Abstract
Release of phosphorothioate antisense oligonucleotides (PS-ASOs) from late endosomes (LEs) is a rate-limiting step and a poorly defined process for productive intracellular ASO drug delivery. Here, we examined the role of Golgi-endosome transport, specifically M6PR shuttling mediated by GCC2, in PS-ASO trafficking and activity. We found that reduction in cellular levels of GCC2 or M6PR impaired PS-ASO release from endosomes and decreased PS-ASO activity in human cells. GCC2 relocated to LEs upon PS-ASO treatment, and M6PR also co-localized with PS-ASOs in LEs or on LE membranes. These proteins act through the same pathway to influence PS-ASO activity, with GCC2 action preceding that of M6PR. Our data indicate that M6PR binds PS-ASOs and facilitates their vesicular escape. The co-localization of M6PR and of GCC2 with ASOs is influenced by the PS modifications, which have been shown to enhance the affinity of ASOs for proteins, suggesting that localization of these proteins to LEs is mediated by ASO-protein interactions. Reduction of M6PR levels also decreased PS-ASO activity in mouse cells and in livers of mice treated subcutaneously with PS-ASO, indicating a conserved mechanism. Together, these results demonstrate that the transport machinery between LE and Golgi facilitates PS-ASO release.
INTRODUCTION
Antisense oligonucleotide (ASO) drugs hybridize with target RNAs and modulate gene expression through different post-RNA binding mechanisms (1–3). Properly designed ASOs can downregulate gene expression via RNase H1-dependent RNA degradation or by triggering nonsense mediated decay or no-go decay (4–6). ASOs can also be designed to increase gene expression by modulating splicing, nonsense-mediated mRNA decay or translation (7–10). Commonly used ASOs are modified with phosphorothioate (PS) backbones and 2′-modifications to increase stability, distribution into tissues and cells, and pharmacological properties. PS-ASOs are active in the cytoplasm and/or the nucleus where the target RNA is localized (11).
PS-ASOs enter cells primarily via the endocytic pathways (12). PS-ASO internalization mediated by cell-surface receptors can direct PS-ASOs to productive pathways through which PS-ASOs can act on target RNAs (13–17). PS-ASOs can also enter cells through non-productive pathways, e.g. macropinocytosis. Previous studies have shown that internalized PS-ASOs are transported into early endosomes (EEs) within 10–20 min, into late endosomes (LE) within 20–50 min and localize to lysosomes within 40–60 min (18). However, PS-ASO activity is observed only after 6–8 h after free uptake, indicating a relatively slow release of PS-ASOs from endocytic pathways (16). Not surprisingly, decreased PS-ASO activity is observed upon inhibition of major endocytic transport pathways by reduction of the expression of Rab5C, an EE protein required for maturation of EEs to LEs or of Rab7, a protein required for LE maturation (19). It appears that LEs or multivesicular bodies (MVBs) are major sites for productive PS-ASO release (12,16,20–22). However, only a small portion of internalized PS-ASOs are released into the cytosol and/or nucleus from the membraned endocytic organelles (23). A better understanding of how PS-ASOs are released from endosomes will facilitate design of ASOs to improve drug performance through enhanced PS-ASO release from these organelles.
Previously we demonstrated that a number of proteins are recruited to LEs in cells incubated with PS-ASOs and that some of these proteins affect PS-ASO activity (24). For example, TCP1 and ANXA2 localize to LEs after cells are treated with PS-ASOs, and reductions in levels of these proteins reduce PS-ASO activity (18,24–25). Furthermore, PS-ASOs localize in intralumenal vesicles (ILVs) inside LE-limiting membranes (20,22), and ANXA2 co-localizes with PS-ASOs in ILVs (18). Based on these and other observations, it has been proposed that PS-ASO release from LEs can occur through multiple pathways that co-exist and that may act in parallel (12,16,26). For example, membrane flip-flop may cause PS-ASOs inside LEs to be exposed to the cytosol (27), protein interactions with LE membranes may trigger membrane deformation and PS-ASO leakage (18), and PS-ASO release may also occur when ILV membranes fuse with the limiting membranes of LEs via a back-fusion process (22).
Coat protein complex II (COPII) vesicles, which are derived from the endoplasmic reticulum (ER), are required for transport of membrane and secreted proteins from the ER to the Golgi (28). In cells incubated with PS-ASOs, COPII vesicles can be re-directed to LEs, where these vesicles facilitate PS-ASO release (26). COPII localization to LEs depends on P115 and STX5, which are normally required for the tethering and fusion of COPII with Golgi membranes (29,30). STX5 can also re-localize to LEs upon PS-ASO incubation, in a process likely mediated by binding of the PS-ASO to STX5 (26). These findings prompted us to evaluate whether other intracellular transport pathways also mediate PS-ASO trafficking and endosomal release. One such important pathway transports cargo between the LEs and the trans-Golgi network (TGN).
Newly synthesized hydrolases are transferred from the ER to the cis-Golgi, where these proteins are modified by mannose-6-phosphate (M6P) addition (31). The M6P-tagged hydrolases are transferred to the trans-Golgi, bound by the M6P receptor, M6PR and then transported to LEs in the form of vesicles (32). After vesicle fusion with LEs, M6PR proteins dissociate from hydrolases due to the low pH in LE lumen. The hydrolases are delivered to lysosomes, whereas M6PR buds off from the LE membrane as vesicles and returns to TGN (33). During this process, GCC2, a TGN-localized peripheral membrane protein, tethers incoming M6PR vesicles to TGN membranes (34–36). Deletion mutations in GCC2 or siRNA-mediated reduction of GCC2 expression impair M6PR tethering to the TGN and lead to scattered localization of M6PR in the cytoplasm (36).
Two distinct M6PR proteins have been identified that exhibit cation-dependent (M6PR-CD) or cation-independent (M6PR-CI) optimal binding to ligands (37,38). Both proteins have a short C-terminal cytosolic domain adjacent to a transmembrane domain. The N-terminal regions are extracytoplasmic and contain M6P-binding sites. M6PR-CI has 15 repetitive units at the extracytoplasmic domain, whereas M6PR-CD has one such unit (39). These two proteins appear to have different functions in transport of substrates, and M6PR-CI seems to handle most of the LE to TGN trafficking (39,40). Most M6PR proteins are present at TGN, but some are localized at plasma membrane (38). However, only M6PR-CI is capable of binding and internalizing extracellular lysosomal enzymes (40). Given the important roles that M6P and M6PR play in cells, we reasoned that it is possible that M6PR-mediated transport influences PS-ASO release from LEs, where PS-ASOs accumulate.
In this study, we characterized the roles of the LE to TGN transport pathway in PS-ASO trafficking and activity. We found that GCC2 and M6PR are required for PS-ASO activity in different human cells. In cells incubated with PS-ASO, GCC2 re-localized to LEs and co-localized with PS-ASOs in ILVs inside LEs. In addition, M6PR-CI localization to LEs was enhanced upon treatment of cells with PS-ASOs, and M6PR-CI co-localized with PS-ASOs both inside and on the membranes of LEs. Co-localization of GCC2 or M6PR with PS-ASO at LEs is most likely mediated by ASO–protein interactions. Importantly, we found that GCC2 and M6PR facilitated PS-ASO release from LEs, likely via vesicular PS-ASO escape and back-fusion-mediated processes. Further, we showed that M6PR-CI was required for PS-ASO activity in mouse cells and in mice. Together our results demonstrate that the LE to TGN trafficking pathway mediates PS-ASO release from LEs, furthering our understanding of the complex mechanisms that influence ASO activity in cells.
MATERIALS AND METHODS
siRNAs and primer probe sets (Supplementary Table S1), antibodies (Supplementary Table S2) and ASOs (Supplementary Table S3) are listed in Supplementary Data.
Cell culture and ASO treatment
HeLa, A431, HepG2 and GFP-Rab7-expressing SVGA cells (a gift from Tomas Kirchhausen's lab) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 0.1 μg/ml streptomycin and 100 units/ml penicillin. ASGR-expressing Hek293 cells were grown in DMEM medium as described above and supplemented with 0.2 mg/ml hygromycin. MHT cells were grown as described previously (20). Cells were seeded at 70% confluency one day before transfection. siRNAs were transfected at 3 nM final concentration into HeLa cells or at 10 nM final concentration for A431 cells using Lipofectamine RNAiMAX (Life Technologies), according to the manufacturer's protocol. At 48 h after siRNA transfection, cells were reseeded into 96-well plates for ASO activity or uptake assays or were seeded into collagen-coated glass-bottom dishes (MatTek) at 70% confluency for immunofluorescent staining.
ASO activity assay
Cells treated with control siRNAs or target-specific siRNAs were trypsinized and reseeded into 96-well plates at about 8000 cells per well for HeLa cells and about 15 000 cells per well for A431 cells. Cells were cultured overnight before ASO treatment. For free uptake, ASOs were directly added to the medium at different final concentrations, and cells were cultured for an additional 16–24 h before collection. For transfection, cells were transfected with ASOs for 4 h using Lipofectamine 2000 (Life Technologies) as per the manufacturer's protocol. Total RNA was prepared, and levels of ASO-targeted RNAs were quantified using qRT-PCR.
RNA preparation and qRT-PCR
Total RNA was prepared using a RNeasy mini kit (Qiagen) from cells grown in 96-well plates using the manufacturer's protocol. qRT-PCR was performed in triplicate using TaqMan primer probe sets as described previously (25). Briefly, ∼50 ng total RNA in 5 μl water was mixed with 0.3 μl primer probe sets containing forward and reverse primers (10 μM of each) and fluorescently labeled probe (3 μM), 0.5 μl RT enzyme mix (Qiagen), 4.2 μl RNase-free water, and 10 μl of 2 × polymerase chain (PCR) buffer in a 20 μl reaction. Reverse transcription was performed at 48°C for 10 min, followed by 94°C for 10 min, and then 40 cycles of PCR were conducted at 94°C for 30 s, and 60°C for 30 s within each cycle using the StepOne Plus RT-PCR system (Applied Biosystems). The mRNA levels were normalized to the amount of total RNA present in each reaction as determined for duplicate RNA samples using the Ribogreen assay (Life Technologies). Statistical analyses were performed from three independent experiments using Prism with either t-test or F-test for curve comparison based on non-linear regression (dose-response curves) for XY analyses, using equation ‘log(agonist) versus normalized response—variable slope’. The Y axis (relative level) was used as the normalized response.
Western analyses
Cell pellets were lysed by incubation at 4°C for 30 min in RIPA buffer (50 mM Tris–HCl, pH 7.4, 1% Triton X-100, 150 mM NaCl, 0.5% sodium deoxycholate and 0.5 mM ethylenediaminetetraacetic acid (EDTA)). Proteins were collected by centrifugation. Approximately 20–40 μg protein were separated on 6–12% NuPAGE Bis-Tris gradient sodium dodecyl sulphate-polyacrylamide gelelectrophoresis gels (Life Technologies), and transferred onto polyvinylidene difluoride (PVDF) membranes using the iBLOT transfer system (Life Technologies). The membranes were blocked with 5% non-fat dry milk in 1 × PBS at room temperature for 30 min. Membranes were then incubated with primary antibodies at room temperature for 2 h or at 4°C overnight. After three washes with 1 × PBS, the membranes were incubated with appropriate HRP-conjugated secondary antibodies (1:2000) at room temperature for 1 h to develop the image using Immobilon Forte Western HRP Substrate (Millipore).
Immunofluorescence staining and confocal imaging
Cells were washed with 1 × phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 0.5–1 h at room temperature, and permeabilized for 4 min with 0.1% Triton in 1 × PBS. After blocking at room temperature for 30 min with 1 mg/ml bovine serum albumin (BSA) in 1 × PBS, cells were incubated with primary antibodies (1:100–1:300) in 1 mg/ml BSA in 1 × PBS for 2–4 h, washed three times (5 min each) with 0.1% nonyl phenoxypolyethoxylethanol-40 (NP-40) in 1 × PBS and incubated for 1 h with secondary antibody conjugated with fluorophores (1:200). After washing three times, cells were mounted with anti-fade reagent containing DAPI (Life Technologies), and images were acquired using confocal microscope (Olympus FV-1000) and processed using FV-10 ASW 3.0 Viewer software (Olympus). Z-stacks were generated from images taken for 20–30 sections at 0.1–0.11 μm depth per section, and 3D images were generated using FV-10 ASW-3.0 viewer. For staining within 2 h of ASO addition, cells were washed three times with acidic buffer (0.1 M acetic acid, 500 mM NaCl) and one time with 1 × PBS, to remove cell surface-associated ASOs before fixation. Quantification of co-localization events was performed manually from 20 cells by counting foci stained with both antibodies that had clearly defined boundaries and that were not saturated. Statistics analysis was performed using an unpaired t-test using Prism software.
For live cell imaging, GFP-Rab7-expressing SVGA cells grown in glass-bottom dishes were incubated with 2 μM PS-ASO 446654 for 6 h. For GFP-M6PR-CD expression, plasmid (RG201277, Origene) was transfected into HeLa cells for 24 h. Cells were reseeded into glass-bottom dish and incubated for ON. Cells were then incubated with 2 μM PS-ASO 446654 for 6 h. Medium was changed to pre-warmed FluoroBrite DMEM Media (Life Technologies), and images were taken at 15-s intervals for 15 min. Movies and images were generated using FV-10 ASW 3.0 Viewer software.
ASO uptake analysis
HeLa cells treated with different siRNAs for 48 h were reseeded into 96-well plates, incubated overnight, and Cy3-labeled PS-ASO 446654 was added to the medium. After 3 h, cells were washed three times with 1 × PBS, trypsinized, and pelleted by centrifugation. Cell pellet was washed once with 1 × PBS supplemented with 3% FBS, to remove residual medium and external ASOs. Cells were resuspended in 1 × PBS supplemented with 3% FBS for analysis by flow cytometry using an Attune NxT Flow Cytometer (ThermoFisher Scientific).
ASO endosomal transport study
Cells were incubated with 2 μM Cy3-labeled PS-ASO 446654 on ice for 20 min and then shifted to 37°C for 25 min to allow the PS-ASOs to enter cells and traffic to EEs or to 37°C for 45 min to allow PS-ASOs to traffic to LEs. Cells were then washed, permeabilized, and fixed as described for immunofluorescent staining. EE and LEs were stained with EEA1 and Rab7, respectively. For quantification, PS-ASO-containing EEs or LEs were counted manually in approximately 20 cells, and the percentages of PS-ASO-containing endosomes relative to total numbers of the corresponding endosomes were calculated.
NanoBRET binding assay
N-terminal NLuc fusions were created using the vector pFN31K Nluc CMV-neo (Promega). Briefly, M6PR-CD was amplified from plasmid RG201277 (Origene) using PCR primers complementary to the full-length cDNA. The forward PCR primer (5′-GCATTCGACTCGAGCATGTTCC CTTTCTACAGCTGCT) and reverse primer (5′- TCGAATGCGAATTCCTACTACATTG GTAATAAATGGTCATCC) were used for PCR reaction. The PCR product was ligated to XhoI and EcoRI sites of the pFN31K Nluc CMV-neo vector, as described previously (26). Fusion proteins were expressed by transfecting the plasmid into 6 × 105 HEK 293 cells using Effectene transfection reagent (Qiagen) according to the manufacturer's protocol. Following a 24-h incubation, cells were removed from the plate by trypsinization, washed with 1 × PBS, and resuspended in 250 μl Pierce IP Lysis Buffer (Thermo Scientific). Lysates were incubated 30 min at 4°C while rotating, then debris was pelleted by centrifugation at 15 000 rpm for 5 min. The fusion protein was purified by adding 20 μl HisPur Ni-NTA Magnetic Beads (Thermo Scientific) and 10 mM imidazole then incubating at 4°C for 2 h. Beads were washed times with 1 × PBS, 10 mM imidazole and 0.01% Tween-20. Fusion protein was eluted from the beads in 100 μl 1 × PBS and 200 mM imidazole, followed by dilution with of 200 μl IP buffer.
BRET assays were performed in white 96-well plates as previously described (41). Alexa-linked ASO 766634 was incubated at room temperature for 15 min in 1× binding buffer (100 mM NaCl, 20 mM Tris–HCl, pH 7.5 or different pH as indicated in Figures, 1 mM EDTA and 0.1% NP-40) with 106 RLU/well of Ni-NTA-purified NLuc fusion protein. Following the incubation, NanoGlo substrate (Promega) was added at 0.1 μl/well. Readings were performed for 0.3 s using a Glomax Discover system using 450/8 nm band pass for the donor filter, and 600 nm long pass for the acceptor filter. BRET was calculated as the ratio of the emission at 600/450 nm (fluorescent excitation emission/RLU).
Animal study
Animal experiments were performed according to American Association for the Accreditation of Laboratory Animal Care guidelines and were approved by the institution's Animal Welfare Committee (Cold Spring Harbor Laboratory′s Institutional Animal Care and Use Committee guidelines). Seven-week-old male BALB/c mice were given 15 mg/kg control PS-ASO 716837, M6PR-CD PS-ASO 1332272 or M6PR-CI PS-ASO 1332273 by subcutaneous injection. After 48 h, mice were given another injection of the same ASOs at the same dose. At 72 h after last dose, animals were given SRB1 PS-ASO 353382 at 0 (saline), 3.75, 7.5, 15 or 30 mg/kg (N = 3 for each dose) by subcutaneous injection. After 24 h, mice were sacrificed, and blood or tissue samples were collected. For PTEN PS-ASO activity assay, animal studies were performed essentially as described above, except that mice were treated with the PTEN PS-ASO 116847 at 0 (saline), 3.7, 11.1, 33.3 or 100 mg/kg (N = 3 for each dose) in animals pre-treated with control PS-ASO, M6PR-CD PS-ASO 1332272 or M6PR-CI PS-ASO 1332273. All animals were included in the analyses. Total RNA or proteins were prepared from mouse liver. The levels of M6PR-CD, M6PR-CI, SRB1 and PTEN mRNAs were determined by qRT-PCR. M6PR-CI protein was detected by western analysis.
RESULTS
Reduction of GCC2 decreases PS-ASO activity upon free uptake
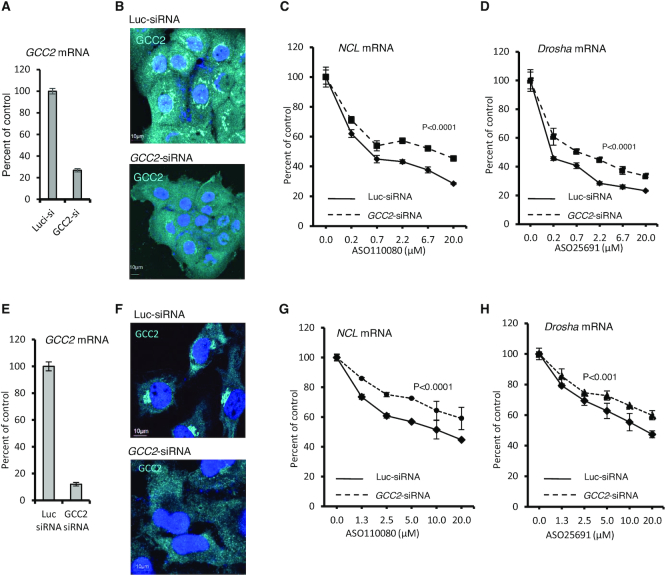
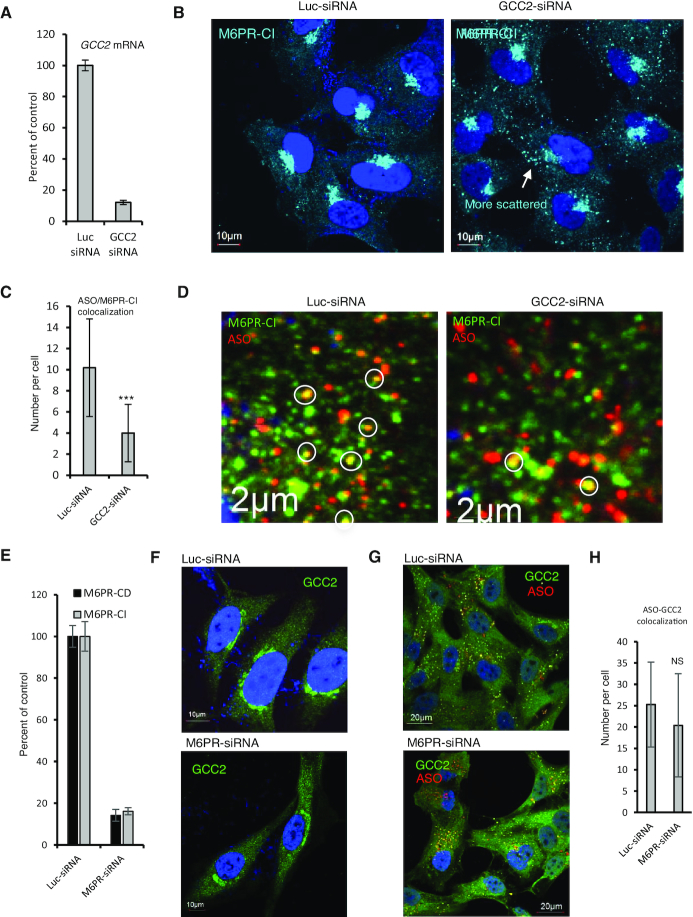
To evaluate whether the transport pathway between LE and TGN influences PS-ASO activity, A431 cells were treated with an siRNA designed to deplete cells of GCC2, a trans-Golgi protein required for transport of M6PR from LEs to the TGN (34). The levels of GCC2 mRNA were dramatically reduced by siRNA treatment as quantified by qRT-PCR (Figure 1A). GCC2 protein was evaluated by immunofluorescence staining (Figure 1B), since we attempted but failed to detect GCC2 protein by western analysis using several different antibodies. In control cells, GCC2 was observed in the Golgi, consistent with a previous report (42). In cells treated with GCC2-specific siRNA, the intensity of staining was dramatically decreased, suggesting that GCC2 protein levels were reduced by the siRNA treatment.
Figure 1.
Reduction of GCC2 decreased ASO activity upon free uptake in different human cells (A) qRT-PCR quantification of GCC2 mRNA levels in A431 cells transfected with control luc-siRNA (targeting luciferase) or GCC2-specific siRNA for 72 h. (B) Immunofluorescent staining of GCC2 protein in A431 cells treated with siRNAs for 72 h. Nuclei were stained with DAPI. Scale bars, 10 μm. (C and D) Levels of C) NCL and D) Drosha mRNAs determined by qRT-PCR in cells treated with siRNAs that were subsequently incubated for 24 h with PS-ASOs 110080 or 25691 targeting NCL or Drosha mRNAs, respectively. (E) qRT-PCR quantification of GCC2 mRNA levels in HeLa cells transfected with control siRNA or GCC2-specific siRNA for 72 h. (F) Immunofluorescent staining of GCC2 protein in HeLa cells treated with siRNAs for 72 h. Nuclei were stained with DAPI. Scale bars, 10 μm. (G and H) qRT-PCR quantification for the levels of (G) NCL and (H) Drosha mRNAs in HeLa cells pretreated with siRNAs and were subsequently incubated for 24 h with PS-ASOs 110080 or 25691 targeting NCL or Drosha mRNAs, respectively. Error bars are standard deviations from three independent experiments. P-values were calculated based on F-test using Prism.
The control or GCC2-depleted cells were treated via free uptake with PS-ASOs designed to reduce NCL or Drosha mRNA through RNase H1 cleavage. Unless specified, the PS-ASOs used in this study are 20-mer gapmers, with 10 deoxynucleotides in the center flanked by 5 nt at each end that are modified by 2′-O-methoxyethyl (2′-MOE). The levels of PS-ASO-targeted mRNAs were determined using qRT-PCR after 24 h of incubation with PS-ASO. The abilities of PS-ASOs to induce degradation of NCL and Drosha mRNAs were reduced in cells depleted of GCC2 compared to control cells (Figure 1C and D). Decreased PS-ASO activity was also observed when GCC2 was reduced using different siRNAs (Supplementary Figure S1). These and subsequent experiments of ASO activity assay were repeated at least three times, and representative results are shown.
To evaluate whether the effects of GCC2 reduction on ASO activity detected in A431 cells was unique to this cell type, we analyzed PS-ASO activity in HeLa cells depleted of GCC2 by siRNA treatment (Figure 1E and F). Reduction of GCC2 in HeLa cells decreased the activity of PS-ASOs targeting NCL and Drosha mRNAs when ASOs were delivered by free uptake (Figure 1G and H). Upon transfection, a method that results in bypass of normal endocytic pathways and delivery of ASOs mostly to the cytosol and nucleus (25,43), PS-ASO activity was not affected by reduction of GCC2 in either A431 or HeLa cells (Supplementary Figure S2). These observations indicate that reduction of GCC2 does not affect PS-ASO activity at steps after ASOs were released to the cytosol or nucleus. Together, our results suggest that GCC2 enhances PS-ASO activity after free uptake by facilitating ASO internalization or endocytic trafficking and that this effect is not unique to a particular cell type.
GCC2 localizes to LEs in PS-ASO-treated cells
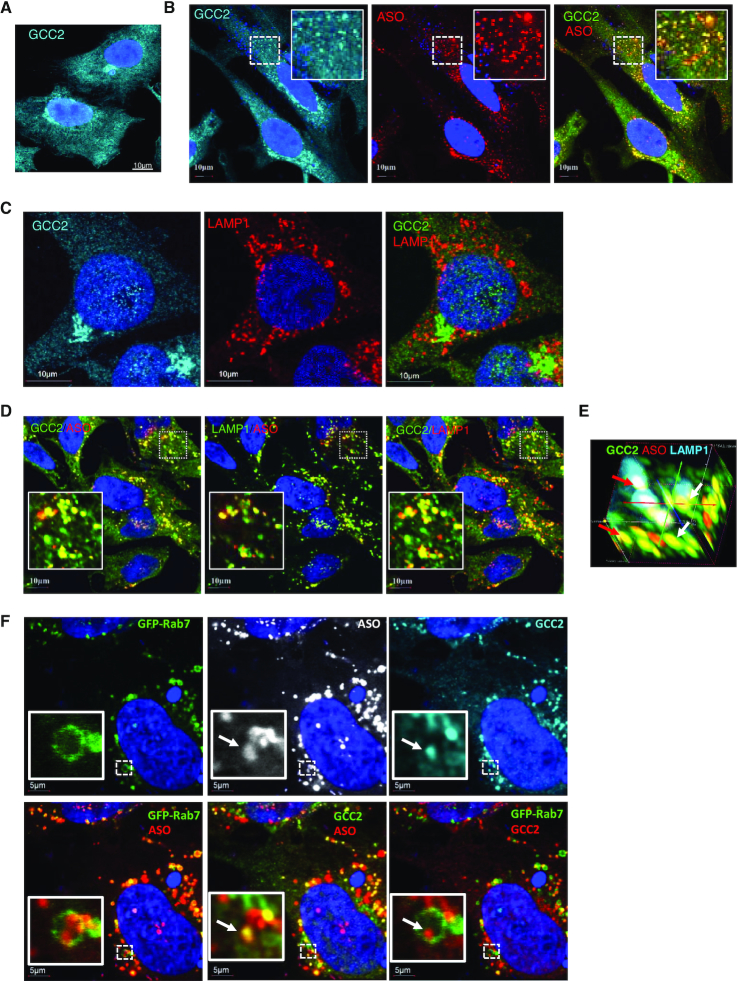
Next, to investigate whether GCC2 protein can co-localize with PS-ASOs upon free uptake, immunofluorescence staining was performed in HeLa cells. (For better resolution, we recommend visualization of confocal images on screen). In cells not treated with PS-ASOs, GCC2 was observed at the Golgi (Figure 2A). After incubation with Cy3-labeled PS-ASOs for 24 h, GCC2 was detected at the Golgi and in small foci scattered across the cytoplasm that co-stained with PS-ASO (Figure 2B).
Figure 2.
GCC2 localizes to LEs in HeLa and SGVA cells treated with PS-ASO. (A) Immunofluorescent staining of GCC2 in HeLa cells. Scale bar, 10 μm. (B) Immunofluorescent staining of GCC2 in HeLa cells incubated with 2 μM Cy3-labeled PS-ASO 446654 for 18 h. Scale bars, 10 μm. The regions boxed with dashed lines are magnified in the insets. (C) Immunofluorescent staining of GCC2 and LAMP1 in HeLa cells. Scale bars, 10 μm. (D) Immunofluorescent staining of GCC2 and LAMP1 in HeLa cells incubated with Cy3-labeled PS-ASO 446654 for 18 h. Scale bars, 10 μm. (E) 3D image of GCC2, PS-ASO and LAMP1 co-localization in HeLa cells incubated with Cy3-labeled PS-ASO 446654 for 18 h. The image was compiled from 25 sections taken at 0.1 μm depth. Two vesicles are marked with red and white arrows. (F) Immunofluorescent staining of GCC2 in GFP-Rab7-expressing SVGA cells incubated with 2 μM Cy3-labeled PS-ASO 446654 for 18 h. Scale bars, 5 μm. The regions boxed with dashed lines are magnified in the insets. The co-localization of GCC2 with PS-ASOs inside an LE is marked with an arrow.
Upon free uptake, PS-ASOs eventually concentrate in LEs and lysosomes in the cytoplasm (16,24). The co-localization of GCC2 with PS-ASOs in the cytoplasmic foci suggests that GCC2 might be relocated to LEs or lysosomes. To evaluate this possibility, cells were co-stained for GCC2 and LAMP1, a marker of LEs and lysosomes. Without PS-ASO incubation, GCC2 was barely detected in scattered cytoplasmic LE and lysosome structures (Figure 2C). Upon treatment of cells with PS-ASO, GCC2 was co-localized with LAMP1 and with PS-ASOs (Figure 2D). This co-localization was confirmed by 3D imaging (Figure 2E). The localization of other proteins known to reside in the Golgi, such as the cis-Golgi protein GM130 and the trans-Golgi protein TGN46 (44,45), were not affected by the presence of PS-ASO, and no significant co-localization with ASOs was detected for these proteins (Supplementary Figure S3). These results suggest that Golgi integrity is not affected by the presence of PS-ASO.
GCC2 co-localized with PS-ASOs of different sequences (Supplementary Figure S4A) and with PS-ASOs containing the constrained ethyl (cEt) modification rather than 2′-MOE (Supplementary Figure S4B). Furthermore, co-localization of GCC2 with LAMP1 was detected in cells incubated with an unlabeled PS-ASO (Supplementary Figure S4C), indicating that the co-localization was not due to the fluorescent dye. ASO-induced GCC2 localization to LEs was also observed in SGVA cells expressing GFP-Rab7, a marker protein for LEs. No LE localization of GCC2 was observed in the absence of PS-ASO (Supplementary Figure S5A). However, upon incubation with PS-ASO, GCC2 was detected to co-localize with Rab7 and with ASOs (Supplementary Figure S5B), confirming that the presence of PS-ASOs led to LE localization of GCC2 protein. Quantification indicated that under these experimental conditions, approximately 90% of LEs contained ASOs, and approximately 60% of PS-ASO-positive LEs contained GCC2 (Supplementary Figure S5C). These results further suggest that GCC2 relocalization to LEs is a dynamic process. GCC2 localization to LEs was also observed in A431 cells treated with PS-ASO (Supplementary Figure S6).
GCC2 localizes to ILVs inside LEs in PS-ASO-treated cells
Previously we showed that in cells treated with PS-ASOs, ANXA2 is observed at distinct foci in the LE lumen (18,22), suggesting that some proteins localize to ILVs in cells that have internalized PS-ASOs. To determine whether GCC2 can also co-localize with PS-ASOs in ILVs inside LEs, Cy3-labeled PS-ASOs were incubated with SVGA cells that express GFP-Rab7. PS-ASOs were observed as distinct foci inside LEs, marked with GFP-Rab7, and some of these foci co-stained with GCC2 (Figure 2F). To confirm that these foci were indeed ILVs, the cells were stained for CD63, a protein found in ILVs (46), or with LBPA, a lipid preferentially located in ILV membranes (47). The results showed that the PS-ASO-containing foci inside LEs are co-stained with both CD63 and LBPA (Supplementary Figure S7), indicating that these foci are membrane-enclosed ILVs.
Reduction of M6PR decreases PS-ASO activity likely by affecting the same pathway as GCC2
As reduction of the Golgi protein GCC2 decreased ASO activity, we evaluated whether reduction of other proteins known to be involved in Golgi function cause similar effects. We first depleted A431 cells of GM130, a Golgi membrane tethering factor that is also involved in maintenance of Golgi structure (48), by siRNA treatment (Supplementary Figure S8A). Reduction of GM130 levels did not significantly affect PS-ASO activity upon free uptake (Supplementary Figure S8B). These results show that not all Golgi proteins affect PS-ASO activity, and that the observed effect of GCC2 reduction on PS-ASO activity is likely related to the specific function of GCC2.
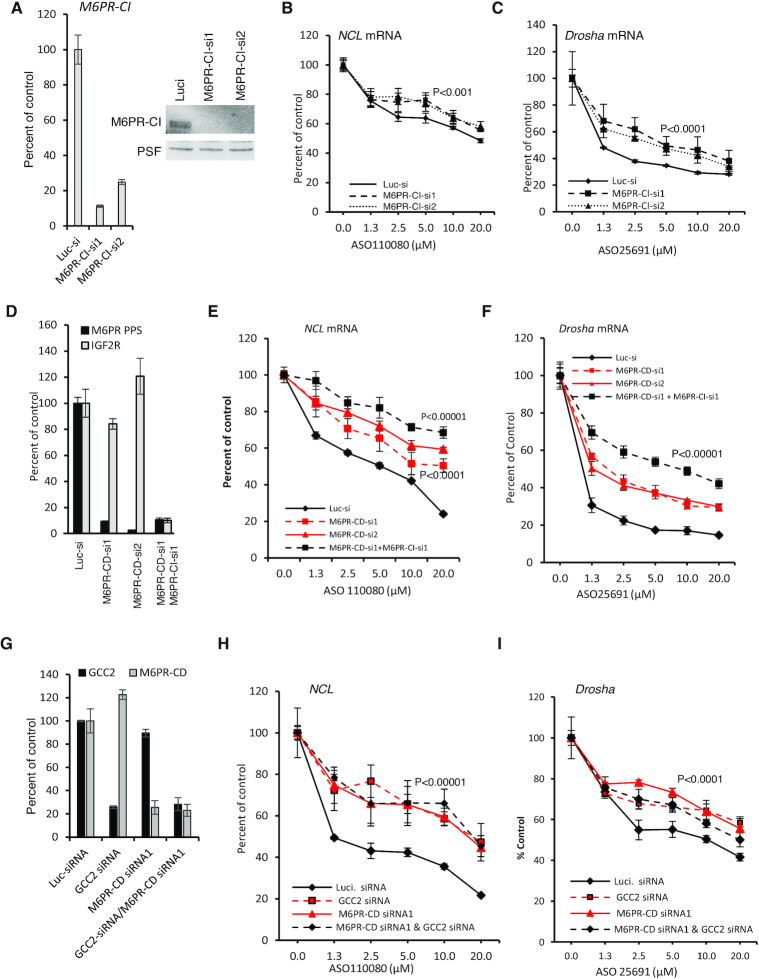
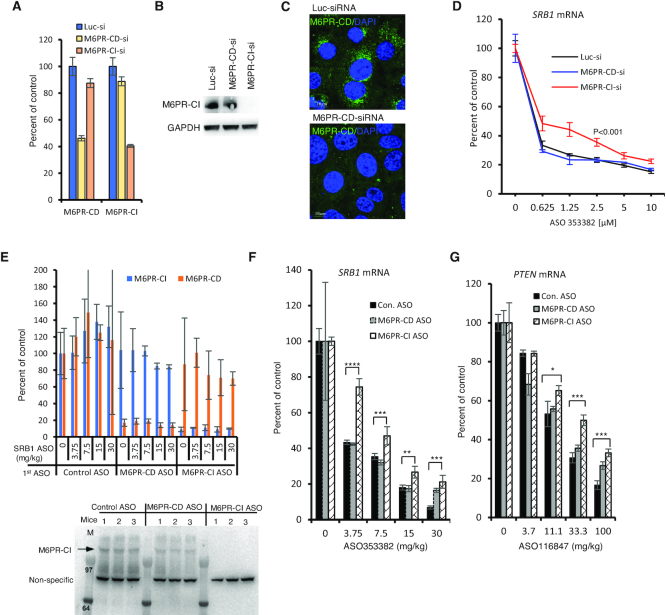
It has been demonstrated that GCC2 is required for LE to TGN shuttling of M6PR, which mediates transport of newly synthesized hydrolases from Golgi to LE and then to lysosomes (31,34). To evaluate the possibility that disruption of M6PR-mediated trafficking would lead to altered PS-ASO activity, M6PR-CI levels were reduced in A431 cells using two different siRNAs (Figure 3A), and the effect of M6PR-CI reduction was evaluated by qRT-PCR quantification of NCL or Drosha mRNAs after incubation of cells with PS-ASOs targeting these mRNAs. Reduction of M6PR-CI levels decreased PS-ASO activity (Figure 3B and C). Similarly, when cells were depleted of M6PR-CD (Figure 3D), PS-ASO activity was also decreased (Figure 3E and F). This indicated that both M6PR-CI and M6PR-CD affect PS-ASO activity in A431 cells. Interestingly, simultaneous reduction of both M6PR-CD and M6PR-CI caused additive effects on PS-ASO activity (Figure 3D–F), indicating that the functions of the two proteins do not entirely overlap, consistent with previous observations (39,40).
Figure 3.
Reduction of M6PR decreases PS-ASO activity in A431 cells. (A) M6PR-CI mRNA levels in A431 cells treated with two different siRNAs for 72 h were quantified using qRT-PCR (left panel) and the protein levels were determined by western analyses (right panel). PSF was probed and served as a control for loading. (B and C) qRT-PCR quantification of NCL and Drosha mRNAs in cells pre-treated with siRNA targeting M6PR-CI and then incubated for 24 h with PS-ASOs 110080 or 25691, respectively. (D) qRT-PCR quantification of M6PR-CD and M6PR-CI mRNAs in A431 cells treated with indicated siRNAs for 72 h. (E and F) qRT-PCR quantification of NCL and Drosha mRNAs in siRNA pretreated cells that were subsequently incubated for 24 h with PS-ASOs 110080 or 25691, respectively. (G) qRT-PCR quantification of M6PR-CD and GCC2 mRNAs in A431 cells treated with indicated siRNAs for 72 h. (H and I) qRT-PCR quantification of NCL and Drosha mRNAs in siRNA pretreated cells that were subsequently incubated for 24 h with PS-ASOs 110080 or 25691, respectively. Error bars are standard deviations from three independent experiments. P-values were calculated based on F-test.
The effects of M6PR reduction on PS-ASO activity were not unique to A431 cells, since reduction of either M6PR-CD or M6PR-CI also decreased PS-ASO activity in HeLa cells (Supplementary Figure S9A–C). Furthermore, over expression of M6PR-CD enhanced ASO activity in HeLa cells (Supplementary Figure S10). As we observed in cells depleted of GCC2, M6PR reduction did not affect PS-ASO activity when PS-ASOs were transfected into cells (Supplementary Figure S9D and E). Interestingly, simultaneous reduction of both GCC2 and M6PR-CD did not cause additive effects on PS-ASO activity (Figure 3G–I). This suggests that GCC2 and M6PR act on the same pathway to influence PS-ASO activity.
M6PR and GCC2 enhance endosomal release of ASOs
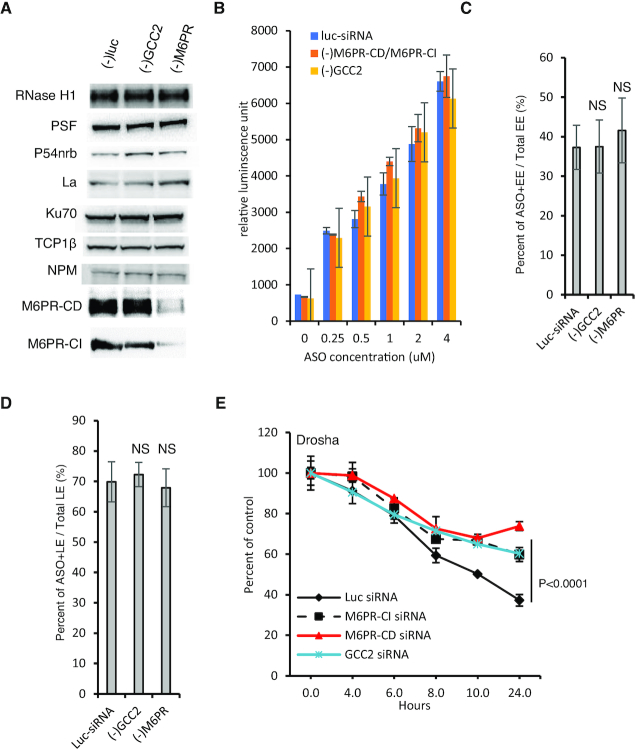
Since reduction of GCC2 and M6PR did not affect activities of PS-ASO transfected into cells, we reasoned that factors required for PS-ASO activity after PS-ASO release into the cytosol or nucleus should not be affected by these proteins. Indeed, levels of proteins known to influence PS-ASO activity, including RNase H1, PSF, P54nrb and others (24–25,49), were not substantially altered by reductions of GCC2 or of both M6PR proteins in HeLa cells (Figure 4A).
Figure 4.
In cells depleted of GCC2 and M6PR PS-ASO release from LEs is impaired. (A) Western analyses of indicated proteins in control siRNA-treated HeLa cells or cells treated with siRNAs targeting GCC2 or M6PR for 72 h. Equal amounts of proteins were loaded in each lane. (B) Flow cytometry analysis of Cy3-labeled PS-ASO 446654 uptake into HeLa cells treated with a control siRNA or siRNAs targeting GCC2 or M6PR for 72 h. Error bars are standard deviation from three independent experiments. (C) Quantification of the EEs containing Cy3-labeled PS-ASO after incubation of HeLa cells pretreated with siRNAs, and subsequently with PS-ASO 446654 on ice for 20 min and then at 37°C for 25 min. The percentages of PS-ASO-containing EEs relative to the number of total EEs were calculated and average values plotted. Error bars are standard deviations of data from 20 cells. P-value was calculated based on unpaired t-test. NS, not significant. (D) Quantification of the PS-ASO-containing LEs in cells pre-treated with control siRNA or siRNAs targeting GCC2 or M6PR and then treated with PS-ASO 446654 on ice for 20 min and then at 37°C for 45 min. The percentage of ASO-containing LEs relative to the number of total LEs was calculated and average values plotted. Error bars are standard deviations of data from 20 cells. P-value was calculated based on unpaired t-test. NS, not significant. (E) qRT-PCR quantification of the levels of Drosha mRNA in HeLa cells. Cells were pretreated with control siRNA or siRNAs targeting GCC2 or M6PR and then with PS-ASO 25691 for 2 h, medium containing PS-ASO was removed, and mRNA was quantified at indicated time points. Error bars are standard deviations from three independent experiments. P-values were calculated based on F-test.
Upon free uptake, PS-ASOs traffic to EEs and then to LEs and then need be released from these endocytic organelles. To identify the trafficking step influenced by GCC2 and M6PR, we first analyzed the effects of GCC2 or M6PR reduction on PS-ASO uptake. HeLa cells were treated with siRNAs targeting GCC2 or both forms of M6PR. The siRNA-treated cells were incubated with Cy3-labeled PS-ASOs at different concentrations for 2 h, and internalized PS-ASOs were analyzed using flow cytometry. Reductions of GCC2 or M6PR did not substantially affect the amounts of PS-ASOs taken into cells (Figure 4B).
Next, PS-ASO trafficking to EEs was analyzed. HeLa cells depleted of GCC2, or M6PR-CD and M6PR-CI together were incubated with Cy3-labeled PS-ASOs for 25 min, a time point at which PS-ASOs are observed in EEs (18). PS-ASO localization to EEs was evaluated by co-staining of EEA1, an EE marker protein (Supplementary Figure S11). Comparable levels of PS-ASO-positive EEs were found in control cells and in cells depleted of GCC2 or M6PR (Figure 4C). These results suggest that PS-ASO transport from plasma membrane to EEs is not regulated by GCC2 or by M6PR proteins.
Consistent with previous observations that a small portion of M6PR is present on the cell surface and can enter cells through the endocytic pathway (31,50), M6PR-CI co-localized with EEs in HeLa cells (Supplementary Figure S12A). However, it appears that M6PR-CI is not required for ASO internalization and EE localization as some PS-ASO-positive EEs, identified by staining with EEA1, did not co-stain with M6PR-CI in cells incubated with PS-ASO for 25 min (Supplementary Figure S12B). In addition, some M6PR-stained EEs did not contain ASOs. Further, incubation of ASOs did not enhance EE localization of M6PR-CI (Supplementary Figure S12C). These observations further support the hypothesis that M6PR is not required for ASO internalization and EE localization.
Similarly, reduction of GCC2 and M6PR proteins did not affect PS-ASO transport into LEs as shown by immunofluorescent staining of Rab7 in cells incubated with Cy3-labeled PS-ASO for 45 min (Supplementary Figure S13), a time at which PS-ASOs are observed in LEs (18). Comparable LE localization of ASOs was observed in cells treated with a control siRNA and in cells depleted of these proteins, as demonstrated by quantification analyses (Figure 4D). These data suggest that GCC2 and M6PR may affect PS-ASO release from endocytic organelles. To evaluate this possibility, cells depleted of GCC2, M6PR-CD or M6PR-CI were incubated with a PS-ASO targeting Drosha mRNA for 2 h to allow ASO internalization. Next, ASOs were removed from the medium to terminate uptake, and cells were incubated in fresh medium for an additional 2–24 h prior to analysis of Drosha mRNA by qRT-PCR. The results showed that reduction of GCC2, M6PR-CD, or M6PR-CI caused slower Drosha mRNA degradation by the PS-ASO as compared with that in control cells (Figure 4E). These observations indicate that GCC2 and M6PR proteins enhance PS-ASO activity by facilitating PS-ASO release, most likely from late endosomes.
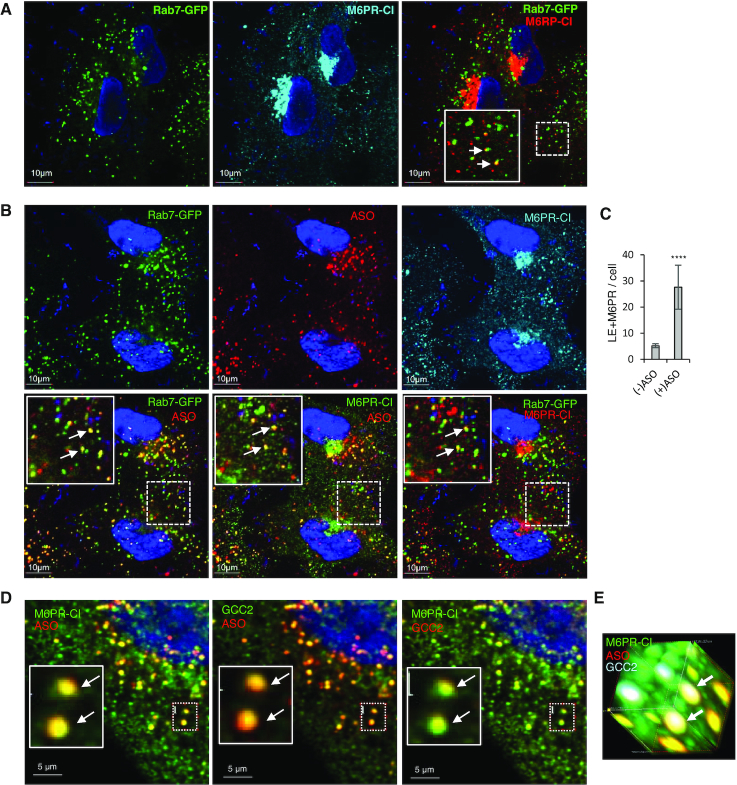
M6PR co-localizes with GCC2 at LEs upon PS-ASO treatment of cells
Next, we sought to understand how M6PR and GCC2 facilitate PS-ASO release from endosomes. M6PR is known to shuttle between LEs and TGN (31), it is possible that PS-ASOs escape from LEs through this intrinsic pathway. We thus analyzed how PS-ASO incubation influences M6PR localization. In SVGA cells not incubated with PS-ASO, M6PR-CI mainly localized to the perinuclear TGN region, as expected, and a few scattered dot-like structures were also detected (Figure 5A and Supplementary Figure S14A). Some of these dot-like structures co-localized with GFP-Rab7 (Figure 5A), consistent with previous reports (51). After incubation of cells with a PS-ASO, the number of scattered cytoplasmic foci that contained M6PR-CI increased (Figure 5B and Supplementary Figure S14B), and these foci were also positive for PS-ASO and Rab7, suggesting increased LE localization of M6PR-CI in the presence of PS-ASO relative to its absence. This was confirmed by quantification of the co-localization between Rab7 and M6PR-CI (Figure 5C). M6PR-CD co-localization with PS-ASOs in LEs was also detected in SVGA cells (Supplementary Figure S15). In addition, ASO was found to co-move with GFP-M6PR-CD protein in distinct foci in live cells (Supplementary Movie S1), further confirming ASO-M6PR-CD colocalization. As the anti-M6PR-CI antibody performed better than the anti-M6PR-CD antibody, the former was used in subsequent studies.
Figure 5.
M6PR co-localizes with PS-ASOs and GCC2 in LEs in SVGA and HeLa cells. (A) Immunofluorescent staining of M6PR-CI in GFP-Rab7-expressing SVGA cells. Scale bars, 10 μm. In the far right panel, the region boxed with a dashed line is magnified in the inset. Co-localization between Rab7 and M6PR-CI is marked by arrows. (B) Immunofluorescent staining of M6PR-CI in GFP-Rab7-expressing SVGA cells incubated with 2 μM Cy3-labeled PS-ASO 446654 for 6 h. Scale bars, 10 μm. The regions boxed with dashed lines are shown at higher magnification in insets, and co-localization is indicated by arrows. (C) Quantification of co-localization between Rab7 and M6PR-CI in cells incubated with or without ASO for 6 h. Error bars are standard deviations. P-value was calculated based on unpaired t-test. ****, P < 0.0001. (D) Immunofluorescent staining of M6PR-CI and GCC2 in HeLa cells incubated with 2 μM Cy3-labeled PS-ASO 446654 for 12 h. An example of co-localization in a cytoplasmic area is boxed. (E) 3D image of the boxed area in panel D. Two foci where M6PR-CI, GCC2, and PS-ASO co-localize are marked by arrows.
We next evaluated whether GCC2 and M6PR co-localize upon PS-ASO incubation. HeLa cells were incubated with PS-ASO, and cells were co-stained for GCC2 and M6PR-CI. Co-localization of GCC2, ASO, and M6PR was readily detected and confirmed by 3D imaging (Figure 5D and E; Supplementary Figure S14C). Interestingly, M6PR-CI and GCC2 co-localization with PS-ASO occurred within 6 h after PS-ASO incubation, and shared similar kinetics, as determined in HeLa cells analyzed after different times of incubation with PS-ASOs (Supplementary Figure S16).
Reduction of GCC2 and M6PR reduces co-localization of ANXA2 with PS-ASOs
The association of GCC2 and M6PR with LEs upon PS-ASO incubation may affect properties of LE membranes. To evaluate this possibility, HeLa cells depleted of GCC2 or both forms of M6PR were incubated with PS-ASO or not, and cells were stained for ANXA2. ANXA2 preferentially binds to negatively charged phospholipids and is enriched in EEs but also present in the cytosol (52,53). Previously we found that, despite the fact that ANXA2 interaction with ASOs is not required, ANXA2 is recruited to LEs when cells are treated with PS-ASOs (18,24). In cells depleted of M6PR or GCC2, co-localization of ANXA2 with PS-ASOs was dramatically reduced (Supplementary Figure S17), suggesting that ANXA2 recruitment to LEs is impaired.
Although changes in the LE membrane might be induced by reduction of GCC2 and M6PR, leading to impaired ANXA2 recruitment to LEs, reduction of M6PR did not affect COPII vesicle recruitment to LEs (Supplementary Figure S18), as determined by staining of Sec31a, a COPII vesicle coat protein that is recruited to LEs upon incubation of cells with PS-ASOs (26). In addition, reduction of STX5, which binds PS-ASOs and is required for COPII localization to LEs (26), did not abolish co-localization of PS-ASO with either GCC2 or M6PR (Supplementary Figure S19). These results suggest that the recruitment of M6PR and COPII vesicles to LEs occur through different mechanisms.
M6PR-CI co-localizes with PS-ASOs to ILVs
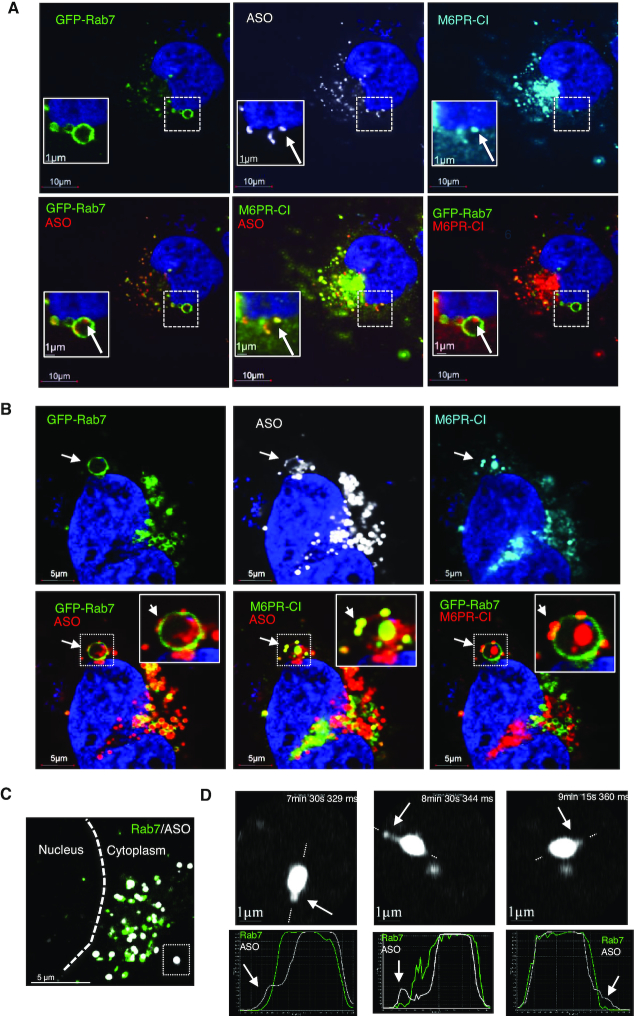
M6PR has been shown to localize to ILVs inside LEs (54). To evaluate whether M6PR co-localizes with PS-ASOs in ILVs, SVGA cells incubated with Cy3-labeled PS-ASO were stained for M6PR-CI (Figure 6A). Like GCC2, M6PR co-localized with PS-ASOs at distinct foci inside LEs (Figure 6A; Supplementary Figure S20 and Movie S2). M6PR traffics back from LEs to the TGN in vesicles that bud from LE membranes (31). Therefore, we next analyzed in more detail whether M6PR can co-localize with PS-ASOs in distinct vesicular structures on the membrane of LEs. At some LEs, PS-ASOs were present in small dot-like structures on the membranes of LEs (Figure 6B). Interestingly, M6PR-CI exactly co-localized with these PS-ASO-containing dots-like structures, which are present at the cytosolic face of the LE membrane and lack of Rab7 staining (Figure 6B).
Figure 6.
M6PR-CI co-localizes with PS-ASOs inside LEs and on the membranes of LEs. (A) Immunofluorescent staining of M6PR-CI in GFP-Rab7-expressing SVGA cells incubated with 2 μM PS-ASO 446654 for 12 h. Scale bars, 10 μm. The regions boxed with dashed lines are shown at higher magnification in insets. M6PR-CI co-localization with a distinct PS-ASO-containing focus inside the LE is marked with an arrow. (B) Immunofluorescent staining of M6PR-CI in GFP-Rab7-expressing SVGA cells incubated with 2 μM PS-ASO 446654 for 12 h. Co-localization of M6PR and PS-ASOs as a distinct structure on the LE membrane is marked by arrows. Scale bars, 5 μm. (C) Live cell imaging of a SGVA cell incubated with PS-ASO 446654 for 6 h. The nuclear border is marked by a dashed line. The boxed PS-ASO-containing LE in the lower right is magnified in panel D. (D) Upper images: Snapshots taken during live cell imaging. Potential PS-ASO release events are marked by arrows. Lower plots: Signal intensity profiles for the LE across the lines as indicated in the upper panels. Arrows indicate ASOs outside the LE body.
This observation raises a possibility that ASOs can escape from LEs in the form of vesicles involved in M6PR trafficking. To determine mechanisms of ASO release from LEs and whether ASOs can escape in the form of small vesicles, we performed live-cell imaging to record PS-ASO movement in SVGA cells that had been incubated with PS-ASO for 16 h (Supplementary Movie S3). From one LE structure (Figure 6C, dashed box), three ASO escape events were captured during a 15-min recording (Supplementary Movie S4 and Figure 6D). In one event, vesicle-like PS-ASO escape was observed (Figure 6D, upper middle panel). In the PS-ASO-containing small vesicular structure leaving the LE, there was little Rab7, as shown by the signal intensity profile (Figure 6D, lower middle panel). Although it is unknown whether these PS-ASO-positive vesicles contain M6PR, it appears clear that ASOs can escape from LEs in vesicular form.
GCC2 acts at a step that precedes the action of M6PR
Our data indicate that GCC2 and M6PR affect PS-ASO activity by acting on the same pathway. To determine which protein acts first in this pathway, GCC2 levels were reduced in HeLa cells by siRNA treatment (Figure 7A). In cells treated with the GCC2 siRNA there was more scattered cytoplasmic distribution of M6PR-CI than in cells treated with control siRNA (Figure 7B). This is consistent with a previous report that GCC2 is required for M6PR shuttling as GCC2 tethers vesicles to the TGN membrane (34). Upon PS-ASO incubation, the number of foci stained for both M6PR and PS-ASO was reduced (Figure 7C and D), suggesting that GCC2 is required for M6PR-CI trafficking and co-localization with PS-ASOs.
Figure 7.
GCC2 affects M6PR distribution and co-localization with PS-ASOs. (A) qRT-PCR quantification of GCC2 mRNA in HeLa cells treated with siRNAs. (B) Immunofluorescent staining of M6PR-CI in siRNA-treated HeLa cells. Scattered cytoplasmic localization of M6PR-CI is indicated by an arrow. Scale bars, 10 μm. (C) Quantification of co-localization of M6PR-CI and PS-ASO in control or GCC2-depleted HeLa cells incubated with 2 μM PS-ASO 446654 for 16 h. Error bars are standard deviations from data obtained from 20 cells. P-value was calculated based on unpaired t-test. ***, P < 0.001. (D) Immunofluorescent staining of M6PR-CI in siRNA-treated HeLa cells incubated with PS-ASO for 16 h. Scale bars, 2 μm. (E) qRT-PCR quantification of M6PR-CD and M6PR-CI in HeLa cells treated with siRNAs. (F) Immunofluorescent staining of GCC2 in HeLa cells treated with control siRNA or cells treated with M6PR-CI and M6PR-CD siRNAs. Scale bars, 10 μm. (G) Immunofluorescent staining of GCC2 in siRNA-treated HeLa cells incubated with 2 μM PS-ASO 446654 for 16 h. (H) Quantification of co-localization of GCC2 with PS-ASO in siRNA-treated HeLa cells. Error bars are standard deviations of data from 20 cells. P-value was calculated based on unpaired t-test. NS, not significant.
Next, HeLa cells were treated with siRNAs to deplete cells of both M6PR-CD and M6PR-CI (Figure 7E). Reduction of M6PR did not affect the Golgi localization pattern of GCC2 (Figure 7F). In cells incubated with PS-ASOs but depleted of M6PR, GCC2 co-localized with PS-ASOs in cytoplasmic foci (Figure 7G). Quantification showed that PS-ASO co-localization with GCC2 was not significantly affected by M6PR reduction (Figure 7H). Together, these results indicate that M6PR is not required for GCC2 localization to LEs in the presence of PS-ASO, whereas GCC2 is involved in M6PR trafficking and co-localization with PS-ASOs.
GCC2 and M6PR co-localization with PS-ASOs in LE requires the phosphorothioate backbone
Compared with ASOs that have phosphodiester (PO) backbones, the PS-backbone containing ASOs used in this study have much higher binding affinity for cellular proteins (25,41). Thus, it is possible that interactions between the PS-ASO and protein may mediate the protein localization. This possibility is supported by our previous findings that PS-ASO–protein interactions can affect protein localization (25) and that COPII vesicles and STX5 localize to LEs upon PS-ASO incubation in a way most likely mediated by PS-ASO–protein interactions (26).
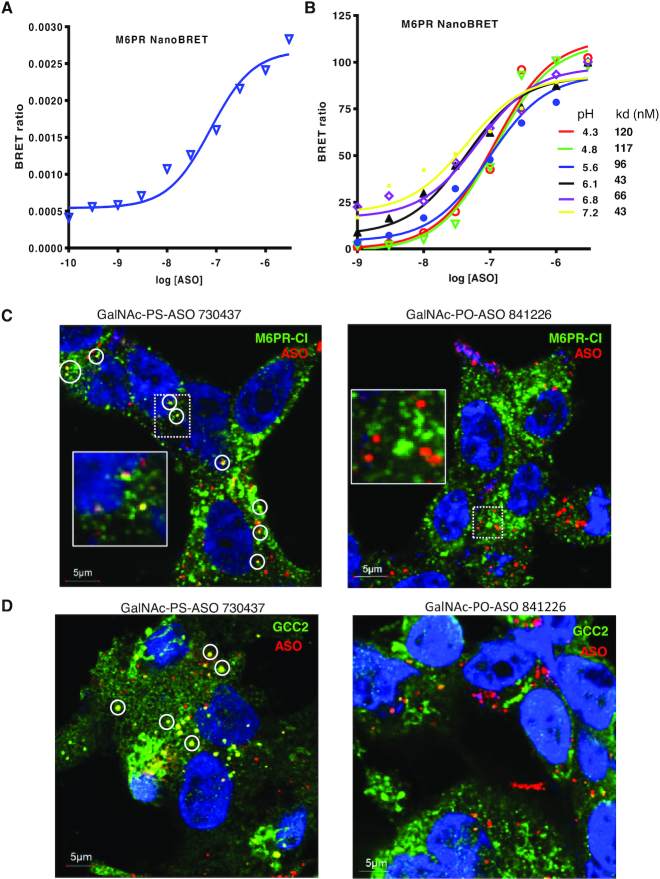
We first analyzed the binding property of M6PR-CD with PS-ASOs using the NanoBRET assay recently developed by our group (41). Due to its relatively small size, M6PR-CD is suitable for the NanoBRET assay. In this assay, the M6PR-CD protein was fused with Nanoluc luciferase, which serves as a donor to transfer light energy to Alex 594 conjugated to the PS-ASO upon binding. M6PR-CD bound to PS-ASO with an affinity of around 75 nM in buffer of pH 7.2 (Figure 8A), whereas no significant binding was detected for a PO-ASO (data not shown). In LEs, the pH is around 5.5 (55). In buffer of pH 5.6, the binding affinity was not substantially different from the affinity at pH 7.2 (Figure 8B), suggesting that this protein could bind PS-ASOs even in LEs.
Figure 8.
Phosphorothioate modification is required for co-localization of PS-ASOs with GCC2 and M6PR. (A) Binding affinity of M6PR-CD for PS-ASO 766634 was determined using the NanoBRET assay. Plotted is the BRET ratio versus log concentration of PS-ASO. (B) Binding affinities of M6PR-CD for PS-ASO 766634 in buffers of different pHs were determined using the NanoBRET assay. The kd values (nM) are shown in the key. (C) Immunofluorescent staining of M6PR-CI in HepG2 cells incubated for 24 h with 3 μM GalNAc-conjugated PS-ASO 730437 (left) or PO-ASO 841226 (right). Co-localization between PS-ASO and M6PR-CI is indicated by circles. Scale bars, 5 μm. The regions boxed with dashed lines are shown at higher magnification in insets. (D) Immunofluorescent staining of GCC2 in HepG2 cells incubated for 24 h with 3 μM GalNAc-conjugated PS-ASO 730437 (left) or PO-ASO 841226 (right). Co-localization between PS-ASO and M6PR-CI is indicated by circles. Scale bars, 5 μm.
Since PO-ASOs have much weaker protein binding affinities than PS-ASOs (16), we reasoned that if a PS-ASO–protein interaction is required for GCC2 and M6PR localization to LEs, the presence of a PO-ASO in cells should not alter localization. To evaluate this hypothesis, we synthesized GalNAc-conjugated PS- or PO-ASOs that have the same sequence and 2′-MOE modification patterns. These ASOs were incubated with HepG2 cells, which express the GalNAc receptor ASGR (56). GalNAc-conjugated oligonucleotides are efficiently internalized by these cells (57). Immunofluorescent staining revealed co-localization between M6PR-CI and the GalNAc-PS-ASO (Figure 8C, left panel). However, no obvious co-localization was detected between M6PR-CI and the GalNAc-PO-ASO, although the PO-ASO was internalized to similar levels as the PS-ASO (Figure 8C, right panel). Similarly, GCC2 protein co-localized with the GalNAc-PS-ASO (Figure 8D, left panel), whereas no substantial co-localization was detected for GCC2 and the GalNAc-PO-ASO (Figure 8D, right panel). Similar observation was also made in ASGR-expressing Hek293 cells (57), in which the GalNAc-PS-ASO, and not the GalNAc-PO-ASO, co-localized with GCC2 in distinct cytoplasmic foci (Supplementary Figure S21A and B). In addition, reduced activity was found for the GalNAc-PO-ASO, as compared with the GalNAc-PS-ASO (Supplementary Figure S21C). Together, these results suggest that GCC2 and M6PR relocalization to LEs requires PS-modification of ASOs, and that the re-localization is most likely mediated by PS-ASO–protein interactions.
Reduction of M6PR-CI in mouse decreases PS-ASO activity
To determine whether the observed effects of M6PR on PS-ASO activity in human cells also occur in other species, we reduced the levels of M6PR-CD and M6PR-CI mRNAs and proteins in mouse MHT cells by siRNA treatment (Figure 9A-C). Depletion of M6PR-CI significantly decreased PS-ASO activity (Figure 9D), consistent with what was observed in human cells. However, the reduction of M6PR-CD levels did not alter PS-ASO activity in the mouse cells, indicative of a species-specific effect of different forms of M6PR on PS-ASO activity.
Figure 9.
M6PR-CI is required for PS-ASO activity in mouse cells and in mice. (A) qRT-PCR quantification of M6PR-CD and M6PR-CI mRNA levels in mouse MHT cells at 72 h after transfection with control siRNA or siRNAs targeting M6PR-CD or M6PR-CI mRNA. Error bars are standard deviations from three independent experiments. (B) Western analysis of M6PR-CI protein in MHT cells transfected with indicated siRNAs. (C) Immunofluorescent staining of M6PR-CD in MHT cells treated with control or M6PR-CD-targeted siRNAs. Scale bars, 10 μm. (D) qRT-PCR quantification of SRB1 mRNA in MHT cells pretreated with siRNAs, followed by incubation with SRB1-targeting PS-ASO 353382. Error bars are standard deviations from three independent experiments. P-values were calculated based on F-test. (E) Upper panel: Quantification of M6PR-CD and M6PR-CI mRNAs in livers of mice treated with control PS-ASO 716837, PS-ASO 1332272 targeting M6PR-CD, or PS-ASO 1332273 targeting M6PR-CI. These mice were subsequently treated with the SRB1 ASO at different doses, as in panel F. Error bars are standard deviations from three animals. Lower panel: M6PR-CI protein levels evaluated by western in livers of mice treated with indicated control or M6PR PS-ASOs. The numbers above lanes represent different animals in each group. (F) qRT-PCR quantification of SRB1 mRNA in livers of mice treated with control PS-ASO 716837, M6PR-CD-targeted PS-ASO 1332272, or M6PR-CI-targeted PS-ASO 1332273 for 5 days followed by treatment with SRB1 PS-ASO 353382 at indicated doses for 24 h. Error bars are standard deviations from three animals. P-values were calculated based on unpaired t-test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (G) qRT-PCR quantification of PTEN mRNA in livers of mice treated with control PS-ASO 716837, PS-ASO 1332272 targeting M6PR-CD or PS-ASO 1332273 targeting M6PR-CI for 5 days, followed by treatment with PTEN PS-ASO 116847 at indicated doses for 24 h. Error bars are standard deviations from three animals. P-values were calculated based on unpaired t-test. *, P < 0.05; ***, P < 0.001.
To confirm this observation and to determine whether M6PR affects PS-ASO activity in vivo, we identified PS-ASOs targeting mouse M6PR-CD and M6PR-CI mRNAs. Mice were given two subcutaneous injections of 15 mg/kg GalNAc-conjugated PS-ASOs targeting individual M6PR mRNAs or a control PS-ASO. Groups of mice were then treated with PS-ASOs targeting either SRB1 or PTEN mRNA. Animals were sacrificed and liver samples were collected for protein or RNA analysis. The levels of serum ALT and serum AST were normal despite depletion of either M6PR protein, and no differences in body weight or organ weights were observed at the time of sacrifice for mice given targeted PS-ASOs compared to mice given the control PS-ASO (data not shown).
As compared with the control PS-ASO treatment, the M6PR PS-ASOs specifically reduced the levels of the targeted M6PR mRNAs in livers of mice (N = 3) that were subsequently treated with different doses of the PS-ASO targeting SRB1 mRNA (Figure 9E, upper panel). Western analyses showed that the levels of M6PR-CI protein were also reduced in livers of mice (N = 3) treated with the ASO targeting M6PR-CI (Figure 9E, lower panel). Reduction of M6PR-CI decreased the activities of PS-ASOs targeting SRB1 (Figure 9F) and PTEN (Figure 9G) in mouse liver as evaluated by qRT-PCR, whereas depletion of M6PR-CD had no substantial effect. This is consistent with the observations made in mouse cells. Together, these results confirmed that M6PR-CI, but not M6PR-CD, is necessary for optimal PS-ASO activity in mice.
DISCUSSION
A better understanding of the intracellular trafficking and endosomal release of PS-ASOs will facilitate the design of PS-ASO drugs that efficiently localize to the site of their targets. Previous studies have shown that productive release of PS-ASOs mainly occurs at LEs or multivesicular bodies, and different potential mechanisms have been proposed for the escape of PS-ASOs from these organelles (12,16,27). Some proteins have been found to localize to LEs and facilitate PS-ASO trafficking and release (18,24). In addition, COPII vesicles, which function as part of the ER-Golgi transport machinery, can be recruited to LEs to ensure efficient PS-ASO release (26). These observations suggest that multiple intracellular machineries can be harnessed to facilitate PS-ASO release. Indeed, here we show that the TGN-LE transport pathway can also facilitate productive PS-ASO release from the endocytic pathway.
The GCC2-mediated LE to TGN retrograde transport pathway is involved in intracellular transport of Shiga toxin and M6PR (34,58). We found that in cells deficient in GCC2, PS-ASO activity was impaired upon free uptake. In addition, depletion of either M6PR-CD or M6PR-CI also reduced PS-ASO activity in different types of human cells, and simultaneous reduction of the two M6PR proteins caused additive effects. These results suggest that the functions of the two M6PR proteins do not entirely overlap and that both proteins facilitate PS-ASO activity in human cells. Consistent with previous reports that GCC2 mediates M6PR trafficking (34), we found that GCC2 and M6PR likely affected PS-ASO activity by acting on the same pathway, as co-depletion of M6PR and GCC2 did not cause cumulative effects. As a control, we reduced levels of another Golgi protein, GM130, in human cells. GM130 has no demonstrated role in M6PR transport and did not affect PS-ASO activity. These observations suggest that a functional M6PR-mediated LE to TGN transport pathway, rather than individual GCC2 and M6PR proteins, is required for optimal PS-ASO activity.
Importantly, we found that reduction of M6PR-CI in mouse cells and in mice also reduced PS-ASO activity, indicating that this pathway might be conserved between human and mouse. However, in contrast to human cells, reduction of M6PR-CD in mouse cells and in mice had no significant effect on PS-ASO activity. This suggests that the two forms of M6PR may have species-specific effects. This is not entirely unexpected, since levels of expression of the two forms of M6PR vary in different cells (40) and M6PR-CI may handle most of the traffic (39).
Depletion of GCC2 or M6PR did not affect PS-ASO activity when PS-ASOs were delivered by transfection. Thus, we analyzed internalization, endocytic transport, and endosomal release steps individually. We found that reduction of GCC2 or M6PR proteins did not substantially affect PS-ASO internalization. Although about 10% of M6PR-CI protein is present on plasma membranes (38), the cell surface M6PR-CI is not required for ASO internalization, as the presence of M6PR-CI in EEs was neither necessary nor sufficient for co-internalization of PS-ASOs. In addition, the extents and kinetics of PS-ASO localization to EE and to LEs were not affected by reduction of GCC2 or M6PR proteins. That the M6PR-mediated LE to TGN transport affects PS-ASO release from LEs was confirmed by the results of a kinetic study of PS-ASO activity.
To better understand how the LE to TGN transport pathway affects PS-ASO release, immunofluorescent staining was performed. Surprisingly, we found that upon incubation of human cells with a PS-ASO, GCC2 was recruited to LEs in a time dependent manner. The kinetics of GCC2 co-localization with PS-ASOs at LEs correlated with the kinetics of PS-ASO-mediated RNA reduction (16), supporting a role of GCC2 in PS-ASO release from LEs. Interestingly, M6PR also co-localized with ASOs in LEs. The LE localization of M6PR-CI increased in a time-dependent manner after PS-ASO addition to cells, similar to the kinetics of GCC2 co-localization with PS-ASOs in LEs, suggesting that PS-ASOs may enhance the shuttling of M6PR between LEs and the TGN.
Consistent with a previous report (34), we observed that reduction of GCC2 caused an increase in cytoplasmic localization of M6PR and reduced PS-ASO co-localization with M6PR in LEs. However, reduction of M6PR did not alter the localization pattern of GCC2 or the co-localization of GCC2 with PS-ASOs at LEs, indicating that GCC2 acts at a step upstream of M6PR during the process that enhances PS-ASO release. This is similar to our previous observations that STX5 is required for COPII vesicle recruitment to LEs and not vice versa (26). However, it appears that the LE recruitment of M6PR and COPII vesicles occurs via different pathways, as reduction of GCC2 or M6PR did not affect co-localization of COPII with PS-ASOs and reduction of STX5 did not disrupt PS-ASO co-localization with either GCC2 or M6PR. These observations further suggest that PS-ASOs can escape from the endocytic pathway through different pathways, and these multiple release pathways may act together to contribute to ASO activity.
Indeed, several different ASO escape events from the same LE were captured by live cell imaging (Supplementary Movie S4), including vesicle-like escape and membrane leakage, suggesting that different release pathways exist even for the same endosome. In addition, we showed previously that although ILV-mediated back fusion process contributes to ASO release, disruption of ILV formation by modulating LBPA levels did not completely inhibit ASO activity and release, and that not all ASOs in LEs are present in ILVs (22). These observations, together with previous findings that reduction of ANXA2, TCP1 or AP2M1 all modestly affect ASO activity (18,20,25), suggest that multiple release pathways coexist. These different release pathways, e.g. protein binding-mediated membrane deformation, back-fusion mediated release from ILVs, and vesicle-mediated escape from LEs, may each contribute to certain degree, independently or cooperatively, generating the pool of cellular productive ASOs. This view is also supported by the observation that no single protein factor (except RNase H1) was found to play a dominant role in ASO activity in a genome-wide screening using shRNAs (Alexey Revenko, personal communication).
Interestingly, reduction of GCC2 or M6PR decreased co-localization of ANXA2 with PS-ASOs at LEs. ANXA2 interacts with negatively charged phospholipids and is recruited to LEs upon PS-ASO incubation (24,52–53), likely by co-transport from EEs to LEs and by recruitment of ANXA2 from the cytosol to the LE membranes (18). Reduced recruitment of ANXA2 to LEs upon depletion of either GCC2 or M6PR suggests that there is a change in the LE membrane when cells are depleted of GCC2 or M6PR, and this may also contribute to the decreased ASO activity.
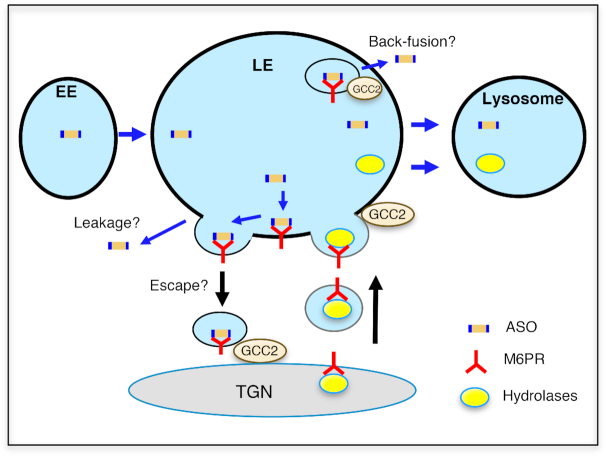
Both GCC2 and M6PR co-localized with PS-ASOs in ILVs inside LEs. It is possible that GCC2 and M6PR facilitate PS-ASO release via a back-fusion process through which ILV membranes fuse with the limiting membranes of LEs (Figure 10). Consistent with the vesicular trafficking of M6PR between LE and TGN, M6PR-CI was also found to co-localize with ASOs in what appear to be vesicles on the LE membrane. This raises the possibility that M6PR on the LE membrane may interact with PS-ASOs present in the lumen of LEs, facilitating PS-ASO escape when M6PR vesicles return to the TGN (Figure 10). This possibility is supported by several observations: First, PS-ASOs can leave LEs in the form of vesicles as shown by live cell imaging; second, PS-ASOs can bind M6PR; and, third, M6PR and GCC2 localization to LEs is most likely mediated by PS-ASO-protein interactions. A better understanding of how M6PR facilitates PS-ASO escape and whether vesicles that contain PS-ASOs and M6PR can reach TGN requires further investigation.
Figure 10.
Proposed model of PS-ASO release from LEs mediated by GCC2 and M6PR. Internalized PS-ASOs traffic from EE to LEs where they accumulated in the lumen of LE or in ILVs. Upon PS-ASO internalization, GCC2 localizes to LEs and M6PR localization to LEs is enhanced. M6PR-containing vesicles fuse with LE membranes, and M6P-tagged hydrolases dissociate from M6PR and are trafficked to lysosomes. In LEs, M6PR may thus be able to interact with PS-ASOs present in LEs, and bud out from LE membrane. PS-ASOs may be released from LEs during membrane fission or may escape together with M6PR-containing vesicles from LEs. In addition, GCC2 and M6PR present in ILVs may facilitate PS-ASO release via a back-fusion-mediated process.
In summary, we identified an intracellular transport pathway that mediates PS-ASO release from LEs. Our observations, together with our previous reports that COPII vesicles, which are part of the ER to Golgi transport machinery, facilitate ASO release (26), suggest that the Golgi apparatus is important for PS-ASO intracellular trafficking and distribution. These findings provide new opportunities to enhance ASO activity. For example, the M6PR-mediated LE to TGN retrograde transport pathway could be manipulated to enhance ASO release. In addition, medicinal chemistry may also be applied to enhance the interaction of ASOs with M6PR and to boost the release of ASOs from this pathway, a research that we are currently conducting.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Eli Scandalis for screening of ASOs targeting mouse M6PR-CD and M6PR-CI mRNAs, Dr Frank Bennett for stimulating discussions, Dr Tomas Kirchhausen (Harvard Medical School) for the kind gift of the GFP-Rab7 SVGA cell line and Dr T.P. Prakash for providing Cy3-labeled, GalNAc-conjugated ASOs and Dr Michael Tanowitz for providing the ASGR-HEK293 cell line.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Internal funding from Ionis Pharmaceuticals. Funding for open access charge: Ionis Pharmaceuticals internal funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F.. RNA-targeted therapeutics. Cell Metab. 2018; 27:714–739. [DOI] [PubMed] [Google Scholar]
- 2. Bennett C.F., Baker B.F., Pham N., Swayze E., Geary R.S.. Pharmacology of antisense drugs. Annu. Rev. Pharmacol. Toxicol. 2017; 57:81–105. [DOI] [PubMed] [Google Scholar]
- 3. Crooke S.T. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017; 27:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crooke S.T., Vickers T.A., Lima W.F., Wu H.-J.. CrookeS T. Mechanisms of antisense drug action, an introduction. Antisense Drug Technology—Principles, Strategies, and Applications. 2008; 2nd ednBoca Raton: CRC Press; 3–46. [Google Scholar]
- 5. Ward A.J., Norrbom M., Chun S., Bennett C.F., Rigo F.. Nonsense-mediated decay as a terminating mechanism for antisense oligonucleotides. Nucleic Acids Res. 2014; 42:5871–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liang X.H., Nichols J.G., Hsu C.W., Vickers T.A., Crooke S.T.. mRNA levels can be reduced by antisense oligonucleotides via no-go decay pathway. Nucleic Acids Res. 2019; 47:6900–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R.. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007; 5:729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liang X.H., Shen W., Sun H., Migawa M.T., Vickers T.A., Crooke S.T.. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotech. 2016; 34:875–880. [DOI] [PubMed] [Google Scholar]
- 9. Liang X.H., Shen W., Crooke S.T.. Specific increase of protein levels by enhancing translation using antisense oligonucleotides targeting upstream open frames. Adv. Exp. Med. Biol. 2017; 983:129–146. [DOI] [PubMed] [Google Scholar]
- 10. Nomakuchi T.T., Rigo F., Aznarez I., Krainer A.R.. Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat. Biotechnol. 2016; 34:164–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang X.H., Sun H., Nichols J.G., Crooke S.T.. RNase H1-dependent antisense oligonucleotides are robustly active in directing RNA cleavage in both the cytoplasm and the nucleus. Mol. Ther. 2017; 25:2075–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Juliano R.L. Intracellular Trafficking and Endosomal Release of Oligonucleotides: What We Know and What We Don’t. Nucleic Acid Ther. 2018; 28:166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Juliano R.L., Carver K., Cao C., Ming X.. Receptors, endocytosis, and trafficking: the biological basis of targeted delivery of antisense and siRNA oligonucleotides. J. Drug Target. 2013; 21:27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prakash T.P., Graham M.J., Yu J., Carty R., Low A., Chappell A., Schmidt K., Zhao C., Aghajan M., Murray H.F. et al.. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014; 42:8796–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang S., Allen N., Vickers T.A., Revenko A.S., Sun H., Liang X.H., Crooke S.T.. Cellular uptake mediated by epidermal growth factor receptor facilitates the intracellular activity of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res. 2018; 46:3579–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H.. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017; 35:230–237. [DOI] [PubMed] [Google Scholar]
- 17. Miller C.M., Donner A.J., Blank E.E., Egger A.W., Kellar B.M., Ostergaard M.E., Seth P.P., Harris E.N.. Stabilin-1 and Stabilin-2 are specific receptors for the cellular internalization of phosphorothioate-modified antisense oligonucleotides (ASOs) in the liver. Nucleic Acids Res. 2016; 44:2782–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T.. Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res. 2016; 44:7314–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller C.M., Wan W.B., Seth P.P., Harris E.N.. Endosomal escape of antisense oligonucleotides internalized by stabilin receptors is regulated by Rab5C and EEA1 During endosomal maturation. Nucleic Acid Ther. 2018; 28:86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F.. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011; 39:4795–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016; 44:6518–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T.. Intra-endosomal trafficking mediated by lysobisphosphatidic acid contributes to intracellular release of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res. 2017; 45:5309–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buntz A., Killian T., Schmid D., Seul H., Brinkmann U., Ravn J., Lindholm M., Knoetgen H., Haucke V., Mundigl O.. Quantitative fluorescence imaging determines the absolute number of locked nucleic acid oligonucleotides needed for suppression of target gene expression. Nucleic Acids Res. 2019; 47:953–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang X.H., Sun H., Shen W., Crooke S.T.. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015; 43:2927–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liang X.H., Shen W., Sun H., Prakash T.P., Crooke S.T.. TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acids Res. 2014; 42:7819–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liang X.H., Sun H., Nichols J.G., Allen N., Wang S., Vickers T.A., Shen W., Hsu C.W., Crooke S.T.. COPII vesicles can affect the activity of antisense oligonucleotides by facilitating the release of oligonucleotides from endocytic pathways. Nucleic Acids Res. 2018; 46:10225–10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Juliano R.L., Carver K.. Cellular uptake and intracellular trafficking of oligonucleotides. Adv. Drug. Deliv. Rev. 2015; 87:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zanetti G., Pahuja K.B., Studer S., Shim S., Schekman R.. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 2011; 14:20–28. [DOI] [PubMed] [Google Scholar]
- 29. Allan B.B., Moyer B.D., Balch W.E.. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000; 289:444–448. [DOI] [PubMed] [Google Scholar]
- 30. Rowe T., Dascher C., Bannykh S., Plutner H., Balch W.E.. Role of vesicle-associated syntaxin 5 in the assembly of pre-Golgi intermediates. Science. 1998; 279:696–700. [DOI] [PubMed] [Google Scholar]
- 31. Coutinho M.F., Prata M.J., Alves S.. Mannose-6-phosphate pathway: a review on its role in lysosomal function and dysfunction. Mol. Genet. Metab. 2012; 105:542–550. [DOI] [PubMed] [Google Scholar]
- 32. Pfeffer S.R. Multiple routes of protein transport from endosomes to the trans Golgi network. FEBS Lett. 2009; 583:3811–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bonifacino J.S., Rojas R.. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 2006; 7:568–579. [DOI] [PubMed] [Google Scholar]
- 34. Reddy J.V., Burguete A.S., Sridevi K., Ganley I.G., Nottingham R.M., Pfeffer S.R.. A functional role for the GCC185 golgin in mannose 6-phosphate receptor recycling. Mol. Biol. Cell. 2006; 17:4353–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hayes G.L., Brown F.C., Haas A.K., Nottingham R.M., Barr F.A., Pfeffer S.R.. Multiple Rab GTPase binding sites in GCC185 suggest a model for vesicle tethering at the trans-Golgi. Mol. Biol. Cell. 2009; 20:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brown F.C., Schindelhaim C.H., Pfeffer S.R.. GCC185 plays independent roles in Golgi structure maintenance and AP-1-mediated vesicle tethering. J. Cell Biol. 2011; 194:779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ghosh P., Dahms N.M., Kornfeld S.. Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 2003; 4:202–212. [DOI] [PubMed] [Google Scholar]
- 38. Klumperman J., Hille A., Veenendaal T., Oorschot V., Stoorvogel W., von Figura K., Geuze H.J.. Differences in the endosomal distributions of the two mannose 6-phosphate receptors. J. Cell Biol. 1993; 121:997–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dahms N.M., Lobel P., Kornfeld S.. Mannose 6-phosphate receptors and lysosomal enzyme targeting. J. Biol. Chem. 1989; 264:12115–12118. [PubMed] [Google Scholar]
- 40. Stein M., Zijderhand-Bleekemolen J.E., Geuze H., Hasilik A., von Figura K.. Mr 46,000 mannose 6-phosphate specific receptor: its role in targeting of lysosomal enzymes. EMBO J. 1987; 6:2677–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vickers T.A., Crooke S.T.. Development of a quantitative BRET affinity assay for nucleic Acid-Protein interactions. PLoS One. 2016; 11:e0161930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luke M.R., Kjer-Nielsen L., Brown D.L., Stow J.L., Gleeson P.A.. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. 2003; 278:4216–4226. [DOI] [PubMed] [Google Scholar]
- 43. Bennett C.F., Chiang M.Y., Chan H., Shoemaker J.E., Mirabelli C.K.. Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides. Mol. Pharmacol. 1992; 41:1023–1033. [PubMed] [Google Scholar]
- 44. Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T.E., Warren G.. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 1995; 131:1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hickinson D.M., Lucocq J.M., Towler M.C., Clough S., James J., James S.R., Downes C.P., Ponnambalam S.. Association of a phosphatidylinositol-specific 3-kinase with a human trans-Golgi network resident protein. Curr. Biol. 1997; 7:987–990. [DOI] [PubMed] [Google Scholar]
- 46. Edgar J.R., Eden E.R., Futter C.E.. Hrs- and CD63-dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic. 2014; 15:197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bissig C., Gruenberg J.. Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb. Perspect. Biol. 2013; 5:a016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakamura N. Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions. J. Pharmacol. Sci. 2010; 112:255–264. [DOI] [PubMed] [Google Scholar]
- 49. Shen W., Liang X.H., Crooke S.T.. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014; 42:8648–8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Prydz K., Brandli A.W., Bomsel M., Simons K.. Surface distribution of the mannose 6-phosphate receptors in epithelial Madin-Darby canine kidney cells. J. Biol. Chem. 1990; 265:12629–12635. [PubMed] [Google Scholar]
- 51. Hirst J., Futter C.E., Hopkins C.R.. The kinetics of mannose 6-phosphate receptor trafficking in the endocytic pathway in HEp-2 cells: the receptor enters and rapidly leaves multivesicular endosomes without accumulating in a prelysosomal compartment. Mol. Biol. Cell. 1998; 9:809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Flower R.J., Perretti M.. ‘Annexins’ themed section. Br. J. Pharmacol. 2015; 172:1651–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morel E., Parton R.G., Gruenberg J.. Annexin A2-dependent polymerization of actin mediates endosome biogenesis. Dev. Cell. 2009; 16:445–457. [DOI] [PubMed] [Google Scholar]
- 54. Griffiths G., Hoflack B., Simons K., Mellman I., Kornfeld S.. The mannose 6-phosphate receptor and the biogenesis of lysosomes. Cell. 1988; 52:329–341. [DOI] [PubMed] [Google Scholar]
- 55. Mellman I. The importance of being acid: the role of acidification in intracellular membrane traffic. J. Exp. Biol. 1992; 172:39–45. [DOI] [PubMed] [Google Scholar]
- 56. Harris R.L., van den Berg C.W., Bowen D.J.. ASGR1 and ASGR2, the genes that encode the asialoglycoprotein receptor (Ashwell Receptor), Are expressed in peripheral blood monocytes and show interindividual differences in transcript profile. Mol. Biol. Int. 2012; 2012:283974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tanowitz M., Hettrick L., Revenko A., Kinberger G.A., Prakash T.P., Seth P.P.. Asialoglycoprotein receptor 1 mediates productive uptake of N-acetylgalactosamine-conjugated and unconjugated phosphorothioate antisense oligonucleotides into liver hepatocytes. Nucleic Acids Res. 2017; 45:12388–12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Derby M.C., Lieu Z.Z., Brown D., Stow J.L., Goud B., Gleeson P.A.. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 2007; 8:758–773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.