Abstract
Intron retention (IR) has been proposed to modulate the delay between transcription and translation. Here, we provide an exhaustive characterization of IR in differentiated white blood cells from both the myeloid and lymphoid lineage where we observed highest levels of IR in monocytes and B-cells, in addition to previously reported granulocytes. During B-cell differentiation, we found an increase in IR from the bone marrow precursors to cells residing in secondary lymphoid organs. B-cells that undergo affinity maturation to become antibody producing plasma cells steadily decrease retention. In general, we found an inverse relationship between global IR levels and both the proliferative state of cells, and the global levels of expression of splicing factors. IR dynamics during B-cell differentiation appear to be conserved between human and mouse, suggesting that IR plays an important biological role, evolutionary conserved, during blood cell differentiation. By correlating the expression of non-core splicing factors with global IR levels, and analyzing RNA binding protein knockdown and eCLIP data, we identify a few splicing factors likely playing an evolutionary conserved role in IR regulation. Our work provides new insights into the role of IR during hematopoiesis, and on the main factors involved in regulating IR.
INTRODUCTION
Hematopoietic cell differentiation gives rise to all blood cell types that carry out diverse functions from oxygen transport to detection and removal of pathogens. It is one of the best studied differentiation systems in human, as samples can easily be acquired. Additionally, cells can be sorted by a plenty of cell type and developmental stage specific surface markers. The investigation of the mechanisms that drive and define differentiation decisions led to the identification of key transcription factors (TFs) that stimulate distinct developmental programs by regulating gene expression as a major driving force (1–3). However, other mechanisms as alternative isoform usage and alternative splicing have been shown to contribute in cell-fate decision making (4).
In general, alternative splicing is assumed to enrich protein diversity in vertebrates, especially through alternative usage of cassette exons (5,6). These may perform regulatory functions, for example when binding or membrane spanning domains are exchanged, removed or added. Compared to other modes of alternative splicing, such as alternative 3′ and 5′ and alternative exon usage, intron retention (IR) has been less investigated (7). Here, introns are not removed by the spliceosome, but remain between their neighbouring exons in the processed transcript (8,9). While the functional mechanism of IR is not yet well understood, it is speculated to halt translation until a signalling response or environmental stimulus is received (10–14). As transcripts with retained introns often contain premature termination codons (PTCs), they may be degraded by nonsense mediated decay (NMD) (15). Since NMD occurs in the cytoplasm, nuclear sequestration allows these transcripts to escape degradation, while preventing them from being translated (16,17).
IR is widespread among almost all eukaryotes from fungi and plants to mammals (8,18,19). Conservation in IR has been reported between human and mouse (15). While differential IR has been found in various cancers (20,21), it appears to play a more important role during differentiation and development. In mammals, IR has been reported predominantly in neuronal development (12,14) and in a variety of differentiation processes in the hematopoietic lineage, especially in the myeloid branch, where IR impacts maturation of erythrocytes (22), megakaryocytes (17) and granulocytes (15). In the lymphoid branch, IR has only been investigated in T-cells, where it declines upon their activation (23).
Here, with the aim of fully characterizing IR during haematopoiesis, we analysed human and mouse RNASeq data from a variety of sources (Supplementary Table S1) obtained in differentiated immune cells, as well as during B-cell and neutrophil/granulocyte differentiation. We found that the global dynamics of intron retention during B-cell differentiation is largely conserved between human and mouse, with IR increasing from precursors in the bone marrow towards lymphoid organs and decreasing when cells undergo affinity maturation. Overall, IR increases towards cellular states with low proliferative potential and with decreased expression levels of splicing factors. This suggest that there is an interplay between IR and splicing at the cellular level, with higher levels of IR partially being a consequence of globally weakened splicing. Finally, by correlating the expression of non-core splicing factors with global IR levels, and analyzing RBP knockdown and eCLIP data, obtained in the framework of the ENCODE project (24), we found a set of non-core splicing factors likely playing a role in the regulation of intron retention. Overall our results uncover a largely unappreciated role of IR in B-cell differentiation.
MATERIALS AND METHODS
Sequencing data
We used different sets of publically available RNASeq samples from cells within the hematopoietic lineage and other blood derived cell lines. First, we used FACS sorted human primary blood cell RNASeq samples, produced by the Blueprint Epigenome Consortium. They encompass the major differentiated human immune cell types (human, total RNA, macrophages were derived from monocytes in vitro) as well as a set of five stages of human B-cell differentiation (human, total RNA) and five stages of human neutrophil differentiation (human, total RNA). Second, we used, murine samples covering seven stages of B-cell development (polyA+) from Brazao et al. (25). Additional murine samples were taken from the ImmGen Consortium (26), also covering seven developmental stages of B-cells (polyA+). Furthermore, we reprocessed RNASeq samples corresponding to three different stages of murine granulocyte development (polyA+) from Wong et al. (15). Finally, we also used polyA+ RNASeq samples of RBP shRNA knockdowns and RBP eCLIP samples from K562 cells generated by the ENCODE Consortium (24,27).
Quantification of intron retention events
Intron retention values for all human and mouse RNASeq data sets were accessed with IRFinder (28). In brief, IRFinder mapped the reads of the FASTQ samples against repeat masked genomes (GRCh38 for human and GRCm38 for mouse) with STAR (29). Using the GENCODE annotations (30) for human v22 and mouse v11, all introns with at least three read pairs overlapping the exon-intron splice junction and a coverage along the intronic sequence of 50% with at least three reads were selected. In a second step, unmapped reads were mapped against the unmasked versions of the genomes. If inclusion of that regions with repetitions or low complexity lead to 90% coverage of the intronic sequence, introns were classified as retained and retention values (between 0 and 1) were computed by the ratio of intronic read coverage in the exon flanking region divided by coverage of neighbouring exons. Introns overlapping exons of other transcripts were removed.
Computation of differential intron retention
Differential intron retention (dIR) values were computed for murine B-cells between the stage with highest and lowest global retention (MZ and GC B-cells, absolute values). The values are based on the median among the five replicates of the stages being compared. In consequence, introns for which IR values could not be determined at a given stage in comparison were omitted. This resulted in values for 47 291 introns from 7786 genes. Then, for each intron, we tested if the differences in inclusion were statistically significant using a t-test. The resulting P-values were corrected for multiple testing. We considered introns with dIR >0.2 and FDR <0.05 as highly differentially retained. This resulted in a set of 630 introns from 520 genes. For all 630 introns, IR was larger for MZ than for GC B-cells.
For computing correlations of IR with gene expression we selected the intron with the highest dIR value between MZ and GC B-cells. We excluded genes if their peak expression was lower than 1 FPKM in any sample.
Gene expression estimation
RNA steady state abundances were taken as proxy for gene expression. Expression values were generated from the raw FASTQ files with the Grape2 pipeline developed in-house (https://github.com/guigolab/grape-nf). In brief, the STAR mapper was used with default settings to map against the human genome GRCh38 and mouse genome mm10. RSEM (31) was used to compute FPKM values from the STAR alignments using the GENCODE annotations human gencode22 and mouse M11. The FPKM values were considered as expression values.
Gene ontology analysis
To determine the function of genes containing retained introns, GO-term enrichment was performed with Gorilla (http://cbl-gorilla.cs.technion.ac.il/). As the gene set of interest, we used all genes with dIR >0.2 for their most differentially retained intron between MZ and GC B-cells. As background, we used all genes expressed with an average expression >0.1 FPKM among all compared stages. Relative enrichment was computed above that background with otherwise default settings of Gorilla. Terms were filtered by FDR <0.05 and GO term enrichment >2. Furthermore, we removed parental GO terms that did not display significant enrichment when evidence provided by their significant child terms was removed.
Assessment of splice site strength
Splice site strength was computed with MaxEntScan developed by Yeo and Burge (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html) The set of introns from mouse B-cells, defined before, with dIR >0.2 and FDR<0.05 (n = 630) was used. Scoring was done with the Maximum Entropy Model for both 3′ and 5′ splice sites.
Motif enrichment
Enrichment of RBP binding sites within intronic sequences was assessed with the MEME Suite 4.12.0. Here, we used AME (32) to find binding motifs enriched in the intronic sequences of interest with default settings for a set of 102 RNA binding protein motifs (33). As background, we used a set of size matched introns with low differential retention values.
Analysis of intron retention in splicing factor knockdown data from ENCODE
RNASeq data of shRNA knockdowns of RNA binding splicing factors were derived from the ENCODE data portal (https://www.encodeproject.org) and processed with IRFinder like the other RNASeq datasets. The analysis was limited to 58 factors overlapping a list of splicing factors provided in Papasaikas et al. (34). The two biological replicates per splicing factor knockdown experiment were processed separately and combined by taking the mean of the IR values. For 42 controls with non-specific targeting, that were processed independently, the median was taken. Those median IR values of the controls were subtracted from the mean IR values of each of the 58 splicing factor knockdowns for each gene. The resulting dIR values were positive for splicing factor knockdowns causing higher IR relative to controls, which indicates a splicing enhancing function. Negative values in contrast indicate a suppressive effect of the splicing factors on IR.
Analysis of splicing factor binding in eCLIP data from ENCODE
BigWig files for RNA-binding splicing factors were derived from the ENCODE data portal (https://www.encodeproject.org). Binding patterns were extracted with bwtool (35). To identify the introns more likely to be mainly regulated by HNRNPK and SRSF1 and not by other splicing factors, for each intron we recomputed dIR for the knockdowns of the factors relative to the average dIR caused by the knockdown of other non-core splicing factors for which eCLIP data is available, excluding the tested factor (FUS, HNRNPA1, HNRNPC, HNRNPL, HNRNPU, ILF3, PTBP1, SRSF1, SRSF7, HNRNPK). We selected the top 100 dIR events for each factor (minimum dIR 0.20 for HNRNPK and 0.14 for SRSF1). We then compared the peak eCLIP signal (average of five nucleotides around the peak) of HNRNPK and SRSF1 in these introns, with the average peak eCLIP signal of the other non-core splicing factors. Significance values were computed with the Kolmogorov-Smirnov test in R.
Sequence conservation of introns
To estimate the sequence conservation of differentially retained introns we downloaded a subsetted bigwig file containing base wise conservation scores of 20 Placental mammals from the UCSC server (http://hgdownload.cse.ucsc.edu/goldenpath/mm10/phastCons60way/). To aggregate the scores over the intronic regions we used bigWigAverageOverBed from KentUtils (https://github.com/ucscGenomeBrowser/kent). For comparison, we selected all introns with an IR value >0.2 against size matched introns with a value <0.02. We tested for a statistically significant difference between the two with a two-tailed t-test.
Correlation of splicing factor expression and IR
As a proxy for the overall impact of IR on murine B-cell and granulocyte development, we took the relative number of genes that had IR values >0.2 for its most retained intron. As the number of expressed genes varies between developmental stages, we assume that the proportions of genes with retained introns reflect the global impact of IR better than the absolute number. The splicing factor set (n = 31) used was defined by the mouse orthologues of the SR-rich and hnRNP genes provided in Papasaikas et al. (34). For each splicing factor, the spearman correlation was computed between the log10 transformed expression values and the proportion of genes with IR >0.2 in each sample.
Statistical analysis
Exploratory statistical analysis and tests were done in R. Correlation were done with the core R function using the Spearman metric. Clustering was done with the pheatmap package (https://CRAN.R-project.org/package = pheatmap). Statistical tests were performed with the core functions implemented in R.
RESULTS
Unprocessed introns are widely present in differentiated human blood cells
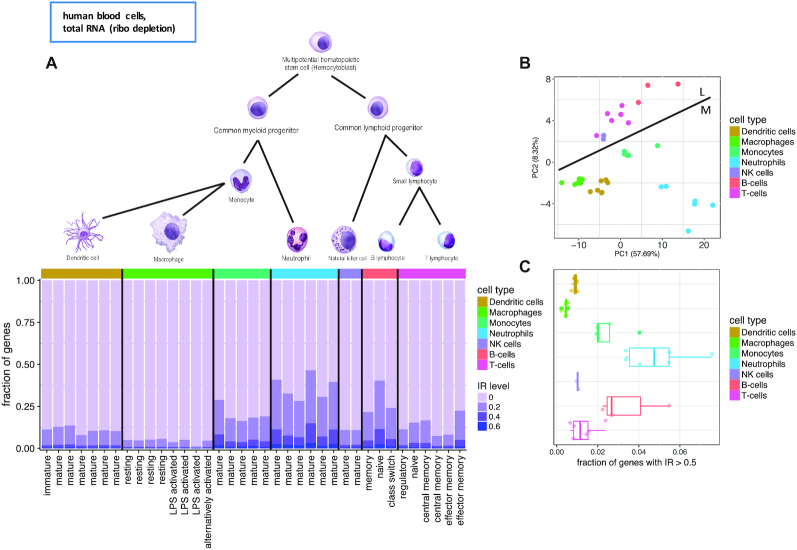
To globally investigate the role of intron retention during hematopoiesis, we first analyzed RNASeq data obtained from a set of Flow Cytometry sorted cell types from the Blueprint Epigenome Project (36). As hematopoietic cells from the Blueprint consortium were taken from different compartments, we focused on a set of 37 samples extracted from peripheral blood and belonging to seven major cell types (Figure 1A and Supplementary Table S1).
Figure 1.
Intron retention (IR) in differentiated primary human blood cells from the Blueprint consortium. (A) Differentiated cells of the hematopoietic tree. Fraction of genes with IR level above displayed threshold (20, 40 and 60%) for the highest retained intron per gene. Cell types identified by color code, subtypes by the X-axis labels. (cell type drawings adapted from OpenStax Anatomy and Physiology Textbook Version 8.25). (B) First two principal components of the PCA based on the IR values from the introns quantified in all samples (i.e. introns with cell type specific IR were omitted). Line indicates separation between the lymphoid (L) and myeloid (M) branch. (C) Fraction of genes with >50% retention of their most retained intron.
To quantify intron retention (IR), we computed the ratios of intronic reads to neighboring exons using IRFinder, a tool developed for assessing intron retention (15,28) (see Methods). Blueprint RNASeq data has been obtained from total RNA, not polyA+ selected, and therefore it potentially contains immature transcripts, which have not been fully processed. Reads, therefore, may originate from introns that will not be retained in the mature transcript. Thus, while we will still use IR to refer to the values obtained by IRFinder in total RNA samples, these values should be understood as the percentage of unprocessed introns, only a fraction of which will be retained in the mature transcripts. Over all samples, we obtained IR values on 148 677 introns from 14 107 genes (on average 35 726 introns from 7730 genes per sample).
IR reveals cell type specific patterns (Figure 1A and Supplementary Figure S1). Principal Component Analysis based on IR of the introns with IR values in all samples (Figure 1B) separated samples according to IR levels along the first principal component. The second component separated cells according to their membership into the myeloid or lymphoid branch of hematopoietic development, suggesting that each of these two differentiation branches has its own specific IR regulation program.
We found that neutrophils exhibit the largest amount of retained introns (Figure 1A). For approximately 5% of all genes, on average amongst the neutrophil samples, at least one intron per gene is retained in >50% of the transcripts (Figure 1C). Granulocytes, which contain a ratio of 70% neutrophils, have previously been reported to show increased intron retention (15). B-cells and monocytes have the highest level of unprocessed introns after neutrophils (Figure 1A and C). Monocytes, that had not been reported to be affected by high levels of unprocessed introns, show higher IR compared to their terminally differentiated stages, macrophages and dendritic cells. Likewise, B-cells display higher IR levels in naive B-cells compared with more mature memory and class switch B-cells. While most previous research (15,17,22) has found differential intron retention occurring during cell differentiation in the myeloid branch, IR has been so far reported only during T-cell activation in the lymphoid branch (23,37). To investigate the relevance of IR in the lymphoid lineage, and more specifically in B-cells, we next focus on IR dynamics during B-cell development and its comparison with neutrophil/granulocyte development.
IR dynamics during B-cell differentiation is conserved between human and mouse
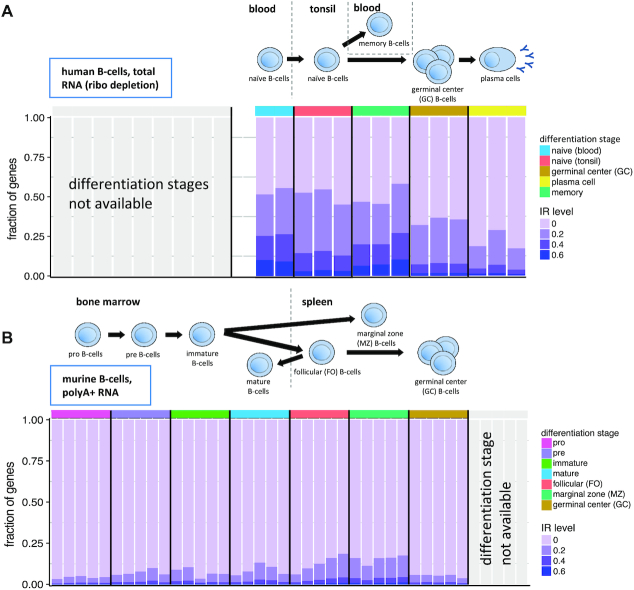
In general, B-cell precursors undergo initial maturation in the bone marrow and migrate via bloodstream to the secondary lymphoid organs where they undergo affinity maturation in the germinal center (GC). GC B-cells can differentiate to antibody secreting cells or give rise to memory B-cells (38).
We have investigated IR in five stages of human B-cell differentiation using RNASeq data produced by the Blueprint consortium ((39), 14 samples from these five stages, Supplementary Table S1). The initial stage sampled, naive B-cells, were taken from blood. A further fraction of naive B-cells, that migrated to tonsils, where they undergo affinity maturation to form either plasma cells or memory B-cells, was extracted from tonsils. Mature memory B-cells were taken from blood. Over all samples, we obtained IR values on 131 943 introns from 12 824 genes; on average, 30 806 introns from 7728 genes per sample (Figure 2A). We observed decreasing levels of IR during B-cell differentiation from naive B-cells in the blood to naive cells in the tonsil, to germinal center B-cells to (antibody producing) plasma cells. Memory cells released to blood after maturation have similar retention values as naive B-cells from blood. This is in contrast to neutrophil differentiation ((40), 15 samples from five stages, Supplementary Table S1), where IR increases towards terminal stages of differentiation (Supplementary Figure S2).
Figure 2.
Intron retention during human and mouse B-cell differentiation. (A) Human B-cell differentiation and migration from blood to tonsil with release of memory cells back into blood. Fraction of genes with IR level above displayed threshold (20, 40 and 60%) for the highest retained intron per gene. Differentiation stages identified by color code. (B) Murine B-cell differentiation and migration from bone marrow to spleen. Fraction of genes with IR level above displayed threshold (20, 40 and 60%) for the highest retained intron per gene. Differentiation stages identified by color code.
The decreasing levels of IR during the late stages of B-cell differentiation appear to have been preserved during evolution, as they can also be observed in mouse. First, we analyzed a set of murine B-cell RNASeq samples ((25), 35 samples from seven different stages). In this data set, differentiating Pro-B-cells that give rise to Pre-B-cells, which in turn give rise to immature B-cells were all taken from the bone marrow. Mature B-cells that migrate back to the bone marrow after maturation were extracted from the bone marrow as well (Figure 2B). Follicular (FO, they are the absolute majority of naive B-cells in the spleen and correspond to the human naive B-cells from tonsil above), marginal zone (MZ, comparable to human memory cells as they respond to pathogens circulating in the blood) and germinal center B-cells were taken from spleen (Figure 2B).
We analyzed IR values obtained on 125 876 introns from 12 709 genes; on average, 51 878 introns from 8633 genes per sample (Supplementary Table S1). Since these are polyA+ enriched samples, our IR values can, in this case, be considered as a measure of ‘bona fide’ intron retention. As expected, in mouse polyA+ samples we observe lower levels of intronic reads compared to human total RNA samples, where reads could stem both from immature transcripts as well as from intron retention. To investigate the magnitude of the effect, we manually inspected four orthologous genes, expressed in human and mouse that contain retained introns in B-cell differentiation (Fus, Ikbkb, Tial1 and Tra2a). As expected, we observed higher intron retention levels for the human total RNA samples in the introns that were also retained in mouse polyA+ samples. In addition, we observed further intronic reads in human introns not retained in mouse, likely stemming from unprocessed introns (Supplementary Figure S3). However, the pattern of intron retention is very clear both in human and mouse. This suggests that, while absolute IR values cannot be compared between polyA+ and total RNA samples, their values relative to other non-retained introns can be compared within the same gene.
As in human, murine MZ B-cells (corresponding to human memory B-cells) show the highest amount of IR, while there is a reduction of IR from FO B-cells (tonsil naive B-cells in human) to GC B-cells. During the early stages of differentiation (for which we do not have human data), we observe, in contrast, an increase in intron retention from pro B-cells to immature B-cells in the bone marrow, as well as in mature B-cells released again from the spleen (Figure 2B).
Because of sequencing depth, murine B-cell differentiation data from Brazao et al. (Figure 2B, Supplementary Table S1 (25)) is the most appropriate to obtain robust IR estimates. However, the correspondence with available human data is limited. Thus, we also analyzed data from the ImmGen Consortium that covers B-cell precursors, splenic FO and MZ cells as well as GC, plasma cells and memory B-cells ((26), unified samples for seven different stages). IR values obtained on 57 594 introns from 8206 genes; on average, 8239 introns from 3089 genes per sample (Supplementary Table S1 and Figure S4). As sequencing for those samples was not very deep, we merged replicates and adjusted to equal read counts. Despite the low number of significantly quantified retention events, we observed the increase of retention towards FO and MZ B-cells seen in the initial murine data set (Supplementary Figure S4). In line with the human data of late developmental stages, we observed a further reduction from FO B-cells to plasma cells, which are among the ones displaying the lowest IR in the two species (Figure 2B and Supplementary Figure S4).
Altogether, thus, during B-cell differentiation, we observe an increase of IR from early precursors in the bone marrow to mature FO and MZ B-cells in secondary lymphoid organs. When FO B-cells undergo the germinal center reaction, IR drops and continues to be low in antibody producing cells, with the exception of memory B-cells that regain the high IR levels of pre GC cells. While direct comparison between human and mouse is difficult because of the different RNA populations being targeted in the RNASeq samples available (total RNA in human, polyA+ in mouse), we have found that not only the general dynamics of IR is conserved between human and mouse, but also the specific events involved in IR do show some level of conservation. We have computed the correlation of IR between human and mouse across orthologous introns at comparable stages during B-cell differentiation and found in all cases significant positive correlations (human naive B-cells from blood vs mouse mature B-cells r = 0.18, P-value <2.2e−16; human naive B-cells from tonsil versus mouse FO B-cells r = 0.27, P-value <2.2e−16; human memory B-cells versus mouse MZ B-cells r = 0.19, P-value <2.2e−16; human GC B-cells versus mouse GC B-cells r = 0.23, P-value <2.2e−16)
Genes with differential intron retention belong to a number of functional categories, notably splicing, cell cycle and NF-kB signalling.
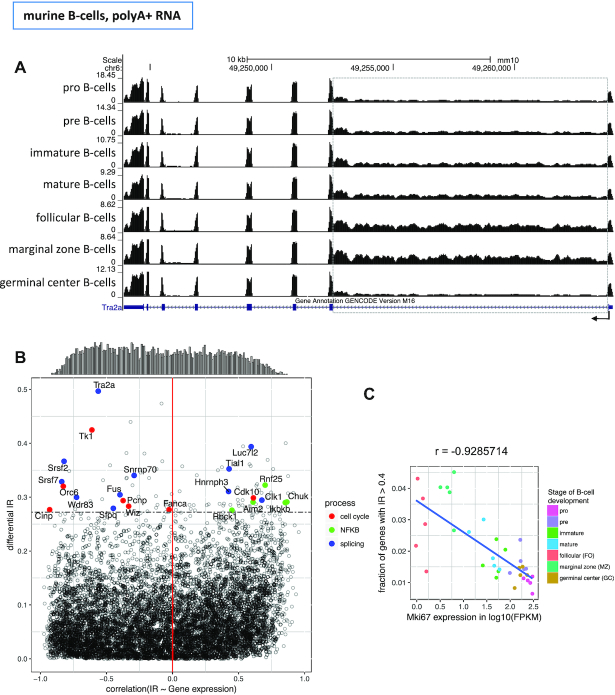
To further understand the biological relevance of IR during B-cell differentiation, we focused on the set of introns with the largest changes in IR during murine B-cell differentiation, using data from Brazao et al. (25). First, we identified the introns with statistically significant differences in intron retention (dIR) between the stages with highest and lowest global retention (MZ and GC B-cells, see Materials and Methods). Then, we considered introns with |dIR| >0.2 and FDR<0.05 as (highly) differentially retained. This resulted in 630 introns from 520 genes with all of them having higher IR in MZ than in GC B-cells (Supplementary Figure S5A, Table S2). These introns are candidates to exhibit regulated retention during B-cell differentiation, and/or to participate in the regulation of this process. Differentially retained introns are shorter than introns overall (2208 nucleotides on average versus 3350). Of the 630 introns, the vast majority (617, 98%), generated, when included, a premature termination codon (PTC), making the resulting transcript a target for non-sense mediated decay (NMD). Moreover, of the remaining 13, only the length of six was a multiple of three, and only in two of those six cases the intron was shorter than 100bp—in these very few cases, the inclusion of an intron with high dIR will not dramatically alter the amino acid sequence of the encoded protein.
Further supporting conservation of the IR program between human and mouse during B-cell differentiation, we have found that the introns from human orthologues of the mouse genes with high dIR, have higher dIR than introns from human genes overall, when measured between human memory and GC B-cells (P-value 1.4e−02, KS test).
Gene Ontology (GO) analysis of these genes revealed terms related to mRNA processing and IkB kinase activity (Supplementary Figure S5C). We further manually curated the literature for the 100 genes containing the most differentially retained introns. About one quarter of the genes belong to three functional categories: eleven genes related to splicing (Tra2a, Srsf2, Srsf7, Snrnp70, Wdr83, Sfpq, Clk1, Tial1, Luc7l2, Hnrnph3, Fus, Supplementary Figure S6), seven genes related to cell cycle (Tk1, Orc6, Cdk10, Pcnp, Wiz, Fanca, Cinp, Supplementary Figure S7), and five genes related to NF-κB regulation (Ikbkb, Chuk, Rbck1, Rnf25, Aim2, Supplementary Figure S8). IκB is a central regulator of rapidly acting transcription factor NF-κB that regulates immune response and cell survival in B-cell differentiation and activation.
Among the introns most affected by IR, we found the first intron of Tra2a (Transformer-2 protein homolog alpha) to be the most differentially retained intron in the B-cell developmental stages analyzed (from IR = 0.65 in MZ B-cells to IR = 0.15 in GC B-cells) (Figure 3A, Supplementary Table S2). With its RNA-binding domain (RBD) Tra2a, like Srsf2 (dIR = 0.37) and Srsf7 (dIR = 0.33), also among the genes most affected by dIR, binds to splicing regulatory elements (SRE) in precursor mRNAs to facilitate or repress splicing in a context dependent manner (41–43). Interestingly, the mRNA of the kinase Clk1, that phosphorylates the SR proteins Srsf2, Srsf7 and Tra2a, is among the most affected by dIR itself (dIR = 0.29). Fus, another gene affected by dIR, couples splicing with transcription and facilitates the splicing of minor introns (44,45). Enriched binding motifs in introns with high IR values had been previously found for Srsf2, Srsf7 and Fus (28). Our data also supports these observations. Known binding motifs exist for five of the eleven splicing related genes with high dIR (Srsf2, Srsf7, Snrnp70, Sfpq and Fus). For each of these factors, we have computed the density of motifs for the very same factor in the differentially retained introns within the factor (dIR > 0.2) and in the introns not differentially retained above the threshold. We have found strong enrichments (>3-fold) for Srsf2 (7.44 motifs per thousand nucleotides in dIR introns compared to 2.32 in other introns), Srsf7 (2.3 over 0.49). For the other factors, we did not find strong enrichments in any direction over 0.50). Overall, these results are in line with previous findings that genes with functions in mRNA processing are affected by intron retention themselves (15,17,22,28).
Figure 3.
Correlation of intron retention with gene expression during mouse B-cell differentiation. (A) RNASeq read distribution along the Tra2a gene in the seven stages of B-cell differentiation (dIR = 0.50 between MZ B-cells and GC B-cells). Read density along intron 7 (dashed grey box) recapitulates the patterns observed globally in Figure 2B (B) Gene-wise Spearman correlation between IR of the highest differential retained intron per gene and the expression of that gene (X-axis) versus dIR (Y-axis). The correlations are computed along 35 samples for 7786 genes. The distribution of the correlation is shown at the top of the plot. The horizontal dashed line identifies the top 100 genes with the highest dIR. Genes involved in cell cycle, splicing and NF-κB are highlighted. (C) Spearman correlation between the expression of the proliferation marker Mki67 and IR along the murine B-cell differentiation samples. Each dot represents Mki67 expression and the fraction of genes that have at least one intron retained with an IR value >0.4 for a given sample.
Since negative correlation between IR and gene expression has been previously reported (8,15,17), we investigate this relation for introns with regulated IR (high dIR) during murine B-cell differentiation. We computed the correlation between IR of the most differentially retained intron in a given gene and the expression of that gene along the 35 murine samples (see Methods), and compared this with dIR (Figure 3B). In contrast with previous reports, we only found a slight bias towards negative correlation (−0.047 median, −0.060 mean). Actually, we observed that the direction of the correlation for genes with high dIR depends largely on gene function. Splicing related genes, for instance, tend to exhibit negative correlations (7 of 11 genes), especially the SR genes Tra2a, Srsf2 and Srsf7, needed for the recruitment of the core splicing complex. For cell cycle related genes, IR is also almost exclusively negatively correlated with expression (6 of 7 genes, Figure 3B and Supplementary Figure S7). The Thymidine Kinase 1 (Tk1, highest dIR (= 0.42) of the cell cycle related genes), that is known for its peak expression in S-phase, has its lowest expression in MZ B-cells. Besides being a marker for cell cycle, Tk1 is also associated with proliferation (46). In line with that, we found the expression of the proliferation marker Mki67 to have a very strong negative correlation with global cellular IR across the murine B-cell samples (r = −0.93, Figure 3C). Consistently, we also observed a sharp decrease in expression of Mki67 during human neutrophil differentiation, when cells lose proliferative potential towards resting neutrophils, which exhibit high levels of IR (Supplementary Figures S2 and S9A). Slightly below the dIR cut-off we found several histone deacetylases with retained introns that all negatively correlated with gene expression: Hdac3 (dIR = 0.19, r = −0.89), Hdac7 (dIR = 0.19, r = −0.89) and Hdac8 (dIR = 0.18, r = −0.71, Supplementary Figure S9B). This suggest a potential role of IR in the regulation of histone deacetylases. Supporting this, an increase of global histone acetylation has been observed towards the resting states of neutrophil differentiation, in which IR is the highest (40), and electron microscopy imaging of MZ B-cells shows a loose coiled structure of the chromatin, in comparison to FO B-cells (47).
In contrast, genes associated with NF-κB regulation were all positively correlated with gene expression, especially Chuk (r = 0.87) and Ikbkb (r = 0.85), coding for the catalytic subunits of the IκB kinase. By ubiquitinating and, thereby, degrading IκB, NF-κB is released to the nucleus to regulate transcription. The physiologically antagonizing positive correlation of IR and gene expression of NF-κB regulators potentially balances the steady state level of active NF-κB in the nucleus. Interestingly, also Fus and Tra2a were found to coactivate NF-κB (48,49).
In summary, we found that during murine B-cell differentiation, IR occurs predominantly in low proliferating states, and that the introns with highly regulated IR belong to a number of functions, among which splicing, cell cycle and Nk-κB signalling are the predominant. We also found that the direction of the relationship between IR and gene expression depends, at least partially, on gene function.
A specific subset of RNA-binding splicing factors contributes to modulate IR
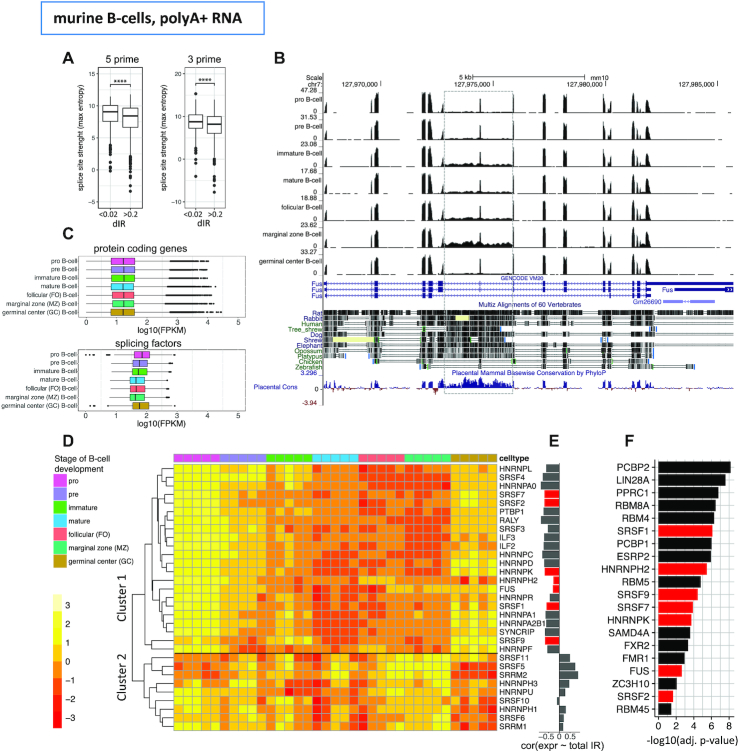
Candidates to participate in the regulation of IR are likely to be among non-core splicing factors. These bind to the RNA sequence in proximity and inside of introns, and participate in the regulation of alternative splicing. In general, splice sites involved in alternative splicing events are weaker than those involved in constitutive splicing, thus allowing for an additional layer of regulation by non-core splicing factors (50–53). Weaker splice sites have also been reported as characteristic of introns affected by IR (8). Consistently, here we found introns with high dIR (dIR > 0.2 in murine B-cell differentiation) to have weaker splice sites compared to the splice sites of other introns (Figure 4A). This strongly suggest that non-core splicing factors are indeed likely to play a role in the regulation of IR. Since IR is conserved between human and mouse, we expect the introns with high dIR to exhibit also some level of sequence conservation. Indeed, we found that average conservation scores among 20 placentalia for introns with high dIR were significantly higher than a set of size matched introns with lower differential retention values (dIR < 0.2, Wilcoxon test, P-value = 1.318e−02). The splicing factor Fus, with introns 6 and 7 retained and a high conservation score all along that introns, is one example (Figure 4B). However, it cannot be completely ruled out that stronger conservation in introns with high dIR may arise from cryptic or unannotated exons as previously indicated for a subset of highly retained introns by Parra et al. (54). We thus, analysed the RefSeq annotation, and for six genes out of 520, we found alternative exons, within and thereby flanked by the retained introns, that were not annotated in GENCODE (Srsf3, Srsf7, Srsf10, Srsf11, Clk1, Snrnp70). With exception of the spliceosomal factor Snrnp70 and the kinase Clk1 that activates SR genes all of them are SR splicing factors. In all cases, the inclusion of these exons leads to a non-coding isoform (Supplementary Figure S10).
Figure 4.
Differential IR during murine B-cell differentiation and splicing factors (A) Splice site strength of introns with high differential retention (dIR>0.2) during murine B-cell differentiation compared with size matched introns with low differential retention (dIR < 0.02). Both 5′ (P-value 1.08e−09, both-sided t-test) and 3′ splice sites (P-value 9.98e−08, both-sided t-test) had significantly lower splice site strength (maximum entropy) in introns with high dIR. (B) RNASeq read distribution and conservation along the Fus transcript. Introns 6 and 7 (dashed grey box) with the highest differential retention also have the highest conservation among vertebrates. (C) Distribution of the expression of 126 murine splicing related genes, including 95 core spliceosomal factors compared to protein coding genes (n = 21 837) along murine B-cell differentiation. Each differentiation stage contains values of five biological replicates. (D) Expression of 31 non-core splicing factors (SR-rich and hnRNP) during murine B-cell differentiation. Genewise z-scores of log10 transformed FPKM values are used. (E) Spearman correlation of the SR-rich and hnRNP gene expression values with the overall fraction of genes that have at least one intron retained per gene with an IR value > 0.2 in the given sample. Splicing factors overlapping with those in panel F) highlighted in red. (F) Enriched RNA Binding Protein (RBP) binding sites (from a set of 102 RBP motifs) in introns with dIR>0.2 between murine MZ and GC B-cells in comparison with size matched introns with low dIR. Splicing factors overlapping within those in panel E) are highlighted in red.
To identify potential candidates to regulate IR, we first analysed the expression levels of 126 splicing factors (curated in Papasaikas et al. (34)) during murine B-cell development, and found that they were expressed at significantly lower levels in stages with higher IR compared to others (GC versus MZ B-cells, paired t-test, P-value 1.95e−14, Figure 4C). During mouse granulocyte differentiation (three samples corresponding to three different stages, (15)), where promyelocytes give rise to myelocytes that in turn differentiate to granulocytes, we observed the same trend of lower splicing factor expression along with the increase of intron retention found by Wong et al. (15) (Supplementary Figure S11A). These results suggest that IR is likely to be, at least partially, just a consequence of globally reduced splicing potential. However, since IR does not occur uniformly across introns, but appears to target a specific set of introns (partially conserved between human and mouse), some form of specific regulation may exist—for instance, by making these introns more prone to inefficient splicing.
As potential candidates for IR regulation, we focused specifically in 31 non-core SR-rich proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs), which are known to modulate splicing by directly binding RNA. SR-rich proteins mostly function as splicing enhancers (55–57), while hnRNPs are described to function as splicing enhancers or silencers in a context specific manner (58,59). The expression patterns of these 31 SR-rich and hnRNP genes in the murine B-cell differentiation samples form two well-defined clusters (Figure 4D), corresponding to genes that are either correlated or anticorrelated with global IR levels (Figure 4E). Most of these factors, 22 (71%), have expression anticorrelated to IR, and are candidates to promote the splicing of retained introns. In neutrophils, the expression of all SR-rich and hnRNP factors is anticorrelated with global levels of IR due to the overall trend of decreasing median expression during maturation of granulocytes (Supplementary Figure S11A, B).
To identify likely candidates to participate in IR regulation, we searched for occurrences of 102 RBP binding motifs (corresponding to 81 RBPs, of which 14 are non-core splicing factors—out of the 31 factors above) within the 630 highly differentially retained introns during B-cell development (since these are the introns more likely to have regulated IR, see above) and compared them with a control set of size matched introns from other genes (Supplementary Table S3). We found a significant enrichment of motifs for seven out of 14 non-core splicing factors in differentially retained introns compared to non-retained introns. Besides the non-core splicing factors, we found enrichment for 13 out of the 74 remaining RBPs in the differentially retained introns (Figure 4E, F). Comparing the number of enriched motifs in differentially retained introns, we found the proportions to be significantly higher for non-core splicing factors compared to all RBPs (hypergeometric test, P-value 1.29e−03). Among these, SRSF1 and SRSF2 binding sites have already been reported to be significantly enriched in retained over non-retained human introns (60). Binding sites for a largely overlapping set of RBPs were enriched in highly differentially retained introns in neutrophils, suggesting a conserved pattern of intron retention within the hematopoietic linage (Supplementary Figure S11C).
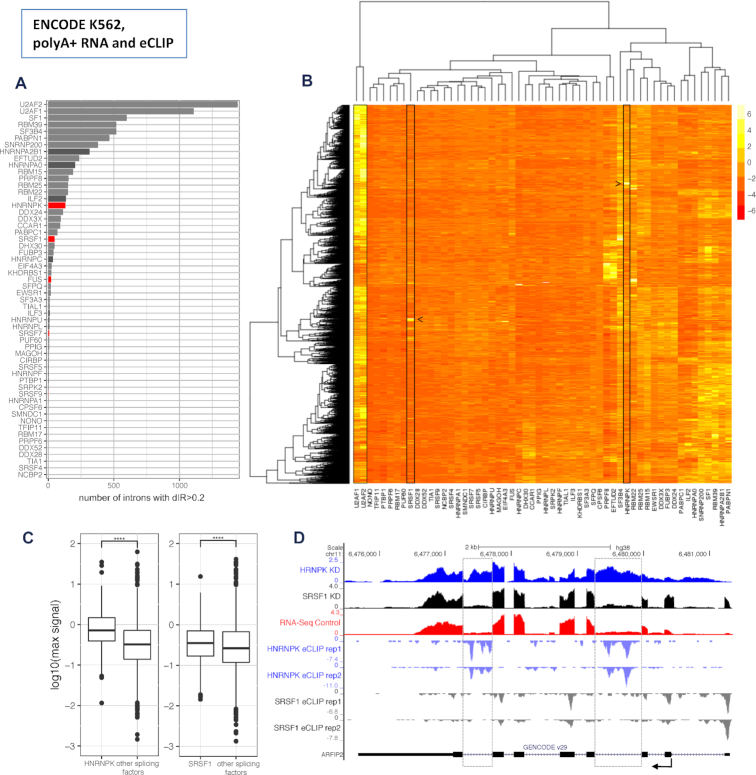
To further validate the potential role of these seven candidates in IR regulation, we investigated polyA+ RNASeq data produced by the ENCODE project after shRNA knockdowns of 58 splicing factors in the human lymphoblast derived K562 cell line (24). These splicing factors included five human orthologues of the seven murine splicing factors candidates to regulate IR (FUS, HNRNPK, SRFS1, SRSF7 and SRSF9). We identified the introns with dIR > 0.2 after knockdown of each of the 58 splicing factors (Figure 5A, B). As expected, we found the knockdown of core splicing factors such as U2AF1, U2AF2, SF3B4 and SF1, essential for splice site and branch point definition, to have the largest impact on intron retention. Notably, however, we found HNRNPK (130 introns) and SRSF1 (48 introns), among the non-core splicing factors with the largest impact (Figure 5A, B). While K562 is a human cell line that does not exactly correspond to any specific mouse B-cell differentiation state, we did find some conservation of the patterns of differential intron retention. Indeed, we computed dIR in the HNRNPK and SRSF1 human knockdown data for the human orthologues of the mouse genes with high dIR during B-cell differentiation, and found that they have significantly higher dIR than genes overall (P-value 4.8e−02 for HNRNPK and 1.9e−02 for SRSF1, KS test). Conversely, the mouse orthologues of the top 500 human genes with highest dIR after HNRNPK knockdown have higher dIR than mouse genes overall (P-value 3.33e−16 for HNRNPK and 9.87e−14 for SRSF1). Overall, these results strongly suggest that HNRNPK and SRSF1 play a (evolutionary conserved) role in the regulation of IR in the hematopoietic lineage.
Figure 5.
Candidate splicing factors to regulate IR (data from the human ENCODE project). (A) Number of introns that increase retention (dIR > 0.2) upon shRNA knockdown of 58 splicing factors in K562 cells from ENCODE. Mean values of two technical replicates per splicing factor normalized by the median of non-targeted control samples. Core splicing factors in grey, non-core splicing factors in black and non-core factors with enriched binding motives in dIR introns 4F) in red. (B) Hierarchical clustering based on IR of the 3102 retained introns with dIR >0.2 for at least one splicing factor knockdown. Mean values of two technical replicates per splicing factor normalized by the median of non-targeted control samples and z-transformed by intron. While core splicing factors like U2AF1 and U2AF2 affect many introns, SR-rich and hnRNP proteins like SRSF1 and HNRNPK (boxed in the plot) have rather distinct sets of intronic targets (identified by arrows). (C) eCLIP derived HNRNPK and SRSF1 strength of binding in the top 100 introns each with their strongest effect on dIR upon their knockdown compared to the strength of binding of nine other non-core splicing factors (see methods) within the same introns in K562 cells. Values are the maximum signal (see Materials and Methods) values within the intronic sequence. (D) RNASeq data of the HNRNPK (blue) and SRSF1 (black) knockdown experiments versus an untreated control (red) in K562 cells from ENCODE for the gene ARFIP2 (top rows). eCLIP binding data for HNRNPK (light blue) and SRSF1 (grey) along the sequence of ARFIP2 (bottom row). Most affected introns are marked by a dashed grey box.
Finally, to investigate direct regulation of IR by HNRNPK and SRSF1, we analyzed human eCLIP data for these two factors produced by the ENCODE Project (24). First, to identify the introns more likely to be mainly regulated by these specific factors and not by others, for each intron we recomputed dIR for the knockdowns of these factors relative to the average dIR caused by the knockdown of nine other non-core splicing factors for which eCLIP data is available (listed in methods). We selected the top 100 dIR events for each factor (minimum dIR 0.20 for HNRNPK and 0.14 for SRSF1, see Materials and Methods). We then compared the peak eCLIP signal of HNRNPK and SRSF1 in these introns, with the average peak eCLIP signal of the other non-core splicing factors. We observed significantly higher HNRNPK and SRSF1 binding signal than the average binding signal for the other splicing factors (P-values 2.69e−06 (HNRNPK) and 1.18e−05 (SRSF1), KS-test, Figure 5C). ARFIP2 is one example where HNRNPK knockdown results in a clear retention pattern compared to SRSF1 knockdown or untreated control cells. eCLIP data reveals binding of HNRNPK, but not of SRSF1 in the affected introns (Figure 5D).
DISCUSSION
While intron retention is a widespread phenomenon among many taxonomic groups, it usually affects a small fraction of the transcriptome in a given cell type (8,14,15,22). In mammals, it has been observed in higher frequencies in brain and blood cells. Here, by analyzing a number of RNASeq datasets from multiple sources, we have provided a comprehensive analysis on intron retention in haematopoiesis.
First, by analyzing the transcriptomes of differentiated human blood cells we found, as previously reported (15), that neutrophils have the highest proportion of genes affected by IR. In addition, we observed similarly high IR values in naive B-cells, which have not been reported previously. During haematopoiesis, we observed a substantial decrease of IR after differentiation of monocytes into either dendritic cells or macrophages. During B-cell differentiation specifically, we observed a continuous decrease of IR towards plasma cells. Memory B-cells, with expression patterns similar to naive B-cells (39), also regained a similar level of IR. All these analyses were based on RNASeq performed by the Blueprint project on total RNA extracts, thus potentially containing immature transcripts. In spite of this, we observed IR levels that were qualitatively consistent with previous findings (15), indicating that total RNA samples could be used as a qualitative proxy when comparing IR changes.
Furthermore, we found the dynamics of intron retention during B-cell differentiation conserved in mouse when analysed in this species using polyA+ RNASeq data—a better RNASeq fraction to monitor IR. The conservation suggests that IR plays a role, possibly regulatory, during haematopoiesis. We found IR increasing from murine precursor B-cells, and decreased in cells undergoing GC reaction towards plasma cells, which is consistent with our observations in human. Genes related to RNA processing and splicing were among the most affected by IR, in agreement with previous studies (15,17,22,28). The role of IR as a self-regulatory mechanism has been previously proposed (15,17). In this regard, we did find enrichment for binding motifs for SRSF2 and SRSF7, most notably, in differentially retained introns from these genes.
Like in Malignant Glioma and for T-cell activation (23,61), we have also found a relation between proliferation and IR. While naive B-cells, with relatively low proliferation potential, and MZ B-cells as well as memory B-cells, that are quiescent, have high levels of intron retention, cells that undergo rapid cycling, like GC B-cells, have significantly reduced global intron retention levels. Likewise, in granulocytes, there is a negative association of intron retention with proliferation activity. Remarkably, we found an intron of the thymidine kinase 1 gene (TK1) to be among the most regulated and its retention level to be negatively correlated with the expression of TK1. With its expression peaking during S-phase, TK1 is a marker for cell cycle, and for proliferation, in general (46). IR in TK1 could potentially be a mechanism contributing to the regulation of proliferation and cell cycle progression during B-cell differentiation.
In electron microscopy images of MZ B-cells, a loose coiled structure of the chromatin was observed in comparison to FO B-cells (47). Interestingly, we detected regulated intron retention during B-cell differentiation in three histone deacetylases with declining expression towards MZ B-cells, which could lead to a decompaction of the chromatin, resulting from higher overall histone tail acetylation.
Directly related to the immunological function of B-cells, we found the three IkB kinase subunits to be among the most affected genes by differential intron retention during murine B-cell differentiation. IkB kinase is crucial for the regulation of NF-κB signalling, essential for B-cell immune response and survival. Contrary to splicing and cell cycle, in which IR is mostly negatively correlated with gene expression, the expression of IκB kinase subunits is, in all cases, positively correlated with IR. This could indicate that IR is involved in fine-tuning the steady state levels of IkB subunits. NF-κB has also been found to be affected by differentially retained introns during T-cell activation (23).
In general, we found only a few introns from a given gene, often only one, affected by IR. We found these introns enriched for RBP binding motifs compared to not retained introns. These RBPs belong significantly often to the hnRNPs and SR-rich families, which are known regulators of alternative splicing. One member of these families, HNRNPLL, has already been found to alter IR of specific introns in T-cells (37), and binding sites for SRSF1, SRSF2 and SRSF7, HNRNPA1 and HNRNPA1L2 have been found enriched in retained introns (28,60). Consistent with the regulatory role of hnRNPs and SR-rich splice factors in IR, we found these factors to be least expressed in B-cell differentiation stages with highest IR. These results suggest that IR could be, at least partially, the consequence of globally inefficient splicing. In this regard, we believe that it is of interest that the kinase Clk1, that activates a number of SR proteins through phosphorylation, contains one of the most differentially retained introns. Interestingly, there is a positive correlation between intron retention and expression of Clk1. It is tempting to speculate that this provides a negative feedback loop in which high levels of IR (which could reveal globally weakened splicing) would lead to the increased expression of Clk1 and thereby higher splicing factor activation, to counteract their lower availability (due to low expression) and partially restore splicing activity in the cell. The regulatory role of Clk1 in intron retention upon stress response has been previously suggested (9).
However, since IR does not occur stochastically, but appears to target a specific set of introns, while the majority undergoes canonical splicing, this suggests an additional layer of specific regulation. In this regard, we have found a number of non-core splicing factors, HNRNPK and SRSF1 in particular, to be potential candidates for IR regulation. All this suggest a specific code for intron retention, in which co-localization of binding sites for specific splicing factors within the sequence of the introns would allow for regulation of retention, explaining the high specificity of intron retention, since the majority of introns undergoes canonical splicing. Additional data on the effect in the transcriptome of the knockdowns of RBPs across multiple cellular conditions, as well as on binding of RBPs, will certainly contribute to deciphering the code involved in the regulation of intron retention. Other mechanisms recently proposed to affect IR are the occupancy of MeCP2 near the splice junction (62), expression of PRMT5 (61) and cryptic exons functioning as splicing decoys (54). In addition a subset of splicing factors that bind RNA can effect gene expression in multifold ways, as it is the case for PABPN1 that can, besides having an active role in splicing, destabilize nuclear transcripts and thereby affect the amount of available transcripts for translation (63).
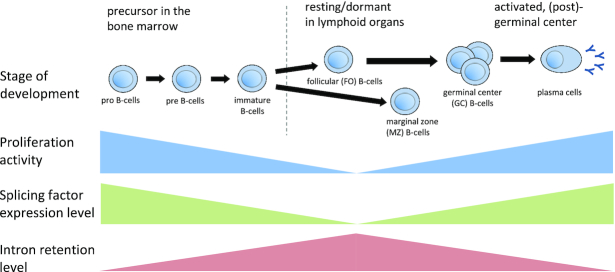
In summary, in our work, we carried out an exhaustive characterization of IR, both during differentiation and in differentiated blood cells. Our results suggest a model where IR increases from B-cell precursors in the bone marrow towards lymphoid organs and decreases when B-cells undergo affinity maturation (Figure 6). Overall, IR increases towards cellular states with low proliferative potential and comparatively inefficient splicing. Our work highlights the important, but largely unappreciated, role of IR in differentiation processes.
Figure 6.
General model of changes in IR, splicing factor expression and proliferation during B-cell differentiation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sílvia Pérez-Lluch and Martin Schaefer for critically reading the manuscript, discussions and advice. We wish to thank those who reviewed the manuscript for their constructive comments.
Notes
Present address: Roderic Guigó, Bioinformatics and Genomics Department, Center for Genomic Regulation, Barcelona, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no 282510; Ministry of Economy, Industry and Competitiveness (MEIC) for the EMBL partnership; Centro de Excelencia Severo Ochoa; CERCA Program/Generalitat de Catalunya.
Conflict of interest statement. None declared.
REFERENCES
- 1. DeKoter R.P., Singh H.. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000; 288:1439–1441. [DOI] [PubMed] [Google Scholar]
- 2. Iwasaki H., Mizuno S., Arinobu Y., Ozawa H., Mori Y., Shigematsu H., Takatsu K., Tenen D.G., Akashi K.. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006; 20:3010–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sasaki H., Kurotaki D., Tamura T.. Regulation of basophil and mast cell development by transcription factors. Allergol. Int. 2016; 65:127–134. [DOI] [PubMed] [Google Scholar]
- 4. Chen L., Kostadima M., Martens J.H.A., Canu G., Garcia S.P., Turro E., Downes K., Macaulay I.C., Bielczyk-Maczynska E., Coe S. et al.. Transcriptional diversity during lineage commitment of human blood progenitors. Science. 2014; 345:1251033–1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J.. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008; 40:1413–1415. [DOI] [PubMed] [Google Scholar]
- 6. Nilsen T.W., Graveley B.R.. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong J.J.-L., Au A.Y.M., Ritchie W., Rasko J.E.J.. Intron retention in mRNA: No longer nonsense: Known and putative roles of intron retention in normal and disease biology. Bioessays. 2016; 38:41–49. [DOI] [PubMed] [Google Scholar]
- 8. Braunschweig U., Barbosa-Morais N.L., Pan Q., Nachman E.N., Alipanahi B., Gonatopoulos-Pournatzis T., Frey B., Irimia M., Blencowe B.J.. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014; 24:1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boutz P.L., Bhutkar A., Sharp P.A.. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015; 29:63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ninomiya K., Kataoka N., Hagiwara M.. Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. J. Cell Biol. 2011; 195:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boothby T.C., Zipper R.S., van der Weele C.M., Wolniak S.M.. Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev. Cell. 2013; 24:517–529. [DOI] [PubMed] [Google Scholar]
- 12. Mauger O., Lemoine F., Scheiffele P.. Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron. 2016; 92:1266–1278. [DOI] [PubMed] [Google Scholar]
- 13. Kalyna M., Simpson C.G., Syed N.H., Lewandowska D., Marquez Y., Kusenda B., Marshall J., Fuller J., Cardle L., McNicol J. et al.. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012; 40:2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yap K., Lim Z.Q., Khandelia P., Friedman B., Makeyev E. V. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012; 26:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wong J.J.L., Ritchie W., Ebner O.A., Selbach M., Wong J.W.H., Huang Y., Gao D., Pinello N., Gonzalez M., Baidya K. et al.. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013; 154:583–595. [DOI] [PubMed] [Google Scholar]
- 16. Raj B., Blencowe B.J.. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015; 87:14–27. [DOI] [PubMed] [Google Scholar]
- 17. Edwards C.R., Ritchie W., Wong J.J.-L., Schmitz U., Middleton R., An X., Mohandas N., Rasko J.E.J., Blobel G.A.. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood. 2016; 127:e24–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.-K., Mockler T.C.. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010; 20:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grützmann K., Szafranski K., Pohl M., Voigt K., Petzold A., Schuster S.. Fungal alternative splicing is associated with multicellular complexity and virulence: a genome-wide multi-species study. DNA Res. 2014; 21:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dvinge H., Bradley R.K.. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015; 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jung H., Lee D., Lee J., Park D., Kim Y.J., Park W.-Y., Hong D., Park P.J., Lee E.. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 2015; 47:1242–1248. [DOI] [PubMed] [Google Scholar]
- 22. Pimentel H., Parra M., Gee S.L., Mohandas N., Pachter L., Conboy J.G.. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2016; 44:838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ni T., Yang W., Han M., Zhang Y., Shen T., Nie H., Zhou Z., Dai Y., Yang Y., Liu P. et al.. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res. 2016; 44:6817–6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brazão T.F., Johnson J.S., Müller J., Heger A., Ponting C.P., Tybulewicz V.L.J.. Long noncoding RNAs in B-cell development and activation. Blood. 2016; 128:e10–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heng T.S.P., Painter M.W., Elpek K., Lukacs-Kornek V., Mauermann N., Turley S.J., Koller D., Kim F.S., Wagers A.J., Asinovski N. et al.. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008; 9:1091–1094. [DOI] [PubMed] [Google Scholar]
- 27. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K. et al.. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Middleton R., Gao D., Thomas A., Singh B., Au A., Wong J.J.-L., Bomane A., Cosson B., Eyras E., Rasko J.E.J. et al.. IRFinder: assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017; 18:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frankish A., Diekhans M., Ferreira A.-M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2018; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McLeay R.C., Bailey T.L.. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010; 11:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A. et al.. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Papasaikas P., Tejedor J.R., Vigevani L., Valcárcel J.. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell. 2015; 57:7–22. [DOI] [PubMed] [Google Scholar]
- 35. Pohl A., Beato M.. bwtool: a tool for bigWig files. Bioinformatics. 2014; 30:1618–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams D., Altucci L., Antonarakis S.E., Ballesteros J., Beck S., Bird A., Bock C., Boehm B., Campo E., Caricasole A. et al.. BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 2012; 30:224–226. [DOI] [PubMed] [Google Scholar]
- 37. Cho V., Mei Y., Sanny A., Chan S., Enders A., Bertram E.M., Tan A., Goodnow C.C., Andrews T.. The RNA-binding protein hnRNPLL induces a T cell alternative splicing program delineated by differential intron retention in polyadenylated RNA. Genome Biol. 2014; 15:R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M.. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015; 15:160–171. [DOI] [PubMed] [Google Scholar]
- 39. Beekman R., Chapaprieta V., Russiñol N., Vilarrasa-Blasi R., Verdaguer-Dot N., Martens J.H.A., Duran-Ferrer M., Kulis M., Serra F., Javierre B.M. et al.. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat. Med. 2018; 24:868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grassi L., Pourfarzad F., Ullrich S., Merkel A., Were F., Carrillo-de-Santa-Pau E., Yi G., Hiemstra I.H., Tool A.T.J., Mul E. et al.. Dynamics of transcription regulation in human bone marrow myeloid differentiation to mature blood neutrophils. Cell Rep. 2018; 24:2784–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tacke R., Tohyama M., Ogawa S., Manley J.L.. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998; 93:139–148. [DOI] [PubMed] [Google Scholar]
- 42. Zhong X.-Y., Wang P., Han J., Rosenfeld M.G., Fu X.-D.. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell. 2009; 35:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ji X., Zhou Y., Pandit S., Huang J., Li H., Lin C.Y., Xiao R., Burge C.B., Fu X.-D.. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013; 153:855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu Y., Reed R.. FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:8608–8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reber S., Stettler J., Filosa G., Colombo M., Jutzi D., Lenzken S.C., Schweingruber C., Bruggmann R., Bachi A., Barabino S.M. et al.. Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. EMBO J. 2016; 35:1504–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. ZHOU J., HE E., SKOG S.. The proliferation marker thymidine kinase 1 in clinical use. Mol. Clin. Oncol. 2013; 1:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sintes J., Gentile M., Zhang S., Garcia-Carmona Y., Magri G., Cassis L., Segura-Garzón D., Ciociola A., Grasset E.K., Bascones S. et al.. mTOR intersects antibody-inducing signals from TACI in marginal zone B cells. Nat. Commun. 2017; 8:1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Uranishi H., Tetsuka T., Yamashita M., Asamitsu K., Shimizu M., Itoh M., Okamoto T.. Involvement of the pro-oncoprotein TLS (translocated in liposarcoma) in nuclear factor-kappa B p65-mediated transcription as a coactivator. J. Biol. Chem. 2001; 276:13395–13401. [DOI] [PubMed] [Google Scholar]
- 49. Matsuda A., Suzuki Y., Honda G., Muramatsu S., Matsuzaki O., Nagano Y., Doi T., Shimotohno K., Harada T., Nishida E. et al.. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003; 22:3307–3318. [DOI] [PubMed] [Google Scholar]
- 50. Xing Y., Lee C.. Alternative splicing and RNA selection pressure — evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006; 7:499–509. [DOI] [PubMed] [Google Scholar]
- 51. ITOH H., Washio T., Tomita M.. Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes. RNA. 2004; 10:1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baek D., Green P.. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:12813–12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. ZHENG C.L., Fu X.-D., Gribskov M.. Characteristics and regulatory elements defining constitutive splicing and different modes of alternative splicing in human and mouse. RNA. 2005; 11:1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Parra M., Booth B.W., Weiszmann R., Yee B., Yeo G.W., Brown J.B., Celniker S.E., Conboy J.G.. An important class of intron retention events in human erythroblasts is regulated by cryptic exons proposed to function as splicing decoys. RNA. 2018; 24:1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tacke R., Manley J.L.. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 1999; 11:358–362. [DOI] [PubMed] [Google Scholar]
- 56. Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000; 6:1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Long J.C., Caceres J.F.. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009; 417:15–27. [DOI] [PubMed] [Google Scholar]
- 58. Smith C.W., Valcárcel J.. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000; 25:381–388. [DOI] [PubMed] [Google Scholar]
- 59. Dreyfuss G., Kim V.N., Kataoka N.. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002; 3:195–205. [DOI] [PubMed] [Google Scholar]
- 60. Sakabe N., de Souza S.. Sequence features responsible for intron retention in human. BMC Genomics. 2007; 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Braun C.J., Stanciu M., Boutz P.L., Patterson J.C., Calligaris D., Higuchi F., Neupane R., Fenoglio S., Cahill D.P., Wakimoto H. et al.. Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant glioma. Cancer Cell. 2017; 32:411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wong J.J.-L., Gao D., Nguyen T. V, Kwok C.-T., van Geldermalsen M., Middleton R., Pinello N., Thoeng A., Nagarajah R., Holst J. et al.. Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment. Nat. Commun. 2017; 8:15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bresson S.M., Hunter O. V., Hunter A.C., Conrad N.K.. Canonical poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genet. 2015; 11:e1005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.