Abstract
Expanding the genetic code to enable the incorporation of unnatural amino acids into proteins in biological systems provides a powerful tool to study protein structure and function. While this technology has been mostly developed and applied in bacterial and mammalian cells, it recently expanded into animals, including worms, fruit flies, zebrafish, and mice. In this review, we highlight recent advances toward the methodology development of genetic code expansion in animal model organisms. We further illustrate the applications, including proteomic labeling in fruit flies and mice, and optical control of protein function in mice and zebrafish. We summarize the challenges of unnatural amino acid mutagenesis in animals, and the promising directions towards broad application of this emerging technology.
Graphical Abstract

Introduction to Genetic Code Expansion
The proteins of all metazoans consist of 20 canonical amino acids, and selenocysteine in some cases,1 as the fundamental building blocks of life. While the number of natural amino acids is small, numerous post-translational modifications, such as methylation, acetylation, glycosylation, and phosphorylation, exist and expand the chemical repertoire and biological function of proteins beyond the limit of the canonical amino acids.2 In fact, many proteins are functional only when post-translationally modified, and this lays the basis for dynamic and robust biological systems. Therefore, the development of methodologies to expand the genetic code to other, unnatural amino acids (UAAs) is very appealing in order to site-specifically add new chemical functionalities to proteins through translation in biological systems. Performing site-specific mutagenesis with UAAs enables precise studies of protein structure and function, and high-resolution external control over protein activity in both cells and, recently, animals (Figure 1A).
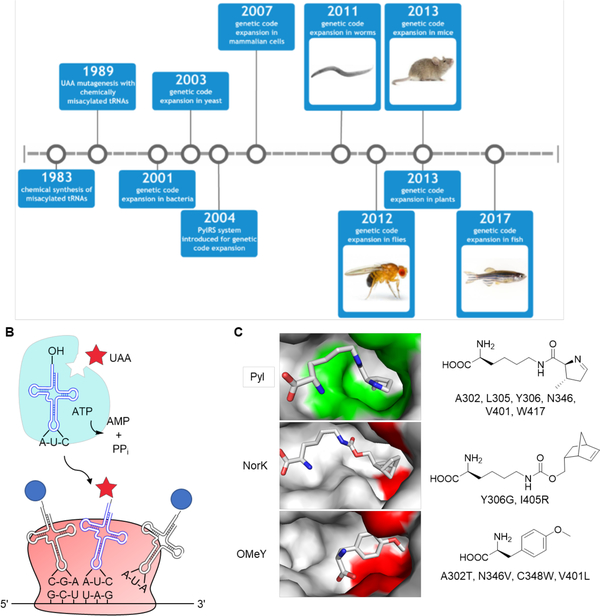
Figure 1.
Overview of genetic code expansion. (A) Major milestones in the development of genetic code expansion and its translation into increasingly complex systems. (B) The tRNA synthetase (aaRS) aminoacylates its cognate tRNA with the unnatural amino acid (UAA). The aminoacylated tRNA is transported to the ribosome, where the UAA is site-specifically incorporated into the growing polypeptide chain in response to the UAG codon. (C) Structure of the M. mazei PylRS reveals key residues for substrate recognition in wild-type (PDB 2ZCE) and mutant (PDB 4BWA and 3QTC) enzymes. Substrate binding pocket residues in the wild-type PylRS are labeled green and mutant residues are shown in red.
In vitro methods for site-specific incorporation of UAAs into dipeptides and proteins were first reported in 19833 and 1989,4 respectively. A chemically acylated suppressor tRNA was used in an in vitro transcription-translation system to incorporate an unnatural amino acid in response to an UAG amber stop codon into a protein of interest. Although this work introduced the concept to express unnaturally mutagenized proteins at the translational level, the difficulty in applying it in cells and the required chemical synthesis of the aminoacylated tRNA limited broad applicability.
A method for UAA incorporation in cells was therefore highly desired, due to the expected larger amounts of protein and the direct application in live biological systems. In initial experiments, an auxotrophic E. coli strain was used for the incorporation of an analogue amino acid in place of its corresponding canonical amino acid throughout the proteome.5–7 However, this method is limited to analogues that are structurally similar to the canonical amino acids and the incorporation is not site-specific.
In 2001, an UAA was site-specifically incorporated into a protein in E. coli using the bacterial translational machinery (Figure 1B).8 To achieve this goal, a tRNA that uniquely recognizes the amber stop codon, UAG, was generated. The amber stop codon was chosen to encode the UAA, because it is the least frequently used among the three stop codons (9% in E. coli).9 Furthermore, this tRNA was engineered to be orthogonal to any endogenous, bacterial aminoacyl-tRNA synthetase (aaRSs). In order to aminoacylate this tRNA with an UAA, its cognate aaRS was engineered to recognize the UAA, but not any of the endogenous amino acids as a substrate. The aaRS also needed to be orthogonal to any endogenous tRNA and its matching UAA should not be recognized by any endogenous aaRS. Additionally, the unnatural amino acid should not be toxic and needs to be transported to the cytoplasm when added to the growth medium.10
Because initial attempts to directly evolve an orthogonal aminoacyl-tRNA synthetase/tRNA pair from the existing E. coli aaRS/tRNA pairs were not successful, a heterologous tyrosyl-tRNA synthetase (TyrRS)/tRNATyr pair from M. jannaschii was chosen.8 An archaea was selected as the source organism, due to inefficient cross-species aminoacylation and the synthetase’s lack of an anticodon-binding motif, allowing for manipulation of the tRNA to recognize an amber stop codon.11 This pair meets the above requirements: both the MjtRNATyr (after sequence optimization through a genetic selection)8 and MjTyrRS are orthogonal to their E. coli counterparts, but are still functional in E. coli. To evolve an MjTyrRS that can accommodate a desired UAA, but not tyrosine (or any other endogenous amino acid), a mutant library was constructed and passed through several rounds of positive and negative selections.8 The positive selection was based on expression of a chloramphenicol acetyltransferase (CAT) gene with an amber codon at a permissive site, thus selecting for MjTyrRS mutants that incorporate the desired UAA and/or endogenous amino acids in media containing chloramphenicol. The negative selection was based on expression of a toxic barnase gene with three amber codons at permissive sites. The cells were grown in the absence of the desired UAA, thus removing clones that utilize endogenous amino acids.
Using this heterologous MjTyrRS/tRNATyr pair, more than 70 UAAs have been genetically incorporated into proteins in E. coli. These UAAs represent distinct structural and functional properties not found in canonical amino acids, including chemically reactive groups for bioconjugation,12 biophysical probes (IR, NMR, and fluorescence) for the study of protein structure and function,13–14 photocaged and photoreactive groups,15 and the products of post-translational modifications.16
Although the MjTyrRS/tRNATyr pair is an effective platform for UAA incorporation in E. coli, it cannot be readily applied to organisms like yeast and mammalian cells because it is not orthogonal in eukaryotes.17 However, the prokaryotic E. coli TyrRS/tRNATyr pair is orthogonal in yeast18 and was used for genetic expansion there.19 To apply the EcTyrRS system in mammalian cells, a Bacillus stearothermophilus tRNATyr, which contains an essential internal promoter sequence for efficient transcription, was paired with the EcTyrRS,20 and yeast was used as a host for synthetase evolution before transfer to the mammalian cell system.21 EcLeuRS was adapted and utilized in a similar fashion, enabling the encoding of hydrophobic and sterically more demanding UAAs.22–23 However, the requirement for separate aaRS/tRNA pairs and separate selection procedures for incorporation of an UAA in prokaryotes and eukaryotes limits rapid application. Therefore, an efficient and facile approach was needed for the incorporation of UAAs in both kingdoms. Ideally, an aaRS/tRNA pair could be evolved for a desired UAA in bacterial cells first, and then be shuttled into single- and multi-cellular eukaryotes directly without loss of orthogonality or functionality.
The Pyrrolysyl-tRNA Synthetase/tRNA Pair and its Application in Genetic Code Expansion
In 2002, a 22nd amino acid – pyrrolysine – was discovered to be genetically encoded in Methanosarcina barkeri.24–25 Pyrrolysine is incorporated into the enzyme methylamine methyltransferase through an in-frame amber UAG codon and acts as a crucial catalytic residue in the catabolism of methylamine substrates.25 The pyrrolysyl-tRNA synthetase (PylRS) and the pyrrolysyl-tRNA (tRNAPyl) were identified in the Methanosarcina barkeri genome24 and, although the complete biosynthetic pathway for pyrrolysine was identified later,26 it was quickly realized that the machinery for the incorporation of pyrrolysine might be a powerful tool for genetic code expansion.27 Similar to Methanosarcina barkeri, the PylRS from Methanosarcina mazei harbors a highly conserved C-terminal catalytic core,27–28 although the length of their N-terminal domain varies. In fact, the residues within the amino acid binding pocket that are randomized to construct a mutant synthetase library are the same between the two strains and have allowed for selection of synthetases that incorporate UAAs. Recently it was shown that a chimeric PylRS from the two strains with the C-terminal catalytic domain from M. mazei and the N-terminal hydrophobic domain from M. barkeri had higher activity for pyrrolysine and UAAs.29 The PylRS/tRNAPyl pair functions in the natural context of the common 20 amino acids and this orthogonality is retained when the PylRS/tRNAPyl pair is transferred from archaea to other organisms. Indeed, in E. coli or mammalian cells, the PylRS/tRNAPyl pair from either M. barkeri or M. mazei does not cross-react with endogenous aaRSs or tRNAs,30 providing several advantages over the previous systems for UAA mutagenesis: First, no directed evolution is needed for the tRNA, because the tRNAPyl is already a perfect substrate for its cognate PylRS, but not any endogenous aaRSs. Second, the anticodon region of the tRNAPyl naturally recognizes the UAG codon, obviating the need for mutation. Third, the mutant PylRS is unlikely to incorporate any endogenous amino acids, as they are structurally very different from pyrrolysine.
The PylRS belongs to the class II aaRS family, but differs from other members of its subfamily, with its deep hydrophobic pocket to harbor pyrrolysine,31 which is significantly larger than any of the 20 canonical amino acids. The crystal structure of the M. mazei PylRS suggests that the pyrroline ring is buried in this pocket (Figure 1C), and that the distance between the ATP-binding domain and the hydrophobic pocket accommodates the long aliphatic lysine chain.31 Moreover, the N346 can hydrogen bond through a water-mediated contact with the primary amine group of Pyl, further stabilizing the substrate. The first application of the PylRS/tRNAPyl system in genetic code expansion in E. coli (with pyrrolysine) was reported in 2004.30 The encoding of an UAA by PylRS/tRNAPyl was subsequently demonstrated in E. coli32–33 and in mammalian cells.34 Surprisingly, the wild-type PylRS exhibits an extraordinarily broad substrate spectrum for many lysine analogues,34–39 including UAAs for bioconjugation reactions 35–36 and photocrosslinking.39 In order to genetically encode other UAAs that are not substrates of the wild-type PylRS, libraries of PylRS mutants were constructed and passed through multiple rounds of positive and negative genetic selections. This strategy has allowed successful engineering of PylRS mutants that enable incorporation of a wide range of lysine analogs (including natural post-translational modifications),40–48 phenylalanine analogs,49–51 histidine analogues,52 tyrosine analogs,53–54 and cysteine analogs.55–56 Structural investigation of, for example, a PylRS triple mutant evolved for incorporation of endo-norbornene-lysine (NorK) showed expansion of the hydrophobic pocket through a Y306G mutation to accommodate the norbornene group. In addition, an I405R mutation shifts the β7-β8 hairpin and arranges the binding pocket so that C348, instead of N346, interacts with the substrate side chain carbonyl (Figure 1C).57 On the other hand, O-methyl tyrosine (OMeY) has a significantly different structure than pyrrolysine and, not surprisingly, mutations allowing for its incorporation both shorten and widen the hydrophobic pocket. Furthermore, an A302T mutation stabilizes OMeY binding through hydrogen bonding with its α-carboxyl group (Figure 1C).58
Due to its activity, versatility, and orthogonality, the PylRS/tRNAPyl system has been recently applied for successful genetic code expansion in animals, including worms,59 flies,60 mice,61 and fish.62 These model organisms are critical tools for the study of cellular and molecular mechanisms in biomedical research,63 and the choice of a model depends on the biological question under study. Caenorhabditis elegans, with its small genome size, is one of the simplest model animals. It is also the easiest of the animal models to maintain in lab, requiring only bacterial lawns on agar for food and allowing for simple freezing and recovery protocols.64 Due to its short life cycle, well-characterized cell lineage and fully defined neural connectivity, C. elegans is frequently used in the study of aging65 and neurodegenerative disease.66 Drosophila melanogaster (fruit fly) is a highly suited model organism for genetic analysis and mutagenesis screens, due to its small size and short life cycle of two weeks. Besides, the regenerative capacity of fruit flies’ imaginal discs have rendered them modern models in studying regenerative biology.67 In contrast to worms and flies, Danio rerio (zebrafish) is a vertebrate organism that is evolutionarily closer to humans. Zebrafish are small, easy to breed, develop ex utero, and are transparent as embryos, making them an ideal model for studying vertebrate development.68 Zebrafish are also an emerging organism for modeling embryologically and genetically tractable diseases,69 as well as a platform for early drug development, including target identification, lead discovery, and toxicology.70 Mus musculus (mouse) is a rodent model organism that shares 99% of genes with humans. Compared with other mammals, the mouse is inexpensive, easy to maintain and breed in large scale. As a result, the mouse is the most widely used model for the study of human biology and human diseases, including cancer, diabetes, cardiovascular disease, and Alzheimer’s disease.71 All four animal models have been used in genetic code expansion studies, enabling distinct applications in biological studies.
Genetic code expansion in Caenorhabditis elegans
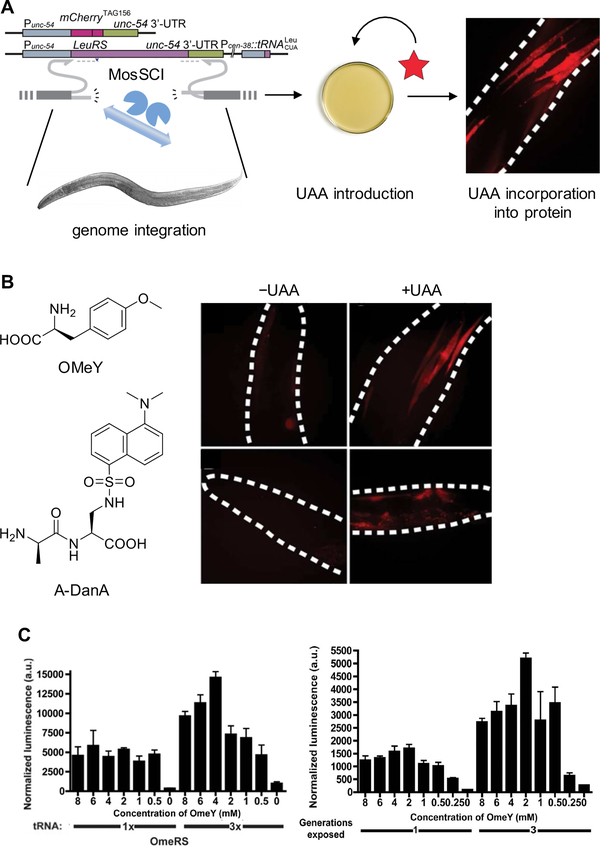
The first multicellular organism with an expanded genetic code was the nematode Caenorhabditis elegans.59 Besides demonstrated tolerance for amber stop codon suppression in previous work,72 C. elegans was an attractive first step for genetic code expansion into the animal kingdom because of their optical transparency throughout all stages of life, making worms amenable to studies with photocrosslinking, photocaging, or fluorescent UAAs. Plasmids encoding the PylRS, the tRNAPyl, and an incorporation reporter were introduced into nonsense mediated decay (NMD) deficient worms (smg-2(e2008) line59 or smg-1 knock-down73), thus effectively removing NMD machinery, which was shown to severely inhibit UAA incorporation. Similar to experiments in mammalian cells,74 the tRNAPyl transcription was driven by a Pol III promoter (here and in the other organisms). Biolistic bombardment of the worms with the plasmids was inefficient, with only 5% of worms showing the desired phenotype. The ectopic expression of these genes also led to mosaic expression of the reporter due to mitosis or silencing of the expression cassette, and read-through of the amber stop codon in the absence of the synthetase/tRNA pair. Enhancement of UAA incorporation was achieved by using the EcLeuRS/tRNALeu system in conjunction with several optimizations; the most important being integration of the reporter gene (mCherry or luciferase), the EcLeuRS, and the tRNA cassette into the host genome (Figure 2A).73 Integration of these into the worm genome was achieved with Mos1-mediated Single Copy gene Insertion (MosSCI).75 This method uses a transposase to insert the expression cassette into a single locus of the genome. This resulted in homogenous expression of all components, higher incorporation efficiency, and 100% transmissible worm lines (Figure 2B). Incorporation of OMeY was achieved through simple addition of the UAA to the media, but for dansyl-alanine (DanA), no incorporation was seen unless the amino acid was coupled to alanine to form a dipeptide, thus allowing uptake via dipeptide transporters in the worm gut (Figure 2B). Incorporation was dependent on UAA concentration, as seen for expression in bacterial and mammalian cells as well (Figure 2C);35 however, at high concentrations, worm viability was reduced. The amount of expressed tRNA also has a major impact on incorporation efficiency, and has been observed as a limiting factor in mammalian cells too (Figure 2C).76 Another interesting observation unique to UAA incorporation in multicellular organisms was the effect of generational exposure to the UAA. Incorporation efficiency increased for worms exposed to the UAA over three generations (Figure 2C), possibly due to enrichment of the UAA in the progeny of exposed worms. Hence, bioavailability of the UAA can be an important and limiting factor for incorporation in multicellular organisms.
Figure 2.
Expanding the genetic code of Caenorhabditis elegans. (A) Diagram of UAA incorporation into mCherry154TAG in C. elegans body-wall muscle using a genome-integrated EcLeuRS/tRNALeu. (B) Expression of the mCherry154TAG reporter in the absence or presence of O-methyl tyrosine (OMeY) or an alanine-dansylalanine dipeptide (A-DanA). Dotted lines indicate the outline of the worms.73 (C) Incorporation of OMeY is dependent on both UAA concentration and tRNA copy number in the genome, as demonstrated by measurement of luciferase activity using the Luc185TAG reporter (left). Incorporation efficiency is also influenced by the number of generations exposed to OMeY (right). Adapted with permission from ACS Chem. Biol. 2012, 7, 1292. Copyright 2012 American Chemical Society.
Genetic code expansion in Drosophila melanogaster
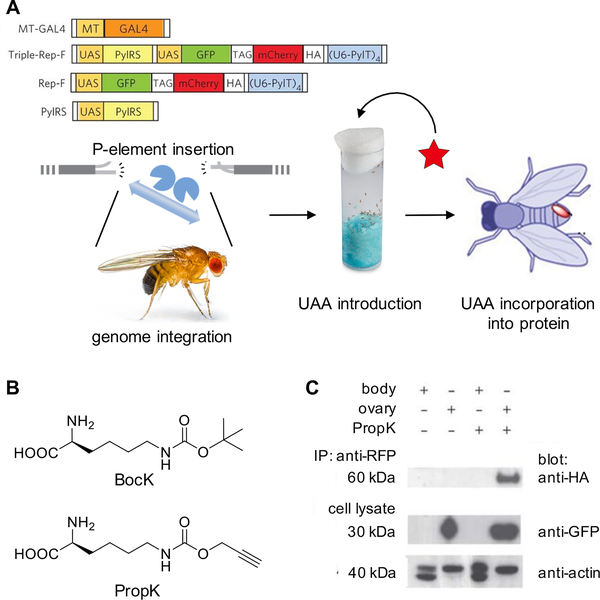
Genetic studies of D. melanogaster have previously revealed tolerance to amber stop codon suppression.77 Genetic code expansion in flies was accomplished using the M. mazei PylRS/tRNAPyl system to incorporate Boc-protected lysine (BocK) and an alkyne containing lysine derivative (PropK),60 both established UAAs that are substrates for the wild-type PylRS (Figure 3A & 3B).35 Several constructs were generated to confirm orthogonality of the PylRS/tRNAPyl pair, first in cultured fly cells (Dmel). A GFP-TAG-mCherry-HA reporter construct was used, which only produces the 60 kDa full-length protein and the HA epitope when the amber stop codon is suppressed. Both PylRS and the reporter genes were placed under transcriptional control by a Gal4 responsive promoter (UAS), and Gal4 itself was under transcriptional control of a metallothionein promoter, providing Cu2+-inducible transcription.
Figure 3.
Expanding the genetic code in Drosophila melanogaster. (A) Diagram of UAA incorporation into proteins in flies via an expression cassette integrated into the genome using P-element insertion. (B) Structures of BocK and PropK. (C) Western blots of lysates from ovary and other body tissue demonstrating tissue-specific incorporation of PropK. The full-length protein, including the HA tag, is only expressed when the amber stop codon is suppressed.60 Adapted with permission from Nat. Chem. Biol. 2012, 8, 748. Copyright 2012 Nature Publishing Group.
For incorporation of UAAs into proteins in fly embryos, transgenic lines with Triple-Rep-F (PylRS, tRNAPyl, and GFP-TAG-mCherry-HA) were generated by P-element insertion, a common method for introducing transgenes into flies, using microinjected DNA and transposase for germline integration, followed by crossing with flies expressing Gal4 under the female germline promoter nos-vp16. Mothers were fed the UAA for two days and embryos were collected for imaging assays. In the presence of the PylRS/tRNAPyl pair, BocK incorporation into GFP-mCherry-HA was detected by immunostaining. It was also shown that PropK incorporation in adult transgenic flies with Gal4 under control of the tissue-specific nos-vp16 promotor (nos-vp16-Gal4, Triple-Ref-F) showed full length GFP-mCherry-HA expression only in the ovaries, but not in other body tissue (Figure 3C). UAA incorporation was amenable to standard transgenic expression techniques in flies, making it an accessible method for further applications (as discussed below).
Genetic code expansion in Danio rerio
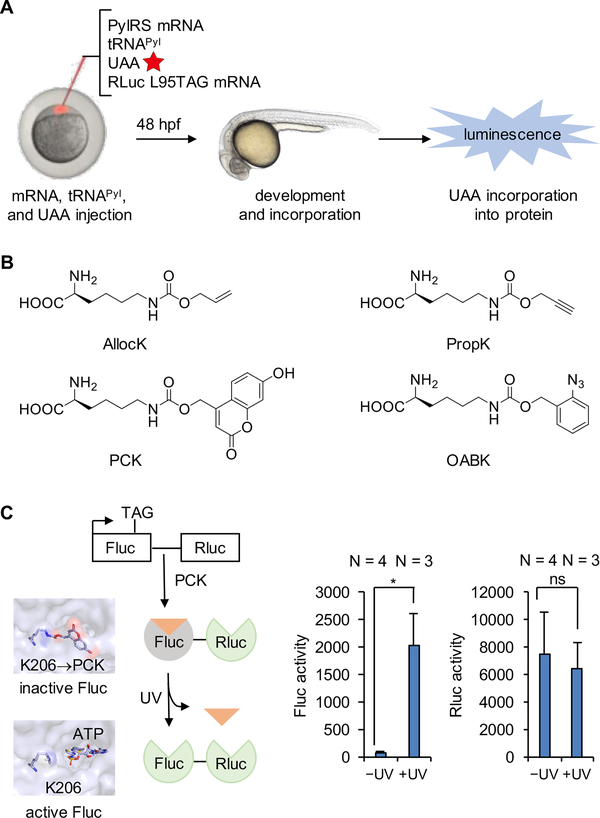
Genetic code expansion has recently been reported in zebrafish embryos.62, 78 A convenient way to generate proteins containing UAAs in live embryos involves the injection of a solution containing mRNA encoding the PylRS, mRNA encoding the protein of interest, tRNAPyl, and the UAA, into zygotes at the 1−2 cell stage (Figure 4A). The Renilla luciferase (Rluc) gene, with an amber codon located at a non-essential L95 position, was used as an initial incorporation reporter. Four UAAs (AllocK, PropK, PCK, and OABK) were tested and luciferase function, as the result of successful stop codon suppression, was only observed in embryos in the presence of the UAA (Figure 4B). No luciferase activity was observed in the absence of the UAA, indicating the fidelity and orthogonality of the PylRS/tRNAPyl system in zebrafish. These four UAAs introduce a diverse set of chemical functions that can be applied to protein labeling, bioconjugation, and conditional – optical or small molecule – control of proteins in live zebrafish. In order to demonstrate optical control of enzyme function in zebrafish through the genetic incorporation of the caged lysine PCK, a firefly luciferase (Fluc) reporter was used as a proof-of-concept. Specifically, PCK incorporation at position K206 blocks luciferase activity until the caging group is removed through light exposure, because K206 resides in close proximity to the luciferase active site.79 Renilla luciferase (Rluc) was fused to the C-terminus of Fluc-K206TAG, serving as an internal control for incorporation efficiency. At 48 hours post fertilization (hpf), injected embryos were either briefly irradiated with 365 nm light or kept in the dark. While virtually no luminescence was observed before UV exposure, a 26-fold increase of Fluc activity was detected after light irradiation (Figure 4C), indicating successful activation of protein function in live zebrafish embryos with an expanded genetic code.
Figure 4.
Expanding the genetic code in Danio rerio embryos. (A) Diagram of UAA incorporation into proteins in zebrafish embryos through injection of the UAA, tRNAPyl, and mRNAs encoding the PylRS and Renilla luciferase (Rluc) L95TAG. (B) Structures of the UAAs incorporated into proteins in zebrafish embryos. (C) Genetic encoding of the photocaged lysine PCK allows for optical control of firefly luciferase activity in zebrafish embryos through blocking of the active site of the enzyme until UV exposure (left), as demonstrated by luminescence recovery after irradiation (right).62 Adapted with permission from J. Am. Chem. Soc. 2017, 139, 9100. Copyright 2017 American Chemical Society.
Genetic code expansion in Mus musculus
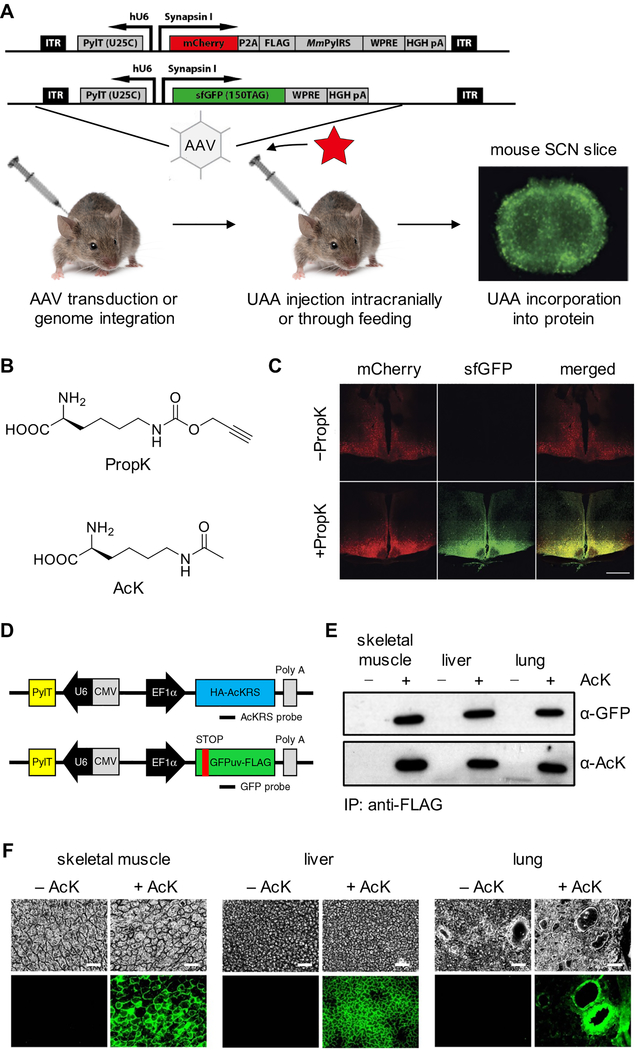
Using the PylRS/tRNAPyl pair, incorporation of a photocaged cysteine into the Kir2.1 ion channel was achieved in dissociated rat hippocampal neuron culture and in the mouse embryo neocortex.23 Recently, this methodology was further expanded to live adult mice61 through application of two adeno-associated virus (AAV) vectors as the delivery tool. These viral vectors efficiently delivered the PylRS/tRNAPyl pair and the reporter DNA into adult mice for transient expression, but do not generate a stable mouse line. One vector delivered the PylRS and the tRNAPyl expressed under control of the neuron-specific human synapsin I promoter and the human U6 promoter, respectively. A second vector provided the gene of interest (containing an amber codon) expressed from the same synapsin promoter. Importantly, another copy of the tRNAPyl was also included in the second AAV vector (Figure 5A). An sfGFP reporter for incorporation with an amber codon at position 150 was used. The viral vectors were injected in the suprachiasmatic nucleus in adult mice, a region of the brain directly above the optic chiasm and near the third ventricle, responsible for the circadian rhythm in mammals. After one week, the UAA (PropK, Figure 5B) was delivered to the third ventricle via an infusion pump that was implanted subcutaneously to allow bypassing of the blood-brain barrier. Imaging of coronal sections after two weeks showed sfGFP expression only in the UAA-treated animals, but not in the control groups (Figure 5C). This result demonstrated successful incorporation of an UAA into proteins in live mice. Moreover, the authors found that delivery of the UAA through the drinking water yielded similar results. In neurons of the suprachiasmatic nucleus, the sfGFP containing PropK could be labeled with an azide containing fluorophore via a Cu(I)-catalyzed cycloaddition.
Figure 5.
Expanding the genetic code in Mus musculus. (A) Diagram of UAA incorporation into proteins in mice via AAV transduction of the expression cassette or generation of stable lines through genome integration, with the UAA introduced either intracranially or through the drinking water. (B) Chemical structure of PropK and AcK. (C) Fluorescence imaging showing incorporation of PropK into sfGFP151TAG in coronal sections of the hypothalamus and suprachiasmatic nucleus. Scale bar: 500 μm.61 (D) DNA constructs used for generation of a transgenic mouse line for UAA incorporation. (E) Western blotting analysis of anti-FLAG-immunoprecipitated proteins from AcK-encoding sfGFP151TAG mice, demonstrating successful genetic code expansion.80 (F) Fluorescence imaging showing incorporation of AcK into GFP39TAG in skeletal muscle, liver, and lung tissues of live mice. Scale bar: 200 μm. Adapted with permission from Nat. Chem. Biol. 2016, 12, 776 and Nat. Commun. 2017, 8, 14568. Copyright 2016 and 2017 Nature Publishing Group.
Genetic code expansion in live mice was also recently achieved by a different approach.80 Two transgenic mice that stably expressed tRNAPyl/PylRS, and tRNAPyl/GFP39TAG, respectively, were generated (Figure 5D), and further crossed to generate double-heterozygous animals carrying the UAA-incorporation machinery and the reporter gene. The generation of the PylRS/tRNAPyl transgenic mouse would allow for crossing with any other mouse containing a transgene with an amber stop codon for future experiments. Daily intraperitoneal injections of the UAA (AcK, Figure 5B) were performed to mice starting at eight weeks of age. Five days later, the tissues were collected and analyzed. GFP expression was observed in the UAA-injected groups, but not the control groups, demonstrating the successful incorporation of UAA in the transgenic animals. Incorporation was confirmed in multiple tissue types (skeletal muscle, liver, and lung) by Western blot with an acetyl lysine-specific antibody (Figure 5E). Fluorescence was detected in these tissues as well, suggesting the ubiquitous expression of mutant protein, as expected under control of the EF1α promotor (Figure 5F). The knock down of Upf2, a component of mammalian NMD, increases UAA incorporation in primary mouse embryonic fibroblasts (MEFs) from double-heterozygous transgenic mice. This result aligns with the strategy in C. elegans. In summary, both approaches established successful UAA-incorporation systems in live mice. The first approach allows facile incorporation of different UAAs through the injection of AAV vectors. However, the expression of PylRS/tRNAPyl is transient, and no stable mouse line was generated. The second approach applies a stable mouse line expressing the necessary components for the incorporation of a specific UAA. While this may provide a more convenient system, the initial generation of stable, transgenic animals for the encoding of different UAAs is time-consuming.
Application I: Proteome Labeling in Flies and Mice Using Stochastic Orthogonal Recoding of Translation with a Chemoselective Modification
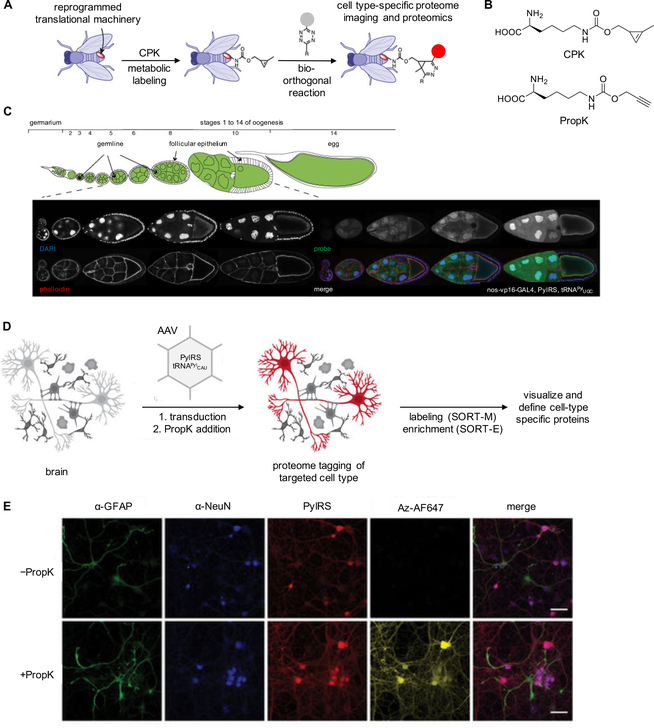
Studying proteome dynamics in living systems can be achieved through random labeling of newly synthesized proteins via incorporation of bioorthogonal handles. For example, an azido-containing analogue of methionine is recognized by the endogenous methionyl-tRNA synthetase (MetRS) and was applied in bioorthogonal noncanonical amino acid tagging (BONCAT).81 Introduction of the azido-methionine results in stochastic incorporation of the azido group at methionine codons within the proteome, followed by copper-catalyzed cycloaddition of the azide with an alkyne containing probe, and enrichment of protein populations and identification via mass spectrometry. Labeling and imaging of cells or in-gel protein visualization has been achieved through noncanonical amino acid tagging (FUNCAT).82 Limitations include low affinity of the MetRS toward the azide-methionine analogue, compared to natural Met, and the introduction of only select chemical handles for proteome labeling. Elliott et al.83 developed a new proteomics technique using an orthogonal aaRS/tRNA pair for incorporation of a bioorthogonal handle in Drosophila. Instead of incorporating UAAs site-specifically in response to introduced amber stop codons, several tRNAPyl recognizing codons that code for canonical amino acids were created (tRNAPylxxx) and effectively competed with their respective natural counterparts for (unnatural) amino acid incorporation. The PylRS system is particularly suited for these studies, since the synthetase does not recognize the anticodon when aminoacylating the tRNAPyl.33 The expression of the PylRS/tRNAPylxxx construct was directed under tissue specific promoters and, combined with the ability to introduce the UAA at defined time points through feeding, allowing for spatial and temporal control over stochastic proteome labeling. This was demonstrated using the nos-vp16 promotor to selectively express the PylRS/tRNAPylxxx pair in fly ovaries. Thus, only ovary cell proteins incorporated cyclopropane-lysine (CPK) (Figure 6A, B) and underwent labeling via a bioorthogonal reaction with a tetrazine-bearing dye. No incorporation was seen in other body tissues, and the protein levels were representative of the mRNA levels for a particular protein, demonstrating the specificity and quantitative potential of this approach (Figure 6C).
Figure 6.
Proteome labeling using Drosophila and mice with an expanded genetic code. (A) Diagram of proteome labeling in fruit flies using cyclopropene-lysine (CPK) incorporation into fly proteins.85 (B) Structure of CPK and PropK. (C) Stochastic incorporation of CPK into proteins was controlled by expression of the PylRS/tRNAPyl under control of nos-vp16, limiting expression of the PylRS/tRNAPyl to germline cells in the ovary. When labeled, fluorescence was restricted to germline cells with no incorporation of CPK in the somatic follicular epithelium.83 (D) Diagram of proteome labeling in the mice brain using PropK. (E) Cell-type specific incorporation of PropK and labeling with an azido-fluorophore. α-GFAP is a glial cell specific protein and α-NeuN is a neuron specific nuclear protein. Adapted with permission from Nat. Biotechnol. 2014, 32, 445, Nat. Biotechnol. 2014, 32, 465, and Nat. Biotechnol. 2017, 36, 156. Copyright 2014 and 2017 Nature Publishing Group.
Krogager et al.84 successfully applied this approach in the mouse brain by injecting AAV containing mCherry-P2A-PylRS under a neuron specific promotor and the tRNAPylCAU (suppressing a lysine codon) under a U6 promoter (Figure 6D). The UAA used in these experiments was PropK (Figure 6B), which requires Cu(I) catalysis for labeling with an azide, in contrast to CPK. Labeling of neuronal proteins with an azide-Alexa-fluor dye (Az-AF647) was first demonstrated in dissociated rat cortex (Figure 6E). Next, Stochastic Orthogonal Recoding of Translation with proteome Enrichment (SORT-E) was performed via stereotactic injection of the AAV vectors and feeding of PropK through the drinking water. The mice brain lysates were then labeled with an azido-diazobenzene-biotin conjugate and the proteins were subsequently enriched via streptavidin pull-down. Tandem MS/MS of this lysate was compared to non-tagged lysate controls to demonstrate enrichment of proteins specific to neurons. The SORT-E method captured 2,119 proteins, including neuronal markers but not many glial cell markers, confirming enrichment of the neuronal proteome. To determine if stochastic incorporation of PropK in neurons had any effect on normal brain function, the mouse brain circadian rhythm was measured using a luciferase reporter and no perturbation was observed.
Validation of SORT for proteomics studies in both flies and mice paves the way for future applications to capture the proteome in specific cell types at specific time points to understand proteome changes during many biological processes such as disease or development, and would also be amenable to proteome-wide labeling with photocrosslinking UAAs to map protein interactions.
Application II: Optical Control of a Potassium Channel in the Mouse Brain
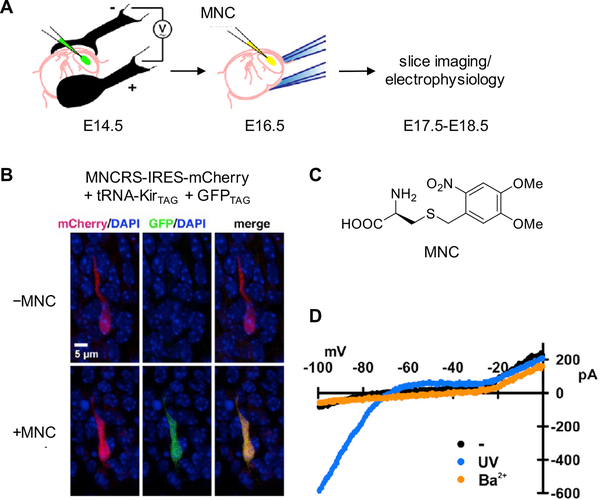
The development of optogenetic tools, such as channelrhodopsins, has greatly facilitated the study of neural circuits and their involvement in animal behavior.86 Unnatural amino acid mutagenesis in animals further expands the optogenetic toolbox through incorporation of photocaged amino acids into proteins. For example, the photocaged cysteine MNC was site-specifically inserted into Kir2.1, a strong inwardly rectifying potassium channel, in the mouse neocortex (Figure 7A–C).23 First, the gene constructs for genetic code expansion were electroporated in utero into the brain of the mouse embryo. The UAA was then injected into the lateral ventricle after two days and, using GFP182TAG as an incorporation reporter, genetic encoding of MNC in embryonic cortical neurons was confirmed (Figure 7B). When the neurons were irradiated with 385 nm light, the inward current rapidly increased, suggesting light-activation of the Kir2.1 potassium channel (Figure 7D). In short, genetic code expansion in mice presents a novel method to trigger neurobiological processes and compared to traditional optogenetic tools, light-regulation of protein function through photocaged amino acids is not limited by the protein type, such as an ion channel.87 Applying this methodology to other proteins will enable the elucidation of highly dynamic processes beyond neuroscience in a system as complex as a mouse.
Figure 7.
Application of genetic code expansion to the optical control of a potassium channel in the mouse neocortex. (A) Electroporation of genetic constructs for UAA mutagenesis into the mouse neocortex and injection of photocaged cysteine (MNC) into the embryonic lateral ventricle after two days. (B) Fluorescence imaging showing incorporation of MNC into GFP182TAG and Kir2.1TAG in mice embryonic cortical neurons. (C) Structure of MNC. (D) I-V plot of currents recorded from mouse neocortical neurons showing light-dependent activation of Kir2.1. Adapted with permission from Neuron 2013, 80, 358. Copyright 2013 Elsevier.
Application III: Optical Control of a Signaling Pathway in Zebrafish Embryos
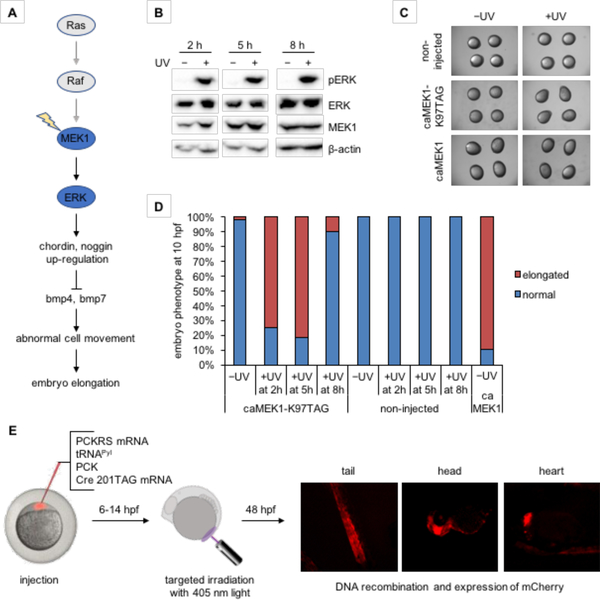
Incorporation of photocaged amino acids allows for spatiotemporal control of a broad range of proteins,87–88 such as the potassium channel discussed above. This non-invasive tool can be further applied to the optical control of signaling pathways in animals, in particular zebrafish due to the transparency of the embryo.62 The regulation of kinase signaling pathways during embryogenesis plays a key role in pathophysiologies of human diseases.89 Understanding the precise mechanisms of regulation and signaling can thus reveal detailed causes of disease and new targets for pharmacological intervention, and various optochemical probes have been developed for this purpose.90 Of the signaling pathways, the MEK/ERK pathway is of particular interest, since it plays a critical role in dorsoventral patterning during early development in zebrafish (Figure 8A).91 The incorporation of a photocaged lysine at the critical K97 position blocks the activity of MEK1, therefore creating a highly specific tool to control the MEK/ERK pathway with light.92 Irradiation at 2 h or 5 h post-injection led to embryos with an elongated phenotype at 10 hpf (Figure 8B–D). In contrast, embryos irradiated at 8 h post-injection showed a normal phenotype (Figure 8D). Light activation of MEK1 increases ERK phosphorylation at all three time points (Figure 8B), and MEK/ERK pathway activity was further confirmed by upregulation of its downstream targets. Altogether, the observed results indicate that active MEK1 was not able to trigger an elongated phenotype when activated at later stages. The temporal triggering of MEK1 therefore revealed a critical time window for the function of the MEK/ERK pathway in early embryo development. Overall, optical control of signaling pathways through genetic code expansion provides a powerful tool to dissect complex signaling networks, and to illustrate spatiotemporal-defined molecular mechanisms of cellular events. Other cell signaling pathways can be interrogated with this methodology as well, since the active-site lysine at position 97 of MEK1 is conserved in most human protein kinases.93
Figure 8.
Optical control of a signaling pathway and of Cre recombinase in zebrafish embryos. (A) In zebrafish, activation of caged MEK1, as part of the Ras/MAPK signaling pathway, induces an elongated phenotype through the secreted bmp inhibitors chordin and noggin. (B) Time-course analysis of ERK phosphorylation by optically activated MEK1.62 (C) Embryos expressing caged MEK1 displayed an elongation phenotype only when activated through light exposure. (D) Temporal control of MEK1 activity reveals a time window when embryo morphogenesis is most susceptible to MEK1 induced elongation. (E) Optical Cre recombinase activation in targeted cell populations in the zebrafish embryo at early developmental periods using a genetically encoded photocaged lysine (PCK, see Figure 4B) allowed for lineage tracing of embryonic cells that formed the tail, head, and heart of the 48 hpf embryo.94 Adapted with permission from J. Am. Chem. Soc. 2017, 139, 9100 and ChemBioChem 2018, 19, 1244. Copyright 2017 American Chemical Society and copyright 2018 John Wiley and Sons.
Application IV: Cell Lineage Tracing in Zebrafish Embryos
Optical control of protein activity using genetically encoded, photocaged amino acids has been extended to Cre recombinase. By replacing a critical lysine residue (K201) with a photocaged lysine analogue (PCK, Figure 4B), Cre activity is suppressed until removal of the photocaging group upon light irradiation.95 After initial experiments in mammalian cell culture, the photocaged Cre recombinase was expressed in zebrafish embryos. Localized decaging and activation of DNA recombination was demonstrated for early embryonic cell populations that would develop into tissue in the tail, head, and heart of the 48 hpf embryo through targeted irradiation at an early developmental period using a 405 nm confocal imaging laser. Using a Cre reporter fish line Tg(Ubi:loxP-EGFP-loxP-mCherry),96 cell lineages could be tracked from early development using mCherry fluorescence with high spatial and temporal resolution (Figure 8E).94
Summary and Outlook
The incorporation of unnatural amino acids into proteins of interest is a powerful tool that is seeing rapid development. Its introduction into common animal models expands accessibility of this technology to many fields of study, while also proving its utility in answering complex biological questions using the unique chemistry installed into the protein of interest. Remarkably, the UAA mutagenesis machinery could be directly transferred from mammalian cells into four, very different animal models without any major adjustment and without major obstacles that could have been caused by differences in codon usage and aaRS/tRNA cross-reactivity with endogenous protein biosynthesis. This methodology conceivably should work in many other animal models as well, owing to the orthogonality of the PylRS/tRNAPyl, EcLeuRS/tRNALeu, and EcTyrRS/tRNATyr systems in eukaryotes. Expressing proteins with UAAs in animals has three requirements: the aaRS/tRNA pair (stably or transiently expressed), the gene of interest with an amber stop codon at the desired residue (potentially inserted into the genome via CRISPR/Cas9 editing), and the UAA. The generation of animal lines stably expressing the aaRS/tRNA pair streamlines the application of this methodology and thus makes it more available to the biological community.78, 80 In many cases, the UAA can be simply introduced through feeding. It is important to note that bioavailability of a UAA through feeding may vary based on what UAA is used, and more direct delivery through injection or the synthesis of actively absorbed dipeptides (as, e.g., applied in C. elegans73) may be required. The structure and function of the UAA is defined by the biological problem to be addressed. For example, the incorporation of bio-orthogonal chemical handles into proteins allowed for tissue specific proteome labeling in flies, enabling the probing of proteomic changes in certain tissues at specific time-points in development. Potential application might also include labeling of distinct (e.g., neoplastic) cells and tissue in live animals through bioconjugation.97 Photocaged amino acids enable investigation of dynamic processes with spatial and temporal resolution, as showcased in the optical control of the MEK1 signaling pathway during zebrafish embryonic development, in particular mapping the time window of hyperactive Ras/MAPK network involvement in birth defects.62 Further applications of optical protein control methodologies may involve measurements of select pathway kinetics in whole animals.92 The light-triggered manipulation of other proteins, such as the Kir2.1 ion channel,23 might allow for control of neuronal activity and behavior in live animals, complementing existing optogenetic approaches.98 While genetic code expansion in multicellular organisms is still in its infancy, further advancement of this methodology can be expected through breakthroughs in synthetase engineering and enhancement of UAA incorporation efficiency.29 One limitation of unnatural amino acid incorporation is the reliance on amber stop codons for site-specific incorporation, which can reduce incorporation efficiency due to competition with release factors at the ribosome and cross-reaction with endogenous amber stop codons. One might suspect that suppression of endogenous amber stop codons would have toxic effects, but in general many prokaryotes and eukaryotes including Drosophila and C. elegans, as mentioned previously, display some natural amber stop codon suppression99 and demonstrate no detriment to viability and growth rates.100 However, use of only the amber stop codon limits the number of UAAs that can be simultaneously encoded. Solutions to this problem may be provided by the use of four-base codons101–102 or the development of fully unnatural DNA/RNA base pairs to create fundamentally new codons that are completely orthogonal.103 Expanding the repertoire of orthogonal codons would allow for incorporation of multiple different UAAs in the same animal. In summary, genetic code expansion in animals provides researchers with a powerful, convenient, and versatile tool to manipulate protein function in vivo, which is largely limited by other techniques. Further application and development of this technology will offer unique and creative ways to study biology with new depth and insight as the toolbox of UAAs with distinct chemical and biological function continues to grow. Having animals with expanded genetic codes by any number of unnatural amino acids with a variety of different chemical functionalities may eventually shed light on nature’s selection of the 20 common amino acids as fundamental building blocks of life.
Acknowledgements
The authors thank the NIH (R01GM112728 and R21HD085206) and the NSF (CBET-1603930) for funding.
Footnotes
Competing Interests
The authors declare no competing financial interests.
References
- 1.Romagne F; Santesmasses D; White L; Sarangi GK; Mariotti M; Hubler R; Weihmann A; Parra G; Gladyshev VN; Guigo R; Castellano S, SelenoDB 2.0: annotation of selenoprotein genes in animals and their genetic diversity in humans. Nucleic Acids Res. 2014, 42, 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chuh Kelly N.; Batt Anna R.; Pratt Matthew R., Chemical methods for encoding and decoding of posttranslational modifications. Cell Chemical Biology 2016, 23, 86–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heckler TG; Zama Y; Naka T; Hecht SM, Dipeptide formation with misacylated tRNAPhes. J. Biol. Chem 1983, 258, 4492–4495. [PubMed] [Google Scholar]
- 4.Noren CJ; Anthonycahill SJ; Griffith MC; Schultz PG, A General-method for site-specific incorporation of unnatural amino-acids into proteins. Science 1989, 244, 182–188. [DOI] [PubMed] [Google Scholar]
- 5.Ibba M; Hennecke H, Relaxing the substrate specificity of an aminoacyl-tRNA synthetase allows in vitro and in vivo synthesis of proteins containing unnatural amino acids. FEBS Lett. 1995, 364, 272–275. [DOI] [PubMed] [Google Scholar]
- 6.Cowie DB; Cohen GN, Biosynthesis by Escherichia coli of active altered proteins containing selenium instead of sulfur. Biochim. Biophys. Acta 1957, 26, 252–261. [DOI] [PubMed] [Google Scholar]
- 7.Sharma N; Furter R; Kast P; Tirrell DA, Efficient introduction of aryl bromide functionality into proteins in vivo. FEBS Lett. 2000, 467, 37–40. [DOI] [PubMed] [Google Scholar]
- 8.Wang L; Brock A; Herberich B; Schultz PG, Expanding the genetic code of Escherichia coli. Science 2001, 292, 498–500. [DOI] [PubMed] [Google Scholar]
- 9.Sharp PM; Cowe E; Higgins DG; Shields DC; Wolfe KH; Wright F, Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens - a review of the considerable within-species diversity. Nucleic Acids Res. 1988, 16, 8207–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu DR; Schultz PG, Progress toward the evolution of an organism with an expanded genetic code. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 4780–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steer BA; Schimmel P, Major anticodon-binding region missing from an archaebacterial tRNA synthetase. J. Biol. Chem 1999, 274, 35601–35606. [DOI] [PubMed] [Google Scholar]
- 12.Chin JW; Santoro SW; Martin AB; King DS; Wang L; Schultz PG, Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc 2002, 124, 9026–9027. [DOI] [PubMed] [Google Scholar]
- 13.Schultz KC; Supekova L; Ryu Y; Xie J; Perera R; Schultz PG, A genetically encoded infrared probe. J. Am. Chem. Soc 2006, 128, 13984–13985. [DOI] [PubMed] [Google Scholar]
- 14.Cellitti SE; Jones DH; Lagpacan L; Hao X; Zhang Q; Hu H; Brittain SM; Brinker A; Caldwell J; Bursulaya B; Spraggon G; Brock A; Ryu Y; Uno T; Schultz PG; Geierstanger BH, In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc 2008, 130, 9268–9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu N; Deiters A; Cropp TA; King D; Schultz PG, A genetically encoded photocaged amino acid. J. Am. Chem. Soc 2004, 126, 14306–14307. [DOI] [PubMed] [Google Scholar]
- 16.Liu CC; Schultz PG, Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat. Biotechnol 2006, 24, 1436–1440. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi T; Nureki O; Ishitani R; Yaremchuk A; Tukalo M; Cusack S; Sakamoto K; Yokoyama S, Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Biol 2003, 10, 425–432. [DOI] [PubMed] [Google Scholar]
- 18.Edwards H; Schimmel P, A bacterial amber suppressor in Saccharomyces cerevisiae is selectively recognized by a bacterial aminoacyl-tRNA synthetase. Mol. Cell. Biol 1990, 10, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin JW; Cropp TA; Anderson JC; Mukherji M; Zhang ZW; Schultz PG, An expanded eukaryotic genetic code. Science 2003, 301, 964–967. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto K; Hayashi A; Sakamoto A; Kiga D; Nakayama H; Soma A; Kobayashi T; Kitabatake M; Takio K; Saito K; Shirouzu M; Hirao I; Yokoyama S, Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002, 30, 4692–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W; Brock A; Chen S; Chen S; Schultz PG, Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods 2007, 4, 239–244. [DOI] [PubMed] [Google Scholar]
- 22.Lemke EA; Summerer D; Geierstanger BH; Brittain SM; Schultz PG, Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat. Chem. Biol 2007, 3, 769–772. [DOI] [PubMed] [Google Scholar]
- 23.Kang JY; Kawaguchi D; Coin I; Xiang Z; O’Leary DD; Slesinger PA; Wang L, In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron 2013, 80, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan G; James CM; Krzycki JA, Pyrrolysine encoded by UAG in archaea: charging of a UAG-decoding specialized tRNA. Science 2002, 296, 1459–1462. [DOI] [PubMed] [Google Scholar]
- 25.Hao B; Gong WM; Ferguson TK; James CM; Krzycki JA; Chan MK, A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 2002, 296, 1462–1466. [DOI] [PubMed] [Google Scholar]
- 26.Gaston MA; Zhang LW; Green-Church KB; Krzycki JA, The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine. Nature 2011, 471, 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan W; Tharp JM; Liu WR, Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta 2014, 1844, 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krzycki JA, The direct genetic encoding of pyrrolysine. Curr. Opin. Microbiol 2005, 8, 706–712. [DOI] [PubMed] [Google Scholar]
- 29.Bryson DI; Fan C; Guo L-T; Miller C; Söll D; Liu DR, Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol 2017, 13, 1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blight SK; Larue RC; Mahapatra A; Longstaff DG; Chang E; Zhao G; Kang PT; Green-Church KB; Chan MK; Krzycki JA, Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature 2004, 431, 333–335. [DOI] [PubMed] [Google Scholar]
- 31.Kavran JM; Gundliapalli S; O’Donoghue P; Englert M; Soell D; Steitz TA, Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 11268–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polycarpo CR; Herring S; Berube A; Wood JL; Soll D; Ambrogelly A, Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006, 580, 6695–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambrogelly A; Gundllapalli S; Herring S; Polycarpo C; Frauer C; Soll D, Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukai T; Kobayashi T; Hino N; Yanagisawa T; Sakamoto K; Yokoyama S, Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun 2008, 371, 818–822. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen DP; Lusic H; Neumann H; Kapadnis PB; Deiters A; Chin JW, Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA Synthetase/tRNA(CUA) pair and click chemistry. J. Am. Chem. Soc 2009, 131, 8720–8721. [DOI] [PubMed] [Google Scholar]
- 36.Lang K; Davis L; Torres-Kolbus J; Chou C; Deiters A; Chin JW, Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem 2012, 4, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torres-Kolbus J; Chou CJ; Liu JH; Deiters A, Synthesis of non-linear protein dimers through a genetically encoded thiol-ene reaction. PloS One 2014, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanagisawa T; Ishii R; Fukunaga R; Kobayashi T; Sakamoto K; Yokoyama S, Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol 2008, 15, 1187–1197. [DOI] [PubMed] [Google Scholar]
- 39.Chou C; Uprety R; Davis L; Chin JW; Deiters A, Genetically encoding an aliphatic diazirine for protein photocrosslinking. Chem. Sci 2011, 480–483. [Google Scholar]
- 40.Chen PR; Groff D; Guo J; Ou W; Cellitti S; Geierstanger BH; Schultz PG, A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chem. Int. Ed 2009, 48, 4052–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gattner MJ; Vrabel M; Carell T, Synthesis of epsilon-N-propionyl-, epsilon-N-butyryl-, and epsilon-N-crotonyl-lysine containing histone H3 using the pyrrolysine system. Chem. Commun 2013, 49, 379–381. [DOI] [PubMed] [Google Scholar]
- 42.Gautier A; Nguyen DP; Lusic H; An W; Deiters A; Chin JW, Genetically encoded photocontrol of protein localization in mammalian cells. J. Am. Chem. Soc 2010, 132, 4086–4088. [DOI] [PubMed] [Google Scholar]
- 43.Groff D; Chen PR; Peters FB; Schultz PG, A genetically encoded epsilon-N-methyl lysine in mammalian cells. ChemBioChem 2010, 11, 1066–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao Z; Song Y; Lin S; Yang M; Liang Y; Wang J; Chen PR, A readily synthesized cyclic pyrrolysine analogue for site-specific protein “click” labeling. Chem. Commun 2011, 47, 4502–4504. [DOI] [PubMed] [Google Scholar]
- 45.Kim CH; Kang M; Kim HJ; Chatterjee A; Schultz PG, Site-specific incorporation of epsilon-N-crotonyllysine into histones. Angew. Chem. Int. Ed 2012, 51, 7246–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neumann H; Peak-Chew SY; Chin JW, Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol 2008, 4, 232–234. [DOI] [PubMed] [Google Scholar]
- 47.Virdee S; Kapadnis PB; Elliott T; Lang K; Madrzak J; Nguyen DP; Riechmann L; Chin JW, Traceless and site-specific ubiquitination of recombinant proteins. J. Am. Chem. Soc 2011, 133, 10708–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang WW; Zeng Y; Wu B; Deiters A; Liu WR, A chemical biology approach to reveal Sirt6-targeted histone H3 sites in nucleosomes. ACS Chem. Biol 2016, 11, 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang YS; Fang X; Wallace AL; Wu B; Liu WR, A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum. J. Am. Chem. Soc 2012, 134, 2950–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tharp JM; Wang YS; Lee YJ; Yang Y; Liu WR, Genetic incorporation of seven ortho-substituted phenylalanine derivatives. ACS Chem. Biol 2014, 9, 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang YS; Fang X; Chen HY; Wu B; Wang ZU; Hilty C; Liu WR, Genetic incorporation of twelve meta-substituted phenylalanine derivatives using a single pyrrolysyl-tRNA synthetase mutant. ACS Chem. Biol 2013, 8, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao H; Peters FB; Yang PY; Reed S; Chittuluru JR; Schultz PG, Genetic incorporation of histidine derivatives using an engineered pyrrolysyl-tRNA synthetase. ACS Chem. Biol 2014, 9, 1092–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arbely E; Torres-Kolbus J; Deiters A; Chin JW, Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. J. Am. Chem. Soc 2012, 134, 11912–11915. [DOI] [PubMed] [Google Scholar]
- 54.Luo J; Torres-Kolbus J; Liu J; Deiters A, Genetic encoding of photocaged tyrosines with improved light-activation properties for the optical control of protease function. ChemBioChem 2017, 18, 1442–1447. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen DP; Mahesh M; Elsasser SJ; Hancock SM; Uttamapinant C; Chin JW, Genetic encoding of photocaged cysteine allows photoactivation of TEV protease in live mammalian cells. J. Am. Chem. Soc 2014, 136, 2240–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uprety R; Luo J; Liu J; Naro Y; Samanta S; Deiters A, Genetic encoding of caged cysteine and caged homocysteine in bacterial and mammalian cells. ChemBioChem 2014, 15, 1793–1799. [DOI] [PubMed] [Google Scholar]
- 57.Schneider S; Gattner MJ; Vrabel M; Flügel V; López-Carrillo V; Prill S; Carell T, Structural insights into incorporation of norbornene amino acids for click modification of proteins. ChemBioChem 2013, 14, 2114–2118. [DOI] [PubMed] [Google Scholar]
- 58.Takimoto JK; Dellas N; Noel JP; Wang L, Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem. Biol 2011, 6, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greiss S; Chin JW, Expanding the genetic code of an animal. J. Am. Chem. Soc 2011, 133, 14196–14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bianco A; Townsley FM; Greiss S; Lang K; Chin JW, Expanding the genetic code of Drosophila melanogaster. Nat. Chem. Biol 2012, 8, 748–750. [DOI] [PubMed] [Google Scholar]
- 61.Ernst RJ; Krogager TP; Maywood ES; Zanchi R; Beranek V; Elliott TS; Barry NP; Hastings MH; Chin JW, Genetic code expansion in the mouse brain. Nat. Chem. Biol 2016, 12, 776–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu J; Hemphill J; Samanta S; Tsang M; Deiters A, Genetic code expansion in zebrafish embryos and its application to optical control of cell signaling. J. Am. Chem. Soc 2017, 139, 9100–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fields S; Johnston M, Whither model organism research? Science 2005, 307, 1885–1886. [DOI] [PubMed] [Google Scholar]
- 64.Corsi AK, A Biochemist’s guide to C. elegans. Anal. Biochem 2006, 359, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopez-Otin C; Blasco MA; Partridge L; Serrano M; Kroemer G, The hallmarks of aging. Cell 2013, 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrington AJ; Hamamichi S; Caldwell GA; Caldwell KA, elegans C as a model organism to investigate molecular pathways involved with Parkinson’s disease. Dev. Dyn 2010, 239, 1282–1295. [DOI] [PubMed] [Google Scholar]
- 67.Jennings BH, Drosophila – a versatile model in biology & medicine. Mater. Today 2011, 14, 190–195. [Google Scholar]
- 68.Grunwald DJ; Eisen JS, Headwaters of the zebrafish - emergence of a new model vertebrate. Nat. Rev. Genet 2002, 3, 717–724. [DOI] [PubMed] [Google Scholar]
- 69.Lieschke GJ; Currie PD, Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet 2007, 8, 353–367. [DOI] [PubMed] [Google Scholar]
- 70.Zon LI; Peterson RT, In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov 2005, 4, 35–44. [DOI] [PubMed] [Google Scholar]
- 71.Rosenthal N; Brown S, The mouse ascending: perspectives for human-disease models. Nat. Cell Biol 2007, 9, 993–999. [DOI] [PubMed] [Google Scholar]
- 72.Hodgkin J, Novel nematode amber suppressors. Genetics 1985, 111, 287–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parrish AR; She X; Xiang Z; Coin I; Shen Z; Briggs SP; Dillin A; Wang L, Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl-tRNA synthetase/tRNA pairs. ACS Chem. Biol 2012, 7, 1292–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chatterjee A; Xiao H; Bollong M; Ai HW; Schultz PG, Efficient viral delivery system for unnatural amino acid mutagenesis in mammalian cells. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 11803–11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frøkjær-Jensen C; Wayne Davis M; Hopkins CE; Newman BJ; Thummel JM; Olesen S-P; Grunnet M; Jorgensen EM, Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet 2008, 40, 1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmied WH; Elsasser SJ; Uttamapinant C; Chin JW, Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J. Am. Chem. Soc 2014, 136, 15577–15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laski FA; Ganguly S; Sharp PA, Construction, stable transformation, and function of an amber suppressor tRNA gene in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A 1989, 86, 6696–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Y; Ma J; Lu W; Tian M; Thauvin M; Yuan C; Volovitch M; Wang Q; Holst J; Liu M; Vriz S; Ye S; Wang L; Li D, Heritable expansion of the genetic code in mouse and zebrafish. Cell Res. 2017, 27, 294–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo J; Uprety R; Naro Y; Chou C; Nguyen DP; Chin JW; Deiters A, Genetically encoded optochemical probes for simultaneous fluorescence reporting and light activation of protein function with two-photon excitation. J. Am. Chem. Soc 2014, 136, 15551–15558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han S; Yang A; Lee S; Lee HW; Park CB; Park HS, Expanding the genetic code of Mus musculus. Nat. Commun 2017, 8, 14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dieterich DC; Link AJ; Graumann J; Tirrell DA; Schuman EM, Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U. S. A 2006, 103, 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ngo JT; Tirrell DA, Noncanonical amino acids in the interrogation of cellular protein synthesis. Acc. Chem. Res 2011, 44, 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Elliott TS; Townsley FM; Bianco A; Ernst RJ; Sachdeva A; Elsasser SJ; Davis L; Lang K; Pisa R; Greiss S; Lilley KS; Chin JW, Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol 2014, 32, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krogager TP; Ernst RJ; Elliott TS; Calo L; Beránek V; Ciabatti E; Spillantini MG; Tripodi M; Hastings MH; Chin JW, Labeling and identifying cell-specific proteomes in the mouse brain. Nat. Biotechnol 2017, 36, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peng T; Hang HC, SORTing out cellular proteomes in vivo. Nat. Biotechnol 2014, 32, 445. [DOI] [PubMed] [Google Scholar]
- 86.Tye KM; Deisseroth K, Optogenetic investigation of neural circuits underlying brain disease in animal models. Nat. Rev. Neurosci 2012, 13, 251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baker AS; Deiters A, Optical control of protein function through unnatural amino acid mutagenesis and other optogenetic approaches. ACS Chem. Biol 2014, 9, 1398–1407. [DOI] [PubMed] [Google Scholar]
- 88.Courtney T; Deiters A, Recent advances in the optical control of protein function through genetic code expansion. Curr. Opin. Chem. Biol 2018, 46, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lahiry P; Torkamani A; Schork NJ; Hegele RA, Kinase mutations in human disease: interpreting genotype-phenotype relationships. Nat. Rev. Genet 2010, 11, 60–74. [DOI] [PubMed] [Google Scholar]
- 90.Kowalik L; Chen JK, Illuminating developmental biology through photochemistry. Nat. Chem. Biol 2017, 13, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anastasaki C; Estep AL; Marais R; Rauen KA; Patton EE, Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development and are sensitive to small molecule inhibitors. Hum. Mol. Genet 2009, 18, 2543–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gautier A; Deiters A; Chin JW, Light-activated kinases enable temporal dissection of signaling networks in living cells. J. Am. Chem. Soc 2011, 133, 2124–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Manning G; Whyte DB; Martinez R; Hunter T; Sudarsanam S, The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- 94.Brown W; Liu J; Tsang M; Deiters A, Cell-lineage tracing in zebrafish embryos with an expanded genetic code. ChemBioChem 2018, 19, 1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luo J; Arbely E; Zhang J; Chou C; Uprety R; Chin JW; Deiters A, Genetically encoded optical activation of DNA recombination in human cells. Chem. Commun 2016, 52, 8529–8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mosimann C; Kaufman CK; Li P; Pugach EK; Tamplin OJ; Zon LI, Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 2011, 138, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Devaraj NK; Upadhyay R; Haun JB; Hilderbrand SA; Weissleder R, Fast and sensitive pretargeted labeling of cancer cells via tetrazine/trans-cyclooctene cycloaddition. Angew. Chem. Int. Ed 2009, 48, 7013–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deisseroth K; Hegemann P, The form and function of channelrhodopsin. Science 2017, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beier H; Grimm M, Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001, 29, 4767–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie J; Schultz PG, An expanding genetic code. Methods 2005, 36, 227–238. [DOI] [PubMed] [Google Scholar]
- 101.Anderson JC; Wu N; Santoro SW; Lakshman V; King DS; Schultz PG, An expanded genetic code with a functional quadruplet codon. Proc. Natl. Acad. Sci. U. S. A 2004, 101, 7566–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee BS; Kim S; Ko BJ; Yoo TH, An efficient system for incorporation of unnatural amino acids in response to the four-base codon AGGA in Escherichia coli. Biochim. Biophys. Acta 2017, 1861, 3016–3023. [DOI] [PubMed] [Google Scholar]
- 103.Zhang Y; Ptacin JL; Fischer EC; Aerni HR; Caffaro CE; San Jose K; Feldman AW; Turner CR; Romesberg FE, A semi-synthetic organism that stores and retrieves increased genetic information. Nature 2017, 551, 644. [DOI] [PMC free article] [PubMed] [Google Scholar]