Abstract
In recent years translation elongation has emerged as an important contributor to the regulation of gene expression. There are multiple quality control checkpoints along the way of producing mature proteins and targeting them to the right cellular compartment, or associating them correctly with their partners. Ribosomes pause to allow co-translational protein folding, protein targeting or protein interactions, and the pausing is dictated by a combination of the mRNA sequence and structure, the tRNA availability and the nascent peptide. However, ribosome pausing can also lead to ribosome collisions and co-translational degradation of both mRNA and nascent chain. Understanding how the translating ribosome tunes the different maturation steps that nascent proteins must undergo, what the timing of these maturation events is, and how degradation can be avoided when pausing is needed, is now possible by the emergence of methods to follow ribosome dynamics in vivo. This review summarizes some of the recent studies that have advanced our knowledge about co-translational events using the power of ribosome profiling, and some of the questions that have emerged from these studies.
INTRODUCTION
Precise control of gene expression is essential for the healthy growth and development of all organisms. At the cellular level, gene expression is the production of the right amount of well-folded protein at the right time, effectively interacting with its cellular partners and targeted to the correct cellular compartment. It entails controlled production of functional proteins, as well as clearance of faulty proteins. Research over the last 20 years has revealed an astonishing level of integration between the different steps of the gene expression pathway. Overall rates of mRNA synthesis and degradation are buffered with reciprocal adjustments (for recent review see (1)). In addition degradation of mRNAs is intimately connected to the process of translation, while clearance of faulty mRNAs and dead-end protein products are coordinated. The ribosome has emerged as an amazing hub at the core of this integrated regulation.
In eukaryotes the steady state pool of mRNAs available for protein synthesis in the cytoplasm is defined by the rates of mRNA synthesis and decay. Cytoplasmic mRNAs can also be partitioned into a variety of different granules (for review, see (2,3)), whose exact roles in defining the pool of mRNAs available for active translation still needs to be clarified. Regulation of gene expression has traditionally been attributed to changes in transcription and translation initiation targeted by signaling pathways, and mRNA degradation has been considered to occur post-translationally because ribosomes protect mRNAs from degradation (discussed in (4)). We now know that mRNA degradation occurs co-translationally, and that translation elongation is not uniform, undergoing multiple quality control checkpoints. Its pattern depends upon the mRNA sequence and structure, the availability of charged tRNAs and translation factors, the nascent chain, as well as upon a myriad of factors interacting with mRNA, ribosome or nascent chains (for review, see (5)). Hence, translation elongation is also an important regulated step that contributes to gene regulation. This review will summarize several recent studies addressing this role of the translating ribosome. In particular, it will focus on those that have benefitted from the development of technologies that allows following the translation elongation process at single codon resolution. Finally open questions will be discussed.
CO-TRANSLATIONAL mRNA DEGRADATION
The processivity of translation elongation defines overall translation efficiency. Decoding is the first step and is a competition between all charged-tRNA complexes until the correct codon–anticodon pairing occurs. The rate will depend upon the relative availability of the right charged tRNA compared to the entire pool of available tRNAs. The ribosome stabilizes the cognate codon–anticodon pair and accelerates the activation of the EF-Tu GTPase and the accommodation of the correct amino-acyl tRNA in the A-site. This process is known as the kinetic discrimination mechanism and is used by the ribosome to maintain a high speed and fidelity during elongation (6,7). Modifications of some tRNAs also impacts the rate of chain elongation. In each kingdom of life some codons are used more and some are used less. In general the amount of tRNA that recognizes rare codons is lower and the elongation rate correlates well with the total tRNA pool, suggesting that the charging of tRNAs is not rate limiting (reviewed in (8)).
In 2015, the Coller laboratory published a study indicating that the overall codon composition of mRNAs, namely the relative percentage of codons above a certain threshold of optimality to the percentage of those below this threshold, correlates with mRNA half-life genome-wide (9). Optimality was defined according to the tRNA adaptive index, itself reflecting the efficiency of tRNA usage, and the recognition of the codon by the translation apparatus. They found that the changes in mRNA stability according to codon optimality were mediated by mRNA deadenylation and decapping, followed by 5′ to 3′ exonucleolytic degradation. Their findings place the ribosome at the core of a regulatory system linking decoding and translation elongation to mRNA degradation.
Co-translational mRNA decay was known to occur in the cases of faulty mRNAs, even though generally mRNA degradation was believed to occur after ribosome removal (10). For instance mRNAs with premature termination codons are turned over by a mechanism called non-sense mediated mRNA decay (NMD) for which the Upf1 protein acts as a hub (Figure 1A) (for review, see (11)). Intriguingly 3–10% of all mRNAs are upregulated in the absence of Upf1, even mRNAs without evident premature termination codons (12,13). This questions whether NMD might not only target faulty mRNAs. Another case of mRNA degradation driven by the ribosome occurs when ribosomes reach the end of an mRNA or a poly(A) tail without encountering a stop codon (NSD or non stop decay), or if the ribosome stalls, either because of stretches of rare codons, higher order mRNA structures, amino acid starvation, tRNA deficiencies or oxidative stress (NGD or no go decay) (Figure 1A) (for review, see (14)). Originally a distinction was made between NSD and NGD. However, it could be that the mechanisms leading to mRNA decay are mostly similar, though additional components may be needed to resolve ribosomes stalling on poly(A) sequences at the mRNA 3′ end. NGD has been extensively studied using reporters with a poly lysine motif or stretches of rare CGA arginine codons inserted between two reporter proteins (for reviews see (14–16)). An endonucleolytic event is triggered by ribosome collision, and factors homologous to the eRF1 and eRF3 termination factors are recruited to split ribosomes with the Rli1 ATPase. This allows recycling of the ribosomes and access of the mRNA to exonucleases (Figure 1A).
Figure 1.
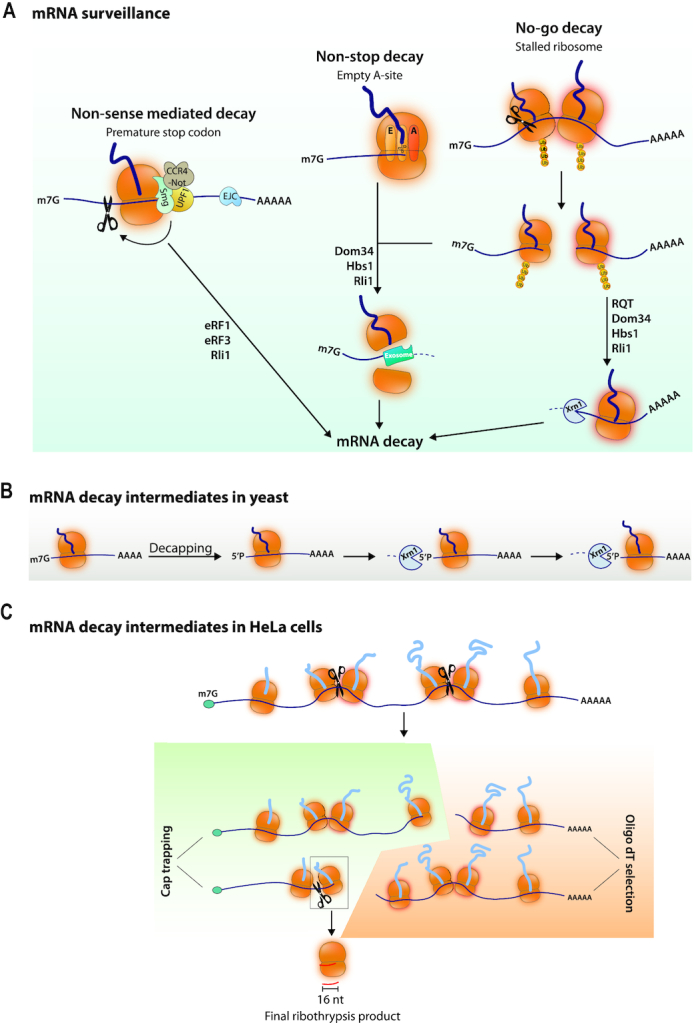
(A) mRNA surveillance induced mRNA decay. NMD occurs when mRNAs have a premature stop codon. In mammalian cells NMD works with Upf1, through the recruitment of the Smg5–7 proteins. Smg5 and Smg6 have a PIN domain associated with endonucleolytic activity and Smg6 has single stranded nuclease activity. Smg7 can recruit the Ccr4-Not deadenylase complex. Recognition of NMD substrates has to do with the distance from the real termination site where termination factors and the poly(A) binding protein are located and/or the presence of a downstream exon-junction complex (EJC). Substrates for NSD and NGD are either mRNAs on which the ribosomes will reach the end of the mRNA with an empty A site or where ribosomes stall. This will trigger ribosome collision that induces ribosomal subunit ubiquitination, endonucleolytic cleavage and exonucleolytic decay of the 5′ and 3′ mRNA fragments generated. The endonuclease has not been identified. The Dom34 and Hbs1 factors in yeast (called Pelota and Hbs1L and Gtpbp2 in mammals) recruited to empty A sites split ribosomes with the help of the Rli1 ATPase (called ABCE1 in human). Then the 5′ mRNA can be degraded by the exosome whereas the 3′ mRNA can be degraded by the Xrn1 5′ to 3′ exonuclease. In yeast an RQT complex contributes to ribosome splitting. (B) mRNA decay intermediates in yeast measure ribosome dynamics. Decapping produces 5′P mRNA ends that are substrates for the Xrn1 5′ to 3′ exonuclease. mRNA decay fragments of different lengths have 5′ ends showing 3-nucleotide periodicity. (C) mRNA decay intermediates in Hela cells. Purification of capped mRNAs and sequencing of their 3′ ends, or purification of poly(A) mRNAs and sequencing of their 5′ ends has revealed 3′ and 5′ ends respectively, within ORFs. The model is that ribosomes pause during translation elongation and this can create a ribosome collision, and then a chain of events similar to NGD called Ribothrypsis. The final product is a 16-nucleotide fragment resulting from the reiterated cleavage events.
As mentioned above co-translational mRNA decay was first connected to faulty mRNAs. However the importance of overall codon composition for mRNA stability indicates that mRNA degradation is generally intimately connected to translation. Two recent studies have confirmed this. In the first study (17), mRNA degradation was profiled in budding yeast by isolating and sequencing 5′ monophosphate mRNAs. 12–14% of the poly(A) mRNA pool was estimated to be in the form of such 5′ decay intermediates, a percentage that could be increased by treatment of cells with cycloheximide (CHX) that increases elongation-paused ribosomes. These mRNA degradation intermediates were detected with size differences showing a 3-nucleotide periodicity and dependent upon the Xrn1 5′ to 3′ exonuclease. This suggested that Xrn1, and hence decay intermediates, measures ribosome dynamics and that Xrn1 follows the last translating ribosome (Figure 1B). Consistent with this model, a structure of Xrn1 associated with ribosomes from yeast shows Xrn1 located at the mRNA exit site, where mRNA can be exposed to the active center of the nuclease (18). In the second study (19), a technique called Akron Seq that captures native 3′ ends of capped mRNAs and 5′ ends of poly(A) mRNAs was developed. Applied to Hela cells it showed that mRNAs undergo co-translational, ribosome-phased, endonucleolytic cuts at the exit site of the mRNA ribosome channel, a mechanism named ribothrypsis (Figure 1C). 63% of capped mRNAs were found to have ends within coding regions, and the mRNA ends showed 3-nucleotide periodicity. Xrn1 was not involved in this profile of cleavages. A similar percentage of polyadenylated RNAs had 5′ ends within coding sequences and also showed the 3-nucleotide periodicity. Final ribothrypsis products from reiterated cleavage events are 16-nucleotide long (Figure 1C) and such fragments have been detected in RNA deep sequencing data from strains where exosome function has been compromised (20). Such 16 nt long products also accumulate in yeast cells lacking Dom34, an ortholog of mammalian eRF1, responsible for ribosome splitting during NGD, and they are presumed to be NGD mRNA truncation products (21).
The mRNA decay patterns or intermediates detected overall in human and yeast are surprisingly reminiscent of those produced upon co-translational mRNA degradation occurring in response to ribosome stalling in yeast. This suggests that co-translational mRNA decay might occur with a certain frequency during translation of normal mRNAs, in the various situations where the ribosomes are made to pause. However, as will be described below, pausing is beneficial and needed for nascent chain folding and protein interactions. Thus cells must have evolved a mechanism that prevents catastrophic collision of ribosomes every time ribosomes pause. Evidence for the existence of such a protective mechanism has been uncovered in a recent study of co-translational association of proteins (see further).
STALLED RIBOSOMES COORDINATE DEGRADATION OF NASCENT CHAINS WITH mRNA DECAY
Synthesis of proteins starts at the ribosome peptidyl transfer center (PTC) and the nascent peptide travels through the ribosome tunnel to the cytosol. It undergoes constraints and multiple interactions already in the tunnel, and as soon as it emerges out of the tunnel it undergoes multiple modifications and folding interactions. Cells have evolved a sophisticated machinery to ensure that the proteins undergo the different maturation steps in the right order. Ribosome stalling interrupts this process and produces aberrant nascent protein products that can be toxic. A mechanism called Ribosome Quality Control (RQC) coordinates the degradation of these aberrant products with mRNA decay and ribosome recycling (for reviews see (14,16)) (Figure 2A). NGD in the context of RQC (NGDRQC+) and RQC start after ribosome stalling, when the following ribosome bumps into the stalled ribosome and collision occurs.
Figure 2.
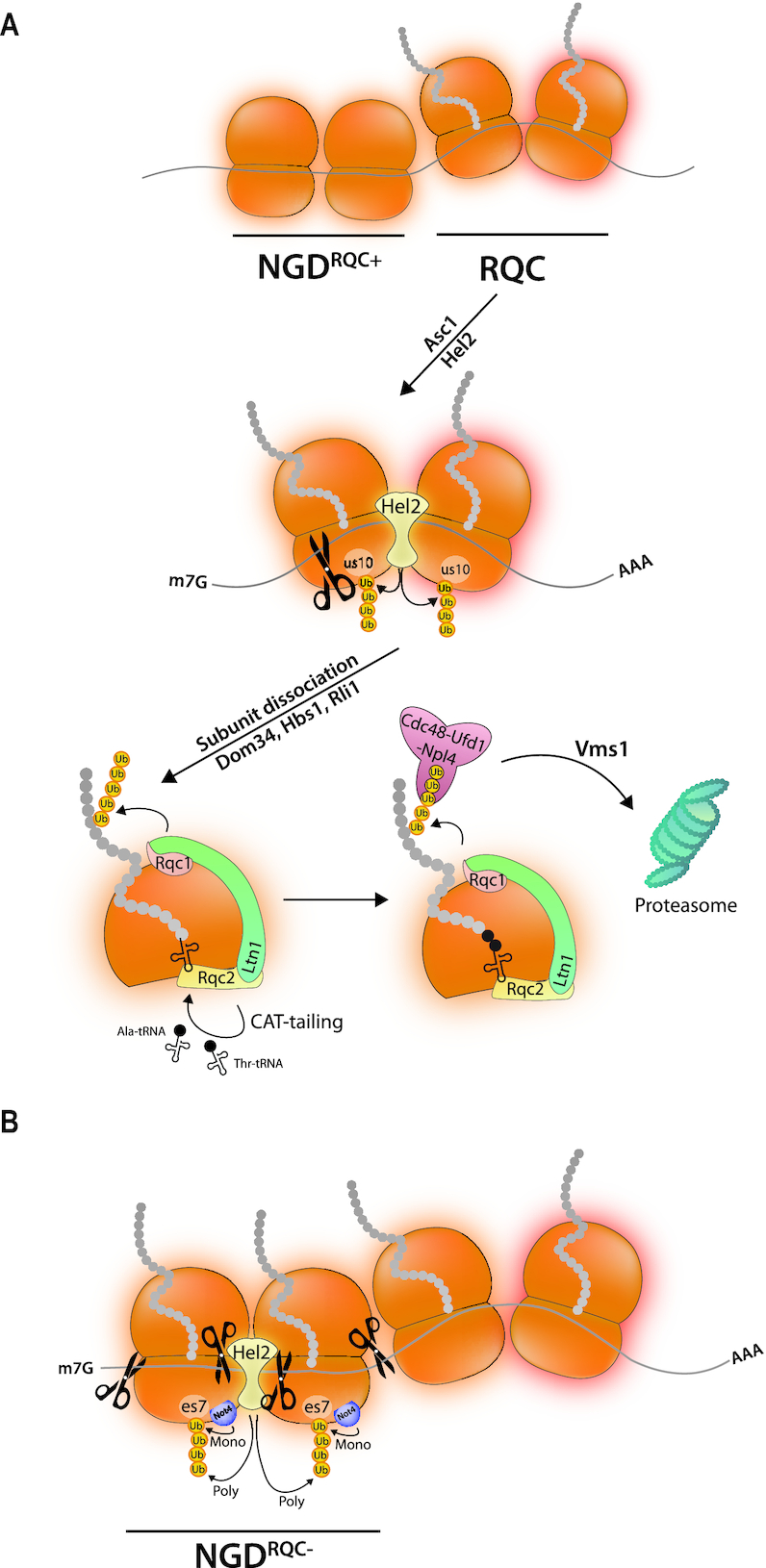
(A) The ribosome tunes nascent chain degradation to mRNA decay. When ribosome collision occurs upon ribosome pausing, RQC is initiated together with NGD by ubiquitination of a ribosomal protein, us10 in yeast, by the Hel2 E3 ligase. This leads to endonucleolytic cleavage and ribosome subunit dissociation involving Dom34, Hbs1, Rli1 and the RQT complex. The released 60S subunit with its peptidyl-tRNA produces a surface recognized by the RQC complex, in particular by its Rqc2 subunit. The Ltn1 E3 ligase ubiquitinates the nascent peptide, which is extracted by Cdc48 and Vms1, and the chain is then targeted to the proteasome. CAT-tails are added to the nascent chain. (B) An alternative pathway occurs when RQC is not functional. The Hel2 E3 ligase poly-ubiquitinates es7 that is first mono-ubiquitinated by the Not4 E3 ligase. This ubiquitination event also leads to endonucleolytic cleavages occuring upstream of the collided ribosome.
RQC happens on the collided ribosomes and requires ubiquitination of a 40S ribosomal protein (us10 in yeast, eS10 in mammalian cells) by an E3 ligase. The latter (Hel2 in yeast and ZNF598 in mammalian cells) recognizes a broad interface of the 40S disome unit of the collided ribosomes (22,23). This recognition occurs with the contribution of Asc1 (RACK1 in mammals), a ribosome-associated protein located on the 40S head. Initiation of the RQC pathway also requires the RQC-trigger complex (RQT), which is composed of the RNA helicase Slh1/Rqt2, the ubiquitin-binding protein Cue3/Rqt3 and yKR023W/Rqt4 (24). Endonucleolytic cleavages within the disome unit happen after ubiquitination of the 40S ribosomal protein and the ATPase activity of Shl1/Rqt2 is essential for the process. Ribosome splitting finally occurs (23) and the mRNA is degraded by exonucleases. The nascent chain is still associated with the 60S subunit in the form of peptidyl-tRNA. A protein complex called RQC will dock and ensure degradation of this stalled nascent chain. The RQC complex includes an E3 ligase (Ltn1 in yeast and Listerin in human) that ubiquitinates the nascent chain as well as Rqc2, a subunit that recognizes the 60S with its peptidyl-tRNA (NEMF in human) and Cdc48 (p97 in human) that with its cofactors will extract the ubiquitinated nascent peptide for proteasomal degradation. This requires an additional factor, Vms1 (ANKZF1 in human), a Cdc48-interacting protein (and either a tRNA hydrolase (25) or tRNA nuclease (26)). Rqc2 additionally stimulates elongation of stalled nascent peptides by multiple alanyl and threonyl residues (CAT-tailing), producing CAT-tailed peptides to safely degrade aberrant peptides. Indeed the addition of residues allows lysines buried in the ribosome tunnel to be pushed out and made accessible for ubiquitination by Ltn1 (27). CAT-tails have been shown to increase protein aggregation in RQC defective strains resulting in proteotoxic stress (28).
An alternative mechanism (NGDRQC−) can occur in yeast when the ribosomal protein us10 cannot be ubiquitinated, for instance due to the absence of Hel2, the absence of Slh1/Rqt2 or because of a point mutation in us10 (23). It is presumed to be as effective as NGDRQC+. NGDRQC− happens on ribosomes upstream of the collided disome unit. It involves mono-ubiquitination of es7 by the Not4 E3 ligase (29) followed by K63-linked polyubiquitination by Hel2. It leads to mRNA cleavage by mechanisms that remain to be clarified (Figure 2B).
Since degradation of nascent chains occurs in parallel to co-translational mRNA degradation, it seems all the more likely that cells must control ribosome collision in response to ribosome pausing. If not, cells spend futile energy synthesizing peptides that will be degraded before they are even useful. Moreover there are many situations where ribosome pausing is important for production of proteins as will be discussed in the following sections.
CO-TRANSLATIONAL CHAPERONE-ASSISTED PROTEIN FOLDING
Proteins are produced from their N-terminus to their C-terminus and emerge vectorially in the cytoplasm. Translation is slow in comparison to folding that can already start within the space constraints of the ribosome tunnel. Then emerging nascent chains can interact with the surface of the ribosome where other factors will contribute to their co-translational folding (for review see (30)). The velocity of elongating ribosomes can define the folding of the nascent peptide. For instance it is presumed that rare codons, overrepresented in the first 90–100 nucleotides of open reading frames (ORFs) of all kingdoms of life and often found at boundaries of protein domains, slow translation elongation to help vectorial folding of proteins. This has been corroborated through direct measurements in vitro and in vivo where synonymous codon changes can change solubility, sensitivity to protease and conformation (reviewed in (8)). Consequently, ribosome pausing can be required for appropriate protein folding.
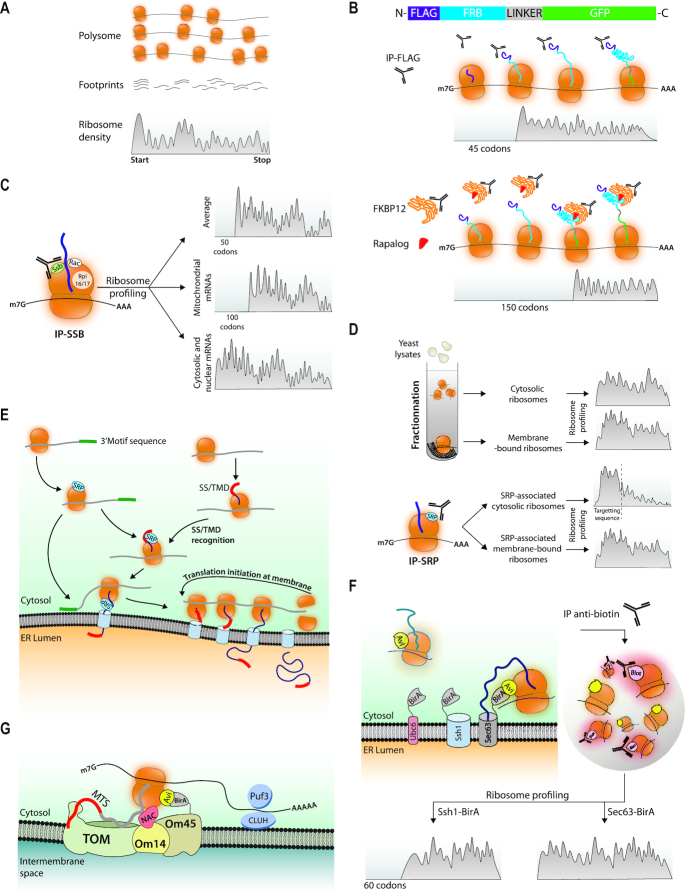
In 2009, a tool to follow translation elongation at single codon resolution in vivo was developed (31). This technique involves the preparation of monosomes from cell extracts digested with RNAse and purifying the ribosome protected mRNA fragments followed by deep sequencing. Then the identified ribosome-protected fragments are aligned with reference sequences. With the size of the protected footprint and the position of the P site within the ribosome-protected fragment, one can get a snap shot of the density of ribosome P sites occupying each codon of the genome (Figure 3A). If translation elongation occurs at a constant rate, ribosome footprints will be evenly distributed throughout an ORF, whereas if ribosomes are slower to translate through specific codons, there will be relatively more ribosome footprints aligning to those particular codons.
Figure 3.
Ribosome profiling associated with selective purification steps determines timing of co-translational folding, chaperone binding and targeting. (A) Ribosome profiling. This is a technique in which total polysomes prepared from a given sample are treated with RNAse to generate monosomes from which the ribosome-protected fragments called footprints are purified and used to make a library that is sequenced. Each fragment is defined by the position of the ribosome P site and the total amount of footprints aligning to each codon normalized to the total complexity of the library (RPKM, y axis) are aligned to reference sequences (x axis). The hypothetical footprints on one ORF are depicted. (B) Ribosome profiling to evaluate co-translational protein folding from cells expressing Flag-FRB-GFP. Upper panel: Ribosome footprints recovered in a Flag immunoprecipitate map to the ORF starting at a position corresponding to ribosomes that had just synthesized enough of the nascent chain to expose the Flag epitope outside of the ribosome tunnel. Lower panel: Ribosome footprints recovered in a FKBP12 immunoprecipitate in presence of a Rapalog map to the ORF starting at a position corresponding to ribosomes that had just synthesized enough of the nascent chain to expose FRB outside of the ribosome tunnel. (C) Ribosome profiling to evaluate co-translational chaperone recruitment. Ssb was immunoprecipitated and the ribosome footprints recovered in the immunoprecipitate were mapped. A large majority, but not all, mRNAs were recovered and the patterns of the ribosome footprints depended upon the category of the proteins encoded by these mRNAs. (D) Ribosome profiling to evaluate Srb targeting to the ER. Upper panel: Cytosol or membranes were fractionated from total extracts and the ribosome footprints recovered in the 2 fractions were mapped. Footprints tended to be distributed throughout the ORFs. Then Srb was immunoprecipitated and the ribosome footprints recovered in the immunoprecipitate were mapped. Lower panel: The Srb-engaged ribosomes from the cytosolic pool showed a gradual depletion of footprints at a position corresponding to exposure of the signal sequence outside of the ribosome tunnel. The Srb-engaged ribosomes from the membrane pool showed mostly distribution throughout the ORF. (E) Proximity labeling at the ER. The Sec61 or Ssh1 transolocon subunits or the C-terminal tail-anchor of Ubc6 were fused to BirA in cells where a ribosomal subunit of either the 40S or of the 60S subunit was fused to an Avi-tag. Ribosomes in proximity to the BirA tagged proteins get biotinylated and can be purified to then map ribosome footprints. (F) Updated model of protein targeting to the ER. The mRNAs encoding proteins with or without a signal sequence recruit SRP before any signal sequence is exposed out of the ribosome tunnel, and 3′UTR sequences can contribute to SRP recruitment as well targeting to the ER (82). SRP is then poised to associate with the signal sequence and dock the RNCs to its receptor and transfer them to the translocon. mRNAs targeted to the ER stay for new rounds of translation. (G) Proximity labeling to investigate targeting of ribosomes to the mitochondrion. BirA was fused to Om45. Om45 interacts with Om14 that acts as a receptor for ribosomes translating mitochondrial proteins via the NAC chaperone, leading to nascent chain transfer to the TOM complex. Targeting to the mitochondria has been attributed on one hand to the nascent chain MTS (mitochondrial targeting sequence) and on the other to the mRNA via RNA binding proteins such as Puf3 in yeast or CLUH in mammalian cells (83–87)).
Ribosome profiling has confirmed the expectation that ribosome density is not uniform across ORFs. There are ribosome pile-ups at certain codons consistent with pausing. So far simple rules to explain the distribution of ribosome reads have not been established (for review, see (32)). In vitro proline acts poorly as a peptidyl donor and acceptor (33). Consistently, ribosome footprints accumulate at proline codons, particularly at stretches of proline codons in vivo (34). Rare codons were originally not found to be an important source of ribosome footprint accumulation (35–38), but several recent studies have instead confirmed that when profiling is performed without pretreatment of cells with the translation elongation inhibitor cycloheximide there is a strong positive correlation (39,40). An important point to underline is that a major difficulty in establishing rules from the translation elongation process that govern ribosome footprint profiles is that the production of the cDNA library from the footprints produces biases. Elements such as sequences at the end of footprints or substrate specificity of the enzymes used during cDNA library preparation and sequencing, bring in variations (41).
The power of ribosome profiling has been used to monitor in vivo the co-translational folding of nascent peptides (42). For instance, in one study a cell line expressing a fusion of the Flag epitope to FRB (FKBP-Rapamycin Binding domain of mTOR) and GFP (Green Fluorescent Protein) with optimal codons placed in the linker region between FRB and GFP was used to isolate nascent chains and monitor when during synthesis the FRB domain actually folded (Figure 3B). The Flag epitope could immunoprecipitate ribosome-nascent chain complexes (RNCs) after 50 codons were translated, so as soon as it was exposed outside of the ribosome tunnel (35–40 amino acids occupy the ribosome tunnel (43) and 10 amino acids correspond to the epitope itself) (Figure 3B, upper panel). Antibodies to FKBP12 that interacts with folded FRB in the presence of a Rapalog could immunoprecipitate RNCs after 150 codons, so as soon as the 100 amino acids of the FRB domain emerged from the ribosome tunnel (Figure 3B, lower panel). This indicated that FRB was folded as soon as it emerged from the ribosome.
Chaperones contribute to effective folding of proteins, and the first chaperones to encounter the nascent chains are the ribosome-associated chaperones. In yeast, the ribosome-associated chaperones are the nascent polypeptide associated complex NAC (44,45) and the Hsp70 Ssb with its RAC (ribosome-associated-complex) co-chaperone, composed of the J-domain protein Zuo1 and the non-canonical Hsp70 Ssz1 (46). These chaperones are not the only ribosome-associated proteins that participate in early interactions with nascent chains. Other players include modifying enzymes such as the N-terminal aminopeptidases (47) and N-acetlytransferases (48), as well as the ER targeting factor SRP (49). All of these factors have overlapping binding sites at the exit of the ribosomal tunnel ((50) and for reviews see (30,51)). This of course raises the question of the timing and specificity of the association of the different factors with the nascent chain in vivo (30). In particular, if protein domains fold as soon as they are exposed from the ribosome tunnel, are the chaperones the first to associate with the nascent chains?
Ssb associates with the 60S ribosomal subunit, and like other Hsp70s it binds linear stretches of hydrophobic residues expected to occur in all unfolded protein sequences (46). Two studies have investigated in budding yeast which nascent chains are actually engaged by Ssb in vivo, and at what time during translation. The Frydman laboratory compared the mRNAs enriched in the Immunoprecipitate (IP) of Ssb relative to the translatome defined by the IP of two different ribosomal proteins (52). They found that Ssb associated with only approximately 65% of the translatome, preferentially with nascent cytosolic and nuclear proteins, and RAC enhances overall Ssb binding and contributes to define its specificity. Bioinformatic analyses indicate that the specificity of Ssb binding correlates with the severity of the challenges facing co-translational folding of nascent chains and with mRNAs displaying a moderate or slow translation rate. In 2017, the Bukau laboratory used the power of ribosome profiling to further characterize the pattern of nascent chain binding pattern by Ssb (53) (Figure 3C). They determined that on average Ssb engaged nascent chains after 50 codons, as soon as 15–20 residues of the nascent chain were exposed out of the ribosome tunnel. The patterns of Ssb engagement to its clients depended upon the destination of the proteins. Ssb-engaged RNCs of cytosolic and nuclear proteins displayed footprints throughout the ORF with patterns suggestive of cycles of binding throughout the synthesis of the protein. Ssb also engaged organellar proteins, notably more than 80% of mitochondrial proteins, where Ssb engaged nascent chains often after ∼100 amino acids. Ssb also engaged ∼40% of ER-targeted proteins showing more complex patterns of binding. Interestingly Ssb-engaged RNCs showed more footprint reads near the translation initiation site in the absence of RAC, suggesting that initiated ribosomes progressed more slowly to the elongation phase without Ssb-RAC.
These experiments have shown that although Ssb can bind the ribosome in vitro, in vivo it is recruited with specificity to ribosome-nascent chain complexes, both in its choice of targets and timing of binding. Moreover Ssb-RAC appears to be important for translation elongation after initiation. A consequence of this observation is that Ssb-RAC not only impacts folding of proteins, but it might also contribute to modulate ribosome collisions at downstream pause sites, and thereby have an impact on productive translation and mRNA turnover rates.
CO-TRANSLATIONAL ORGANELLAR TARGETING
Cytoplasmic ribosomes translate proteins that must be targeted to different compartments. One third of the cell's mRNAs synthesize proteins that will transit or be directed to the ER. The co-translational targeting of nascent chains to the ER has been intensely studied in vitro and the successive steps are well established (for review see (54)). SRP first recognizes the linear hydrophobic signal sequences or the transmembrane domains, then docks onto its receptor on the ER membrane and finally starts transferring the nascent chain-ribosome complex to the translocon channel. However, as mentioned above, in vivo SRP might be in competition with other factors to associate with nascent chains. Several studies have investigated the selectivity and timing of SRP association with ribosomes in vivo. Surprisingly the in vivo targets of SRP in budding yeast are not only mRNAs bearing a targeting sequence, but also some nuclear and mitochondrial mRNAs (55). Moreover ribosome footprints of membrane-associated RNCs isolated by fractionation methods are distributed throughout ORFs (56) (Figure 3D). This indicates that RNCs are close to the ER before synthesis of any signal sequence, and that de novo initiation must occur on mRNAs at the membrane, possibly after a first round of translation targets them there. SRP-associated RNCs are both membrane-bound and cytosolic, though the majority is membrane-bound, and all have ribosome footprints throughout the ORFs. This argues against the canonical model proposing that SRP recognizes the nascent chain only after the targeting signal is exposed. Nevertheless the cytosolic pool of SRP-associated RNCs show a gradual loss of ribosome footprints at a point corresponding to the exposure of the signal sequence from the ribosome tunnel. This indicates that when the signal sequence is exposed there is a competition between continued elongation in the cytosol and SRP-dependent targeting to the membrane. If in vivo SRP can engage nascent chains without a signal sequence, but yet shows very few off-targets, this is because specificity is defined by combined contributions of non-coding mRNA sequences, ongoing translation and the nascent chain (see model on Figure 3E).
In 2014, the Weissman group (57) established a protocol to analyze global translation in defined cellular locations that avoids cell fractionation. This method involves expressing a spatially restricted biotin ligase (BirA) to mark ribosomes containing a biotin acceptor peptide (Avi-Tag), and purification of biotinylated ribosomes followed by ribosome profiling with the purified RNCs. It was applied to the ER (Figure 3F) (57). In both yeast and human, ER-enriched gene sets were exclusively from the secretome and defined most of the secretome, indicating that most secretory proteins are co-translationally translocated in vivo, even proteins that can be inserted post-translationally into microsomes in vitro and proteins that are independent of SRP (58). Moreover this work showed that transfer of RNCs to the different translocons show different ribosome footprint patterns and hence timing of targeting (Figure 3F).
In vitro proteins can be imported post-translationally into mitochondria and most of our knowledge about mitochondrial import stems from in vitro studies (for reviews see (59,60)) but several reports indicate that nuclear-encoded mitochondrial mRNAs are targeted to the mitochondrial surface (for review see (61)). Consistently, mitochondrial RNCs could be strongly enriched using a fusion of BirA to the mitochondrial outer membrane protein Om45 (62) (Figure 3G). Enrichment of the annotated mitochondrial proteome (mitoP2) was only 27% but it could be increased to 68% with the translation elongation inhibitor cycloheximide, suggesting that there is a competition between translation elongation in the cytosol and targeting to the mitochondrion.
The comparison of ER and mitochondrial proximity labeling experiments revealed few proteins with dual ER and mitochondrial localization. However a recent study in yeast suggests that during their production mitochondrial proteins can transit by the ER where an ER- localized J protein called Djp1 safeguards their import to the right compartment via a retrieval pathway (63). We do not know whether in these situations RNCs may have first been targeted to the ER, but one consideration to keep in mind with the proximity labeling approach is the possibility that not all tethered translating ribosomes are accessible to the BirA proteins used.
CO-TRANSLATIONAL COMPLEX ASSEMBLY
Proteins often do not work alone, but assemble into homo- or hetero-oligomeric complexes to form ‘cellular machines’. The assembly process contributes in an important way to shape the cellular proteome. Some proteins can assemble post-translationally. For this, they must be available in appropriate quantities at the same time and adopt assembly-competent forms with half-lives sufficient to provide them time to meet by diffusion (Figure 4A). Alternatively, proteins can associate co-translationally, if one fully synthesized subunit is recruited to the nascent chain of its partner subunit as it is still being synthesized by the ribosome, and as soon as the domain of interaction folds (Figure 4B, upper panel). This mechanism can concern not only subunits of protein complexes but also dedicated chaperones recruited to their clients (Figure 4B, lower panel). Finally two nascent chains still being synthesized can interact if their mRNAs are translated in close proximity (Figure 4C). This can occur between nascent chains of partner subunits (upper panel), but also between nascent chains of the same protein and in this case maybe also between nascent chains being synthesized by two ribosomes on the same mRNA molecule (lower panel) (for reviews, see (64,65)).
Figure 4.
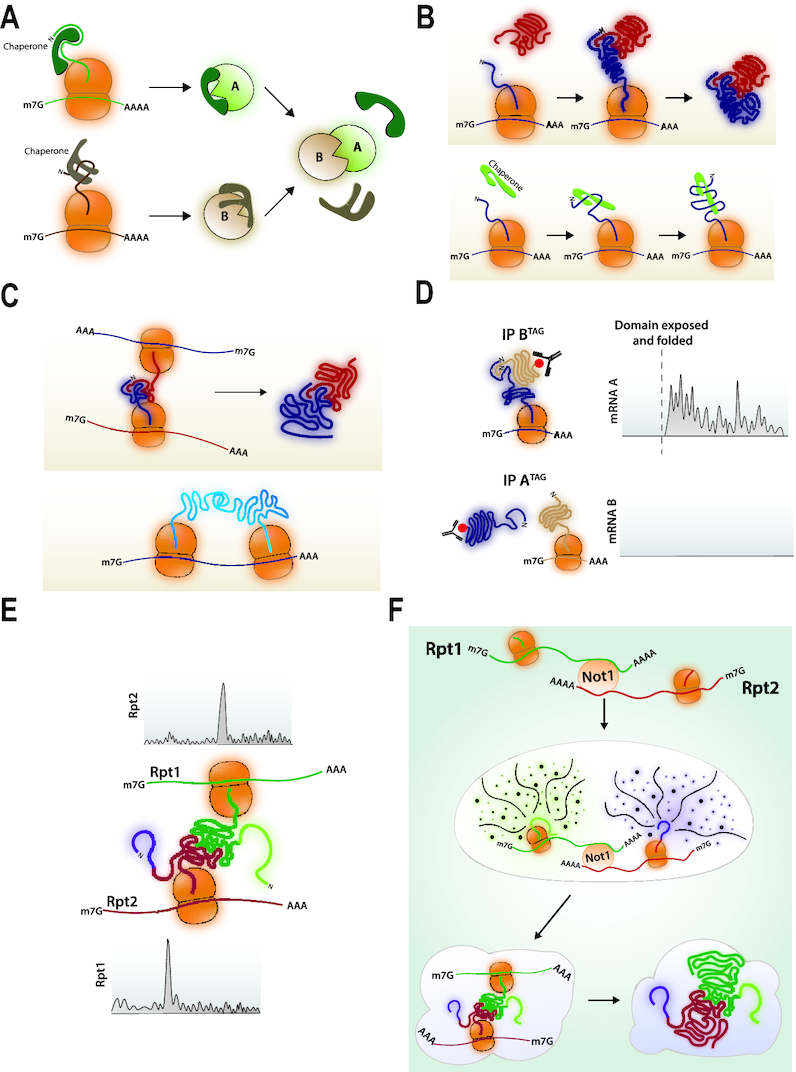
(A) Post-translational protein association. Proteins A and B are synthesized separately and kept soluble and assembly competent with dedicated chaperones that are released upon association of A and B. (B) Asymetric co-translational protein interaction. Upper panel: Protein B is produced, folded, released and recruited to the site of protein A translation. It associates with its domain of interaction in the nascent chain of protein A as soon as this domain folds. Lower panel: A chaperone is recruited to the site of translation of its client protein. (C) Co-translational assembly promoted by mRNA co-localization. Upper panel: The mRNAs encoding proteins A and B are tethered during translation so that the nascent chains of A can B can associate as soon as the interaction domains have folded and are exposed out of the tunnel. Lower panel: In the case of homo-oligomers, interaction of the nascent chains can occur from neighboring ribosomes on the same mRNA. (D) Asymetric co-translational protein interaction detected by ribosome profiling. Protein pairs known to interact are C-terminally tagged and immunoprecipitated. The ribosome footprints in the immunoprecipitate are recovered and aligned to the genome. They are detected with a sharp onset on the ORF of the mRNA encoding the partner subunit. This is asymmetric. (E) Co-translational association revealed by ribosome profiling. Ribosome pauses are detected by ribosome footprint accumulation on the RPT1 and RPT2 mRNAs that expose nascent chain-interacting domains outside of the ribosome tunnel. (F) Model for co-translational interaction of Rpt1 and Rpt2. mRNAs are first co-localized with the Not1 protein and dependent upon Not1. Then nascent chains emerging from the ribosome tunnel contribute to the formation of higher order granules in which paused ribosomes will be stable and assembly of nascent chain interacting domains can occur. After interaction, translation elongation resumes in the heavy particles.
In prokaryotes, transcription and translation occur in the same cellular compartment and genes encoding proteins that participate in the same cellular function or assemble into protein complexes, are often organized and co-expressed in operons. This results in high local concentrations of nascent proteins that need to associate. In contrast, in eukaryotic cells mRNAs are produced in the nucleus and exported to the cytoplasm where they will be translated. Thus, the entire process of complex assembly needs specific mechanisms in the cytoplasm to be made efficient. There is evidence that genes encoding subunits of protein complexes are co-expressed ((66) and references therein). There are also specific examples of co-localized mRNAs (67–70). In some cases, the co-localization of the mRNAs does not depend upon the nascent chain, indicating that it is an RNA-mediated process. This is for instance the case of the co-localization of proteasome mRNAs driven together to distinct granules by CNOT1 (70), the scaffold of the Ccr4-Not complex (71) (see further).
In 2013, Duncan and Mata took a genomic approach to address how frequent co-translational assembly might occur in fission yeast (72). They immunoprecipitated 31 proteins (not RNA binding proteins) and identified mRNAs in the IP by sequencing. In 38% of the cases, proteins could immunoprecipitate the mRNA encoding a partner protein. This was dependent upon expression of the partner and polysome integrity, indicating that the interaction occurred during translation. The interactions were usually asymmetric, namely one protein was recruited to the nascent chain of its partner, but not the other way around. The Bukau laboratory tackled this question more recently in budding yeast, using the power of ribosome profiling (73). C-terminally tagged proteins from 12 established different hetero-oligomeric complexes were immunopreciptated and the RNCs in the IP were characterized. A majority of the protein pairs showed co-translational assembly that was often asymmetric. The co-translational assembly nature of the interactions was fully supported by the sharp onset of the ribosome footprints in the RNCs that were immunoprecipitated and that corresponded to exposure of the interaction domain of the nascent chain outside of the ribosome tunnel (Figure 4D). Interestingly, Ssb engages the same RNCs before the co-translational interaction of the partner subunits, during the synthesis of the interaction domains, most likely thereby ensuring their folding in good coordination with the co-translational process.
The approaches described so far evaluated the timing of co-translation events by relying on the accessibility of a nascent chain to antibodies or to partner proteins, and defining where associated RNCs start displaying ribosome footprints on the mRNAs. A different approach consists of analyzing ribosome footprinting data to determine where ribosome footprints might accumulate because of ribosome pausing (70). This approach was used to investigate the assembly of the proteasome. Important peaks of ribosome footprints indicative of ribosome pausing were detected on two of the mRNAs encoding partner subunits of the proteasome base, Rpt1 and Rpt2, at a position corresponding to a DP codon pair, through which translation elongation is particularly dependent upon the translation factor eIF5A (74,75). Ribosomes paused at these positions expose a nascent chain out of the ribosome tunnel that includes the interaction domains for both subunits. This led to the model that ribosomes were pausing during translation to enable the interaction of the nascent chains to engage each other before translation was completed (Figure 4E) (70).
RNA GRANULES TO PROTECT REGULATORY RIBOSOME PAUSING FROM NGD AND RQC
As mentioned above, ribosome pausing is expected to provoke ribosome collision and induce NGD and RQC. Thus, it was surprising to detect important peaks of ribosome footprints on the proteasome mRNAs, unless a mechanism to prevent ribosome collision and NGD was occurring. Indeed, the proteasome RNCs were shown to form heavy particles in which the RNCs were very stable, in which there was no evidence for the presence of multiple ribosomes translating the mRNA with a stalled ribosome (hence no risk for collision) compatible with a repression of new translation initiation events. The formation of these particles was dependent upon the nascent chains, in particular upon the disordered most N-terminal regions of the nascent chains, since when they were deleted, RNCs no longer assembled into soluble heavy particles enabling the co-translational process.
The formation of these heavy particles was a two-step procedure, since it was preceded by the co-localization of the two mRNAs, dependent upon the Not1 protein but not upon translation elongation (Figure 4F). These heavy particles or granules were named ‘Not1-Containing Assemblysomes’ since they stabilize RNCs to enable co-translational association of nascent chains. They were effectively enabling rapid degradation of subunits that had been fully translated without associating co-translationally and would be dead-end protein products likely to aggregate. Interestingly after interaction of the nascent chains, translational pause was relieved and translation elongation continued in the heavy particles, suggesting that further steps of assembly might continue in the same localized manner (70). According to these findings, Not1 plays a key role for the protection of the proteome. It promotes the formation of granules that enable assembly of the proteasome, and protects the cytoplasm from the accumulation of unassembled proteins that would aggregate.
The mechanism by which these granules stabilize RNCs still remains to be clarified. As mentioned above, there was no evidence for nascent peptides of different lengths in the granules with paused ribosomes, suggesting that there were no collided ribosomes, hence no substrates for NGD. Thus RNCs might be protected from NGD because new translation initiation events are inhibited on the mRNAs with paused ribosomes. Alternatively, or in addition, components of NGD/RQC may have limited access to paused ribosomes. Further work will be needed to determine this. In this context it is interesting to note that in cells lacking Dom34 necessary for recycling of ribosomes during NGD, there was no major change in ribosome footprints on open reading frames (ORFs) genome-wide (21). The exception was a 30-fold increase of a footprint on the HAC1 mRNA that encodes the transcription factor for the unfolded protein response and whose translation is regulated by splicing in the cytoplasm. An interaction between the 5′ untranslated region and the intron stalls translation of the unspliced mRNA associated with polyribosomes (76). Hence, substrates of this mRNA with paused ribosomes are specifically not protected from NGD. Translation of XBP1, the human ortholog of HAC1 mRNA, also occurs with ribosome pausing. As soon as a domain of the nascent protein from the unspliced mRNA is exposed at the ribosome surface, ribosome pausing due to a conserved motif sequence found in the C-terminal region of the protein occurs, and the paused mRNA-RNC complex is recruited to the ER membrane. This pausing is essential for efficient recruitment of the complex to the ER membrane and mRNA cytoplasmic splicing upon ER stress (77).
There are other cases of translation attenuation that rely upon ribosome pausing, such as that of the arg-2 mRNA when arginine is supplied and ribosomes stall at the termination codon of an upstream ORF, preventing scanning of ribosomes to reach the initiation codon (78). Repression of the human antizyme inhibitor 1 (AZIN1) mRNA under high polyamines also involves ribosome stalling on an upstream ORF and ribosome queuing, ultimately preventing scanning to the start of the AZIN1 ORF (79). There is also a mechanism in vertebrates to limit translation from a single adenosylmethionine decarboxylase I (AMD1) mRNA. It involves stalling of ribosomes that read through the stop codon at the next in frame stop codon, provoking ribosome queuing that halts translation of the main ORF (80). One can question whether these different translational controls based upon ribosome pausing and queuing rely on specific granules to avoid co-translational quality control mechanisms.
In summary, assemblysomes are granules that serve the purpose of tethering mRNAs, limiting ribosome collision events when ribosomes pause for co-translational processes or translation regulation, and/or protecting the mRNAs from NGD in cases of ribosome collision. They may or may not be similar to the recently described translation factor mRNA granules (81). Indeed, we still have to understand how diverse such types of granules are, how frequently granules are relevant during translation, and finally whether they are or not phase-separation granules similar to stress granules that are translationally silent.
CONCLUSION
In conclusion, in this review we have described how cells cope with aberrant ribosome stalling but also needs ribosome pausing for co-translational processes. We have summarized several examples of studies that have used new methods to follow the translation elongation process at single codon resolution to understand how nascent chains are folded, matured and targeted, and how factors are recruited to meet nascent peptides during their production in vivo, situations likely to need ribosome pausing. We described the identification of granules, Not1-containing assemblysomes, that protect paused ribosomes from co-translational quality control mechanisms. Finally we have underlined that while we now have tools to follow global ribosome dynamics in cells we still have to learn how to read and interpret the information that is available. Indeed, the profiles of ribosome footprints reflect the net result of at least ribosome velocity, the mRNA code, co-translational mRNA decay, the influence of the nascent chains themselves and the availability of interacting co-translational partners. Importantly they also result from biases in the generation of the data that we should be able to account for better as the amount of data generated expands.
ACKNOWLEDGEMENTS
We thank William Kelly, Olesya Panasenko and Zoya Ignatova for a critical reading of the manuscript.
FUNDING
Swiss National Science Foundation [31003A_172999 to M.A.C.]. Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Timmers H.T.M., Tora L.. Transcript buffering: a balancing act between mRNA synthesis and mRNA degradation. Mol. Cell. 2018; 72:10–17. [DOI] [PubMed] [Google Scholar]
- 2. Guzikowski A.R., Chen Y.S., Zid B.M.. Stress-induced mRNP granules: form and function of processing bodies and stress granules. Wiley Interdiscip. Rev. RNA. 2019; 10:e1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Standart N., Weil D.. P-bodies: cytosolic droplets for coordinated mRNA storage. Trends Genet. 2018; 34:612–626. [DOI] [PubMed] [Google Scholar]
- 4. Kurosaki T., Maquat L.E.. Molecular autopsy provides evidence for widespread ribosome-phased mRNA fragmentation. Nat. Struct. Mol. Biol. 2018; 25:299–301. [DOI] [PubMed] [Google Scholar]
- 5. Stein K.C., Frydman J.. The stop-and-go traffic regulating protein biogenesis: how translation kinetics controls proteostasis. J. Biol. Chem. 2019; 294:2076–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodnina M.V., Fischer N., Maracci C., Stark H.. Ribosome dynamics during decoding. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017; 372:20160182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gromadski K.B., Rodnina M.V.. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004; 13:191–200. [DOI] [PubMed] [Google Scholar]
- 8. Rodnina M.V. The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci. 2016; 25:1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R. et al.. Codon optimality is a major determinant of mRNA stability. Cell. 2015; 160:1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu W., Sweet T.J., Chamnongpol S., Baker K.E., Coller J.. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009; 461:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karousis E.D., Muhlemann O.. Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb. Perspect. Biol. 2019; 11:a032862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He F., Li X., Spatrick P., Casillo R., Dong S., Jacobson A.. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell. 2003; 12:1439–1452. [DOI] [PubMed] [Google Scholar]
- 13. Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C.. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004; 36:1073–1078. [DOI] [PubMed] [Google Scholar]
- 14. Ikeuchi K., Izawa T., Inada T.. Recent progress on the molecular mechanism of quality controls induced by ribosome stalling. Front. Genet. 2018; 9:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuda R., Ikeuchi K., Nomura S., Inada T.. Protein quality control systems associated with no-go and nonstop mRNA surveillance in yeast. Genes Cells. 2014; 19:1–12. [DOI] [PubMed] [Google Scholar]
- 16. Joazeiro C.A.P. Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu. Rev. Cell Dev. Biol. 2017; 33:343–368. [DOI] [PubMed] [Google Scholar]
- 17. Pelechano V., Wei W., Steinmetz L.M.. Widespread Co-translational RNA decay reveals ribosome dynamics. Cell. 2015; 161:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tesina P., Heckel E., Cheng J., Fromont-Racine M., Buschauer R., Kater L., Beatrix B., Berninghausen O., Jacquier A., Becker T. et al.. Structure of the 80S ribosome-Xrn1 nuclease complex. Nat. Struct. Mol. Biol. 2019; 26:275–280. [DOI] [PubMed] [Google Scholar]
- 19. Ibrahim F., Maragkakis M., Alexiou P., Mourelatos Z.. Ribothrypsis, a novel process of canonical mRNA decay, mediates ribosome-phased mRNA endonucleolysis. Nat. Struct. Mol. Biol. 2018; 25:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lubas M., Andersen P.R., Schein A., Dziembowski A., Kudla G., Jensen T.H.. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 2015; 10:178–192. [DOI] [PubMed] [Google Scholar]
- 21. Guydosh N.R., Green R.. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014; 156:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Juszkiewicz S., Chandrasekaran V., Lin Z., Kraatz S., Ramakrishnan V., Hegde R.S.. ZNF598 Is a quality control sensor of collided ribosomes. Mol. Cell. 2018; 72:469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ikeuchi K., Tesina P., Matsuo Y., Sugiyama T., Cheng J., Saeki Y., Tanaka K., Becker T., Beckmann R., Inada T.. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 2019; 38:e100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsuo Y., Ikeuchi K., Saeki Y., Iwasaki S., Schmidt C., Udagawa T., Sato F., Tsuchiya H., Becker T., Tanaka K. et al.. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017; 8:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verma R., Reichermeier K.M., Burroughs A.M., Oania R.S., Reitsma J.M., Aravind L., Deshaies R.J.. Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature. 2018; 557:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuroha K., Zinoviev A., Hellen C.U.T., Pestova T.V.. Release of ubiquitinated and non-ubiquitinated nascent chains from stalled mammalian ribosomal complexes by ANKZF1 and Ptrh1. Mol. Cell. 2018; 72:286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kostova K.K., Hickey K.L., Osuna B.A., Hussmann J.A., Frost A., Weinberg D.E., Weissman J.S.. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science. 2017; 357:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choe Y.J., Park S.H., Hassemer T., Korner R., Vincenz-Donnelly L., Hayer-Hartl M., Hartl F.U.. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature. 2016; 531:191–195. [DOI] [PubMed] [Google Scholar]
- 29. Panasenko O.O., Collart M.A.. Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase. Mol. Microbiol. 2012; 83:640–653. [DOI] [PubMed] [Google Scholar]
- 30. Gloge F., Becker A.H., Kramer G., Bukau B.. Co-translational mechanisms of protein maturation. Curr. Opin. Struct. Biol. 2014; 24:24–33. [DOI] [PubMed] [Google Scholar]
- 31. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hanson G., Coller J.. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018; 19:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Melnikov S., Mailliot J., Rigger L., Neuner S., Shin B.S., Yusupova G., Dever T.E., Micura R., Yusupov M.. Molecular insights into protein synthesis with proline residues. EMBO Rep. 2016; 17:1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Artieri C.G., Fraser H.B.. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 2014; 24:2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Charneski C.A., Hurst L.D.. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013; 11:e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qian W., Yang J.R., Pearson N.M., Maclean C., Zhang J.. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012; 8:e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gardin J., Yeasmin R., Yurovsky A., Cai Y., Skiena S., Futcher B.. Measurement of average decoding rates of the 61 sense codons in vivo. Elife. 2014; 3:doi:10.7554/eLife.03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dana A., Tuller T.. The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res. 2014; 42:9171–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hussmann J.A., Patchett S., Johnson A., Sawyer S., Press W.H.. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet. 2015; 11:e1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weinberg D.E., Shah P., Eichhorn S.W., Hussmann J.A., Plotkin J.B., Bartel D.P.. Improved Ribosome-Footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 2016; 14:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O’Connor P.B., Andreev D.E., Baranov P.V.. Comparative survey of the relative impact of mRNA features on local ribosome profiling read density. Nat. Commun. 2016; 7:12915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han Y., David A., Liu B., Magadan J.G., Bennink J.R., Yewdell J.W., Qian S.B.. Monitoring cotranslational protein folding in mammalian cells at codon resolution. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:12467–12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y., Wolfle T., Rospert S.. Interaction of nascent chains with the ribosomal tunnel proteins Rpl4, Rpl17, and Rpl39 of Saccharomyces cerevisiae. J. Biol. Chem. 2013; 288:33697–33707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reimann B., Bradsher J., Franke J., Hartmann E., Wiedmann M., Prehn S., Wiedmann B.. Initial characterization of the nascent polypeptide-associated complex in yeast. Yeast. 1999; 15:397–407. [DOI] [PubMed] [Google Scholar]
- 45. Rospert S., Dubaquié Y., Gautschi M.. Nascent-polypeptide-associated complex. Cell Mol. Life Sci. 2002; 59:1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., Sinning I., Rospert S.. Two chaperones locked in an embrace: structure and function of the ribosome-associated complex RAC. Nat. Struct. Mol. Biol. 2017; 24:611–619. [DOI] [PubMed] [Google Scholar]
- 47. Chen S., Vetro J.A., Chang Y.H.. The specificity in vivo of two distinct methionine aminopeptidases in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2002; 398:87–93. [DOI] [PubMed] [Google Scholar]
- 48. Starheim K.K., Gevaert K., Arnesen T.. Protein N-terminal acetyltransferases: when the start matters. Trends Biochem. Sci. 2012; 37:152–161. [DOI] [PubMed] [Google Scholar]
- 49. Zhang X., Shan S.O.. Fidelity of cotranslational protein targeting by the signal recognition particle. Annu. Rev. Biophys. 2014; 43:381–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nyathi Y., Pool M.R.. Analysis of the interplay of protein biogenesis factors at the ribosome exit site reveals new role for NAC. J. Cell Biol. 2015; 210:287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deuerling E., Gamerdinger M., Kreft S.G.. Chaperone interactions at the ribosome. Cold Spring Harb. Perspect. Biol. 2019; a033977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Willmund F., del Alamo M., Pechmann S., Chen T., Albanese V., Dammer E.B., Peng J., Frydman J.. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013; 152:196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doring K., Ahmed N., Riemer T., Suresh H.G., Vainshtein Y., Habich M., Riemer J., Mayer M.P., O’Brien E.P., Kramer G. et al.. Profiling Ssb-Nascent chain interactions reveals principles of Hsp70-assisted folding. Cell. 2017; 170:298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mandon E.C., Trueman S.F., Gilmore R.. Protein translocation across the rough endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013; 5:a013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. del Alamo M., Hogan D.J., Pechmann S., Albanese V., Brown P.O., Frydman J.. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011; 9:e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chartron J.W., Hunt K.C., Frydman J.. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016; 536:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jan C.H., Williams C.C., Weissman J.S.. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014; 346:1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ast T., Cohen G., Schuldiner M.. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell. 2013; 152:1134–1145. [DOI] [PubMed] [Google Scholar]
- 59. Hansen K.G., Herrmann J.M.. Transport of proteins into mitochondria. Protein J. 2019; 38:330–342. [DOI] [PubMed] [Google Scholar]
- 60. Pfanner N., Warscheid B., Wiedemann N.. Mitochondrial proteins: from biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019; 20:267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lesnik C., Golani-Armon A., Arava Y.. Localized translation near the mitochondrial outer membrane: An update. RNA Biol. 2015; 12:801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Williams C.C., Jan C.H., Weissman J.S.. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014; 346:748–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hansen K.G., Aviram N., Laborenz J., Bibi C., Meyer M., Spang A., Schuldiner M., Herrmann J.M.. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science. 2018; 361:1118–1122. [DOI] [PubMed] [Google Scholar]
- 64. Williams N.K., Dichtl B.. Co-translational control of protein complex formation: a fundamental pathway of cellular organization. Biochem. Soc. Trans. 2018; 46:197–206. [DOI] [PubMed] [Google Scholar]
- 65. Wells J.N., Bergendahl L.T., Marsh J.A.. Co-translational assembly of protein complexes. Biochem. Soc. Trans. 2015; 43:1221–1226. [DOI] [PubMed] [Google Scholar]
- 66. Zhang W., Xu J., Li Y., Zou X.. Integrating network topology, gene expression data and GO annotation information for protein complex prediction. J. Bioinform. Comput. Biol. 2019; 17:1950001. [DOI] [PubMed] [Google Scholar]
- 67. Mingle L.A., Okuhama N.N., Shi J., Singer R.H., Condeelis J., Liu G.. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 2005; 118:2425–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu F., Jones D.K., de Lange W.J., Robertson G.A.. Cotranslational association of mRNA encoding subunits of heteromeric ion channels. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:4859–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kassem S., Villanyi Z., Collart M.A.. Not5-dependent co-translational assembly of Ada2 and Spt20 is essential for functional integrity of SAGA. Nucleic Acids Res. 2017; 45:1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Panasenko O.O., Somasekharan S.P., Villanyi Z., Zagatti M., Bezrukov F., Rashpa R., Cornut J., Iqbal J., Longis M., Carl S.H. et al.. Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct. Mol. Biol. 2019; 26:110–120. [DOI] [PubMed] [Google Scholar]
- 71. Collart M.A. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA. 2016; 7:438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Duncan C.D., Mata J.. Widespread cotranslational formation of protein complexes. PLos Genet. 2011; 7:e1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shiber A., Doring K., Friedrich U., Klann K., Merker D., Zedan M., Tippmann F., Kramer G., Bukau B.. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature. 2018; 561:268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schuller A.P., Wu C.C., Dever T.E., Buskirk A.R., Green R.. eIF5A functions globally in translation elongation and termination. Mol. Cell. 2017; 66:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pelechano V., Alepuz P.. eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 2017; 45:7326–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ruegsegger U., Leber J.H., Walter P.. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001; 107:103–114. [DOI] [PubMed] [Google Scholar]
- 77. Yanagitani K., Kimata Y., Kadokura H., Kohno K.. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011; 331:586–589. [DOI] [PubMed] [Google Scholar]
- 78. Wang Z., Fang P., Sachs M.S.. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol. Cell Biol. 1998; 18:7528–7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ivanov I.P., Shin B.S., Loughran G., Tzani I., Young-Baird S.K., Cao C., Atkins J.F., Dever T.E.. Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol. Cell. 2018; 70:254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yordanova M.M., Loughran G., Zhdanov A.V., Mariotti M., Kiniry S.J., O’Connor P.B.F., Andreev D.E., Tzani I., Saffert P., Michel A.M. et al.. AMD1 mRNA employs ribosome stalling as a mechanism for molecular memory formation. Nature. 2018; 553:356–360. [DOI] [PubMed] [Google Scholar]
- 81. Pizzinga M., Bates C., Lui J., Forte G., Morales-Polanco F., Linney E., Knotkova B., Wilson B., Solari C.A., Berchowitz L.E. et al.. Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth. J. Cell Biol. 2019; 218:1564–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Loya A., Pnueli L., Yosefzon Y., Wexler Y., Ziv-Ukelson M., Arava Y.. The 3′-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. RNA. 2008; 14:1352–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Saint-Georges Y., Garcia M., Delaveau T., Jourdren L., Le Crom S., Lemoine S., Tanty V., Devaux F., Jacq C.. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 2008; 3:e2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gerber A.P., Herschlag D., Brown P.O.. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004; 2:E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kershaw C.J., Costello J.L., Talavera D., Rowe W., Castelli L.M., Sims P.F., Grant C.M., Ashe M.P., Hubbard S.J., Pavitt G.D.. Integrated multi-omics analyses reveal the pleiotropic nature of the control of gene expression by Puf3p. Sci. Rep. 2015; 5:15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cox R.T., Spradling A.C.. Clueless, a conserved Drosophila gene required for mitochondrial subcellular localization, interacts genetically with parkin. Dis. Model. Mech. 2009; 2:490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fields S.D., Conrad M.N., Clarke M.. The S. cerevisiae CLU1 and D. discoideum cluA genes are functional homologues that influence mitochondrial morphology and distribution. J. Cell Sci. 1998; 111:1717–1727. [DOI] [PubMed] [Google Scholar]