Abstract
Welding generates and releases fumes that are hazardous to human health. Welding fumes (WFs) are a complex mix of metallic oxides, fluorides and silicates that can cause or exacerbate health problems in exposed individuals. In particular, WF inhalation over an extended period carries an increased risk of cancer, but how WFs may influence cancer behaviour or growth is unclear. To address this issue we employed a quantitative analytical framework to identify the gene expression effects of WFs that may affect the subsequent behaviour of the cancers. We examined datasets of transcript analyses made using microarray studies of WF-exposed tissues and of cancers, including datasets from colorectal cancer (CC), prostate cancer (PC), lung cancer (LC) and gastric cancer (GC). We constructed gene-disease association networks, identified signaling and ontological pathways, clustered protein-protein interaction network using multilayer network topology, and analyzed survival function of the significant genes using Cox proportional hazards (Cox PH) model and product-limit (PL) estimator. We observed that WF exposure causes altered expression of many genes (36, 13, 25 and 17 respectively) whose expression are also altered in CC, PC, LC and GC. Gene-disease association networks, signaling and ontological pathways, protein-protein interaction network, and survival functions of the significant genes suggest ways that WFs may influence the progression of CC, PC, LC and GC. This quantitative analytical framework has identified potentially novel mechanisms by which tissue WF exposure may lead to gene expression changes in tissue gene expression that affect cancer behaviour and, thus, cancer progression, growth or establishment.
Subject terms: Cancer genomics, Machine learning
Introduction
Welding processes join rigid material pieces (usually metal) at their contact interface by using high temperatures to cause fusion. This process can be hazardous because it exposes the operator to extremely toxic fumes and to radiant energy1. The International Agency for Research on Cancer (IARC) has recognized WFs and UV radiation from welding as Group 1 carcinogens2. WFs are mainly composed of metallic oxides, silicates and fluorides, including those of magnesium, manganese, zinc, aluminum, beryllium, copper, chromium, cadmium, lead, iron, nickel and vanadium3.
Welders inhaling WFs in large quantities over a long period run a significantly elevated risk of developing certain types of cancer1,2. These metastatic diseases involve uncontrolled or neoplastic growth of cancer cells that arise after the accumulation of genomic mutations, but other factors with powerful effects on cancer behaviour and growth include genetic factors and environmental factors the suffer is exposed to4. Environmental factors include inhaled toxic fumes that affect the lungs and enter the circulation to reach many tissues, and which can affect cellular gene expression of cancer cells and thereby their behaviour, survival, growth and invasiveness. Thus, influences such as WF inhalation affects the progression of many types of cancers, including those focused on in this study, specifically CC, PC, LC and GC, which are among the cancers most commonly linked with WF exposure5–7. The aim of this study is therefore to identify mechanisms through which WFs may increase cancer incidence.
LC is one of the most lethal types of cancer and globally is a leading cause of death1,2,8. WFs contain toxic metallic oxides and silicates that directly affect the sensitive tissues of the lung when inhaled, the manner of exposure (by inhalation) makes this the cancer with the highest risk for welders9. CC arises in the colon and the rectum and has a typical 5-year survival rate of about 60%. It damages colon or rectum by uncontrollable and invasive cell growth10. Iron, aluminum and magnesium oxide of the welding fumes are known to affect the incidence of CC9, although this is not well understood. PC affects prostate, the gland which produces seminal fluid and controls the transportation of sperm11. Nitrogen oxides, carbon dioxide and phosgene are risk factors for prostate neoplasms that are found in WFs9. GC (gastric or stomach cancer)12 is linked to exposures to nickel, beryllium and cobalt oxides which are all present in WFs9.
In this study, we developed a systematic and quantitative network-based approach to investigate the effects of WFs on gene expression and how these effects may give a clue as to how they encourage the incidence and progression of cancers through affecting pathways and pathway genes that are also altered in these cancers. Thus, we compared gene expression effects of WF exposure with the altered pattern of gene expression seen in CC, PC, LC and GC. This involved, firstly, analyzing differentially expressed gene profiles, then filtering these genes through gene-disease association networks, signaling and ontological pathways, and protein-protein interaction networks. We also investigated the importance of genes and pathways thus identified by using the gold benchmark databases dbGaP and OMIM to identify evidence to support the involvement of these genes in pathological processes such as cancer development. Moreover, we analysed patient survival and its association with the genes that are dysregulated in both the WF-exposed tissue and the four types of cancers. The influence on cancer patient survival of these identified genes provides evidence for their involvement in WF effect on cancer progression.
Methods and Materials
Overview of the analytical approach
We applied an analytical approach to identify links between WF exposure and the incidence of the cancers by employing selected microarray datasets shown in the block diagram of the applied analytical approach shown in Fig. 1. This quantitative approach used genes differentially expressed in WF exposure, and identifies those that are also common to the differentially expressed genes observed in each cancer study. Further, these shared or common differentially expressed genes were used to construct gene-disease (diseasome) association network, identify signaling and ontological pathways, protein-protein interaction (PPI) network and survival function analysis. This approach also used gold benchmark databases OMIM and dbGaP validate genes and pathways identified in our study as showing possible disease associations.
Figure 1.
Flow-diagram of the analytical approach used in this study.
Datasets employed in this study
To identify the gene expression dysregulation that is common to WFs and the four types of cancers under investigation, we analyzed gene expression microarray datasets from the National Center for Biotechnology Information (NCBI). We examined five different microarray datasets with accession numbers GSE62384, GSE25071, GSE55945, GSE10072 and GSE268513–17. Dataset GSE62384 was produced using human upper airway epithelial cells (RPMI 2650) exposed to spark generated WFs. These data were generated from cells exposed to WFs for 6 hours continuously at low (85 μg/m3) and high (760 μg/m3) concentrations. The CC dataset (GSE25071) consists of microarray data taken from 17 colorectal cancer sufferers who had late-onset CC (mean age 79 years) and 24 patients with early-onset CC (mean age 43 years). The PC dataset (GSE55945) is a microarray data on RNA taken from radical prostatectomy tissue from prostate cancer patients at the Beth Israel Deaconess Medical Center which compared tissue from PC sufferers (Gleason score 6 or 7) with normal prostate tissue. The LC dataset (GSE10072) contained microarray data comparing normal lung tissue and lung adenocarcinoma tissue collected from 26 former smokers, 20 non-smokers (who never smoked) and 28 current smokers; gene expression data are reported by comparing 49 non-tumor and 58 tumor lung tissues. The GC dataset (GSE2685) contains microarray data from 22 gastric cancer and 8 non-cancerous gastric tissues.
To analyze the patient survival association of the altered genes that are common to WFs and the four types of cancers under investigation, we retrieved clinical and RNAseq data for CC, PC, LC and GC from the cBioPortal 18,19. In the clinical dataset of CC (Colorectal Adenocarcinoma, TCGA, Nature 2012) there are 585 samples with 24 features. The samples of CC have RNAseq gene expression data included 224 cases with 224 mutated genes20. The clinical dataset of PC (Prostate Adenocarcinoma, TCGA, Cell 2015) includes 333 samples with 86 features. The RNAseq gene expression data of PC has 333 cases with 333 mutated genes21. The LC clinical dataset (Lung Adenocarcinoma, TCGA, PanCancer Atlas) consists of 566 samples with 81 features. The samples of LC have RNAseq gene expression data included 510 cases with 566 genes22. The clinical dataset of GC (Stomach Adenocarcinoma, TCGA, Nature 2014) contains 295 samples with 52 features. The samples of GC have RNAseq gene expression data included 265 cases with 295 mutated genes23. We employed six clinical factors (ethnicity, anatomical site of cancer, histological grade of cancer, primary tumour site, and neoplasm status with tumour) to analyze the survival of the altered genes that are common to WFs and the four types of cancers under investigation. The summarized description of the datasets is shown in Tables 1 and 2.
Table 1.
Summarized description of the datasets used for gene expression and enrichment analysis.
Table 2.
Summarized description of the datasets used for survival prediction.
| Sl. | Disease name | Datasets name in the cBioPortal | Number of samples | |||
|---|---|---|---|---|---|---|
| Patients | Clinical features | RNA-Seq | Mutated genes | |||
| 2 | Colorectal Cancer (CC) | Colorectal Adenocarcinoma (TCGA, Nature 2012) | 585 | 24 | 224 | 224 |
| 3 | Prostate Cancer (PC) | Prostate Adenocarcinoma (TCGA, Cell 2015) | 333 | 86 | 333 | 333 |
| 4 | Lung cancer (LC) | Lung Adenocarcinoma (TCGA, PanCancer Atlas) | 566 | 81 | 510 | 566 |
| 5 | Gastric Cancer (GC) | Stomach Adenocarcinoma (TCGA, Nature 2014) | 295 | 52 | 265 | 295 |
Analysis methods
Microarray-based gene expression analysis is a global and sensitive method to identify and quantify possible molecular mechanisms that underlie human disorders24. We used these approaches to analyze the gene expression profiles of CC, PC, LC and GC to find the genetic effects of WFs that may influence the development of these cancers. To allow comparisons of the mRNA expression data generated using different platforms and to avoid complications arising from the different experimental systems employed in the original studies, we normalized the gene expression data by means of Z-score transformation (Zij) for each type of cancer tissue gene expression profile using , where SD denotes the standard deviation, gij denotes the value of the gene expression i in sample j. After this transformation gene expression values of different diseases at different platforms can be directly compared. We applied unpaired t-tests to find differentially expressed genes of each disease over control data and selected significantly dysregulated genes. We have chosen a threshold of at least 1 log2 fold change and a p-value for the t-tests of . We employed the neighborhood-based benchmark and the multilayer topological methods to find gene-disease associations. We constructed a gene-disease network (GDN) using the gene-disease associations, where the nods in the network represent either gene or disease. This network can also be recognized as a bipartite graph. The primary condition for a disease to be connected with other diseases in GDN is they should share at least one or more significant dysregulated genes. Let is a specific set of diseases and G is a set of dysregulated genes, gene-disease associations attempt to find whether gene is associated with disease . If and , the sets of significantly dysregulated genes associated with diseases Di and Dj respectively, then the number of shared dysregulated genes associated with both disorders Di and Dj is as follows25:
| 1 |
The common neighbours are the based on the Jaccard Coefficient method, where the edge prediction score for the node pair is as26:
| 2 |
where G is the set of nodes and E is the set of all edges. We used R software packages “comoR”27 and “POGO”28 to cross check their genes-disease associations.
To investigate how molecular determinants from the WF exposed tissues relate gene expression alterations in the cancers, we analyzed pathway and gene ontology using Enrichr 29,30. We used KEGG, WikiPathways, Reactome and BioCarta databases for analyzing signaling pathway31–34. We used GO Biological Process and Human Phenotype Ontology databases for ontological analysis35,36. We also constructed a protein-protein interaction sub-network for each CD, using the STRING database, a biological database and web resource of known and predicted protein-protein interactions37. Furthermore, we examined the validity of our study by employing two gold benchmark databases OMIM and dbGaP.
To determine the patient survival association of the altered genes that are common to WFs and the four types of cancers under investigation, we employed Cox PH model for univariate and multivariate analysis38,39. The Cox PH model can be written as follows:
| 3 |
Here is the hazard function conditioned on a subject with covariate information given as the vector , is the baseline hazard function which is independent of covariate information, and β represents a vector of regression coefficients to the covariates correspondingly. We have calculated the hazard ratio (HR) based on the estimated regression coefficients from the fitted Cox PH model to determine whether a specific covariate affects patient survival. The HR for a covariate can be expressed by the following simple formula exp . Thus, the HR for any covariate can be calculated by applying an exponential function to the corresponding coefficient.
The survival status of a patient can be estimated by calculating PL estimator40 of the survival function can be defined as follows:
| 4 |
Here is estimated survival function at time tj, dj is the number of events occurred at tj, and nj is the number of subjects available at tj. After estimating survival function, two or more groups can be compared using a log-rank test. We used Log-rank tests to detect the most significant genes in the case of patient’s survival time in altered versus normal (non-altered) groups in context of gene expression. The null hypothesis for this test can be symbolically explained as follows:
| 5 |
| 6 |
Here H0 is survival functions that are the same for altered and normal gene and HA is survival functions that are not the same for these two groups.
If the survival function of a specific gene is different among altered and normal groups then we include it to the combined Cox PH model. This approach is efficient for learning the effect of a specific gene of interest on patient survival in the presence of the clinical factors.
Results
Gene expression analysis
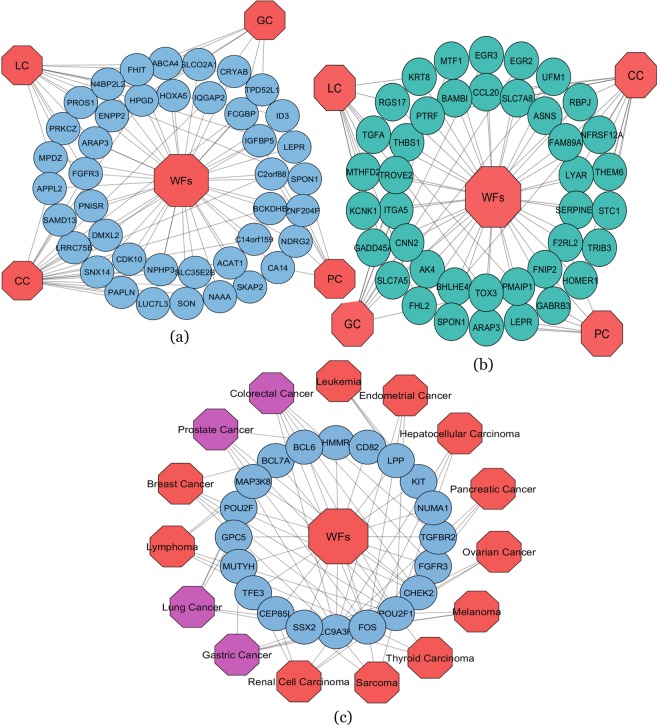
To identify and investigate the gene expression effects of WFs that may influence the behaviour of various types of cancer, we analyzed the gene expression microarray data collected from the National Center for Biotechnology Information (NCBI). We observed that WFs have 903 differentially expressed genes obtained by adjusted and . The differentially expressed genes of WFs contain 392 up and 511 down-regulated genes relative to controls. Similarly, the statistical analysis identified the most significant genes with altered expression in each cancer type. The number of differentially expressed genes we identified was 939 (503 up and 436 down) in CC, 553 (323 up and 230 down) in PC, 890 (673 up and 217 down) in LC and 691 (463 up and 228 down) in GC. We also employed a cross-comparative analysis to find the common genes with altered expression between WFs and each CD. We found that WF treated cells share a number of differentially expressed genes with for CC (36 dysregulated genes), PC (13 genes), LC (25 genes) and GC (17 genes). To identify the significant associations among these cancer types with the effects of WF exposure, we constructed two separate gene-disease association-ship networks for up and down-regulated genes using Cytoscape plugins41, centered on the WF data as shown in Fig. 2(a,b). The necessary condition for two diseases to be associated is they must have at least one or more common differentially expressed genes in between them. Notably, two particular significant genes, C2orf88 and IGFBP5 were differentially expressed among WF exposure, CC and PC; and three significant genes, FCGBP, IQGAP2 and HPGD are common among WF exposure, CC and GC. One gene, FGFR3, is commonly dysregulated among WF exposure, CC and LC.
Figure 2.
(a) Up-regulated gene-disease association network of welding fumes (WFs) exposure with colorectal cancer (CC), prostate cancer (PC), lung cancer (LC) and gastric cancer (GC). Octagon-shaped red-colored nodes represent different cancer types and sky-blue colored round-shaped nodes represent commonly up-regulated genes for WFs with the cancers examined. (b) Down-regulated gene-disease association network of welding fumes (WFs) exposure with colorectal cancer (CC), prostate cancer (PC), lung cancer (LC) and gastric cancer (GC). Octagon-shaped red colored nodes represent different cancer types and dark-cyan colored round-shaped nodes represent commonly down-regulated genes for WFs exposure with the different types of cancer examined. (c) Diseasome network showing validation of our study. Red colored octagon-shaped nodes represent different cancer types, pink-colored octagon-shaped nodes represent our selected four CDs and round-shaped sky-blue colored nodes represent differentially expressed genes for WFs exposure. A link is placed between a disease and a gene if mutations in that gene lead to the specific disease.
Pathway and functional association analysis
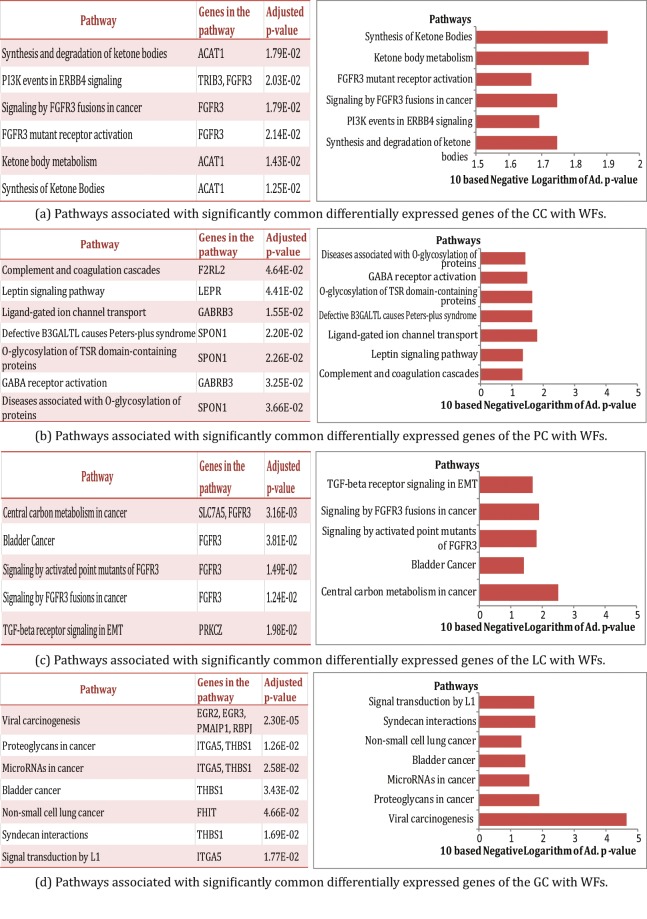
Pathways are constituted by a series of interactions at the molecular level in a cell, and are a vital key to understand the internal changes of an organism. Pathway-based analysis can be used to identify molecular or biological mechanisms that underlie the development of complex diseases42,43. We analyzed pathways of the commonly altered expression genes seen in WF exposure and in the cancers using Enrichr, a comprehensive web-based gene set enrichment analyzing tool29,44. Signaling pathways of the commonly altered expression genes of WF exposure and each type of cancer examined were analyzed using four globally recognized databases includes KEGG, WikiPathways, Reactome and BioCarta. We considered signaling pathways from the selected four databases and identified the most significant signaling pathways of each CD after applying several statistical analysis. Notably, we found 6, 7, 5 and 7 signaling pathways are associated with CC, PC, LC and GC, respectively, as shown in Fig. 3.
Figure 3.
Pathway analysis for identifying the most significant signaling pathways common to the WF exposed cells and the cancer types revealed by the common differentially expressed genes. These include significant signaling pathways common to WFs exposed cells and (a) CC (b) PC (c) LC and (d) GC.
Gene ontological analysis
The Gene Ontology (GO) refers to a universal conceptual model for representing gene functions and their relationship in the domain of gene regulation. It is constantly expanded by accumulating the biological knowledge to cover the regulation of gene functions and the relationship of these functions in terms of ontology classes and semantic relations between classes45. We analyzed ontological pathways of the commonly altered expression genes seen in WFs exposed cells and each cancer type using two recognized databases including GO Biological Process and Human Phenotype Ontology. We considered ontological pathways from selected two databases and identified the most significant ontological pathways for each cancer type after applying several statistical analysis. We found 10, 11, 14 and 14 ontological pathways are associated with the CC, PC, LC and GC, respectively, as shown in Tables 3–6.
Table 4.
The most significant ontological pathways common to the WFs exposed cells and PC.
| GO Term | Pathway | Genes in the pathway | Adjusted p-value |
|---|---|---|---|
| GO:0071420 | Cellular response to histamine | GABRB3 | 4.79E-03 |
| GO:0044321 | Response to leptin | LEPR | 5.39E-03 |
| GO:0035024 | Negative regulation of Rho protein signal transduction | ARAP3 | 8.97E-03 |
| GO:2000369 | Regulation of clathrin-dependent endocytosis | ARAP3 | 9.56E-03 |
| GO:0071417 | Cellular response to organonitrogen compound | GABRB3, IGFBP5 | 1.52E-03 |
| GO:2000146 | Negative regulation of cell motility | IGFBP5, ARAP3 | 1.52E-03 |
| GO:0030336 | Negative regulation of cell migration | IGFBP5, ARAP3 | 2.34E-03 |
| GO:0071407 | Cellular response to organic cyclic compound | GABRB3, IGFBP5 | 2.90E-03 |
| GO:0014912 | Negative regulation of smooth muscle cell migration | IGFBP5 | 8.97E-03 |
| HP:0000823 | Delayed puberty | LEPR | 1.67E-02 |
| HP:0000824 | Growth hormone deficiency | LEPR | 1.67E-02 |
Table 5.
The most significant ontological pathways common to the WFs exposed cells and LC.
| GO Term | Pathway | Genes in the pathway | Adjusted p-value |
|---|---|---|---|
| GO:1903708 | Positive regulation of hemopoiesis | N4BP2L2, HOXA5 | 4.18E-05 |
| GO:0009132 | Nucleoside diphosphate metabolic process | AK4 | 9.96E-03 |
| GO:0045647 | Negative regulation of erythrocyte differentiation | HOXA5 | 8.72E-03 |
| GO:2000665 | Regulation of interleukin-13 secretion | PRKCZ | 8.72E-03 |
| GO:0044320 | Cellular response to leptin stimulus | LEPR | 9.96E-03 |
| GO:0045837 | Negative regulation of membrane potential | PMAIP1 | 8.72E-03 |
| GO:2000394 | Positive regulation of lamellipodium morphogenesis | ENPP2 | 9.96E-03 |
| GO:0050730 | Regulation of peptidyl-tyrosine phosphorylation | ENPP2, PRKCZ | 5.14E-03 |
| GO:0032754 | Positive regulation of interleukin-5 production | PRKCZ | 9.96E-03 |
| HP:0000975 | Hyperhidrosis | SLCO2A1, FGFR3 | 5.38E-03 |
| HP:0000522 | Alacrima | FGFR3 | 1.24E-02 |
| HP:0010662 | Abnormality of the diencephalon | LEPR | 1.86E-02 |
| HP:0000495 | Recurrent corneal erosions | FGFR3 | 1.49E-02 |
| HP:0001413 | Micronodular cirrhosis | KRT8 | 1.86E-02 |
Table 3.
The most significant ontological pathways common to the WFs exposed cells and CC.
| GO Term | Pathway | Genes in the pathway | Adjusted p-value |
|---|---|---|---|
| GO:0009083 | Branched-chain amino acid catabolic process | BCKDHB, ACAT1 | 7.11E-04 |
| GO:0009081 | Branched-chain amino acid metabolic process | BCKDHB, ACAT1 | 7.78E-04 |
| GO:0009063 | Cellular amino acid catabolic process | BCKDHB, ACAT1 | 8.48E-04 |
| GO:0051924 | Regulation of calcium ion transport | HOMER1, STC1 | 4.10E-03 |
| GO:0032092 | Positive regulation of protein binding | BAMBI, TRIB3 | 5.39E-03 |
| GO:0051726 | Regulation of cell cycle | SON, HPGD, CDK10 | 6.82E-03 |
| GO:0009966 | Regulation of signal transduction | IGFBP5, HOMER1,FGFR3 | 8.49E-03 |
| GO:0051099 | Positive regulation of binding | BAMBI, TRIB3 | 9.56E-03 |
| HP:0001643 | Patent ductus arteriosus | HPGD, NPHP3, FGFR3 | 1.46E-03 |
| HP:0001946 | Ketosis | BCKDHB, ACAT1 | 1.16E-03 |
Table 6.
The most significant ontological pathways common to the WFs exposed cells and GC.
| GO Term | Pathway | Genes in the pathway | Adjusted p-value |
|---|---|---|---|
| GO:0072331 | Signal transduction by p53 class mediator | PMAIP1, FHIT | 5.20E-04 |
| GO:2001235 | Positive regulation of apoptotic signaling pathway | PMAIP1, TPD52L1 | 1.92E-03 |
| GO:0097193 | Intrinsic apoptotic signaling pathway | PMAIP1, FHIT | 3.33E-03 |
| GO:0042981 | Regulation of apoptotic process | UFM1, EGR3, PMAIP1, THBS1 | 4.28E-03 |
| GO:0010634 | Positive regulation of epithelial cell migration | ENPP2, THBS1 | 1.82E-03 |
| GO:0045766 | Positive regulation of angiogenesis | ITGA5, THBS1 | 3.46E-03 |
| GO:0001936 | Regulation of endothelial cell proliferation | EGR3, THBS1 | 2.07E-03 |
| GO:0010038 | Response to metal ion | MTF1, THBS1 | 1.72E-03 |
| GO:0043066 | Negative regulation of apoptotic process | UFM1, EGR3, ITGA5, THBS1 | 6.37E-04 |
| GO:0051094 | Positive regulation of developmental process | ENPP2, RBPJ | 4.14E-03 |
| GO:0034976 | Response to endoplasmic reticulum stress | UFM1, THBS1 | 3.66E-03 |
| GO:0043069 | Negative regulation of programmed cell death | UFM1, EGR3, THBS1 | 4.66E-03 |
| HP:0003577 | Congenital onset | EGR2, HPGD | 6.35E-03 |
| HP:0000890 | Long clavicles | HPGD | 7.63E-03 |
Protein-protein interaction analysis
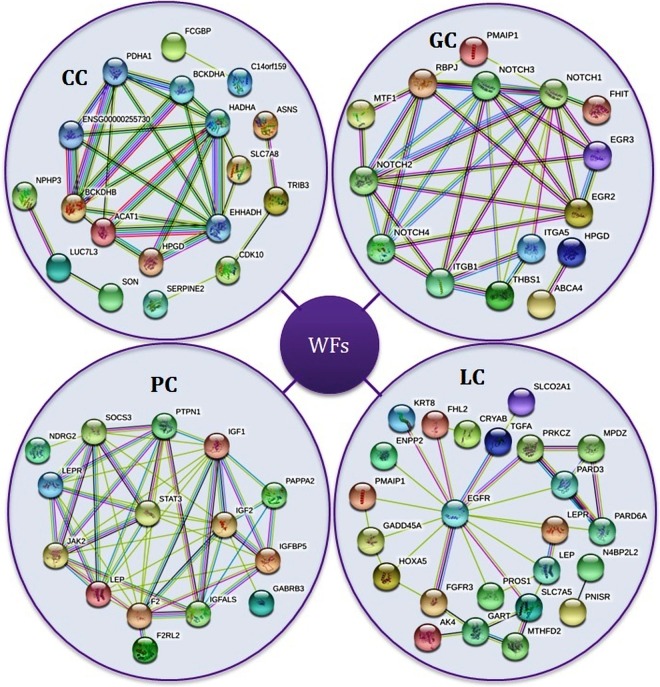
A protein-protein interaction network refers to the binding of proteins in the cell formed by biochemical or complex biological functions. Protein-protein interactions are essential to understand the cell physiology in health and disease states. We constructed and analyzed protein-protein interaction networks of the significantly altered expression genes of each CD using the STRING database. We clustered protein-protein interactions of cancer types into four different groups as shown in Fig. 4.
Figure 4.
Protein-protein interaction network of the four types of cancer using STRING.
Survival analysis
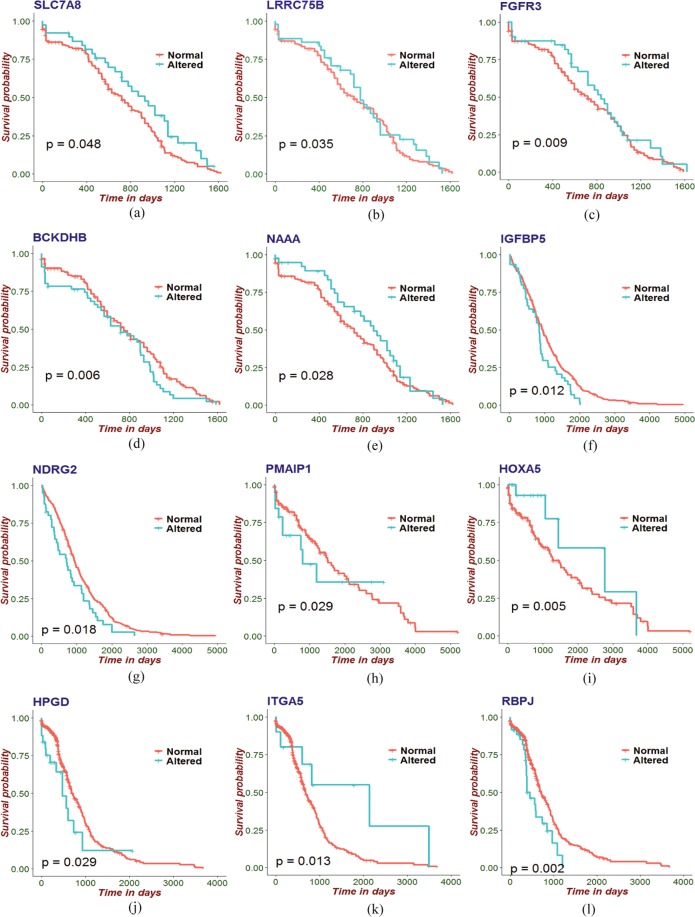
Patient survival analysis using both gene expression and clinical data is a popularly used feature in research to predict and characterize gene signatures in cancer46. In this study, we estimated survival function for altered and normal groups of the significant genes that are common to WFs and the four types of cancers under investigation by employing Cox PH model and PL estimator analysis. We fitted both univariate and multivariate analysis of the Cox PH regression model. The significant genes of the four selected cancers with estimated coefficients (β), hazard ratios (HR) and p-values from those analyses are shown in Tables 7–10. After these analyses we selected the most significant genes for the four types of cancers by choosing a threshold () of the p-value. The survival curves of the most significant genes, comparing altered and normal groups had been obtained by using the PL estimator as shown in Fig. 5. Note that, from Fig. 5, we can see that those with altered expression of genes show lower survival compared to the normal group.
Table 8.
β coefficient, hazard ratio and p-values in univariate, multivariate and combined models of the identified genes that are common between WFs and PC.
| Gene symbol | Univariate | Multivariate | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β | HR | p-value | β | HR | p-value | β | HR | p-value | |
| ARAP3 | 1.12E-01 | 1.12E+00 | 4.48E-01 | 1.81E-01 | 1.20E+00 | 2.57E-01 | 2.38E-01 | 1.27E+00 | 1.39E-01 |
| CA14 | 1.49E-01 | 1.16E+00 | 3.74E-01 | 1.64E-01 | 1.18E+00 | 3.59E-01 | 2.11E-01 | 1.24E+00 | 2.45E-01 |
| F2RL2 | −4.70E-02 | 9.54E-01 | 7.53E-01 | −8.16E-02 | 9.22E-01 | 6.00E-01 | −5.31E-02 | 9.48E-01 | 7.41E-01 |
| FNIP2 | 3.27E-03 | 1.00E+00 | 9.84E-01 | −6.86E-03 | 9.93E-01 | 9.68E-01 | 1.20E-01 | 1.13E+00 | 4.93E-01 |
| GABRB3 | −9.83E-02 | 9.06E-01 | 4.13E-01 | −1.21E-01 | 8.86E-01 | 3.28E-01 | −1.14E-01 | 8.93E-01 | 3.67E-01 |
| IGFBP5 | 3.63E-01 | 1.44E+00 | 2.28E-02 | 3.85E-01 | 1.47E+00 | 1.86E-02 | 4.17E-01 | 1.52E+00 | 1.23E-02 |
| LEPR | −1.95E-02 | 9.81E-01 | 8.78E-01 | −2.36E-02 | 9.77E-01 | 8.57E-01 | −9.40E-02 | 9.10E-01 | 4.84E-01 |
| NDRG2 | 4.03E-01 | 1.50E+00 | 1.61E-02 | 3.80E-01 | 1.46E+00 | 2.74E-02 | 4.16E-01 | 1.52E+00 | 1.85E-02 |
| SPON1 | −8.50E-02 | 9.19E-01 | 6.38E-01 | −2.19E-01 | 8.04E-01 | 2.66E-01 | −3.41E-01 | 7.11E-01 | 8.93E-02 |
| TOX3 | −2.11E-02 | 9.79E-01 | 8.95E-01 | 8.41E-03 | 1.01E+00 | 9.60E-01 | −5.66E-02 | 9.45E-01 | 7.40E-01 |
Table 9.
β coefficient, hazard ratio and p-values in univariate, multivariate and combined models of the identified genes that are common between WFs and LC.
| Gene symbol | Univariate | Multivariate | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β | HR | p-value | β | HR | p-value | β | HR | p-value | |
| PROS1 | −5.57E-02 | 9.46E-01 | 8.12E-01 | 2.56E-02 | 1.03E+00 | 9.28E-01 | 2.42E-01 | 1.27E+00 | 4.09E-01 |
| SPON1 | 1.76E-01 | 1.19E+00 | 5.24E-01 | 4.52E-01 | 1.57E+00 | 1.72E-01 | 4.30E-01 | 1.54E+00 | 2.48E-01 |
| KCNK1 | −2.05E-01 | 8.15E-01 | 5.02E-01 | −1.35E-01 | 8.73E-01 | 7.04E-01 | −2.63E-01 | 7.68E-01 | 4.76E-01 |
| KRT8 | 1.19E-01 | 1.13E+00 | 6.81E-01 | 2.11E-01 | 1.23E+00 | 5.35E-01 | 2.88E-01 | 1.33E+00 | 4.21E-01 |
| ID3 | 7.21E-02 | 1.07E+00 | 8.34E-01 | 1.62E-01 | 1.18E+00 | 7.43E-01 | 1.13E-01 | 1.12E+00 | 8.39E-01 |
| CRYAB | −1.58E-01 | 8.54E-01 | 5.87E-01 | −8.98E-03 | 9.91E-01 | 9.79E-01 | −1.44E-01 | 8.66E-01 | 6.85E-01 |
| FGFR3 | 4.95E-01 | 1.64E+00 | 1.49E-01 | 5.82E-01 | 1.79E+00 | 1.71E-01 | 4.73E-01 | 1.60E+00 | 2.80E-01 |
| ENPP2 | −3.64E-01 | 6.95E-01 | 1.99E-01 | −5.45E-01 | 5.80E-01 | 1.26E-01 | −5.27E-01 | 5.90E-01 | 1.44E-01 |
| PMAIP1 | 2.78E-01 | 1.32E+00 | 4.37E-01 | 6.49E-01 | 1.91E+00 | 1.31E-01 | 1.02E+00 | 2.78E+00 | 2.91E-02 |
| RGS17 | −1.95E-01 | 8.23E-01 | 4.77E-01 | −2.73E-01 | 7.61E-01 | 4.00E-01 | −4.14E-02 | 9.59E-01 | 8.99E-01 |
| MTHFD2 | 2.14E-01 | 1.24E+00 | 4.29E-01 | 4.27E-01 | 1.53E+00 | 2.89E-01 | 5.09E-01 | 1.66E+00 | 2.13E-01 |
| ARAP3 | 3.37E-01 | 1.40E+00 | 2.32E-01 | 3.25E-01 | 1.38E+00 | 3.89E-01 | 5.24E-01 | 1.69E+00 | 1.83E-01 |
| TGFA | 3.10E-01 | 1.36E+00 | 3.66E-01 | 3.53E-01 | 1.42E+00 | 4.32E-01 | 4.43E-01 | 1.56E+00 | 3.62E-01 |
| HOXA5 | −6.82E-01 | 5.06E-01 | 1.41E-01 | −1.59E+00 | 2.03E-01 | 9.55E-03 | −1.86E+00 | 1.56E-01 | 5.18E-03 |
| PNISR | −1.38E-01 | 8.71E-01 | 6.23E-01 | −6.86E-01 | 7.56E-01 | 4.19E-01 | −4.15E-01 | 6.60E-01 | 2.44E-01 |
| GADD45A | 2.66E-01 | 1.31E+00 | 3.07E-01 | −2.80E-01 | 1.07E+00 | 8.28E-01 | 2.42E-01 | 1.27E+00 | 4.78E-01 |
| FHL2 | −3.39E-02 | 9.67E-01 | 9.02E-01 | 7.20E-02 | 6.50E-01 | 2.89E-01 | −2.87E-01 | 7.51E-01 | 4.61E-01 |
| TOX3 | −1.10E-01 | 8.96E-01 | 6.91E-01 | −4.30E-01 | 5.89E-01 | 1.65E-01 | −7.57E-01 | 4.69E-01 | 6.12E-02 |
| LEPR | 1.51E-01 | 1.16E+00 | 5.71E-01 | −5.29E-01 | 1.12E+00 | 7.35E-01 | 2.02E-01 | 1.22E+00 | 5.45E-01 |
| SLCO2A1 | −2.08E-01 | 8.13E-01 | 5.45E-01 | 1.12E-01 | 8.36E-01 | 6.82E-01 | −1.20E-01 | 8.87E-01 | 7.90E-01 |
| SLC7A5 | 1.42E-01 | 1.15E+00 | 5.88E-01 | −1.79E-01 | 1.23E+00 | 5.05E-01 | −2.26E-02 | 9.78E-01 | 9.44E-01 |
| MPDZ | 2.37E-01 | 1.27E+00 | 3.00E-01 | 2.11E-01 | 1.34E+00 | 2.58E-01 | 4.03E-01 | 1.50E+00 | 1.42E-01 |
Table 7.
β coefficient, hazard ratio and p-values in univariate, multivariate and combined models of the identified genes that are common between WFs and CC.
| Gene symbol | Univariate | Multivariate | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β | HR | p-value | β | HR | p-value | β | HR | p-value | |
| NPHP3 | −1.74E-01 | 8.40E-01 | 3.27E-01 | −2.52E-01 | 7.77E-01 | 2.34E-01 | −5.21E-02 | 9.49E-01 | 8.38E-01 |
| SAMD13 | 8.58E-02 | 1.09E+00 | 6.66E-01 | 1.24E-02 | 1.01E+00 | 9.58E-01 | 1.97E-01 | 1.22E+00 | 4.55E-01 |
| STC1 | 9.02E-02 | 1.09E+00 | 6.19E-01 | 1.08E-01 | 1.11E+00 | 6.06E-01 | −1.08E-01 | 8.98E-01 | 6.56E-01 |
| THEM6 | 9.20E-02 | 1.10E+00 | 5.89E-01 | 8.11E-02 | 1.08E+00 | 6.97E-01 | 2.95E-01 | 1.34E+00 | 2.10E-01 |
| TNFRSF12A | 3.16E-01 | 1.37E+00 | 8.94E-02 | 6.14E-01 | 1.85E+00 | 5.27E-03 | 8.19E-01 | 2.27E+00 | 6.48E-01 |
| ASNS | 1.35E-01 | 1.14E+00 | 5.23E-01 | 1.66E-01 | 1.18E+00 | 4.86E-01 | 2.38E-01 | 1.27E+00 | 4.17E-01 |
| FAM89A | 2.19E-01 | 1.24E+00 | 2.12E-01 | 3.28E-01 | 1.39E+00 | 1.12E-01 | 1.80E-01 | 1.20E+00 | 4.13E-01 |
| ACAT1 | −3.24E-02 | 9.68E-01 | 8.49E-01 | −9.12E-02 | 9.13E-01 | 6.55E-01 | 1.41E-01 | 1.15E+00 | 5.78E-01 |
| HPGD | 3.90E-01 | 1.48E+00 | 6.10E-02 | 3.63E-01 | 1.44E+00 | 1.25E-01 | 2.60E-01 | 1.30E+00 | 3.36E-01 |
| SLC7A8 | −3.99E−01 | 6.71E-01 | 5.01E-02 | −2.38E-01 | 7.88E-01 | 3.46E-01 | −5.80E-01 | 5.60E-01 | 4.87E-02 |
| TRIB3 | 1.30E-02 | 1.01E+00 | 9.39E-01 | −9.81E-02 | 9.07E-01 | 6.12E-01 | −3.09E-01 | 7.34E-01 | 1.58E-01 |
| APPL2 | −1.63E-01 | 8.50E-01 | 4.50E-01 | −1.29E-01 | 8.79E-01 | 5.98E-01 | 5.43E-02 | 1.06E+00 | 8.45E-01 |
| LRRC75B | −1.42E-01 | 8.67E-01 | 4.52E-01 | −3.28E-01 | 7.20E-01 | 1.35E-01 | −5.23E-01 | 5.93E-01 | 3.58E-02 |
| BAMBI | 4.46E-01 | 1.56E+00 | 1.39E-02 | 5.17E-01 | 1.68E+00 | 1.53E-02 | 2.39E-01 | 1.27E+00 | 3.32E-01 |
| FGFR3 | −2.06E-01 | 8.14E-01 | 3.04E-01 | −6.86E-01 | 5.03E-01 | 4.39E-03 | −8.88E-01 | 4.11E-01 | 9.54E-03 |
| DMXL2 | 5.36E-02 | 1.06E+00 | 7.46E-01 | 3.93E-02 | 1.04E+00 | 8.45E-01 | 1.93E-01 | 1.21E+00 | 3.98E-01 |
| CCL20 | 2.16E-01 | 1.24E+00 | 2.62E-01 | 4.86E-02 | 1.05E+00 | 8.24E-01 | 1.03E-02 | 1.01E+00 | 9.67E-01 |
| SERPINE2 | −3.53E-01 | 7.03E-01 | 6.55E-02 | −4.57E-01 | 6.33E-01 | 4.42E-02 | −5.14E-01 | 5.98E-01 | 4.31E-01 |
| CDK10 | 3.49E-02 | 1.04E+00 | 8.50E-01 | 1.02E-01 | 1.11E+00 | 6.21E-01 | 4.02E-01 | 1.50E+00 | 9.56E-02 |
| SON | 4.35E-01 | 1.54E+00 | 1.48E-02 | 2.89E-01 | 1.33E+00 | 1.44E-01 | 3.43E-01 | 1.41E+00 | 1.78E-01 |
| FCGBP | 1.18E-01 | 1.12E+00 | 5.39E-01 | 2.44E-01 | 1.28E+00 | 2.91E-01 | 2.33E-01 | 1.26E+00 | 3.44E-01 |
| IQGAP2 | −6.58E-02 | 9.36E-01 | 7.27E-01 | 1.66E-01 | 1.18E+00 | 4.71E-01 | 2.61E-01 | 1.30E+00 | 2.94E-01 |
| LUC7L3 | −9.57E-03 | 9.90E-01 | 9.57E-01 | −2.53E-01 | 7.77E-01 | 2.32E-01 | −2.38E-02 | 9.77E-01 | 9.22E-01 |
| BCKDHB | 2.77E-01 | 1.32E+00 | 1.01E-01 | 4.12E-01 | 1.51E+00 | 3.43E-02 | 5.91E-01 | 1.81E+00 | 6.52E-03 |
| LYAR | 2.14E-01 | 1.24E+00 | 2.71E-01 | 1.13E-01 | 1.12E+00 | 6.14E-01 | −1.11E-01 | 8.95E-01 | 6.57E-01 |
| NAAA | −2.33E-01 | 7.92E-01 | 2.44E-01 | −2.19E-01 | 8.03E-01 | 4.02E-01 | −5.92E-01 | 5.53E-01 | 2.86E-02 |
| SKAP2 | −1.80E-01 | 8.35E-01 | 3.12E-01 | −2.83E-01 | 7.54E-01 | 1.70E-01 | −1.09E-01 | 8.97E-01 | 6.46E-01 |
| BHLHE40 | 4.44E-03 | 1.00E+00 | 9.81E-01 | −1.13E-01 | 8.94E-01 | 6.30E-01 | −1.50E-01 | 8.61E-01 | 5.75E-01 |
| IGFBP5 | −4.24E-02 | 9.59E-01 | 8.31E-01 | 9.18E-02 | 1.10E+00 | 6.98E-01 | 2.18E-01 | 1.24E+00 | 3.99E-01 |
| SNX14 | 2.05E-02 | 1.02E+00 | 9.08E-01 | 6.87E-02 | 1.07E+00 | 7.37E-01 | −1.35E-01 | 8.74E-01 | 5.59E-01 |
| HOMER1 | −1.84E-01 | 8.32E-01 | 3.60E-01 | −2.37E-01 | 7.89E-01 | 2.83E-01 | −3.02E-01 | 7.39E-01 | 2.03E-01 |
| PAPLN | −3.29E-01 | 7.20E-01 | 1.23E-01 | −4.48E-01 | 6.39E-01 | 8.79E-02 | −2.94E-01 | 7.45E-01 | 3.21E-01 |
Table 10.
β coefficient, hazard ratio and p-values in univariate, multivariate and combined models of the identified genes that are common between WFs and GC.
| Gene symbol | Univariate | Multivariate | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β | HR | p-value | β | HR | p-value | β | HR | p-value | |
| ABCA4 | 1.44E-01 | 1.16E+00 | 6.89E-01 | 2.86E-01 | 1.33E+00 | 4.53E-01 | 3.51E-01 | 1.42E+00 | 3.60E-01 |
| CNN2 | −9.03E-02 | 9.14E-01 | 6.45E-01 | −2.10E-01 | 8.10E-01 | 3.12E-01 | −3.64E-01 | 6.95E-01 | 9.73E-02 |
| EGR2 | −2.19E-01 | 8.03E-01 | 6.64E-01 | −9.34E-02 | 9.11E-01 | 8.58E-01 | −7.03E-02 | 9.32E-01 | 8.96E-01 |
| EGR3 | −2.51E-01 | 7.78E-01 | 4.41E-01 | −1.95E-01 | 8.23E-01 | 5.64E-01 | −5.92E-02 | 9.43E-01 | 8.65E-01 |
| ENPP2 | −6.84E-02 | 9.34E-01 | 8.04E-01 | −6.85E-02 | 9.34E-01 | 8.10E-01 | 6.74E-02 | 1.07E+00 | 8.20E-01 |
| FCGBP | −5.25E-02 | 9.49E-01 | 8.66E-01 | −2.88E-02 | 9.72E-01 | 9.29E-01 | −6.37E-03 | 9.94E-01 | 9.84E-01 |
| FHIT | −2.65E-02 | 9.74E-01 | 9.38E-01 | 3.03E-01 | 1.35E+00 | 3.98E-01 | 3.37E-01 | 1.40E+00 | 3.51E-01 |
| HPGD | 4.71E-01 | 1.60E+00 | 8.91E-02 | 6.97E-01 | 2.01E+00 | 2.22E-02 | 6.83E-01 | 1.98E+00 | 2.90E-02 |
| IQGAP2 | −4.14E-01 | 6.61E-01 | 1.82E-01 | −7.08E-01 | 4.93E-01 | 3.29E-02 | −6.01E-01 | 5.48E-01 | 7.69E-02 |
| ITGA5 | −8.11E-01 | 4.45E-01 | 5.39E-02 | −9.77E-01 | 3.77E-01 | 2.95E-02 | −1.16E+00 | 3.15E-01 | 1.35E-02 |
| MTF1 | −9.49E-02 | 9.10E-01 | 6.40E-01 | −1.21E-01 | 8.86E-01 | 5.76E-01 | −6.43E-02 | 9.38E-01 | 7.71E-01 |
| PMAIP1 | −5.46E-02 | 9.47E-01 | 8.44E-01 | −1.10E-01 | 8.96E-01 | 7.18E-01 | −6.08E-02 | 9.41E-01 | 8.45E-01 |
| RBPJ | 5.92E-01 | 1.81E+00 | 6.64E-03 | 7.14E-01 | 2.04E+00 | 2.41E-03 | 7.31E-01 | 2.08E+00 | 2.37E-03 |
| THBS1 | −2.71E-01 | 7.63E-01 | 4.52E-01 | −2.75E-01 | 7.60E-01 | 4.62E-01 | −2.04E-01 | 8.15E-01 | 5.85E-01 |
| TPD52L1 | 2.90E-03 | 1.00E+00 | 9.92E-01 | −1.43E-01 | 8.67E-01 | 6.54E-01 | −1.79E-01 | 8.36E-01 | 5.80E-01 |
| TROVE2 | −1.26E-02 | 9.88E-01 | 9.54E-01 | −1.33E-01 | 8.75E-01 | 5.78E-01 | −1.54E-01 | 8.57E-01 | 5.25E-01 |
| UFM1 | −1.47E-01 | 8.63E-01 | 3.50E-01 | −1.61E-01 | 8.51E-01 | 3.52E-01 | −2.06E-01 | 8.14E-01 | 2.46E-01 |
Figure 5.
Survival function for an altered and normal group of the most significant genes that are common to WFs and the four types of cancers under investigation. These include significant genes common to WFs exposed cells and CC (a–e), PC (f,g), LC (h,i) and GC (j–l). Here, the cyan colored line in the survival graphs indicates the altered and the red indicates the normal gene expression group.
Discussion
In this study we investigated how WF exposure may influence a number of types of cancer whose development and growth is greater with exposure to WFs or the components of WFs. We compared the gene expression alterations that result from WF exposure in cells with that of the genes that have dysregulated expression in several cancer types. The idea behind this is similar to studies of comorbidities, where dysregulated genes (or more usually gene pathways) that are common to two diseases give clues to how those diseases interact when co-occurring in the same individual, even if we are unclear as to the reason for the altered expression of individual genes or pathways is unclear. Thus, genes or gene pathways altered in response to WF exposure and the cancers of interest can be means by which WF exposure encourages those cancers to develop. Note that WFs included components such as metal fumes that are absorbed by the lungs into the bloodstream, to expose many tissues around the body. Many of these fumes are carcinogenic, but cancer initiation is only one of a number of stages of cancer development and progression, and welders commonly have regular exposure to fumes over long periods. Unlike in other morbidities, some altered gene expression may arise in individual cancer cells due to mutations which will affect survival of those cells; if such altered expression the is detected in whole cancer tissue across many individuals (as in our studies) then the alteration may be affecting pathways that encourage survival and growth. Thus we have applied a systematic approach to identify pathways that WFs may affect the cancer behaviours.
For our analysis we employed gene regulation analysis, gene-disease association networks, signaling and ontological pathways, and protein-protein interaction networks. To identify pathways and genes that are important in WF interactions in the cellular processes that influence cancer progression, we examined gene expression microarray data from WF exposed cells, CC, PC, LC and GC, each with control datasets. This identified a large number of significant genes that were commonly dysregulated between WF-exposure and cancer profiles, and evident by simple gene expression comparisons. There were a number of dysregulated genes that were common between WF exposure responses and cancer types, which suggests that WF exposure may cause gene expression changes that could affect the behaviour of cancers. It should be noted that the cancer transcriptome datasets, such as those employed here, contain transcripts from both cancer cells and the supporting stromal cells found in the tumors themselves. Thus, it should be noted that WFs may exert their effects on cancers either indirectly (through tumor stroma) or on the cancer cells themselves.
We constructed two separate gene-disease association networks for up- and down-regulated genes showed strong evidence that WFs may indeed influence these cancers as indicated in Fig. 2(a,b). The pathway-based analysis is a technique to better understand the molecular or biological mechanisms underlying different complex diseases by determining common pathways that a stimulus (such as WFs) may influence cells of interest. We identified significant signaling and ontological pathways of the commonly dysregulated genes of each cancer. These identified pathways indicated how WFs may affect these cancer types. Similarly, protein-protein interaction sub-networks of the commonly altered genes suggest that WFs affect several types of cancers. Note that if a pathway is a conduit for the effects of an important risk factor for a disease, this points to that pathway being particularly important to the pathogenesis of the disease and that reducing that pathways effects could be a way to attack the disease progression itself. It should be noted that these findings only point to possible ways that WF exposure may affect the cancers and cannot prove causation. However, when we investigated whether the gene expression patterns that we have observed could be associated with reduced survival of the patients (pointing to the importance of those gene expression levels either directly or indirectly) that is what we observed for several of significant genes that are common WF the cancer profiles under investigation as shown in Fig. 5.
It should be noted that the datasets employ a number of different cell types, which is commonly the case in this type of study. While gene expression patterns are, by definition, different in different cell types, here we were only concerned with expression alterations; certain responses to WFs may not occur in all cells so, while our approach cannot identify all pathways affected by WFs in nascent tumour cells, it will find some. Indeed, our data provides evidence to suggest the involvement of a number of genes in cancer behaviours that are linked to the noxious effect of WFs on cancer.
We used the gold benchmark databases OMIM and dbGaP for cross checking the validity of our outcome and found that there were some shared genes in between the WF exposure and cancer types as shown in Fig. 2(c). For validation purposes, we collected disease with associated genes from the dbGaP, OMIM Disease and OMIM Expanded databases using differentially expressed genes of WFs. After several steps of statistical analysis we selected only cancer related diseases. Interestingly, we found our selected four cancers among the list of cancers collected from the mentioned databases as shown in Fig. 2(c).
Moreover, we found our identified genes in Fig. 2(c) had been shown in other studies to be associated with disease progression in cancers. Specifically, Vázquez-ArreguÃn K. et al., Cybulski C. et al. and Wang L. et al. shown RAB4B, CHEK2 and FOS to be associated with CC incidence47–50; Biswas S. et al. found a link between TGFBR2 and CC51. Lijovic M. et al. showed CD82 to be linked to PC incidence52; Wang Y. et al. shown the association between CHEK2 and PC progression53; Ouyang X. et al. identified a link between FOS and PC54; Gruosso T. et al. showed MAP3K8 to be associated with LC55; Vallejo A. et al. found a link between FOS and LC incidence56; Yuan S. et al. showed an association between GPC5 and LC progression57. Kim CJ. et al. found MUTYH to be associated to GC incidence58; Myllykangas S. found an association between FOS and GC59; Teodorczyk U. et al. found CHEK2 to be linked to GC progression60. Therefore, it suggested that WFs may have a strong interaction with CC, PC, LC and GC.
Conclusions
In this study, we considered gene expression microarray data from WFs exposure, CC, PC, LC, GC and control datasets to analyze and investigate the genetic links between WF exposure and the effects that they have on cancers. We analyzed gene expression, constructed gene-disease association networks, identified signaling and ontological pathways, analyzed protein-protein interaction networks and survival function of WFs exposed cells and cancers. The outcome of our study indicated that WFs can exert a strong influence on cancers. This kind of study will be useful for making more accurate disease prediction, and identifyi potentially better therapeutic approaches. This study will also be useful for assessing the dangerous effects of welding on the human body.
Acknowledgements
This study was supported by funds from the Sydney Medical School - K1329 and University of Sydney - U2347.
Author contributions
H.K. Rana and M.R. Akhtar performed all experiments, analyzed the results and prepared the manuscript; M.B. Islam and M.B. Ahmed performed some statistical analysis; J.M.W. Quinn reviewed the manuscript; M.A. Moni supervised and reviewed the manuscript; F. Huq and P. Lió approved the final draft of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Humayan Kabir Rana and Mst. Rashida Akhtar.
References
- 1.Antonini JM. Health effects of welding. Critical reviews toxicology. 2003;33:61–103. doi: 10.1080/713611032. [DOI] [PubMed] [Google Scholar]
- 2.Guha N, et al. Carcinogenicity of welding, molybdenum trioxide, and indium tin oxide. The Lancet Oncol. 2017;18:581–582. doi: 10.1016/S1470-2045(17)30255-3. [DOI] [PubMed] [Google Scholar]
- 3.Rana HK, et al. Genetic effects of welding fumes on the progression of neurodegenerative diseases. Neurotoxicology. 2019;71:93–101. doi: 10.1016/j.neuro.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 5.MacLeod JS, Harris MA, Tjepkema M, Peters PA, Demers PA. Cancer risks among welders and occasional welders in a national population-based cohort study: canadian census health and environmental cohort. Saf. health at work. 2017;8:258–266. doi: 10.1016/j.shaw.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oddone E, Modonesi C, Gatta G. Occupational exposures and colorectal cancers: a quantitative overview of epidemiological evidence. World J. Gastroenterol. WJG. 2014;20:12431. doi: 10.3748/wjg.v20.i35.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parent M-É, Siemiatycki J. Occupation and prostate cancer. Epidemiol. reviews. 2001;23:138–43. doi: 10.1093/oxfordjournals.epirev.a000779. [DOI] [PubMed] [Google Scholar]
- 8.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat. Rev. Cancer. 2017;17:725. doi: 10.1038/nrc.2017.87. [DOI] [PubMed] [Google Scholar]
- 9.Hazardous substances in welding fumes: How they affect the human body - safe welding, https://safe-welding.com/hazardous-substances-in-welding-fumes-how-they-affect-the-human-body/. (Accessed on 11/23/2018).
- 10.Siegel RL, et al. Colorectal cancer statistics, 2017. CA: a cancer journal for clinicians. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 11.Prostate cancer - symptoms and causes - mayo clinic, https://www.mayoclinic.org/diseases-conditions/prostate-cancer/symptoms-causes/syc-20353087. (Accessed on 11/23/2018).
- 12.Stomach cancer: Symptoms, treatment, and possible causes, https://www.webmd.com/cancer/stomach-gastric-cancer#1. (Accessed on 11/23/2018).
- 13.Geo accession viewer, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62384. (Accessed on 11/23/2018).
- 14.Ågesen T, et al. Clc and ifnar1 are differentially expressed and a global immunity score is distinct between early-and late-onset colorectal cancer. Genes immunity. 2011;12:653. doi: 10.1038/gene.2011.43. [DOI] [PubMed] [Google Scholar]
- 15.Arredouani, M. S. et al. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin. Cancer Res. 1078–0432 (2009). [DOI] [PMC free article] [PubMed]
- 16.Landi MT, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PloS one. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hippo Y, et al. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer research. 2002;62:233–240. [PubMed] [Google Scholar]
- 18.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal. 2013;6:pl1–pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami, E. et al. The cbio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data (2012). [DOI] [PMC free article] [PubMed]
- 20.Network CGA, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nat. 2012;487:330. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abeshouse A, et al. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoadley KA, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Network CGAR, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nat. 2014;513:202. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rana HK, et al. Genetic effects of welding fumes on the development of respiratory system diseases. Comput. biology medicine. 2019;108:142–149. doi: 10.1016/j.compbiomed.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Moni MA, Lio’ P. Genetic profiling and comorbidities of zika infection. The J. infectious diseases. 2017;216:703–712. doi: 10.1093/infdis/jix327. [DOI] [PubMed] [Google Scholar]
- 26.Moni MA, et al. A computational approach to identify blood cell-expressed parkinson’s disease biomarkers that are coordinately expressed in brain tissue. Comput. biology medicine. 2019;113:103385. doi: 10.1016/j.compbiomed.2019.103385. [DOI] [PubMed] [Google Scholar]
- 27.Moni MA, Liò P. comor: a software for disease comorbidity risk assessment. J. clinical bioinformatics. 2014;4:8. doi: 10.1186/2043-9113-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moni MA, Liò P. How to build personalized multi-omics comorbidity profiles. Front. cell developmental biology. 2015;3:28. doi: 10.3389/fcell.2015.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman MR, et al. The influence of depression on ovarian cancer: Discovering molecular pathways that identify novel biomarkers and therapeutic targets. Informatics Medicine Unlocked. 2019;16:100207. doi: 10.1016/j.imu.2019.100207. [DOI] [Google Scholar]
- 31.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. Kegg: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research. 2016;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutmon M, et al. Wikipathways: capturing the full diversity of pathway knowledge. Nucleic acids research. 2015;44:D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabregat A, et al. Reactome pathway analysis: a high-performance in-memory approach. BMC bioinformatics. 2017;18:142. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura D. Biocarta. Biotech Softw. & Internet Report: The Comput. Softw. J. for Sci. 2001;2:117–120. doi: 10.1089/152791601750294344. [DOI] [Google Scholar]
- 35.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. genetics. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Köhler S, et al. The human phenotype ontology in 2017. Nucleic acids research. 2016;45:D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franceschini A, et al. String v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2012;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox DR. Regression models and life-tables. J. Royal Stat. Soc. Ser. B (Methodological) 1972;34:187–202. [Google Scholar]
- 39.Xu H, Moni MA, Liò P. Network regularised cox regression and multiplex network models to predict disease comorbidities and survival of cancer. Comput. biology chemistry. 2015;59:15–31. doi: 10.1016/j.compbiolchem.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Hossain, M. A., Islam, S. M. S., Quinn, J. M., Huq, F. & Moni, M. A. Machine learning and bioinformatics models to identify gene expression patterns of ovarian cancer associated with disease progression and mortality. J. biomedical informatics 103313 (2019). [DOI] [PubMed]
- 41.Moni MA, Xu H, Lio P. Cytocom: a cytoscape app to visualize, query and analyse disease comorbidity networks. Bioinforma. 2014;31:969–971. doi: 10.1093/bioinformatics/btu731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin L, et al. Pathway-based analysis tools for complex diseases: a review. Genomics, proteomics & bioinformatics. 2014;12:210–220. doi: 10.1016/j.gpb.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hossain, M. A., Asa, T. A., Huq, F., Quinn, J. M. & Moni, M. A. A network-based approach to identify molecular signatures and comorbidities of thyroid cancer. In Proceedings of International Joint Conference on Computational Intelligence, 235–246 (Springer, Singapore, 2020).
- 44.Hossain, M. A., Asa, T. A., Rahman, M. R. & Moni, M. A. Network-based approach to identify key candidate genes and pathways shared by thyroid cancer and chronic kidney disease (2019).
- 45.Gene regulation ontology - summary | ncbo bioportal, https://bioportal.bioontology.org/ontologies/GRO. (Accessed on 11/05/2019).
- 46.Hossain, M. E., Khan, A., Moni, M. A. & Uddin, S. Use of electronic health data for disease prediction: A comprehensive literature review. IEEE/ACM Transactions on Comput. Biol. Bioinforma. (2019). [DOI] [PubMed]
- 47.Vázquez-Arreguín K, et al. Oct1/pou2f1 is selectively required for colon regeneration and regulates colon malignancy. PLoS genetics. 2019;15:e1007687. doi: 10.1371/journal.pgen.1007687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cybulski C, et al. Germline chek2 mutations and colorectal cancer risk: different effects of a missense and truncating mutations? Eur. J. Hum. Genet. 2007;15:237. doi: 10.1038/sj.ejhg.5201734. [DOI] [PubMed] [Google Scholar]
- 49.Wang L, Sun Y, Jiang M, Zhang S, Wolfl S. Fos proliferating network construction in early colorectal cancer (crc) based on integrative significant function cluster and inferring analysis. Cancer investigation. 2009;27:816–824. doi: 10.1080/07357900802672753. [DOI] [PubMed] [Google Scholar]
- 50.Rahman M, et al. Identification of prognostic biomarker signatures and candidate drugs in colorectal cancer: Insights from systems biology analysis. Medicina. 2019;55:20. doi: 10.3390/medicina55010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biswas S, et al. Mutational inactivation of tgfbr2 in microsatellite unstable colon cancer arises from the cooperation of genomic instability and the clonal outgrowth of transforming growth factor β resistant cells. Genes, Chromosom. Cancer. 2008;47:95–106. doi: 10.1002/gcc.20511. [DOI] [PubMed] [Google Scholar]
- 52.Lijovic M, Somers G, Frauman AG. Kai1/cd82 protein expression in primary prostate cancer and in bph associated with cancer. Cancer Detect. Prev. 2002;26:69–77. doi: 10.1016/S0361-090X(02)00012-0. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Dai B, Ye D. Chek2 mutation and risk of prostate cancer: a systematic review and meta-analysis. Int. journal clinical experimental medicine. 2015;8:15708. [PMC free article] [PubMed] [Google Scholar]
- 54.Ouyang X, et al. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer research. 2008;68:2132–2144. doi: 10.1158/0008-5472.CAN-07-6055. [DOI] [PubMed] [Google Scholar]
- 55.Gruosso T, et al. Map3k8/tpl-2/cot is a potential predictive marker for mek inhibitor treatment in high-grade serous ovarian carcinomas. Nat. communications. 2015;6:8583. doi: 10.1038/ncomms9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vallejo A, et al. An integrative approach unveils fosl1 as an oncogene vulnerability in kras-driven lung and pancreatic cancer. Nat. communications. 2017;8:14294. doi: 10.1038/ncomms14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan S, et al. Gpc5, a novel epigenetically silenced tumor suppressor, inhibits tumor growth by suppressing wnt/β-catenin signaling in lung adenocarcinoma. Oncogene. 2016;35:6120. doi: 10.1038/onc.2016.149. [DOI] [PubMed] [Google Scholar]
- 58.Kim CJ, et al. Genetic alterations of the myh gene in gastric cancer. Oncogene. 2004;23:6820. doi: 10.1038/sj.onc.1207574. [DOI] [PubMed] [Google Scholar]
- 59.Myllykangas S, Monni O, Nagy B, Rautelin H, Knuutila S. Helicobacter pylori infection activates fos and stress-response genes and alters expression of genes in gastric cancer-specific loci. Genes, Chromosom. Cancer. 2004;40:334–341. doi: 10.1002/gcc.20047. [DOI] [PubMed] [Google Scholar]
- 60.Teodorczyk U, et al. The risk of gastric cancer in carriers of chek2 mutations. Fam. cancer. 2013;12:473–478. doi: 10.1007/s10689-012-9599-2. [DOI] [PubMed] [Google Scholar]