Abstract
During canonical translation, the ribosome moves along an mRNA from the start to the stop codon in exact steps of one codon at a time. The collinearity of the mRNA and the protein sequence is essential for the quality of the cellular proteome. Spontaneous errors in decoding or translocation are rare and result in a deficient protein. However, dedicated recoding signals in the mRNA can reprogram the ribosome to read the message in alternative ways. This review summarizes the recent advances in understanding the mechanisms of three types of recoding events: stop-codon readthrough, –1 ribosome frameshifting and translational bypassing. Recoding events provide insights into alternative modes of ribosome dynamics that are potentially applicable to other non-canonical modes of prokaryotic and eukaryotic translation.
INTRODUCTION
Ribosomes produce proteins by translating the sequence of an mRNA into the amino acid sequence of a protein. To make a protein that is encoded by a given open reading frame (ORF) of an mRNA, the ribosome has to select the correct AUG codon to start translation, ensure the collinearity of the mRNA and the protein sequences during translation elongation, and terminate translation at a stop codon marking the end of the ORF. Cells have evolved sophisticated control mechanisms that ensure fidelity of each translation phase. However, in special cases, signals encoded in an mRNA reprogram the ribosome to read the message in an alternative way, a phenomenon called translational recoding. In this review, we will focus on three types of recoding: (i) stop-codon readthrough; (ii) ribosome frameshifting and (iii) translational bypassing (Figure 1).
Figure 1.
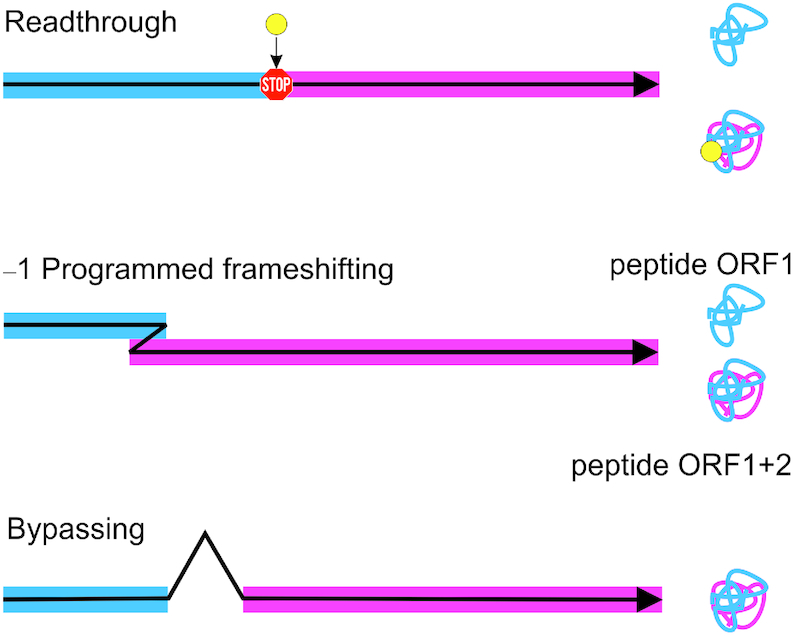
Three types of recoding events. Translational readthrough extends the polypeptide C-terminally allowing the production of two protein isoforms from the same transcript. Frameshifting produces typically two functional polypeptides from different reading frames of the same mRNA. Bypassing is a recoding event that synthesizes one protein from two open discontinuous reading frames.
During translation elongation, the mRNA is decoded with the help of aminoacyl-tRNAs (aa-tRNA) that are delivered to the ribosome in complex with an elongation factor (EF-Tu in bacteria or eEF1 in eukaryotes) and GTP. The ribosome selects the tRNAs according to the match between the mRNA codon and the tRNA anticodon. Failing to discriminate against incorrect aa-tRNA results in missense errors of translation. Generally, the fidelity of decoding is very high, with a frequency of missense errors in the range from <10−7 to 10−4 per codon depending on the type of mismatch and the position of the amino acid in the protein (1–4). At the end of the open reading frame, stop codons (UAA, UAG and UGA) are recognized by termination (release) factors (RF1 and RF2 in bacteria or eRF1 in eukaryotes). The frequency of occasional readthrough is low, <10−4 per stop codon (5,6), which can increase dramatically, up to 0.1–0.3, when induced by sequence and structural elements in the mRNA and by trans factors (7–9). Missense and nonsense errors are mistakes of decoding. After peptide bond formation, the ribosome moves along the mRNA to read the next codon in a tightly orchestrated process of translocation. In order to produce a correct protein, the ribosome must be translocated by exactly one codon at a time. Failing to maintain the correct reading frame results in ribosome frameshifting in – or + direction. Depending on the conditions, frameshifting errors can occur during decoding or translocation. The frequency of spontaneous frameshifting is rather low, i.e. <10−5 (10–12). Signals in the mRNA provide a context in which frameshifting is greatly enhanced, which is referred to as programmed ribosome frameshifting (PRF). The efficiency of PRF can vary in a wide range between 0.5% and 80%, depending on the organism and the frameshifting sequence (for reviews, see (13–16)). Finally, translational bypassing is a recoding phenomenon that produces a single protein from a discontinuous reading frame. Bypassing is a post-decoding event that requires multiple signals in the mRNA. In the following, we will discuss the mechanisms of each of these recoding events in light of the recent progress of biochemical, kinetic and structural studies.
TRANSLATIONAL READTHROUGH
Stop codon readthrough can result from decoding of a stop codon as a sense codon by a near-cognate tRNA. Natural tRNAs that are prone to readthrough usually have an anticodon that has a single mismatch upon pairing to a stop codon, such as tRNAGln, tRNATyr, tRNACys or tRNATrp (17). Translational readthrough is widely employed by viruses to expand the coding potential of their limited genome (7,18–20). Readthrough does not alter the translational reading frame, but rather extends the polypeptide C-terminally allowing the production of two protein isoforms from the same transcript. The C-terminal extension can carry cellular localization signals or homo/heterodimerization domains or alter the function of the protein such as its ligand-binding properties (Table 1).
Table 1.
Examples of translational readthrough (RT) in genes from different kingdoms of life
| Protein | RT % | Function of RT isoform | References | |
|---|---|---|---|---|
| Viruses | ||||
| Coliphage Qβ | Minor coat protein A1 | 5% | Formation of infectious particles | (20,27) |
| Tobacco mosaic virus | Replicase | 10–35% | RNA polymerase domain | (18,142) |
| Sindbis virus | nsP4 | 10% | Viral replication | (28,143–145) |
| Luteoviruses (BYDV, BWYV) | Coat protein | Virus transmission | ||
| MuLV | Gag-pol | 5% | Replication by gag-pol fusion protein | (146,147) |
| Bacteria | ||||
| B. subtilis | SacB levansucrase | Modification of enzymatic properties | (148) | |
| Eukaryotes | ||||
| S. cerevisae | PDE2 | 0.5–2.2% | Proteasome dependent degradation | (149) |
| U. maydis | PGK | Peroxisomal Targeting Signal 1 | (150) | |
| A. nidulans | GAPDH | Peroxisomal Targeting Signal 1 | ||
| Rabbit | β-globin | (151–153) | ||
| Vertebrates | MPZ | 14% | Role in myelination | (154,155) |
| Mammals | VEGFA | 7–25% | Antiangiogenic activity | (8) |
| MTCH2 | 13% | |||
| AGO1 | 24% | |||
| OPRL1 | 31% | (21) | ||
| OPRK1 | 13% | |||
| MAPK10 | 14% | |||
| AQP4 | 7% | |||
| Human | LDHB MDH1 | 1.5–5% | Peroxisomal Targeting Signal 1 | (57,156) |
| VDR | 6.7% | Reduced transcriptional response to calcitriol | (157) | |
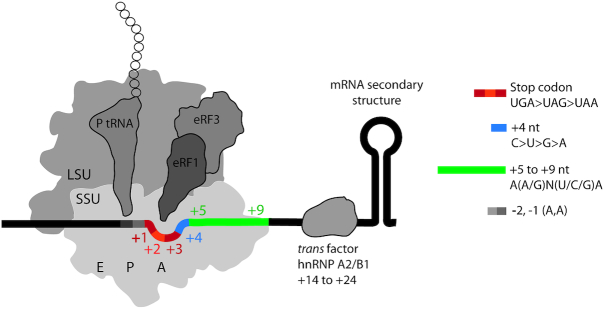
The minimal mRNA sequence motif that modulates readthrough is comprised of the stop codon (nt +1, +2, +3) and its context from nt –2 to +9 (Figure 2). The propensity for readthrough is lowest on the UAA and highest on the UGA codon (21–24). The 5′ context of stop codons shows a non-random distribution of nucleotides in Escherichia coli and in humans (25). The presence of two adenines at positions –1 and –2 favors readthrough (26). The presence of a cytidine at position +4 (C+4) is associated with leaky termination in various organisms, in particular on UGA codons (20,27–31); notably, UGA C or UAG C are rare in mammals (32). The effect of bases other than C+4 varies between the three stop codons (6,32,33). The nucleotides +4 to +6 in the context of UGA-CUA or UGA-CGG induce readthrough in a number of viral and eukaryotic genes (21,22,34,35). In several cases, the mRNA context up to nt +9 can modulate readthrough (9,36). For example, in the Tobacco Mosaic Virus (TMV) replicase gene, the consensus sequence CARYYA (R, purines; Y, pyrimidines) triggers readthrough at all stop codons (37).
Figure 2.
Factors affecting translational readthrough in eukaryotes. Cis factors that affect readthrough include sequences upstream of the stop codon (light gray), the identity of the stop codon (red-orange), the +4 nucleotide (blue) and the downstream sequences that occupy the mRNA channel (green). Distal cis element includes downstream mRNA secondary structure. Among several trans factors that affect readthrough, the specific case of hnRNP A2/B1 is depicted. hnRNP A2/B1 promotes readthrough by binding to a cis element in the 3′ UTR of mammalian gene VEGFA. A, P and E depict the three stable tRNA-binding sites. SSU, small ribosomal subunit; LSU, large ribosomal subunit.
The structural basis for sequence effects in readthrough is unclear. Recognition of stop codons by RFs is achieved by sequence- and shape-specific recognition of the three nucleotides of the stop codons (nt +1 to +3) and, in eukaryotes, of the adjacent nucleotide +4 (38,39). Nucleotides +4 and +5 are involved in stacking interactions with rRNA bases around the decoding center, which are more stable with purines than pyrimidines (38,39). This might suggest that C or U at positions +4 and/or +5 decrease the stability of the decoding complex and interfere with the compaction of the mRNA in the A site, which is a hallmark of stop-codon recognition by eRF1 in eukaryotes (38). Although the details of stop codon recognition differ between bacteria and eukaryotes, there are indications that adenosines in positions +4 and +5 interact with the 16S rRNA, which might account for the reported context bias in prokaryotes (40).
In addition to the immediate context, more distal stimulatory 3′ cis elements involving mRNA structures regulate readthrough in several viral and eukaryotic mRNAs (7,41–44). For example, an 80-nucleotide sequence downstream of the stop codon in the Drosophila hdc gene forms a stem–loop (SL) structure that stimulates readthrough (45). Cis-acting RNA structures can modulate readthrough by (i) interfering with release factor recruitment to the ribosome; (ii) modulating ribosome function by interacting with ribosomal proteins or rRNAs; (iii) inducing ribosome stalling or (iv) recruiting trans factors (7,8,46). We note that the sequences downstream of stop codons evolved to limit the negative consequences of leaky termination, as in-frame stop codons are significantly over-represented immediately downstream of the primary stop signal, which ensures termination in close proximity of the correct end of the ORF (36).
In addition to elements in the mRNA, several trans factors may influence the efficiency of termination by various mechanisms. For example, readthrough of the mammalian vascular endothelial growth factor A (VEGFA) mRNA is facilitated by the heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1 that binds the hnRNP A2/B1 recognition element (A2RE) in the termination region (8) (Figure 2). Recently, eIF3 was proposed to promote readthrough at all three stop codons in leaky context by preventing eRF1 from recognizing the third position of the stop codon (47). Depletion of termination factors eRF1 and/or eRF3 results in increased levels of readthrough in humans independent of the codon context (48,49). The [Psi+] strain of Saccharomyces cerevisiae exhibits the epigenetically inherited prion state of termination factor eRF3 where translation termination is compromised. In these strains, eRF3 forms amyloid fibrils that sequester a part of the release factor pool (50–52). The abundance and properties of tRNAs also influence readthrough efficiency (17,53,54). For example, the relative abundance of the major tRNAGln isoacceptor with the 5′-UUG-3′ anticodon compared to the minor tRNAGln with 5′-CUG-3′ in S. cerevisiae explains why glutamine is preferentially incorporated at UAA compared to UAG, despite the same non-conventional G-U base pairing that forms upon decoding. The modification of the tRNA bases within the anticodon or in its vicinity affects its ability to read stop codons (34,55).
The prevalence of readthrough varies between organisms. Analysis of the stop codon contexts of 12 Drosophila species and ribosome profiling studies suggested potential readthrough in several hundred Drosophila genes (44,56). However, similar genomic analyses and profiling studies of human genes have so far found only a few candidate genes (44,56,57). Computational analysis of readthrough protein isoforms suggests that these are mostly long, modular proteins with intrinsically disordered C-termini of low sequence complexity (58,59). The lack of a structurally ordered C-terminus might provide conformational flexibility that allows the readthrough extensions to perform functions without distorting the native protein. The majority of readthrough genes identified in D. melanogaster have regulatory roles, and appending a functional C-terminal extension may confer conditional advantage to protein function.
In addition to readthrough by near-cognate aa-tRNAs, stop codons can be recoded by the specialized cognate tRNAs with an anticodon that is complementary to the stop codon, such as tRNAPyl or tRNASec (60,61). Pyrrolysin and selenocysteine are natural proteinogenic non-canonical amino acids that are not encoded by a sense codon. Pyl-specific tRNAPyl reads the UAG stop codon, whereas Sec-specific tRNASec reads the UGA codon (61–64). The Pyl trait is restricted to several microbes, mostly methanogenic archaea, which encode a tRNAPyr (pylT) and the dedicated aa-tRNA synthetase (pylS). Pyl-tRNAPyl is recognized by EF-Tu. Genome analysis of Pyl-containing organisms suggested that UAG is not a typical stop signal in Pyl-utilizing archaea and that Pyl insertion can effectively compete with translation termination for UAG codons obviating the need for specific mRNA structures that recruit tRNAPyl to a specific stop codon (61). In contrast to tRNAPyl, tRNASec is found in bacteria, archaea and eukaryotes. Sec is required for synthesis of a specialized group of proteins, selenoproteins. Sec-tRNASec is delivered to the ribosome by the specialized elongation factor SelB (EFSec in eukaryotes), a GTP-binding protein that belongs to the family of translational GTPases (65,66). The key element for recruitment of the SelB–GTP–Sec-tRNASec to the stop codon on bacterial ribosomes is a selenocysteine insertion sequence (SECIS) in the mRNA, a SL structure located immediately downstream of the in-frame UGA codon at which Sec is incorporated (67).
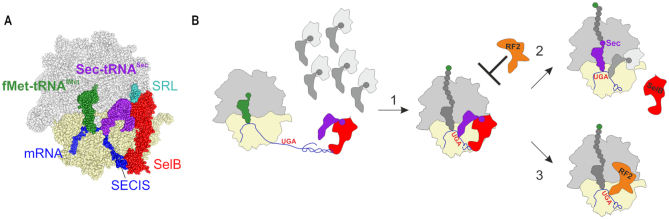
Recent cryo-EM structures revealed how Sec-tRNASec–SelB–GTP is recognized by the ribosome (68) (Figure 3A). Because tRNASec is cognate for the UGA codon, the codon–anticodon recognition initiates the same ribosome rearrangements as the canonical aa-tRNA–EF-Tu–GTP complex (69–71). This includes the domain movements of the SSU, GTPase activation of the factor by the interaction with the sarcin–ricin loop on the LSU and the accommodation of aa-tRNA on the LSU upon dissociation of SelB–GDP (68). However, some details of the interaction are Sec-specific. SECIS recruits SelB domain 4. The specific recognition of Sec-tRNASec by SelB is achieved by interactions between unique regions in SelB with the extra-long variable arm of tRNASec and the acceptor- and T-stems of tRNASec (68). These elements distinguish tRNASec from canonical tRNAs (72). Finally, the amino acid-binding pocket of SelB is lined with positively charged residues, allowing SelB to specifically recognize the negatively charged selenol group and to discriminate against Ser-tRNASec (68,73).
Figure 3.
UGA recoding by Sec-tRNASec. (A) Structure of the SelB–GTP–Sec-tRNASec complex on the ribosome during recoding (modified from (68)). The GTPase of SelB is activated by the sarcin–ricin loop (SRL) of 23S rRNA. (B) SECIS-mediated Sec insertion versus RF2-dependent termination at UGA. The Sec-tRNASec–SelB–GTP complex is rapidly recruited to the SECIS while still distant from the ribosome. Step 1: while the ribosome moves along the mRNA toward the UGA codon, the lower part of the SECIS becomes unwound and the Sec-tRNASec–SelB–GTP complex occupies the entry to the A site, thereby hindering the recruitment of RF2 to the stop codon. Step 2: after delivery of Sec-tRNASec to the A site and Sec insertion into the growing peptide, the ribosome can recruit the next EF-Tu–GTP–aa-tRNA complex (gray) and continue translation. Alternatively (step 3), if Sec incorporation fails, the A site becomes accessible for RF2, which promotes termination and peptide release.
The affinity of SelB–GTP for Sec-tRNASec is very high, with a Kd in the picomolar range (73). Also, SelB binding to the SECIS is in the nanomolar range and is rapid (kon = 108 M−1s−1) (74). This implies that in the cell the Sec-tRNASec–SelB–GTP complex can bind to the SECIS before it enters the ribosome, thereby facilitating the recruitment of Sec-tRNASec to the UGA codon preceding the SECIS. Although tRNASec is recognized by the ribosome as a cognate aa-tRNA (68,71), the efficiency of Sec incorporation is only about 40%, whereas 60% of the ribosomes terminate translation with the help of RF2 (75). Why some translating ribosomes incorporate Sec and others do not, remains unclear. Surprisingly, RF2 does not act as a direct competitor of Sec, but rather terminates translation on the ribosomes that failed to incorporate Sec. It is possible that when the ribosome arrives at the UGA, the SECIS-bound Sec-tRNASec–SelB–GTP blocks the entrance of RF2 to the A site (Figure 3B). However, if the attempt to deliver Sec is unsuccessful, the interaction of SelB with the SECIS will be lost eventually, thereby freeing the access for RF2 to the stop codon. Alternatively, conformational heterogeneity of translating ribosomes and the folding-unfolding dynamics of the SECIS may define the preference for Sec binding on one fraction of ribosome complexes, whereas the other fraction favours RF2 (75).
SPONTANEOUS AND PROGRAMMED RIBOSOME FRAMESHIFTING
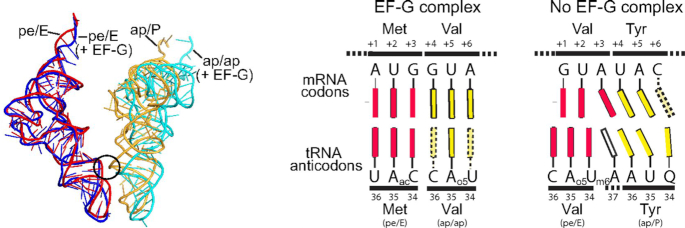
The propensity of the ribosome for spontaneous frameshifting depends on the stability of the codon–anticodon complexes. Early studies suggested that in solution even fully matched codon–anticodon complexes dissociate very rapidly, at 3–6 s−1 (76). In the A site of the ribosome, the dissociation is much slower, about 0.2 s−1 (77). However, when these stabilizing ribosome interactions are released during translocation, the tRNA may unpair from the mRNA within the time of translocation and thus the inherent stability of the codon–anticodon complex may be insufficient to hold the tRNA in frame. At mRNA sequences where tRNA pairing with its 0-frame codon is favored over –1 or +1 alternative frames, transient loss of base pairing may be unimportant, because even if the anticodon dissociates from the anticodon, the 0-frame codon is the most likely target for it to rebind. However, when the mRNA sequence is ‘slippery’, i.e., allows tRNA base pairing with the codon in the –1- or +1-frame, the loss of interactions with the codon, together with the movements of the elements of the SSU that occur during translocation, may result in frameshifting. A recent crystal structure of a translocation intermediate formed in the absence of EF-G indeed shows that the interactions of the ribosome with the codon–anticodon complex are disrupted and the A-site tRNA in the complex is shifted by one nucleotide toward the –1-frame of the mRNA (78) (Figure 4). In comparison, in crystal structures obtained in the presence of EF-G, residues at the tip of domain 4 of EF-G interact with the A-site tRNA and prevent it from shifting (79). The interacting residues at the tip of EF-G domain 4, H583 and Q507 (E. coli numbering), are known to play a key role in translocation (80).
Figure 4.
Reading frame maintenance during translocation. Positions of the P- and A-site tRNAs in the intermediate state of translocation in the presence and absence of EF-G (left panel) and the schematics illustrating the movement of the tRNA anticodons toward the –1-frame in the absence of EF-G (right panel) (reproduced from Zhou, J., Lancaster, L., Donohue, J.P. and Noller, H.F. (2019) Spontaneous ribosomal translocation of mRNA and tRNAs into a chimeric hybrid state. Proc. Natl. Acad. Sci. U.S.A., 116, 7813–7818 (78) with permission). Complexes depicted in the schematics are from (79) with EF-G and from (78) without EF-G and contain a different sets of tRNAs in the A and P sites.
In contrast to spontaneous frameshifting, which produces non-functional polypeptides, PRF typically leads to the synthesis of a functional polypeptide from an altered frame. PRF was initially identified in viral genomes, where it plays an important role in viral propagation by modulating synthesis of viral proteins in specific stoichiometric ratios (81,82). Examples of –1PRF were found in all three domains of life (83–88). In eukaryotes, frameshifting can regulate the stability of an mRNA. After a frameshifting event, the translating ribosome soon encounters an out-of-frame stop codon, causing premature termination of translation and thereby recruiting the machinery of the nonsense-mediated decay pathway (86).
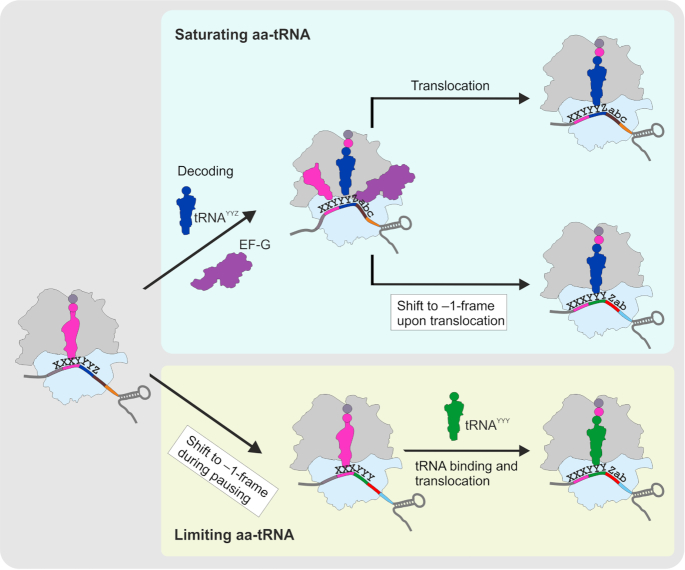
In most cases, –1PRF is facilitated by two regulatory elements in the mRNA sequence, a slippery site and a secondary structure element (a pseudoknot, a SL or a kissing loop) at a precisely defined distance of 5 to 9 nt from the slippery site (15,89–91). The mRNA structure element stalls the ribosome, which facilitates slippage (14,92). –1PRF can be also facilitated by binding of miRNAs (86) or proteins (93–96) to the sequence following the slippery site. Recent mechanistic studies suggested that despite the great variety of the frameshifting sequences, –1 frameshifting follows one of two main pathways (97–105) (Figure 5). One route is predominant under translation conditions where the tRNAs that read the slippery sequence codons are abundant. In this case, frameshifting occurs at the late stage of translocation, with two tRNAs moving through the ribosome, and requires the presence of the stimulatory element within the mRNA sequence. The other route is favored at conditions of aa-tRNA limitation and occurs via one-tRNA slippage of the P-site tRNA when the A site is vacant; its efficiency is independent of the downstream mRNA stimulators. The latter mechanism is often called ‘hungry’ frameshifting, because it can be triggered by aa-tRNA limitation due to starvation (106–108).
Figure 5.
Mechanism of –1PRF. Under conditions where aa-tRNAs are abundant (blue box), –1PRF takes place during the late stage of translocation by two-tRNA slippage (the P- and A-site tRNAs are shown in magenta and blue, respectively). Aa-tRNA limitation (yellow box) causes translational pausing that leads to the one-tRNA slippage of the P-site tRNA. X XXY YYZ is the sequence of the slippery site; abc and Zab are codons following the slippery site in 0- and –1-frames, respectively.
The detailed insights into the kinetic mechanism of translocation-dependent –1PRF came from ensemble and single molecule kinetic studies on 1a/1b mRNA of the avian infectious bronchitis virus (IBV) and dnaX mRNA from E. coli (97,98,102,104). Despite differences in sequence and structure in those mRNAs, frameshifting proceeds by a very similar mechanism. The frameshifting motif of the 1a/1b mRNA consists of a slippery site U1 UUA4 AAG7 encoding Leu (UUA) and Lys (AAG) in 0-frame followed by a pseudoknot (109). The dnaX frameshifting motif has the slippery site A1 AAA4 AAG7 encoding two Lys (AAA and AAG) in 0-frame preceded by a Shine-Dalgarno-like sequence and followed by a SL (110). In both cases, the role of the downstream secondary structure element is to slow down the late stages of translocation (97,98,102,104). At this point the ribosome is stalled in a rotated or even hyper-rotated state in which the stabilizing contacts between the ribosome and the codon–anticodon complexes are disrupted, which allows the tRNA to sample alternative reading frames (78,102,111). Both the dissociation of the E-site tRNA and the backward rotation of the ribosomal subunits are slow, but the E-site tRNA is released before the ribosome rotates backwards (102). EF-G, which usually restricts the A-site tRNA in the 0-frame position (78), can also dissociate prior to the completion of translocation (102). When both EF-G and the deacylated tRNA have been released, a single tRNA in transit from the A to the P site may be particularly prone to frameshifting (78). There are two ways to resolve the metastable stalled state, either by spontaneous unwinding of the mRNA secondary structure element that hinders the progression of the ribosome, which would allow the ribosome to resume its progression in the 0-frame, or by slippage in the –1 direction (97). The latter scenario may be kinetically advantageous because this would move the base of the pseudoknot to the entrance of the mRNA tunnel where the helicase center of the ribosome can actively unwind the mRNA secondary structure (112,113).
The choice of frameshifting pathway on the dnaX mRNA is dictated by environmental conditions, i.e. the availability of nutrients (Figure 5). However, there are cases where both pathways are constitutive. One prominent example is the gag-pol mRNA of human immunodeficiency virus type 1 (HIV-1). Here, the function of –1PRF is to produce viral structural proteins (Gag, 0-frame) and enzymes (Gag-Pol, –1-frame) at a defined ratio (114). The gag-pol mRNA contains the slippery sequence U1 UUU4 UUA7 encoding Phe (UUU) and Leu (UUA) in 0-frame followed by a SL (114). The –1 frameshifting efficiency in HIV-1 is modulated by the availability of the Leu-tRNALeu(UAA) isoacceptor that is rare in CD4+ T-lymphocytes—cells infected by the virus in the human host (99). When tRNALeu is abundant, it is rapidly accommodated at its cognate codon UUA, and –1PRF takes place during the late stage of translocation by two-tRNA slippage of tRNAPhe and tRNALeu. The frameshifting scenario changes markedly when Leu-tRNALeu(UAA) is limiting. During the translation pausing due to the ‘hungry’ UUA codon in the A site, the P-site tRNAPhe can slip into the –1-frame, which exposes a UUU Phe codon in the A site and bypasses the limitation for Leu-tRNA. Taking into account the low level of Leu-tRNALeu(UAA) in HIV-1 target cells and potential changes in tRNA profiles upon viral infection and interferon signaling activation (115–117), the alternative mechanism could act as a rescue pathway to allow for frameshifting under the limitation of the key tRNA (99). Most likely, HIV-1 has evolved to use both mechanisms to maintain the efficiency of –1PRF at the constant value, which is critical for viral replication and infectivity (118,119). Rescue pathways regulated by tRNA availability may be operational in other viruses, as recent studies of –1PRF on the 6K mRNA of the alphavirus Semliki Forest virus (SFV) identified a very similar switch between frameshifting pathways, also operated by tRNALeu(UAA) (100).
Manipulation of the frameshifting efficiency opens new perspectives in developing antiviral therapies and controlling gene expression of cellular mRNAs (120). An intriguing example is the interferon-stimulated cellular protein Shiftless. –1PRF in retroviruses (HIV) and alphaviruses (SFV) seems to be suppressed by this protein, which is thought to bind to both the translating ribosome and the frameshifting mRNA motif by a mechanism that is not fully understood (93). Multiple attempts have been made to design synthetic drugs targeting the frameshifting motif of HIV-1 (121–125) and SARS coronavirus (126). Recently, Matsumoto et al. have developed a small-molecule tool that can induce pseudoknot formation and activate –1PRF both in vitro and in vivo in human cells (127). Such inducible –1PRF was previously reported for HIV-1 using PRF stimulation by antisense nucleotides (91) and can serve to control viral propagation and gene expression using small synthetic molecules.
TRANSLATIONAL BYPASSING
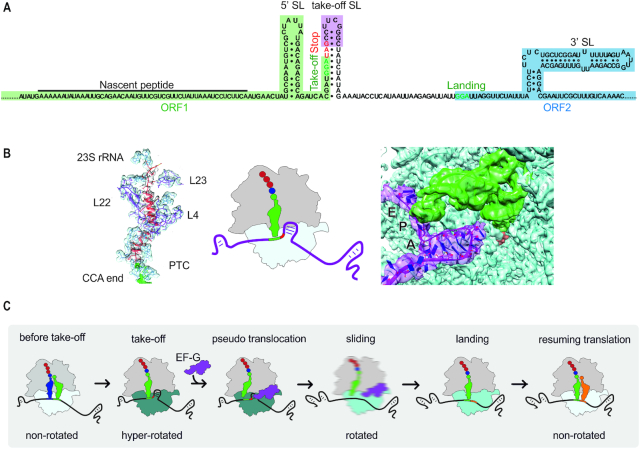
Another remarkable example of recoding is translational bypassing, which involves skipping of a portion of the mRNA by the translating ribosome, leading to the production of one polypeptide from a discontinuous frame. Translational bypassing was first identified in gene 60 of bacteriophage T4 (128), which remains the best-studied example of bypassing, and was later found in the mitochondrial genome of the yeast Magnusiomycetes (129). The mRNA of gene 60 contains two open reading frames (ORF1 and ORF2) separated by a non-coding gap (Figure 6A). Chemical and enzymatic probing of the mRNA structure suggested that mRNA of both ORFs are highly structured, whereas the gap is largely unfolded and forms a module that is structurally independent of the two ORFs (130). The gap appears to represent a mobile genetic element inserted into the gene 60 mRNA to inhibit cleavage by homing endonuclease MobA (131). The ribosome translates the first 46 mRNA codons of ORF1 up to a GGA triplet coding for amino acid glycine. The subsequent codon is a stop codon UAG, but instead of terminating protein synthesis, the ribosome slides over a 50 nt-long non-coding gap, lands at a distal GGA codon and resumes translation to the end of ORF2 (132). Gene 60 mRNA elements that stimulate bypassing are located 5′ of the take-off site, in the take-off SL and 3′ of the landing site (132–136). Remarkably, the key bypassing signals, such as the take-off SL element and the matching take-off and landing codons, are present also in yeast mitochondrial bypassing mRNAs (129), suggesting a similar mechanism of bypassing to that in bacteriophage T4.
Figure 6.
Translational bypassing. (A) Schematic of gene 60 mRNA. The nascent peptide, the SL element upstream (5′ SL) and downstream (3′ SL) of the take-off site, as well as the take-off SL are key elements facilitating bypassing. (B) Structure of the nascent peptide in the exit tunnel of the ribosome (left panel) and of the short A-site SL (right panel) obtained by cryo-EM (133). Middle panel represents a cartoon of the ribosome at the take-off site. (C) Schematic of bypassing. For details, see text. Rotation of the SSU relative to the LSU is indicated by different shades of blue. The blurred cartoon represents the ribosome in motion.
Recent biochemical, single molecule and structural work suggests how translational bypassing works. Translation of ORF1 is a non-uniform process: at the beginning, translation of ORF1 is rapid but then gradually slows down (136), probably because the ribosome has to unwind the secondary structure elements on its way along the mRNA. The ribosome pauses at the take-off GGA codon (136). To start bypassing, the ribosome requires the action of EF-G accompanied with GTP hydrolysis and a rotation of the ribosomal subunits relative to each other into an unusual hyper-rotated conformation (137).
The cryo-EM structure of the take-off complex reveals that the nascent peptide, which is known to be a key determinant for bypassing (135), forms numerous interactions with the polypeptide exit tunnel of the ribosome (133) (Figure 6B). These contacts help to hold the peptidyl-tRNA on the ribosome during sliding and likely contribute to the slow down at the take-off codon. In addition, the interactions of the nascent peptide residues with the ribosome lock an inactive conformation of the peptidyl transferase center, thus preventing the premature termination and readthrough at the take-off site.
Another remarkable feature of the take-off complex is a short dynamic SL formed by the mRNA in the decoding site of the SSU (133) (Figure 6B). The short SL hinders access of the translation termination factor or near-cognate aa-tRNAs into the A site (133). In addition, the SL serves as a mimic of an A-site tRNA to help EF-G to promote a pseudo-translocation event (Figure 6C). This displaces the P-site peptidyl-tRNA from its codon and starts ribosome sliding. As the ribosome moves forward, the mRNA upstream of the take-off site starts to emerge from the ribosome and can re-fold, thereby preventing backward sliding of the ribosome (135). The directionality of the ribosome movement may be also facilitated by cycles of EF-G binding and GTP hydrolysis (137). In fact, the kinetics of GTP hydrolysis by EF-G and bypassing are identical. EF-G appears to hydrolyze, on average, about 90 molecules of GTP for each ribosome that completes bypassing. Considering the length of the non-coding gap (50 nt), EF-G hydrolyzes on average 1.8 molecules of GTP per nucleotide of the sliding sequence. This GTP expenditure may be required to maintain the ribosome conformation that is prone to sliding or to facilitate the forward direction of sliding, similarly to the power-stroke action of EF-G in translocation (138). Although all ribosomes disengage from the take-off GGA codon and start sliding, only 50–60% of them synthesize the full-length protein, while the remaining ribosomes stop translation due to termination or spontaneous drop-off of the peptidyl-tRNAGly (135,136,139). At the end of the non-coding mRNA gap, the ribosome lands at the GGA codon guided by the 3′ SL in the mRNA downstream of the landing codon (135). The ribosome adopts a rotated conformation into which the next aa-tRNA accommodates (135,136). After peptide bond formation and subsequent translocation, the ribosome returns into a canonical non-rotated state and resumes translation of ORF2.
CONCLUSIONS
Although at the first glance recoding events seem to be a heterogeneous group of different phenomena facilitated by specific regulatory elements, collectively they provide insights into the dynamic modes of translation. Comparison of aa-tRNA recognition during canonical decoding and UGA recoding by Sec-tRNASec shows that the major key steps on the ribosome are identical (68). Specific recognition of Sec-tRNASec and the discrimination against all other similar aa-tRNAs occur at the preceding, pre-ribosomal steps of Sec-tRNASec recruitment to SelB and SECIS. This probably reflects the evolution of the ribosome as a universal decoder for all different tRNAs and mRNA codons, which relies on the geometry of the codon–anticodon complex, rather than on the structural specifics of each tRNA–codon pair. The mechanism of programmed readthrough is remarkably unclear, except for the fact that the near-cognate aa-tRNA and the RF must compete with each other. In contrast, comparison between canonical translocation, spontaneous and PRF and translational bypassing show common mechanisms underlying these processes. One general theme is the importance of ribosome dynamics. For example, the hyper-rotated state is found not only in ribosomes starting bypassing, but also during frameshifting (111) or in complexes stalled by the SecM peptide (137), suggesting that a hyper-rotated state may be a hallmark for stalled ribosomes resuming translation. Ribosome stalling is another important factor that defines the outcome of translation, as it regulates the efficiency of spontaneous and PRF, as well as bypassing. In these three cases, EF-G has a key role by either holding and escorting the tRNA or facilitating a pseudo-translocation of a tRNA-like A-site SL. Formation of the short dynamic SL in the A site may regulate ribosome pausing. In contrast to normal translation where ribosomes move by one codon at a time, during bypassing the ribosome slides over the mRNA. Similarly, ribosomes can move along the 3′ untranslated regions of eukaryotic mRNAs (140,141). Ribosome sliding exploits conserved elements of the translational machinery, such as the decoding center of the ribosome and EF-G. Thus, bypassing may explain how the ribosome changes from canonical decoding to unconventional EF-G–promoted movement through noncoding regions on the mRNA and suggests several new modes of ribosome dynamics that are potentially applicable in prokaryotic and eukaryotic translation.
FUNDING
Deutsche Forschungsgemeinschaft [SFB860 to M.V.R.]; Boehringer Ingelheim Fonds (to N.K.). Funding for open access charge: Max Planck Institute for Biophysical Chemistry.
Conflict of interest statement. None declared.
REFERENCES
- 1. Garofalo R., Wohlgemuth I., Pearson M., Lenz C., Urlaub H., Rodnina M.. Broad range of missense error frequencies in cellular proteins. Nucleic Acids Res. 2019; 47:2932–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Manickam N., Nag N., Abbasi A., Patel K., Farabaugh P.. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014; 20:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kramer E., Vallabhaneni H., Mayer L., Farabaugh P.. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010; 16:1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joshi K., Cao L., Farabaugh P.. The problem of genetic code misreading during protein synthesis. Yeast. 2019; 36:35–42. [DOI] [PubMed] [Google Scholar]
- 5. Schueren F., Thoms S.. Functional translational readthrough: A systems biology perspective. PLoS Genet. 2016; 12:e1006196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dabrowski M., Bukowy-Bieryllo Z., Zietkiewicz E.. Translational readthrough potential of natural termination codons in eucaryotes–The impact of RNA sequence. RNA Biol. 2015; 12:950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Firth A., Wills N., Gesteland R., Atkins J.. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011; 39:6679–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eswarappa S., Potdar A., Koch W., Fan Y., Vasu K., Lindner D., Willard B., Graham L., DiCorleto P., Fox P.. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 2014; 157:1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Namy O., Hatin I., Rousset J.. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001; 2:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kurland C. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992; 26:29–50. [DOI] [PubMed] [Google Scholar]
- 11. Drummond D., Wilke C.. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009; 10:715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hardin A., Villalta C., Doan M., Jabri M., Chockalingham V., White S., Fowler R.. A molecular characterization of spontaneous frameshift mutagenesis within the trpA gene of Escherichia coli. DNA Repair. 2007; 6:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Atkins J., Loughran G., Bhatt P., Firth A., Baranov P.. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016; 44:7007–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caliskan N., Peske F., Rodnina M.. Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting. Trends Biochem. Sci. 2015; 40:265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brierley I., Gilbert R., Pennell S.. Atkins J.F., Gesteland R.F.. Recoding: expansion of decoding rules enrichesgene expression. 2010; NY: Springer; 149–174. [Google Scholar]
- 16. Advani V., Dinman J.. Reprogramming the genetic code: the emerging role of ribosomal frameshifting in regulating cellular gene expression. Bioessays. 2016; 38:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blanchet S., Cornu D., Argentini M., Namy O.. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res. 2014; 42:10061–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pelham H. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature. 1978; 272:469–471. [DOI] [PubMed] [Google Scholar]
- 19. Felsenstein K., Goff S.. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J. Virol. 1988; 62:2179–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hofstetter H., Monstein H., Weissmann C.. The readthrough protein A1 is essential for the formation of viable Q beta particles. Biochim. Biophys. Acta. 1974; 374:238–251. [DOI] [PubMed] [Google Scholar]
- 21. Loughran G., Chou M., Ivanov I., Jungreis I., Kellis M., Kiran A., Baranov P., Atkins J.. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014; 42:8928–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cridge A., Crowe-McAuliffe C., Mathew S., Tate W.. Eukaryotic translational termination efficiency is influenced by the 3′ nucleotides within the ribosomal mRNA channel. Nucleic Acids Res. 2018; 46:1927–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manuvakhova M., Keeling K., Bedwell D.. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000; 6:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Howard M., Shirts B., Petros L., Flanigan K., Gesteland R., Atkins J.. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 2000; 48:164–169. [PubMed] [Google Scholar]
- 25. Arkov A., Korolev S., Kisselev L.. 5′ contexts of Escherichia coli and human termination codons are similar. Nucleic Acids Res. 1995; 23:4712–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tork S., Hatin I., Rousset J., Fabret C.. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res. 2004; 32:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weiner A., Weber K.. A single UGA codon functions as a natural termination signal in the coliphage Qβ coat protein cistron. J. Mol. Biol. 1973; 80:837–855. [DOI] [PubMed] [Google Scholar]
- 28. Li G., Rice C.. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J. Virol. 1993; 67:5062–5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown C., Stockwell P., Trotman C., Tate W.. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990; 18:6339–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pedersen W., Curran J.. Effects of the nucleotide 3′ to an amber codon on ribosomal selection rates of suppressor tRNA and release factor-1. J. Mol. Biol. 1991; 219:231–241. [DOI] [PubMed] [Google Scholar]
- 31. Tate W., Poole E., Horsfield J., Mannering S., Brown C., Moffat J., Dalphin M., McCaughan K., Major L., Wilson D.. Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem. Cell Biol. 1995; 73:1095–1103. [DOI] [PubMed] [Google Scholar]
- 32. McCaughan K., Brown C., Dalphin M., Berry M., Tate W.. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bonetti B., Fu L., Moon J., Bedwell D.. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995; 251:334–345. [DOI] [PubMed] [Google Scholar]
- 34. Beier H., Grimm M.. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001; 29:4767–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urban C., Zerfass K., Fingerhut C., Beier H.. UGA suppression by tRNACmCATrp occurs in diverse virus RNAs due to a limited influence of the codon context. Nucleic Acids Res. 1996; 24:3424–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williams I., Richardson J., Starkey A., Stansfield I.. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2004; 32:6605–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Skuzeski J., Nichols L., Gesteland R., Atkins J.. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol. 1991; 218:365–373. [DOI] [PubMed] [Google Scholar]
- 38. Shao S., Murray J., Brown A., Taunton J., Ramakrishnan V., Hegde R.. Decoding mammalian Ribosome-mRNA states by translational GTPase complexes. Cell. 2016; 167:1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown A., Shao S., Murray J., Hegde R., Ramakrishnan V.. Structural basis for stop codon recognition in eukaryotes. Nature. 2015; 524:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laurberg M., Asahara H., Korostelev A., Zhu J., Trakhanov S., Noller H.. Structural basis for translation termination on the 70S ribosome. Nature. 2008; 454:852–857. [DOI] [PubMed] [Google Scholar]
- 41. Feng Y., Yuan H., Rein A., Levin J.. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J. Virol. 1992; 66:5127–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wills N., Gesteland R., Atkins J.. Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J. 1994; 13:4137–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wills N., Gesteland R., Atkins J.. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:6991–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jungreis I., Lin M., Spokony R., Chan C., Negre N., Victorsen A., White K., Kellis M.. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011; 21:2096–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steneberg P., Samakovlis C.. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2001; 2:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Napthine S., Yek C., Powell M., Brown T., Brierley I.. Characterization of the stop codon readthrough signal of Colorado tick fever virus segment 9 RNA. RNA. 2012; 18:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beznoskova P., Wagner S., Jansen M., von der Haar T., Valasek L.. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015; 43:5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chauvin C., Salhi S., Le Goff C., Viranaicken W., Diop D., Jean-Jean O.. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol. Cell Biol. 2005; 25:5801–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carnes J., Jacobson M., Leinwand L., Yarus M.. Stop codon suppression via inhibition of eRF1 expression. RNA. 2003; 9:648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paushkin S., Kushnirov V., Smirnov V., Ter-Avanesyan M.. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996; 15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 51. Liebman S., Sherman F.. Extrachromosomal psi+ determinant suppresses nonsense mutations in yeast. J. Bacteriol. 1979; 139:1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wickner R., Masison D., Edskes H.. [PSI] and [URE3] as yeast prions. Yeast. 1995; 11:1671–1685. [DOI] [PubMed] [Google Scholar]
- 53. Beznoskova P., Gunisova S., Valasek L.. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA. 2016; 22:456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Roy B., Leszyk J., Mangus D., Jacobson A.. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:3038–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blanchet S., Cornu D., Hatin I., Grosjean H., Bertin P., Namy O.. Deciphering the reading of the genetic code by near-cognate tRNA. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:3018–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dunn J., Foo C., Belletier N., Gavis E., Weissman J.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schueren F., Lingner T., George R., Hofhuis J., Dickel C., Gartner J., Thoms S.. Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. Elife. 2014; 3:e03640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pancsa R., Macossay-Castillo M., Kosol S., Tompa P.. Computational analysis of translational readthrough proteins in Drosophila and yeast reveals parallels to alternative splicing. Sci. Rep. 2016; 6:32142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kleppe A., Bornberg-Bauer E.. Robustness by intrinsically disordered C-termini and translational readthrough. Nucleic Acids Res. 2018; 46:10184–10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hoffman K., Crnkovic A., Soll D.. Versatility of synthetic tRNAs in genetic code expansion. Genes (Basel). 2018; 9:E537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Y., Baranov P., Atkins J., Gladyshev V.. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J. Biol. Chem. 2005; 280:20740–20751. [DOI] [PubMed] [Google Scholar]
- 62. Blight S., Larue R., Mahapatra A., Longstaff D., Chang E., Zhao G., Kang P., Green-Church K., Chan M., Krzycki J.. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature. 2004; 431:333–335. [DOI] [PubMed] [Google Scholar]
- 63. Bock A., Forchhammer K., Heider J., Leinfelder W., Sawers G., Veprek B., Zinoni F.. Selenocysteine: the 21st amino acid. Mol. Microbiol. 1991; 5:515–520. [DOI] [PubMed] [Google Scholar]
- 64. Forchhammer K., Boesmiller K., Bock A.. The function of selenocysteine synthase and SELB in the synthesis and incorporation of selenocysteine. Biochimie. 1991; 73:1481–1486. [DOI] [PubMed] [Google Scholar]
- 65. Forchhammer K., Leinfelder W., Bock A.. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989; 342:453–456. [DOI] [PubMed] [Google Scholar]
- 66. Fagegaltier D., Hubert N., Yamada K., Mizutani T., Carbon P., Krol A.. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000; 19:4796–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zinoni F., Heider J., Bock A.. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:4660–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fischer N., Neumann P., Bock L., Maracci C., Wang Z., Paleskava A., Konevega A., Schroder G., Grubmuller H., Ficner R. et al.. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature. 2016; 540:80–85. [DOI] [PubMed] [Google Scholar]
- 69. Loveland A., Demo G., Grigorieff N., Korostelev A.. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature. 2017; 546:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ogle J., Murphy F., Tarry M., Ramakrishnan V.. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002; 111:721–732. [DOI] [PubMed] [Google Scholar]
- 71. Ogle J., Brodersen D., Clemons W. Jr, Tarry M., Carter A., Ramakrishnan V.. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001; 292:897–902. [DOI] [PubMed] [Google Scholar]
- 72. Rudinger J., Hillenbrandt R., Sprinzl M., Giege R.. Antideterminants present in minihelix(Sec) hinder its recognition by prokaryotic elongation factor Tu. EMBO J. 1996; 15:650–657. [PMC free article] [PubMed] [Google Scholar]
- 73. Paleskava A., Konevega A., Rodnina M.. Thermodynamic and kinetic framework of selenocysteyl-tRNASec recognition by elongation factor SelB. J. Biol. Chem. 2010; 285:3014–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Thanbichler M., Bock A., Goody R.. Kinetics of the interaction of translation factor SelB from Escherichia coli with guanosine nucleotides and selenocysteine insertion sequence RNA. J. Biol. Chem. 2000; 275:20458–20466. [DOI] [PubMed] [Google Scholar]
- 75. Kotini S., Peske F., Rodnina M.. Partitioning between recoding and termination at a stop codon-selenocysteine insertion sequence. Nucleic Acids Res. 2015; 43:6426–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rodnina M., Wintermeyer W.. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 2001; 70:415–435. [DOI] [PubMed] [Google Scholar]
- 77. Gromadski K., Rodnina M.. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004; 13:191–200. [DOI] [PubMed] [Google Scholar]
- 78. Zhou J., Lancaster L., Donohue J., Noller H.. Spontaneous ribosomal translocation of mRNA and tRNAs into a chimeric hybrid state. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:7813–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou J., Lancaster L., Donohue J., Noller H.. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014; 345:1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Savelsbergh A., Matassova N., Rodnina M., Wintermeyer W.. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J. Mol. Biol. 2000; 300:951–961. [DOI] [PubMed] [Google Scholar]
- 81. Jacks T., Varmus H.. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985; 230:1237–1242. [DOI] [PubMed] [Google Scholar]
- 82. Plant E., Rakauskaite R., Taylor D., Dinman J.. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 2010; 84:4330–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tsuchihashi Z., Kornberg A.. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cobucci-Ponzano B., Conte F., Benelli D., Londei P., Flagiello A., Monti M., Pucci P., Rossi M., Moracci M.. The gene of an archaeal alpha-L-fucosidase is expressed by translational frameshifting. Nucleic Acids Res. 2006; 34:4258–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Luthi K., Moser M., Ryser J., Weber H.. Evidence for a role of translational frameshifting in the expression of transposition activity of the bacterial insertion element IS1. Gene. 1990; 88:15–20. [DOI] [PubMed] [Google Scholar]
- 86. Belew A., Meskauskas A., Musalgaonkar S., Advani V., Sulima S., Kasprzak W., Shapiro B., Dinman J.. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature. 2014; 512:265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Manktelow E., Shigemoto K., Brierley I.. Characterization of the frameshift signal of Edr, a mammalian example of programmed -1 ribosomal frameshifting. Nucleic Acids Res. 2005; 33:1553–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wills N., Moore B., Hammer A., Gesteland R., Atkins J.. A functional -1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J. Biol. Chem. 2006; 281:7082–7088. [DOI] [PubMed] [Google Scholar]
- 89. Atkinson J., Dodge M., Gallant J.. Secondary structures and starvation-induced frameshifting. Mol. Microbiol. 1997; 26:747–753. [DOI] [PubMed] [Google Scholar]
- 90. Brierley I., Jenner A., Inglis S.. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992; 227:463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lin Z., Gilbert R., Brierley I.. Spacer-length dependence of programmed -1 or -2 ribosomal frameshifting on a U6A heptamer supports a role for messenger RNA (mRNA) tension in frameshifting. Nucleic Acids Res. 2012; 40:8674–8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Plant E., Dinman J.. Comparative study of the effects of heptameric slippery site composition on -1 frameshifting among different eukaryotic systems. RNA. 2006; 12:666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang X., Xuan Y., Han Y., Ding X., Ye K., Yang F., Gao P., Goff S., Gao G.. Regulation of HIV-1 Gag-Pol Expression by Shiftless, an Inhibitor of Programmed -1 Ribosomal Frameshifting. Cell. 2019; 176:625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Napthine S., Ling R., Finch L., Jones J., Bell S., Brierley I., Firth A.. Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat. Commun. 2017; 8:15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li Y., Treffers E., Napthine S., Tas A., Zhu L., Sun Z., Bell S., Mark B., van Veelen P., van Hemert M. et al.. Transactivation of programmed ribosomal frameshifting by a viral protein. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2172–E2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kobayashi Y., Zhuang J., Peltz S., Dougherty J.. Identification of a cellular factor that modulates HIV-1 programmed ribosomal frameshifting. J. Biol. Chem. 2010; 285:19776–19784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Caliskan N., Katunin V., Belardinelli R., Peske F., Rodnina M.. Programmed -1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014; 157:1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Caliskan N., Wohlgemuth I., Korniy N., Pearson M., Peske F., Rodnina M.. Conditional switch between frameshifting regimes upon translation of dnaX mRNA. Mol. Cell. 2017; 66:558–567. [DOI] [PubMed] [Google Scholar]
- 99. Korniy N., Goyal A., Hoffmann M., Samatova E., Peske F., Pohlmann S., Rodnina M.. Modulation of HIV-1 Gag/Gag-Pol frameshifting by tRNA abundance. Nucleic Acids Res. 2019; 47:5210–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Korniy N., Samatova E., Anokhina M., Peske F., Rodnina M.. Mechanisms and biomedical implications of –1 programmed ribosome frameshifting on viral and bacterial mRNAs. FEBS Lett. 2019; 593:1468–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chen C., Zhang H., Broitman S., Reiche M., Farrell I., Cooperman B., Goldman Y.. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 2013; 20:582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chen J., Petrov A., Johansson M., Tsai A., O’Leary S., Puglisi J.. Dynamic pathways of -1 translational frameshifting. Nature. 2014; 512:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Namy O., Moran S., Stuart D., Gilbert R., Brierley I.. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006; 441:244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kim H., Liu F., Fei J., Bustamante C., Gonzalez R. Jr, Tinoco I. Jr. A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yan S., Wen J., Bustamante C., Tinoco I. Jr. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015; 160:870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gallant J., Lindsley D.. Leftward ribosome frameshifting at a hungry codon. J. Mol. Biol. 1992; 223:31–40. [DOI] [PubMed] [Google Scholar]
- 107. Olubajo B., Taylor E.. A -1 frameshift in the HIV-1 env gene is enhanced by arginine deficiency via a hungry codon mechanism. Mutat. Res. 2005; 579:125–132. [DOI] [PubMed] [Google Scholar]
- 108. Temperley R., Richter R., Dennerlein S., Lightowlers R., Chrzanowska-Lightowlers Z.. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science. 2010; 327:301. [DOI] [PubMed] [Google Scholar]
- 109. Brierley I., Digard P., Inglis S.. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989; 57:537–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Tsuchihashi Z., Brown P.. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 1992; 6:511–519. [DOI] [PubMed] [Google Scholar]
- 111. Qin P., Yu D., Zuo X., Cornish P.. Structured mRNA induces the ribosome into a hyper-rotated state. EMBO Rep. 2014; 15:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Qu X., Wen J., Lancaster L., Noller H., Bustamante C., Tinoco I. Jr. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011; 475:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Takyar S., Hickerson R., Noller H.. mRNA helicase activity of the ribosome. Cell. 2005; 120:49–58. [DOI] [PubMed] [Google Scholar]
- 114. Jacks T., Power M., Masiarz F., Luciw P., Barr P., Varmus H.. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988; 331:280–283. [DOI] [PubMed] [Google Scholar]
- 115. van Weringh A., Ragonnet-Cronin M., Pranckeviciene E., Pavon-Eternod M., Kleiman L., Xia X.. HIV-1 modulates the tRNA pool to improve translation efficiency. Mol. Biol. Evol. 2011; 28:1827–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Smith B., Chen G., Wilke C., Krug R.. Avian influenza virus PB1 gene in H3N2 viruses evolved in humans to reduce interferon inhibition by skewing codon usage toward interferon-altered tRNA pools. MBio. 2018; 9:e01222-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Pavon-Eternod M., David A., Dittmar K., Berglund P., Pan T., Bennink J., Yewdell J.. Vaccinia and influenza A viruses select rather than adjust tRNAs to optimize translation. Nucleic Acids Res. 2013; 41:1914–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Biswas P., Jiang X., Pacchia A., Dougherty J., Peltz S.. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 2004; 78:2082–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Karacostas V., Wolffe E., Nagashima K., Gonda M., Moss B.. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993; 193:661–671. [DOI] [PubMed] [Google Scholar]
- 120. Dinman J. Translational recoding signals: Expanding the synthetic biology toolbox. J. Biol. Chem. 2019; 294:7537–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ofori L., Hilimire T., Bennett R., Brown N. Jr, Smith H., Miller B.. High-affinity recognition of HIV-1 frameshift-stimulating RNA alters frameshifting in vitro and interferes with HIV-1 infectivity. J. Med. Chem. 2014; 57:723–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Brakier-Gingras L., Charbonneau J., Butcher S.. Targeting frameshifting in the human immunodeficiency virus. Exp. Opin. Ther. Targets. 2012; 16:249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hung M., Patel P., Davis S., Green S.. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 1998; 72:4819–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Brakier-Gingras L., Charbonneau J., Miller B.. Drugs targeting the− 1 ribosomal frameshifting that generates the enzymes of the human immunodeficiency virus. Front. Clin. Drug Res. 2014; 1:67–82. [Google Scholar]
- 125. Anokhina V., McAnany J., Ciesla J., Hilimire T., Santoso N., Miao H., Miller B.. Enhancing the ligand efficiency of anti-HIV compounds targeting frameshift-stimulating RNA. Bioorg. Med. Chem. 2019; 27:2972–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ahn D., Lee W., Choi J., Kim S., Plant E., Almazan F., Taylor D., Enjuanes L., Oh J.. Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antiviral Res. 2011; 91:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Matsumoto S., Caliskan N., Rodnina M., Murata A., Nakatani K.. Small synthetic molecule-stabilized RNA pseudoknot as an activator for -1 ribosomal frameshifting. Nucleic Acids Res. 2018; 46:8079–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Huang W., Ao S., Casjens S., Orlandi R., Zeikus R., Weiss R., Winge D., Fang M.. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science. 1988; 239:1005–1012. [DOI] [PubMed] [Google Scholar]
- 129. Lang B., Jakubkova M., Hegedusova E., Daoud R., Forget L., Brejova B., Vinar T., Kosa P., Fricova D., Nebohacova M. et al.. Massive programmed translational jumping in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5926–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Todd G., Walter N.. Secondary structure of bacteriophage T4 gene 60 mRNA: implications for translational bypassing. RNA. 2013; 19:685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bonocora R., Zeng Q., Abel E., Shub D.. A homing endonuclease and the 50-nt ribosomal bypass sequence of phage T4 constitute a mobile DNA cassette. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:16351–16356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Weiss R., Huang W., Dunn D.. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990; 62:117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Agirrezabala X., Samatova E., Klimova M., Zamora M., Gil-Carton D., Rodnina M., Valle M.. Ribosome rearrangements at the onset of translational bypassing. Sci. Adv. 2017; 3:e1700147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Herr A., Atkins J., Gesteland R.. Coupling of open reading frames by translational bypassing. Annu. Rev. Biochem. 2000; 69:343–372. [DOI] [PubMed] [Google Scholar]
- 135. Samatova E., Konevega A., Wills N., Atkins J., Rodnina M.. High-efficiency translational bypassing of non-coding nucleotides specified by mRNA structure and nascent peptide. Nat. Commun. 2014; 5:4459. [DOI] [PubMed] [Google Scholar]
- 136. Chen J., Coakley A., O’Connor M., Petrov A., O’Leary S., Atkins J., Puglisi J.. Coupling of mRNA structure rearrangement to ribosome movement during bypassing of Non-coding regions. Cell. 2015; 163:1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Klimova M., Senyushkina T., Samatova E., Peng B., Pearson M., Peske F., Rodnina M.. EF-G-induced ribosome sliding along the noncoding mRNA. Sci. Adv. 2019; 5:eaaw9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Chen C., Cui X., Beausang J., Zhang H., Farrell I., Cooperman B., Goldman Y.. Elongation factor G initiates translocation through a power stroke. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7515–7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Herr A., Wills N., Nelson C., Gesteland R., Atkins J.. Drop-off during ribosome hopping. J. Mol. Biol. 2001; 311:445–452. [DOI] [PubMed] [Google Scholar]
- 140. Miettinen T., Bjorklund M.. Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3′ untranslated regions. Nucleic Acids Res. 2015; 43:1019–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Guydosh N., Green R.. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014; 156:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Beier H., Barciszewska M., Krupp G., Mitnacht R., Gross H.. UAG readthrough during TMV RNA translation: isolation and sequence of two tRNAs with suppressor activity from tobacco plants. EMBO J. 1984; 3:351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Brault V., van den Heuvel J., Verbeek M., Ziegler-Graff V., Reutenauer A., Herrbach E., Garaud J., Guilley H., Richards K., Jonard G.. Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J. 1995; 14:650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Brown C., Dinesh-Kumar S., Miller W.. Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J. Virol. 1996; 70:5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Filichkin S., Lister R., McGrath P., Young M.. In vivo expression and mutational analysis of the barley yellow dwarf virus readthrough gene. Virology. 1994; 205:290–299. [DOI] [PubMed] [Google Scholar]
- 146. Yoshinaka Y., Katoh I., Copeland T., Oroszlan S.. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:1618–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Csibra E., Brierley I., Irigoyen N.. Modulation of stop codon read-through efficiency and its effect on the replication of murine leukemia virus. J. Virol. 2014; 88:10364–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Chambert R., Rain-Guion M., Petit-Glatron M.. Readthrough of the Bacillus subtilis stop codon produces an extended enzyme displaying a higher polymerase activity. Biochim. Biophys. Acta. 1992; 1132:145–153. [DOI] [PubMed] [Google Scholar]
- 149. Namy O., Duchateau-Nguyen G., Rousset J.. Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol. Microbiol. 2002; 43:641–652. [DOI] [PubMed] [Google Scholar]
- 150. Freitag J., Ast J., Bolker M.. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature. 2012; 485:522–525. [DOI] [PubMed] [Google Scholar]
- 151. Chittum H., Lane W., Carlson B., Roller P., Lung F., Lee B., Hatfield D.. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998; 37:10866–10870. [DOI] [PubMed] [Google Scholar]
- 152. Geller A., Rich A.. A UGA termination suppression tRNATrp active in rabbit reticulocytes. Nature. 1980; 283:41–46. [DOI] [PubMed] [Google Scholar]
- 153. Hatfield D., Thorgeirsson S., Copeland T., Oroszlan S., Bustin M.. Immunopurification of the suppressor tRNA dependent rabbit beta-globin readthrough protein. Biochemistry. 1988; 27:1179–1183. [DOI] [PubMed] [Google Scholar]
- 154. Yamaguchi Y., Hayashi A., Campagnoni C., Kimura A., Inuzuka T., Baba H.. L-MPZ, a novel isoform of myelin P0, is produced by stop codon readthrough. J. Biol. Chem. 2012; 287:17765–17776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Yamaguchi Y., Baba H.. Phylogenetically conserved sequences around myelin P0 stop codon are essential for translational readthrough to Produce L-MPZ. Neurochem. Res. 2018; 43:227–237. [DOI] [PubMed] [Google Scholar]
- 156. Stiebler A., Freitag J., Schink K., Stehlik T., Tillmann B., Ast J., Bolker M.. Ribosomal readthrough at a short UGA stop codon context triggers dual localization of metabolic enzymes in Fungi and animals. PLoS Genet. 2014; 10:e1004685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Loughran G., Jungreis I., Tzani I., Power M., Dmitriev R., Ivanov I., Kellis M., Atkins J.. Stop codon readthrough generates a C-terminally extended variant of the human vitamin D receptor with reduced calcitriol response. J. Biol. Chem. 2018; 293:4434–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]