61. There are numerous publications on the potential use of Actinobacteria as probiotics4,62–65, but they have all focused on the genera Streptomyces and Lactococcus. Meanwhile, Xiong et al., 201654 suggest that the selection of probiotics should favor the specific microbiota of each species to provide a greater probability of intestinal colonization. Given the results obtained here, potential probiotics belonging to the genera Actinomadura, Fodinicola and Agromyces should be evaluated for their abundance and association with MOS (Fig. 7). On the other hand, it has been reported that the presence of Verrucomicrobiaceae is related to the recovery of the functionality of the intestinal epithelial barrier, since these bacteria use complex oligosaccharides as substrates for fermentation66. The limitation of this group may be that isolates are difficult to obtain.
The primers used in the study (341F, 518R) correspond to conserved regions that have been evaluated in silico for Illumina and Ion Torrent sequencing67. This study revealed coverage of 96.7% in Bacteria, 44.6% in Archaea and 0.2% in Eucarya while not detecting the candidate divisions OP11 and WS6 or Armatimonadetes. Those undetected phyla have not been mentioned in any previous report about the intestinal microbiota of the L. vannamei. Therefore, we consider that with the filtering of the sequences assigned to Archaea and Eucarya, we obtained a widely representative library. However, the detection of Archaea may open a new field of exploration in the description of shrimp microbiota. The abundance of Archaea (10%) may imply that these organisms play important roles within the microbiota of shrimp. For example, one of the detected archaeal taxa corresponded to Thaumarchaeota, which includes microorganisms involved in the nitrogen cycle, a critical parameter for shrimp health68. However, this observation must be verified using the proper primers to describe this domain.
In 2010, Daniels et al.36 stated that the action of MOS stabilizes the composition of the microbiota and partly suppresses the variations and influxes of new bacterial strains from the environment. This statement is fully consistent with the results of our diversity analysis and may be extended to say that the action of the MOS also controls the influx of bacterial strains with potential pathogenicity for L. vannamei. The key innovation of this research can be summarized as the first approach through NGS to the management of the L. vannamei’s microbiota modification under commercial cultivation conditions.
Materials and Methods
Experimental animals and environmental conditions
The study was performed at the Santa Ana intensive shrimp farm, located at 3° 31′ 10.7′′S, 80°10′ 29.74′′W, belonging to canton Huaquillas, El Oro Province, Ecuador. Santa Ana is located in the dry thorn scrub zone of the province of El Oro, between Arenillas and Huaquillas, in the altitudinal range 0–50 meters above sea level69. Approximately 1,780,000 juvenile L. vannamei with an average initial weight of 2.2 ± 0.53 g (calculated by routine weekly population sampling in culture procedures, n = 4000) from a breeding pond at the shrimp farm were distributed in 4 pools of 0.5 ha each with the temperatures controlled between 32 °C and 34 °C, salinity at 5‰, oxygen saturation above 60%, over 60 days of cultivation. To minimize the factors that could influence the results of the experiment, we standardized conditions such as maturation stage and the conditions of the ponds in order to avoid interfering with the culture population, since a significantly representative periodic monitoring of the microbiota of culture population would require intervention in the biomass with the periodic slaughter of a large number of individuals cultivated for market. The ponds are only linked hydrologically by a common water source, with independent inputs. The ponds were constructed under greenhouse conditions, including the use of a geotextile liner for impermeabilization. Environmental variables have been parameterized throughout the culture production tracking protocol in intensive culture systems. Under commercial production conditions, there is less control of environmental variables than under laboratory conditions or experimental production, so the management protocol seeks to standardize the conditions in the production pools. We used the most common commercial feed formula, whose standardized proximal composition has been published by the manufacturer (Nicovita Classic Camarón Vitapro, Callao, Peru; Bio-Mos®, Alltech Inc., Nicholasville, KY, US).
Food and treatment
The shrimps were fed 4 times per day (08:00, 11:00, 14:00 and 17:00), cultured for 59 days, until they reached their harvest average weight (total body weight W ± 14 g; calculated by routine weekly sampling, n = 1000), with feed adjustments according to demand at the trough. A 2.0 mm commercial feed from Nicovita Classic Camarón (Vitapro, Callao, Peru) was used; the composition reported by the manufacturer was 35% protein, 5% fat, 10% ash, fiber 5% and 12% moisture. The inclusion of 0.5% (w/w) of MOS (Bio-Mos®, Alltech Inc., Nicholasville, KY, US) to the feed was through the dilution of MOS in distilled water (0.125 g/mL) plus commercial gelatin (0.125 g/mL), sprayed onto the feed (0.04 mL/g) in the mechanical hopper for homogenization. The food for the two control pools was prepared with the same protocol with the addition of commercial gelatin without MOS.
Sampling procedure
Complete harvesting is usually undertaken by using a bag net installed at the drainage gate of the pond. Taking advantage of the positive phototactic behavior of the species during the night, shrimps are attracted by a lamp to the drained water stream and collected at the bag net, until the pond is dry. At 60 days of cultivation, based on synchronization with the ecdysis period, the total capture in the ponds was conducted, and the total biomass harvested per pond was quantified (B). Characterization of the intestinal tract microbiota was based on the sampling performed during the harvest that included 200 shrimp were collected per test pond, 1 Control pond and 2 replicate ponds of the MOS condition; the weight (W) was recorded for each individual, and the digestive tracts were dissected under aseptic conditions for all individuals in the samples. The removed digestive tracts were divided into 3 groups per pond, homogenized for the extraction of DNA and immediately preserved at −40 °C. In our study, we chose to increase the number of individuals because this is commercial culture with a total of 1.7 x 106 shrimps. Hence, pools included 200 shrimps to obtain a more comprehensive representation of the microbiota and its variations. The extracted digestive tracts were pooled and homogenized by pond, according to the observations described in previous publications; in which pooled samples and individual samples were similar in terms of microbiota profiles70. It is thus a common practice to study the gut microbiota in fish using pooled samples70–73.
Statistical analysis
Specific weight gain was determined as WG = (WF − WI)/t, where WF represents the average final weight, WI represents the average initial weight, and t represents the time in days of cultivation. The survival rate (S) of shrimp in each pond was determined by the following formula: S = 100 × (nT/N0), where S is the survival rate, nT is the number of shrimp harvested at the time t, and N0 is the number of shrimp at the beginning of the experiment. The conversion of food to biomass ratio was calculated using the following equation: FCA = AT/B, where AT represents the total food supplied, and B represents the biomass harvested.
To compare the productivity indexes for each treatment, we used Welch’s t-test74.
The survival (S) per condition was studied via an analysis of contingency tables using Pearson’s X2 test75.
DNA extraction
For the DNA extraction, 100 mg of each homogenized subsample was taken. The sample was incubated with 0.8 mg/ml lysozyme (Merck, Germany) and (0.8 mg/ml) lyticase (Sigma-Aldrich Corp, US) for 1 hour at 37 °C. The sample was later incubated with (0.1 mg/ml) proteinase K (Ambion/Life Technologies, CA, USA) for another hour at 37 °C. Next, the protocol for the PowerSoil ® DNA Isolation Kit from MoBio (Mo Bio Laboratories Inc., Qiagen, Carlsbad, CA, USA) was used according to the manufacturer’s instructions.
PCR amplification
To analyze the composition of the microbiota, part of the hypervariable V2-V3 region of the 16S rRNA gene was amplified. The PCR was performed using the primers 341F (5′ GCCTACGGGAGGCAGCAG 3′) and 518R (5′ CGTATTACCGCGGCTGCTGG 3′)67. The DNA concentration was determined using the Qubit dsDNA BR Assay Kit (Life Technologies, Grand Island, NY, US). Each reaction took place in a 30-μl solution consisting of 1 μl ≈ 1 ng of DNA, 18.5 μl of sterile, deionized deoxyribonuclease-free water, 6 µl of 5X buffer, 1.5 mM of MgCl2, 0.5 mM dNTPs, 0.84 μM each of forward and reverse primers and 0.5U of GoTaq (Promega, US). The PCR was performed in a Swift MiniPro thermocycler (Esco, China). The PCR program included an initial denaturation at 94 °C for 10 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 53 °C for 1 min and 72 °C for 1 min for extension, with a final incubation of 72 °C for 10 min. The resulting amplicons were visualized using PAGE76 and then purified using a QIAquick PCR Purification kit (Qiagen, Crawley, UK) according to the manufacturer’s protocol.
High-performance mass sequencing
A Qubit Fluorimeter (Invitrogen, CA, USA) was used to quantify the purified PCR products. The amplicons were then evaluated for fragment concentration using Ion Library Quantitation Kit (Life Technologies, CA, USA). The concentration was adjusted to 26 pM. The amplicons were attached to ion sphere particles (ISPs) using the Ion PGM kit Template OT2 400 (Life Technologies, CA, USA) according to the manufacturer’s protocol. Multiplexed sequencing was conducted using the 318 chip (Life Technologies, CA, USA) on the Ion Torrent Personal Genome platform (Life Technologies, CA, USA). Sequences were sorted by sample and filtered within the PGM software to remove low-quality reads. Finally, the data for each sample were exported to an individual Fastq file.
Data processing of the high-performance mass sequencing results
The quality of the Fastq sequences obtained was analyzed by using prescreening in the program FastQC from Babraham Bioinformatics77. Then, the parameters retrieved (p: minimum number of consecutive high-quality base calls to retain read; q: last quality score considered low quality; r: maximum number of consecutive low-quality base calls allowed before truncating a read; and n: maximum number of ambiguous (N) characters) were used to filter our reads using the UPARSE pipeline according to the recommendations of Bokulich et al.78 and Edgard79. The sequences were merged in a single FASTQ file using the script Split_libraries_fastq.py from QIIME (version 1.9.1-20150604)79. Then, the UPARSE pipeline was used to perform chimera checking and OTU clustering, using the default parameters of 97% identity with the biological sequence as described Edgar 201379. Taxonomic designations by RDP80 and sequence alignment with Pynast81, with a minimum threshold length of 170 bp, were performed in the QIIME pipeline82, using the 123 SILVA open source version as a reference83 with a threshold of 0.55 assigned by QIIME. The remaining sequences were considered unclassified, and the archaeal sequences were removed from the table and located in a separated table (see Suppl. Data). QIIME was also used to calculate a phylogenetic tree using the default parameters84 and 123 Silva as a reference database. Alpha diversity metric was obtained in the rarefied OTU tables. Beta diversity was evaluated using the Bray-Curtis method85 and is presented using principal component analysis (PCoA) based on the QIIME pipeline. The identification of OTUs with differential abundance between treatments was performed using MetagenomeSeq’s fitZIG method86, in QIIME. The comparison of the composition of the microbiota between treatments with the ANOSIM test87 were the last steps in the QIIME pipeline. To compare the microbiota associated with the treatments in terms of both statistical significance and biological relevance, we used the Linear Discriminant Analysis (LDA) Effect Size (LEfSe)88.
Supplementary information
Acknowledgements
The authors wish to thank SENESCYT Ecuador for their contribution (O.G. fellowship) and also to Avila owners of the shrimp farm. Fondecyt 1171129 supported this work.
Author contributions
O.G. develops the bioassay, performed bioinformatics and work and wrote draft version of the manuscript. J.R contributed with experimental design, sample preparation, laboratory support, critical revision of the article and wrote the version submitted.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-59587-y.
References
- 1.Schawarz, L. Visión general del sector acuícola nacional, Ecuador. Departamento de Pesca y Acuicultura de la FAO, http://www.fao.org/fishery/countrysector/naso_ecuador/es#tcN70044 (2005).
- 2.FAO (Food and Agriculture Organization of the United Nations). The state of world Fisheries and Aquaculture. FAO Fisheries and Aquaculture Department, Rome, Italia (2012).
- 3.FAO (Food and Agriculture Organization of the United Nations). The state of world Fisheries and Aquaculture. FAO Fisheries and Aquaculture Department, Rome, Italia (2014).
- 4.Tuong, N. C. T., Ha, L. T. T. & Thuan, N. A. H. Characterization of Actinomycetes Antagonistic to Vibrio parahaemoliticus Isolated from Shrimp Pond Sediment. VNU Journal of Science: Earth and Environmental Studies, 32 (2016).
- 5.Tran L, et al. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Diseases of aquatic organisms. 2013;105:45–55. doi: 10.3354/dao02621. [DOI] [PubMed] [Google Scholar]
- 6.Holmström K, et al. Antibiotic use in shrimp farming and implications for environmental impacts and human health. International Journal of Food Science & Technology. 2003;38:255–266. doi: 10.1046/j.1365-2621.2003.00671.x. [DOI] [Google Scholar]
- 7.Gainza RO. Terapia antimicrobiana con enrofloxacina en camaronicultura. Aqua Cultura. 2009;76:44–45. [Google Scholar]
- 8.Hoa PTP, et al. Antibiotic contamination and occurrence of antibiotic-resistant bacteria in aquatic environments of northern Vietnam. Science of the Total Environment. 2011;409:2894–2901. doi: 10.1016/j.scitotenv.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 9.Ringø, E., Olsen, R. E., Jensen, I., Romero, J. & Lauzon, H. L. Application of vaccines and dietary supplements in aquaculture: possibilities and challenges. Reviews in Fish Biology and Fisheries, 1–28 (2014).
- 10.Nayak SK. Role of gastrointestinal microbiota in fish. Aquaculture Research. 2010;41:1553–1573. doi: 10.1111/j.1365-2109.2010.02546.x. [DOI] [Google Scholar]
- 11.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izvekova GI. Hydrolytic activity of enzymes produced by symbiotic microflora and its role in digestion processes of bream and its intestinal parasite Caryophyllaeus laticeps (Cestoda, Caryophyllidea) Biology Bulletin. 2006;33:287–292. doi: 10.1134/S1062359006030125. [DOI] [PubMed] [Google Scholar]
- 13.Roeselers G, et al. Evidence for a core gut microbiota in the zebrafish. The ISME journal. 2011;5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullam KE, et al. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta‐analysis. Molecular ecology. 2012;21:3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semova I, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell host & microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torrecillas S, Montero D, Izquierdo M. Improved health and growth of fish fed mannan oligosaccharides: potential mode of action. Fish & shellfish immunology. 2014;36:525–544. doi: 10.1016/j.fsi.2013.12.029. [DOI] [PubMed] [Google Scholar]
- 17.Sanz Y, Collado MC, Haros M, Dalmau J. Funciones metabólico-nutritivas de la Microbiota intestinal y su modulación a través de la dieta: probióticos y prebióticos. Acta Pediatrica Española. 2004;62:520–526. [Google Scholar]
- 18.Guarner F. Papel de la flora intestinal en la salud y en la enfermedad. Nutrición Hospitalaria. 2007;22:14–19. [PubMed] [Google Scholar]
- 19.Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nature Reviews Microbiology. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 20.Frei R, Akdis M, O’Mahony L. Prebiotics, probiotics, synbiotics, and the immune system: experimental data and clinical evidence. Current opinion in gastroenterology. 2015;31:153–158. doi: 10.1097/MOG.0000000000000151. [DOI] [PubMed] [Google Scholar]
- 21.Merrifield DL, et al. Assessment of the effects of vegetative and lyophilized Pediococcus acidilactici on growth, feed utilization, intestinal colonization and health parameters of rainbow trout (Oncorhynchus mykiss Walbaum) Aquaculture Nutrition. 2011;17(1):73–79. doi: 10.1111/j.1365-2095.2009.00712.x. [DOI] [Google Scholar]
- 22.Standen BT, et al. Probiotic Pediococcus acidilactici modulates both localised intestinal and peripheral immunity in tilapia (Oreochromis niloticus) Fish & shellfish immunology. 2013;35(4):1097–1104. doi: 10.1016/j.fsi.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Abid A, et al. Dietary synbiotic application modulates Atlantic salmon (Salmo salar) intestinal microbial communities and intestinal immunity. Fish & shellfish immunology. 2013;35(6):1948–1956. doi: 10.1016/j.fsi.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, et al. Dietary supplementation of Bacillus subtilis and fructooligosaccharide enhance the growth, non-specific immunity of juvenile ovate pompano, Trachinotus ovatus and its disease resistance against Vibrio vulnificus. Fish & shellfish immunology. 2014;38(1):7–14. doi: 10.1016/j.fsi.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Wu DX, et al. Effects of a probiotic (Bacillus subtilis FY99-01) on the bacterial community structure and composition of shrimp (Litopenaeus vannamei, Boone) culture water assessed by denaturing gradient gel electrophoresis and hig-troughput sequencing. Aquaculture research. 2014;47(3):857–869. doi: 10.1111/are.12545. [DOI] [Google Scholar]
- 26.Pham KC, et al. Protection of Penaeus monodon against white spot syndrome by continuous oral administration of a low concentration of Bacillus subtilis spores expressing the VP28 antigen. Letters in Applied Microbiology. 2017;64(3):184–191. doi: 10.1111/lam.12708. [DOI] [PubMed] [Google Scholar]
- 27.Azevedo RVD, et al. Economic evaluation of prebiotics, probiotics and symbiotics in juvenile Nile tilapia. Revista Ciência Agronômica. 2015;46:72–79. doi: 10.1590/S1806-66902015000100009. [DOI] [Google Scholar]
- 28.Merrifield DL, et al. The current status and future focus of probiotic and prebiotic applications for salmonids. Aquaculture. 2010;302(1):1–18. doi: 10.1016/j.aquaculture.2010.02.007. [DOI] [Google Scholar]
- 29.Bindels LB, Delzenne NM, Cani PD, Walter J. Towards a more comprehensive concept for prebiotics. Nature reviews Gastroenterology &. hepatology. 2015;12(5):303–310. doi: 10.1038/nrgastro.2015.47. [DOI] [PubMed] [Google Scholar]
- 30.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. The Journal of nutrition. 1995;125(6):1401. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 31.Dildey D, et al. Effect of mannan oligosaccharide supplementation on performance and health of Holstein calves. Journal of Dairy Science. 1997;80(1):188. [Google Scholar]
- 32.Ringø E, et al. Prebiotics in aquaculture: a review. Aquaculture Nutrition. 2010;16:117–136. doi: 10.1111/j.1365-2095.2009.00731.x. [DOI] [Google Scholar]
- 33.Genc MA, Aktas M, Genc E, Yilmaz E. Effects of dietary mannan oligosaccharide on growth, body composition and hepatopancreas histology of Penaeus semisulcatus (de Haan 1844) Aquaculture Nutrition. 2007;13(2):156–161. doi: 10.1111/j.1365-2095.2007.00469.x. [DOI] [Google Scholar]
- 34.Sang HM, Ky LT, Fotedar R. Dietary supplementation of mannan oligosaccharide improves the immune responses and survival of marron, Cherax tenuimanus (Smith, 1912) when challenged with different stressors. Fish & shellfish immunology. 2009;27(2):341–348. doi: 10.1016/j.fsi.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Van Hai N, Fotedar R. Comparison of the effects of the prebiotics (Bio-Mos® and β-1, 3-D-glucan) and the customized probiotics (Pseudomonas synxantha and P. aeruginosa) on the culture of juvenile western king prawns (Penaeus latisulcatus Kishinouye, 1896) Aquaculture. 2009;289(3):310–316. [Google Scholar]
- 36.Daniels CL, et al. Effect of dietary Bacillus spp. and mannan oligosaccharides (MOS) on European lobster (Homarus gammarus L.) larvae growth performance, gut morphology and gut microbiota. Aquaculture. 2010;304:49–57. doi: 10.1016/j.aquaculture.2010.03.018. [DOI] [Google Scholar]
- 37.Sang HM, Fotedar R. Effects of mannan oligosaccharide dietary supplementation on performances of the tropical spiny lobsters juvenile (Panulirus ornatus, Fabricius 1798) Fish & Shellfish Immunology. 2010;28:483–489. doi: 10.1016/j.fsi.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Mazlum Y, Yılmaz E, Genç MA, Guner O. A preliminary study on the use of mannan oligosaccharides (MOS) in freshwater crayfish, Astacus leptodactylus Eschscholtz, 1823 juvenile diets. Aquaculture International. 2011;19(1):111–119. doi: 10.1007/s10499-010-9345-4. [DOI] [Google Scholar]
- 39.Sang HM, Fotedar R, Filer K. Effects of dietary mannan oligosaccharide on the survival, growth, immunity and digestive enzyme activity of freshwater crayfish, Cherax destructor Clark (1936) Aquaculture Nutrition. 2011;17(2):e629–e635. doi: 10.1111/j.1365-2095.2010.00812.x. [DOI] [Google Scholar]
- 40.Zhang J, et al. Effects of dietary mannan oligosaccharide on growth performance, gut morphology and stress tolerance of juvenile Pacific white shrimp, Litopenaeus vannamei. Fish & shellfish immunology. 2012;33:1027–1032. doi: 10.1016/j.fsi.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Genc MA, Ebeoglu B. The Effects of Different Salinity and Supplemented Mannan Oligosaccharides (MOS) on Growth of Litopenaeus varmamei (Penaeus: Decapoda) Journal of Animal and Veterinary Advances. 2013;12:942–947. [Google Scholar]
- 42.Hoang DH, Jones CM. Effects of dietary mannan oligosaccharides supplementation on juvenile spiny lobster (Panulirus homarus) (Palinuridae) Aquaculture. 2014;432:258–264. doi: 10.1016/j.aquaculture.2014.05.013. [DOI] [Google Scholar]
- 43.Sang HM, Kien NT, Thanh Thuy NT. Effects of dietary mannan oligosaccharide on growth, survival, physiological, immunological and gut morphological conditions of black tiger prawn (Penaeus monodon Fabricius 1798) Aquaculture Nutrition. 2014;20(3):341–348. doi: 10.1111/anu.12083. [DOI] [Google Scholar]
- 44.Zhang M, et al. Characterization of the intestinal microbiota in Pacific white shrimp, Litopenaeus vannamei, fed diets with different lipid sources. Aquaculture. 2014;434:449–455. doi: 10.1016/j.aquaculture.2014.09.008. [DOI] [Google Scholar]
- 45.Qiao, F. et al. Influence of different dietary carbohydrate sources on the growth and intestinal microbiota of Litopenaeus vannamei at low salinity. Aquaculture Nutrition (2016).
- 46.Sha Y, et al. Bacterial Population in Intestines of Litopenaeus vannamei Fed Different Probiotics or Probiotic Supernatant. Journal of Microbiology and Biotechnology. 2016;26(10):1736–1745. doi: 10.4014/jmb.1603.03078. [DOI] [PubMed] [Google Scholar]
- 47.Zhang M, et al. Response of gut microbiota to salinity changes in two euryhaline aquatic animals with reverse salinity preference. Aquaculture. 2016;454:72–80. doi: 10.1016/j.aquaculture.2015.12.014. [DOI] [Google Scholar]
- 48.Gainza O, Ramírez C, Ramos AS, Romero J. Intestinal microbiota of white shrimp Penaeus vannamei under intensive cultivation conditions in Ecuador. Microbial ecology. 2018;75(3):562–568. doi: 10.1007/s00248-017-1066-z. [DOI] [PubMed] [Google Scholar]
- 49.Chan SM, Rankin SM, Keeley LL. Characterization of the molt stages in Penaeus vannamei: setogenesis and hemolymph levels of total protein, ecdysteroids, and glucose. The Biological Bulletin. 1988;175(2):185–192. doi: 10.2307/1541558. [DOI] [Google Scholar]
- 50.Griffith DRW, Wigglesworth JM. Growth rhythms in the shrimp Penaeus vannamei and P. schmitti. Marine biology. 1993;115(2):295–299. doi: 10.1007/BF00346347. [DOI] [Google Scholar]
- 51.Luquet G. Biomineralizations: insights and prospects from crustaceans. Zookeys. 2012;176:103–121. doi: 10.3897/zookeys.176.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson MJ, et al. Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist. Ecology Letters. 2011;14(1):19–28. doi: 10.1111/j.1461-0248.2010.01552.x. [DOI] [PubMed] [Google Scholar]
- 53.Huang Z, Li X, Wang L, Shao Z. Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei. Aquaculture Research. 2014;6(47):1737–1746. [Google Scholar]
- 54.Xiong J, et al. Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Applied microbiology and biotechnology. 2015;99:6911–6919. doi: 10.1007/s00253-015-6632-z. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, J., et al Contrasting Ecological Processes and Functional Compositions Between Intestinal Bacterial Community in Healthy and Diseased Shrimp. Microbial Ecology, 1–11 (2016). [DOI] [PubMed]
- 56.Rungrassamee W, Klanchui A, Maibunkaew S, Karoonuthaisiri N. Bacterial dynamics in intestines of the black tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. Journal of Invertebrate Pathology. 2016;133:12–19. doi: 10.1016/j.jip.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Dierckens KR, et al. Aeromonas hydrophila causes black disease in fairy shrimps (Anostraca; Crustacea) Journal of fish diseases. 1998;21:113–119. doi: 10.1046/j.1365-2761.1998.00085.x. [DOI] [PubMed] [Google Scholar]
- 58.Balcázar JL, et al. In vitro competitive adhesion and production of antagonistic compounds by lactic acid bacteria against fish pathogens. Veterinary microbiology. 2007;122(3):373–380. doi: 10.1016/j.vetmic.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 59.Opazo R, Gajardo F, Ruiz M, Romero J. Genome sequence of a Lactococcus lactis strain isolated from salmonid intestinal microbiota. Genome announcements. 2016;4(4):e00881–16. doi: 10.1128/genomeA.00881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahajan GB, Balachandran L. Antibacterial agents from actinomycetes-a review. Frontiers in bioscience (Elite edition) 2011;4:240–253. doi: 10.2741/373. [DOI] [PubMed] [Google Scholar]
- 61.Manivasagan P, Venkatesan J, Sivakumar K, Kim SK. Marine actinobacterial metabolites: current status and future perspectives. Microbiological research. 2013;168:311–332. doi: 10.1016/j.micres.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Das S, Ward LR, Burke C. Prospects of using marine actinobacteria as probiotics in aquaculture. Applied Microbiology and Biotechnology. 2008;81:419–429. doi: 10.1007/s00253-008-1731-8. [DOI] [PubMed] [Google Scholar]
- 63.Kamarudheen N, George CS, Pathak S, George SL, Rao KV. Antagonistic activity of Marine Streptomyces sp. on Fish pathogenic Vibrio species isolated from Aquatic Environment. Research Journal of Pharmacy and Technology. 2015;8:1529–1533. doi: 10.5958/0974-360X.2015.00273.5. [DOI] [Google Scholar]
- 64.Velmurugan S, et al. Isolation of Actinomycetes from Shrimp Culture Pond and Antagonistic to Pathogenic Vibrio spp. and WSSV. Int. J. Curr. Microbiol. App. Sci. 2015;4:82–92. [Google Scholar]
- 65.Tan, L. T. H., Chan, K. G., Lee, L. H. & Goh, B. H. Streptomyces bacteria as potential probiotics in aquaculture. Frontiers in microbiology, 7 (2016). [DOI] [PMC free article] [PubMed]
- 66.Ottman N, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One, 2017;12(3):e0173004. doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic acids research, 2012;41(1):e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sauder LA, et al. Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters. PLoS One, 2011;6(8):e23281. doi: 10.1371/journal.pone.0023281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lozano, P. “Los tipos de bosque en el sur de Ecuador.” Bótanica Austroecuatoriana. Estudios sobre los recursos vegetales en las provincial de El Oro, Loja y Zamora-Chinchipe. Abya-Yala, Quito, 29–49 (2002).
- 70.Romero J, Navarrete P. 16S rDNA-based analysis of dominant bacterial populations associated with early life stages of coho salmon (Oncorhynchus kisutch) Microbial ecology. 2006;51(4):422–430. doi: 10.1007/s00248-006-9037-9. [DOI] [PubMed] [Google Scholar]
- 71.Hovda MB, Lunestad BT, Fontanillas R, Rosnes JT. Molecular characterisation of the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.) Aquaculture. 2007;272(1):581–588. doi: 10.1016/j.aquaculture.2007.08.045. [DOI] [Google Scholar]
- 72.Navarrete P, Espejo RT, Romero J. Molecular analysis of microbiota along the digestive tract of juvenile Atlantic salmon (Salmo salar L.) Microbial ecology. 2009;57(3):550–561. doi: 10.1007/s00248-008-9448-x. [DOI] [PubMed] [Google Scholar]
- 73.Navarrete P, et al. Short-term effects of dietary soybean meal and lactic acid bacteria on the intestinal morphology and microbiota of Atlantic salmon (Salmo salar) Aquaculture Nutrition. 2013;19(5):827–836. doi: 10.1111/anu.12047. [DOI] [Google Scholar]
- 74.Welch BL. The generalization ofstudent’s’ problem when several different population variances are involved. Biometrika. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 75.Pearson K. X. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science. 1900;50:157–175. doi: 10.1080/14786440009463897. [DOI] [Google Scholar]
- 76.Espejo RT, Feijoo CG, Romero J, Vásquez M. PAGE analysis of the heteroduplexes formed between PCR-amplified 16S rRNA genes: estimation of sequence similarity and rDNA complexity. Microbiology. 1998;144(6):1611–1617. doi: 10.1099/00221287-144-6-1611. [DOI] [PubMed] [Google Scholar]
- 77.Andrews, S. FastQC: a quality Control tool for high throughput sequence data. Cambridge, UK Babraham Institute. http://www. bioinformatics. babraham. ac. uk/projects/fastqc/. Accessed, 5 (2016).
- 78.Bokulich NA, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 80.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caporaso J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS one. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecological monographs. 1957;27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 86.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nature methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology. 1993;18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 88.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12:1. doi: 10.1186/1465-6906-12-S1-P1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





