Abstract
The human apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3 (APOBEC3, A3) family member proteins can deaminate cytosines in single-strand (ss) DNA, which restricts human immunodeficiency virus type 1 (HIV-1), retrotransposons, and other viruses such as hepatitis B virus, but can cause a mutator phenotype in many cancers. While structural information exists for several A3 proteins, the precise details regarding deamination target selection are not fully understood. Here, we report the first parallel, comparative analysis of site selection of A3 deamination using six of the seven purified A3 member enzymes, oligonucleotides having 5′TC3′ or 5′CT3′ dinucleotide target sites, and different flanking bases within diverse DNA secondary structures. A3A, A3F and A3H were observed to have strong preferences toward the TC target flanked by A or T, while all examined A3 proteins did not show a preference for a TC target flanked by a G. We observed that the TC target was strongly preferred in ssDNA regions rather than dsDNA, loop or bulge regions, with flanking bases influencing the degree of preference. CT was also shown to be a potential deamination target. Taken together, our observations provide new insights into A3 enzyme target site selection and how A3 mutagenesis impacts mutation rates.
INTRODUCTION
Human apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3 (APOBEC3, A3) family proteins are composed of seven members that are all encoded on human chromosome 22 (1). All seven members are cytidine deaminases that target cytidines in ssDNA, and generally differ in their preferred sites of deamination, which is a trend seen in A3s from other species (2–4). Specifically, APOBEC3G prefers to deaminate cytidine within 5′CC3′ target sites, but can also deaminate cytidines within 5′TC3′ target sites (5). The other six APOBEC3 members preferentially deaminate cytidines within a 5′TC3′ target site (2–4). Deamination of cytidine, which results in a cytidine to uridine transition, generates mutations within the target DNA (6–9). In the context of somatic cell division, such mutations can facilitate oncogenesis, whereas in the context of viral replication, C to U mutations can generate lethal mutations, thereby inhibiting replication of a wide variety of viruses, including human immunodeficiency virus (HIV) (10–12). In HIV as well as cellular genomic DNA, C to U mutations in the minus-strand DNA appear as G to A mutations in the plus-strand DNA; high G to A mutation frequencies can be lethal for HIV, and in human genomic DNA, such mutations are associated with cancer (2–4,6,13,14).
Of the seven APOBEC3 proteins, A3D, A3G, A3F and A3H have been shown to restrict HIV replication when the virally encoded Vif protein is absent (2,15). In the presence of Vif, APOBEC3 is targeted for degradation, which prevents it from inhibiting HIV replication (16,17). During HIV replication, the viral RNA genome is reverse transcribed from ss(+)RNA to ss(−)DNA and then finally to a dsDNA, which is integrated into the human genome (18–20). APOBEC3-mediated inhibition has been attributed to cytidine deamination within ssDNA that is generated during reverse transcription (21). During deamination, the amine group of the cytidine is replaced by a carbonyl group, which transforms the cytidine with a uridine (C to U). This transition replaces the guanine-pairing cytidine by adenine pairing uridine, resulting in a G to A mutation in the plus strand DNA. The frequency of G to A mutations in the HIV provirus can exceed 10% of all G residues leading to a lethal level of mutations (22). The high frequency of mutations prevent the virus from replicating with enough fidelity to remain infectious (23). This G to A hypermutation eliminates virus infectivity. Previous studies have suggested that sub-lethal levels of mutations could contribute to viral evolution (24–26), but recent studies argue that the contribution of APOBEC3 proteins to HIV mutation, recombination and virus evolution is minimal (27,28). In addition to inhibition of viral replication through mutagenesis, APOBEC3 proteins have also been shown to restrict HIV through direct inhibition of reverse-transcriptase (RT) activity (29,30), although this may be a secondary mechanism of inhibiting viral infectivity (31).
Not only do the APOBEC3 proteins differ in their preferred sites of deamination and their ability to restrict HIV (3,32), they also differ in their subcellular localization (33), tissue expression patterns (34), and the number of domains that they contain. Three of the APOBEC3 proteins (A3A, A3C, A3H) contain a single domain while the other four APOBEC3 proteins (A3B, A3D, A3F, A3G) have two domains (35–37). Each of the single domain APOBEC3 proteins has a single, zinc finger containing catalytic domain. In contrast, the double domain APOBEC3 proteins have two zinc finger containing domains, both of which contain similar three-dimensional folding. However, only the C terminal domain of the double domain APOBEC3 proteins is catalytically active (38–41).
While residues within a loop of the catalytic domain that are responsible for the APOBEC3 proteins preferred dinucleotide deamination sites, the presence of the TC dinucleotide target site (or CC in the case of A3G) alone, does not ensure that a site will be deaminated. In particular, some of the TC dinucleotide target sites do not appear to be APOBEC3 targets while others appear to be preferentially targeted and are referred to as APOBEC3 ‘hotspots’ based on the frequency at which these sites are deaminated (7,10,42). Some target sites are heavily deaminated while others are not, suggesting that the APOBEC3 proteins have preferences beyond the dinucleotide target site that are not yet well characterized. One previous study focused on individual APOBEC3 proteins and identified a propensity to deaminate within areas of single-stranded DNA (21). Furthermore, preferences for the sequences flanking the CC dinucleotide target site have been elucidated for A3G (44). However, an extensive analysis of all seven APOBEC3 family members has not been performed to date and as a result, it is not known if there are differences among the family members in their propensity to deaminate cytidines located within different regions of DNA sequence and secondary structures. In particular, the catalytic properties of A3D have been a challenge to fully characterize, and the target site preferences not fully established (43).
Several previous studies have provided some insights into the nature of target site specificity. In one study, cytosine deamination with A3G using stem-loop DNA substrates showed that A3G cytosine deamination hotspots are defined by both the sequence context of the cytosine dinucleotide target as well as ssDNA secondary structure (44). Analysis of the co-crystal structure of A3G-NTD in complex with ssDNA revealed that flanking bases (i.e. −1 and +1) form specific binding interactions with loop-1, 3, 5 and 7 of A3G (45). Nucleic acid residues adjacent to the target sequence affect A3F-CTD deamination activity, with TTCA/G being the most preferred sequences (46). The sequence ATCG has been found to be a deamination target for A3A, with (T/C) TC (A/G) being important for A3A binding (47). Analysis of co-crystal structures of either A3A or the A3B-CTD with ssDNA revealed that the loop regions of DNA stem–loop structures may serve as hotpots for APOBEC-mediated mutagenesis (48). To date, there is a knowledge gap in understanding the nature of target site preferences (i.e., nucleotide context, secondary structure) among the APOBEC3 proteins.
Here, we report the first parallel, comparative analysis of site selection of APOBEC3 deamination using six of the seven purified APOBEC3 proteins (i.e. A3A, A3B, A3C, A3F, A3G and A3H haplotype II) and oligonucleotides having 5′TC3′ or 5′CT3′ dinucleotide target sites and variable flanking bases within diverse DNA secondary structures. In particular, observations were made which implicated that A3A, A3F and A3H have strong preferences towards TC targets flanked by A or T, while all examined APOBEC3 proteins were observed not to have preferences with a TC target flanked by G. Additionally, it was also observed that the APOBEC3 enzymes could recognize a TC target site in a stem structure, and deaminate a CT target. In coincidence with the expectation that A3 enzymes preferentially act on ssDNA templates, the majority of the A3 proteins were observed to have a higher preference for the TC target in a ssDNA region than in other regions, such as a dsDNA, loop and bulge. Notably, the secondary structure of ‘−1’ T of TC target was shown to influence the A3 deamination preference. Intriguingly, A3 enzymes that possess closer sequence and/or structural similarities were observed to follow comparable rules for target selection, e.g. A3A and A3H. Collectively, the findings of this study provide new insights into target site selection of A3 enzyme and aid in understanding how A3 mutagenesis impacts mutation rates.
MATERIALS AND METHODS
Expression and purification of APOBEC3 proteins
Recombinant baculovirus for expression of GST-A3A, GST-A3B, GST-A3C, GST-A3F, GST-A3G and GST-A3H haplotype II (referred to as A3H) was constructed using either the pAcG2T (BD Biosciences) or pFAST-bac1 (Life Technologies) vectors as previously described (49–54). Recombinant baculovirus infected Sf9 cells were harvested after 72 h of infection. Cells were lysed, and the GST-tagged proteins were purified as described previously to obtain protein that was cleaved from the GST tag. Cleaved protein fractions were stored at −80°C. Enzymes were assessed by SDS-PAGE (Supplemental Figure S1).
Oligonucleotide design and synthesis
Each oligonucleotide (Table 1) was synthesized to contain both a 5′-FAM and a 3′-TAMRA modification. Each oligonucleotide was also designed to have a TC or CT context in a given DNA secondary structure, which are listed in Table 1 by category. The Mfold program (http://unafold.rna.albany.edu/?q=mfold/dna-folding-form) was used to design oligonucleotides that were predicted to have only one secondary structure at 37°C.
Table 1.
Sequences of oligonucleotides used in FRET assay for detecting APOBEC3 enzyme activitya
| Sequence context | predicted secondary structure at deamination site | Oligonucleotide name | ΔG | 5′-3′ sequence |
|---|---|---|---|---|
| XTCX | Open | ATCA open | −2.44 | ATTGAATCAGCATGGTGGCATGGAATATA |
| TTCT open | −2.44 | ATTGATTCTGCATGGTGGCATGGTGTATA | ||
| GTCG open | −2.44 | ATTGAGTCGGCATGGTGGCATGGTATATA | ||
| CTCC open | −2.44 | ATTGACTCCGCATGGTGGCATGGTATATA | ||
| Stem | ATCA stem | −6.22 | AAATCACGTGAGAGAACGTGATAAT | |
| TTCT stem | −4.64 | AATTCTTGTGAGAGAACAAGAAAAT | ||
| GTCG stem | −7.60 | AAGTCGCAAGAGAGATTGCGACAAT | ||
| CTCC stem | −6.35 | AACTCCAAAGAGAGATTTGGAGAAT | ||
| Loop | ATCA 10-loop | −4.90 | ATTGATGCACACGAATCAAGATGTGCAAATATA | |
| ATCA 3-loop | −3.67 | ATTGAACAACATCATGTTGAAATATA | ||
| GTCG 10-loop | −4.80 | ATTGATGCACATGAGTCGAGGTGTGCAAATATA | ||
| GTCG 3-loop | −4.44 | ATTGAACAACGTCGCGTTGAAATATA | ||
| Bulge | ATCA bulge | −2.18 | AGTGAAAATCAACGCAACGTTTTTTGTATA | |
| GTCG bulge | −3.46 | AGTGAATGTCGTACGAAGTACCATTGTATA | ||
| XCTX | Open | ACTA open | −2.44 | ATTGAACTAGCATGGTGGCATGGAATATA |
| TCTT open | −2.44 | ATTGATCTTGCATGGTGGCATGGTGTATA | ||
| GCTG open | −2.44 | ATTGAGCTGGCATGGACGCATGGGATATA | ||
| CCTC open | −2.44 | ATTGACCTCGCATGGTGGCATGATATATA | ||
| Stem | ACTA stem | −5.89 | AAACTACGTGAGAGAACGTAGTAAT | |
| TCTT stem | −4.64 | AATCTTTGTGAGAGAACAAAGAAAT | ||
| GCTG stem | −7.66 | AAGCTGCAAGAGAGATTGCAGCAAT | ||
| CCTC stem | −6.35 | AACCTCAAAGAGAGATTTGAGGAAT | ||
| XTCY | Open | GTCA open | −2.44 | ATTGAGTCAGCATGGTGGCATGGAATATA |
| CTCA open | −2.44 | ATTGACTCAGCATGGTGGCATGGTATATA | ||
| CTCT open | −2.44 | ATTGACTCTGCATGGTGGCATGGTATATA | ||
| CTCG open | −2.44 | ATTGACTCGGCATGGTGGCATGGTATATA | ||
| Half open | Half open C in ss region ACTT (ACTT) | −0.92 | ATGGAACTTCAATAATGAATACGAATATA | |
| Half open T in ss region ATCT (ATCT) | −2.16 | ATGGAATCTCAATAATGAGTTGGAATATA | ||
| Half open C in ss region ACTG (ACTG) | −2.43 | TAAGAACTGCAATAATGCAAACGAATATA | ||
| Half open T in ss region ATCG (ATCG) | −3.99 | TAAGAATCGCAATAATGCGTACGAATATA |
aThe predicted target cytidine for deamination is indicated by an underline in each oligonucleotide. The X or Y nucleotides in the sequence context represent a variable nucleotide. The X and Y in the same oligonucleotide indicates different variable nucleotides.
Fluorescence energy transfer (FRET) assay for detecting purified APOBEC3 activity
Cytosine deaminase activity for each APOBEC3 protein was detected through a fluorescence resonance energy transfer (FRET) based assay, which was performed as previously described with minor modifications (44,55). DNA oligonucleotides labeled with 5′FAM and 3′TAMRA were used as substrates. Intact oligonucleotides maintain FAM and TAMRA in close proximity that allows TAMRA to quench the FAM signal (490 nm). When the oligonucleotide is targeted by APOBEC3, a uridine substrate is generated and the oligonucleotide is subjected to uracil DNA glycosylase digestion that results in its cleavage of the oligonucleotide. As a result, TAMRA no longer quenches the FAM signal and an increase in the FAM signal at 490 nm (ΔRFU) is generated, which corresponds to APOBEC3 activity. Briefly, 200 pmol (for A3B/A3C/A3F/A3G) or 20 pmol (for A3A/A3H) oligonucleotide and 0.8 μM of one kind of APOBEC3 (0.05 μM for A3F) were incubated in deamination buffer (50 mM Tris–HCl pH 8.0, 5 mM MgCl2 and 1 mM DTT) at 37°C for 2 h using 96 white-well assay plates (Bio-Rad). The standard assay reaction volume used was 20 ul. Then, 0.04 U of uracil DNA glycosylase (NEB) was added to each assay and the plate was incubated at 37°C for 45 min. Next, 2 M Tris–Acetate (7.9) was added to each assay. After incubating at 37°C for 20 min, the plate was incubated at 95°C for 2 min and at 4°C for 2.5 min with a CFX96 real-time PCR system (Bio-Rad). The fluorescence was then measured at 4°C. In order to calculate a relative change in fluorescence (ΔRFU) due to APOBEC3 activity, the endpoint fluorescence from buffer without APOBEC3 proteins was measured for all oligonucleotides as negative controls, and was subtracted from all the endpoint fluorescence of experimental samples. Experiments were conducted in three independent replicates.
Generation of APOBEC3 protein-expressing cell lines
HEK-293 cells that stably express A3C, A3D, A3F, A3G or A3H (haplotype II) were established by transfecting HEK-293 cells with APOBEC3-pcDNA3 (2). The plasmids were kindly provided by Reuben Harris (University of Minnesota). HEK-293 cells (American Type Culture Collection) were cultured in DMEM containing 10% FC3 (HyClone) and 1% penicillin/streptomycin (Invitrogen). Transfected HEK-293 cells were selected with 1000 μg/ml G418 (Gibco) for 7 days. The cells were then diluted to yield approximately 1 cell per well of a 96-well plate to form single cell colonies. Cell lines stably expressing the APOBEC3 were maintained in DMEM containing 10% FC3 and 250 μg/ml G418.
Transient transfection of A3A and A3B
HEK-293 cells were transiently transfected with 10 μg APOBEC3-pcDNA3.1 using 30 μg of polyethylenimine (PEI; Polysciences, Inc.) (56). Transfected cells were maintained in DMEM containing 10% FC3 and cells were harvested 48 h after transfection.
Immunoblot and cell line verification
Cell lysates were made by lysing cells with 1% Triton X-100 in PBS. Protein concentration was determined by using a BCA kit (Thermo Fisher). 10% SDS-PAGE (Bio-Rad) was used for protein separation. An anti-HA antibody (Biolegend, 1:800) and goat anti-mouse antibody (Thermo Fisher, 1:8000) were used for A3 protein detection by immunoblot analysis.
Fluorescence energy transfer (FRET) assay for detecting APOBEC3 enzyme activity from cell lysates
The FRET assay used to assess APOBEC3 activity from cell lysates was performed as previously described with minor modifications (44,57). Briefly, 20 pmol oligonucleotide, 10 ug RNase A (Sigma), 100 μl cell lysate of APOBEC3 cell lines and 0.04 U of uracil DNA glycosylase (NEB) were used for each experiment. Cell lysates were made by lysing 5 000 000 cells with 250 μl of lysis buffer (0.626% NP40, 10 mM tris-acetate pH 7.4, 50 mM potassium acetate, 100 mM NaCl and 10 mM EDTA). In order to calculate a relative change in fluorescence (ΔRFU) due to APOBEC3 activity, the endpoint fluorescence from the 293-vector cell lysate was measured for all oligonucleotides as negative controls and was subtracted from all the endpoint fluorescence of experimental samples (293-APOBEC cell lysates).
Statistical analysis
All graphs were created in GraphPad Prism, version 5.0 (GraphPad Software, Inc.). Data were analyzed by calculating the mean ± standard deviation (SD). Differences between groups were analyzed by either one-way or two-way ANOVA test as indicated. A P-value of <0.05 was considered statistically significant. Statistical analysis was not shown in Figure 2B–G because of multiple comparisons, but listed in Supplemental Tables S1–S6, and the results were summarized in Tables 2, 3 and 5.
Figure 2.
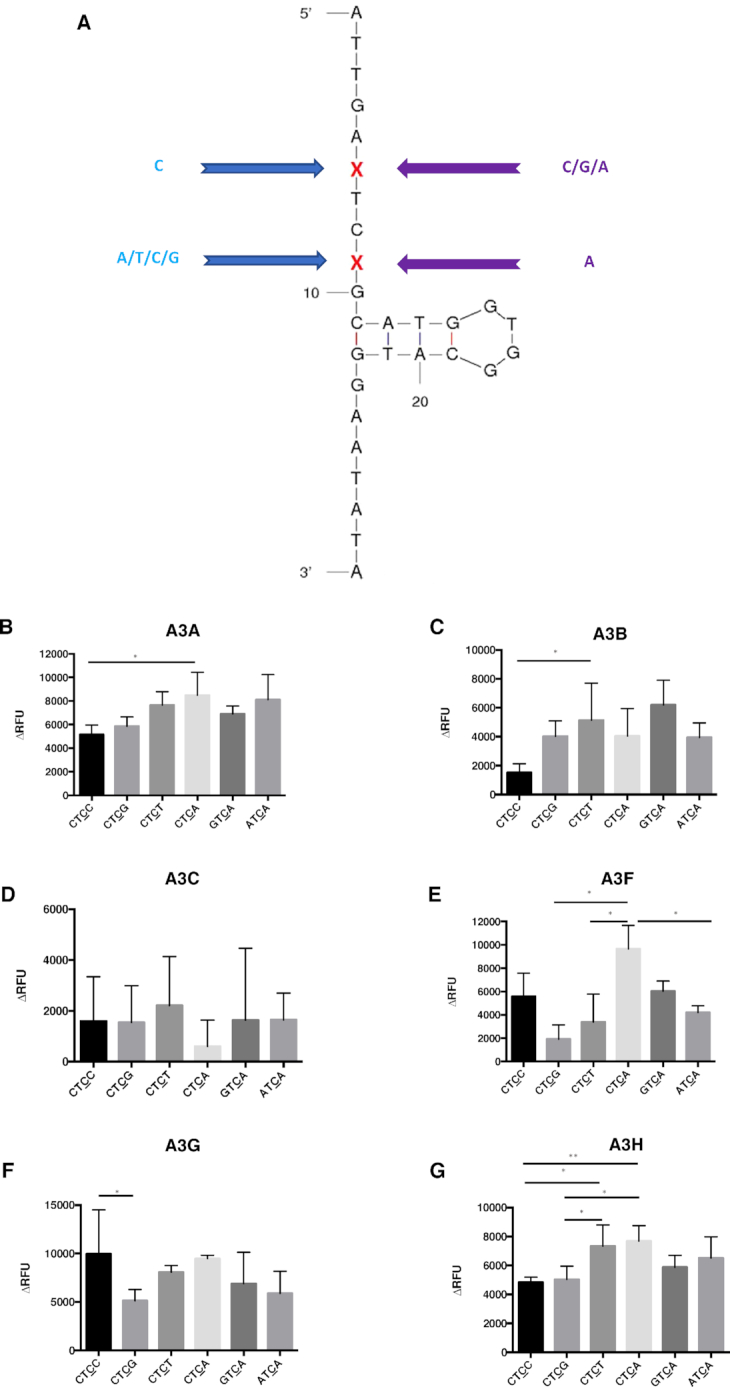
APOBEC3 proteins have different preferences with oligonucleotides having different flanking bases on either side of the target site. (A) Oligonucleotide sequence and predicted secondary structure. Cytidine was chosen for the 5′ flanking base of ‘TC’ motif, while the 3′ flanking base is either adenine, cytidine, guanine, or thymidine. In contrast, adenine was chosen for the fixed 3′ flanking base of ‘TC’ motif, while the 5′ flanking base is either adenine, cytidine or guanine. The same sequence (backbone) as the ‘open’ oligonucleotides was used for all the substrates besides the modifications of the flanking bases. (B–G) APOBEC3 protein activity with different oligonucleotide substrates. Comparison of APOBEC3 activity was determined among the oligonucleotides CTCC, CTCG, CTCT, CTCA, GTCA and the ATCA open oligonucleotide. Results are shown as the average ± standard deviation of 3 independent experiments. The asterisks and brackets above any two bars indicate the significant difference between experiments. Significance was analyzed by using a one-way ANOVA with significance defined as P ≤ 0.05. Experiments that were not significant or significant are indicated as follows: ‘ns’ = not significant; ‘**’ = P ≤ 0.01; ‘*’ = P ≤ 0.05.
Table 2.
Comparison of the APOBEC3 protein deamination preferences in the XTCX sequence contexta
| Protein\preference | Nucleotides flanking the XTCX open | Nucleotides flanking the XTCX stem | ‘+1’ position X for CTCX open | ‘−2’ position X for XTCA open |
|---|---|---|---|---|
| A3A | A = T>C = G | T>C≧A = G | A≧T = G = C | C = A = G |
| A3B | A = C = T = G | A = T = C = G | T≧A = G = C | G = C = A |
| A3C | T = A = C = G | T = A = C = G | T = C = G = A | G = A = C |
| A3F | T≧G = C = A | A = T = G = C | A = C≧T = G | C = G≧A |
| A3G | C>A = T>G | C>T≧A = G | C = A = T≧G | G = C = A |
| A3H | A = T≧C>G | T>A = C = G | A = T>G = C | C = A = G |
aThe predicted target cytidine for deamination is indicated by an underline in each oligonucleotide. The X nucleotide in the sequence context represents a variable nucleotide. ‘>’ = greater than; ‘<’ = smaller than; ‘ = ’ = equal; ‘≧’ = greater or equal; ‘≦’ = smaller or equal. Statistical differences between groups were analyzed by using either a one-way or two-way ANOVA test, with significance having a P-value of P < 0.05.
Table 3.
Comparison of APOBEC3 protein deamination preferences with oligonucleotides containing TC targetsa
| Protein\preference | Open versus stem | Open versus bulge | Open versus loop | 10-loop versus 3-loop |
|---|---|---|---|---|
| A3A | > | > | >(ATCA); ≦ (GTCG 3-loop) | ≦ |
| A3B | = | = | = | = |
| A3C | = | = | = | = |
| A3F | ≧ | ≧ | = (ATCA); > (GTCG) | = |
| A3G | = | >(ATCA); < (GTCG) | ≧(ATCA); ≦ (GTCG) | ≧ |
| A3H | > | ≧ | ≧ | < |
aThe predicted target cytidine for deamination is indicated by an underline in each oligonucleotide. ‘>’ = greater than; ‘<’ = smaller than; ‘ = ’ = equal; ‘≧’ = greater or equal; ‘≦’ = smaller or equal. Statistical differences between groups were analyzed by using a two-way ANOVA test, with significance having a P-value of P < 0.05.
Table 5.
Comparison of the APOBEC3 protein deamination preferences with TC or CT targets in oligonucleotides with different predicted DNA secondary structuresa
| Protein\preference | Open versus ‘−1’ T open & target C stem versus stem (TC target) | Open versus ‘+1’ T stem & target C open versus stem (CT target) |
|---|---|---|
| A3A | open ≧ ‘−1’ T open & target C stem ≧ stem | stem > open = ‘+1’ T stem & target C open |
| A3F | open = stem = ‘−1’ T open & target C stem | open = stem = ‘+1’ T stem & target C open |
| A3G | open = stem ≧ ‘−1’ T open & target C stem | stem > open = ‘+1’ T stem & target C open |
| A3H | open ≧ ‘−1’ T open & target C stem > stem | ‘+1’ T stem & target C open ≧ open > stem |
aThe predicted target cytidine for deamination is indicated by an underline in each oligonucleotide. ‘>’ = greater than; ‘<’ = smaller than; ‘ = ’ = equal; ‘≧’ = greater or equal; ‘≦’ = smaller or equal. Statistical differences between groups were analyzed by using a one-way ANOVA test, with significance having a P-value of P < 0.05.
RESULTS
The activity of APOBEC3 proteins within ssDNA is dictated, in part, by the nucleotides that flank the TC target site
Our previous studies implicated that the nucleotides immediately adjacent to 5′CC3′ targets influenced the activity of A3G (44). In those experiments, cell lysates from cells stably expressing A3G were used, which cannot exclude the possibility that other cell factors could influence the results. In this present study, in order to more directly assess APOBEC3 enzyme activity, we have conducted in vitro deaminase experiments with purified APOBEC3 enzymes for A3A, A3B, A3C, A3F, A3G and A3H (Supplemental Figure S1). In particular, we sought to determine whether enzyme activity among APOBEC3 enzymes was influenced by the nucleotides flanking the 5′TC3′ target site, since six of seven APOBEC3 members preferentially deaminate the 5′TC3′ target site.
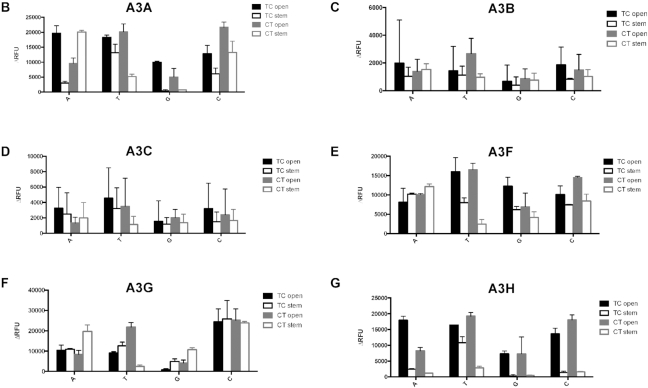
A single parent oligonucleotide sequence was used to create derivative oligonucleotides (Table 1). We designed oligonucleotides where the 5′TC3′ target site was flanked on both sides by an A, C, G or T to examine the effect of the nucleotides that flank the 5′TC3′ target site (XTCX open, Table 1 and Figure 1A). It was found that A3A and A3H had a demonstrated preference for A and T as the flanking bases. G was found to be the least preferred flanking base for A3G and A3H when recognizing a TC target. A3A exhibited the least enzymatic activity with a TC target that was flanked by either a C or a G. A3A, A3F and A3H were found to have selection site preferences of either ATCA or TTCT. In contrast, all examined A3 enzymes were observed to have minimal activity with a GTCG target (Figure 1B–G, Table 2 and Supplemental Tables S1–S6a). A3G was observed to exhibit the highest deamination activities with CTCC substrates (Figure 1F, Table 2 and Supplemental Table S5a).
Figure 1.
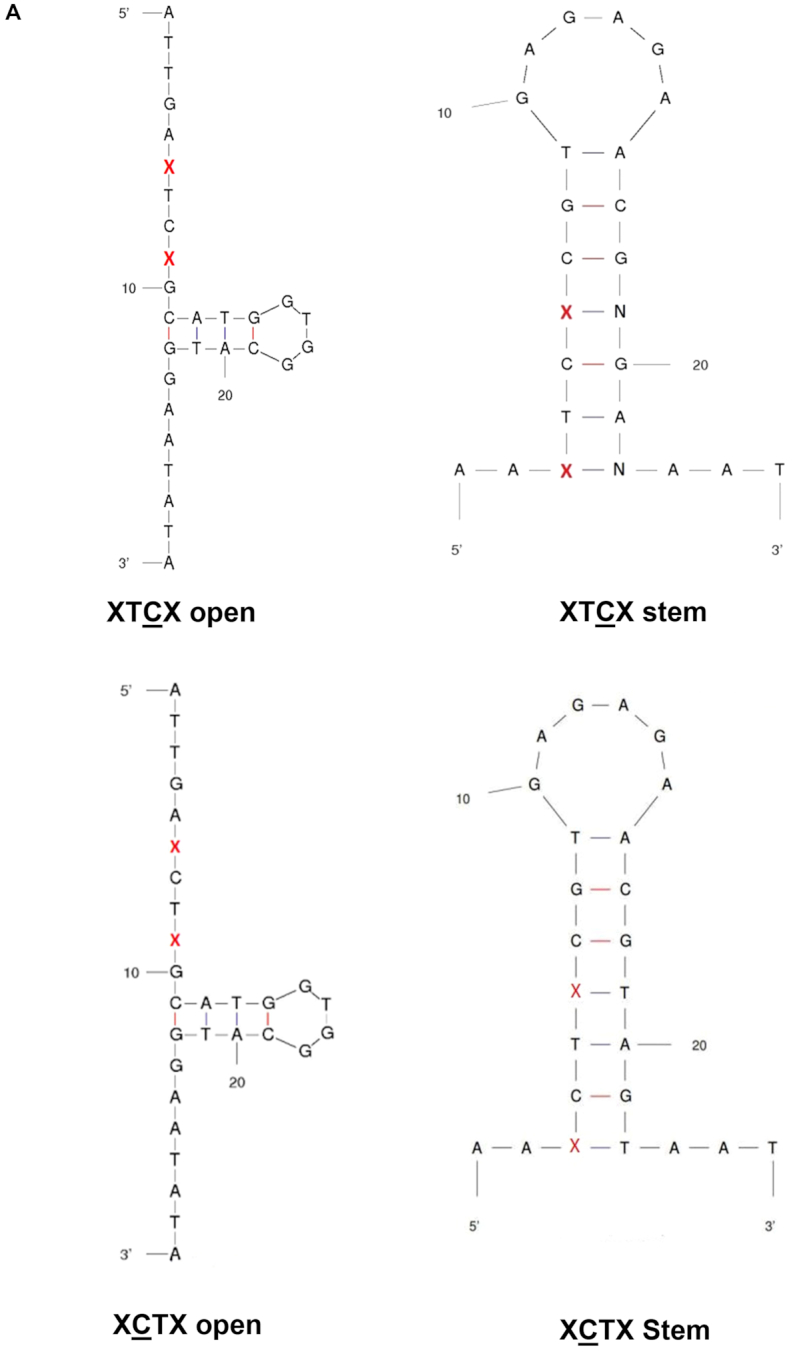
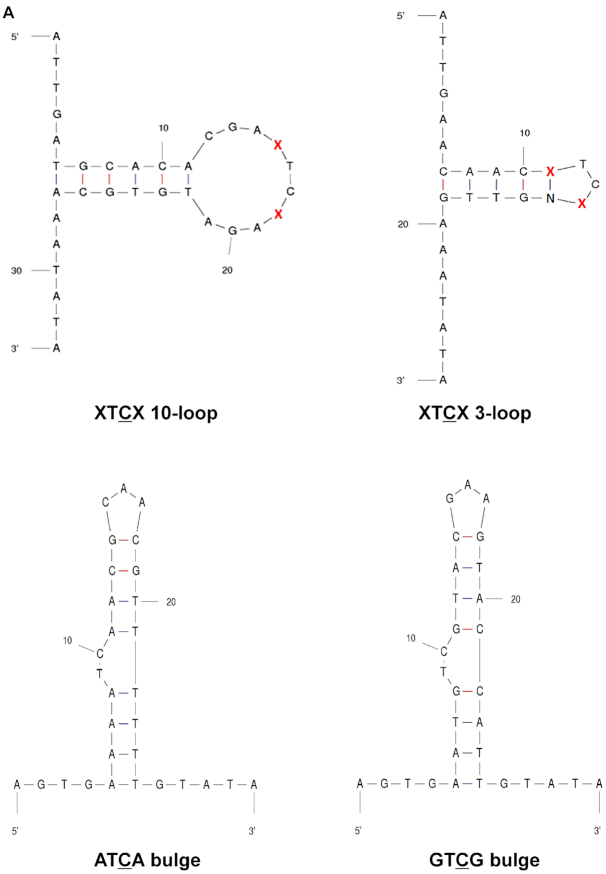
APOBEC3 activity has a preference for TC in an open versus a stem DNA structure and is influenced by nucleotides flanking the 5′TC3′/5′CT3′ site. (A) Oligonucleotide sequences and predicted secondary structures. The ‘open’ oligonucleotides contain the TC/CT target site in a ssDNA region, i.e. XTCX/XCTX open. The ‘stem’ oligonucleotide contains the TC/CT target site in a dsDNA region, i.e., XTCX/XCTX stem. The red ‘X’ represents the bases that are immediately adjacent to the TC target site, which can be A, T, G or C. For the stem oligonucleotide, the ‘N’ represents the base that is complementary to the changeable base ‘X.’ (B–G) Comparison of purified APOBEC3 activity using open oligonucleotides (target TC/CT is in ssDNA) and stem oligonucleotides (target TC/CT is in dsDNA) with different flanking regions. Results are shown as the average ± standard deviation of three independent experiments. Significance was analyzed by using a two-way ANOVA, P ≤ 0.05.
We next examined oligonucleotides with different TC flanking bases on two sides. As shown in Figure 2A, C was chosen for the 5′ flanking base of TC motif (i.e., −2 position), while the 3′ flanking base (i.e. +1 position) is either an A, C, G or T. An A was chosen for the fixed 3′ flanking base for the TC motif (i.e. +1 position), while the 5′ flanking base (i.e. −2 position) was either A, C, or G (Figure 2A). The oligonucleotide with T as the 5′ flanking base in the TCA motif was predicted to have a different DNA secondary structure. Therefore, this oligonucleotide was not used. The data indicates that the A3 enzymes exhibited a preference, albeit mild, for such substrates (Figure 2 and Table 2). A3A and A3B were observed to have a mild preference for a C at the +1 position (Figure 2B and C). Furthermore, A3H had the least activity with substrates when either G or C were at the +1 position (Figure 2G), and A3G had low activity when G was at the +1 position (Figure 2F). In contrast, A3F had a preference of A and C over that of T and G at the +1 position (Figure 2E). For A3F, the ordered nucleotide preference at the −2 position was observed to be C = G≧A, which is the opposite to that observed previously with the A3F-CTD (46) (Figure 2E and Table 2). This discrepancy suggests substrate differences may exist between full-length and truncated A3F. Taken together, these observations indicate that the +1 position X in the CTCX target site is one of the variables that determines the target preferences of A3 proteins, while the −2 position X for XTCA does not represent a significant variable in defining target site preference.
APOBEC3 proteins can recognize the TC target in a stem structure, but prefers the TC target in an open structure
The target site in a stem region was previously observed to be a less preferred target of A3G in assays using cell lysates from cells stably expressing A3G (44). In order to determine if the TC target is deaminated only in ssDNA (i.e., an open, ssDNA structure), oligonucleotide derivatives were designed to contain the TC target site within a region of dsDNA (XTCX stem, Figure 1A) compared to derivatives that have the TC target site within a region of ssDNA (XTCX open, Figure 1A). The data indicates that A3A and A3H strongly prefer the TC target in open than stem structure, irregardless of the flanking base (Figure 1B and G, Table 3, Supplemental Tables S1b and S6b). A3F was also shown to prefer TTCT open, but had similar preferences between ATCA/GTCG/CTCC open and stem (Figure 1E, Table 3, Supplemental Table S4b). No significant differences were observed in regards to deamination preferences between the TC target sites within the open and stem regions for A3G (Figure 1F, Table 3, Supplemental Table S5b). Furthermore, the flanking base preference of A3A, A3F and A3H for the TC target site was altered when the target site and flanking bases were changed from an open to a stem structure, demonstrating that the secondary structure of the TC target influences the preference of flanking bases of the target site (Table 2). These observations, taken together, imply that A3 enzymes can recognize a TC target site in a stem structure. However, a more likely explanation for this observation is DNA ‘breathing’—i.e., transient departure of base-paired DNA sequences from their most stable structures, creating an open configuration that would allow for cytosine deamination to occur (58).
APOBEC3 proteins can recognize both the XCTX open and the stem target as well as the XTCX open/stem target, with a preference for targets with special flanking bases
A3G is known to deaminate C with a 3′ to 5′ polarity (53,59). It is formally possible that other APOBEC3 enzymes may be able to deaminate C with a 3′ to 5′ polarity, and could deaminate C when T is in +1 versus the −1 position. It should be noted that the increase in APOBEC3 enzymatic activity observed when the TC target site was flanked by Ts and Cs (i.e., TTCT and CTCC; Figure 1) could be attributed to the presence of more than one target site rather than a preference of a TC target site that is flanked by these bases. These extra target sites would only be present if APOBEC3 could recognize both the 5′CT3′ and the 5′TC3′ sites. In order to discriminate the ability of APOBEC3 enzymes to deaminate either a 5′TC3′ (TC) or a 5′CT3′ (CT) site, oligonucleotides derivatives were designed to have the same secondary structure as TC oligos, but the 5′TC3′ was replaced with 5′CT3′ (i.e. XCTX open, Figure 1A). While the canonical target site was 5′TC3′, each APOBEC3 enzyme was able to recognize the 5′CT3′ target site. A3B, A3C and A3F were equally capable of deaminating the 5′CT3′ or 5′TC3′ target site (Figure 1C–E, Table 4 and Supplemental Tables S2–S4b). The target preference between XTCX and XCTX of A3A, A3G and A3H was affected by the flanking bases. A3A was shown to have no preference between TTCT and TCTT, as predicted. A3A preferred XTCX when X = A/G; while it preferred XCTX with X = C (Figure 1B, Table 4 and Supplemental Table S1b). When the flanking base was A/G/C, A3G showed the same activity to deaminate 5′CT3′ as the 5′TC3′ target site as well. When the flanking base was changed to a T, A3G was found to prefer the 5′CT3′ site more than the 5′TC3′ target site (Figure 1F, Table 4 and Supplemental Table S5b). When the flanking base was T/G/C, A3H also showed the same activity to deaminate 5′CT3′ or the 5′TC3′ target site. When the flanking base was changed to an A, A3H was found to prefer the 5′TC3′ site more than the 5′CT3′ target site (Figure 1G, Table 4 and Supplemental Table S6b). These observations indicated that the flanking base of the TC target site affected the deamination preference of APOBEC3 enzymes, and suggests that APOBEC3 activity detected with oligonucleotide derivatives having a TC target site flanked by either a T or a C (i.e. TTCT, CTCC) was due, in part, to the ability of APOBEC3 enzymes to recognize both the 5′TC3′ and 5′CT3′ target sites.
Table 4.
Comparison of APOBEC3 protein deamination preferences between the XTCX and XCTX sequence contextsa
| Protein\preference | TC open versus CT open | TC stem versus CT stem | CT open versus CT stem |
|---|---|---|---|
| A3A | X = T, =; X = A/G, >; X = C, < | X = G, =; X = A/C, <;X = T, > | X = T/C, >; X = A, <; C = G, = |
| A3B | = | = | X = A/G/C, =; X = T, > |
| A3C | = | = | = |
| A3F | = | X = A/G/C, =; X = T, > | X = T/C, >; X = A/G, = |
| A3G | X = A/G/C, =; X = T, < | X = G/C, =; X = A, <; X = T, > | X = T, >; X = A/G, <; X = C, = |
| A3H | X = T/G/C, =; X = A,> | X = A/G/C, =; X = T, > | > |
aThe predicted target cytidine for deamination is indicated by an underline in each oligonucleotide. The X nucleotide in the sequence context represents a variable nucleotide. ‘>’ = greater than; ‘<’ = smaller than; ‘ = ’ = equal; ‘≧’ = greater or equal; ‘≦’ = smaller or equal. Statistical differences between groups were analyzed by using a two-way ANOVA test, with significance having a P-value of P < 0.05.
For most APOBEC3 enzymes, no preferences have been observed for deamination of TC and CT target sites when the target site is in a ssDNA region. We next tested if locating the target site to a stem region would affect target site recognition and deamination. We observed that when the target site was located within a stem region, A3B and A3C had no propensity to deaminate TC over CT target sites (Figure 1C and D, Table 4, Supplemental Tables S2b and S3b). A3A, A3F, A3G and A3H all preferred TTCT over TCTT in a stem, and all showed no preference between GTCG and GCTG in the stem. A3F, A3G and A3H showed no preference between CTCC and CCTC in a stem as well. A3F and A3H also showed no preference between ATCA and ACTA in a stem. A3A and A3G preferred ACTA than ATCA in a stem, and A3A also preferred CCTC than CTCC in a stem. As shown in Table 4, APOBEC3 proteins can recognize both a XCTX open and a stem target primarily as the same as XTCX open/stem target, but in some instances had a preference for either one with special flanking bases.
We next compared the XCTX open with the XCTX stem to see if the preference of an open target was changed with a CT target. As shown in Table 4, the preference of XCTX open was either larger or equal to that of the XCTX stem. In contrast, A3A and A3G preferred ACTA in a stem than ACTA open, and A3G preferred GCTG in a stem stem than GCTG open as well. Taken together, these observations lead to the conclusion that the preference of APOBEC3 proteins for the CT target was affected by both the secondary structure and flanking bases.
Location of the ‘−1’ T of the TC target in an open or stem structure may affect the APOBEC 3 deamination preference but not that of the ‘+1’ T of the CT target
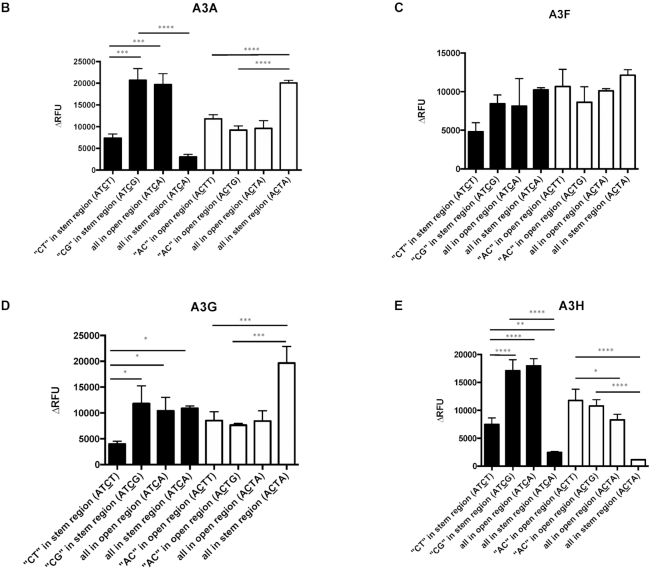
Given the observed recognition of the TC target site either in an open or stem region, we next examined whether the TC target site needs to be entirely in an open or a stem region to be recognized as a deamination target site. To test this, we designed 1) oligonucleotides that had the C of the TC target site in an area of stem region, while the T of the TC target site was located in an open region (ATCT and ATCG), and 2) oligonucleotide derivatives that have the C of the CT target site in an open region and the T of the CT target site in a stem region (ACTT and ACTG) (Figure 3A). We observed that A3A deaminated in the context of ATCT to the same efficiency as that of the ATCA stem (Figure 3B). In contrast, A3A and A3H deaminated in the context of ATCG to the same efficiency as that of the ATCA open, which indicated that the secondary structure of ‘−1’ T of the TC target played a role in target selection. Furthermore, A3G deaminated ATCT with a much lower efficiency than ATCA stem. This effect was not obvious for A3F, perhaps because A3F did not have strong preference between TC/CT in the open versus the stem (Figure 3C). However, A3A and A3G deaminated in the context of ACTT or ACTG to the same efficiency as that of the ACTA open, but much lower to that of ACTA stem, which indicated minimal impact of the secondary structure of ‘+1’ T in the deaminase preference (Figure 3B and D). This was also seen with A3H, but in an opposite manner (Figure 3E). In particular, A3H deaminated in the context of ACTT or ACTG almost to the same efficiency as that of the ACTA open, but much higher to that of ACTA stem. As summarized in Table 5, the secondary structure of ‘−1’ T of the TC target may affect the deamination preferences of the APOBEC3 proteins, but not the ‘+1’ T of the CT target.
Figure 3.
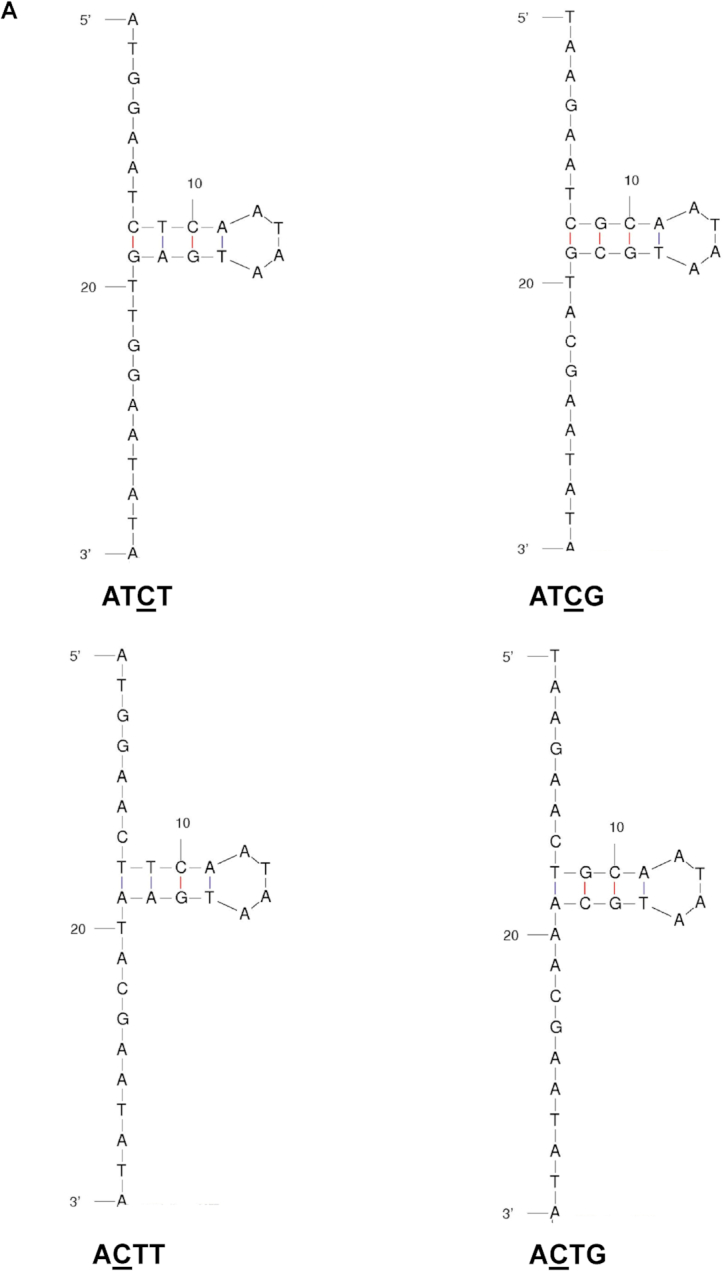
Influence of the ‘−1’ T in the TC target but not the ‘+1’ T in the CT target on APOBEC3 protein activity in an open or stem structure. (A) Oligonucleotide sequence and predicted secondary structure. The ‘half open’ oligonucleotide was designed to contain a T in the TC target site in the ssDNA region and a C in the TC target site of the dsDNA region. The ‘half open’ oligonucleotide contains a C in the CT target site of the ssDNA region and a T in the CT target site of the dsDNA region. (B–E) APOBEC3 protein activity with different oligonucleotide substrates. APOBEC3 activity was analyzed in the presence of half open oligonucleotides with different flanking nucleotides (ATCT VS. ATCG) compared to that of oligonucleotides with an ATCA/ACTA open oligonucleotide or an ATCA/ACTA stem oligonucleotide. Data are shown as the average ± standard deviation from three independent experiments. The asterisks and brackets above any two experiments indicate statistical significance. Significance was analyzed by using a one-way ANOVA with significance defined as P ≤ 0.05. Experiments that were significant or not significant are indicated as follows: ‘ns’ = not significant; ‘****’ = P ≤ 0.0001; ‘***’ = P ≤ 0.001; ‘**’ = P ≤ 0.01; ‘*’ = P ≤ 0.05.
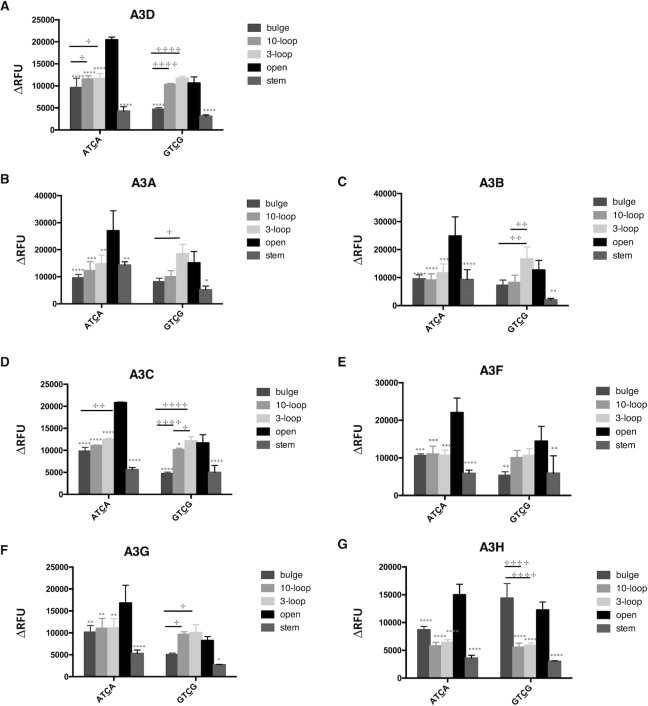
APOBEC3 proteins prefer a TC target in a ssDNA region over other structures, such as loop and bulge structures
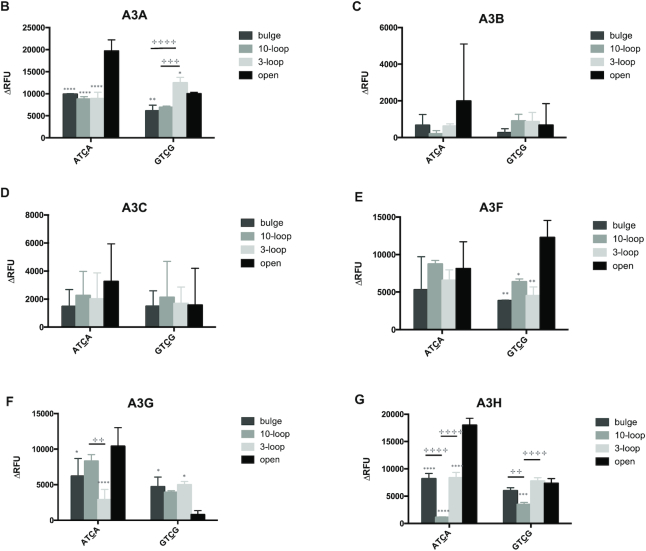
Although the target site and adjacent flanking base of oligonucleotides examined were either in an open or a stem region, the host genomic and proviral DNAs can complex secondary structures. As alluded to above, co-crystal-structures of A3A or A3B-CTD with ssDNA indicated that the loop regions of stem–loop structures in ssDNA can be a hotspot for deamination (48). Furthermore, A3A was shown to prefer a binding target sequence in the loop of structured hairpins (47). Therefore, we sought to investigate if other secondary structures of oligonucleotides can affect the deamination preference of APOBEC3 enzymes. First, we tested oligonucleotides in which the TC target site and the flanking region was either located within a long stretch of ssDNA (XTCX 10-loop, Figure 4A) or within a short stretch of ssDNA (XTCX 3-loop, Figure 4A). Second, we tested oligonucleotides that contained the TC target site within an area of ssDNA with flanking residues (either A or G residues) located within a region of dsDNA (bulge, Figure 4A). A3H more readily deaminated the TC target when the target and flanking region were in a 3-base loop rather than in a 10-base loop; A3B, A3C and A3F had no preference (Figure 4C, D, E, G and Table 3). A3A preferred GTCG in a 3-loop than in a 10-loop, but showed the same preference between ATCA in a 3-loop and in a 10-loop (Figure 4B and Table 3). A3G preferentially deaminated the TC target with an A flanking base in a 10-base loop compared to an A flanking base in a 3-base loop, and A3G exhibited the same preference for GTCG in a 3-loop versus that in a 10-loop (Figure 4F and Table 3). A3G was found to also prefer ATCA in an open configuration than in either a bulge or a loop, but preferred GTCG in a bulge or a loop configuration rather than in an open configuration. A3A was observed to have higher deaminase activity with XTCX in a 3-loop configuration only when the flanking base was a G. In other instances, A3A, A3B, A3C, A3F and A3H had either lower or equal deamination activities with oligonucleotides that had the TC target located in a bulge or a loop rather than the TC located in an ssDNA region.
Figure 4.
APOBEC3 proteins prefer a TC target site in ssDNA rather than in the context of a loop or a bulge. (A) Oligonucleotide sequence and predicted secondary structure. The ‘10-loop’ oligonucleotide contains the TC target site in a 10nt ssDNA loop. The ‘3-loop’ oligonucleotide contains the TC target site in a 3nt ssDNA loop. The ‘Bulge’ oligonucleotide contains TC target site in a ssDNA region with flanking bases in a dsDNA region. The ‘X’ represents the flanking bases (‘A’ or ‘G’). (B–G) APOBEC3 protein activity with different oligonucleotide substrates. APOBEC3 activity with oligonucleotides containing the target site either within a large region of ssDNA (10-loop), within a small region of ssDNA (3-loop) or a bulge was compared to that of the target site in an open TC. Data is shown as the average ± standard deviation of three independent experiments. Significance was analyzed by using a two-way ANOVA with significance defined as P ≤0.05. Significant differences between experiments of an open structure compared to another secondary structure is indicated by a ‘*’ above relative histogram bar. The ‘+’ symbol and brackets above any two bars indicates the significant difference between analyses. Experiments that were significantly different are indicated as follows: ‘****’/ ‘+ + + +’ = P ≤ 0.0001; ‘***’/ ‘+ + +’ = P ≤ 0.001; ‘**’/ ‘+ +’ = P ≤ 0.01; ‘*’/ ‘+’ = P ≤ 0.05.
Low activity of purified A3B and A3C could help explain the lack of significance when target sites are associated with either open or other secondary structures. To help explore this possibility, HEK 293 cells that either stably or transiently (i.e. A3A, A3B) expressed each of the A3 enzymes were generated (Supplemental Figure S2). Since overexpression of A3A and A3B can result in cell toxicity, these were transiently expressed. Cell lysates were then used in an in vitro deaminase assay (44). Using this assay, the deamination preferences were tested with oligonucleotide substrates having an ATCA or GTCG target in an open, stem, bulge or loop context (Figure 5). Results are summarized in Table 6 and revealed that all APOBEC3 proteins preferred the TC target in an open structure rather than a stem structure. Additionally, lysates from APOBEC3 protein-expressing cells were observed to have higher or equal deamination preferences with target sites in an open context rather than a bulge or a loop (Figure 5 and Table 6). Taken together, the results shown in Table 3 and Table 6 demonstrate similar trends, with the noted differences likely being due to the nature of the cell lysates that are not represented in assays using purified A3 enzymes. In sum, the secondary structure of the target TC and its flanking base were further confirmed to affect the deamination preference of APOBEC3 enzymes, with the TC target in the ssDNA being the most preferred target for most all APOBEC3 proteins.
Figure 5.
Preference of APOBEC3 proteins from cell lysates for the TC target in the ssDNA region rather than in a stem, loop and bulge secondary structure. APOBEC3 protein activity from cell lysates with different oligonucleotide substrates. The deaminase activity of cell lysates from cells expressing APOBEC3 proteins with oligonucleotides containing the target site either within a 10-loop, 3-loop, bulge or stem was compared to that of of an oligonucleotide with open TC. Data is shown as the average ± standard deviation of three independent experiments. Significance was analyzed by using a two-way ANOVA with significance defined as P ≤ 0.05. Significant differences between experiments of an open structure compared to another secondary structure is indicated by a ‘*’ above relative histogram bar. The ‘+’ symbol and brackets above any two bars indicates the significant difference between analyses. Experiments having statistical significance are indicated as follows: ‘****’/ ‘+ + + +’ = P ≤ 0.0001; ‘***’/ ‘+ + +’ = P ≤ 0.001; ‘**’/ ‘+ +’ = P ≤ 0.01; ‘*’/ ‘+’ = P ≤ 0.05.
Table 6.
Comparison of APOBEC3 protein deamination preference activities from cell lysates in oligonucleotides with TC targets having different DNA secondary structuresa
| Protein/preference | Open versus stem | Open versus bulge | Open versus loop | 10-loop versus 3-loop |
|---|---|---|---|---|
| A3A | > | ≧ | > | = |
| A3B | > | ≧ | ≧ | ≦ |
| A3C | > | > | ≧ | ≦ |
| A3D | > | > | ≧ | = |
| A3F | > | > | ≧ | = |
| A3G | > | > | ≧ | = |
| A3H | > | ≧ | > | = |
aThe predicted target cytidine for deamination is indicated by an underline in each oligonucleotide. ‘>’ = greater than; ‘<’ = smaller than; ‘ = ’ = equal; ‘≧’ = greater or equal; ‘≦’ = smaller or equal. Statistical differences between groups were analyzed by using a two-way ANOVA test, with significance having a P-value of P < 0.05.
Analysis of oligonucleotide secondary structures
The oligonucleotides used in these studies were predicted to have only one highly energetically favorable secondary structure based upon analysis by using the Mfold program server (http://unafold.rna.albany.edu/?q=mfold). To provide an experimental support for these predictions, we compared the initial fluorescence of the oligonucleotides used in these studies. Based upon theoretical predictions, the further the fluorophore is at a distance from the quencher, the higher the predicted fluorescence is for each of the labeled oligonucleotides. For example, the 5′ end of the open oligonucleotides is 19 bases away from the 3′ end, while 5′ end of the stem oligonucleotides is only 7 bases away from the 3′ end – and this predicts that the quencher in the stem oligonucleotides will function more efficiently. As anticipated, most all of the oligonucleotides with a TC in the ssDNA region (open) had a significantly higher fluorescence than those with a TC target site in a dsDNA (stem), which provides an independent line of experimental evidence that the oligonucleotides used in these studies have the predicted secondary structure (Supplemental Figure S3).
DISCUSSION
APOBEC3 proteins serve as an important arm of the innate immune system through their ability to inhibit retrotransposons and exogenous viruses such as hepatitis B virus (11,60,61), human T-cell leukemia virus type 1 (62,63), and HIV (2). Restriction of viral replication is attributed to APOBEC3-mediated deamination of cytidines within single-stranded DNA. Deamination converts cytidines to uridines which in the case of HIV generates G to A mutations in the plus strand of the dsDNA provirus (64). The frequency at which some of the APOBEC3 proteins can deaminate cytidines within the target genome can reach a lethal level of mutations resulting in an inhibition of viral replication (2,31,32,62). While previous studies have examined the structural and sequence determinants on dinucleotide preference, less is known about what makes a dinucleotide target site a ‘hot spot.’
We previously reported the effect of DNA secondary structure and the nucleotides that flank the target site on A3G activity (44). In the current study, we have conducted a comparative, parallel investigation of the target sequence selection for purified A3A, A3B, A3C, A3F, A3G and A3H (Tables 2–5). Key observations made included (i) confirmation of the critical importance of both the flanking bases and their secondary structures on deamination sequence selection; (ii) A3A, A3F and A3H were observed to have a strong preferences with a TC target flanked by either A or T; (iii) all A3 enzymes were found to not have a preference for a TC target flanked by a G. Intriguingly, deamination of the TC target site occurred within either an open or stem region, but APOBEC3 proteins mainly preferred a TC target in ssDNA rather than in dsDNA region, presumably because the APOBEC3 proteins can only access their substrate when the dsDNA ‘breaths’ and exposes ssDNA bases (58). APOBEC3 proteins were found to readily act at either 5′CT3′ or 5′TC3′ target sites, but in some instances had a preference for either one with special flanking bases. The secondary structure of ‘−1’ T in the TC target was shown to play a role in target selection of deamination. Almost all the APOBEC3 proteins preferred to deaminate a TC target in a ssDNA region rather than in the context of a loop or a bulge region. These observations indicate that target site selection of the APOBEC3 enzymes is multivariable and complex.
Due to the role of the APOBEC3 proteins in oncogenesis and virus restriction, there is significant interest in understanding the relationship(s) among APOBEC3 sequence, structure, and dinucleotide preference sites. Sequence and structural analysis of the APOBEC3 proteins have revealed that the APOBEC3 proteins either have one (A3A, A3C, A3H) or two (A3B, A3D, A3F, A3G) zinc-coordinating domains (13,35,65,66). Based on sequence similarities, the zinc coordinating domains have been divided into three groups (67): The Z1 group includes A3A and the C terminal domains of A3B and A3G; the Z2 group includes A3C, the C terminal domains of A3D and A3F, and the N terminal domain of all double-domain proteins. APOBEC3H is the sole member of the Z3 group, and in general, has the most sequence divergence among the APOBEC3 family members. Both solution and crystal structures have shown that the secondary and tertiary structure of the zinc coordinating domains are highly conserved with a five-stranded beta sheet flanked by six alpha helices (38–40,65,66,68,69). The beta strands and alpha helices that make up the zinc-coordinating domains are connected by several loops that have been shown to influence APOBEC3 dinucleotide preferences (38,70). For example, residue swaps within Loop 7 between APOBEC3 proteins switches the enzyme's dinucleotide target site preference. In particular, for A3G, the preference for the 5′CC3′ dinucleotide target site was mapped to D317, a specific residue within Loop 7 (70).
Our data provides an independent experimental confirmation of predictions made from analyses of co-crystal structures of A3A, A3B-CTD or the A3G-NTD with ssDNA (45,48,71). While ssDNA was reported to be the target for all APOBEC3 proteins, we cannot exclude the possibility that the APOBEC3 proteins can deaminate dsDNA, at least a TC target in a dsDNA stem region. It was previously reported that ssDNA that forms during DNA lagging strand synthesis can be a major substrate for APOBEC3 catalysis, which implies that transcription and DNA repair intermediates can also serve as substrates (72). A3A has also been shown to deaminate dsDNA undergoing transcription (49). According to the ‘U-shaped’ ssDNA-binding mechanism revealed by co-crystal structures of A3A, A3B-CTD with ssDNA, loop regions of a DNA stem–loop structure may help to define a hotspot for APOBEC mutagenesis. Our results have also revealed that APOBEC3 proteins deaminate at some stem TC target sites, though have a preference for open TC target sites. These observations do not exclude the possibility of transient ssDNA in stem regions. Co-crystal structures have implicated the impact of the flanking base to cytidine on APOBEC3 deamination activity. For example, co-crystal structures of A3A or A3B-CTD with ssDNA have revealed that base stacking or hydrophobic interactions between the +1 to +3 nucleotides may be relevant for ssDNA deamination. Additionally, the co-crystal structure of A3G-NTD with ssDNA has revealed that one dT residue inserted into the zinc center and the two flanking residues form specific binding interactions with loops 1, 3, 5 and 7. This could help explain our observations that flanking bases can influence deamination activity. Taken together, these observations indicate that the interaction between flanking bases or between flanking bases and loops connected via beta strands and alpha helices of the catalytic domain is one of the factors that can determine APOBEC3 catalytic target site selection.
Analysis of the co-crystal structures of APOBEC3 proteins with ssDNA has implicated the function of T residue at the −1 position in target selection, which is consistent with our finding that the secondary structure of ‘−1’ T of the TC target affected the deamination preference of APOBEC3 proteins. The 5′TC3′ dinucleotide is known as a universal dinucleotide target site for all APOBEC3 proteins, except for A3G. The fact that A3G catalyzes C to U deamination with 3′ to 5′ deamination polarity implies that other APOBEC3 proteins may be able to recognize 3′TC’ as well, which is supported by the findings of our study. Most APOBEC3 proteins readily recognize both the 5′TC’ and 3′TC’ dinucleotides. However, it is important to note that the selection on TC over CT is also affected by the flanking base and whether C is in a stem structure, as observed with A3A and A3H.
APOBEC3 proteins, particularly A3A and A3B, have been implicated in the mutagenesis of many cancer types (72–74). For example, A3B has been reported to be the major source of mutagenesis in breast cancer cell lines (6). C to T and C to G base substitution mutations were commonly observed as signature mutations (75,76). The target C is usually within either a 5′TC dinucleotide motif or in either 5′TCA or 5′TCT trinucleotide contexts (77). Our observations imply that A3A has a preference for A at the +1 position, which is consistent with a preference for the 5′TCA trinucleotide context.
Recent studies of HIV-1 replication in the context of specific APOBEC3 protein overexpression help to provide evidence in support of the biological relevance of the secondary structures and nucleotide base neighbor contexts of the TC target representing related sequence contexts in the HIV-1 genome. In particular, Illumina sequencing revealed 17 deamination hotspots in HIV-1 pol (2574–3301) related to A3F or A3G overexpression (28). We used the Mfold program to analyze the predicted DNA secondary structure of HIV-1 pol (1792–3536) (Supplemental Figure S4). The characteristics of the deamination hotspots resembled the ones used in our study (Supplemental Table S7). For example, 41% of the hotspots were found to have an open structure (Supplemental Table S7b). Furthermore, while 35% were found to be in a loop structure, those were primarily in large loops, which likely behave more like an open structure. A likely target was also observed in a predicted stem structure (e.g., structure number 12). In this case, the T was preferred at the −2 position, while the A was preferred at the +1 position. No evidence for a preference of either C or G was observed. These observations are consistent with our findings using oligonucleotide targets and purified enzymes. Notably, the changing of the −2 and +1 positions together with the same nucleotide resulted in target sites observed in HIV-1 during APOBEC3 overexpression—i.e., 5 of 15 TC targets (Supplemental Table S7a) and 6 of 14 TC targets (Supplemental Table S8a). Our findings are also consistent with Sanger sequencing results obtained from the sequences that encode the HIV-1 protease (2250–2631) (52). The predicted DNA sequence secondary structures and characteristics of the deaminated TC or CC targets are shown in Supplemental Figure S5 and Supplemental Table S8, respectively. In another study, Desimmie and colleagues sequenced HIV-1 pol (RT region) following virus replication in the presence of A3D, A3F, A3G, or A3H (78). These authors reported that the nucleotide at the +2 position affected APOBEC3-induced G-to-A mutation frequencies. Our analysis of their findings indicates the importance of the nucleotide at −2 position on APOBEC3-induced C-to-T mutation frequencies (Supplemental Table S9). This analysis of the published data is fairly consistent with our data presented in this study. For example, the preference of A3H for the −2 position of the TC target is C ≧ A > G, which was also observed in this study (Table 2). However, the preferences of −2 position nucleotide was not fully consistent. For instance, the published data indicated that A was preferred over G at the −2 position by either A3F or A3G (Supplemental Table S9), but our data indicates that G was preferred over A (Table 2). The discrepancies may be due, in part, to the fact that the data in our study was based only on the TC target in an open structure. Taken together, comparison of the data reported in our study with available published data from analyses done in the context of HIV-1 replication help support the conclusion that the data generated here in this study with oligonucleotides and purified APOBEC3 enzymes are biologically relevant in predicting potential TC targets in a known sequence that are deaminated by APOBEC3 enzymes.
The parallel, comparative analysis of APOBEC3 site selection using six of the seven purified APOBEC3 deaminases demonstrates that enzyme target site selection is multivariable and complex. The correlation between nucleotide sequence and preference sites provides greater insights into the specific residues that confer preference site specificity beyond just the dinucleotide target site, and helps provide an improved understanding into what defines a ‘hot spot’. Future biochemical and structural studies will help fill the current knowledge gap of the enzyme-nucleic acid interactions involved in the recognition of deamination target sites as well as better defining what nucleotides/sequence contexts/secondary structures contribute substrate specificity and optimal target site identification.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Paul J. Jardine for constructive comments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01 GM105876 to L.M.M.]; Canadian Institutes of Health Research [PJT162-407 to L.C.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
References
- 1. Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N.. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002; 79:285–296. [DOI] [PubMed] [Google Scholar]
- 2. Hultquist J.F., Lengyel J.A., Refsland E.W., LaRue R.S., Lackey L., Brown W.L., Harris R.S.. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 2011; 85:11220–11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zennou V., Bieniasz P.D.. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006; 349:31–40. [DOI] [PubMed] [Google Scholar]
- 4. Jern P., Stoye J.P., Coffin J.M.. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 2007; 3:2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simon V., Zennou V., Murray D., Huang Y., Ho D.D., Bieniasz P.D.. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005; 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burns M.B., Lackey L., Carpenter M.A., Rathore A., Land A.M., Leonard B., Refsland E.W., Kotandeniya D., Tretyakova N., Nikas J.B. et al.. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013; 494:366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liddament M.T., Brown W.L., Schumacher A.J., Harris R.S.. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 2004; 14:1385–1391. [DOI] [PubMed] [Google Scholar]
- 8. Refsland E.W., Hultquist J.F., Luengas E.M., Ikeda T., Shaban N.M., Law E.K., Brown W.L., Reilly C., Emerman M., Harris R.S.. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014; 10:e1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hache G., Liddament M.T., Harris R.S.. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 2005; 280:10920–10924. [DOI] [PubMed] [Google Scholar]
- 10. Bishop K.N., Holmes R.K., Sheehy A.M., Davidson N.O., Cho S.J., Malim M.H.. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004; 14:1392–1396. [DOI] [PubMed] [Google Scholar]
- 11. Janahi E.M., McGarvey M.J.. The inhibition of hepatitis B virus by APOBEC cytidine deaminases. J. Viral Hepat. 2013; 20:821–828. [DOI] [PubMed] [Google Scholar]
- 12. Sasada A., Takaori-Kondo A., Shirakawa K., Kobayashi M., Abudu A., Hishizawa M., Imada K., Tanaka Y., Uchiyama T.. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology. 2005; 2:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Langlois M.A., Beale R.C., Conticello S.G., Neuberger M.S.. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005; 33:1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jern P., Russell R.A., Pathak V.K., Coffin J.M.. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009; 5:e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H.. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002; 418:646–650. [DOI] [PubMed] [Google Scholar]
- 16. Stopak K., de Noronha C., Yonemoto W., Greene W.C.. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003; 12:591–601. [DOI] [PubMed] [Google Scholar]
- 17. Malim M.H. Natural resistance to HIV infection: the Vif-APOBEC interaction. C. R. Biol. 2006; 329:871–875. [DOI] [PubMed] [Google Scholar]
- 18. Peliska J.A., Benkovic S.J.. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992; 258:1112–1118. [DOI] [PubMed] [Google Scholar]
- 19. Charneau P., Mirambeau G., Roux P., Paulous S., Buc H., Clavel F.. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 1994; 241:651–662. [DOI] [PubMed] [Google Scholar]
- 20. Dyda F., Hickman A.B., Jenkins T.M., Engelman A., Craigie R., Davies D.R.. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994; 266:1981–1986. [DOI] [PubMed] [Google Scholar]
- 21. Suspene R., Sommer P., Henry M., Ferris S., Guetard D., Pochet S., Chester A., Navaratnam N., Wain-Hobson S., Vartanian J.P.. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004; 32:2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gillick K., Pollpeter D., Phalora P., Kim E.Y., Wolinsky S.M., Malim M.H.. Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination. J. Virol. 2013; 87:1508–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holtz C.M., Mansky L.M.. Variation of HIV-1 mutation spectra among cell types. J. Virol. 2013; 87:5296–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sadler H.A., Stenglein M.D., Harris R.S., Mansky L.M.. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J. Virol. 2010; 84:7396–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Janini M., Rogers M., Birx D.R., McCutchan F.E.. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J. Virol. 2001; 75:7973–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kieffer T.L., Kwon P., Nettles R.E., Han Y., Ray S.C., Siliciano R.F.. G→A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J. Virol. 2005; 79:1975–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delviks-Frankenberry K.A., Nikolaitchik O.A., Burdick R.C., Gorelick R.J., Keele B.F., Hu W.S., Pathak V.K.. Minimal Contribution of APOBEC3-Induced G-to-A Hypermutation to HIV-1 Recombination and Genetic Variation. PLoS Pathog. 2016; 12:e1005646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mohammadzadeh N., Love R.P., Gibson R., Arts E.J., Poon A.F.Y., Chelico L.. Role of co-expressed APOBEC3F and APOBEC3G in inducing HIV-1 drug resistance. Heliyon. 2019; 5:e01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adolph M.B., Webb J., Chelico L.. Retroviral restriction factor APOBEC3G delays the initiation of DNA synthesis by HIV-1 reverse transcriptase. PLoS One. 2013; 8:e64196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iwatani Y., Chan D.S., Wang F., Maynard K.S., Sugiura W., Gronenborn A.M., Rouzina I., Williams M.C., Musier-Forsyth K., Levin J.G.. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007; 35:7096–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anderson J.L., Hope T.J.. APOBEC3G restricts early HIV-1 replication in the cytoplasm of target cells. Virology. 2008; 375:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chaipan C., Smith J.L., Hu W.S., Pathak V.K.. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J. Virol. 2013; 87:444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lackey L., Law E.K., Brown W.L., Harris R.S.. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013; 12:762–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koning F.A., Newman E.N., Kim E.Y., Kunstman K.J., Wolinsky S.M., Malim M.H.. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 2009; 83:9474–9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Navarro F., Bollman B., Chen H., Konig R., Yu Q., Chiles K., Landau N.R.. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005; 333:374–386. [DOI] [PubMed] [Google Scholar]
- 36. Zheng Y.H., Irwin D., Kurosu T., Tokunaga K., Sata T., Peterlin B.M.. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 2004; 78:6073–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harris R.S., Liddament M.T.. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004; 4:868–877. [DOI] [PubMed] [Google Scholar]
- 38. Shandilya S.M., Bohn M.F., Schiffer C.A.. A computational analysis of the structural determinants of APOBEC3′s catalytic activity and vulnerability to HIV-1 Vif. Virology. 2014; 471–473:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bohn M.F., Shandilya S.M., Albin J.S., Kouno T., Anderson B.D., McDougle R.M., Carpenter M.A., Rathore A., Evans L., Davis A.N. et al.. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013; 21:1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shandilya S.M., Nalam M.N., Nalivaika E.A., Gross P.J., Valesano J.C., Shindo K., Li M., Munson M., Royer W.E., Harjes E. et al.. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010; 18:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holden L.G., Prochnow C., Chang Y.P., Bransteitter R., Chelico L., Sen U., Stevens R.C., Goodman M.F., Chen X.S.. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008; 456:121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beale R.C., Petersen-Mahrt S.K., Watt I.N., Harris R.S., Rada C., Neuberger M.S.. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 2004; 337:585–596. [DOI] [PubMed] [Google Scholar]
- 43. Adolph M.B., Love R.P., Chelico L.. Biochemical basis of APOBEC3 deoxycytidine deaminase activity on diverse DNA substrates. ACS Infect Dis. 2018; 4:224–238. [DOI] [PubMed] [Google Scholar]
- 44. Holtz C.M., Sadler H.A., Mansky L.M.. APOBEC3G cytosine deamination hotspots are defined by both sequence context and single-stranded DNA secondary structure. Nucleic Acids Res. 2013; 41:6139–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xiao X., Li S.X., Yang H., Chen X.S.. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat. Commun. 2016; 7:12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wan L., Nagata T., Katahira M.. Influence of the DNA sequence/length and pH on deaminase activity, as well as the roles of the amino acid residues around the catalytic center of APOBEC3F. Phys. Chem. Chem. Phys. 2018; 20:3109–3117. [DOI] [PubMed] [Google Scholar]
- 47. Silvas T.V., Hou S., Myint W., Nalivaika E., Somasundaran M., Kelch B.A., Matsuo H., Kurt Yilmaz N., Schiffer C.A.. Substrate sequence selectivity of APOBEC3A implicates intra-DNA interactions. Sci. Rep. 2018; 8:7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shi K., Carpenter M.A., Banerjee S., Shaban N.M., Kurahashi K., Salamango D.J., McCann J.L., Starrett G.J., Duffy J.V., Demir O. et al.. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 2017; 24:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Love R.P., Xu H., Chelico L.. Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A. J. Biol. Chem. 2012; 287:30812–30822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adolph M.B., Love R.P., Feng Y., Chelico L.. Enzyme cycling contributes to efficient induction of genome mutagenesis by the cytidine deaminase APOBEC3B. Nucleic Acids Res. 2017; 45:11925–11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adolph M.B., Ara A., Feng Y., Wittkopp C.J., Emerman M., Fraser J.S., Chelico L.. Cytidine deaminase efficiency of the lentiviral viral restriction factor APOBEC3C correlates with dimerization. Nucleic Acids Res. 2017; 45:3378–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ara A., Love R.P., Chelico L.. Different mutagenic potential of HIV-1 restriction factors APOBEC3G and APOBEC3F is determined by distinct single-stranded DNA scanning mechanisms. PLoS Pathog. 2014; 10:e1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chelico L., Prochnow C., Erie D.A., Chen X.S., Goodman M.F.. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J. Biol. Chem. 2010; 285:16195–16205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Feng Y., Love R.P., Ara A., Baig T.T., Adolph M.B., Chelico L.. Natural polymorphisms and oligomerization of human APOBEC3H contribute to Single-stranded DNA scanning ability. J. Biol. Chem. 2015; 290:27188–27203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nair S., Rein A.. In vitro assay for cytidine deaminase activity of APOBEC3 protein. Biol. Protoc. 2014; 4:e1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Godbey W.T., Barry M.A., Saggau P., Wu K.K., Mikos A.G.. Poly(ethylenimine)-mediated transfection: a new paradigm for gene delivery. J. Biomed. Mater. Res. 2000; 51:321–328. [DOI] [PubMed] [Google Scholar]
- 57. Thielen B.K., Klein K.C., Walker L.W., Rieck M., Buckner J.H., Tomblingson G.W., Lingappa J.R.. T cells contain an RNase-insensitive inhibitor of APOBEC3G deaminase activity. PLoS Pathog. 2007; 3:1320–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. von Hippel P.H., Johnson N.P., Marcus A.H.. Fifty years of DNA “breathing”: reflections on old and new approaches. Biopolymers. 2013; 99:923–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chelico L., Pham P., Calabrese P., Goodman M.F.. APOBEC3G DNA deaminase acts processively 3′ → 5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 2006; 13:392–399. [DOI] [PubMed] [Google Scholar]
- 60. Vartanian J.P., Henry M., Marchio A., Suspene R., Aynaud M.M., Guetard D., Cervantes-Gonzalez M., Battiston C., Mazzaferro V., Pineau P. et al.. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 2010; 6:e1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Suspene R., Guetard D., Henry M., Sommer P., Wain-Hobson S., Vartanian J.P.. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:8321–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ooms M., Krikoni A., Kress A.K., Simon V., Munk C.. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J. Virol. 2012; 86:6097–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mahieux R., Suspene R., Delebecque F., Henry M., Schwartz O., Wain-Hobson S., Vartanian J.P.. Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J. Gen. Virol. 2005; 86:2489–2494. [DOI] [PubMed] [Google Scholar]
- 64. Yu Q., Konig R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J.M., Landau N.R.. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004; 11:435–442. [DOI] [PubMed] [Google Scholar]
- 65. Byeon I.J., Byeon C.H., Wu T., Mitra M., Singer D., Levin J.G., Gronenborn A.M.. Nuclear magnetic resonance structure of the APOBEC3B catalytic domain: structural basis for substrate binding and DNA deaminase activity. Biochemistry. 2016; 55:2944–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shaban N.M., Shi K., Li M., Aihara H., Harris R.S.. 1.92 Angstrom zinc-free APOBEC3F catalytic domain crystal structure. J. Mol. Biol. 2016; 428:2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. LaRue R.S., Andresdottir V., Blanchard Y., Conticello S.G., Derse D., Emerman M., Greene W.C., Jonsson S.R., Landau N.R., Lochelt M. et al.. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 2009; 83:494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kitamura S., Ode H., Nakashima M., Imahashi M., Naganawa Y., Kurosawa T., Yokomaku Y., Yamane T., Watanabe N., Suzuki A. et al.. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 2012; 19:1005–1010. [DOI] [PubMed] [Google Scholar]
- 69. Stauch B., Hofmann H., Perkovic M., Weisel M., Kopietz F., Cichutek K., Munk C., Schneider G.. Model structure of APOBEC3C reveals a binding pocket modulating ribonucleic acid interaction required for encapsidation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:12079–12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rathore A., Carpenter M.A., Demir O., Ikeda T., Li M., Shaban N.M., Law E.K., Anokhin D., Brown W.L., Amaro R.E. et al.. The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J. Mol. Biol. 2013; 425:4442–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Maiti A., Myint W., Kanai T., Delviks-Frankenberry K., Sierra Rodriguez C., Pathak V.K., Schiffer C.A., Matsuo H.. Crystal structure of the catalytic domain of HIV-1 restriction factor APOBEC3G in complex with ssDNA. Nat. Commun. 2018; 9:2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hoopes J.I., Cortez L.M., Mertz T.M., Malc E.P., Mieczkowski P.A., Roberts S.A.. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 2016; 14:1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. King J.J., Larijani M.. A novel regulator of Activation-Induced cytidine Deaminase/APOBECs in immunity and cancer: Schrodinger's CATalytic pocket. Front. Immunol. 2017; 8:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang Y., Schmitt K., Guo K., Santiago M.L., Stephens E.B.. Role of the single deaminase domain APOBEC3A in virus restriction, retrotransposition, DNA damage and cancer. J. Gen. Virol. 2016; 97:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G. et al.. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013; 45:970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L. et al.. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Refsland E.W., Harris R.S.. The APOBEC3 family of retroelement restriction factors. Curr. Top. Microbiol. Immunol. 2013; 371:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Desimmie B.A., Burdick R.C., Izumi T., Doi H., Shao W., Alvord W.G., Sato K., Koyanagi Y., Jones S., Wilson E. et al.. APOBEC3 proteins can copackage and comutate HIV-1 genomes. Nucleic Acids Res. 2016; 44:7848–7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.