Abstract
Precise noncoding RNA (ncRNA)-based network prediction is necessary to reveal ncRNA functions and pathological mechanisms. Here, we established a systemic pipeline to identify prognostic ncRNAs, predict their functions and explore their pathological mechanisms in lung adenocarcinoma (LUAD). After in silico and experimental validation based on evaluations of prognostic value in multiple LUAD cohorts, we selected the PTTG3P pseudogene from among other prognostic ncRNAs (MIR497HG, HSP078, TBX5-AS1, LOC100506990 and C14orf64) for mechanistic studies. PTTG3P upregulation in LUAD cells shortens the metaphase to anaphase transition in mitosis, increases cell viability after cisplatin or paclitaxel treatment, facilitates tumor growth that leads to poor survival in orthotopic lung models, and is associated with a poor survival rate in LUAD patients in the TCGA cohort who received chemotherapy. Mechanistically, PTTG3P acts as an ncRNA that interacts with the transcription factor FOXM1 to regulate the transcriptional activation of the mitotic checkpoint kinase BUB1B, which augments tumor growth and chemoresistance and leads to poor outcomes for LUAD patients. Overall, we established a systematic strategy to uncover prognostic ncRNAs with functional prediction methods suitable for pan-cancer studies. Moreover, we revealed that PTTG3P, due to its upregulation of the PTTG3P/FOXM1/BUB1B axis, could be a therapeutic target for LUAD patients.
INTRODUCTION
According to the annotations generated by the GENCODE and ENCODE consortia, 75% of the human genome has been estimated to be transcribed into RNAs in different cell types and tissues (1,2). Approximately one-third of these complex transcripts are categorized as coding RNAs with various splicing isoforms, and the remaining two-thirds are categorized as noncoding RNAs (ncRNAs), which comprise long noncoding RNAs (lncRNAs), pseudogenes and other structural and regulatory small RNAs (3). Emerging evidence suggests that ncRNAs play critical roles in a broad range of biological functions to regulate transcription, gene expression and protein signaling via participation in histone modification and transcriptional protein complexes, the remodeling of 3D nuclear organelles, the efficacy of protein translation via direct binding to mRNAs, and the modulation of cellular signaling via scaffolding protein complexes (4,5). In comparison to coding genes that are well annotated with the biological functions and pathological connections to human diseases and symptoms, the molecular functions and pathological mechanisms of abundant expressed ncRNAs remain elusive. Since alterations of some ncRNAs were found to be associated with disease progression and patient outcomes, especially in cancers, aberrant ncRNAs might serve as biomarkers and therapeutic targets for potential clinical interventions (6). Nevertheless, the lack of systematic studies that predict the biological functions and prognostic values of ncRNAs has hampered progress in the development of mechanistic understanding and clinical interventions utilizing ncRNAs to prolong the lives of cancer patients.
Lung cancer is the most common cancer in humans, and there were an estimated 2 million cases and 1.7 million deaths due to lung cancer worldwide in 2018 (7). Lung adenocarcinoma (LUAD) is the predominant subtype and accounts for 40% of lung cancers for which there are abundant resources including genomic information accompanied with patient clinical features in the public domains for integrated genomic analysis (8). With the conventional focus on mutated coding genes in LUAD for target therapies, some bioinformatic efforts were conducted to identify prognostic coding and non-coding gene signatures but lack of mechanistic and experimental validations for potential translational applications (9–11).
To comprehensively explore prognosis-associated ncRNAs by utilizing functional prediction and mechanistic validations, we established a systematic pipeline to identify prognostic ncRNAs and their functions and mechanisms in LUAD. We hypothesized that the co-expression of coding genes with ncRNAs could precisely predict the unknown functions of the ncRNAs based on the presence of the same biological condition in a given dataset. After determining the association of aberrantly expressed genes with LUAD patient prognosis, the networks containing prognostic ncRNAs with highly correlated coding genes that existed in at least half of the LUAD datasets were selected for functional and mechanistic predictions. We identified six prognostic ncRNA-associated networks and selected the most consistent prognostic ncRNA, PTTG3P (pituitary tumor-transforming gene 3 pseudogene), for experimental and mechanistic validations of its correlation with poor prognosis in LUAD patients.
PTTG3P is located in chromosomal region 8q13 and has been annotated as a processed, intronless pseudogene with very high RNA sequence similarity to its ancestral genes PTTG1 and PTTG2, which are located in chromosomal regions 5q33 and 4p12, respectively (12,13). PTTG1 is an aberrantly expressed oncogenic securin protein that has a negative regulatory effect on p53 in modulating chromosome stability and DNA repair in cancers (14–16). In contrast, although a few studies have noted differences in the expression and function of PTTG family members in different cancers (17–19), neither PTTG2 nor PTTG3P has been shown to participate in tumor progression in LUAD. In our study, consistent with the functional and pathological predictions that PTTG3P participates in mitosis and drug resistance, PTTG3P ncRNA forms a complex with the FOXM1 transcription factor to target the promoter of BUB1B (mitotic checkpoint serine/threonine kinase B). The activation of the PTTG3P ncRNA/FOXM1/BUB1B axis shortens the metaphase-anaphase transition, which increases cell proliferation, tumor growth and drug resistance and leads to poor survival in LUAD patients. Overall, we established a comprehensive and systematic pipeline that is applicable to other cancer types to identify prognostic driver ncRNAs, to predict their biological functions, and to validate and reveal the molecular pathological mechanisms that underlie the involvement of ncRNAs in tumor progression, which could aid in the development of potential clinical interventions.
MATERIALS AND METHODS
Dataset collection
The cohort datasets of transcriptomes of LUAD patients were downloaded from Genome Expression Omnibus (GEO). The identification of differentially expressed probes (DEPs) was conducted by using four datasets (GSE27262, GSE31210, GSE30219 and GSE19188) containing both the tumor and normal tissue samples. Six datasets (GSE31210, GSE50081, GSE37745, GSE30219, GSE3141 and GSE19188) with 10-year survival information of LUAD patients were selected to conduct the survival analysis. The microarray datasets were all normalized and downloaded from GEO, which were generated using Affymetrix Human Genome U133 Plus 2.0 Arrays containing 39455 coding and 2038 non-coding probes, and summarized in Supplementary Table S1. The level 3 gene expression data of LUAD RNA-Seq was downloaded from The Cancer Genome Atlas (TCGA).
Identification of prognostic differential expressed probes in LUAD
The DEPs in tumor samples were selected independently in each of four datasets via F-test and t-test with FDR (false discovery rate) <0.05 and cross-validated in between datasets deposited from different laboratories for reducing the false positive possibility. For prognostic DEPs, we conducted the cox-regression test on six datasets with 10-year survival information by using the function coxph in survival R package (20). We identified 627 prognostic DEPs with criterion of each possible candidate in the log-rank test P-value < 0.05 by computing univariate cox analyses and were presented in at least three different datasets.
Identification of ncRNA-associated networks with a functional prediction method
Modules were constructed from co-expression networks with the weighted gene co-expression network analysis (WGCNA) package by using the standard procedure (21). A strong correlation was obtained by weighting the correlation based on power β derived from the power adjacency function. The approximate scale-free topological overlap of each dataset was represented by an adjacency matrix by using a well-defined formula for weighted networks and applied to average linkage hierarchical clustering through topological overlap dissimilarity (1-topological overlap). According to the hierarchical clustering, the probes were grouped as a module with a height cut-off for the dynamic tree of 0.95 and the minimum number of probes in a module was 5.
For precise functional prediction, the significance of a gene identified by the WGCNA was calculated by the –log(P-value of the log-rank test) and the kWithin value to demonstrate the connectivity of the probes within the modules. In this study, the top 15 correlated DEPs in each dataset were ranked on the basis of the kWithin values to identify the downstream targets of the specific ncRNAs. After extracting each prognostic ncRNA with its co-expressed coding genes from specific modules in at least three datasets, the prognostic ncRNA networks were applied to pathway enrichment analysis by WebGestalt (a web-based gene set analysis toolkit).
LUAD Tissue arrays and RNA-ISH
Tumors tissues of Taiwan LUAD patients were collected and spotted onto microarray by Dr Teh-Ying Chou from Taipei Veterans General Hospital (VGH) (22). LUAD patients enrolled from 2002 to 2006 at Taipei VGH in the surgical pathology archives were included in this study. Preparation of tissue arrays and patient demography were provided in the Supplementary Methods and Supplementary Table S2, respectively.
Probes for PTTG3P RNA were designed through LNATM Probe Designer (https://www.exiqon.com/mRNA-probes), and the sequences of probes and the competition RNAs for assays are provided in the Supplementary Table S3. After dewaxing, the tissue arrays were treated with protease K at 37°C for 10 min, fixed by 4% paraformaldehyde at room temperature for 15 min and then hybridized by LNA probes at 60°C overnight. Arrays were washed with 0.2× SSC buffer with 2% BSA at 4°C for 10 min to remove non-specific probes by following the manufactural protocol of IsHyb In Situ Hybridization (ISH) Kit (BC-K2191050, Blossom Biotechnologies). After arrays dehydration and sealing, expression intensity of PTTG3P was determined by the staining intensity with H-score (intensity scores multiplied by the number of percentage of cells with positive PTTG3P staining) by two pathologists. The high and low expression of PTTG3P were grouped based on high and low tertiles of total PTTG3P H-score for survival analysis.
LUAD cell lines
Eight lung cell lines including immortal lung epithelial NL-20 and LUAD cell lines H1299, A549, CL1-0, CL1-5, H23, H1435 and H1437 were described previously (23,24). All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS and incubated in 5% CO2 at 37°C.
Transfections with knockdown or overexpression reagents
Knockdown of PTTG3P in H1299 and CL1-5 was conducted by the CRISPR interference CRISPRi (RNAi Core of Academia Sinica in Taiwan), and the sequence of sgRNA are provided in Supplementary Table S3. At first, each sgRNA vector (PTTG3P sgRNA-1, PTTG3P sgRNA-2 and sgCtrl) were transfected with p5w-dCas9-KRAB.pBsd plasmid into H1299 and CL1-5 cell lines by standard protocol of jetPRIME (PolyPlus). After 2 days transfection, we obtained the specific clonal population by single cell dilution in 96-well plates with selection of 2 μg/ml puromycin and 10 μg/ml Blastidin (Thermo Fisher Scientific™) for at least 2 weeks.
H1299 cells were infected with luciferase-expressing lentivirus for an lung orthotopic model and selected in the presence of 600 μg/ml G418 (Thermo Fisher Scientific™). Luciferase expression vector of psPAX2 and pMD2.G were transfected into 293T by jetPRIME for virus production. After 2 days, the virus containing supernatant was collected by passing through the 0.22 μm filters with polybrene ready for H1299 infection. Plasmids for overexpression were transfected into cells by following the standard protocol of jetPRIME, and the details for plasmid construction are provided in the Supplementary Methods.
Lung orthotopic mouse models
The 5-week old male NOD-SCID/CB.17 mice were purchased from LASCO, Taiwan. Cells (H1299: 1 × 106/50 μl; A549: 3 × 106/50 μl) mixed with Matrigel (BD) were injected into the left lung of anesthetized mice (6–8-week old) as described in the previous studies (25). Tumor tissue blocks and slides of orthotopic lung were prepared from left lung of NOD/SCID mice by the Pathology Core of IBMS (Academia Sinica) for staining with PTTG3P RNA-ISH probe or H&E.
Subcellular fractionation, RNA extraction and RT-qPCR
Subcellular fractionation was conducted by following the standard protocol (26). Total RNA samples were extracted by Trizol (Thermo Fisher Scientific™) based on the recommended protocol and cDNA were generated by Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific™) followed by the manufacturer's instructions. For detecting RNA expression, equal amounts of cDNA mixed with SYBR Green Master Mix (Thermo Fisher Scientific™) and primers was performed in Applied Biosystems 7500 Real-Time PCR System. 18S ribosomal RNA (rRNA) served as an internal control for real-time qPCR analyses. See Supplementary Tables S3 for sequences of RT-qPCR primers.
Cell proliferation and cell viability assays
Cell proliferation assay was performed by xCELLigence RTCA system and followed the standard protocol of xCELLigenece instrument. In general, 2500 cells were seeded in each well of a 16-well E-plate and monitored the cell index of each hour. The cell viability assay detected by Alamar blue reagent (Thermo Fisher Scientific™) according to manufacturer's instruction were provided in the Supplementary Methods. Cisplatin is gifts kindly provided by Dr Tang Tang of our institute.
Flow cytometry
Cells (1 × 106) were washed by phosphate buffered saline (PBS) twice, and then fixed with 70% ice-cold ethanol at −20°C overnight. The fixed cells were washed by PBS and incubated in the dark place containing 400 μg/ml RNase A, 20 μg/ml propidium iodide (PI), 0.1% Triton X-100 in PBS at room temperature for at least 90 minutes. After PBS washes, cells were visualized by BD LSR II flow cytometer.
Live cell time-lapse imaging
Cancer cells were seeded into the 12-well plate with 80% confluence and replaced medium containing 65 ng/ml nocodazole in the next day. After treated with nocodazole for 18 h, cells were treated with Hoechst 33342 (Sigma-Aldrich), washed by PBS in at least three times and replaced with fresh medium to monitor live cells by Leica DMI 6000B Live-Cell Fluorescence Observation System.
Western blot
Cells were lysed with RIPA buffer containing protease inhibitor cocktail (Roche) and phosphatase inhibitor (Millipore). Equal amount of lysates were loaded onto a 10% SDS-PAGE gel and blotted onto PVDF membranes. After treated with 5% blocking buffer (non-fat dry milk dissolved in PBST with 0.1% Tween-20) for 30 min in room temperature, western blotting was incubated with primary antibodies against HA-tag (Covance) or β-actin, or GAPDH (proteintech). After 2-hour incubation with primary antibodies, membranes were washed by 0.1% PBST three times followed by incubated with peroxidase-conjugated rabbit/mouse secondary antibodies for detection. The chemoluminescense was detected by ImageQuant LAS 4000 mini.
RNA-seq
RNA samples were extracted by NucleoSpin RNA (MN) kit and the transcriptome were performed by next generation sequencing (NGS) and analyzed by Novogene (BIOTOOLS). In brief, libraries were generated by using the Agilent Sure Select Strand Specific RNA Library Prep (Illumina) kit after rRNA removed from RNA samples. NGS sequencing were performed in HiSeq4000 (Illumina) for 150 bp paired-end reads. The detailed versions of software and analysis are provided in the Supplementary Table S4. The transcriptome data was deposited into GEO database with accession number GSE114826.
Reporter constructs and luciferase assays
The BUB1B promoter fragment (–3000/+39, −585/+39, −236/+39) were PCR amplified from A549 genomic DNA incorporating NheI and XhoI cutting sites, and then PCR products were inserted into pGL3 Luciferase Reporter Vectors (Promega). The primers of promoter fragments were provided in Supplementary Table S2.
Reporter constructs containing BUB1B promoter fragment were co-transfected with pRK5 Renilla reporter vector as control of transfection efficiency. After 36–48 h, the luciferase assay was followed the standard protocol of Dual-Glo® Luciferase Assay System (Promega), and detected by the recommended program of (TECAN)-Infinite® M1000 PRO. The luciferase activity was normalized to the Renilla activities.
Chromatin-immunoprecipitation (ChIP) and RNA-immunoprecipitation (RIP)
DNA samples were prepared by following the standard protocol of EpiTect ChIP OneDay Kit (Qiagen), and the RIP assay was based on the protocol as previous study (27). The ChIP-grade primary antibodies against FOXM1 or IgG were ordered from Cell Signaling Technology. The primer sequences for cloning the promoter regions of BUB1B were provided in Supplementary Table S3.
RNA fluorescence in situ hybridization (RNA-FISH)
Cells were cultured on the silane-coating slides for 48 hours with 50% confluence. After washing cells with PBS, cells were fixed in 4% paraformaldehyde for 20 min at room temperature. After washing with DEPC-treated PBS, cells were incubated with 0.2 M HCl for 20 min at room temperature. Cells were further treated with protease K at 37°C for 10 min after washing with DEPC-treated PBS, and then further hybridized by LNA probes at 60°C overnight. Slides were washed with 0.2xSSC buffer with 2% BSA at 4°C for 10 min to remove non-specific probes, and further incubated with anti-DIG-HRP (NEF832001EA, Blossom Biotechnologies) for 1 h. After washing cells by DEPC-PBS, fluorescence were amplified by following the manufactural protocol of TSA Plus Fluorescence Kits (PK-NEL705A, Blossom Biotechnologies). After slides were counterstained with Hoechst dye 33342 and mounted, we examined the intensity of PTTG3P signaling by confocal microscope LSM510.
RNA pull-down
Cells were synchronized to G2/M by 65 ng/ml nocodazole, and RNA pull-down assay was performed by following the previous study (28). In vitro transcription of biotinylated T7-based or SP6-based RNA were prepared by AmpliScribeTMT7-FlashTMBiotin-RNA Transcription Kit (Lucigen) and SP6 RNA polymerase with Biotin RNA labeling Mix (Roche).
Chromatin Isolation by RNA purification (ChIRP)
ChIRP was performed by following the previous protocol (29). In the beginning, we designed anti-sense oligo probes of PTTG3P and LacZ with biotin-labeled at 3′-prime end by ChIRP Probe Designer. Forty million H1299 cells were treated with 65 ng/ml nocodazole for G2/M synchronization followed by 1% glutaraldehyde for 10 minutes at room temperature for crosslinking. After lysing crosslinking, DNA was sheared to 200–500 bp at 4°C and hybridized with biotinylated target probes in 37°C. Chromatin bound with probes were isolated by Streptavidin Mag Sepharose (GE Healthcare) and individually extracted DNA, RNA, and protein for further detection as previously described (29). Sequences of biotinylated probes used in ChIRP were listed in Supplementary Table S3
Statistical analysis
Differentially expressed probes (DEPs) were selected through Student's t-test, F-test and adjust P-value by Benjamini–Hochberg (BH) correction. Statistical significance of triplicates for Student's t-test, one-way or two-way ANOVA, and survival analysis was calculated by Graphic Prism version 8.
RESULTS
Identification of prognostic ncRNAs in LUAD patients with predicted biological functions and drug responses
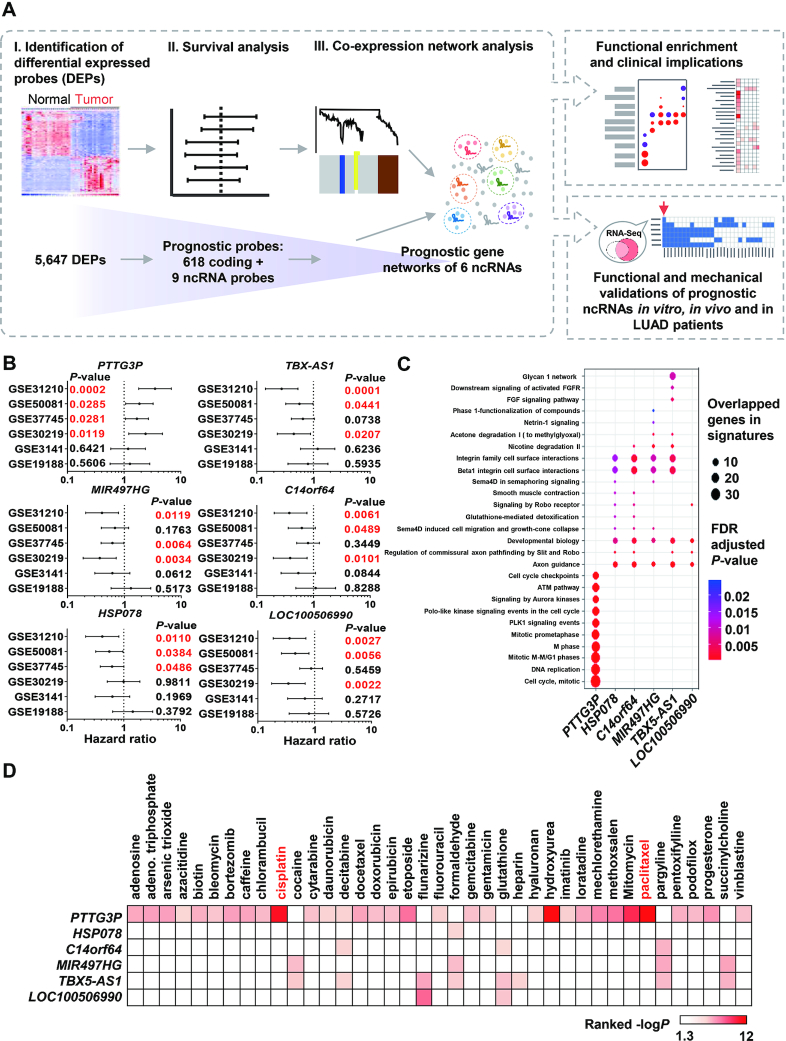
To discover prognostic ncRNAs with their biological functions and possible clinical applications in LUAD patients, we applied a systematic approach to identify and predict the functions and drug sensitivities of ncRNAs that were highly correlated with patient survival in LUAD (Figure 1A). We identified 5647 differentially expressed probes (DEPs) in the tumor and normal samples (Supplementary Table S1), including 627 DEPs that were associated with patient survival. These 627 DEPs were examined the enrichment analyses and were highly enriched in the reported gene signature of poor survival in NSCLC (Supplementary Figure S1A) (30), and then 627 prognostic DEPs, including 618 coding and 9 noncoding DEPs, were further applied to identify the prognostic gene networks by constructing co-expression networks with the weighted gene co-expression network analysis (WGCNA) (Supplementary Figure S1B–D) (see details in the Materials and Methods section) (21). We found six ncRNA-containing networks, including modules containing PTTG3P, MIR497HG, HSP078, TBX5-AS1, C14orf64 and LOC100506990 that were associated with patient survival. Each ncRNA and its associated module maintained their association with prognosis in at least half of the datasets, as indicated by a statistically significant P-value based on the log-rank test (Figure 1B; Supplementary Figure S1E; Tables S5 and S6). We noticed that two datasets, GSE3141 and GSE19188, showed no statistical association between the six ncRNAs and LUAD patient survival, and these two datasets had relatively small sample sizes compared to those of the other datasets. Our results indicated that the upregulation of PTTG3P and its associated network was correlated with poor prognosis in LUAD patients, while the rest of the ncRNAs tended to be associated with a good prognosis in LUAD patients.
Figure 1.
Identification of prognostic ncRNAs in LUAD with predicted biological functions and drug responses. (A) A systematic pipeline was used to discover 5,647 differentially expressed probes (DEPs) and 627 prognostic probes, including nine noncoding probes, with log-rank tests based on multiple LUAD transcriptome datasets containing 10-year patient survival information. We identified six prognostic ncRNA-containing networks by WGCNA and then predicted their biological functions through WebGestalt. The validated and most promising ncRNAs were selected for functional and mechanistic studies of their roles in LUAD tumor progression and clinical intervention. (B) Survival analysis of ncRNAs in six LUAD transcriptomic datasets; a P-value lower than 0.05 is highlighted in red. (C) Pathway prediction in ncRNA modules (networks) by WebGestalt, and the enrichment results was generated using the clusterProfiler R package. The dot size indicates the number of overlapping genes, and the color indicates the significance of the adjusted P-value after scaling. (D) Drug association analysis of ncRNA-containing modules was performed in WebGestalt to determine their clinical effects. The color scale indicates the ranked –log P-value. The white box indicates that the P-value was over 0.05.
After identifying prognostic ncRNAs and their highly correlated networks according to stringent criteria, gene enrichment analysis was applied to each ncRNA-containing network by the web-based gene set analysis toolkit (WebGestalt) to investigate their potential biological functions and involvement in drug responses (Figure 1C, D) (31). Consistently, although only in a relatively small number of prognostic probes were used, several predicted biological processes, such as the nicotine degradation II pathway, axon guidance, the Slit-Robo pathway, sema4D-related and integrin-related cell migration, and developmental biology were demonstrated to contribute to lung cancer tumor progression (Figure 1C) (32–35). Interestingly, our statistical analysis showed that the PTTG3P-containing network was significantly enriched in cell cycle-related pathways, especially those related to mitosis. This implied that aberrant PTTG3P expression might lead to a poor prognosis by disturbing the process of mitosis in LUAD. Moreover, drug association analysis showed that the PTTG3P-containing network was significantly enriched in genes associated with first-line clinical chemotherapeutic drugs such as cisplatin and paclitaxel in LUAD patients (Figure 1D).
In silico and experimental validations of ncRNAs associated with poor survival in LUAD patient data retrieved from TCGA
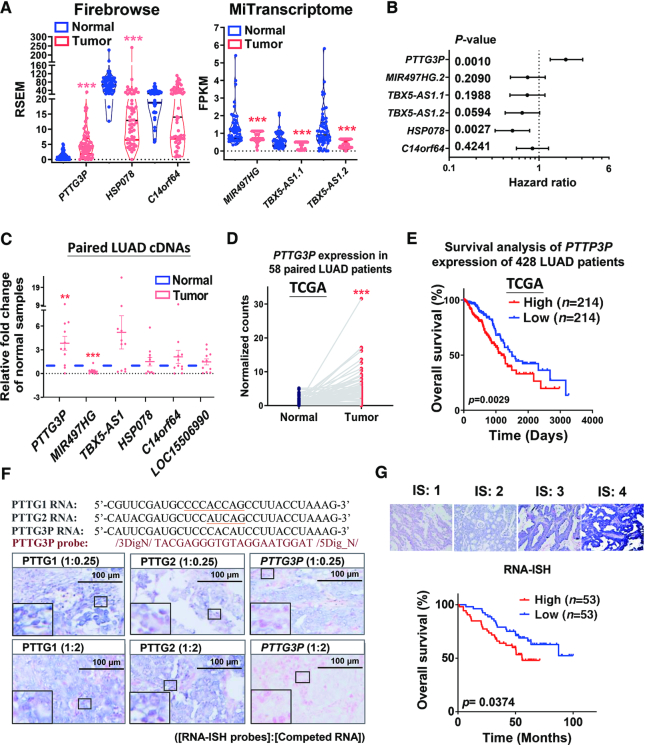
To validate the aberrant expression and prognostic value of the six prognostic ncRNAs in silico, we examined the association of their expression with LUAD patient survival with RNA-Seq data from TCGA that was normalized by either Firebrowse or MiTranscriptome (36,37). Except for the lack of annotation of LOC100506990 in both datasets, we found consistently aberrant expression and similar trends in the hazard ratios for the remaining five ncRNAs in LUAD patient data collected from TCGA (Figure 2A, B). Experimentally, we also validated the altered expression of the six ncRNAs with cDNAs generated from paired LUAD samples by RT-qPCR (Figure 2C; Supplementary Table S2). Notably, PTTG3P showed the most consistent aberrant expression and statistical association with poor prognosis in LUAD, which was validated in the independent and LUAD datasets in the TCGA (Figure 2C–E), suggesting that PTTG3P could serve as a prognostic oncogenic marker in LUAD. Consistently, increasing expression of PTTG3P is also predicted in association with poor patient survival of LUAD independently analyzed by The Atlas of Noncoding RNAs in Cancer (TANRIC) (38).
Figure 2.
Upregulation of PTTG3P contributed to poor survival in LUAD patients based on data from TCGA and tissue arrays from LUAD tumors. (A) Data showing the expression of selected ncRNAs in paired lung adenocarcinoma patients were collected from Firebrowse (n = 58) and MiTranscriptome (n = 53), which were reanalyzed to determine gene expression in cancer patients from TCGA. Notably, there were no annotated expression data obtained from the RNA-seq transcriptomes of LUAD patients for the ncRNA LOC100506990 in the public datasets. (B) Survival analysis in LUAD patients showing the contribution of ncRNAs based on data from Firebrowse (n = 428) and MiTranscriptome (n = 316), which shows the hazard ratios and P-values based on tertile criteria. (C) The expression of selected ncRNAs was examined in cDNAs obtained from paired normal and LUAD tumor samples by real-time qPCR (n = 11). 18S rRNA served as an internal control. (D) PTTG3P expression was upregulated in tumor samples compared to that in adjacent normal samples from 58 paired LUAD patients collected from TCGA. (E) Upregulation of PTTG3P expression is associated with poor overall survival rates in LUAD patients according to data collected from TCGA (n = 428). (F) Sequence alignments and specificity determination via competition of PTTG3P RNA-ISH probes with RNAs from the paralogs PTTG1, PTTG2, and PTTG3P in LUAD tissues collected from surgical patients in Taipei VGH. Concentration ratios of [probes]: [competed RNA] are shown in parentheses. Underlined nucleotides indicated the differences of RNA sequences in between members of PTTG family. The large rectangle at left/bottom corner is the 9X magnified image of the small rectangle in each representative image. (G) Representative intensity scores of RNA-ISH results in LUAD patients in Taipei VGH were used for Kaplan–Meier survival analysis of PTTG3P expression. PTTG3P expression was grouped into tertile (n = 53 in each high and low group) according to overall survival time. IS: intensity score; *P< 0.05, **P< 0.005, ***P< 0.0005.
PTTG3P was shown to be upregulated and associated with poor survival by using RNA-ISH in tumors from LUAD patients in Taiwan
After in silico validation of the six prognostic ncRNAs, we selected the PTTG3P ncRNA for biological and pathological validations based on its consistency and the stringent criteria described above. Owing to the high sequence identity among members of the PTTG family and the clarification of the role of PTTG3P RNA expression in LUAD, we found that both PTTG1 and PTTG3P but not PTTG2 were upregulated in LUAD tissues in the TCGA dataset (Supplementary Figure S2A). The expression of upregulated PTTG3P showed no correlation with the different stages of LUAD in tissues (Supplementary Figure S2B). Since PTTG1 is upregulated in LUAD tissues, as shown in previous reports (15,16), we examined the aberrant expression and prognostic value of PTTG3P with RNA in situ hybridization (RNA-ISH) in LUAD patients from Taiwan with tissue arrays (Supplementary Table S2). We designed a PTTG3P-specific RNA-ISH probe and competing PTTG1, PTTG2, and PTTG3P RNAs (Figure 2F, upper panel). After competition with different concentrations of competing RNAs (Figure 2F, lower panels; Supplementary Figure S3), we demonstrated that the PTTG3P-specific probe could specifically recognize PTTG3P expression with high specificity in tissue arrays of LUAD. Based on the RNA-ISH experiments, we consistently showed that PTTG3P is indeed upregulated and is not correlated with LUAD stages (Supplementary Figure S2C), but it is associated with poor survival of LUAD patients in Taiwan (Figure 2G).
Inhibition of PTTG3P perturbed cell mitosis, proliferation and drug sensitivity in LUAD
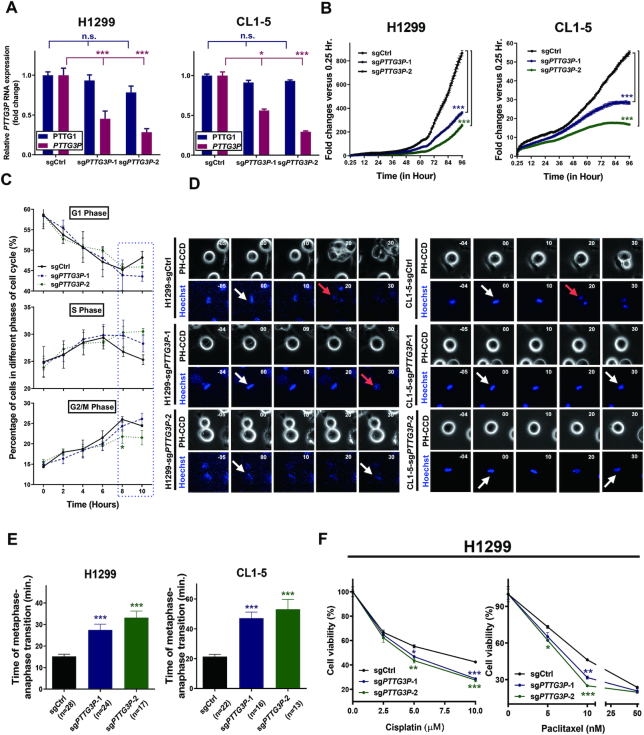
Since PTTG3P could play a role in modulating the cell cycle (Figure 1C), we next examined the predicted functions of PTTG3P in LUAD cell lines. We investigated the RNA expression of PTTG3P in LUAD cell lines with RT-qPCR assays. We found that PTTG3P had relatively low expression levels in the immortalized lung cell line NL-20 and in the cancer cell lines A549 and H23; moderate expression levels in the H1435, H1437 and CL1-0 cell lines; and high expression levels in the CL1-5 and H1299 LUAD cell lines (Supplementary Figure S2D). Based on these results, we selected the H1299 and CL1-5 cells for PTTG3P knockdown experiments and the A549 and CL1-0 cell lines for PTTG3P overexpression experiments for functional assays.
Because of the high sequence identity of PTTG family members but their different chromosomal locations, we performed PTTG3P-specific knockdown via CRISPR interference (CRISPRi) (39). Two PTTG3P-specific single guide RNAs (sgRNAs) (sgPTTG3P-1 and sgPTTG3P-2) were designed and cotransfected with dCas9-KRAB into H1299 and CL1-5 cells to repress PTTG3P expression at the transcriptional level. Since PTTG2 was shown to be expressed at low levels in normal lung and cancer tissues without activation during LUAD progression in the TCGA dataset (Supplementary Figure S2A), we compared the knockdown specificity of sgPTTG3P-1 and sgPTTG3P-2 by verifying the RNA expression of PTTG1 and PTTG3P in LUAD cell lines. Our results showed that PTTG3P knockdown significantly decreased PTTG3P expression with no significant interference in the expression of PTTG1 in the H1299 and CL1-5 cell lines (Figure 3A).
Figure 3.
Knockdown of PTTG3P with CRISPR/dCas9-KRAB decreased cell proliferation, prolonged metaphase to anaphase transition, and sensitized LUAD cell lines to chemotherapy. (A) Specificity and efficiency of PTTG3P RNA knockdown in H1299 and CL1-5 cell lines compared to that of the knockdown of PTTG1 by with sgRNAs, as determined by a real-time qPCR assay. 18S rRNA served as an internal control; mean , n = 3. (B) PTTG3P knockdown in H1299 and CL1-5 cell lines decreased cell proliferation, and the statistics was performed by two-way ANOVA at the final time point. (C) PTTG3P knockdown delayed the progression from the G2/M phase to the G1 phase, especially 8 hours after entering the cell cycle, as indicated by the dashed rectangle for the H1299 cell line. (D) PTTG3P knockdown prolonged the process of metaphase to anaphase transition when compared to that produced by the control sgRNA, as shown by live-cell time-lapse imaging in H1299 and CL1-5 cell lines. White and red arrows represent metaphase and anaphase, respectively, as defined by Hoechst dye staining. (E) Summary of metaphase to anaphase transition, based on a comparison between the treatment of H1299 and CL1-5 with PTTG3P and control sgRNAs, and the statistics was performed by One-way ANOVA with the multiple comparison test. (F) Viability assays of PTTG3P and control sgRNA-transfected H1299 cells treated with the LUAD clinical drugs paclitaxel and cisplatin; mean
, n = 3. (B) PTTG3P knockdown in H1299 and CL1-5 cell lines decreased cell proliferation, and the statistics was performed by two-way ANOVA at the final time point. (C) PTTG3P knockdown delayed the progression from the G2/M phase to the G1 phase, especially 8 hours after entering the cell cycle, as indicated by the dashed rectangle for the H1299 cell line. (D) PTTG3P knockdown prolonged the process of metaphase to anaphase transition when compared to that produced by the control sgRNA, as shown by live-cell time-lapse imaging in H1299 and CL1-5 cell lines. White and red arrows represent metaphase and anaphase, respectively, as defined by Hoechst dye staining. (E) Summary of metaphase to anaphase transition, based on a comparison between the treatment of H1299 and CL1-5 with PTTG3P and control sgRNAs, and the statistics was performed by One-way ANOVA with the multiple comparison test. (F) Viability assays of PTTG3P and control sgRNA-transfected H1299 cells treated with the LUAD clinical drugs paclitaxel and cisplatin; mean , n = 3; *P< 0.05, **P< 0.005, ***P< 0.0005.
, n = 3; *P< 0.05, **P< 0.005, ***P< 0.0005.
To investigate the involvement of the aberrant upregulation of PTTG3P, as predicted by the mitotic cell cycle, in LUAD progression, we examined the downregulation of PTTG3P expression in the processes involved in mitosis, cell proliferation and drug sensitivity. We found that cell proliferation was decreased in PTTG3P-knockdown H1299 and CL1-5 cells compared with that in cells treated with control sgRNA (Figure 3B). When examining the effects of PTTG3P knockdown on the percentages of cells distributed in different phases of the cell cycle after serum-free synchronization, we found that most of the H1299 cells entered the G2/M phase after approximately 8 hours when treated with control sgRNA and entered the G2/M phase after approximately 10 hours upon PTTG3P knockdown after replacement with fresh medium (Figure 3C). Moreover, the population of cells in the G1 phase increased after 10 hours in cells transfected with control sgRNA but not in cells transfected with sgRNA targeting PTTG3P, indicating that the depletion of PTTG3P affected cell cycle progression.
To dissect the mechanism involved in mitotic cell cycle interference in PTTG3P knockdown cells, we monitored the transition from metaphase to anaphase in mitosis with Hoechst dye staining and fluorescence microscopy. Our results indicated that PTTG3P knockdown in H1299 and CL1-5 cells significantly lengthened the time required for the metaphase to anaphase transition by two-fold compared with that in cells treated with control sgRNA (Figure 3D, E).
To further investigate the effect of mitotic interference by upregulated PTTG3P on drug sensitivity, we examined the effect of PTTG3P knockdown on viability in H1299 cells treated with the common LUAD chemotherapeutic drugs paclitaxel and cisplatin. We found that the knockdown of PTTG3P in H1299 cells significantly decreased cell viability after paclitaxel or cisplatin treatment in a dose-dependent manner compared with control sgRNA (Figure 3F). Overall, our results suggested that the aberrant upregulation of PTTG3P ncRNA in LUAD might perturb the process of mitosis, accelerate cell proliferation, as was predicted, and increase resistance to common LUAD drug treatments in H1299 cells. Our results indicated that the aberrant activation of PTTG3P not only contributed to LUAD tumor progression but also decreased drug sensitivity to common LUAD chemotherapeutic drugs, which led to a reduction in therapeutic efficacy.
Knockdown of PTTG3P suppressed LUAD tumor growth and prolonged survival time in an orthotopic lung model
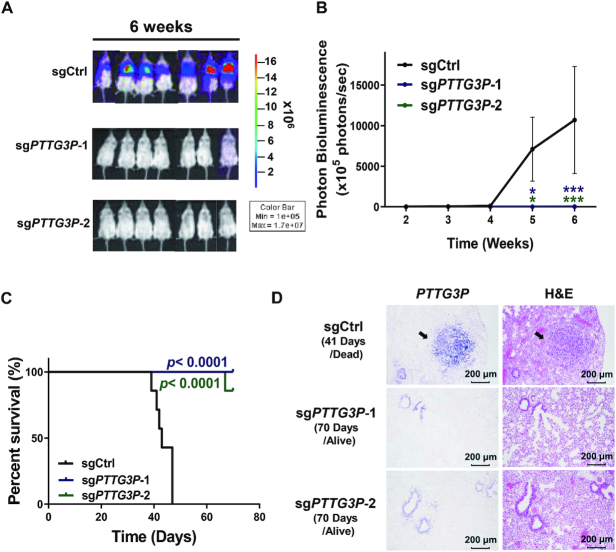
To investigate whether PTTG3P expression was involved in modulating LUAD tumor growth in vivo, we implanted PTTG3P-knockdown H1299 cells into the lungs of mice, which served as orthotopic LUAD models, and tracked tumor formation by measuring bioluminescence (BLI) using an IVIS spectrum imaging system. The intensity of luciferase staining as an indicator of tumor growth was detected in the control sgRNA group in 3 weeks after implantation (Supplementary Figure S4A). However, only three mice had traceable BLI signaling in the sgPTTG3P knockdown groups, and the majority of the mice in the sgPTTG3P-1 and sgPTTG3P-2 knockdown groups showed no signs of tumor growth in 6 weeks after implantation (Figure 4A, B; Supplementary Figure S4A). To further interpret the significant tumor suppressive feature, we examine whether PTTG3P knockdown could trigger cell death to enhance tumor suppression. Our results indicate that the knockdown of PTTG3P enhanced the cleavages of PARP and caspase-3 in H1299 and CL1-5 cells (Supplementary Figure S4B), implying that PTTG3P knockdown not only delays cell cycle progression but also induces the cancer cell death led to tumor suppression in an orthotopic lung tumor model.
Figure 4.
Knockdown of PTTG3P inhibited tumor growth and prolonged survival time in an orthotopic model of lung cancer. (A) Representative bioluminescence images of mice bearing 6-week-old tumors inoculated with sgCtrl-Luc+ (sgCtrl)- or PTTG3P-sgRNA-1/2-Luc+ (sgPTTG3P-1/2)-transfected H1299 cells obtained by IVIS; n = 7 in each group, mean ± SEM. (B) Normalized photon bioluminescence images from three groups of mice monitored by IVIS. The error bars show the S.E.M. (C) Kaplan–Meier plot analysis was performed by comparing the survival rates in the three groups of mice. P-values were provided by log-rank tests. (D) Representative images of PTTG3P expression during RNA-ISH experiments in tissues from orthotopic lung tumors. PTTG3P RNA-ISH and H&E staining were performed on 1-month and 3-month tumor tissues isolated during control and PTTG3P knockdown experiments, respectively. *P< 0.05, **P< 0.005, ***P< 0.0005.
To validate the association between the upregulation of PTTG3P and poor survival in LUAD, we monitored the survival rate of mice for 10 weeks after the implantation of PTTG3P-knockdown H1299 cells in orthotopic lung models. Consistent with the minimal tumor growth in the PTTG3P sgRNA knockdown groups, the suppression of PTTG3P significantly improved the survival of mice after 10 weeks of implantation compared with that of control sgRNA (Figure 4C). We further confirmed PTTG3P expression in orthotopic lung tissues using RNA-ISH and hematoxylin and eosin (H&E) staining. Our results showed that higher PTTG3P RNA expression was detected in lung tissues of the control sgRNA group compared with expression in the PTTG3P knockdown groups (Figure 4D). Overall, our data suggested that PTTG3P expression was highly correlated with tumor growth and survival rates in LUAD.
PTTG3P enhances cell proliferation and drug resistance at the RNA level
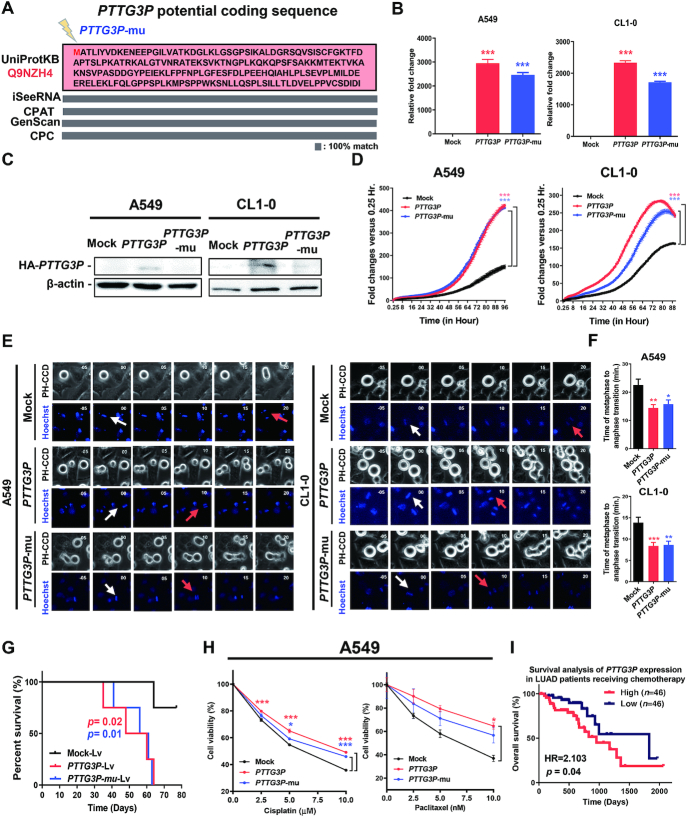
Because a very low percentage of pseudogenes are transcribed and translated into polypeptides that participate in biological functions (40), we would like to clarify firstly whether the PTTG3P pseudogene functions at the RNA and/or protein level as an oncogene to drive LUAD tumorigenesis. Based on polypeptide prediction tools including GenScan (41), CPC (42), iSeeRNA (43) and CPAT (44), PTTG3P pseudogene is universally predicted to encode a 22 kDa polypeptide with 202 amino acids in a single exon identical to an unknown protein, Q9NZH4, in the UniProt database (Figure 5A). We established constructs of PTTG3P and PTTG3P-mu (the ATG codon mutated to the stop codon TAG of the PTTG3P predicted translation start site) with HA-tags and conducted assays of in vitro cell-free transcription/translation and in vivo translation in A549 cells (45). Our results indicated that the PTTG3P construct translated a 25 kDa HA-tagged polypeptide but not in the mock and the PTTG3P-mu constructs in vitro by western blotting analysis (Supplementary Figure S5A). To determine the essential role of PTTG3P polypeptides or RNA in LUAD tumorigenesis, we expressed these constructs in A549 and CL1-0 LUAD cell lines to examine their tumorigenic effects. We found that the RNAs of both PTTG3P and PTTG3P-mu were overexpressed in comparison with those in mock-transfected cells (Figure 5B) and that PTTG3P but not PTTG3P-mu expressed a protein detected by the corresponding anti-HA tag antibody that was located at the predicted molecular weight, as shown by western blotting analysis (Figure 5C; Supplementary Figure S5B). The expression of the PTTG3P and PTTG3P-mu constructs in the A549 and CL1-0 LUAD cell lines induced oncogenic features, including elevated cell proliferation (Figure 5D) and reduced transition time from metaphase to anaphase in mitosis (Figure 5E, F). In contrast to the knockdown of PTTG3P in an orthotopic lung tumor model, both overexpressed-PTTG3P and PTTG3P-mu cells enhanced the tumor growth and reduced the survival rate in comparison to mock in LUAD orthotopic mice models (Figure 5G; Supplementary Figure S5C–E). Furthermore, consistent with the results in knockdown cells treated with cisplatin and paclitaxel, both the overexpressed PTTG3P and PTTG3P-mu cells showed increased cell viability during treatment with cisplatin and paclitaxel in a dose-dependent manner (Figure 5H). These results suggested that PTTG3P ncRNA per se, regardless of its translation into a polypeptide, is oncogenic in LUAD tumorigenesis. Moreover, we found that the high expression of PTTG3P in LUAD patients in the TCGA cohort resulted in a poor survival rate during treatment with chemotherapy, in contrast to the improved survival rate in LUAD patients with low PTTG3P expression (Figure 5I). Collectively, our results implied that the upregulation of PTTG3P ncRNA but not the predicted polypeptide in LUAD cells could induce oncogenesis and resistance to common LUAD drug treatments that might lead to an association with poor patient prognosis.
Figure 5.
PTTG3P ncRNA, but not the corresponding polypeptide, enhanced cell proliferation and drug resistance. (A) The predicted polypeptide sequence of PTTG3P produced by multiple prediction tools aligned it with that of the Q9NZH4 protein from the UniProtKB database. The PTTG3P-mu construct was generated by mutating the start codon (ATG) into a stop codon (TAG) in the HA-tagged PTTG3P construct. (B) RNA expression of PTTG3P or PTTG3P-mu constructs was detected by real-time qPCR in A549 or CL1-0 cells, and (C) protein expression was detected by western blotting analysis. (D) Ectopic expression of PTTG3P increased A549 and CL1-0 cell proliferation, and the statistics was performed by two-way ANOVA at the final time point; mean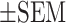 , n = 3. (E) PTTG3P or PTTG3P-mu expression shortened the process of metaphase to anaphase in comparison to that observed in the mock-transfected cells, as shown by live-cell time-lapse imaging in A549 and CL1-0 cell lines. White and red arrows represent metaphase and anaphase, respectively, as defined by Hoechst dye staining. (F) Summary of the comparison of the metaphase to anaphase transition in PTTG3P-, PTTG3P-mu and mock-transfected A549 and CL1-0 cells, and the statistics was performed by One-way ANOVA with the multiple comparison test. (G) Kaplan–Meier plot analysis was performed by comparing the survival rates in the lung orthotopic model bearing the PTTG3P-overexpressed A549 cells; Lv: Lentivirus-infected cells; n = 4 in each group. P-values were provided by log-rank tests. (H) Expression of PTTG3P or PTTG3P-mu increased resistance to clinical drugs used in LUAD patients in A549 and CL1-0 cells; mean
, n = 3. (E) PTTG3P or PTTG3P-mu expression shortened the process of metaphase to anaphase in comparison to that observed in the mock-transfected cells, as shown by live-cell time-lapse imaging in A549 and CL1-0 cell lines. White and red arrows represent metaphase and anaphase, respectively, as defined by Hoechst dye staining. (F) Summary of the comparison of the metaphase to anaphase transition in PTTG3P-, PTTG3P-mu and mock-transfected A549 and CL1-0 cells, and the statistics was performed by One-way ANOVA with the multiple comparison test. (G) Kaplan–Meier plot analysis was performed by comparing the survival rates in the lung orthotopic model bearing the PTTG3P-overexpressed A549 cells; Lv: Lentivirus-infected cells; n = 4 in each group. P-values were provided by log-rank tests. (H) Expression of PTTG3P or PTTG3P-mu increased resistance to clinical drugs used in LUAD patients in A549 and CL1-0 cells; mean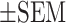 , n = 3. (I) Kaplan–Meier plot of data from LUAD patients treated with chemotherapy in the TCGA cohort. The survival analysis was examined with the Gehan-Breslow–Wilcoxon test with tertile criteria. *P< 0.05, **P< 0.005, ***P< 0.0005.
, n = 3. (I) Kaplan–Meier plot of data from LUAD patients treated with chemotherapy in the TCGA cohort. The survival analysis was examined with the Gehan-Breslow–Wilcoxon test with tertile criteria. *P< 0.05, **P< 0.005, ***P< 0.0005.
PTTG3P, as a chromatin-associated ncRNA, increased cell proliferation and drug resistance in LUAD cells by enhancing FOXM1-BUB1B oncogenic signaling
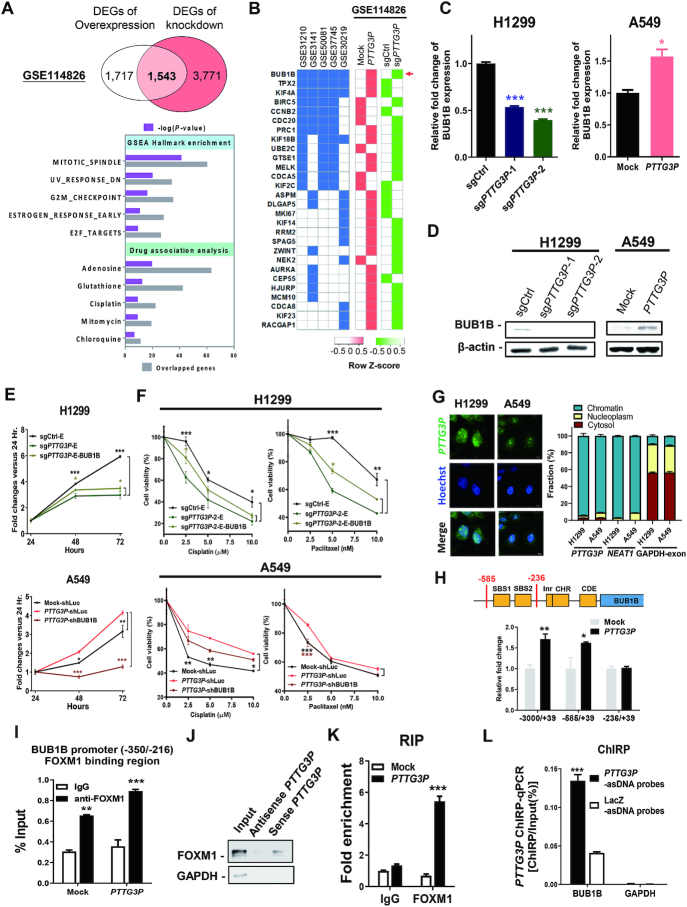
To uncover the oncogenic mechanisms of PTTG3P ncRNA in LUAD, we performed RNA sequencing (RNA-seq) in PTTG3P-overexpressing A549 and PTTG3P-knockdown H1299 cells and their corresponding control cells, followed by gene enrichment analyses (GSE114826). Because of the high consistency of the two transcriptomic datasets, we uncovered 1543 differentially expressed genes (DEGs) and conducted hallmark pathway analysis with GSEA and drug association analysis with WebGestalt (Figure 6A, upper panel; Supplementary Table S7). Consistent with the prediction shown in Figure 1, not only were 1543 DEGs significantly enriched in the cell cycle-associated gene lists, especially in hallmark pathways involved in the mitotic-spindle and G2/M checkpoints, but these DEGs also showed a strong association with the chemotherapeutic drugs cisplatin (Figure 6A, lower panel) and paclitaxel (P-value = 0.0145).
Figure 6.
Interaction of PTTG3P ncRNA with FOXM1 augments BUB1B transcriptional activity to enhance cell proliferation and drug resistance. (A) 1543 DEGs were shared between the upregulated genes in PTTG3P-overexpressing A549 cells and the downregulated genes in PTTG3P-knockdown H1299 cells (GSE114826), which were further analyzed with GSEA and WebGestalt. (B) Genes in the PTTG3P-associated networks in five LUAD datasets that were ranked according to their kWithin value from the WGCNA were aligned with the common DEGs in the deposited transcriptomes in GSE114826 dataset. The GSE19188 dataset was excluded because no PTTG3P-associated module was constructed. Heat maps were individually generated on the basis of the Z-score by Heatmapper and show the upregulated (red) and downregulated (green) genes. A red arrow indicates the highest concordant gene in each dataset. (C) BUB1B expression was examined at the RNA level by real-time qPCR, and (D) its protein level was determined by western blotting analysis in H1299 and A549 cells. (E) Proliferation assays and (F) cell viability assays were performed after treatment with clinical drugs in PTTG3P knockdown H1299 cells overexpressing BUB1B with an EGFP-tag (sgPTTG3P-E-BUB1B) or in PTTG3P-overexpressing A549 cells with shBUB1B knockdown (PTTG3P-shBUB1B). (G) The cellular distribution of PTTG3P ncRNA detected by fluorescence RNA-ISH and the RNA expression of PTTG3P in the cytoplasmic, nucleoplasmic, and chromatin fractions were examined by real-time qPCR in both of H1299 and A549 cell lines. GAPDH-exon and NEAT1 RT-qPCR amplicons served as RNA markers for the cellular and nuclear fractions, respectively. (H) The transcriptional activities of different promoter regions of BUB1B were detected in A549 transfectants with luciferase reporter assays. (I) The binding region of PTTG3P ncRNA/FOXM1 versus FOXM1 alone on the BUB1B promoter was determined by a chromatin immunoprecipitation assay with an anti-FOXM1 antibody and real-time qPCR. (J) Western blotting analysis of FOXM1 obtained from the RNA pull-down assay was performed in nocodazole-synchronized H1299 cells. (K) PTTG3P ncRNA was immunoprecipitated with an anti-FOXM1 antibody and examined by real-time qPCR for an RNA-IP assay. (L) Chromatin isolation by RNA purification (ChIRP) in the presence of FOXM1 was conducted with an antisense PTTG3P DNA probe (PTTG3P-asDNA probe) and a LacZ antisense probe (LacZ-asDNA probe) for a comparison of their binding to the promoters of BUB1B and GAPDH with a ChIRP-qPCR assay. GAPDH and a LacZ-asDNA probe served as negative controls; mean ± SEM, n = 3. *P< 0.05, **P< 0.005, ***P< 0.0005.
To identify the PTTG3P-mediated downstream mitosis-related genes, we examined the association of the 1543 DEGs with the genes in the aforementioned PTTG3P-associated networks in each LUAD transcriptome dataset. We found that BUB1B showed the most consistent association in the PTTG3P-associated networks and in the LUAD transcriptomes which was upregulated in PTTG3P-overexpressing cells and downregulated in PTTG3P knockdown cells (Figure 6B). Interestingly, BUB1B is a mitotic checkpoint protein that plays a critical role in regulating the spindle-assembly checkpoint and that shows aberrant expression in several cancers (46). Therefore, we selected BUB1B as a possible PTTG3P downstream target that may contribute to aberrant mitosis and poor prognosis.
We found that the expression of PTTG3P in A549 cells upregulated BUB1B expression at both the RNA and protein levels (Figure 6C, D). In contrast, the knockdown of PTTG3P expression in H1299 cells reduced BUB1B expression in terms of its RNA and protein levels. To confirm that BUB1B is the downstream target of PTTG3P, we compared the impacts of cell viability by conducting the PTTG3P knockdown (sgPTTG3P) with BUB1B overexpression in H1299 cells and PTTG3P overexpression with BUB1B knockdown (shBUB1B) in A549 cells (Supplementary Figure S6A). Interestingly, cell proliferation was slightly rescued by BUB1B overexpression upon PTTG3P depletion, conversely, the overexpression of PTTG3P failed to rescue the cessation of cell proliferation under BUB1B depletion (Figure 6E). Moreover, the cell resistance to cisplatin and paclitaxel showed a similar trend with the effect of cell proliferation (Figure 6F). Together, we concluded that BUB1B is the downstream of PTTG3P-driven LUAD tumorigenesis.
To clarify how PTTG3P regulated BUB1B, we examined the subcellular localization of PTTG3P since the localization of the ncRNA might affect RNA biological functions (47,48). Our results showed that PTTG3P is more highly expressed in chromatin than in the cytoplasm and nucleoplasm fractions, as detected by RNA fluorescence in situ hybridization (RNA-FISH) and RNA subcellular localization (Figure 6G). Because of the likely role played by nuclear chromatin ncRNAs in the modulation of gene transcription, we speculated that the upregulation of PTTG3P ncRNA might facilitate mitotic catastrophe via regulating BUB1B transcriptional activation by modulating its promoter region (49). Indeed, the expression of PTTG3P directly modulated the transcription of BUB1B by targeting the (–350/–216) promoter region of BUB1B, which was not observed in the mock control, as shown by luciferase reporter assays (Figure 6H).
Next, we speculated that FOXM1 is one of the transcription factors that binds to the (–350/–216) promoter region of BUB1B along with PTTG3P ncRNA because the FOXM1/ BUB1B signaling pathway is essential for the tumorigenicity of glioblastoma and rhabdomyosarcoma, especially in terms of mitotic progression (50,51). First, we examined BUB1B expression during FOXM1 knockdown in PTTG3P-overexpressing cells (Supplementary Figure S6B) and found that BUB1B expression was significantly decreased, suggesting that PTTG3P might modulate BUB1B expression via FOXM1. Second, we examined whether upregulated PTTG3P ncRNA might interact with FOXM1 to target the BUB1B promoter to affect transcriptional activation during aberrant mitosis in tumorigenesis. Indeed, the expression of PTTG3P potentiated FOXM1 binding to the BUB1B promoter region in A549 cells as detected by chromatin immunoprecipitation (ChIP) assays (Figure 6I).
Third, we revealed that sense- but not antisense-strand PTTG3P interacted with FOXM1 with an RNA pull-down assay (Figure 6J). In addition, FOXM1 was shown to interact with PTTG3P and was also detected by an RNA immunoprecipitation (RIP) assay (Figure 6K), strongly suggesting that the upregulated PTTG3P ncRNA interacted with the FOXM1 transcription factor to target the BUB1B promoter, which resulted in aberrant transcriptional activation and facilitated mitosis. Finally, we performed chromatin isolation by using RNA purification (ChIRP) to confirm that PTTG3P ncRNA and FOXM1 form a complex on the BUB1B promoter with 3-fold stronger intensity than the LacZ control (Figure 6L; Supplementary Figure S6C, D). Collectively, we established a systematic approach to reveal prognostic ncRNAs networks that could predict potential biological functions and pathological mechanisms involved in LUAD. Moreover, we identified PTTG3P as a functional ncRNA that guides and complexes with FOXM1 on the BUB1B promoter to augment BUB1B transcriptional activities to facilitate mitosis, tumor growth and drug resistance, resulting in poor prognosis in LUAD patients.
DISCUSSION
To systematically determine the biological functions, clinical relevance and therapeutic targets of ncRNAs, we established a pipeline for the identification of prognostic ncRNAs and their associated networks. The prognostic ncRNA-containing co-expressed gene module allowed us to produce precise predictions of the functions and drug responses of ncRNAs with gene enrichment analysis. By performing validation in multiple LUAD cohorts, we identified the upregulation of PTTG3P ncRNA in LUAD and revealed its involvement in functional mechanisms associated with drug sensitivity and prognosis. We demonstrated that PTTG3P ncRNA interacts with FOXM1 to target the downstream BUB1B promoter, leading to aberrant transcriptional activation that facilitates the mitotic metaphase to anaphase transition and tumor growth and results in poor prognosis in LUAD animal models and human patients. Moreover, LUAD cells with PTTG3P upregulation acquired resistance to treatment with paclitaxel and cisplatin in cell models and in LUAD patients from TCGA cohorts, which further suggested that the detection of upregulated PTTG3P ncRNA might be crucial for therapeutic intervention in LUAD patients.
Several strategies were applied for the functional prediction of ncRNAs, including the prediction of ncRNA-protein interactions, the determination of subcellular localization, the analysis of disease gene clustering and networks, and the analysis of the enrichment of transcriptional factor targets to reveal the downstream signaling pathways of ncRNAs (52–54). In lung cancers, several prognostic coding and non-coding RNAs were identified based on different bioinformatic approaches, especially focusing on prognostic signature gene sets for predicting patient's therapeutic responses and survival at the early stage (9,30,55–57). With increasing evidence of ncRNAs participating in tumor progression, more efforts conducted for exploring their prognostic value in LUAD. For instance, the lncRNA-mRNA-miRNA competing endogenous RNA networks from LUAD differential expressed RNAs were established to reveal prognostic RNAs in association with LUAD patient survival (58,59). Two lncRNAs (DKFZP434 L187 and LOC285548) were discovered for their prognostic properties in LUAD based on their differential expression (10), and another two lncRNA (C1orf132 and TMPO-AS1) were identified to predict survival of stage I-II LUAD patients without adjuvant therapies (60). We noticed that these predicted prognostic LUAD coding genes and ncRNAs based on transcriptome analysis are not identical owing to heterogeneity of LUAD samples and analytic algorisms. Additional endeavors on functional validations and mechanistic actions of identified ncRNAs in LUAD should be critical for revealing useful prognostic ncRNAs to improve LUAD therapeutic interventions.
In comparison to other similar approaches (55–57,61), several features of our study are important for identifying robust prognostic ncRNAs in heterogeneous LUAD transcriptomes that are suitable for functional prediction in other cancer types. First, we collected more than 604 LUAD transcriptomic datasets along with 10-year survival information from different ethnic groups to reveal the common etiology in LUAD (Supplementary Table S1). Second, we applied highly stringent criteria to the already restricted prognostic noncoding gene networks, including requirements for highly correlated genes with cutoff heights of 0.95 and minimal module sizes of 5 genes in multiple datasets, to obtain promising prognostic ncRNAs. Finally, we conducted comprehensive in silico, pathological and experimental validation with mechanistic understanding in mouse cancer models and human patients to obtain ncRNAs that could be used as innovative LUAD therapeutic markers.
In addition to PTTG3P ncRNA in LUAD, the remaining five prognostic ncRNAs including MIR497HG, HSP078, TBX5-AS1, LOC100506990, C14orf64 and their WGCNA networks were predicted to participate in biological functions and pathways involved in developmental biology, axon guidance, the nicotine degradation II pathway, Slit/Robo signaling and Sema4D- and integrin-related cell migration during progression in LUAD patients. Except for MIR497HG, the remaining 4 ncRNAs HSP078 (LOC101928517), TBX5-AS1, LOC100506990 and C14orf64 (LINC01550) have been rarely studied in cancers. One of the mature forms of the MIR497HG cluster host gene, hsa-miR-195, was shown previously to function as a negative regulator of lung cancer growth and metastasis by targeting the oncogenic CHEK1, HDGF, IGF1R, MYB genes and other genes, which gave it prognostic value (62–66). Interestingly, hsa-miR-195 was found to be one of the core schizophrenia genes participating in axonal guidance signaling that is consistent with our WGCNA and functional prediction (67). Notably, several predicted biological pathways involved in axon guidance, the Slit-Robo pathway, sema4D-related and integrin-related cell migration, and developmental biology that were identified with these prognostic ncRNA networks were previously shown to contribute to lung cancer tumor progression and to be related to guidance molecules in the nervous system that control the direction of axon outgrowth (33). Owing to the relatively small number of overlapping probes, the level of statistical significance was relatively low, therefore further exploration of the pathological mechanisms of guidance molecules in LUAD progression is needed to warrant the development of novel therapeutic markers.
Our results also clarify that PTTG3P functions as a FOXM1-interacting ncRNA to target and activate the mitotic checkpoint kinase BUB1B without the need to translate the predicted polypeptide, leading to PTTG3P-mediated LUAD tumorigenesis and drug resistance. Notably, PTTG3P is labeled an ‘uncertain protein’ in the UniProt database, and no predicted peptide fragments of PTTG3P were detected in the database of the human proteome map (www.humanproteomemap.org). These indications further support our results that PTTG3P functions as a ncRNA. Moreover, we also found that upregulated PTTG3P is associated with patients with LUAD that is classified as the AD.1 subtype, which is a genomic and molecular subtyping group of LUAD based on the clustering of multiomics data in TCGA (68) (Supplementary Figure S7). Consistently, the AD.1 subtype in LUAD patients conferred poor differentiation and malignant tumor features based on the enrichment of cell-cycle associated genes, leading to a poor prognosis in LUAD. Furthermore, our analysis of aberrant PTTG3P expression in 36 cancer types in TCGA datasets indicated that PTTG3P is upregulated in more than half of the cancer types examined, and 10 out of 36 cancer types, including LUAD, exhibited a high and statistically significant hazard ratio for patient survival (Supplementary Figure S8). Overall, all of these results provide lines of evidence that show that the upregulation of PTTG3P ncRNA at minimum activates FOXM1/BUB1B oncogenic signaling to facilitate cancer cell proliferation and tumor growth along with malignant progression, leading to poor prognosis in cancer patients. In conclusion, our study provides a highly stringent pipeline that can be used to reveal prognostic ncRNAs with weighted co-expression networks for accurate mechanical prediction and the determination of their association with drug responses. Moreover, we demonstrated that the biological function and pathological mechanisms involved in the upregulation of the PTTG3P ncRNA/ FOXM1/BUB1B axis resulted in poor prognosis in LUAD patients. Notably, the activation of PTTG3P ncRNA during cancer progression increased the resistance of LUAD cells to paclitaxel and cisplatin treatment, and validation in TCGA patients who received chemotherapy might reveal an important biomarker that could be used for dosage determination when evaluating the optimal therapeutic intervention for LUAD. Importantly, our approach would be suitable for use in other cancer types to develop new prognostic ncRNAs as novel therapeutic targets to benefit cancer patients.
DATA AVAILABILITY
To review GEO accession GSE114826: Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114826.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Common Equipment Core of IBMS and Academia Sinica including Light Microscopy Core Facility, DNA sequencing, Flow Cytometry, and Pathology core of Institute of Biomedical Sciences for supporting our experiments (AS-CFII-108-103, AS-CFII-108-113 and AS-CFII-108-115).
Author contributions: Conception and experimental designs: J.H.S., T.Y.C., C.Y.C., R.H.C. and Y.S.J.; Tissue arrays experiments and pathological interpretations: S.C.L., Y.C.Y. and T.Y.C.; Computational analysis, animal and molecular biology experiments: J.H.S., H.Y.C., R.S., D.C.W. and Y.D.L.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academia Sinica and MOST [106-0210-01-15-02]; MOST of Taiwan [MOST1042320B001009MY3]. Funding for open access charge: Academia Sinica and MOST [106-0210-01-15-02]; MOST of Taiwan [MOST1042320B001009MY3]
Conflict of interest statement. None declared.
REFERENCES
- 1. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al.. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang J.X., Rastetter R.H., Wilhelm D.. Non-coding RNAs: an introduction. Adv. Exp. Med. Biol. 2016; 886:13–32. [DOI] [PubMed] [Google Scholar]
- 4. Garraway L.A., Lander E.S.. Lessons from the cancer genome. Cell. 2013; 153:17–37. [DOI] [PubMed] [Google Scholar]
- 5. Calore F., Lovat F., Garofalo M.. Non-coding RNAs and cancer. Int. J. Mol. Sci. 2013; 14:17085–17110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sas-Chen A., Srivastava S., Yarden Y.. The short and the long: non-coding RNAs and growth factors in cancer progression. Biochem. Soc. Trans. 2017; 45:51–64. [DOI] [PubMed] [Google Scholar]
- 7. Cheng T.Y., Cramb S.M., Baade P.D., Youlden D.R., Nwogu C., Reid M.E.. The international epidemiology of lung cancer: latest trends, disparities, and tumor characteristics. J. Thorac. Oncol. 2016; 11:1653–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Siegel R.L., Miller K.D., Jemal A.. Cancer statistics, 2017. CA Cancer J. Clin. 2017; 67:7–30. [DOI] [PubMed] [Google Scholar]
- 9. Shukla S., Evans J.R., Malik R., Feng F.Y., Dhanasekaran S.M., Cao X., Chen G., Beer D.G., Jiang H., Chinnaiyan A.M.. Development of a RNA-Seq based prognostic signature in lung adenocarcinoma. J. Natl. Cancer Inst. 2017; 109:djw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li L., Feng T., Qu J., Feng N., Wang Y., Ma R.N., Li X., Zheng Z.J., Yu H., Qian B.. LncRNA expression signature in prediction of the prognosis of lung adenocarcinoma. Genet. Test. Mol. Biomarkers. 2018; 22:20–28. [DOI] [PubMed] [Google Scholar]
- 11. van Dam S., Vosa U., van der Graaf A., Franke L., de Magalhaes J.P.. Gene co-expression analysis for functional classification and gene-disease predictions. Brief. Bioinform. 2018; 19:575–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marques A.C., Dupanloup I., Vinckenbosch N., Reymond A., Kaessmann H.. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005; 3:e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen L., Puri R., Lefkowitz E.J., Kakar S.S.. Identification of the human pituitary tumor transforming gene (hPTTG) family: molecular structure, expression, and chromosomal localization. Gene. 2000; 248:41–50. [DOI] [PubMed] [Google Scholar]
- 14. Zou H., McGarry T.J., Bernal T., Kirschner M.W.. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999; 285:418–422. [DOI] [PubMed] [Google Scholar]
- 15. Zhang W., Gong W., Ai H., Tang J., Shen C.. Gene expression analysis of lung adenocarcinoma and matched adjacent non-tumor lung tissue. Tumori. 2014; 100:338–345. [DOI] [PubMed] [Google Scholar]
- 16. Li H., Yin C., Zhang B., Sun Y., Shi L., Liu N., Liang S., Lu S., Liu Y., Zhang J. et al.. PTTG1 promotes migration and invasion of human non-small cell lung cancer cells and is modulated by miR-186. Carcinogenesis. 2013; 34:2145–2155. [DOI] [PubMed] [Google Scholar]
- 17. Huang J.L., Cao S.W., Ou Q.S., Yang B., Zheng S.H., Tang J., Chen J., Hu Y.W., Zheng L., Wang Q.. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol. Cancer. 2018; 17:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mendez-Vidal C., Gamez-Del Estal M.M., Moreno-Mateos M.A., Espina-Zambrano A.G., Torres B., Pintor-Toro J.A.. PTTG2 silencing results in induction of epithelial-to-mesenchymal transition and apoptosis. Cell Death. Dis. 2013; 4:e530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weng W., Ni S., Wang Y., Xu M., Zhang Q., Yang Y., Wu Y., Xu Q., Qi P., Tan C. et al.. PTTG3P promotes gastric tumour cell proliferation and invasion and is an indicator of poor prognosis. J. Cell Mol. Med. 2017; 21:3360–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Therneau T.M., Grambsch P.M.. Modeling Survival Data: Extending the Cox Model. 2000; NY: Springer; 39–77. [Google Scholar]
- 21. Langfelder P., Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008; 9:599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kao H.-L., Yeh Y.-C., Lin C.-H., Hsu W.-F., Hsieh W.-Y., Ho H.-L., Chou T.-Y.. Diagnostic algorithm for detection of targetable driver mutations in lung adenocarcinomas: comprehensive analyses of 205 cases with immunohistochemistry, real-time PCR and fluorescence in situ hybridization methods. Lung Cancer. 2016; 101:40–47. [DOI] [PubMed] [Google Scholar]
- 23. Lin K.T., Wang Y.W., Chen C.T., Ho C.M., Su W.H., Jou Y.S.. HDAC inhibitors augmented cell migration and metastasis through induction of PKCs leading to identification of low toxicity modalities for combination cancer therapy. Clin. Cancer Res. 2012; 18:4691–4701. [DOI] [PubMed] [Google Scholar]
- 24. Wang Y.W., Tu P.H., Lin K.T., Lin S.C., Ko J.Y., Jou Y.S.. Identification of oncogenic point mutations and hyperphosphorylation of anaplastic lymphoma kinase in lung cancer. Neoplasia. 2011; 13:704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okimoto R.A., Breitenbuecher F., Olivas V.R., Wu W., Gini B., Hofree M., Asthana S., Hrustanovic G., Flanagan J., Tulpule A. et al.. Inactivation of Capicua drives cancer metastasis. Nat. Genet. 2017; 49:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Conrad T., Ørom U.A.. Ørom UA. Enhancer RNAs: Methods and Protocols. 2017; NY: Springer; 1–9. [Google Scholar]
- 27. Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E. et al.. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007; 129:1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Panda A.C., Martindale J.L., Gorospe M.. Affinity pulldown of biotinylated RNA for detection of Protein-RNA complexes. Bio-protocol. 2016; 6:e2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chu C., Quinn J., Chang H.Y.. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. 2012; 3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shedden K., Taylor J.M., Enkemann S.A., Tsao M.S., Yeatman T.J., Gerald W.L., Eschrich S., Jurisica I., Giordano T.J., Misek D.E. et al.. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med. 2008; 14:822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang J., Duncan D., Shi Z., Zhang B.. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013; 41:W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dallol A., Da Silva N.F., Viacava P., Minna J.D., Bieche I., Maher E.R., Latif F.. SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 2002; 62:5874–5880. [PubMed] [Google Scholar]
- 33. Nasarre P., Potiron V., Drabkin H., Roche J.. Guidance molecules in lung cancer. Cell Adh. Migr. 2010; 4:130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grando S.A. Connections of nicotine to cancer. Nat. Rev. Cancer. 2014; 14:419–429. [DOI] [PubMed] [Google Scholar]
- 35. Basile J.R., Castilho R.M., Williams V.P., Gutkind J.S.. Semaphorin 4D provides a link between axon guidance processes and tumor-induced angiogenesis. Proc. Natl Acad. Sci. U.S.A. 2006; 103:9017–9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tomczak K., Czerwinska P., Wiznerowicz M.. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Pozn.). 2015; 19:A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al.. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li J., Han L., Roebuck P., Diao L., Liu L., Yuan Y., Weinstein J.N., Liang H.. TANRIC: An interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015; 75:3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dominguez A.A., Lim W.A., Qi L.S.. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016; 17:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Poliseno L., Marranci A., Pandolfi P.P.. Pseudogenes in Human Cancer. Front. Med. 2015; 2:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burge C., Karlin S.. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997; 268:78–94. [DOI] [PubMed] [Google Scholar]
- 42. Kong L., Zhang Y., Ye Z.Q., Liu X.Q., Zhao S.Q., Wei L., Gao G.. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007; 35:W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun K., Chen X., Jiang P., Song X., Wang H., Sun H.. iSeeRNA: identification of long intergenic non-coding RNA transcripts from transcriptome sequencing data. BMC Genomics. 2013; 14(Suppl. 2):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W.. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013; 41:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y., Fan X., Lin F., He G., Terzaghi W., Zhu D., Deng X.W.. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10359–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ricke R.M., van Deursen J.M.. Aneuploidy in health, disease, and aging. J. Cell Biol. 2013; 201:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Long Y., Wang X., Youmans D.T., Cech T.R.. How do lncRNAs regulate transcription. Sci. Adv. 2017; 3:eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Noh J.H., Kim K.M., McClusky W.G., Abdelmohsen K., Gorospe M.. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA. 2018; 9:e1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Myslinski E., Gerard M.A., Krol A., Carbon P.. Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143. Nucleic Acids Res. 2007; 35:3453–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ma Q., Liu Y., Shang L., Yu J., Qu Q.. The FOXM1/BUB1B signaling pathway is essential for the tumorigenicity and radioresistance of glioblastoma. Oncol. Rep. 2017; 38:3367–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wan X., Yeung C., Kim S.Y., Dolan J.G., Ngo V.N., Burkett S., Khan J., Staudt L.M., Helman L.J.. Identification of FoxM1/Bub1b signaling pathway as a required component for growth and survival of rhabdomyosarcoma. Cancer Res. 2012; 72:5889–5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang Y., Han L., Yuan Y., Li J., Hei N., Liang H.. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat. Commun. 2014; 5:3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Signal B., Gloss B.S., Dinger M.E.. Computational approaches for functional prediction and characterisation of long noncoding RNAs. Trends Genet. 2016; 32:620–637. [DOI] [PubMed] [Google Scholar]
- 54. Li Y., Li L., Wang Z., Pan T., Sahni N., Jin X., Wang G., Li J., Zheng X., Zhang Y. et al.. LncMAP: Pan-cancer atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Res. 2018; 46:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Botling J., Edlund K., Lohr M., Hellwig B., Holmberg L., Lambe M., Berglund A., Ekman S., Bergqvist M., Ponten F. et al.. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin. Cancer Res. 2013; 19:194–204. [DOI] [PubMed] [Google Scholar]
- 56. Li Y., Tang H., Sun Z., Bungum A.O., Edell E.S., Lingle W.L., Stoddard S.M., Zhang M., Jen J., Yang P. et al.. Network-based approach identified cell cycle genes as predictor of overall survival in lung adenocarcinoma patients. Lung Cancer. 2013; 80:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Feng L., Tong R., Liu X., Zhang K., Wang G., Zhang L., An N., Cheng S.. A network-based method for identifying prognostic gene modules in lung squamous carcinoma. Oncotarget. 2016; 7:18006–18020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li X., Li B., Ran P., Wang L.. Identification of ceRNA network based on a RNA-seq shows prognostic lncRNA biomarkers in human lung adenocarcinoma. Oncol. Lett. 2018; 16:5697–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang X., Ding Y., Da B., Fei Y., Feng G.. Identification of potential prognostic long noncoding RNA signatures based on a competing endogenous RNA network in lung adenocarcinoma. Oncol. Rep. 2018; 40:3199–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peng F., Wang R., Zhang Y., Zhao Z., Zhou W., Chang Z., Liang H., Zhao W., Qi L., Guo Z. et al.. Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol. Cancer. 2017; 16:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang J., Lin J., Liu T., Chen T., Pan S., Huang W., Li S.. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer. 2014; 85:110–115. [DOI] [PubMed] [Google Scholar]
- 62. Zhang H., Su Y., Xu F., Kong J., Yu H., Qian B.. Circulating microRNAs in relation to EGFR status and survival of lung adenocarcinoma in female non-smokers. PLoS One. 2013; 8:e81408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guo H., Li W., Zheng T., Liu Z.. MiR-195 targets HDGF to inhibit proliferation and invasion of NSCLC cells. Tumour Biol. 2014; 35:8861–8866. [DOI] [PubMed] [Google Scholar]
- 64. Wang X., Wang Y., Lan H., Li J.. MiR-195 inhibits the growth and metastasis of NSCLC cells by targeting IGF1R. Tumour Biol. 2014; 35:8765–8770. [DOI] [PubMed] [Google Scholar]
- 65. Yongchun Z., Linwei T., Xicai W., Lianhua Y., Guangqiang Z., Ming Y., Guanjian L., Yujie L., Yunchao H.. MicroRNA-195 inhibits non-small cell lung cancer cell proliferation, migration and invasion by targeting MYB. Cancer Lett. 2014; 347:65–74. [DOI] [PubMed] [Google Scholar]
- 66. Liu B., Qu J., Xu F., Guo Y., Wang Y., Yu H., Qian B.. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015; 6:9445–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guo A.Y., Sun J., Jia P., Zhao Z.. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst. Biol. 2010; 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen F., Zhang Y., Parra E., Rodriguez J., Behrens C., Akbani R., Lu Y., Kurie J.M., Gibbons D.L., Mills G.B. et al.. Multiplatform-based molecular subtypes of non-small-cell lung cancer. Oncogene. 2017; 36:1384–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
To review GEO accession GSE114826: Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114826.