Abstract
Multifractionated irradiation is the mainstay of radiation treatment in cancer therapy. Yet, little is known about the cellular DNA repair processes that take place between radiation fractions, even though understanding the molecular mechanisms promoting cancer cell recovery and survival could improve patient outcome and identify new avenues for targeted intervention. To address this knowledge gap, we systematically characterized how cells respond differentially to multifractionated and single-dose radiotherapy, using a combination of genetics-based and functional approaches. We found that both cancer cells and normal fibroblasts exhibited enhanced survival after multifractionated irradiation compared with an equivalent single dose of irradiation, and this effect was entirely dependent on 53BP1-mediated NHEJ. Furthermore, we identified RIF1 as the critical effector of 53BP1. Inhibiting 53BP1 recruitment to damaged chromatin completely abolished the survival advantage after multifractionated irradiation and could not be reversed by suppressing excessive end resection. Analysis of the TCGA database revealed lower expression of 53BP1 pathway genes in prostate cancer, suggesting that multifractionated radiotherapy might be a favorable option for radio-oncologic treatment in this tumor type. We propose that elucidation of DNA repair mechanisms elicited by different irradiation dosing regimens could improve radiotherapy selection for the individual patient and maximize the efficacy of radiotherapy.
INTRODUCTION
Radiotherapy, which is usually applied in multiple fractions over a period of days or weeks, is a key component of cancer treatment. Several different irradiation (IR) schedules with or without systemic therapy are currently in clinical use. The majority of patients are irradiated with a conventional fractionated schedule consisting of 2-Gy fraction per day up to a total dose of 60–90 Gy, although for some tumor types there is currently a preference toward the use of hypofractionation applying one or a few larger fractions (1). This trend is in part due to technological advances that have enabled more accurate targeting of malignant tissue, which allows the application of higher doses without significant side-effects on normal tissue (1).
In general, both tumor and normal cells show better survival when radiotherapy is delivered in multiple fractions. Several factors may contribute to this effect including DNA repair and repopulation processes between fractions as well as changes in cell cycle distribution and tissue oxygenation (2). We have previously demonstrated that IR-induced activation of pro-survival pathways is affected by the fractionation regimen of radiotherapy (3,4). In this report, we sought to address whether cellular DNA repair mechanisms also differ following a single dose of IR compared to the same total dose delivered over multiple fractions.
Besides its eponymous role of interacting with the cell cycle regulator p53, tumor suppressor p53 binding protein 1 (53BP1) is a DNA repair protein that is rapidly recruited to DNA double-strand breaks (DSBs) (5). Localization of 53BP1 to DSBs is dependent on multiple chromatin changes occurring in the vicinity of DNA lesions. The Tudor domain of 53BP1 binds to di-methylated histone H4 (H4-K20me2) (6,7). This interaction is regulated by bromodomain proteins (BET) and therefore can be suppressed by treatment with BET inhibitors such as JQ1 (8,9). In addition, histone ubiquitylation at DSB sites by the RING finger E3 ubiquitin ligases RNF8 and RNF168 are crucial for the recruitment of 53BP1 (10,11).
53BP1 protects DNA ends from nuclease-mediated 5′ end resection, which favors DNA repair by non-homologous end-joining (NHEJ), such as during immunoglobulin class switch recombination (CSR) (12). Recently, Rap1-interacting factor 1 (RIF1) and Pax transactivation domain-interacting protein (PTIP) were identified as key downstream effectors of 53BP1. RIF1 and PTIP bind distinct DNA damage- and ATM-inducible phospho-epitopes on 53BP1 and both factors contribute to limiting end resection at DNA DSB sites (13–15). By contrast, BRCA1 promotes end resection by antagonizing 53BP1, at least partly by inducing dephosphorylation of 53BP1 and thereby shift DNA repair to homologous recombination (HR) (16–18). Unrestrained 53BP1 activity is highly toxic in cells lacking BRCA1 and causes profound sensitivity to chemotherapeutic agents that normally trigger HR, such as inhibitors of Poly(ADP-ribose) polymerase (PARP) (19). Conversely, loss of 53BP1 largely restores HR in BRCA1-deficient cells and represents an important mechanism by which tumors become resistant to PARP inhibition (20). While both NHEJ and HR are known to participate in the repair of DNA lesions produced by radiotherapy, it is unclear how each may contribute to the adaptive response observed during multifractionated IR regimens and if mutual antagonism between these two pathways exists also in this setting. Here, we demonstrate that 53BP1 and RIF1, but not BRCA1, are specifically required for the efficient repair of DNA DSBs that lead to enhanced survival following multifractionated radiotherapy. Moreover, deleting BRCA1 in 53BP1-deficient cells does not diminish the survival benefit of fractionation, suggesting that limiting end resection cannot readily restore NHEJ in the absence of 53BP1.
MATERIALS AND METHODS
Antibodies
Antibodies for western blotting included 53BP1 (Cell Signaling), β-actin (Millipore), IRDye 800CW donkey anti-mouse and IRDye 680RD Donkey anti-rabbit antibodies (LI-COR). Antibodies for immunofluorescence staining included 53BP1 (BD), γH2AX (Millipore), RIF1 (21), AlexaFluor 488 anti-rabbit and AlexaFlour 594 anti-mouse antibodies (Invitrogen).
Cell culture and radiation exposure
DU145, PC3 and H1299 were obtained from the NCI tumor bank and LNCaP and A549 from ATCC. A passage number of 15 was not exceeded. Atm−/− (22), Brca1Δ11 (20), 53bp1−/− (23), 53bp1−/− Brca1Δ11 (20), Rnf8−/− (24), Rnf168−/− (25), Ku80−/− (26), 53BP1 S25A (15) and Rif1fl/fl (21,27) MEF have been previously described. Ptip−/− MEF were a kind gift from Dr Kai Ge (NIDDK/NIH). Asynchronous exponentially growing cells were used in all experiments. DU145, PC3, LNCaP and H1299 were cultured in RPMI 1640 containing GlutaMAX (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen); A549 cells in Dulbecco's modified Eagle's medium (DMEM) containing GlutaMAX supplemented with 10% FBS, and MEF in DMEM containing GlutaMAX supplemented with 15% FBS and 1% penicillin/streptomycin (Invitrogen). Irradiation was delivered at room temperature as a single dose or multiple fractions of 320 kV X-rays (Precision X-Ray Inc., North Branford, CT, USA). The dose-rate was ∼2.3 Gy/min, and the total doses ranged from 0 to 10 Gy. Multifractionated irradiation was carried out in fractions of 2 Gy per day.
Conditional deletion of Rif1 by Cre-mediated recombination
BOSC23 cells were transfected with pMX-Cre (20) and the packaging plasmid pCL-Eco (Addgene plasmid #12371) using Lipofectamine2000 (Invitrogen). After 48 h, filtered (0.45 μM PVDF syringe filter) viral supernatant was harvested and spiked with 8 μg/ml polybrene (Sigma) to transduce Rif1fl/fl MEF. Selection of Cre-expressing cells was accomplished with puromycin (Invivogen). Additionally, cells were sorted two times for GFP expression. RIF1 knockout was confirmed with immunofluorescence staining and western blotting.
Ctip shRNA retroviral transduction
BOSC23 cells were transfected with pMX-IRES-GFP empty vector or pMX-IRES-GFP containing a Ctip shRNA construct (5′-TGCTGTTGACAGTGAGCGAACAGAG AATCTCTTTGGTGATTAGTGAAGCCA CAGATGTAATCACCAAAGAGATTCTC TGTCTGCCTACTGCCTCGGA-3′) using FuGENE 6 (Promega) as previously described (20). pCL-Eco was used as the packaging plasmid. WT and Rnf168−/− MEF were incubated with the viral supernatant spiked with 10 μg/ml polybrene for 48 h. Successful transduction was confirmed by qPCR.
CRISPR/Cas9 gene editing
Deletion of 53BP1 was accomplished by CRISPR–CAS9 gene editing using an established protocol (3,28). LentiCRISPR v2 was acquired from Addgene (plasmid # 52961). Design of target-specific oligonucleotides was performed with the CHOPCHOP algorithm tool (https://chopchop.rc.fas.harvard.edu/). The oligonucleotides were annealed and cloned into the linearized LentiCRISPR v2 vector (FastDigest Esp3I; ThermoFisher Scientific). 293T cells were transfected with the 53BP1-targeting LentiCRISPR plasmid along with the psPAX2 and pMD2.VSVG packaging constructs using Lipofectamine 2000. The virus-containing media was harvested, filtered and spiked with 8 μg/ml polybrene before it was added to PC3 and H1299 cells. Selection was performed with puromycin. Knockdown of 53BP1 expression was confirmed by Western blotting. Initially, five different oligonucleotide sequences were used. Construct 1 (F: GCACAAGAACTTATGGAAAG, R: CTTTCCATAAGTTCTTGTGC) had the highest knockdown efficiency and was chosen for further experiments.
Colony formation assay
Colony formation assays were performed as previously published (29). In brief, cells were trypsinized, counted and seeded in six-well plates. Irradiation was started at 24 h after plating. Cells were treated with the BET inhibitor JQ1 (Selleckchem) 30 min before the start of irradiation. JQ1-containing media was renewed every 24 h for 3 days. DMSO treated cells were used as control. Cells were cultured for 7–14 days depending on the cell line. After fixation and staining with 0.4% crystal violet, cell clusters with >50 cells were counted with a stereomicroscope (Cambridge Instruments). Surviving fractions were calculated as follows: (irradiated colony number/unirradiated colony number).
Western blotting
Asynchronous exponentially growing cell cultures were harvested and lysed in modified RIPA buffer supplemented with protease inhibitors (Complete; Roche) and phosphatase inhibitors as previously described (30). Homogenization of lysates was accomplished by four passages through a 25-gauge needle followed by centrifugation at 16000×g for 20 min. Samples were stored at −80°C. SDS-PAGE, transfer of proteins onto nitrocellulose membranes (BioRad, Hercules, CA, USA) and probing with indicated primary antibodies was carried by standard procedures. Detection was achieved with fluorescently-labeled secondary antibodies on the Odyssey CLx imager (LI-COR).
Real-time PCR
1 μg of total RNA was reverse transcribed using RT2 First Strand synthesis kit (Qiagen as previously described (4). qPCR assays were performed using RT2 SYBR Green ROX qPCR Mastermix and RT² qPCR Primer Assays (Qiagen) for Ctip and Cdkn1a. Actb was used as the normalizing gene. Real-time PCR reactions were performed using an Applied Biosystems’ thermal cycler (Quant Studio 3). PCR steps included a 15-min holding stage at 95°C, 40 cycles of alternate denaturation at 95°C for 15 s, and annealing/extension at 60°C for 1 min. For specificity control of the corresponding RT-PCR reactions, a melt curve analysis was performed. Fold change in gene expression was calculated using the 2-ddCt method where ddCt = dCt (test) - dCt (control); dCt = Ct (gene) – Ct (Actb); and Ct is the threshold cycle number.
Immunofluorescence staining
Immunofluorescence staining was carried out as described (31). At 24 h after irradiation, cells were fixed with 3% formaldehyde/PBS for 15 min. Permeabilization was performed with 0.25% Triton X-100/PBS for 10 min. After washing with PBS, samples were blocked with 3% BSA/PBS for 30 min. Samples were stained with 53BP1, γH2AX or RIF1 antibody overnight at 4°C. A 1-h incubation with secondary antibodies was performed after washing with PBS. Samples were mounted with Vectashield/DAPI mounting medium (Alexis, Farmingdale, NY, USA). Images were acquired using an AxioImager.Z1/ApoTome microscope (Zeiss, Peabody, MA, USA). The number of foci per nuclei was quantified for at least 50 cells. Nuclei showing signs of apoptosis were excluded from the analysis.
Cell cycle analysis
Cell cycle distribution of MEF and tumor cells was analyzed as published (4). After cells were incubated with 10 mM BrdU for 10 min, samples were fixed with 80% EtOH and incubated with 2 N HCl and 0.1 M Na2B4O7. Staining was performed with anti-BrdU antibody (BD PharMingen) and anti-mouse AlexaFlour 647 (Invitrogen) overnight at 4°C. Total DNA staining was carried out with propidium iodide (PI, Invitrogen) solution containing RNAse (Invitrogen). Data for 10000 events were acquired on an LSR Fortessa flow cytometer (Becton Dickinson) with DIVA software. Cell cycle distribution was analyzed using the FlowJo software (Tree Star).
Ex vivo immunoglobulin class switch recombination assay
Resting primary B lymphocytes were isolated from mouse spleens using anti-CD43 microbeads (Miltenyi Biotec). One million cells were stimulated to proliferate with a cytokine cocktail containing 25 μg/ml lipopolysaccharide (LPS, Sigma-Aldrich), 5 ng/ml interleukin-4 (IL-4, Sigma-Aldrich) and 0.5 μg/ml anti-CD180 (RP105, BD PharMingen). CSR to IgG1 was detected on day 3 using biotinylated anti-IgG1 followed by PE-conjugated streptavidin (BD Biosciences). Anti-B220 was used to confirm the purity of B cell samples.
Gene expression analysis in human patient samples
Expression of DDR genes in prostate cancer and normal prostate was analyzed using the ‘The Cancer Genome Atlas’ (TCGA) database as recently published (3). Normalized mRNA expression data for prostate cancer and normal prostate tissue and for NSCLC and normal lung tissue were retrieved using the R package ‘TCGAbiolinks’ (32). The following genes were preselected for analysis: TP53BP1, RNF8, RNF168, RIF1 and KU80. Unpaired, two-sided Student's t-test was used to test for significant differences in gene expression between cancer and normal tissue.
Data and statistical analysis
Analyses of experimental data were performed with Microsoft Excel or R. Fold change was calculated by normalization of measured values to the corresponding control. To test for statistical significance, the unpaired, two-sided Student's t-test was used. Results were considered statistically significant if a P value of <0.05 was reached.
RESULTS
Efficacy of multifractionated versus single-dose radiotherapy differs between cancer cell lines
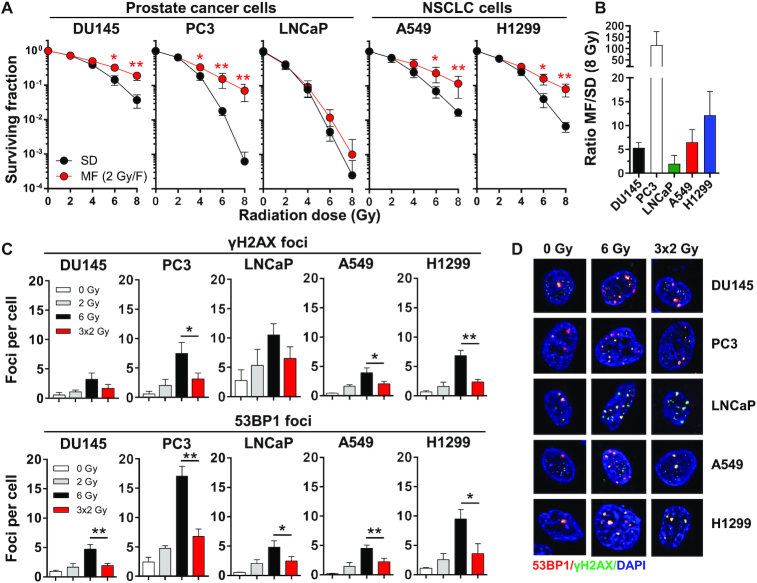
Different types of cancer or even individual tumors of the same cancer type can differ strongly in their responses to IR. Therefore, we first determined the clonogenic survival of three prostate carcinoma cell lines (DU145, PC3, LNCaP) and two non-small cell lung cancer (NSCLC) cell lines (A549, H1299) after radiotherapy, given either as a single dose (SD) or in multiple 2-Gy fractions (MF) amounting to the same total dose (Figure 1A). Additionally, we calculated the survival ratio for each cell line enabling a direct comparison between the efficacy of the SD and MF regimens (Figure 1B). We found that all of the tested cell lines had a higher survival rate after fractionated irradiation but the MF/SD ratios differed in a cell line-dependent manner ranging from 114.4 for PC3 to 2.0 for LNCaP (Figure 1A, B). Consistent with the increased survival rates, significantly lower numbers of residual γH2AX and 53BP1 foci were detected in cells after MF than in cells that had received the corresponding SD irradiation (Figure 1C,D). In general, cells with larger differences in residual γH2AX/53BP1 foci tended to show higher MF/SD ratios (Figure 1B–D), suggesting that DNA repair could play a key role in conferring survival benefits after MF. Indeed, the 3 × 2-Gy MF regimen produced similar amounts of residual γH2AX/53BP1 foci as a single 2-Gy dose (Figure 1C), supporting the notion that efficient repair of DNA damage occurred between each radiation fraction. Cell cycle analyses revealed G1 arrest in MF-irradiated p53 wildtype cells (LNCaP, A549) and G2 arrest in p53 deleted or mutated cells (DU145, PC3, H1299) (Supplementary Figure S1). Thus, the type of checkpoint engagement alone did not appear to predict response to MF irradiation. Overall, these results clearly demonstrated that cancer cells differ in their susceptibility to SD and MF regimens. Therefore, identifying the most appropriate dosing schemes (standard 2-Gy MF versus hypofractionation/stereotactic irradiation with higher single doses) for any individual tumor may significantly impact patient survival and outcome.
Figure 1.
Survival after single-dose and fractionated radiotherapy is cell line-dependent. (A) Clonogenic survival of human prostate (DU145, PC3, LNCaP) and non-small cell lung carcinoma (NSCLC) cells (A549, H1299) after irradiation with a single dose (SD) of X-ray (2–8 Gy) or with a multifractionated (MF) regimens (1 fraction of 2 Gy per day, 2–8 Gy total dose). Unirradiated cell cultures were used as control. (B) Efficacy of MF versus SD radiotherapy. Ratio of MF to SD survival was calculated as follows: surviving fraction after four 2 Gy-fractions (4 × 2 Gy)/surviving fraction after a single dose of 8 Gy. (C) Number of residual γH2AX (green) and 53BP1 (red) foci and (D) representative images in tumor cells at 24 h after the final irradiation dose of SD and MF radiotherapy. Results show mean ± STDEV (n = 3, *P < 0.05, **P < 0.01, Student's t-test).
53BP1 is crucial for efficient DNA DSB repair after multifractionated radiotherapy
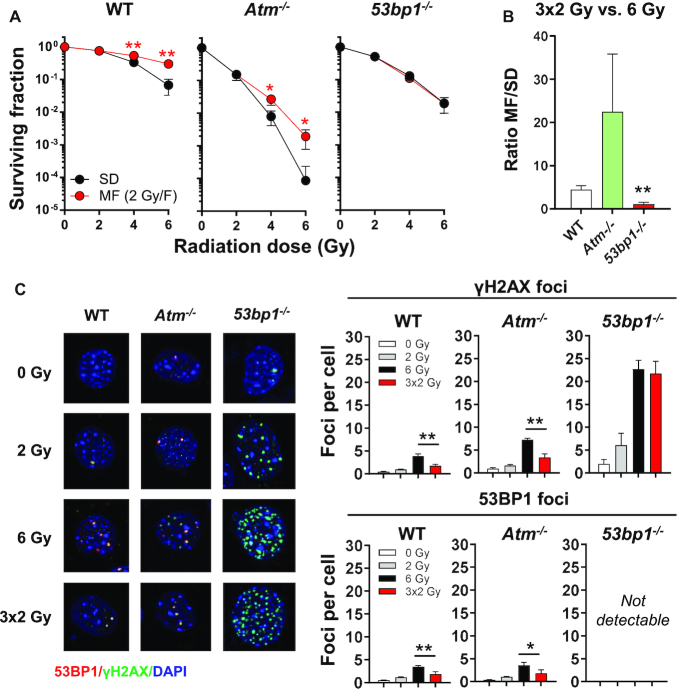
Our initial data suggested that DNA damage incurred during MF irradiation may be more readily repaired than lesions produced by the corresponding SD irradiation. To clarify the role of DNA damage response (DDR) proteins in cell survival after multifractionated radiotherapy, we used different knockout mouse embryonic fibroblast (MEF) lines and irradiated the cells with single doses of radiation (2, 4, 6 Gy) as well as the corresponding fractionated regimens (2 × 2 Gy, 3 × 2 Gy). Similar to human cancer cells, wild type (WT) MEF had significantly better survival after MF irradiation compared to SD irradiation, resulting in an MF/SD survival ratio of ∼5 (Figure 2A, Supplementary Figure S2A, B). The effect was most pronounced with a 24-h time interval between fractions but also occurred when fractions were applied at intervals as short as 30 min (Supplementary Figure S2C). While a significant checkpoint response was elicited in WT MEF when cells were irradiated at a 24-h time interval, no cell cycle changes were observed when the time interval between fractions was reduced to 2 h (Supplementary Figure S3). Thus, robust checkpoint-mediated cell cycle arrest between radiation fractions is not strictly required for the enhanced survival of cells following MF IR. Compared to WT, MEF deficient in ATM were significantly more radiosensitive to both SD and MF irradiation. Nevertheless, Atm−/− MEF still exhibited a significantly higher survival rate after MF irradiation compared to SD irradiation (Figure 2A, B). By contrast, loss of 53BP1 completely abolished the survival benefit from fractionation (Figure 2A, B, Supplementary Figure S2C) but only moderately sensitized cells to SD irradiation (Supplementary Figure S4A). Consistent with this, residual γH2AX foci in 53bp1−/− MEF were comparable after MF and SD irradiation (3 × 2-Gy versus 6-Gy), suggesting a key role of 53BP1 in promoting enhanced survival after MF irradiation (Figure 2C). Notably, loss of 53BP1 resulted in significantly elevated residual γH2AX foci regardless of the IR schedule, suggesting that 53BP1 function is required for the repair of radiation-induced DNA damage in general (Figure 2C). Although 53BP1 has also been reported to regulate checkpoint responses (33,34), we found that differences in cell cycle arrest did not correlate with MF/SD ratio (Supplementary Figure S4B). Taken together, our results indicate that 53BP1-mediated DSB repair during multifractionated radiotherapy is crucial for enhanced cell survival.
Figure 2.
53BP1 is essential for DNA DSB repair after fractionated radiotherapy. (A) Clonogenic survival of wildtype (WT), Atm−/− and 53bp1−/− MEF after irradiation with single doses (2, 4, 6 Gy) or multiple fractions (2 or 3 fractions of 2 Gy, 1 fraction per day). Cells were plated 24 h prior to irradiation. Fold change was calculated by normalization to unirradiated controls (0 Gy). (B) Ratio of MF to SD survival was calculated as follows: surviving fraction after three 2 Gy-fractions (3 × 2 Gy)/surviving fraction after a single dose of 6 Gy. (C) Representative images and numbers of γH2AX (green) and 53BP1 (red) foci in cells irradiated with a single dose of 6 Gy or 3 fractions of 2 Gy. Nuclei were counterstained with DAPI. Cells were fixed at 24 h after the end of radiotherapy. Results show mean ± STDEV (n = 3, *P < 0.05, **P < 0.01, Student's t-test).
Decreasing 53BP1 binding to chromatin reduces the survival benefit from multifractionated radiotherapy
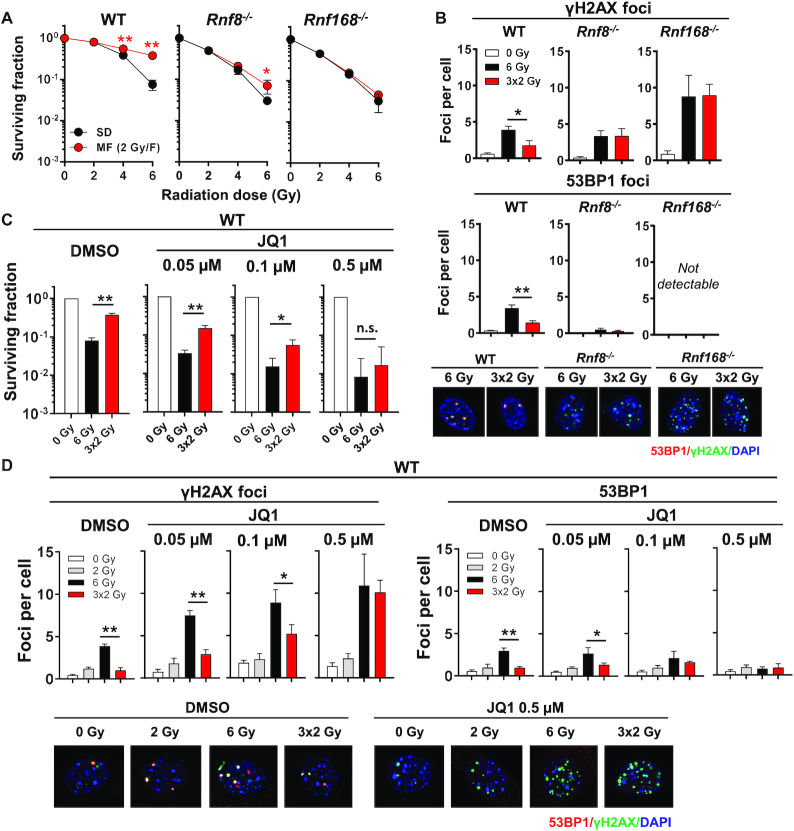
Chromatin modifications promote 53BP1 accumulation at DSB sites. Stable binding of 53BP1 to nucleosomes necessitates dual recognition of ubiquitylated H2A (H2A-K13/K15) and di-methylated H4 (H4-K20me2). In response to DSBs, the E3 ubiquitin ligases RNF8 and RNF168 catalyze H2A ubiquitylation while the bromodomain (BET) protein BRD2 facilitates access to H4-K20me2 (8,9). Corroborating the key role of 53BP1 in promoting enhanced survival after MF irradiation, we found that loss of RNF8 or RNF168 strongly decreased the survival benefit of fractionation and in parallel diminished the difference in residual γH2AX foci after SD and MF irradiation (Figure 3A, B). Similarly, inhibition of BET proteins by JQ1 resulted in concentration-dependent reductions in both 53BP1 foci formation and survival benefit after MF, such that identical survival rates were observed when cells were irradiated in the presence of 0.5 μM JQ1 (Figure 3C). Consistent with this, co-treatment with 0.5 μM JQ1 resulted in comparably high levels of residual γH2AX foci following both MF and SD irradiation (Figure 3D), which was reminiscent of the effect seen upon 53BP1 deletion (Figure 2C). Thus, 53BP1 chromatin binding is essential for enhanced cell survival after multifractionated radiotherapy.
Figure 3.
Inhibition of 53BP1 recruitment to damaged chromatin diminishes the survival benefit of multifractionated radiotherapy. (A) Clonogenic survival of Rnf8−/− and Rnf168−/− MEF irradiated with different single doses (2, 4, or 6 Gy) or with multiple fractions (2 × 2 Gy or 3 × 2 Gy). Wild type cells (WT) served as control. (B) Number of residual γH2AX and 53BP1 foci in MEF 24 h after irradiation with 6 Gy or 3 × 2 Gy. (C) Clonogenic survival in wild type MEF (WT) treated with different concentrations (0.05–0.5 μM) of the BET inhibitor JQ1. DMSO was used as control. Inhibitor or DMSO was applied 30 min before the start of irradiation and was renewed daily. After a total treatment time of 72 h (24 h after the last irradiation dose of the multifractionated regimen), the inhibitor was removed, and the cells were cultured for an additional 4 days. (D) Residual γH2AX and 53BP1 foci numbers and representative images at 24 h after irradiation in WT MEF treated with indicated concentrations of JQ1 or DMSO. DAPI was used to stain the nucleus. Results show mean ± STDEV (n = 3, *P < 0.05, **P < 0.01, Student's t-test).
53BP1 uses RIF1 to channel DNA repair into NHEJ after multifractionated radiotherapy
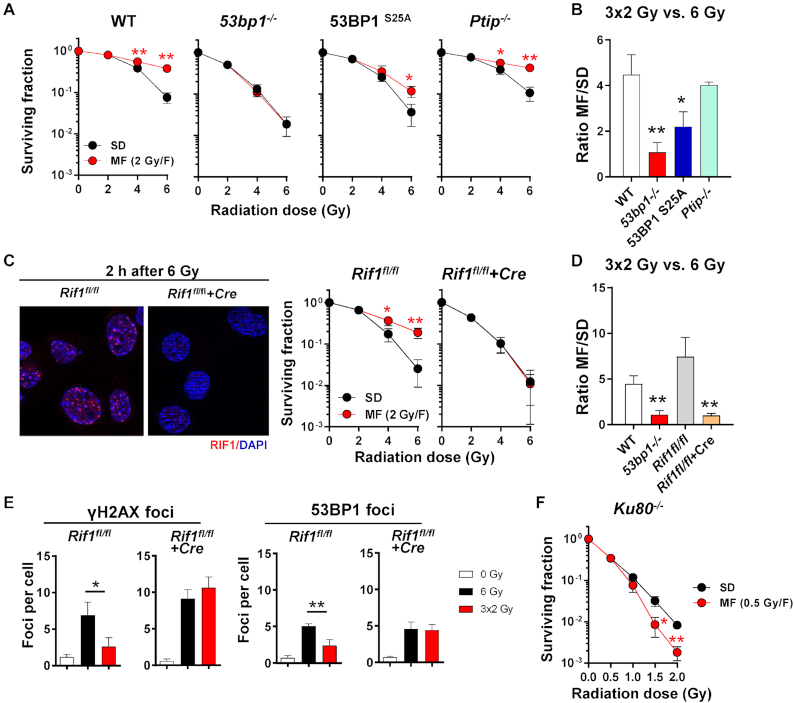
53BP1 engages its downstream effectors PTIP and RIF1 through phosphorylation-mediated binding (15). The phospho-epitope for PTIP interaction has been unequivocally mapped to serine 25, while no single phospho-site has been assigned for RIF1 (15,35). Unlike complete loss of 53BP1, mutating 53BP1 serine 25 to alanine (S25A) did not abolish the survival benefit after multifractionated radiotherapy (Figure 4A, B). Similar observations were made in PTIP-deficient MEF (Figure 4A, B). By contrast, conditional deletion of RIF1 by Cre recombinase completely abolished the survival benefit of fractionation (Figure 4C, D) and increased residual γH2AX foci after MF to levels similar to SD irradiation (Figure 4E). MF irradiation-induced G2 accumulation at the expense of S-phase in both Rif1fl/fl and Rif1fl/fl+Cre MEF (Supplementary Figure S5), suggesting that cell cycle redistribution alone is not sufficient to explain why loss of RIF1 negated the survival benefit seen in MF irradiated RIF1-proficient cells. Importantly, survival benefit after MF irradiation was also absent in cells lacking Ku80 (Figure 4F). Thus, 53BP1-dependent survival after multifractionated radiotherapy is mediated through RIF1 and utilizes NHEJ as the predominant DSB repair mechanism.
Figure 4.
RIF1 but not PTIP is the downstream effector of 53BP1 responsible for promoting DNA repair after multifractionated radiotherapy. (A) Colony formation assays of WT, 53bp1−/−, 53BP1 serine-25 phospho-mutant (53BP1 S25A) and Ptip−/− MEFs. (B) Ratio of MF to SD survival was calculated as follows: surviving fraction after three 2 Gy-fractions (3 × 2 Gy)/surviving fraction after a single dose of 6 Gy. (C) Immunofluorescence staining of RIF1 at 2 h after irradiation and clonogenic survival of RIF1 knockout (Rif1fl/fl+Cre) versus control MEF (Rif1fl/fl) after SD or MF irradiation. (D) Ratio of MF to SD survival was calculated as in (B). (E) Number of γH2AX and 53BP1 foci in Rif1fl/fl and Rif1fl/fl+Cre MEF after irradiation with SD (6 Gy) or MF (3 × 2 Gy) radiotherapy. Nuclei were counterstained with DAPI. (F) Clonogenic survival of Ku80−/− MEF after SD (0.5–2 Gy) or MF (1–4 fractions of 0.5 Gy) irradiation. Results show mean ± STDEV (n = 3, * P < 0.05, ** P < 0.01, Student's t-test).
Inhibiting end resection in 53BP1-deficient cells does not restore survival benefit after multifractionated radiotherapy
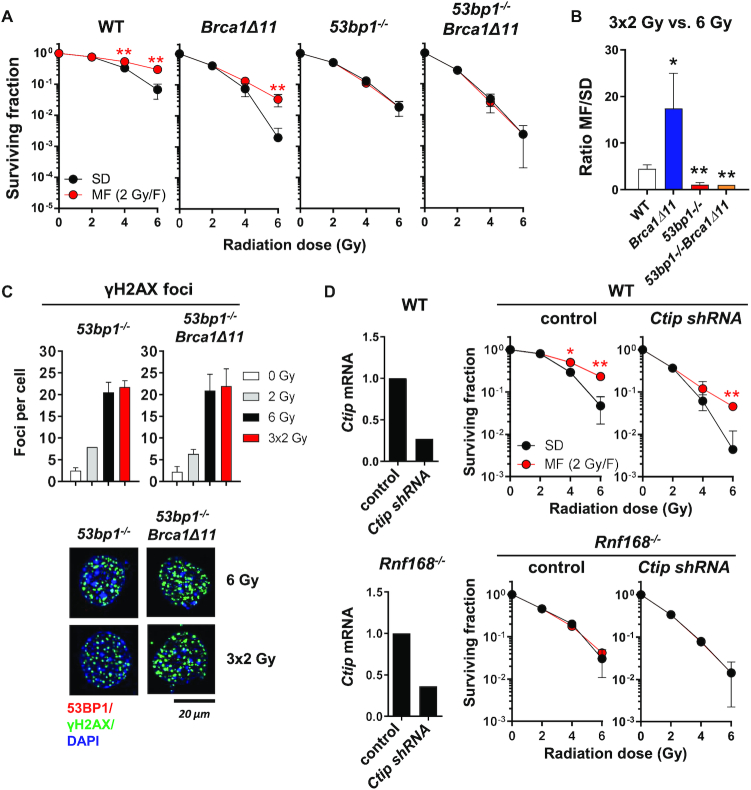
53BP1 and BRCA1 have been shown to reciprocally modulate end resection (12,20). While deletion of 53BP1 largely rescues end resection and HR in BRCA1 deficient cells, it is not clear whether loss of BRCA1 function would restore NHEJ in cells lacking 53BP1. As shown in Figure 5A, although MEF expressing mutant BRCA1 (Brca1Δ11) exhibited a general increase in radiosensitivity, their survival after MF irradiation was nevertheless elevated compared to SD irradiation, with an MF/SD ratio even higher than WT MEF (Figure 5A, B). This is likely due to increased 53BP1-dependent DNA repair in cells lacking functional BRCA1 (20). Interestingly, cells deficient in both BRCA1 and 53BP1 showed no survival benefit after MF irradiation, behaving identically to 53bp1−/− MEF (Figure 5A–C). To note, Brca1Δ11 and 53bp1−/− MEF showed similar cell cycle profiles after MF irradiation (Supplementary Figure S6A), which were quite different from either WT or 53bp1−/−Brca1Δ11 MEF (Supplementary Figures S4B, S6B). These data reinforced the notion that cell cycle control likely played no major role in the observed survival effects. Abolition of 53BP1/RIF1 function enables enhanced end resection driven by CtIP (36). However, depletion of CtIP also failed to restore survival benefit cells with impaired 53BP1 signaling after MF irradiation, although it did increase cellular radiosensitivity to both MF and SD irradiation (Figure 5D). Finally, we found that inactivating BRCA1 did not ameliorate the severe CSR defect in 53bp1−/− and Rnf168−/− B cells (Supplementary Figure S6C). Thus, reducing end resection, cannot rescue 53BP1-dependent NHEJ.
Figure 5.
Inhibition of end resection in 53BP1 deficient cells fails to restore survival benefit after multifractionated radiotherapy. (A) Clonogenic survival of WT, BRCA1-deficient (Brca1Δ11), 53BP1-deficient (53bp1−/−) and 53BP1/BRCA1 doubly deficient (53bp1−/−Brca1Δ11) MEF after single-dose (SD) and multifractionated (MF) radiotherapy. Unirradiated cells were used as control. (B) Ratio of MF to SD survival was calculated as follows: surviving fraction after three 2 Gy-fractions (3 × 2 Gy)/surviving fraction after a single dose of 6 Gy. (C) Number of residual γH2AX foci in 53bp1−/− and 53bp1−/− Brca1Δ11 at 24 h after SD (6 Gy) or MF (3 × 2 Gy) irradiation. (D) Ctip transcript levels and clonogenic survival of WT and Rnf168−/− MEF after shRNA-mediated knockdown of Ctip. An empty vector was used as control. Results show mean ± STDEV (n = 3, *P < 0.05, **P < 0.01, Student's t-test).
53BP1 loss reduces the survival of cancer cells after multifractionated radiotherapy independently of cellular p53 status
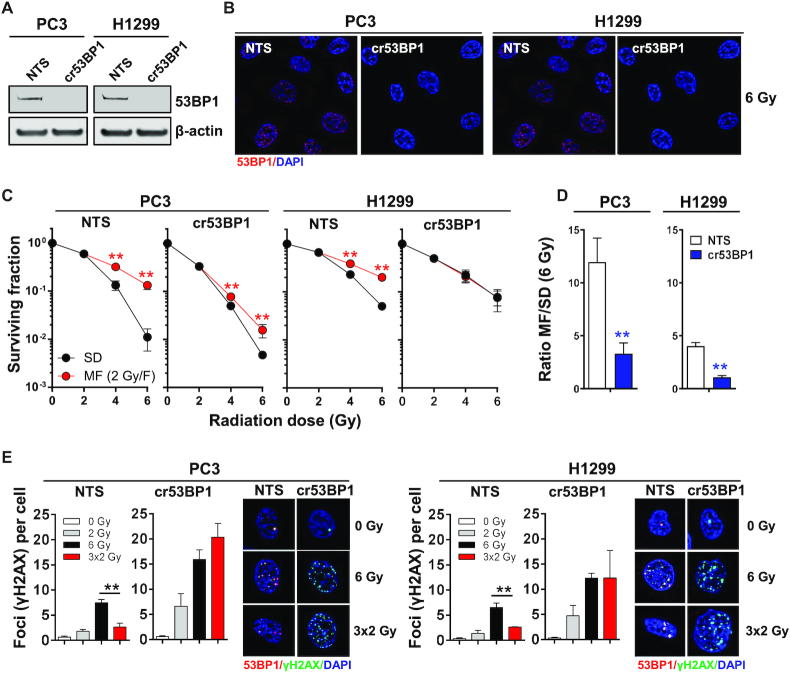
53BP1 was originally identified as a cellular factor that binds to the master tumor suppressor p53 (37). While the precise functional significance of 53BP1/p53 interaction is debated, it is widely thought that 53BP1 can modulate apoptosis through p53 (38). Since p53 mutations and deletion are frequently found in cancer, we next evaluated whether loss of 53BP1 also diminishes the survival benefit from radiation fractionation in cells with impaired p53 function. Using CRISPR/Cas9-mediated gene editing, we deleted TP53BP1 in PC3 and H1299 (both p53-deficient) cells (Figure 6A, B). Similar to our observations in MEF, which are p53-proficient (Supplementary Figure S7A, B), loss of 53BP1 in cells lacking functional p53 preferentially reduced survival after MF irradiation and significantly decreased MF/SD ratios (Figure 6C, D). Further, the number of residual γH2AX foci after MF irradiation increased strongly to levels similar to those produced by SD irradiation (Figure 6E). Thus, the impact of 53BP1 on DNA repair and survival after multifractionated radiotherapy is not mediated by p53.
Figure 6.
53BP1 gene ablation decreases cancer cell survival after multifractionated radiotherapy independently of p53. (A) Western blot and (B) immunofluorescence staining of 53BP1 in PC3 (p53-deleted) and H1299 (p53-mutated) cells after CRISPR/Cas9-mediated 53BP1 gene knockout (cr53BP1). A non-targeting sequence (NTS) was used as control. β-Actin expression was evaluated to ensure equal sample loading. (C) Clonogenic survival of PC3 and H1299 NTS or cr53BP1 cells after single-dose (SD) or multifractionated (MF) irradiation with fractions of 2 Gy per day. Cells were plated 24 h prior to the start of radiotherapy. (D) Ratio of MF to SD survival was calculated as follows: surviving fraction after three 2 Gy-fractions (3 × 2 Gy)/surviving fraction after according single dose (6 Gy). (E) Staining of γH2AX (green) and 53BP1 (red) in cells irradiated with a single dose of 6 Gy or three fractions of 2 Gy. Nuclei were counterstained with DAPI. Cells were fixed at 24 h after the end of radiotherapy. Results show mean ± STDEV (n = 3, *P < 0.05, **P < 0.01, Student's t-test).
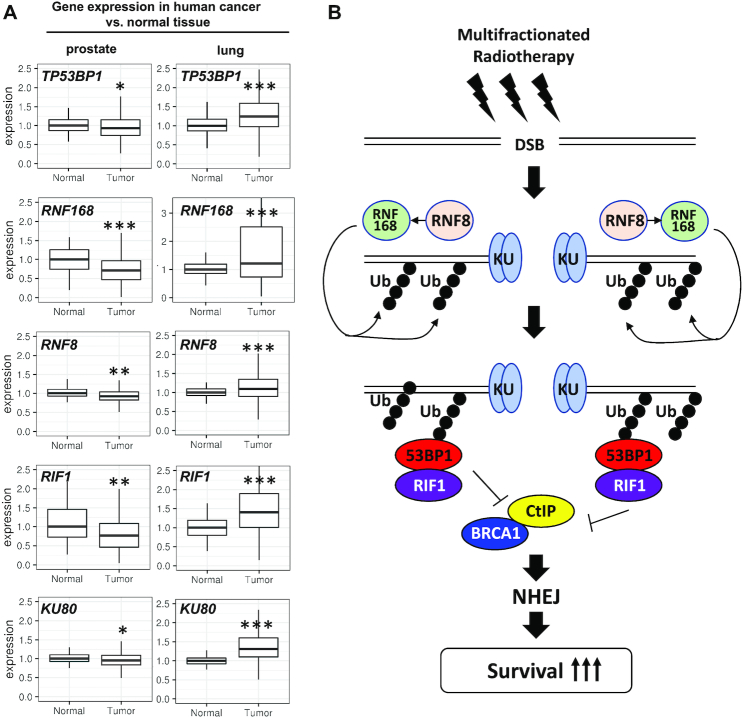
Expression of 53BP1 pathway genes is altered in human cancer
The importance of 53BP1 to cell survival following MF irradiation suggested that any tumor-associated alterations in this pathway may significantly impact the efficacy of multifractionated radiotherapy. To evaluate the expression of DDR genes in human cancer and normal tissues, we queried the TCGA database. As shown in Figure 7A, expression of TP53BP1, RNF8, RNF168 and RIF1, all of which have been shown to promote survival after fractionated irradiation, were significantly lower in prostate cancer than in normal prostate epithelium. In contrast, NSCLC tumors showed increased expression of 53BP1-associated genes (Figure 7A). Although for both prostate cancer and NSCLC, multifractionated irradiation is the standard radiotherapy regimen, some patients receive stereotactic body radiotherapy (SBRT) in which an ablative radiation dose of one or few fractions is applied. Our data suggest that application of multifractionated irradiation regimens could be beneficial for the treatment of prostate cancer but may be less effective against NSCLC. Overall, we propose that differential expression of DDR genes in cancer and normal tissues can be exploited to select the most effective fractionation regimen for individual tumors.
Figure 7.
TP53BP1 and associated genes are downregulated in prostate cancer. (A) Expression of indicated 53BP1 pathway genes in 52 normal prostate gland and 497 prostate cancer samples and in 110 normal lung tissue and 1018 non-small cell lung cancer samples (NSCLC; including squamous cell carcinoma: 501 samples and adenocarcinoma: 517 samples) obtained from the TCGA data bank and shown as box plots. For statistical analysis, the Student's t-test was used (*P < 0.05, **P < 0.01, ***P < 0.001). (B) Schematic depicting how 53BP1 pathway activation contributes to efficient DNA repair and enhanced cell survival after multifractionated radiotherapy.
DISCUSSION
Radiotherapy can be administered as a single large dose or in a few high-dose fractions (hypofractionation) as well as over multiple lower dose fractions (multifractionation). Both tumor and normal cells generally survive better when radiotherapy is delivered in multiple fractions. Thus, MF radiotherapy regimens may reduce collateral damages to healthy tissues, but this could come at the expense of anti-tumor efficacy. Several processes have been proposed to contribute to enhanced cellular tolerance to MF irradiation, including cell cycle checkpoint responses and activation of pro-survival signaling pathways (2). Nevertheless, how mammalian cells repair DNA damage in response to MF irradiation remains largely unexplored.
Radiotherapy produces potentially lethal DNA DSBs, which can be repaired by either NHEJ or HR. These two pathways normally counteract each other through the mutual antagonism between 53BP1-mediated end protection and BRCA1-facilitated end resection. Our results revealed that HR-associated repair factors such as BRCA1 and CtIP were equally important for cell survival after both MF and SD irradiation. On the contrary, we now provide evidence supporting 53BP1-dependent NHEJ as a key pathway cells utilize to efficiently repair DSBs during an MF irradiation regimen, leading to enhanced cell survival compared to SD irradiation (Figure 7B). We further demonstrated that 53BP1 mediates this effect through RIF1, but not its other known binding partners PTIP or p53. Surprisingly, we found that while loss of ATM resulted in pronounced radiosensitization to either irradiation regimens, it did not specifically affect the enhanced ability of cells to survive MF irradiation. This was in contrast to previous findings in human ataxia–telangiectasia (A–T) fibroblasts, which were shown to exhibit no or very limited split-dose recovery when two irradiation doses were applied 12 h apart (39). In addition, it may seem counterintuitive that cells rely on 53BP1 and RIF1, but not ATM, to enhance their survival after MF irradiation given that ATM is known to promote 53BP1–RIF1 interaction by phosphorylation (40). Several factors may cause these seemingly discrepant results. First, human A-T fibroblasts are significantly more radiosensitive than their Atm−/− MEF counterparts (39). Second, the current study used a much longer 24 h interval to mimic clinically relevant irradiation schedules, which might have allowed A–T cells sufficient time to repair potentially lethal damages between fractions. In this regard, we note that ATM has been reported to be specifically required for cellular response to DSBs within heterochromatin, which are generally repaired much more slowly than euchromatic DSBs (41). Finally, redundancy between ATM and DNA-PK could allow the latter to functionally compensate (42). Additional experiments will be needed to address these questions.
In general, our findings agree well with the observations of Somaiah et al., who reported that NHEJ-deficient Chinese hamster ovary (CHO) cells are equally susceptible to a few large doses of radiation (hypofractionation) compared to a multifractionated regimen of 1 Gy per fraction (43). By contrast, CHO cells deficient in HR exhibited higher tolerance to multifractionated irradiation. However, it should be pointed out that NHEJ-deficient CHO cells eventually became resistant after about seven fractions of irradiation, while their HR-deficient counterparts never became fully resistant. These data therefore suggested that HR could also contribute to split-dose recovery, albeit at a later point in time. In line with this idea, split-dose recovery in the chicken B-cell lymphoma line DT40 has been shown to be highly dependent on the HR-associated gene RAD54 (44,45). A possible caveat here is that HR is known to be hyperactive in DT40 owing to its participation in avian immunoglobulin gene conversion (46). In the same vein, we note that PTIP was reported to facilitate HR in DT40 (47), while in mammals it was clearly shown to promote 53BP1-dependent NHEJ (15). As such, differences in repair pathway usage could be biased by cell-intrinsic properties. Moreover, unlike RAD54, which acts very late during HR (48), loss of factors that function at earlier steps during HR, including RAD52, XRCC2, XRCC3, RAD51C and RAD51D, did not abolish the split-dose recovery of the DT40 lymphocytes (45). Similarly, we found that MEF lacking upstream HR factors such as BRCA1 or CtIP still exhibited enhanced survival after MF compared to SD irradiation. These results indicated that while loss of 53BP1 can restore CtIP-dependent end resection and HR in BRCA1-deficient cells and represents a known mechanism for acquired resistance to PARP inhibition, suppression of end resection generally cannot rescue NHEJ when the 53BP1 pathway is impaired. Thus, 53BP1 likely has specialized functions during NHEJ that go beyond simply suppressing end resection.
The cellular DNA repair systems appear to have a finite capacity. While the exact capacity may vary between cell lines, exceeding this capacity could lead to repair factor exhaustion and system failure, as was demonstrated for RPA (49). One possible explanation for how 53BP1 augments survival after MF irradiation may be that 53BP1-dependent DNA repair between fractions helps keep the overall damage level below the exhaustion limit, whereas a single large dose of irradiation may overwhelm the cellular repair system. Indeed, a finite capacity for 53BP1-mediated end protection has been suggested (50). Nevertheless, while it is possible that this mechanism could potentially contribute to loss of survival benefit after MF irradiation in 53BP1-deficient cells, it is important to note that NHEJ-deficient Ku80-/- MEF are hypersensitive to MF irradiation compared to SD irradiation. These findings instead indicate a more prominent role for NHEJ in DSB repair after MF irradiation.
Besides its function in DNA repair, 53BP1 modulates radiation-induced cell cycle checkpoint responses and apoptosis through its interactions with the tumor suppressor p53 and the ubiquitin-specific protease USP28 (38,51). Recently, it was reported that 53BP1 compartmentalizes at DNA damage sites through phase separation, which is important not only to shield DNA ends against resection but also for p53 stabilization (52). However, the current study showed that 53BP1-dependent survival after MF irradiation can be separated from its roles in stimulating p53 function. In general, our data indicated that enhanced cell survival after MF irradiation is not associated with any particular type of cell cycle checkpoint response. Moreover, decreasing the time between fractions eliminated cell cycle redistribution but not the survival benefit from fractionation. These results support the notion that 53BP1-mediated survival after MF irradiation cannot be adequately explained by its role in regulating checkpoint or pro-apoptotic signaling. Nevertheless, even with an enhanced survival rate, the number of colonies formed after MF irradiation represented only a subset of the original cells (typically 10–50%). As such, the possibility that any MF irradiation-induced cell cycle change within this subpopulation might be obscured in the bulk analysis cannot be completely excluded.
From a therapeutic point of view, the instrumental role of 53BP1 in the adaptive response to MF irradiation suggests that pharmacological targeting of the 53BP1 pathway can potentially augment the efficacy of fractionated radiotherapy. Indeed, we demonstrated that abrogating 53BP1 chromatin recruitment by deleting RNF8/RNF168 or treating cells with the BET inhibitor JQ1 phenocopied 53BP1 loss and abolished the survival benefit after fractionation. Notably, RNF8/RNF168 activity can also be effectively suppressed with clinically approved proteasome inhibitors (e.g. bortezomib) while multiple BET inhibitors are currently being tested in clinical trials (53). This approach of targeting factors upstream of 53BP1 recruitment may therefore hold considerable promise, but additional studies are necessary to test whether normal and malignant tissues would respond differently to the inhibition of 53BP1 signaling.
In summary, our study provides strong evidence that different DNA repair mechanisms are activated by MF and SD irradiation and that enhanced cellular survival after multifractionated radiotherapy is dependent on 53BP1/RIF1 signaling and NHEJ. These findings are clinically highly relevant for two main reasons. First, analyzing the expression status of the 53BP1 pathway in tumors can aid physicians in choosing the most effective radiotherapy regimen and thereby maximize therapeutic benefits while minimizing collateral damages to normal tissue. Second, suppression of 53BP1 pathway activation with proteasome- and/or BET inhibitors offers a tantalizing opportunity to augment the efficacy of multifractionated radiotherapy. These possibilities, if strengthened by further evidence, may ultimately enable clinicians to tailor radiotherapy to the tumor genotype and individualize treatment to optimize cancer therapy outcomes.
DATA AVAILABILITY
The Cancer Genome Atlas (TCGA) data is available through the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). The R package ‘TCGAbiolinks’ is an open source software available in the Bioconductor repository (http://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html).
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by the NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research. The authors thank Adeola Makinde (NIH/NCI), Katherine Wilsdon (NIH/NCI), Shannon Martello (NIH/NCI) and Matthew Brown (NIH/NCI) for excellent technical assistance and Amanda Day (NIH/NCI) for generating Ku80−/− MEF. The results shown here are in part based upon data generated by the TCGA Research Network. We would like to thank the TCGA Research Network and the specimen donors.
Notes
Present address: Iris Eke, Department of Radiation Oncology, Stanford University School of Medicine, Center for Clinical Sciences Research (CCSR), 269 Campus Dr., Room 1260, Stanford, CA 94305, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Intramural Research Program, National Cancer Institute, Center for Cancer Research [ZIA BC 010670 to C.N.C]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Martin A.G.R., Thomas S.J., Harden S.V., Burnet N.G.. Evaluating competing and emerging technologies for stereotactic body radiotherapy and other advanced radiotherapy techniques. Clin. Oncol. (R. Coll. Radiol). 2015; 27:251–259. [DOI] [PubMed] [Google Scholar]
- 2. Denekamp J. Cell kinetics and radiation biology. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1986; 49:357–380. [DOI] [PubMed] [Google Scholar]
- 3. Eke I., Makinde A.Y., Aryankalayil M.J., Sandfort V., Palayoor S.T., Rath B.H., Liotta L., Pierobon M., Petricoin E.F., Brown M.F. et al.. Exploiting radiation-induced signaling to increase the susceptibility of resistant cancer cells to targeted drugs: AKT and mTOR inhibitors as an example. Mol. Cancer Ther. 2018; 17:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eke I., Makinde A.Y., Aryankalayil M.J., Reedy J.L., Citrin D.E., Chopra S., Ahmed M.M., Coleman C.N.. Long-term tumor adaptation after radiotherapy: therapeutic implications for targeting integrins in prostate cancer. Mol. Cancer Res. 2018; 16:1855–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zimmermann M., de Lange T.. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014; 24:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Botuyan M.V., Lee J., Ward I.M., Kim J.E., Thompson J.R., Chen J., Mer G.. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006; 127:1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pei H., Zhang L., Luo K., Qin Y., Chesi M., Fei F., Bergsagel P.L., Wang L., You Z., Lou Z.. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011; 470:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gursoy-Yuzugullu O., Carman C., Price B.D.. Spatially restricted loading of BRD2 at DNA double-strand breaks protects H4 acetylation domains and promotes DNA repair. Sci. Rep. 2017; 7:12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li X., Baek G., Ramanand S.G., Burma S., De Bono J. Correspondence, R.S.M. . BRD4 promotes DNA repair and mediates the formation of TMPRSS2-ERG gene rearrangements in prostate cancer. Cell Rep. 2018; 22:796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J.. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007; 131:887–900. [DOI] [PubMed] [Google Scholar]
- 11. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J. et al.. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009; 136:435–446. [DOI] [PubMed] [Google Scholar]
- 12. Bothmer A., Robbiani D.F., Feldhahn N., Gazumyan A., Nussenzweig A., Nussenzweig M.C.. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 2010; 207:855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jowsey P., Morrice N.A., Hastie C.J., McLauchlan H., Toth R., Rouse J.. Characterisation of the sites of DNA damage-induced 53BP1 phosphorylation catalysed by ATM and ATR. DNA Repair (Amst.). 2007; 6:1536–1544. [DOI] [PubMed] [Google Scholar]
- 14. Callen E., Zong D., Wu W., Wong N., Stanlie A., Ishikawa M., Pavani R., Dumitrache L.C., Byrum A.K., Mendez-Dorantes C. et al.. 53BP1 enforces distinct Pre- and Post-resection blocks on homologous recombination. Mol. Cell. 2020; 77:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Callen E., Di Virgilio M., Kruhlak M.J., Nieto-Soler M., Wong N., Chen H.-T., Faryabi R.B., Polato F., Santos M., Starnes L.M. et al.. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013; 153:1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schlegel B.P., Jodelka F.M., Nunez R.. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006; 66:5181–5189. [DOI] [PubMed] [Google Scholar]
- 17. Isono M., Niimi A., Oike T., Hagiwara Y., Sato H., Sekine R., Yoshida Y., Isobe S.-Y., Obuse C., Nishi R. et al.. BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep. 2017; 18:520–532. [DOI] [PubMed] [Google Scholar]
- 18. Feng L., Li N., Li Y., Wang J., Gao M., Wang W., Chen J.. Cell cycle-dependent inhibition of 53BP1 signaling by BRCA1. Cell Discov. 2015; 1:15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farmer H., McCabe N., Lord C.J., Tutt A.N.J., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C. et al.. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005; 434:917–921. [DOI] [PubMed] [Google Scholar]
- 20. Bunting S.F., Callén E., Wong N., Chen H.-T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L. et al.. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010; 141:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A.D., Feldhahn N., Resch W., Oliveira T.Y., Chait B.T. et al.. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013; 339:711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pellegrini M., Celeste A., Difilippantonio S., Guo R., Wang W., Feigenbaum L., Nussenzweig A.. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 2006; 443:222–225. [DOI] [PubMed] [Google Scholar]
- 23. Ward I.M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J.S., Cascalho M., Chen L., Nussenzweig A., Livak F. et al.. 53BP1 is required for class switch recombination. J. Cell Biol. 2004; 165:459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Santos M.A., Huen M.S.Y., Jankovic M., Chen H.-T., López-Contreras A.J., Klein I.A., Wong N., Barbancho J.L.R., Fernandez-Capetillo O., Nussenzweig M.C. et al.. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J. Exp. Med. 2010; 207:973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zong D., Adam S., Wang Y., Sasanuma H., Callén E., Murga M., Day A., Kruhlak M.J., Wong N., Munro M. et al.. BRCA1 haploinsufficiency is masked by RNF168-Mediated chromatin Ubiquitylation. Mol. Cell. 2019; 73:1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nussenzweig A., Chen C., da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M.C., Li G.C.. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996; 382:551–555. [DOI] [PubMed] [Google Scholar]
- 27. Buonomo S.B.C., Wu Y., Ferguson D., de Lange T.. Mammalian Rif1 contributes to replication stress survival and homology-directed repair. J. Cell Biol. 2009; 187:385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanjana N.E., Shalem O., Zhang F.. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014; 11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eke I., Schneider L., Förster C., Zips D., Kunz-Schughart L.A., Cordes N.. EGFR/JIP-4/JNK2 signaling attenuates cetuximab-mediated radiosensitization of squamous cell carcinoma cells. Cancer Res. 2013; 73:297–306. [DOI] [PubMed] [Google Scholar]
- 30. Eke I., Zscheppang K., Dickreuter E., Hickmann L., Mazzeo E., Unger K., Krause M., Cordes N.. Simultaneous β1 integrin-EGFR targeting and radiosensitization of human head and neck cancer. J. Natl. Cancer Inst. 2015; 107:dju419. [DOI] [PubMed] [Google Scholar]
- 31. Eke I., Leonhardt F., Storch K., Hehlgans S., Cordes N.. The small molecule inhibitor QLT0267 Radiosensitizes squamous cell carcinoma cells of the head and neck. PLoS One. 2009; 4:e6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I. et al.. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016; 44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meitinger F., Anzola J. V, Kaulich M., Richardson A., Stender J.D., Benner C., Glass C.K., Dowdy S.F., Desai A., Shiau A.K. et al.. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 2016; 214:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lambrus B.G., Daggubati V., Uetake Y., Scott P.M., Clutario K.M., Sluder G., Holland A.J.. A USP28–53BP1–p53–p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J. Cell Biol. 2016; 214:143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munoz I.M., Jowsey P.A., Toth R., Rouse J.. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res. 2007; 35:5312–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bakr A., Köcher S., Volquardsen J., Petersen C., Borgmann K., Dikomey E., Rothkamm K., Mansour W.Y.. Impaired 53BP1/RIF1 DSB mediated end-protection stimulates CtIP-dependent end resection and switches the repair to PARP1-dependent end joining in G1. Oncotarget. 2016; 7:57679–57693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iwabuchi K., Bartel P.L., Li B., Marraccino R., Fields S.. Two cellular proteins that bind to wild-type but not mutant p53. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:6098–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cuella-Martin R., Oliveira C., Lockstone H.E., Snellenberg S., Grolmusova N., Chapman J.R.. 53BP1 integrates DNA repair and p53-Dependent cell fate decisions via distinct mechanisms. Mol. Cell. 2016; 64:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alsbeih M.G.A., Fertil B., Arlett C.F., Malaise E.P.. High split-dose recovery in hypersensitive human fibroblasts: a case of induced radioresistance. Int. J. Radiat. Biol. 1996; 69:225–239. [DOI] [PubMed] [Google Scholar]
- 40. Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T.F., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D. et al.. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013; 49:872–883. [DOI] [PubMed] [Google Scholar]
- 41. Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P.A.. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008; 31:167–177. [DOI] [PubMed] [Google Scholar]
- 42. Callén E., Jankovic M., Wong N., Zha S., Chen H.-T., Difilippantonio S., Di Virgilio M., Heidkamp G., Alt F.W., Nussenzweig A. et al.. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol. Cell. 2009; 34:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Somaiah N., Yarnold J., Lagerqvist A., Rothkamm K., Helleday T.. Homologous recombination mediates cellular resistance and fraction size sensitivity to radiation therapy. Radiother. Oncol. 2013; 108:155–161. [DOI] [PubMed] [Google Scholar]
- 44. Utsumi H., Tano K., Takata M., Takeda S., Elkind M.M.. Requirement for repair of DNA double-strand breaks by homologous recombination in split-dose recovery. Radiat. Res. 2001; 155:680–686. [DOI] [PubMed] [Google Scholar]
- 45. Rao B.S.S., Tano K., Takeda S., Utsumi H.. Split dose recovery studies using homologous recombination deficient gene knockout chicken B lymphocyte cells. J. Radiat. Res. 2007; 48:77–85. [DOI] [PubMed] [Google Scholar]
- 46. Buerstedde J.-M., Takeda S.. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991; 67:179–188. [DOI] [PubMed] [Google Scholar]
- 47. Wang X., Takenaka K., Takeda S.. PTIP promotes DNA double-strand break repair through homologous recombination. Genes Cells. 2010; 15:243–254. [DOI] [PubMed] [Google Scholar]
- 48. Mazin A. V, Mazina O.M., Bugreev D.V., Rossi M.J.. Rad54, the motor of homologous recombination. DNA Repair (Amst.). 2010; 9:286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Toledo L.I., Altmeyer M., Rask M.-B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J.. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013; 155:1088–1103. [DOI] [PubMed] [Google Scholar]
- 50. Ochs F., Somyajit K., Altmeyer M., Rask M.-B., Lukas J., Lukas C.. 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol. 2016; 23:714–721. [DOI] [PubMed] [Google Scholar]
- 51. Zhang D., Zaugg K., Mak T.W., Elledge S.J.. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006; 126:529–542. [DOI] [PubMed] [Google Scholar]
- 52. Kilic S., Lezaja A., Gatti M., Bianco E., Michelena J., Imhof R., Altmeyer M.. Phase separation of 53 BP 1 determines liquid‐like behavior of DNA repair compartments. EMBO J. 2019; 38:e101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stathis A., Bertoni F.. BET proteins as targets for anticancer treatment. Cancer Discov. 2018; 8:24–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Cancer Genome Atlas (TCGA) data is available through the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). The R package ‘TCGAbiolinks’ is an open source software available in the Bioconductor repository (http://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html).