Abstract
Ribosomopathies are diseases caused by defects in ribosomal constituents or in factors with a role in ribosome assembly. Intriguingly, congenital ribosomopathies display a paradoxical transition from early symptoms due to cellular hypo-proliferation to an elevated cancer risk later in life. Another association between ribosome defects and cancer came into view after the recent discovery of somatic mutations in ribosomal proteins and rDNA copy number changes in a variety of tumor types, giving rise to somatic ribosomopathies. Despite these clear connections between ribosome defects and cancer, the molecular mechanisms by which defects in this essential cellular machinery are oncogenic only start to emerge. In this review, the impact of ribosomal defects on the cellular function and their mechanisms of promoting oncogenesis are described. In particular, we discuss the emerging hallmarks of ribosomopathies such as the appearance of ‘onco-ribosomes’ that are specialized in translating oncoproteins, dysregulation of translation-independent extra-ribosomal functions of ribosomal proteins, rewired cellular protein and energy metabolism, and extensive oxidative stress leading to DNA damage. We end by integrating these findings in a model that can provide an explanation how ribosomopathies could lead to the transition from hypo- to hyper-proliferation in bone marrow failure syndromes with elevated cancer risk.
INTRODUCTION
The ribosome is one of the most ancient primordial molecular machines, posited to have originated 4 billion years ago. The ancient ribosomal heart still beats with the same purpose today: executing a key role in the central dogma of molecular biology by translating messenger RNA (mRNA) into proteins. The human ribosome is composed of a small 40S subunit consisting of the 18S rRNA chain and 33 RPS proteins and a large 60S subunit encompassing the 28S, 5S and 5.8S rRNA chains, and 47 RPL proteins. The accuracy of sequential ribosome assembly followed by functional quality checks are crucial aspects of the ribosomal biorhythm. It has been estimated that thousands of ribosomes are being manufactured and functionally checked every minute in a growing eukaryotic cell (1).
A defect in even one of the components of this essential cellular apparatus can have a major impact on cellular function. A group of diseases—ribosomopathies—are characterized by defects in RPs, rRNA processing or ribosome assembly factors (2). An intriguing characteristic of ribosomopathies is the remarkable tissue-specificity of the phenotypic abnormalities. Despite the fact that every cell in the body relies on ribosomes to translate mRNA into proteins, the disease-associated abnormalities in ribosomopathy patients are restricted to particular tissues, for example the frequently affected hematopoietic system. We refer to other recent reviews that have provided insights and potential explanations for this observation (3–5). A second peculiarity of ribosomopathies is the evolution of the disease phenotype. Early in life, ribosomopathy patients present symptoms such as bone marrow failure and anemia, broadly fitting into the category of cellular hypo-proliferation phenotypes. Whereas the consequences of these phenotypes used to be lethal, supportive treatments now allow patients to survive this initial disease phase. However, the improved life-span has illuminated a paradoxical second disease phase, as these patients have an elevated risk of progressing to a hyper-proliferative cellular state and ultimately cancer later in life (6). Overall, ribosomopathy patients have a 2.5- to 8.5-fold higher risk to develop cancer throughout their life, and for particular cancer types these risks can be up to 200-fold higher (7–9). For a recent review providing an overview on the cancer risks in ribosomopathies, we refer to (10). The question thus arises of how ribosomopathies can undergo a transition from an illness founded on a lack of cell proliferation to cancer, a disease of uncontrolled growth? The recent discovery of somatic mutations in RPs in a variety of tumor types provides a second link between ribosome defects and cancer. In this review, we discuss the most recent findings and perspectives with respect to ribosomopathies, with a main focus on the molecular mechanisms driving ribosomopathy-associated cancer development. Defining the ‘hallmarks of ribosomopathies’ is inspired by the landmark paper by Hanahan and Weinberg on the ‘hallmarks of cancer’, in which the molecular biological cellular changes that are at the basis of cancer are described (11).
RIBOSOMOPATHIES
Mammalian ribosome assembly, also referred to as ribosome biogenesis, is a complex and incompletely understood process (reviewed in (12)). Mature mammalian ribosomes consist of ∼6000 rRNA bases divided over 4 rRNA molecules as well as 80 RPs. Hundreds of accessory trans-acting biogenesis factors facilitate the eukaryotic ribosome assembly process. These proteins are not incorporated into the mature ribosome, but instead guide the maturation process. This process starts in the nucleolus, where three out of four rRNA molecules are transcribed as a long precursor rRNA (pre-rRNA). These pre-rRNAs undergo a series of cleavages (known as ‘processing’) along with modifications, such as methylation and pseudouridylation mediated by hundreds of small nucleolar RNAs and protein co-factors, to become the mature 18S, 28S, 5.8S and 5S rRNAs (13). Ribosomal protein (RP) encoding mRNAs are transcribed in the nucleoplasm, translated in the cytoplasm, after which RPs are shuttled back to the nucleolus to associate with the maturing ribosome. A series of ribosome assembly intermediates are formed before the pre-60S and pre-40S particles are exported from the nucleolus into the nucleus and subsequently to the cytoplasm for final rRNA processing and protein associations. Finally, the fully assembled and mature 60S and 40S subunits bind to form translationally active 80S ribosomes.
Given the essential cellular function of ribosomes as protein production factories, non-lethal alterations in ribosome assembly and/or function are expected to cause cellular dysfunction and potentially disease. Ribosomopathies can be defined as diseases associated with a mutation in a RP, in rRNA, in a biogenesis factor, or with a defect in rDNA transcription that is linked to disease causality (6). The term ribosomopathy was historically used to refer to disease syndromes caused by congenital mutations in the ribosome or a biogenesis factor. The definition above may however also apply to cancers with somatic RP gene mutations, as the data supporting causality of these mutations in tumor pathogenesis cannot be ignored as discussed below. However, cancers are caused by the accumulation of multiple genetic events, which can include somatic RP mutations. A somatic RP mutation may thus promote transformation but will not be sufficient to cause cancer. On the other hand, mutations in RPs and ribosome biogenesis factors are likely sufficient to cause the hypo-proliferative phenotypes observed in the early phase of the congenital ribosomopathy syndromes. We provide an overview of congenital and somatic ribosomopathies in the following section. For several of these diseases, relevant animal models have been established, which have been reviewed elsewhere (14).
Congenital ribosomopathies
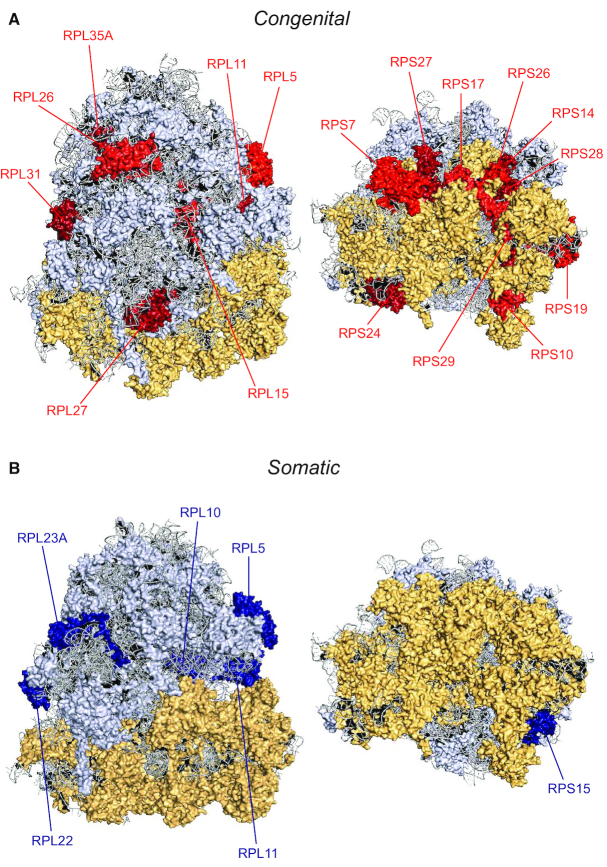
In 1999, the first recurrent RP mutations were described in RPS19 (also known as eS19 according to the new nomenclature (15)), in patients with the congenital bone marrow failure syndrome Diamond-Blackfan anemia (DBA) (16). Since then, RP mutations have been identified in ∼50% of DBA patients, with loss-of-function mutations in RPS19, RPL5 (uL18), RPL11 (uL5) and RPS10 (eS10) being the most frequent (17) (Figure 1A). Other congenital syndromes have been linked to defects in ribosome biogenesis factors. Each of these genetic abnormalities disrupts a specific step in ribosome biogenesis. The most studied ribosomopathies besides DBA include Shwachman-Diamond syndrome (SDS), X-linked dyskeratosis congenita (DC), cartilage hair hypoplasia (CHH) and Treacher Collins syndrome (TCS) (2). In SDS, 90% of patients display inactivating mutations in the SBDS gene (18). SBDS encodes a trans-acting factor involved in late cytoplasmic maturation of 60S subunits by promoting the release of eukaryotic initiation factor 6 (EIF6) from pre-60S subunits (19). EIF6 keeps the nascent 60S subunit inactive during cytoplasmic 60S assembly by preventing premature association with the 40S subunit. In SDS patients, efficient EIF6 release does not take place, thereby stalling 60S maturation (20). CHH is caused by mutations in RMRP, a long non-coding RNA (lnc-RNA) component of the RNase mitochondrial RNA processing complex. RMRP mutations or knock-down affect rRNA processing by inhibiting cleavage of pre-rRNA in the internal transcribed spacer 1 (ITS1), limiting the maturation of 18S and 5.8S rRNAs (21,22). DBA-associated mutations in several RPs in both subunits also have a direct impact on pre-rRNA. These specific processing defects in DBA cells are even utilized to diagnose the disease (23–25). Twenty five percent of the patients suffering from DC carry mutations in DKC1, encoding dyskerin—the enzyme executing rRNA pseudouridylation. DKC1 also has a role in telomere maintenance and shares this function with other genes such as TINF2, TERC or TERT that are recurrently mutated in DC (26). DBA, SDS and DC are characterized by bone marrow failure phenotypes early in life, followed by an elevated risk to develop cancer later in life. The fact that TCS is not associated with increased cancer incidence or bone marrow failure is of interest. In contrast to other RP defects that are linked to a specialized translational program of subsets of genes controlling hematopoiesis, this has not been described for TCS. TCS is caused by a lack of polymerase I/III activity due to a deletion or mutation in TCOF1, POLR1C and POLR1D (27–29). These defects lead to reduced levels of mature ribosomes and lower overall translation, thereby weakening the cells and promoting a quiescent state. More rare and less studied ribosomopathies include isolated congenital asplenia and North American Indian childhood cirrhosis. More recently, a germline truncating mutation in RPS20 (uS10) was identified as a colon cancer predisposing mutation, and knockdown of RPS20 was also shown to impair pre-ribosomal RNA maturation (30).
Figure 1.
Structural model of the ribosome with indication of RPs affected in ribosomopathies. Structural model of the human 80S ribosome with indication of RPs with recurrent mutations and/or deletions in ribosomopathies. The 60S large subunit, 40S small subunit and ribosomal RNA are indicated in yellow, light blue and gray, respectively. In panel (A), RPs with congenital defects are indicated in red. RPs with somatic mutations and deletions are marked in blue in panel (B). Each panel shows two different viewpoints of the solvent side of the ribosome. This figure was generated in PyMOL and is based on the human cryo-EM structure with a resolution of 3.9Å (PDM entry: 6IP5) (140). New RP nomenclature: RPL5 (uL18), RPL10 (uL16), RPL11 (uL5), RPL15 (eL15), RPL22 (eL22), RPL23A (uL23), RPL26 (uL24), RPL27 (eL27), RPL31 (eL31), RPL35A (eL33), RPS7 (eS7), RPS10 (eS10), RPS14 (uS11), RPS15 (uS19), RPS17 (eS17), RPS19 (eS19), RPS24 (eS24), RPS26 (eS26), RPS27 (eS27), RPS28 (eS28), RPS29 (uS14).
Somatic ribosomopathies
RP mutations and copy number changes
Genomic analyses on 10 744 cancer patient samples and cell lines revealed that about 43% of cancers display RP deletions (31). Over 95% of these RP deletions are heterozygous, as homozygous loss of most RPs is lethal for a cell, with the exception of homozygous loss of non-essential RPs such as RPL22. The deletions in cancer patients encompassing RP genes sometimes also affect established tumor suppressors such as TP53 and CDKN2A/B. In the past, it was therefore often assumed that RP gene deletions were a meaningless side effect of larger deletions affecting these tumor suppressors. However, additional genetic data support a causative role for these mutations in cancer: besides deletions, somatic heterozygous inactivating mutations are recurrently observed in RP genes in large deletion regions such as RPL5, RPL11 and RPL22 (eL22) (32–37). Moreover, the RPL5 and RPL22 genes on chromosome bands 1p22 and 1p36 respectively are located within the common minimally deleted region across patients (34,38), further supporting specific clonal selection of loss of these RP alleles throughout tumor development. Finally, experiments in animal models support the oncogenic role of heterozygous loss of RP genes. Development of peripheral nerve sheet tumors have been observed in 17 different heterozygous RP gene mutant zebrafish lines, and transgenic mouse lines modeling heterozygous loss of Rpl22 or Rpl11 display tumor acceleration phenotypes (34,39–41).
Haploinsufficiency for RPS14 (uS11) in 5q− myelodysplastic syndrome (MDS) leads to an erythroid differentiation defect highly similar to DBA. This disorder is characterized by a somatically acquired deletion of the entire 5q chromosomal region, and other genes besides RPS14 in the deleted 5q region may therefore also contribute to the disease phenotype (42,43).
Somatic recurrent RP mutations and deletions have also been detected in cancer samples (Figures 1B and 2). While low frequency mutations in RPL11 are observed in melanoma and relapsed T-cell acute lymphoblastic leukemia (T-ALL), heterozygous loss of RPL5 by deletions or inactivating mutations occurs in 2% of T-ALL samples and in up to 30% of multiple myeloma, melanoma, glioblastoma and breast cancers (32,33,38,44–46). These RPs, which are also recurrently deleted and mutated in DBA, share certain extra-ribosomal functions as discussed below. A tumor suppressor role for RPL5 is supported by the fact that its downregulation accelerates tumor formation in xenografted breast cancer cell lines (32). In contrast, loss of RPL5 rather inhibits cell proliferation of normal cells (47). Breast cancer cell lines carry a high mutational load as opposed to normal cells. These contradictory observations may thus suggest that, in addition to the tumor promoting actions of RPL5 as discussed below, RPL5 can only function as a tumor suppressor in cooperation with other genetic defects already present, as in the case of breast cancer cells.
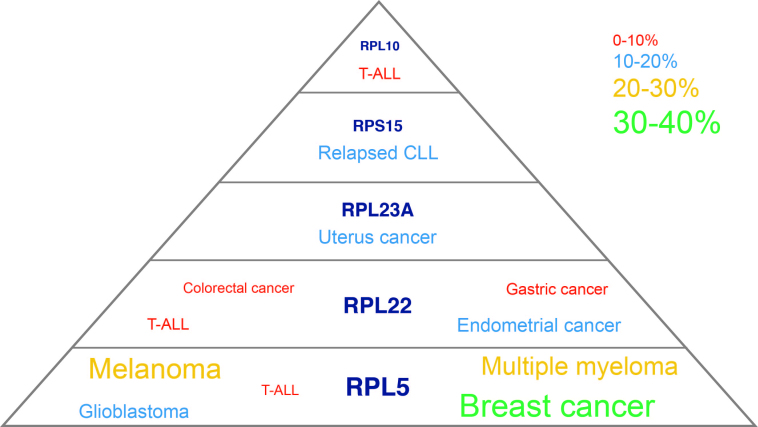
Figure 2.
Scheme illustrating the RP genes with recurrent somatic mutations and/or deletions in cancer. Font sizes in this figure are proportional to incidence of the RP mutations and deletions in the cancer types where they have been described, and incidence percentages of the RP defects are indicated by the color legend; CLL, chronic lymphocytic leukemia; T-ALL, T-cell acute lymphoblastic leukemia.
Mutations or deletions in RPL22 were described in 4% of T-ALL samples (34,46). Moreover, truncating RPL22 mutations are present in ∼10% of gastric, endometrial and colorectal cancer samples (35–37). In mice, Rpl22 directly represses expression of its paralog Rpl22l1. Therefore, Rpl22 null mice have only subtle phenotypes with no significant translation defects because of a compensatory increase in Rpl22-like1 (Rpl22l1) expression and its incorporation into ribosomes (48). The relevance and implications of this in the context of cancers with RPL22 loss remains to be determined.
The genomic region encoding RPL23A (uL23) is amplified in 12.5% of uterine cancers where it is part of a distinct amplification peak. These amplifications occur more frequent in serous endometroid tumors, a more rare and aggressive subtype of uterine cancer (32).
For RPS15 (uS19) and RPL10 (uL16), no copy number changes have been described, but these genes are targeted by clustered mutations. RPS15 mutations concentrate in a region encoding 7 conserved C-terminal amino acids (131–138) and are detected in up to 20% of patients with relapsed chronic lymphoid leukemia (CLL) (49,50). The mutational clustering is even more striking for RPL10: 8% of pediatric T-ALL patients display the same R98S missense mutation in this RP in their leukemia cells (45,46). Interestingly, the P-site loop of RPL10, which harbors this mutated R98 residue, interacts with the N-terminal domain of SBDS. SBDS mutations interfere with this RPL10 interaction, causing similar ribosome biogenesis defects with impaired 60S subunit maturation due to defective EIF6 release from the ribosome (19).
Copy number changes in ribosomal DNA
In addition to the RPs, the rRNA is essential for ribosome functioning, and variations in the rDNA may contribute to ribosomopathies. So far, the studies on the role of rDNA variations in human disease are sparse. This is a consequence of the complex genetic structure of the rDNA, with hundreds of copies of the rDNA operon organized in tandem arrays. This is further complicated by the absence of rDNA sequences in mammalian genome assemblies and the inherent difficulty of sequence assembly and variant discovery in highly repetitive regions such as the rDNA loci. Recent work revealed that rDNA copy numbers vary between 50 and 1500 copies among individuals. Moreover, rDNA operons were found to encode thousands of single nucleotide variants in the rRNA components of assembled and actively translating ribosomes. Furthermore, rDNA variants are evolutionarily conserved between mouse and humans and map to functional centers of the ribosome, including defined interaction sites with translation factors. These findings support the concept that genomic variants in the rDNA can give rise to heterogeneous ribosomes (51). Genomic instability of the rDNA loci has been reported in congenital diseases that are linked to an elevated cancer risk, such as Bloom syndrome and ataxia-teleangiectasia (52). Somatic copy number losses of the 45S rDNA have recently been described in a variety of tumor types (53–55). Loss of 45S rDNA copy numbers in cancer are commonly observed together with hyperactivity of mTOR, an activator of ribosome biogenesis (54), and rDNA loss may thus act as a mechanism to rebalance ribosome biogenesis. Furthermore, tumors with mutations in ATRX, a chaperone of histone H3, are enriched for rDNA copy number loss, supporting a role for ATRX in rDNA maintenance (55). Finally, tumors cells with rDNA copy loss display elevated sensitivity to DNA damaging agents and RNA polymerase I inhibitors (54,55).
ONCOGENIC MECHANISMS IN RIBOSOMOPATHIES
Research on the effects of ribosomal defects on ribosome function and cell physiology has provided insights into the oncogenic molecular mechanisms in ribosomopathies. These mechanisms can be broadly divided into three categories. The first category concerns the direct effect of ribosomal gene defects on the ribosomal protein synthesis function. Ribosomal defects not only lead to ribosome insufficiency due to ribosome mis-assembly, but also alter the translational output of the mis-assembled, structurally distinct ribosomes. As a consequence, the resulting translatome can be shifted toward growth-promoting and oncogenic protein expression signatures. Second, extra-ribosomal moonlighting functions of RPs involved in ribosomopathies may contribute to oncogenic transformation, as some RPs regulate major cancer proteins in a translation-independent manner. The third category entails the influence of ribosome defects on cellular protein and energy metabolism, which can result in cellular stress conditions that can promote acquisition of secondary mutations (Figure 3).
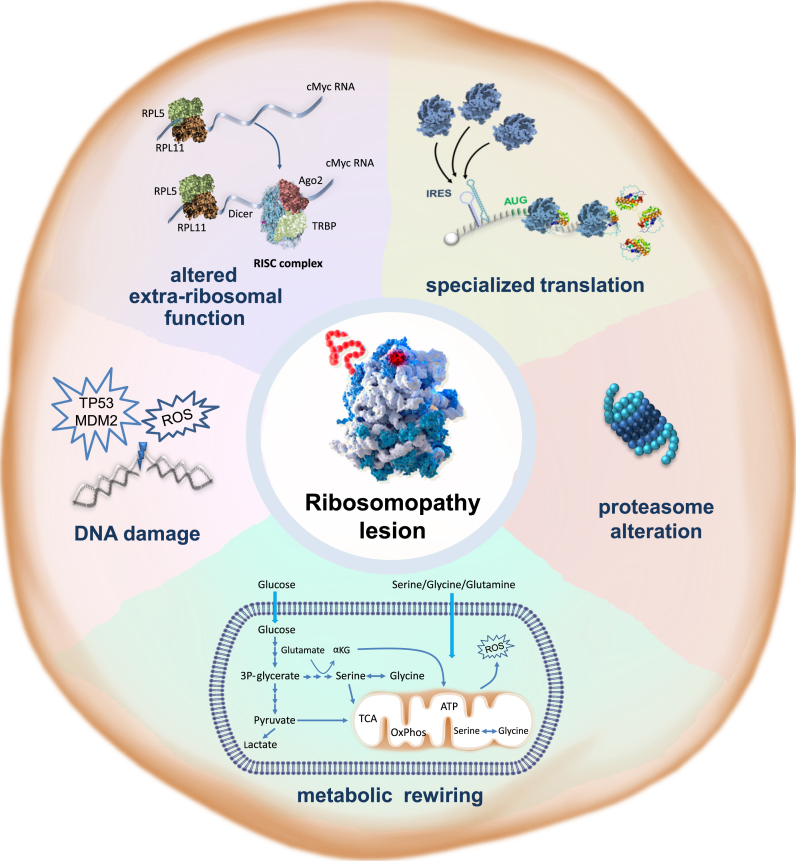
Figure 3.
Hallmarks of ribosomopathies. Ribosomopathies are characterized by a collection of cellular phenotypes that promote cancer as summarized in this figure that is inspired by the figure from the landmark ‘Hallmarks of Cancer’ review by Hanahan and Weinberg (11). (i) Ribosomopathy lesions reprogram translation, affecting cellular translation of a subset of hematopoietic and cancer-promoting mRNAs. (ii) Many ribosomopathies display an altered proteasome function, which can lead to stabilization or increased degradation of subsets of proteins, including oncogenes and tumor suppressors. (iii) Ribosomopathies display metabolic rewiring. The implications for translational reprogramming of specific subsets of proteins or supporting an alternative metabolic requirement of RP-defective cells is currently unclear. (iv) Ribosomapthy cells display elevated levels of oxidative DNA damage that can promote acquisition of secondary mutations with a key role in cancer transformation. (v) Lesions can influence the extra-ribosomal function of the mutated RPs.
Altered mRNA translation by specialized onco-ribosomes
Many consider the ribosome as a conserved machine with the same composition in all cells and tissues of the body that translates available mRNA equally. Plenty of studies however support ribosomal heterogeneity at the level of core RPs, rRNA and of proteins interacting with the ribosome (56–59). Importantly, variations in ribosome composition have functional implications: ribosomes with different composition display specialized functions with preferential translation of particular mRNAs. Within the context of congenital and somatic RP mutations in ribosomopathies and cancer, there are data supporting that, in addition to negatively impacting ribosome assembly, these mutations change the intrinsic ribosomal preferences for particular mRNAs. The resulting specialized proteome could be instrumental for enabling a pre-oncogenic state, and we discuss various aspects of this possibility below.
Speed and accuracy are critical properties of translation. Many RP-mutant cell models display altered translational speed and fidelity, suggesting that such defects have a role in the pathogenesis of ribosomopathies. The RPS15 mutations in CLL are one example: three of the recurrent mutations in the mutational hotspot region (P131S, T136A and S139F) cause a general reduction of translation, two other recurrent mutations (H137Y and S138F) promote near-cognate amino acid misincorporation and the majority of tested mutations promotes stop-codon read-through (50). Similarly, the T-ALL associated RPL10-R98S mutation increases near-cognate amino acid misincorporation and stop-codon read-through in a yeast model (60).
In addition to these more general translation fidelity phenotypes, very specific translation defects only affecting particular mRNAs have been described in some instances. For example, erythroid cells of Rps19 and Rpl11 mutant zebrafish lines show a reduced production of globin proteins, which partially explains their anemic phenotype (61). Moreover, cells derived from nerve sheath tumors in zebrafish display a specific defect in Tp53 translation (62). Frequently, the reshaping of the translational output of mutant ribosomes can be explained by altered interactions with translation regulatory elements. For example, internal ribosomal entry site (IRES) elements in mRNAs can recruit ribosomes independent of the canonical cap-driven translation initiation. These elements are frequently found on mRNAs encoding stress-response genes, enabling the rapid activation of their translation in conditions of cellular stress, when cap-dependent translation is repressed. In a physiological setting, specific RPs can promote translation of IRES-containing mRNAs. An example is Rpl38, which is required for IRES-mediated translation of Hoxa genes (63). In a disease context, mutations in ribosomes can influence IRES-mediated translation rates, independent of stress conditions. For example, the RPL10-R98S mutation drives specific and constitutive IRES-mediated overexpression of the anti-apoptotic factor BCL2 in leukemia cells. This enables ribosome-mutant cells to survive high levels of oxidative stress associated with the RPL10-R98S mutation (64). With respect to inactivation of RPL22, it may be of interest that RPL22 protein levels have been shown to influence rates of translation driven by a hepatitis C virus IRES sequence, an observation that requires further investigation in the context of cancer development (65). DBA-associated mutations in RPS19 and RPL11 reduce IRES-mediated translation of erythroid differentiation factors BAG1 and CSDE1 in DBA mouse models and patient samples (66). Whereas this most likely cannot explain the cancer predisposition in DBA, it may provide an explanation for the erythroid defect in this disease. In addition to RP mutations, defective RNA modifications can also influence IRES-mediated translation. DKC1 mutations linked to DC inhibit the translation of particular IRES-containing mRNAs, such as those encoding the tumor suppressors TP53 and CDKN1B and the anti-apoptotic factors BCL2L1 and XIAP and they enhance IRES-translation of the mRNA encoding the angiogenic growth factor VEGF (67–69). Furthermore, p53-inactivated cancer cells display elevated expression of the rRNA methyl-transferase fibrillarin (FBL) leading to altered 2′-O-methylation of the rRNA and increased IRES-dependent translation of cancer genes (70,71). These studies highlight that altered IRES-dependent translation by defective ribosomes may contribute to cancer transformation in ribosomopathies by shifting the balance to pro-oncogenic proteins at the expense of tumor suppressor proteins.
Another class of cis-acting mRNA control elements include programmed −1 ribosomal frameshift (-1 PRF) signals. In mammalian cells, such signals induce translating ribosomes to move by one nucleotide in the −1 direction on an mRNA, leading to translation in a new −1 reading frame and termination at a premature stop codon (72). Increased incidences of −1 PRF on an mRNA thus lead to lower rates of translation of that particular mRNA. Depletion of DKC1 broadly reduces translational fidelity and increases the frequency of −1 PRF in yeast and human cells (73). Furthermore, ribosomes carrying the T-ALL associated RPL10-R98S mutation display lower rates of −1 PRF on mRNAs encoding proteins of the JAK-STAT signaling cascade, causing elevated expression of JAK-STAT proteins (74).
Upstream open reading frames (uORFs) represent another class of mRNA regulons in the 5′ untranslated regions (5′ UTRs) that can influence translation rates of the downstream ORF. An example of altered translation mediated by an uORF has been described in SDS. SBDS mutations in SDS specifically affect translation of the C/EBPα and β proteins, which are important regulators of hematopoietic granulocyte differentiation. SBDS is required for efficient translation re-initiation on the C/EBPα and β mRNAs, which is controlled by a single uORF in the 5′ UTR of these mRNAs (75). Besides the impaired hematopoiesis as observed in SBDS, the inability to translate C/EBP α and β properly may have a role in the 202-fold elevated risk to develop acute myeloid leukemia (AML) in SDS patients (8), as loss-of-function mutations in C/EBPα have a known role in AML pathogenesis (76).
Several additional types of mRNA regulatory elements have recently been discovered that can direct selective cap-dependent translation of specific pro-tumorigenic mRNAs (reviewed in (77)). It remains to be determined if translation of transcripts containing such secondary structures is also altered in ribosomopathies. New insights here may further broaden the repertoire of specialized ribosome functions in ribosomopathies and their potential to contribute to transformation and cancer progression.
Finally, reduced expression of RPS19, RPL5, RPL11 or RPS24 (eS24) in DBA cells leads to a specific decrease in GATA1 translation (78,79). Here, no particular mRNA regulatory element has yet been identified that may explain this translational reduction, and the phenotype has been attributed to the short and unstructured nature of the 5′ UTR of the GATA1 mRNA. GATA1 is an essential transcription factor for erythroid development, and inactivating GATA1 mutations have been described in DBA in addition to the GATA1 translation defect in this disease (78,80).
Changes in translation-independent moonlighting functions of RPs
Translational dysregulation contributes to the observed disease phenotypes in ribosome-mutant diseases, but other data indicate that additional factors are also involved. For example, extensive characterization of the transcriptome and translatome changes induced by the leukemia-associated RPL10-R98S mutation revealed that half of the 246 detected protein level changes could be explained by transcriptional rather than translation efficiency changes (81). Translation-independent extra-ribosomal moonlighting functions have been described for certain RPs (82), of which several entail regulation of established oncogenes and tumor suppressors. In this context, it is important to consider dysregulation of these extra-ribosomal functions of RPs as a source of oncogenesis.
Several RPs act as suppressors of c-MYC. This factor enhances ribosome biogenesis by inducing rRNA and RP transcription, and certain RPs in turn repress c-MYC expression and function (83). RPL11 binds to the promoter regions of c-MYC target genes, thereby reducing c-MYC–dependent transcription (83,84). Moreover, RPL5 and RPL11 can jointly bind to the c-MYC mRNA and guide it to the RNA-induced silencing complex (RISC) for degradation (85). Similar mechanisms of repressing c-MYC expression and function have also been described for RPS14 (86). In addition, c-MYC activation has been described upon RPL22 inactivation via an indirect NF-kB–Lin28B–Let7 miRNA mechanism (34). Mutations or deletions of these RPs might thus promote transformation by oncogenic c-MYC overexpression. In support of this notion, c-MYC upregulation has been described in mouse lymphoma models that were accelerated by heterozygous Rpl11 or Rpl22 deletions (34,41).
A second well-established extra-ribosomal function of some RPs entails TP53 regulation. The causes of the hypo-proliferative clinical symptoms of most ribosomopathies have long been linked to TP53 activation (87). However, TP53-independent mechanisms of cell-cycle arrest after ribosomal stress have also been described (88). The MDM2 protein is a central regulator of TP53, functioning as an ubiquitin ligase that guides TP53 degradation. Ribosome assembly defects result in freely available RPs, some of which (e.g. RPL5/RPL11) can bind and sequester MDM2, preventing MDM2-induced TP53 degradation and thereby inducing TP53 activity (24,89). Consistent with this, RPL5 and RPL11 mutations in DBA cells are associated with defects in ribosome biogenesis and cell cycle progression (24,85,89). Furthermore, RPL5 or RPL11 loss-of-function might predispose DBA patients to cancer by loss of capacity to activate the TP53 tumor suppressor. While this extra-ribosomal function is most established for RPL5 and RPL11, many other RPs have been described to regulate TP53 by binding MDM2 (82).
RPL10 interferes with the oncogenic transcription factor c-JUN by inhibiting its DNA binding and transactivation (90–92). c-JUN can dimerize with c-FOS to form the AP-1 oncogenic transcription factor complex. In particular, RPL10 was reported to compete with c-FOS for the same binding domain on c-JUN (92). As described above, expression of the RPL10-R98S mutation in lymphoid cells induces extensive transcriptional changes. Intriguingly, the majority of downregulated transcripts in RPL10-R98S cells are predicted c-FOS target genes (81). These results may indicate an impact of the R98S mutation on the extra-ribosomal regulation of c-JUN by RPL10.
Cell metabolism reprogramming
The role of defective ribosomes in the transition to cancer might also be indirect. Besides inducing specific translational changes, multiple lines of evidence support interaction between ribosomes and the composition and functioning of proteasomes, thereby controlling protein metabolism. Additionally, proper ribosomal functioning is also required for controlling cellular redox homeostasis. In this regard, excessive levels of reactive oxygen species (ROS) and the resulting acceleration of secondary mutations have been linked to several ribosomopathies. Finally, ribosomal defects can alter glycolysis and serine/glycine metabolism through direct or indirect regulation. In the sections below, we discuss the ribosome-dependent and independent mechanisms of cell metabolism dysregulation in ribosomopathies.
Effects on the proteasome
Proteasomes fulfill the opposite cellular function of ribosomes: the degradation of proteins that have been tagged with an ubiquitin mark. A number of observations indicate that the functioning of ribosomes and proteasomes is interconnected. For example, 15 proteasomal protein components were identified in the set of protein interaction partners of the ribosome (ribo-interactome) obtained from mouse embryonic fibroblasts (58). This observation suggests physical interactions and potential mutual regulation between ribosomes and proteasomes. The Biogrid database supports RP-proteasome interactions between RPL5 and PSMB1 and between RPL11 and PSMD4 in human cells (93). Various ribosomopathy models strengthen that dysregulation of ribosomes also has implications on the proteasome. RPS19 is affected in 25% of DBA patients and interacts with proteasomal subunits, PSMC5 and PSMC6. Intriguingly, this interaction is lost in the DBA-associated R62W and R101H RPS19 mutants (94,95). Furthermore, proteome comparison of erythrocytes from DBA patients versus healthy controls revealed that immunoproteasome components PSMB8, PSM9 and PSMB10 were overexpressed in DBA erythrocytes (96). Proteome analysis of Hek293T cells expressing WT or CLL-associated RPS15 mutants also showed a significant enrichment of proteasome components among the differentially expressed proteins (50). The same applies to proteomes from isogenic lymphoid cell models expressing RPL10-WT or R98S mutant ribosomes (74). The RPL10-R98S cell model showed up- and downregulation of various proteasomal units, including upregulation of Psmb10. This altered proteasomal unit expression in RPL10-R98S cells is associated with reduced chymotrypsin-like and caspase-like proteasomal activities, and with elevated sensitivity of RPL10-R98S expressing cells for the clinically used proteasome inhibitors, bortezomib and carfilzomib (74). These observations may indicate the expression of a distinct type of proteasomes in RPL10-R98S cells, and is consistent with previously described ‘mixed type’ proteasomes, consisting of constitutive as well as immuno-subunits (97,98). Different proteasome varieties show quantitative differences in cleavage efficiency of particular epitopes, which might provide a certain degree of protein specificity (99,100). In the context of RPL10-R98S, data support that the altered composition and activity of the proteasome influence stability of particular cellular proteins and that this may help in reshaping the cellular proteome toward a more oncogenic one. Indeed, JAK1, an oncogenic kinase with a known pathogenic role in T-ALL showed an enhanced stability in RPL10-R98S cells. RPL5 haploinsufficiency occurs in various cancer types (Figure 2) including multiple myeloma, for which proteasome inhibitors are part of the standard of care. Analysis of clinical trial data revealed that multiple myeloma patients with low RPL5 mRNA expression levels respond better to the proteasome inhibitor bortezomib as compared to patients with high RPL5 expression (38). For the CLL-associated RPS15 mutants, reduced protein stability has been observed as compared to WT RPS15, and these mutants could be partially restabilized by bortezomib (50). It remains to be determined whether proteasome inhibitors also have a stronger effect on the viability of RPS15 mutant CLL cells as compared to WT cells. Finally, proteasome inhibitors may also have therapeutic potential for certain RPS19-mutant DBA patients. Two distinct classes of RPS19 mutants have been described: a first class with normal nucleolar localization and slightly decreased protein expression, and a second class characterized by a stronger reduction in protein expression on top of a failure to localize to the nucleolus. Proteasome inhibitor treatment of cells expressing the second class of RPS19 mutants leads to RPS19 protein restabilization and restoration of its nucleolar localization (101). Altogether, these observations underscore the effects of ribosomal defects on the proteasome and support that proteasome inhibitors may have potential for the treatment of RP-mutant diseases.
Excessive ROS levels
Reactive oxygen species (ROS) accumulate when cells increase the processing of very long-chain fatty acids in the peroxisomes or the mitochondrial electron chain reactions for ATP production, or upon limited availability of cellular anti-oxidants such as glutathione. Either of these processes can be affected by transformation-associated metabolic reprogramming of cells. Changes in RP expression levels, RP mutations or assembly factors can lead to dynamic adaptations in cellular metabolic preferences.
It is becoming clear that RP-mutant diseases are associated with high cellular oxidative stress due to increased cellular ROS levels. The mechanisms by which defective ribosomes induce elevated ROS levels are however poorly understood. In the case of the leukemia-associated RPL10-R98S mutation, this may arise from enhanced peroxisome activity (64). Furthermore, wild-type RPL10 regulates the expression of proteins related to ROS production and it controls mitochondrial ROS production in pancreatic cancer (102). RPL10-R98S may also alter these ROS regulatory functions. Other RPs have also been associated with oxidative stress responses. For example, ROS-inducing agents can cause RPS3 (uS3) translocation to the mitochondria, where it can protect cells from ROS-induced mitochondrial DNA damage (103).
Enhanced ROS levels can induce different cellular outcomes: low ROS levels stimulate cell proliferation by activating the PI3K and MAPK signaling pathways (104–107), whereas higher levels of ROS-associated oxidative stress become toxic and inhibit proliferation. In the context of RP mutations, the latter seems to apply: lowering cellular ROS levels by antioxidants can rescue the proliferation defects in RPL10-R98S cells (64), indicating that RPL10-R98S associated ROS production impairs cell proliferation. Elevated ROS levels have been linked to increased DNA damage and genomic instability in cancer (108). Leukemia-associated RP defects were shown to be connected to elevated DNA damage, with a mutational signature consistent with oxidative stress (64,109). Models of DBA also display enhanced oxidative stress and DNA damage. RPL5- and RPS19-deficient mouse erythroleukemia-DBA clones and DBA patient samples contained increased ROS levels and displayed higher levels of DNA double-strand breaks and 8-oxoguanine oxidative DNA damage (110). Additionally, TF-1 myeloid cells with knockdown of SBDS and SDS patient lymphocytes showed elevated cellular oxidative stress (111,112). In a mouse model with specific Sbds deletion in the mesenchymal cells of the hematopoietic niche, inflammation was induced, resulting in ROS accumulation and genotoxic stress in neighboring wild-type hematopoietic stem and progenitor cells (113). This observation supports that within the context of SDS, the microenvironment induces oxidative stress in hematopoietic stem cells. Also T-lymphocytes from DC patients display elevated ROS levels, and transient suppression of DKC1 induced both oxidative stress and expression of antioxidant enzymes in HeLa cells (73,114). While no changes in γH2A.X DNA double-strand break foci were observed, DNA lesions defined by protein poly ADP-ribosylation were increased. Finally, CHH patient samples display 2.4-fold higher expression of the ROS scavenger catalase (115), pointing toward elevated oxidative stress in this disease as well. Upregulation of ROS scavengers is however not a generalized phenotype in ribosomopathies: expression of genes that involved in ROS detoxification, such as superoxide dismutase 2, is decreased in DBA models such as Rpl11 zebrafish mutants and RPS19 (eS19)-deficient human cell lines (116,117). Besides a role for ROS in causing DNA damage, alternative pathways likely support the enhanced DNA damage that is observed in RP deficiencies, such as alterations in DNA repair mechanisms or failure of DNA damage recognition via the MDM2/TP53 axis.
High oxidative stress can also induce mitochondrial dysfunction, interfering with adequate oxidative phosphorylation, mitochondrial respiration and consequently TCA cycle intermediates and end products like ATP (64,111). This can contribute to the hypo-proliferative nature of early stage ribosomopathies (64,111,112,118). Furthermore, restricted availability of TCA-derived substrates might increase the pool of metabolic enzymes that are available to fulfill their additional roles as mRNA-binding proteins (58,119) as discussed below. Furthermore, rRNA is a target for oxidative nucleobase damage and, consequently, enhanced oxidative stress levels can interfere with ribosome assembly and different substeps of the translation elongation cycle (120), as well as reduce the fidelity of protein synthesis (121). This can further augment ROS production, leading to an oxidative and translation-defective cellular feedback loop that might even further advance a mutagenic phenotype.
Altered glycolysis and serine/glycine synthesis
Metabolic reprogramming occurs very frequently in cancer, as transformed cells depend on augmented nucleotide and protein synthesis and ATP. In addition, cellular protein synthesis is one of the most energy-consuming processes in the cell, estimated to consume 30% of cellular ATP (122). Therefore, it is not surprising that ribosomopathies affecting the cellular protein translation apparatus can have profound impacts on cellular energy metabolism.
Glycolytic changes have been observed in diseases associated with ribosomopathies. Leukocytes from CHH patients display elevated mRNA expression of glycolysis enzymes, such as fructose-1,6-bisphosphatase 1 (FBP1), glucokinase (GK) and hexokinase 2 (HK2) (115). SDS patient lymphoblasts contain low levels of pyruvate and present impaired oxidative phosphorylation, accompanied by elevated lactate levels, indicating that SBDS-deficient cells undergo high levels of glycolysis (111). In DBA, however, Rpl11-deficient zebrafish and RPS19-deficient mouse fetal liver cells downregulate genes encoding glycolytic enzymes and increase the expression of genes involved in aerobic respiration (117). The key glycolytic enzymes pyruvate kinase isozyme 2 (PKM2), fructose-bisphosphate aldolase A (ALDOA) and lactate dehydrogenase A (LDHA) directly bind to ribosomes (58), making it tempting to speculate that altered ribosome availability in ribosomopathies may directly impact glycolytic enzyme availability and activity.
Modulation of serine synthesis, a glycolysis-diverting pathway, is well-described in breast cancer (123). This however may also be relevant for ribosome-mutant disorders. Combined transcriptome and translatome analysis on cells expressing the RPL10-R98S mutation revealed that one of the key enzymes in serine synthesis—phosphoserine phosphatase (PSPH)—is more efficiently transcribed and translated in RPL10-R98S mutant cells. This mutant ribosome-dependent increase in PSPH translation resulted in enhanced serine/glycine synthesis, which in turn supported purine synthesis (81). Similarly, fibroblasts of DBA patients present elevated levels of serine/glycine synthesis enzymes phosphoglycerate dehydrogenase (PHGDH), phosphoserine aminotransferase 1 (PSAT1) and the mitochondrial serine hydroxymethyltransferase 2 (SHMT2) (124). This is further supported by the observation of increased PHGDH and PSAT1 mRNA levels in erythroid TF-1 cells upon knockdown of DBA-associated ribosomal proteins RPL5, RPS19 and RPL11 (116). Several studies thus support that ribosome-defective cells undergo metabolic reprogramming to benefit from glycolysis and serine synthesis. These intriguing observations however require further investigation to define whether metabolic rewiring is a direct consequence of particular ribosomal defects, as in the case of RPL10 R98S.
To add another layer of complexity, metabolic reprogramming and altered levels of metabolic intermediates and enzymes in ribosomopathies might in turn cause translational reprogramming. Besides their catalytic function to convert metabolites, many metabolic enzymes have been identified as mRNA-binding proteins (58,119). Why these proteins bind mRNAs and how this affects mRNA translation is still unknown. The enzymes for which these functions have been described cover much of the metabolomic landscape and do not cluster into particular pathways. Many of these enzymes share the ability to simultaneously bind ATP and a substrate such as succinate, aspartate or pyruvate. The influence of restricted availability of these metabolic intermediates on the RNA-binding capacity of the enzymes also requires further investigation. In addition, many metabolic enzymes form dimers or oligomers and, therefore, enzyme availability might affect RNA-binding ability or capacity. Furthermore, besides an important role as RNA-binding protein, a recent study highlights the requirement of SHMT2-driven serine catabolism for maintaining formylmethionyl-tRNAs, associated with proper mitochondrial translation initiation (125), providing another connection between cell metabolism and translational regulation.
TRANSITION FROM HYPO- TO HYPER-PROLIFERATION IN RIBOSOMOPATHIES AND RELATED BONE MARROW FAILURE SYNDROMES
In the sections above, cellular changes induced by ribosome defects were discussed. Here, we propose a model that integrates several of these findings and that can explain the paradoxical evolution from hypo- to hyper-proliferative disease phenotypes in ribosomopathy patients (Figure 4). Recent examples illustrate that a ribosome defect can generate specialized ‘onco-ribosome’ species with a shifted translational output, resulting in hypo-translation of tumor suppressors such as CDKN1B and TP53, and hyper-translation of oncogenes such as BCL-2, JAK-STAT proteins or VEGF (64,67–69,74). Altered extra-ribosomal regulation of proteins such as TP53, MYC or c-FOS/c-JUN represents a second means by which cancer-associated proteins can be affected. Despite these tumor-promoting changes in the cells, introduction of a RP or biogenesis defect typically impairs cell proliferation at first. This can be explained by the fact that dysregulation of the cellular oncogene/tumor suppressor balance is not the only cellular change that occurs: defective ribosomes also induce extensive oxidative stress and generate high ROS levels, which are toxic and impair cellular growth. The hypo-proliferative phenotypes associated with the RPL10-R98S mutation or with SBDS or DKC1 inactivation are entirely rescued by reduction of ROS levels with an antioxidant (64,109,112,114). Thus, as long as cells cannot lower these excessive ROS levels, hypo-proliferation persists. ROS damages multiple cellular components, including the DNA, causing high mutation rates. Alternatively, selection of MDM2- or TP53-deficient cells may bypass the DNA damage checkpoints and their repair, increasing the mutational load. At a certain moment, some of these newly acquired mutations may function in reducing cellular oxidative stress levels and relieve the hypo-proliferative pressure. In the context of the RPL10-R98S mutation, secondary NOTCH1 mutations have been shown to fulfill this function: acquisition of a NOTCH1 mutation was able to rescue the RPL10-R98S associated proliferation defect and reduced elevated ROS and DNA double-strand break levels. Other secondary mutations such as inactivating lesions in TP53 may exert this function in other ribosomopathies (109). As such, these secondary mutations can alleviate RP mutation associated toxicity, leading to transition into a hyper-proliferative state, with specialized translational and/or extra-ribosomal functions preserved. Other mutations caused by the initial acceleration of DNA damage can further promote cell growth and are selected in a continuous process, strengthening the hyper-proliferation phenotype yet again (Figure 4).
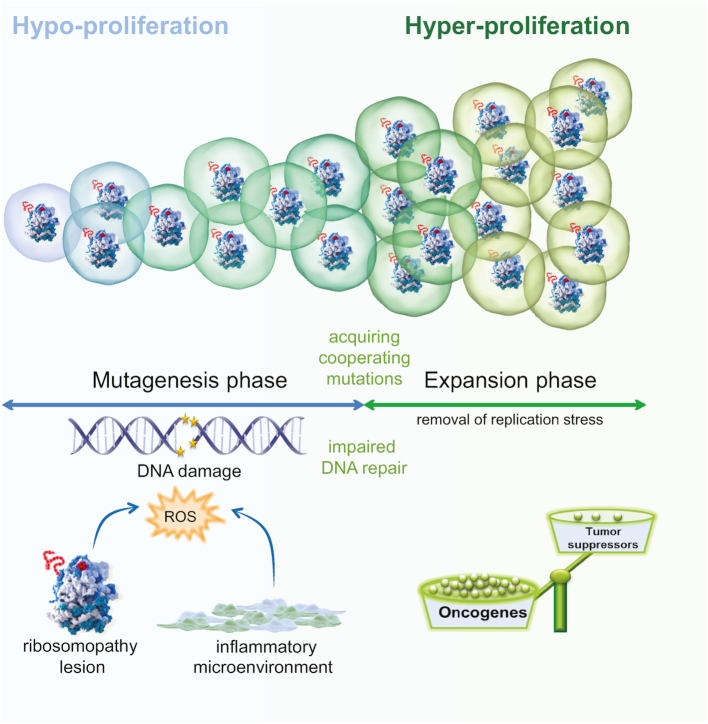
Figure 4.
Cellular hypo- to hyper-proliferation transition model. Intracellular oxidative stress caused by alterations in the assembly and/or function of ribosomes or by micro-environmental factors such as inflammation, impairs cell proliferation and promotes DNA damage. Defective DNA repair mechanisms can also induce or further aggravate this DNA damage. After a certain latency, this elevated DNA damage results in the acquisition of rescuing mutations that inhibit ROS production, thereby removing the block on cellular proliferation. At this stage, the expansion phase starts. This phase is then further accelerated by overexpression of oncogenes and/or downregulation of tumor suppressor genes caused by the excessive DNA damage that occurred in the hypoproliferative phase, as well as by reprogramming of the cellular protein translation landscape in the case of RP defects.
MDS with elevated risk to AML progression also occurs in patients that do not display ribosome defects, and causes of MDS are not always understood. The pathogenic role of elevated ROS levels and oxidative DNA damage may be similar in some non-ribosomopathy MDS patients, since increased oxidative DNA damage has been observed in hematopoietic progenitors from MDS patients (126). In contrast to the high ROS levels induced by a ribosome defect in the cell, ROS levels can originate from a microenvironment dependent inflammatory response (127). If our model explaining transition from hypo- to hyper-proliferation is correct, it implies that reducing oxidative stress levels in hematopoietic cells by means of antioxidants would alleviate the hypo-proliferative disease phase. It would prevent DNA damage and thus progression to the hyper-proliferative phase in bone marrow failure syndromes where excessive ROS levels are accumulating in the cells. This is an attractive idea that requires further investigation. Such a strategy will however not be efficient in all MDS syndromes, because the excessive genomic instability in these diseases can also be caused by defective DNA repair, as for instance in patients with the congenital bone marrow failure syndrome Fanconia Anemia.
MITORIBOSOMES
The vast majority of the human proteome is synthesized by cytoplasmic or endoplasmic reticulum associated ribosomes. However, in humans 13 proteins of the mitochondrial oxidative phosphorylation machinery are translated exclusively by mitochondrial ribosomes (mitoribosomes) that are built from nuclear genome-encoded mito-RPs and mitochondrial DNA-encoded rRNA. Mitochondria are generally accepted to be descendant of ancient bacteria that started to cooperate with primordial single-cell organisms. As a result, mitochondrial ribosomes structurally more closely resemble ribosomes found in modern day bacteria rather than those in the cytoplasm of eukaryotic cells. Mitoribosomes, however, display a much higher protein-to-rRNA ratio. It is widely believed that mitoribosomes lost segments of rRNA and replaced it with proteins as a part of a devolutionary process, as parasites and symbionts devolved by losing non-core components (128). However, we speculate that an alternate theory is also possible: because mitoribosomes function in the ROS-intensive environment of the mitochondria, they evolved a protein ‘shield’ to protect the rRNA core from ROS attack, which is particularly sensitive to oxidative nucleobase damage (120,121). As such, they have become ‘specialized’ to function in this predominantly harsh environment. This is particularly relevant in cancer cells, which generally display elevated levels of ROS and higher rates of mitochondrial biogenesis (129).
While mitoribosomes have been less intensively studied, their central role in cancer progression is becoming clear. For example, cancer cell fitness is improved through a concerted increase in both cytosolic and mitochondrial translation (130), and mitochondrial translation inhibition was proposed as an attractive therapeutic strategy for some cancers (129,131). The clinical relevance of mitochondrial translation is further highlighted by the occurrence of mutations in mitochondrial RP genes, associated with mitochondrial dysfunction disorders (reviewed in (128)). For example, a recent case report highlights a common familiar mitochondrial mutation that sensitizes the affected individuals to the ototoxicity of aminoglycoside antibiotics due to the mutation-induced structural changes in the mitoribosome (132). Somatic mutations in mito-RP genes regularly appear in cancer genomics datasets, but their significance in cancer pathogenesis has not been established. We speculate that these mutations may impact the functionality of cellular oxidative phosphorylation and may thus have a role in the well-known Warburg effect, whereby tumors switch from oxidative phosphorylation to anaerobic glycolysis as the main energy source.
CONCLUSIONS AND FUTURE PERSPECTIVES
As outlined in this review article, great progress has been made in understanding the ribosomopathies since the description on RPS19 mutations in DBA over 20 year ago. Several findings have also resulted in novel therapeutic approaches that can already be explored, or are finding their implementation into the clinic. An overview on the potential therapeutic options for ribosomopathies has been provided elsewhere (14). More recently, a novel therapeutic opportunity was described to target the leukemic addiction to BCL-2, provided by the IRES-mediated translation in RPL10-R98S mutant leukemias (64). Furthermore, since high ROS levels seem to be a general consequence of ribosomal defects, it can be hypothesized that ribosomopathy patients may benefit from antioxidant supplements during the hypo-proliferative period to control cellular oxidative stress and its associated DNA damage.
Despite recent advances, a number of questions still require investigation. First, the concept of ribosome specialization needs further attention. As described in detail above, there are now plenty of examples, both in congenital and somatic ribosomopathies, supporting a differential translational output of ribosomes with RP or ribosome biogenesis factor defects. However, the exact composition of these specialized ribosomes has not been elucidated. In-depth structural studies by cryogenic electron microscopy (cryo-EM), combined with analysis of the protein composition, the rRNA modification status and the spectrum of proteins interacting with these ribosomes (ribo-interactome), would be required to characterize these specialized ribosomes in more detail. This might in turn encourage the development of ‘specialized’ translation inhibitors. Indeed, new classes of translation inhibitors are emerging that selectively target translation of small subsets of mRNAs, with few off-target effects (133,134).
A second open question concerns the spectrum of mutations in ribosomopathies. For example, in 25% of DBA patients, the causal mutation for their disease remains unidentified. Furthermore, somatic mutations have been described in genes to which multiple functions, including regulating ribosome biogenesis, have been associated. Examples here are the mutations in the NPM1 gene, observed in one-third of AML patients, as well as mutations in PHF6, present in 20% of T-ALL and 3% of AML patients (135–137). It is currently however unclear whether the mutations in these genes are promoting cancer by dysregulating the ribosome. Also mutations in factors affecting tRNA biology can alter translation, leading to ribosomopathy phenotypes. In this regard, loss of PUS7, an enzyme involved in tRNA pseudouridinylation, has recently been described in MDS and has been shown to drive elevated protein translation (138).
While many of the described cellular changes in ribosomopathies are cell autonomous, non-cell autonomous effects can also assist in transformation of precancerous ribosome defective cells. Supportive mesenchymal cells affect ribosome-defective cells by exposure to genotoxic stress and inducing a pro-inflammatory response that enhances oxidative stress and DNA damage (113). Activation of the innate immune system is also observed in Rps14-deficient mouse model of MDS (139). In addition, due to high ROS and metabolic rewiring, both oxygen and nutrient diffusion from dynamic autocrine and paracrine pathways can assist ribosome-defective cells and their interacting supportive niches, potentially contributing to cancerous transformation (81).
To quote the Nobel laureate Albert Szent-Gyorgyi: ‘Discovery consists of looking at the same thing as everyone else and thinking something different.’ In regards to the ribosomes field, such thinking enabled to centrally position the translational machinery in cancer research. We are confident that expecting the unexpected will continue to lead to novel discoveries and insights to ultimately improve the quality of life for patients suffering from ribosomopathies and prevent the progression to cancer.
ACKNOWLEDGEMENTS
We thank all researchers and clinicians for their contributions to the field and apologize to those whose work we did not describe or cite.
FUNDING
Postdoctoral Fellowship, Kom Op Tegen Kanker(to K.R.K.); Leukemia Research Grant, ‘Me To You’ Foundation (to K.R.K.); SB PhD Fellowship, FWO (n° 1S49817N) (to S.V.); Research Grant,FWO [G067015N to K.D.K.]; Research Grant, Stichting Tegen Kanker [2016-775, 2016-801 to K.D.K.]; Research Grant, KU Leuven Research Council [C14/18/104 to K.D.K.]. Funding for open access charge: FWO [G067015N].
Conflict of interest statement. None declared.
REFERENCES
- 1. Warner J.R., Vilardell J., Sohn J.H.. Economics of ribosome biosynthesis. Cold Spring Harb. Symp. Quant. Biol. 2001; 66:567–574. [DOI] [PubMed] [Google Scholar]
- 2. Narla A., Ebert B.L.. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010; 115:3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mills E.W., Green R.. Ribosomopathies: there’s strength in numbers. Science. 2017; 358:eaan2755. [DOI] [PubMed] [Google Scholar]
- 4. Sulima S.O., De Keersmaecker K.. Bloody mysteries of ribosomes. Hemasphere. 2018; 2:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerst J.E. Pimp my Ribosome: Ribosomal protein paralogs specify translational control. Trends Genet. 2018; 34:832–845. [DOI] [PubMed] [Google Scholar]
- 6. De Keersmaecker K., Sulima S.O., Dinman J.D.. Ribosomopathies and the paradox of cellular hypo- to hyperproliferation. Blood. 2015; 125:1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vlachos A., Rosenberg P.S., Atsidaftos E., Kang J., Onel K., Sharaf R.N., Alter B.P., Lipton J.M.. Increased risk of colon cancer and osteogenic sarcoma in Diamond-Blackfan anemia. Blood. 2018; 132:2205–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alter B.P., Giri N., Savage S.A., Rosenberg P.S.. Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica. 2018; 103:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taskinen M., Ranki A., Pukkala E., Jeskanen L., Kaitila I., Makitie O.. Extended follow-up of the Finnish cartilage-hair hypoplasia cohort confirms high incidence of non-Hodgkin lymphoma and basal cell carcinoma. Am. J. Med. Genet. A. 2008; 146A:2370–2375. [DOI] [PubMed] [Google Scholar]
- 10. Sulima S.O., Kampen K.R., De Keersmaecker K.. Cancer biogenesis in ribosomopathies. Cells. 2019; 8:E229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanahan D., Weinberg R.A.. The hallmarks of cancer. Cell. 2000; 100:57–70. [DOI] [PubMed] [Google Scholar]
- 12. Klinge S., Woolford J.L. Jr. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019; 20:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma S., Lafontaine D.L.J.. ‘View from a Bridge’: A new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015; 40:560–575. [DOI] [PubMed] [Google Scholar]
- 14. Sulima S.O., Hofman I.J.F., De Keersmaecker K., Dinman J.D.. How ribosomes translate cancer. Cancer Discov. 2017; 7:1069–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ban N., Beckmann R., Cate J.H.D., Dinman J.D., Dragon F., Ellis S.R., Lafontaine D.L.J., Lindahl L., Liljas A., Lipton J.M. et al.. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014; 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Draptchinskaia N., Gustavsson P., Andersson B., Pettersson M., Willig T.N., Dianzani I., Ball S., Tchernia G., Klar J., Matsson H. et al.. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999; 21:169–175. [DOI] [PubMed] [Google Scholar]
- 17. Aspesi A., Ellis S.R.. Kupfer GM, Reaman GH, Smith FO. Bone Marrow Failure. 2018; Cham: Springer International Publishing; 99–110. [Google Scholar]
- 18. Woloszynek J.R., Rothbaum R.J., Rawls A.S., Minx P.J., Wilson R.K., Mason P.J., Bessler M., Link D.C.. Mutations of the SBDS gene are present in most patients with Shwachman-Diamond syndrome. Blood. 2004; 104:3588–3590. [DOI] [PubMed] [Google Scholar]
- 19. Weis F., Giudice E., Churcher M., Jin L., Hilcenko C., Wong C.C., Traynor D., Kay R.R., Warren A.J.. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015; 22:914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Finch A.J., Hilcenko C., Basse N., Drynan L.F., Goyenechea B., Menne T.F., Gonzalez Fernandez A., Simpson P., D'Santos C.S., Arends M.J. et al.. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011; 25:917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thiel C.T., Horn D., Zabel B., Ekici A.B., Salinas K., Gebhart E., Ruschendorf F., Sticht H., Spranger J., Muller D. et al.. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am. J. Hum. Genet. 2005; 77:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goldfarb K.C., Cech T.R.. Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev. 2017; 31:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flygare J., Aspesi A., Bailey J.C., Miyake K., Caffrey J.M., Karlsson S., Ellis S.R.. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007; 109:980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gazda H.T., Sheen M.R., Vlachos A., Choesmel V., O’Donohue M.-F., Schneider H., Darras N., Hasman C., Sieff C.A., Newburger P.E. et al.. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 2008; 83:769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quarello P., Garelli E., Carando A., Mancini C., Foglia L., Botto C., Farruggia P., De Keersmaecker K., Aspesi A., Ellis S.R. et al.. Ribosomal RNA analysis in the diagnosis of Diamond-Blackfan Anaemia. Br. J. Haematol. 2016; 172:782–785. [DOI] [PubMed] [Google Scholar]
- 26. Mason P.J., Bessler M.. The genetics of dyskeratosis congenita. Cancer Genet. 2011; 204:635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dauwerse J.G., Dixon J., Seland S., Ruivenkamp C.A.L., van Haeringen A., Hoefsloot L.H., Peters D.J.M., Boers A.C.-d., Daumer-Haas C., Maiwald R. et al.. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat. Genet. 2011; 43:20–22. [DOI] [PubMed] [Google Scholar]
- 28. Bowman M., Oldridge M., Archer C., O’Rourke A., McParland J., Brekelmans R., Seller A., Lester T.. Gross deletions in TCOF1 are a cause of Treacher-Collins-Franceschetti syndrome. Eur. J. Human Genetics. 2012; 20:769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Splendore A., Jabs E.W., Passos-Bueno M.R.. Screening of TCOF1 in patients from different populations: confirmation of mutational hot spots and identification of a novel missense mutation that suggests an important functional domain in the protein treacle. J. Med. Genet. 2002; 39:493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nieminen T.T., O’Donohue M.-F., Wu Y., Lohi H., Scherer S.W., Paterson A.D., Ellonen P., Abdel-Rahman W.M., Valo S., Mecklin J.-P. et al.. Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology. 2014; 147:595–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ajore R., Raiser D., McConkey M., Joud M., Boidol B., Mar B., Saksena G., Weinstock D.M., Armstrong S., Ellis S.R. et al.. Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol. Med. 2017; 9:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fancello L., Kampen K.R., Hofman I.J.F., Verbeeck J., De Keersmaecker K.. The ribosomal protein gene RPL5 is a haploinsufficient tumor suppressor in multiple cancer types. Oncotarget. 2017; 8:14462–14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tzoneva G., Perez-Garcia A., Carpenter Z., Khiabanian H., Tosello V., Allegretta M., Paietta E., Racevskis J., Rowe J.M., Tallman M.S. et al.. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat. Med. 2013; 19:368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rao S., Lee S.-Y., Gutierrez A., Perrigoue J., Thapa R.J., Tu Z., Jeffers J.R., Rhodes M., Anderson S., Oravecz T. et al.. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012; 120:3764–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferreira A.M., Tuominen I., van Dijk-Bos K., Sanjabi B., van der Sluis T., van der Zee A.G., Hollema H., Zazula M., Sijmons R.H., Aaltonen L.A. et al.. High frequency of RPL22 mutations in microsatellite-unstable colorectal and endometrial tumors. Hum. Mutat. 2014; 35:1442–1445. [DOI] [PubMed] [Google Scholar]
- 36. Nagarajan N., Bertrand D., Hillmer A.M., Zang Z.J., Yao F., Jacques P.-E., Teo A.S.M., Cutcutache I., Zhang Z., Lee W.H. et al.. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol. 2012; 13:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Novetsky A.P., Zighelboim I., Thompson D.M. Jr, Powell M.A., Mutch D.G., Goodfellow P.J.. Frequent mutations in the RPL22 gene and its clinical and functional implications. Gynecol. Oncol. 2013; 128:470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hofman I.J.F., van Duin M., De Bruyne E., Fancello L., Mulligan G., Geerdens E., Garelli E., Mancini C., Lemmens H., Delforge M. et al.. RPL5 on 1p22.1 is recurrently deleted in multiple myeloma and its expression is linked to bortezomib response. Leukemia. 2017; 31:1706–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Amsterdam A., Sadler K.C., Lai K., Farrington S., Bronson R.T., Lees J.A., Hopkins N.. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004; 2:E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rao S., Cai K.Q., Stadanlick J.E., Greenberg-Kushnir N., Solanki-Patel N., Lee S.-Y., Fahl S.P., Testa J.R., Wiest D.L.. Ribosomal protein Rpl22 controls the dissemination of T-cell Lymphoma. Cancer Res. 2016; 76:3387–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morgado-Palacin L., Varetti G., Llanos S., Gomez-Lopez G., Martinez D., Serrano M.. Partial loss of Rpl11 in adult mice recapitulates Diamond-Blackfan Anemia and Promotes Lymphomagenesis. Cell Rep. 2015; 13:712–722. [DOI] [PubMed] [Google Scholar]
- 42. Boultwood J., Fidler C., Strickson A.J., Watkins F., Gama S., Kearney L., Tosi S., Kasprzyk A., Cheng J.-F., Jaju R.J. et al.. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood. 2002; 99:4638–4641. [DOI] [PubMed] [Google Scholar]
- 43. Hosono N., Makishima H., Mahfouz R., Przychodzen B., Yoshida K., Jerez A., LaFramboise T., Polprasert C., Clemente M.J., Shiraishi Y. et al.. Recurrent genetic defects on chromosome 5q in myeloid neoplasms. Oncotarget. 2017; 8:6483–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G.. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014; 505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. De Keersmaecker K., Atak Z.K., Li N., Vicente C., Patchett S., Girardi T., Gianfelici V., Geerdens E., Clappier E., Porcu M. et al.. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 2013; 45:186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Y., Easton J., Shao Y., Maciaszek J., Wang Z., Wilkinson M.R., McCastlain K., Edmonson M., Pounds S.B., Shi L. et al.. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 2017; 49:1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Teng T., Mercer C.A., Hexley P., Thomas G., Fumagalli S.. Loss of tumor suppressor RPL5/RPL11 does not induce cell cycle arrest but impedes proliferation due to reduced ribosome content and translation capacity. Mol. Cell Biol. 2013; 33:4660–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O’Leary M.N., Schreiber K.H., Zhang Y., Duc A.-C.E., Rao S., Hale J.S., Academia E.C., Shah S.R., Morton J.F., Holstein C.A. et al.. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 2013; 9:e1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ljungstrom V., Cortese D., Young E., Pandzic T., Mansouri L., Plevova K., Ntoufa S., Baliakas P., Clifford R., Sutton L.-A. et al.. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood. 2016; 127:1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bretones G., Alvarez M.G., Arango J.R., Rodriguez D., Nadeu F., Prado M.A., Valdes-Mas R., Puente D.A., Paulo J.A., Delgado J. et al.. Altered patterns of global protein synthesis and translational fidelity in RPS15-mutated chronic lymphocytic leukemia. Blood. 2018; 132:2375–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Parks M.M., Kurylo C.M., Dass R.A., Bojmar L., Lyden D., Vincent C.T., Blanchard S.C.. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 2018; 4:eaao0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Killen M.W., Stults D.M., Adachi N., Hanakahi L., Pierce A.J.. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum. Mol. Genet. 2009; 18:3417–3428. [DOI] [PubMed] [Google Scholar]
- 53. Wang M., Lemos B.. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 2017; 13:e1006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu B., Li H., Perry J.M., Singh V.P., Unruh J., Yu Z., Zakari M., McDowell W., Li L., Gerton J.L.. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017; 13:e1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Udugama M., Sanij E., Voon H.P.J., Son J., Hii L., Henson J.D., Chan F.L., Chang F.T.M., Liu Y., Pearson R.B. et al.. Ribosomal DNA copy loss and repeat instability in ATRX-mutated cancers. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:4737–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Genuth N.R., Barna M.. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genetics. 2018; 19:431–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Genuth N.R., Barna M.. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell. 2018; 71:364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simsek D., Tiu G.C., Flynn R.A., Byeon G.W., Leppek K., Xu A.F., Chang H.Y., Barna M.. The mammalian Ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell. 2017; 169:1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shi Z., Fujii K., Kovary K.M., Genuth N.R., Rost H.L., Teruel M.N., Barna M.. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs Genome-wide. Mol. Cell. 2017; 67:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sulima S.O., Patchett S., Advani V.M., De Keersmaecker K., Johnson A.W., Dinman J.D.. Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5640–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Y., Ear J., Yang Z., Morimoto K., Zhang B., Lin S.. Defects of protein production in erythroid cells revealed in a zebrafish Diamond-Blackfan anemia model for mutation in RPS19. Cell Death Dis. 2014; 5:e1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. MacInnes A.W., Amsterdam A., Whittaker C.A., Hopkins N., Lees J.A.. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:10408–10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xue S., Tian S., Fujii K., Kladwang W., Das R., Barna M.. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015; 517:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kampen K.R., Sulima S.O., Verbelen B., Girardi T., Vereecke S., Rinaldi G., Verbeeck J., Op de Beeck J., Uyttebroeck A., Meijerink J.P.P. et al.. The ribosomal RPL10 R98S mutation drives IRES-dependent BCL-2 translation in T-ALL. Leukemia. 2019; 33:319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wood J., Frederickson R.M., Fields S., Patel A.H.. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol. 2001; 75:1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Horos R., Ijspeert H., Pospisilova D., Sendtner R., Andrieu-Soler C., Taskesen E., Nieradka A., Cmejla R., Sendtner M., Touw I.P. et al.. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012; 119:262–272. [DOI] [PubMed] [Google Scholar]
- 67. Bellodi C., Kopmar N., Ruggero D.. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010; 29:1865–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yoon A., Peng G., Brandenburger Y., Brandenburg Y., Zollo O., Xu W., Rego E., Ruggero D.. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006; 312:902–906. [DOI] [PubMed] [Google Scholar]
- 69. Rocchi L., Pacilli A., Sethi R., Penzo M., Schneider R.J., Trere D., Brigotti M., Montanaro L.. Dyskerin depletion increases VEGF mRNA internal ribosome entry site-mediated translation. Nucleic Acids Res. 2013; 41:8308–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Marcel V., Ghayad S.E., Belin S., Therizols G., Morel A.-P., Solano-Gonzalez E., Vendrell J.A., Hacot S., Mertani H.C., Albaret M.A. et al.. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013; 24:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sharma S., Marchand V., Motorin Y., Lafontaine D.L.J.. Identification of sites of 2′-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci. Rep. 2017; 7:11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Advani V.M., Dinman J.D.. Reprogramming the genetic code: The emerging role of ribosomal frameshifting in regulating cellular gene expression. Bioessays. 2016; 38:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jack K., Bellodi C., Landry D.M., Niederer R.O., Meskauskas A., Musalgaonkar S., Kopmar N., Krasnykh O., Dean A.M., Thompson S.R. et al.. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell. 2011; 44:660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Girardi T., Vereecke S., Sulima S.O., Khan Y., Fancello L., Briggs J.W., Schwab C., de Beeck J.O., Verbeeck J., Royaert J. et al.. The T-cell leukemia-associated ribosomal RPL10 R98S mutation enhances JAK-STAT signaling. Leukemia. 2018; 32:809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. In K., Zaini M.A., Muller C., Warren A.J., von Lindern M., Calkhoven C.F.. Shwachman-Bodian-Diamond syndrome (SBDS) protein deficiency impairs translation re-initiation from C/EBPalpha and C/EBPbeta mRNAs. Nucleic Acids Res. 2016; 44:4134–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pabst T., Mueller B.U.. Transcriptional dysregulation during myeloid transformation in AML. Oncogene. 2007; 26:6829–6837. [DOI] [PubMed] [Google Scholar]
- 77. Truitt M.L., Ruggero D.. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer. 2016; 16:288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ludwig L.S., Gazda H.T., Eng J.C., Eichhorn S.W., Thiru P., Ghazvinian R., George T.I., Gotlib J.R., Beggs A.H., Sieff C.A. et al.. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat. Med. 2014; 20:748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Khajuria R.K., Munschauer M., Ulirsch J.C., Fiorini C., Ludwig L.S., McFarland S.K., Abdulhay N.J., Specht H., Keshishian H., Mani D.R. et al.. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell. 2018; 173:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sankaran V.G., Ghazvinian R., Do R., Thiru P., Vergilio J.-A., Beggs A.H., Sieff C.A., Orkin S.H., Nathan D.G., Lander E.S. et al.. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J. Clin. Invest. 2012; 122:2439–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kampen K., Fancello L., Girardi T., Rinaldi G., Planque M., Sulima S.O., Loayza-Puch F., Verbelen B., Vereecke S., Verbeeck J. et al.. Translatome analysis reveals altered serine/glycine metabolism in T-cell acute lymphoblastic leukemia. Nat. Comm. 2019; 10:2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Molavi G., Samadi N., Hosseingholi E.Z.. The roles of moonlight ribosomal proteins in the development of human cancers. J. Cell Physiol. 2018; 234:8327–8341. [DOI] [PubMed] [Google Scholar]
- 83. Dai M.-S., Sears R., Lu H.. Feedback regulation of c-Myc by ribosomal protein L11. Cell Cycle. 2007; 6:2735–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dai M.-S., Sun X.-X., Lu H.. Ribosomal protein L11 associates with c-Myc at 5 S rRNA and tRNA genes and regulates their expression. J. Biol. Chem. 2010; 285:12587–12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liao J.M., Zhou X., Gatignol A., Lu H.. Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. Oncogene. 2014; 33:4916–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhou X., Hao Q., Liao J.-M., Liao P., Lu H.. Ribosomal protein S14 negatively regulates c-Myc activity. J. Biol. Chem. 2013; 288:21793–21801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Golomb L., Volarevic S., Oren M.. p53 and ribosome biogenesis stress: the essentials. FEBS Lett. 2014; 588:2571–2579. [DOI] [PubMed] [Google Scholar]
- 88. Donati G., Montanaro L., Derenzini M.. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res. 2012; 72:1602–1607. [DOI] [PubMed] [Google Scholar]
- 89. Bursac S., Brdovcak M.C., Pfannkuchen M., Orsolic I., Golomb L., Zhu Y., Katz C., Daftuar L., Grabusic K., Vukelic I. et al.. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:20467–20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Monteclaro F.S., Vogt P.K.. A Jun-binding protein related to a putative tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:6726–6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Inada H., Mukai J., Matsushima S., Tanaka T.. QM is a novel zinc-binding transcription regulatory protein: its binding to c-Jun is regulated by zinc ions and phosphorylation by protein kinase C. Biochem. Biophys. Res. Commun. 1997; 230:331–334. [DOI] [PubMed] [Google Scholar]
- 92. Imafuku I., Masaki T., Waragai M., Takeuchi S., Kawabata M., Hirai S., Ohno S., Nee L.E., Lippa C.F., Kanazawa I. et al.. Presenilin 1 suppresses the function of c-Jun homodimers via interaction with QM/Jif-1. J. Cell Biol. 1999; 147:121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Oughtred R., Stark C., Breitkreutz B.-J., Rust J., Boucher L., Chang C., Kolas N., O’Donnell L., Leung G., McAdam R. et al.. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019; 47:D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Orro S., Aspesi A., Armiraglio M., Caterino M., Loreni F., Ruoppolo M., Santoro C., Dianzani I.. Analysis of the ribosomal protein S19 interactome. Mol. Cell Proteom. 2007; 6:382–393. [DOI] [PubMed] [Google Scholar]
- 95. Caterino M., Aspesi A., Pavesi E., Imperlini E., Pagnozzi D., Ingenito L., Santoro C., Dianzani I., Ruoppolo M.. Analysis of the interactome of ribosomal protein S19 mutants. Proteomics. 2014; 14:2286–2296. [DOI] [PubMed] [Google Scholar]
- 96. Pesciotta E.N., Lam H.-S., Kossenkov A., Ge J., Showe L.C., Mason P.J., Bessler M., Speicher D.W.. In-Depth, Label-Free analysis of the erythrocyte cytoplasmic proteome in Diamond Blackfan Anemia identifies a unique inflammatory signature. PLoS One. 2015; 10:e0140036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Klare N., Seeger M., Janek K., Jungblut P.R., Dahlmann B.. Intermediate-type 20 S proteasomes in HeLa cells: “asymmetric” subunit composition, diversity and adaptation. J. Mol. Biol. 2007; 373:1–10. [DOI] [PubMed] [Google Scholar]
- 98. Dahlmann B., Ruppert T., Kuehn L., Merforth S., Kloetzel P.M.. Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J. Mol. Biol. 2000; 303:643–653. [DOI] [PubMed] [Google Scholar]
- 99. Huber E.M., Basler M., Schwab R., Heinemeyer W., Kirk C.J., Groettrup M., Groll M.. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012; 148:727–738. [DOI] [PubMed] [Google Scholar]
- 100. Mishto M., Liepe J., Textoris-Taube K., Keller C., Henklein P., WeberruSs M., Dahlmann B., Enenkel C., Voigt A., Kuckelkorn U. et al.. Proteasome isoforms exhibit only quantitative differences in cleavage and epitope generation. Eur. J. Immunol. 2014; 44:3508–3521. [DOI] [PubMed] [Google Scholar]
- 101. Cretien A., Hurtaud C., Moniz H., Proust A., Marie I., Wagner-Ballon O., Choesmel V., Gleizes P.-E., Leblanc T., Delaunay J. et al.. Study of the effects of proteasome inhibitors on ribosomal protein S19 (RPS19) mutants, identified in patients with Diamond-Blackfan anemia. Haematologica. 2008; 93:1627–1634. [DOI] [PubMed] [Google Scholar]
- 102. Yang J., Chen Z., Liu N., Chen Y.. Ribosomal protein L10 in mitochondria serves as a regulator for ROS level in pancreatic cancer cells. Redox Biol. 2018; 19:158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kim Y., Kim H.D., Kim J.. Cytoplasmic ribosomal protein S3 (rpS3) plays a pivotal role in mitochondrial DNA damage surveillance. Biochim. Biophys. Acta. 2013; 1833:2943–2952. [DOI] [PubMed] [Google Scholar]
- 104. Schieber M., Chandel N.S.. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014; 24:R453–R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sullivan L.B., Martinez-Garcia E., Nguyen H., Mullen A.R., Dufour E., Sudarshan S., Licht J.D., Deberardinis R.J., Chandel N.S.. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol. Cell. 2013; 51:236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ishikawa K., Takenaga K., Akimoto M., Koshikawa N., Yamaguchi A., Imanishi H., Nakada K., Honma Y., Hayashi J.-I.. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008; 320:661–664. [DOI] [PubMed] [Google Scholar]
- 107. Woo D.K., Green P.D., Santos J.H., D'Souza A.D., Walther Z., Martin W.D., Christian B.E., Chandel N.S., Shadel G.S.. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/+) mice. Am. J. Pathol. 2012; 180:24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tubbs A., Nussenzweig A.. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017; 168:644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sulima S.O., Kampen K.R., Vereecke S., Pepe D., Fancello L., Verbeeck J., Dinman J.D., De Keersmaecker K.. Ribosomal lesions promote oncogenic mutagenesis. Cancer Res. 2019; 79:320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kapralova K., Saxova Z., Kralova B., Lanikova L., Pospisilova D., Divoky V., Horvathova M.. Inflammatory signature, oxidative stress, and DNA damage response in DBA pathogenesis. Blood. 2017; 130:2452. [Google Scholar]
- 111. Ravera S., Dufour C., Cesaro S., Bottega R., Faleschini M., Cuccarolo P., Corsolini F., Usai C., Columbaro M., Cipolli M. et al.. Evaluation of energy metabolism and calcium homeostasis in cells affected by Shwachman-Diamond syndrome. Sci. Rep. 2016; 6:25441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ambekar C., Das B., Yeger H., Dror Y.. SBDS-deficiency results in deregulation of reactive oxygen species leading to increased cell death and decreased cell growth. Pediatr. Blood Cancer. 2010; 55:1138–1144. [DOI] [PubMed] [Google Scholar]
- 113. Zambetti N.A., Ping Z., Chen S., Kenswil K.J.G., Mylona M.A., Sanders M.A., Hoogenboezem R.M., Bindels E.M.J., Adisty M.N., Van Strien P.M.H. et al.. Mesenchymal inflammation drives genotoxic stress in hematopoietic stem cells and predicts disease evolution in human Pre-leukemia. Cell Stem Cell. 2016; 19:613–627. [DOI] [PubMed] [Google Scholar]
- 114. Pereboeva L., Westin E., Patel T., Flaniken I., Lamb L., Klingelhutz A., Goldman F.. DNA damage responses and oxidative stress in dyskeratosis congenita. PLoS One. 2013; 8:e76473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hermanns P., Bertuch A.A., Bertin T.K., Dawson B., Schmitt M.E., Shaw C., Zabel B., Lee B.. Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Hum. Mol. Genet. 2005; 14:3723–3740. [DOI] [PubMed] [Google Scholar]
- 116. Aspesi A., Pavesi E., Robotti E., Crescitelli R., Boria I., Avondo F., Moniz H., Da Costa L., Mohandas N., Roncaglia P. et al.. Dissecting the transcriptional phenotype of ribosomal protein deficiency: implications for Diamond-Blackfan Anemia. Gene. 2014; 545:282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Danilova N., Sakamoto K.M., Lin S.. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br. J. Haematol. 2011; 152:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Calamita P., Gatti G., Miluzio A., Scagliola A., Biffo S.. Translating the Game: ribosomes as active players. Front. Genet. 2018; 9:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al.. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 120. Willi J., Kupfer P., Evequoz D., Fernandez G., Katz A., Leumann C., Polacek N.. Oxidative stress damages rRNA inside the ribosome and differentially affects the catalytic center. Nucleic Acids Res. 2018; 46:1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Netzer N., Goodenbour J.M., David A., Dittmar K.A., Jones R.B., Schneider J.R., Boone D., Eves E.M., Rosner M.R., Gibbs J.S. et al.. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009; 462:522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Buttgereit F., Brand M.D.. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995; 312:163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.-K., Jang H.G., Jha A.K. et al.. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011; 476:346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Avondo F., Roncaglia P., Crescenzio N., Krmac H., Garelli E., Armiraglio M., Castagnoli C., Campagnoli M.F., Ramenghi U., Gustincich S. et al.. Fibroblasts from patients with Diamond-Blackfan anaemia show abnormal expression of genes involved in protein synthesis, amino acid metabolism and cancer. BMC Genomics. 2009; 10:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Minton D.R., Nam M., McLaughlin D.J., Shin J., Bayraktar E.C., Alvarez S.W., Sviderskiy V.O., Papagiannakopoulos T., Sabatini D.M., Birsoy K. et al.. Serine catabolism by SHMT2 is required for proper mitochondrial translation initiation and maintenance of Formylmethionyl-tRNAs. Mol. Cell. 2018; 69:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Novotna B., Bagryantseva Y., Siskova M., Neuwirtova R.. Oxidative DNA damage in bone marrow cells of patients with low-risk myelodysplastic syndrome. Leuk. Res. 2009; 33:340–343. [DOI] [PubMed] [Google Scholar]
- 127. Sallman D.A., List A.. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood. 2019; 133:1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Greber B.J., Ban N.. Structure and function of the mitochondrial ribosome. Annu. Rev. Biochem. 2016; 85:103–132. [DOI] [PubMed] [Google Scholar]
- 129. Skrtic M., Sriskanthadevan S., Jhas B., Gebbia M., Wang X., Wang Z., Hurren R., Jitkova Y., Gronda M., Maclean N. et al.. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011; 20:674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Vendramin R., Verheyden Y., Ishikawa H., Goedert L., Nicolas E., Saraf K., Armaos A., Delli Ponti R., Izumikawa K., Mestdagh P. et al.. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat. Struct. Mol. Biol. 2018; 25:1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lamb R., Ozsvari B., Lisanti C.L., Tanowitz H.B., Howell A., Martinez-Outschoorn U.E., Sotgia F., Lisanti M.P.. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: treating cancer like an infectious disease. Oncotarget. 2015; 6:4569–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ou Y.-H., Chen A.W.-G., Fan J.-Y., Cheng W.-L., Lin T.-T., Chen M.-K., Liu C.-S.. Aminoglycoside-associated nonsyndromic deafness and speech disorder in mitochondrial A1555G mutation in a family: a case report. Medicine (Baltimore). 2018; 97:e12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Lintner N.G., McClure K.F., Petersen D., Londregan A.T., Piotrowski D.W., Wei L., Xiao J., Bolt M., Loria P.M., Maguire B. et al.. Selective stalling of human translation through small-molecule engagement of the ribosome nascent chain. PLoS Biol. 2017; 15:e2001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Petersen D.N., Hawkins J., Ruangsiriluk W., Stevens K.A., Maguire B.A., O’Connell T.N., Rocke B.N., Boehm M., Ruggeri R.B., Rolph T. et al.. A Small-Molecule Anti-secretagogue of PCSK9 targets the 80S ribosome to inhibit PCSK9 protein translation. Cell Chem. Biol. 2016; 23:1362–1371. [DOI] [PubMed] [Google Scholar]
- 135. Falini B., Mecucci C., Tiacci E., Alcalay M., Rosati R., Pasqualucci L., La Starza R., Diverio D., Colombo E., Santucci A. et al.. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005; 352:254–266. [DOI] [PubMed] [Google Scholar]
- 136. Van Vlierberghe P., Palomero T., Khiabanian H., Van der Meulen J., Castillo M., Van Roy N., De Moerloose B., Philippe J., Gonzalez-Garcia S., Toribio M.L. et al.. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat. Genet. 2010; 42:338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Van Vlierberghe P., Patel J., Abdel-Wahab O., Lobry C., Hedvat C.V., Balbin M., Nicolas C., Payer A.R., Fernandez H.F., Tallman M.S. et al.. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011; 25:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Guzzi N., Ciesla M., Ngoc P.C.T., Lang S., Arora S., Dimitriou M., Pimkova K., Sommarin M.N.E., Munita R., Lubas M. et al.. Pseudouridylation of tRNA-Derived fragments steers translational control in stem cells. Cell. 2018; 173:1204–1216. [DOI] [PubMed] [Google Scholar]
- 139. Schneider R.K., Schenone M., Ferreira M.V., Kramann R., Joyce C.E., Hartigan C., Beier F., Brummendorf T.H., Germing U., Platzbecker U. et al.. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat. Med. 2016; 22:288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Yokoyama T., Machida K., Iwasaki W., Shigeta T., Nishimoto M., Takahashi M., Sakamoto A., Yonemochi M., Harada Y., Shigematsu H. et al.. HCV IRES captures an actively translating 80S ribosome. Mol. Cell. 2019; 74:1205–1214. [DOI] [PubMed] [Google Scholar]