Abstract
In metazoan cell nuclei, heterochromatin constitutes large chromatin domains that are in close contact with the nuclear lamina. These heterochromatin/lamina-associated domains (LADs) domains are difficult to profile and warrants a simpler and direct method. Here we report a new method, Protect-seq, aimed at identifying regions of heterochromatin via resistance to nuclease degradation followed by next-generation sequencing (NGS). We performed Protect-seq on the human colon cancer cell line HCT-116 and observed overlap with previously curated LADs. We provide evidence that these protected regions are enriched for and can distinguish between the repressive histone modification H3K9me3, H3K9me2 and H3K27me3. Moreover, in human cells the loss of H3K9me3 leads to an increase in chromatin accessibility and loss of Protect-seq signal. For further validation, we performed Protect-seq in the fibrosarcoma cell line HT1080 and found a similar correlation with previously curated LADs and repressive histone modifications. In sum, Protect-seq is an efficient technique that allows rapid identification of nuclease resistant chromatin, which correlate with heterochromatin and radial positioning.
INTRODUCTION
Heterochromatin domains are linked to a number of chromosomal structures and behaviors including chromosomal topology, replication timing, transcriptional repression, and lamina-association (1). Histone H3 lysine 9 methylation (H3K9me) is a hallmark of heterochromatin and has been shown to be necessary for chromatin to associate with the nuclear periphery suggesting an interplay between histone modifications and nuclear localization/LAD formation (2–4). However, recent work suggests chromosome architecture is maintained by heterochromatin attraction to drive phase separation independent of LAD formation (5). Although the function of LADs remains unclear, LADs are conserved across cell types and species and constitute more than one-third of the genome suggesting these domains play an important role in genome organization (4,6,7). However, detecting such changes using current NGS approaches has proved challenging. We set out to design a direct technique that measures heterochromatin on the periphery and can contribute addition layers of information which will allow for a greater understanding of chromosome organization.
Chromatin accessibility is often measured by enzyme accessibility. DNase-seq (8), ATAC-seq (9), MNase-seq (10) and NicE-seq (11) all require an enzyme to cleave DNA in order to define accessible chromatin. DNase-seq, ATAC-seq and NicE-seq have a strong preference towards nucleosome free chromatin (termed open chromatin). A similar technique, DIVA, use viral integration to distinguish between accessible and inaccessible chromatin (12,13). ATAC-seq and DIVA both directly insert exogenous sequences into accessible chromatin. For unknown reasons, DIVA seems to have less bias towards open chromatin compared to ATAC-seq and therefore demarcates accessible chromatin. Alternatively, MNase-seq identifies both euchromatin and heterochromatin, suggesting the entire genome is accessible to nucleases (14–16). However, the degree of bias towards euchromatin remains less unclear. Sono-seq (17), FAIRE-seq (18) and Gradient-seq (19) use sonication to detect chromatin accessibility of crosslinked chromatin. Gradient-seq fractionates sonicated chromatin using a sucrose gradient. Fractions enriched for larger/heavier fragments are enriched for heterochromatin suggesting that heterochromatin is compacted and more resistant to perturbation. However, multiple fractions need to be assayed to find the sonication resistant heterochromatin (srHC) fraction. Taken together, chromatin accessibility is a spectrum with open chromatin as the most accessible and sonication resistant chromatin as the most inaccessible chromatin.
Here, we describe a novel in situ sequencing technique (termed Protect-seq) in which a cocktail of nucleases degrades chromatin that is accessible to nucleases while either failing to degrade inaccessible chromatin or sequestration through tight association with the nuclear lamina. Our approach finds that chromatin near the nuclear periphery is enriched for nuclease resistant chromatin. To validate our approach, we applied Protect-seq to human HCT116 and HT1080 cells and demonstrated that our approach identified known heterochromatin domains. Protect-seq is a simple, reliable, and cost-and-time effective method to quantify heterochromatin domains using NGS. Importantly, Protect-seq is a direct readout of chromatin accessibility, which does not require multiple rounds of cell division or ectopic transgene expression.
MATERIALS AND METHODS
Cell culture
HCT116 and DKO cells were cultured in McCoy5A media. DKO cells were grown in the presence of G418, geneticin. HT1080 cells were cultured in DMEM media plus L-glutamine. All media was supplemented with 10% fetal bovine serum (FBS) at 37°C and 5% CO2.
Crosslinking and Nuclei Preparation
Cells were grown to ∼75% confluency, harvested with trypsin, washed in 1× PBS, and frozen/stored at −80°C. Thawed cells were fixed in 1% formaldehyde and quenched in 0.125 M glycine, then washed twice in 1× PBS. Fixed cells were then resuspended in 500 μl lysis buffer (50 mM Tris–HCl pH 8.0, 10 mM NaCl, 0.2% NP40, 1× PITC) for 30 min on ice with periodic resuspension. Lysed cells were spun 3500 RPM for 3 min and resuspended in 300 μl 1× NEB buffer 2, spun and resuspended in 198 μl 1× NEB buffer 2. 2μl of 10% SDS was added and incubated at 65°C for 10 min. After, 400 μl 1× NEB buffer 2 and 60 μl 10% Triton X-100 were added to quench the SDS. Samples were incubated at 37°C for 15 min. Nuclei were spun 3500 RPM for 3 min and resuspended in 300 μl 1× NEB buffer 2, repeat wash step.
Protect-seq protocol
Pelleted nuclei were resuspended in 183 μl DNaseI Buffer then 2 μl 100 mM Ca2+ (1 mM final), 5 μl DNase I (10 U), 5 μl MNase (10 000 U) and 5 μl RNase A (20 mg/ml) were added (200 μl final volume). Cells plus enzyme cocktail were incubated at room temperature (RT) (also works at 37°C) for 30 min. Digested cells were spun 3500 RPM for 3 min and resuspended in 400 μl of 1× NEB buffer 2, then rotated at RT for 15 min. Digested/Wash#1 cells were spun 5000 RPM for 3 min and resuspended in same 200 μl cocktail mix and incubated again at RT (or 37°C) for 30 min. Digested cells#2 were spun 10 000 RPM for 3 min and resuspended in 400 μl of 1× NEB buffer 2, then rotate at RT for 15 min (save aliquot for microscopy). Spin digested cells#2 10 000 RPM for 3 min and resuspended in 200 μl of 1× NEB buffer 2, 20 μl Proteinase K (SDS optional). Digest overnight at 65°C then purify using phenol/chloroform and ethanol precipitation (compatible with silica-bead purification). Note: Nuclei clumping prevents nuclease digestion. Lower temperatures helped prevent clumping.
Illumina library preparation
DNA was quantified with Qubit (high-sensitivity) and sonicated using Covaris 50 μl 300 bp protocol. Illumina libraries were prepared using NEB Ultra II DNA library kit using the manufacturer's protocol. 4–5 PCR cycles were used to amplify NGS libraries and index samples. Note: overcycling can introduce GC-bias and other unwanted artifacts.
Oxford nanopore library prep
Libraries were prepared using manufacturer's protocol with slight modifications. Protect-seq purified DNA (not fragmented) was treated using NEB FFPE-repair mix. Samples were purified using (0.9×) SPRI beads, washed twice with 70% ethanol, resuspended in 25 μl water. End Prep: 25 μl DNA, 3.5 μl UltraII End-prep Buffer, 1.5 μl UltraII End-prep enzyme mix; incubate RT for 20 min then 65°C for 20 min. Adaptor Ligation: 30 μl DNA from end-prep, 20 μl AMX adaptor, 24 μl UltraII ligation master mix, 1 μl UltraII ligation enhancer; incubate RT 20 min.
Chromatin immunoprecipitation (ChIP) experiments
SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005 from Cell Signaling Technologies was used for all ChIP-seq experiments using manufacturer's recommended protocol. Four million cells per IP. Digested chromatin was pooled into single tube for brief sonication to lyse nuclei. Supernatant was then split evenly between IPs (minus 2% input). Antibodies and chromatin were incubated overnight at 4°C, rotating. DNA was purified using spin columns and prepared using NEB Ultra II DNA library kit.
Repli-seq
Repli-seq experiments were conducted as previously described (20). Cells were sorted on a SONY SH800 FACS machine.
Data analysis
FASTQ files were trimmed and assessed for quality with fastp (21). Trimmed reads were mapped to hg38 (UCSC) using bwa mem (22). Alignment files (BAM format) were filtered using encode_filter.py (https://github.com/ENCODE-DCC/chip-seq-pipeline2). Replicate filtered BAM files were merged using sambamba (23). Signal tracks were generated using 100 bp bins and normalized to reads per genomic content (RPGC) using deeptools bamCoverage (24–26) with the following parameters: -of bigwig -effectiveGenomeSize 2913022398 -normalizeUsing RPGC -e. Differential signal tracks (log2ratio) were generated using deeptools bamCompare. Published data (Fastq files) were downloaded from GEO and processed as described above. Pearson and spearman correlation were performed using deeptools.
HMM domain calls and analysis
A 25 kb binned pandas dataframe was generated using cooler (27). Log2ratio signal tracks were imported into the binned dataframe using pybbi (https://github.com/nvictus/pybbi). Missing values were represented as NaN. Enriched Protect-seq domains were identified with HMMs using Pomegranate (28). Viterbi state calls were made on a per bin bases and used for downstream analysis. Neighboring states were merged to create domains then converted to bed files (https://github.com/gspracklin/hmm_bigwigs).
chromHMM
Single-end (or read1 if paired-end) ChIP-seq data was merged for replicates from ENCODE, this study, and PolII data (29). Merged and sorted bam files (via sambamba) were used as input for the binarization step with default parameters. LearnModel was used with 15-states (default parameters) and genome version hg38, in addition LaminB1 DamID-seq domains and Protect-seq domains were included with the precompiled refSeq TSS/TES/Exons/Genes and CpG islands.
RESULTS
Protect-seq assay and design
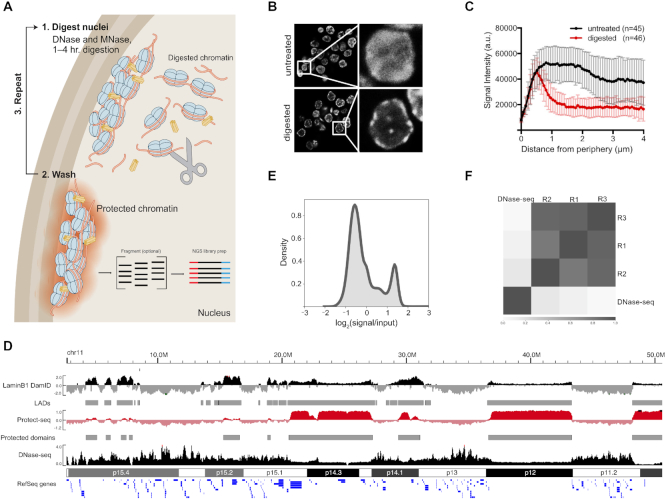
We hypothesized that heterochromatic DNA, which localizes near the nuclear periphery, might be resistant to nuclease digestion (30) and that this nuclease resistance might allow such chromatin to be identified quickly using next-generation sequencing. We performed our initial experiments in the human colon cancer cell line HCT-116. HCT-116 cells are near-diploid, easy to culture, and extensively characterized by the ENCODE and 4DN consortium. Protect-seq relies on the degradation of accessible chromatin by a combination of DNase I and micrococcal nuclease (MNase) via two sequential digestion steps (Figure 1A). The addition of a second nuclease increased digestion efficiency (data not shown). First, HCT-116 cells are crosslinked with formaldehyde, nuclei are extracted with mild detergent, then incubated twice with a cocktail of nucleases and washes (see Materials and Methods, Supplemental File 1). The remaining DNA can be purified (via phenol/chloroform or silica-bead genomic isolation kits) and prepared for NGS.
Figure 1.
Protect-seq principle and assay. (A) Schematic of Protect-seq. DNase I and MNase (scissors) degrade accessible chromatin. The remaining ‘protected’ chromatin is isolated for sequencing. (B) DAPI stain of HCT-116 nuclei with and without nuclease treatment, white boxes represent zoomed image (on right) (C) Spatial quantification of DAPI signal with and without nuclease treatment (D) Next-generation sequencing coverage track of DNase-seq (ENCSR000ENM) (46) (black; accessible chromatin), Protect-seq (red; inaccessible chromatin), and LaminB1 DamID-seq (4DNFIFKMR1J8) (47) (black; LADs). Representative chromosome used (chr11). LaminB1 DamID-seq and Protect-seq data are normalized using Reads Per Genome Content (RPGC) and displayed as log2ratio (signal/input). (E) Kernel density estimation plot (KDE) of Protect-seq signal. (F) Pearson correlation of Protect-seq replicates and DNase-seq in HCT116.
Next, we wanted to confirm accessible/euchromatin was being degraded during the nuclease digestion steps. To address this issue, we imaged nuclei using DAPI staining before and after performing Protect-seq (Figure 1B). As expected, the DAPI signal in the untreated cells is evenly distributed throughout the nucleus whereas, in the treated cells the DAPI signal is predominantly around the periphery with dense punctate occasionally in the center (Figure 1B, C), consistent with published heterochromatin and lamin immunofluorescence (3,4). These data suggest that after Protect-seq digestion the remaining chromatin may be protected via sequestration through tight association with the nuclear lamina and other bodies and surfaces, or by being in a compacted state or phase which prevents nuclease accessibility.
In order to identify the DNA remaining after nuclease digestion, Illumina libraries were prepared and sequenced. The resulting reads were mapped to the human genome (hg38), normalized to reads per genomic coverage and represented as log2(signal/input) (Figure 1D and E and Supplemental Figure S1) (see Materials and Methods). Protect-seq replicates displayed a high degree of similarity via Pearson correlation suggesting the technique is reproducible (Figure 1F and Supplemental Figure S1). In addition to short-read sequencing, Protect-seq has the potential to determine the exact size of the protected region using long-read sequencing approaches. We took purified DNA and ligated adaptors without fragmentation and sequenced using Oxford nanopore. We obtained ∼2 million reads with an average read length of 488 bp and a median length of 399 bp (2–3 nucleosomes) (Supplemental Figure S2). We infer from these data that DNaseI and MNase are able to access both euchromatin and heterochromatin. This observation is consistent with MNase-seq experiments in both human and drosophila cell lines (15,16). As expected, the enriched regions were similar between Illumina libraries and nanopore libraries (r = 0.76). Taken together, prolonged nuclease digestion is an easy and effective method to identify protected regions of the genome and is compatible with both long and short-read platforms.
Protect-seq reveals inaccessible chromatin domains
Next, we aimed to identify and assess the reproducibility of Protect-seq enriched domains (hereafter referred to as protected domains). Hidden Markov Models (HMMs) have been used to identify LADs (31,32). Using a similar approach, we identified 390, 375, 284 domains in each Protect-seq replicate, respectively (Supplemental Figures S1 and S3). To compare our domain calls between replicates, we first quantified the number of overlapping domains in all three samples (n = 249). To calculate the degree of overlap we used the Jaccard similarity coefficient (J) (or intersection over union) which ranges from 0 (no overlap) to 1 (perfect overlap). For this, we observed a Jaccard similarity coefficient of 0.80, 0.76, 0.75 for pairwise interactions (Rep1–Rep2, Rep1–Rep3, Rep2–Rep3) suggesting our replicate domains have a high degree of overlap (Supplemental Figure S1). Another tool for comparisons is the fraction of the genome covered by the domains. Consistent with domain overlap, protected domains covered a similar fraction of the genome in each replicate (30%, 22%, 26%, respectively) (Supplemental Figure S1). Moving forward, we pooled our biological replicates and recalled domains (n = 456). In sum, these data suggest that our method and HMM domain calls are robust and reproducible.
DNase-seq and ATAC-seq identify open chromatin (8,9,33). Open chromatin regions, defined as DNaseI Hypersensitive Sites (DHS), are on average 2–3 nucleosomes wide and have been reported to occupy ∼2% of the genome (8). An alternative approach, DIVA, leverages viral integration to map accessible chromatin (13). In HeLa cells, DIVA-enriched accessible chromatin constitutes ∼50% of the genome (12). If Protect-seq indeed identities inaccessible chromatin then Protect-seq and DNase-seq/DIVA should identify mutually exclusive chromatin. To compare the techniques, we visualized signal tracks on a genome browser (Figure 1D and Supplemental Figure S5). Indeed, Protect-seq enriched regions are very broad and depleted of DNase-seq and DIVA enriched regions. In support of this, DNase-seq and DIVA do not share a high degree of similarity with Protect-seq via Pearson's correlation (DNase-seq r ≤ 0.14, DIVA r = −0.39) (Figure 1D and F). Taken together, we have developed a novel technique to sequence inaccessible chromatin which provides orthogonal information to accessible chromatin techniques.
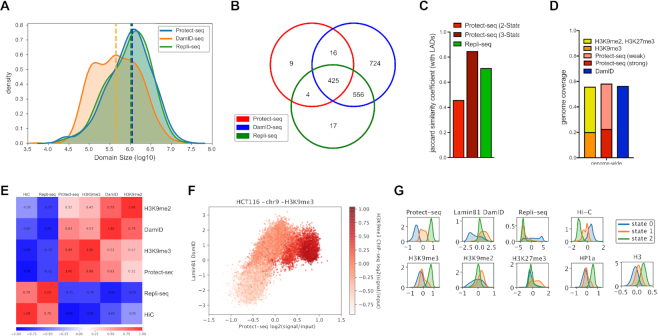
If Protect-seq identifies inaccessible chromatin then we predict that Protect-seq will identify similar chromatin as Lamin DamID-seq and Repli-seq, which are known to identify inaccessible chromatin. LADs (via Lamin DamID-seq) and late-replicating domains (via Repli-seq) are largely overlapping, very broad-ranging in size from ∼80 kb to 30 Mb and invariant across cell types suggesting little plasticity in chromosome organization (6,32,34,35). For comparative analysis between LADs, late-replicating domains, and protected domains, we examined the distribution of domain sizes, domain overlap between techniques, and Pearson's correlation of signal tracks represented as log2(signal/input) in 25-kb genomic windows. As expected, LADs and late-replicating domains are strongly correlated using Pearson's correlation (r = 0.85) and their domains are highly overlapping (J = 0.71). However, late replicating domains are larger than LADs (median: DamID = 445 kb, Repli-seq = 1 Mb). Taken together, LADs and late replicating domains are the same genomic regions (Figure 2A–C, Supplemental Table S2 and Figure S3). When compared to protected domains, LADs and late-replicating domains are also highly correlated via Pearson's correlation (r: LADs = 0.63, late-replicating domains = 0.72) and contained a high degree of overlapping domains (93% overlap) (Figure 2B and E). However, protected domains cover a lower percentage of the genome (23%) when compared to LADs (56%) and the two sets of domains have a low Jaccard similarity coefficient (J = 0.45) (Figure 2C and D). These data suggest that protected domains, although overlapping, do not fully encompass or are a subdomain within LADs. Upon visual examination, we noticed that protected domains can be classified as having a weak or strong signal intensity. To capture weak and strong protected domains based on signal intensity we used a three-state HMM (Supplemental Figure S3). When weak and strong protected domains were joined, the degree of overlap with LADs increased (Jaccard similarity coefficient: two-state = 0.45 versus three-state = 0.84) and genomic coverage of protected domains (58%) was now proportional to LADs (56%) (Figure 2C and D and Supplemental Figure S3). Taken together, these data suggest protected domains are identifying similar genomic regions as LADs and late replicating DNA, which is known to be localized to the nuclear periphery.
Figure 2.
Comparison of Protect-seq domains with LADs, late-replicating domains, and heterochromatin domains in HCT116 cells (A) Kernel density estimation plot (KDE) of domain size in Protect-seq, LaminB1 DamID-seq, and Repli-seq (B) Venn diagram of domain overlap between Protect-seq (two-state HMM), LaminB1 DamID-seq and Repli-seq (C) Jaccard similarity coefficient comparing LADs with Protect-seq domains called using a two-state and three-state HMM and Repli-seq. (D) Fraction of genome covered by histone modifications (H3K9me2, H3K9me3, H3K27me3), Protect-seq domains (strong and weak) and LaminB1 DamID-seq. (E) Pearson correlation of Hi-C eigenvectors, Repli-seq, Protect-seq, LaminB1 DamID-seq, H3K9me3, and H3K9me2 log2ratio bigwigs (25 kb bins) in HCT116. (F) Scatter Plot of Protect-seq signal and LaminB1 DamID-seq, shading in red indicates the strength of H3K9me3 ChIP-seq signal. (G) KDE plots of various genomic features separated by HMM state (three-state)
Protect-seq reveals two types of heterochromatin domains
The above data support a model in which Protect-seq is a simple and robust way to identify inaccessible chromatin. The histone modification H3K9me3 is linked to compacted chromatin in many organisms. We predicted that Protect-seq identified chromatin would therefore correspond to H3K9me3 enriched chromatin. We performed Chromatin immunoprecipitation followed by sequencing (ChIP-seq) using antibodies against H3K9me2, H3K9me3, HP1 α/β (H3K9me3 binding proteins), and H3 (Supplemental Table S1). Then, by combining our data and data available from ENCODE for HCT116, we examined 20 histone and chromatin factors in a combinatorial epigenomic map using chromHMM (36). This approach revealed an enrichment for the repressive histone modifications H3K9me-2/-3, HP1 proteins, and H3K27me3 (Supplemental Figure S4). These data are consistent with previous reports on the histone modifications enriched within LADs (6). To further refine the contribution of repressive histone modifications we used Pearson's correlation. Protect-seq signal tracks have the strongest correlation with H3K9me3 (r = 0.87) and HP1α/β (r = 0.85/0.87) and to a lesser extent H3K9me2 (r = 0.34) and H3K27me3 (r = 0.15), whereas DamID-seq displayed the inverse (Figure 2E and 3A and Supplemental Figure S4). Moreover, Protect-seq is able to quantitatively distinguish H3K9me3, unlike LaminB1 DamID-seq (Figure 2F and G). Identifying an enrichment with H3K9me3 is consistent with previous reports that demonstrated heterochromatin domains are sonication resistant (srHC) and tightly compacted (19). Moreover, Protect-seq domains overlap and correlate with srHC gradient-seq domains and chromatin domains refractory to viral integration using DIVA, despite being from different cell types (Supplemental Figure S5). Interestingly, a three-state HMM is able to separate histone modifications and other genomic features based on the strength of the Protect-seq signal, consistent with the existence of weak and strong domains (Figure 2G and Supplemental Figure S4). In support of these data, k-means clustering of histone modifications within protected domains suggests the existence of an H3K9me3/HP1-enriched cluster and H3K27me3-enriched cluster (Figure 3B). In sum, the data show that Protect-seq identifies regions of chromatin containing high levels of H3K9me3 and HP1 proteins. Interestingly, the data also show that, unlike Lamin DamID-seq, Protect-seq can distinguish between H3K9me3-containing chromatin and H3K9me2/H3K27me3-containing chromatin.
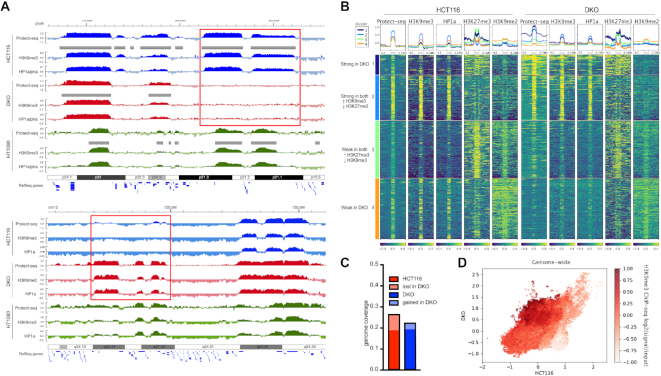
Figure 3.
Protect-seq domains are dependent on the histone modification H3K9me3 and HP1 proteins in colon cancer cells. (A) Genome browser of HCT116 (blue), DKO (red), and HT1080 (green) cells displaying Protect-seq, H3K9me3 and HP1alpha ChIP-seq for cell line. Grey boxes denote domain calls by HMM. Representative example of region that lost Protect-seq and H3K9me3/HP1a signal in DKO cells (top, red box) or gained Protect-seq and H3K9me3/HP1a signal in DKO cells (bottom, red box). (B) Heatmap centered around Protect-seq domains clustered using k-means. Each row represents one scaled Protect-seq domain that includes ± 500 kb of flanking region. Each column is a different signal track with a profile plot above the heatmap (Protect-seq signal, H3K9me3, HP1a, H3K27me3 and H3K9me2 ChIP-seq signal). (C) Genome coverage of protected domains in HCT116 and DKO cells (D) Scatter Plot of Protect-seq signal in HCT116 and DKO cells, shading in red indicates the strength of H3K9me3 ChIP-seq signal.
As further support that Protect-seq is identifying heterochromatin, we asked if Protect-seq correlates with inactive chromosome compartments as measured by high-throughput chromosome conformation capture techniques (Hi-C) which are known to be enriched for repressive histone modifications. A recent report found that heterochromatin, not LAD formation, is the driver behind chromosome compartmentalization (5). Therefore, we predict that Protect-seq will have a higher correlation with inactive compartments than either Lamin DamID-seq or Repli-seq. To address this question, we compared Protect-seq, Lamin DamID-seq, and Repli-seq signal tracks with publicly available Hi-C data in HCT-116 (Supplemental Table S1). Using Pearson's correlation, we observed that Protect-seq has a higher correlation with inactive/B-compartments (r = 0.91), as measured by Hi-C eigenvectors, than DamID-seq (r = 0.62) or Repli-seq (r = 0.70) (Figure 2E and Supplemental Figure S4). Moreover, Protect-seq is able to quantitatively separate A/B-compartments based on signal intensity using HMMs (Figure 2G and Supplemental Figure S4). Taken together, our results suggest that Protect-seq is a simple and reliable tool to identify heterochromatin domains.
Protect-seq domains are dependent on the histone modification H3K9me3 and HP1 proteins in colon cancer cells
If Protect-seq domains correspond to heterochromatin then Protect-seq domains should be dependent on the histone modification H3K9me3. To test this idea, we chose a mutant cell line that has been previously found to have regional defects in H3K9me3 (37). These cells are derived from HCT-116 and contain deletions in DNMT1 and DNMT3b (termed double-knockout or DKO) (38). We performed Protect-seq in DKO cells in triplicate and observed a high correlation amongst replicates (Supplemental Figure S1). When compared to HCT-116, DKO cells have similar numbers of protected domain, similar size distribution, and a high degree of domain overlap suggesting that the defects are not global (Figure 3C and Supplemental Figure S6). However, we observed 168 regions missing in DKO when compared to HCT-116. To examine the chromatin states in these regions, we performed ChIP-seq using antibodies against H3K9me2, H3K9me3 and HP1α/β in HCT116 and DKO cells. We also observed regional defects in H3K9me3 similar to previous reports (37) (Figure 3A and Supplemental Figure S6). Not surprisingly, the regions that lost H3K9me3 and HP1 had less Protect-seq signal suggesting those loci are becoming more accessible (Figure 3A, B and D and Supplemental Figure S6). Conversely, regions that gained H3K9me3/HP1 became less accessible as measured by Protect-seq (Figure 3B and D and Supplemental Figure S6). Interestingly, the domains that lost H3K9me3 were enriched for H3K9me2 suggesting that the chromatin defect in DKO cells is specific to the H3K9 dimethyl to trimethyl conversion (Figure 3A and B and Supplemental Figure S6). These data support our previous observation in HCT116 cells that H3K9me2-containing chromatin does not strongly contribute to Protect-seq enriched chromatin. These data also show that Protect-seq enriched chromatin is dependent on the repressive histone modification H3K9me3 and HP1 proteins in colon cancer cells. Together, from these experiments we conclude that Protect-seq is a new method that can be used to efficiently and quickly identify heterochromatin domains.
Protect-seq is adaptable to additional cell types
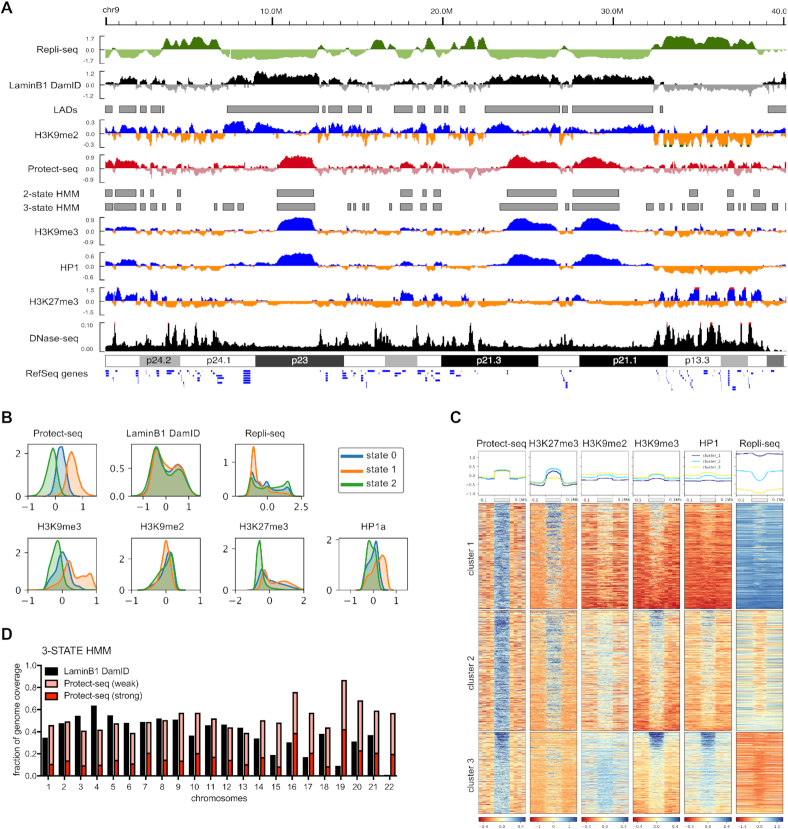
The above data show that Protect-seq is a simple and robust technique for identifying heterochromatin. To address generality and to expand the usage of Protect-seq beyond HCT-116 and DKO cells, we assayed Protect-seq domains in HT1080 cells. HT1080 is a human cell line derived from a fibrosarcoma and consists of immature fibroblasts (39). We performed Protect-seq in HT1080 and observed consistency among replicates, again demonstrating the reproducibility of our technique (Supplemental Figure S1). Protected domains in HT1080 (median = 350 kb) are much smaller than HCT-116 (median = 1.1 Mb) suggesting that protected domains are dynamic and vary between cell types. To characterize the chromatin landscape of protected domains in HT1080 cells, we performed ChIP-seq using antibodies against H3K9me2, H3K9me3, HP1α and H3K27me3 (Supplemental Table S1). In HT1080 cells, protected domains are strongly associated with H3K9me3/HP1α, as expected, but also strongly enriched for H3K27me3 (Figure 4A–C and Supplemental Figures S4 and S7). These data are not surprising given H3K27me3 is known to play a significant role in gene repression during differentiation (40). K-means clustering identified discrete patterns of histone modifications similar to those described above for HCT-116 cells (Figure 4C). The cluster analysis supports the existence of an H3K9me3/HP1 cluster and H3K27me3 cluster, similar to HCT116, but also evidence for a third cluster enriched in both H3K9me3/HP1 and H3K27me3 (Figure 4C). The biological significance of these clusters remains unclear. To ask if protected domains correlate with genomic features associated with the nuclear periphery, we again compared protected domains to LADs and replication timing domains. For this analysis, we performed Repli-seq in duplicates (see Materials and Methods) and used previously curated LADs (31). Again, similar to HCT116, protected domains cover similar percentages of the genome as LADs when weak and strong HMM state are joined (Figure 4D and Supplemental Figure S7) and protected domains overlap with, but do not fully encompass, late replicating domains and LADs (Figure 4A and Supplemental Figure S7). Interestingly, the H3K27me3 cluster is enriched for early replicating DNA (Figure 4A and D). This observation is consistent with a distinct subclass of nucleolar-associated heterochromatin in mouse cells that also lacks lamin association, and tends to be early replicating and associated with H3K27me3 (41). From these data, we conclude that our Protect-seq protocol is compatible with multiple cell types and that protected domains are composed of high levels of H3K9me3-containing and H3K27me3-containing chromatin in HT1080 cells.
Figure 4.
Protect-seq domains are enriched for repressive histone modifications in HT1080 cells (A) Genome browser track showing Log2ratio (signal/input) for Repli-seq, LaminB1 DamID-chip (Meuleman et al., 2015), Protect-seq, H3K9me2/H3K9me3/H3K27me3 ChIP-seq and RPGC for DNase-seq (ENCSR000FDI). Grey boxes below denote domain calls by HMM. Data are pooled from two biological replicates (B) KDE plots for Protect-seq, DamID, Repli-seq and H3K9me2/H3K9me3/H3K27me3/HP1alpha ChIP-seq signal separated based on HMM state. (C) Heatmap centered around Protect-seq domains clustered using k-means. Each row represents one scaled Protect-seq domain that includes ±100 kb of flanking region. Each column is a different signal track with a profile plot above the heatmap (Protect-seq signal, H3K27me3, H3K9me2, H3K9me3 and HP1 ChIP-seq signal, and Repli-seq). (D) Fraction of chromosome coverage by DamID and Protect-seq using a three-state HMM (C) KDE plots of various genomic features separated by HMM state (three-state)
DISCUSSION
In summary, we developed Protect-seq to measure heterochromatin near the nuclear periphery. Protect-seq enables the identification of inaccessible chromatin domains by incubating nuclei with nucleases. Protected domains correlate with lamina-associated domains (LADs) and late-replicating domains. Interestingly, the underlying histone modifications that constitute strong protected domains vary between cell types. This observation suggests Protect-seq could potentially be used to define heterochromatin domains in an unbiased way.
Protect-seq has the potential to distinguish repressive chromatin states. One potential mechanism is that repressive histone modifications could have differential degrees of accessibility and potentially different helical structures (42). Strong protect-seq domains are enriched for the conserved repressive histone modification H3K9me3. These domains are also enriched for HP1 proteins which bind H3K9me3 and condense chromatin (43,44). Perhaps, the compaction of HP1-bound chromatin could lead to nuclease resistance. In support of this, Protect-seq is sensitive to changes in repressive chromatin modifications (i.e. H3K9me3) and HP1 localization suggesting an intimate link between histone modifications and chromatin accessibility. Likewise, weak protect-seq domains correlate with H3K9me2 and H3K27me3 chromatin domains. In Drosophila melanogaster, H3K27me3 domains are densely packed and occupy a smaller volume compared to transcriptionally active regions (45). Then, what is the biological meaning behind weak domains? One possibility is that weak Protect-seq domains could represent heterogeneous domains in the population. In other words, weak domains could be fully accessible in some cells and fully inaccessible in others. Whereas strong Protect-seq domains would be more consistently inaccessible in a population of cells. Future studies using microscopy and single cell approaches will be necessary to interrogate the biological significance of weak protect-seq domains.
Alternatively, Protect-seq enriched regions could be the result of sequestration on the nuclear periphery, not chromatin accessibility. In such a model, Protect-seq enriched regions are tethered/crosslinked to the nuclear lamina and thus insoluble during the wash steps. One way to test this model would be to perform Protect-seq on inverted nuclei. Inverted nuclei contain heterochromatin concentrated in the center of the nucleus, opposed to the typical positioning on the nuclear periphery. Cells with inverted nuclei have normal heterochromatin and 3D structure (5). If heterochromatin, now located in the center, is susceptible to nuclease degradation then radial positioning and sequestration would be favored over chromatin accessibility. In support of this model, MNase can access to both euchromatin and heterochromatin suggesting all chromatin is accessible to nucleases (14–16). Moreover, this model is also consistent with reports that H3K9 methylation is necessary for positioning on the nuclear lamina (2–4). Thus, domains that lose H3K9 methylation, could potentially lose lamina positioning and thus sequestration during the wash steps in Protect-seq. Taken together, we are proposing two potential models (sequestration v chromatin accessibility) to explain the mechanism(s) behind protected domains. Future studies will be necessary to determine the exact mechanism each epigenetic mark contributes with respect to nuclear tethering and chromatin accessibility.
Future development of Protect-seq should expand the types of cells used (i.e. tissue sections, FFPE samples, mouse, fly, worm nuclei). In addition, Protect-seq is performed in nucleo so single-cell (sc)Protect-seq is feasible, given the ability to accurately sort nuclei and create high-quality NGS libraries with adequate genome coverage from low input DNA. And importantly, Protect-seq does not require actively dividing cells, unlike current approaches, thus making the technique amenable to non-model organisms and clinical samples. In sum, Protect-seq is a simple, easy-to-use tool that can be readily adopted without the need for specialized equipment and/or reagents to identify inaccessible chromatin, heterochromatin domains, and potentially identify information about radial position.
DATA AVAILABILITY
The references and accession numbers of published data used and analyzed in this work are indicated in Supplementary Table S1. All data sets generated in this study are deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE135580.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pierre-Oliver Estève, Nezar Abdennur, Anton Goloborodko, and Leonid Mirny for helpful suggestions during the experiments and analysis and Brandon Fields and Scott Kennedy for critical reading of the manuscript. We thank Tasha Josè for help with graphics design. We thank Donald Comb, Rich Roberts, Tom Evans, William Jack and Clotilde Carlow at New England Biolabs Inc. for research support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New England Biolabs, Inc. Funding for open access charge: New England Biolabs.
Conflict of interest statement. None declared.
REFERENCES
- 1. Grewal S.I.S., Jia S.. Heterochromatin revisited. Nat. Rev. Genet. 2007; 8:35–46. [DOI] [PubMed] [Google Scholar]
- 2. Towbin B.D., González-Aguilera C., Sack R., Gaidatzis D., Kalck V., Meister P., Askjaer P., Gasser S.M.. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012; 150:934–947. [DOI] [PubMed] [Google Scholar]
- 3. Solovei I., Wang A.S., Thanisch K., Schmidt C.S., Krebs S., Zwerger M., Cohen T.V., Devys D., Foisner R., Peichl L. et al.. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013; 152:584–598. [DOI] [PubMed] [Google Scholar]
- 4. Kind J., Pagie L., Ortabozkoyun H., Boyle S., de Vries S.S., Janssen H., Amendola M., Nolen L.D., Bickmore W.A., van Steensel B.. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013; 153:178–192. [DOI] [PubMed] [Google Scholar]
- 5. Falk M., Feodorova Y., Naumova N., Imakaev M., Lajoie B.R., Leonhardt H., Joffe B., Dekker J., Fudenberg G., Solovei I. et al.. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature. 2019; 570:395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W. et al.. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008; 453:948–951. [DOI] [PubMed] [Google Scholar]
- 7. Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S.W.M., Solovei I., Brugman W., Gräf S., Flicek P., Kerkhoven R.M., van Lohuizen M. et al.. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 2010; 38:603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E.. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008; 132:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schones D.E., Cui K., Cuddapah S., Roh T.-Y., Barski A., Wang Z., Wei G., Zhao K.. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008; 132:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ponnaluri V.K.C., Zhang G., Estève P.-O., Spracklin G., Sian S., Xu S.-Y., Benoukraf T., Pradhan S.. NicE-seq: high resolution open chromatin profiling. Genome Biol. 2017; 18:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tchasovnikarova I.A., Timms R.T., Douse C.H., Roberts R.C., Dougan G., Kingston R.E., Modis Y., Lehner P.J.. Hyperactivation of HUSH complex function by Charcot-Marie-Tooth disease mutation in MORC2. Nat. Genet. 2017; 49:1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Timms R.T., Tchasovnikarova I.A., Lehner P.J.. Differential viral accessibility (DIVA) identifies alterations in chromatin architecture through large-scale mapping of lentiviral integration sites. Nat. Protoc. 2019; 14:153–170. [DOI] [PubMed] [Google Scholar]
- 14. Mieczkowski J., Cook A., Bowman S.K., Mueller B., Alver B.H., Kundu S., Deaton A.M., Urban J.A., Larschan E., Park P.J. et al.. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 2016; 7:11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chereji R.V., Bryson T.D., Henikoff S.. Quantitative MNase-seq accurately maps nucleosome occupancy levels. Genome Biol. 2019; 20:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwartz U., Németh A., Diermeier S., Exler J.H., Hansch S., Maldonado R., Heizinger L., Merkl R., Längst G.. Characterizing the nuclease accessibility of DNA in human cells to map higher order structures of chromatin. Nucleic Acids Res. 2019; 47:1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Auerbach R.K., Euskirchen G., Rozowsky J., Lamarre-Vincent N., Moqtaderi Z., Lefrançois P., Struhl K., Gerstein M., Snyder M.. Mapping accessible chromatin regions using Sono-Seq. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:14926–14931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D.. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007; 17:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Becker J.S., McCarthy R.L., Sidoli S., Donahue G., Kaeding K.E., He Z., Lin S., Garcia B.A., Zaret K.S.. Genomic and proteomic resolution of heterochromatin and its restriction of alternate fate genes. Mol. Cell. 2017; 68:1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marchal C., Sasaki T., Vera D., Wilson K., Sima J., Rivera-Mulia J.C., Trevilla-García C., Nogues C., Nafie E., Gilbert D.M.. Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat. Protoc. 2018; 13:819–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen S., Zhou Y., Chen Y., Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P.. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015; 31:2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczyński B., Riddell A., Furlong E.E.M.. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012; 44:148–156. [DOI] [PubMed] [Google Scholar]
- 25. Bulut-Karslioglu A., De La Rosa-Velázquez I.A., Ramirez F., Barenboim M., Onishi-Seebacher M., Arand J., Galán C., Winter G.E., Engist B., Gerle B. et al.. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014; 55:277–290. [DOI] [PubMed] [Google Scholar]
- 26. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abdennur N., Mirny L.A.. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 2019; btz540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schreiber J. Pomegranate: fast and flexible probabilistic modeling in python. J. Mach. Learn. Res. 2017; 18:1–6. [Google Scholar]
- 29. Chen F., Gao X., Shilatifard A.. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev. 2015; 29:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilkie G.S., Schirmer E.C.. Purification of nuclei and preparation of nuclear envelopes from skeletal muscle. Methods Mol. Biol. 2008; 463:23–41. [DOI] [PubMed] [Google Scholar]
- 31. Meuleman W., Peric-Hupkes D., Kind J., Beaudry J.-B., Pagie L., Kellis M., Reinders M., Wessels L., van Steensel B.. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013; 23:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kind J., Pagie L., de Vries S.S., Nahidiazar L., Dey S.S., Bienko M., Zhan Y., Lajoie B., de Graaf C.A., Amendola M. et al.. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015; 163:134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crawford G.E., Davis S., Scacheri P.C., Renaud G., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S.. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006; 3:503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peric-Hupkes D., van Steensel B.. Role of the Nuclear Lamina in Genome Organization and Gene Expression. Cold Spring Harb. Symp. Quant. Biol. 2010; 75:517–524. [DOI] [PubMed] [Google Scholar]
- 35. Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K. et al.. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014; 515:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lay F.D., Liu Y., Kelly T.K., Witt H., Farnham P.J., Jones P.A., Berman B.P.. The role of DNA methylation in directing the functional organization of the cancer epigenome. Genome Res. 2015; 25:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rhee I., Bachman K.E., Park B.H., Jair K.-W., Yen R.-W.C., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W. et al.. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002; 416:552–556. [DOI] [PubMed] [Google Scholar]
- 39. Rasheed S., Nelson-Rees W.A., Toth E.M., Arnstein P., Gardner M.B.. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer. 1974; 33:1027–1033. [DOI] [PubMed] [Google Scholar]
- 40. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al.. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 41. Vertii A., Ou J., Yu J., Yan A., Pagès H., Liu H., Zhu L.J., Kaufman P.D.. Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res. 2019; 29:1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Risca V.I., Denny S.K., Straight A.F., Greenleaf W.J.. Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping. Nature. 2017; 541:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hiragami-Hamada K., Xie S.Q., Saveliev A., Uribe-Lewis S., Pombo A., Festenstein R.. The molecular basis for stability of heterochromatin-mediated silencing in mammals. Epigenet. Chromatin. 2009; 2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., Narlikar G.J.. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017; 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boettiger A.N., Bintu B., Moffitt J.R., Wang S., Beliveau B.J., Fudenberg G., Imakaev M., Mirny L.A., Wu C.-T., Zhuang X.. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016; 529:418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dekker J., Belmont A.S., Guttman M., Leshyk V.O., Lis J.T., Lomvardas S., Mirny L.A., O’Shea C.C., Park P.J., Ren B. et al.. The 4D nucleome project. Nature. 2017; 549:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The references and accession numbers of published data used and analyzed in this work are indicated in Supplementary Table S1. All data sets generated in this study are deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE135580.