Abstract
Background
Proprotein convertase subtilisin/kexin type 9 (PCSK9) is a secreted protein that down-regulates hepatic low-density lipoprotein receptor (LDLR) by binding and shuttling LDLR to lysosomes for degradation. The development of therapy that inhibits PCSK9 has attracted considerable attention for the management of cardiovascular disease risk. However, only monoclonal antibodies of PCSK9 have reached the clinic use. Oral administration of small-molecule transcriptional inhibitors has the potential to become a therapeutic option.
Methods
Here, we developed a cell-based small molecule screening platform to identify transcriptional inhibitors of PCSK9. Through high-throughput screening and a series of evaluation, we found several active compounds. After detailed investigation on the pharmacological effect and molecular mechanistic characterization, 7030B-C5 was identified as a potential small-molecule PCSK9 inhibitor.
Findings
Our data showed that 7030B-C5 down-regulated PCSK9 expression and increased the total cellular LDLR protein and its mediated LDL-C uptake by HepG2 cells. In both C57BL/6 J and ApoE KO mice, oral administration of 7030B-C5 reduced hepatic and plasma PCSK9 level and increased hepatic LDLR expression. Most importantly, 7030B-C5 inhibited lesions in en face aortas and aortic root in ApoE KO mice with a slight amelioration of lipid profiles. We further provide evidences suggesting that transcriptional regulation of PCSK9 by 7030B-C5 mostly depend on the transcriptional factor HNF1α and FoxO3. Furthermore, FoxO1 was found to play an important role in 7030B-C5 mediated integration of hepatic glucose and lipid metabolism.
Interpretation
7030B-C5 with potential suppressive effect of PCSK9 expression may serve as a promising lead compound for drug development of cholesterol/glucose homeostasis and cardiovascular disease therapy.
Fund
This work was supported by grants from the National Natural Science Foundation of China (81473214, 81402929, and 81621064), the Drug Innovation Major Project of China (2018ZX09711001-003-006, 2018ZX09711001-007 and 2018ZX09735001-002), CAMS Innovation Fund for Medical Sciences (2016-I2M-2-002, 2016-I2M-1-011 and 2017-I2M-1-008), Beijing Natural Science Foundation (7162129).
Keywords: PCSK9, Small-molecule inhibitor, FoxO1/3, HNF1α, Lipid and glucose metabolism, Atherosclerosis
Research in context.
Evidence before this study
To date, PCSK9 inhibitors are novel therapeutics for the treatment of cardiovascular disease in addition to statins. Two monoclonal antibodies alirocumab and evolocumab targeting PCSK9 have been approved by FDA, but they are expensive and requiring subcutaneous administration, limiting their widespread use. As small-molecule inhibitors targeting protein-protein interaction of PCSK9 and LDLR are difficult to develop, more options are needed.
Added value of this study
Here, we have developed a novel cell-based screening platform to discover inhibitors targeting PCSK9 transcription. Using a robust and pragmatic approach, we identified an effective small-molecule PCSK9 inhibitor 7030B-C5. Our data showed that 7030B-C5 down-regulated the expression of PCSK9 at both mRNA and protein levels and increased the cellular LDLR protein level and its mediated cellular LDL-C uptake. In both C57BL/6 J mice and ApoE KO mice, oral administration of 7030B-C5 significantly reduced hepatic PCSK9 expression/secretion and increased LDLR expression. Moreover, the compound profoundly reduced atherosclerosis progression, and showed dual benefits in lipid and glucose metabolism in an HNF1α and FoxO1/3-responsive elements-dependent manner.
Implications of all the available evidence
A small-molecule compound, 7030B-C5, with PCSK9 inhibitory and anti-atherosclerosis activity may potentially serve as a drug lead compound for cholesterol/glucose modulation and cardiovascular disease.
Alt-text: Unlabelled box
1. Introduction
Increases in hepatic low-density lipoprotein receptor (LDLR) play a pivotal role in enhancing the clearance of plasma low-density lipoprotein cholesterol (LDL-C) and are associated with reducing the risk of cardiovascular disorder [1]. Proprotein convertase subtilisin/kexin type 9 (PCSK9) is a critical player in LDL-C metabolism through regulating the degradation of hepatic LDLRs [2,3]. PCSK9 is a liver-derived plasma protease which is initially synthesized as a 75 kDa precursor protein and converted into a 62 kDa mature form undergoing the autocatalytic cleavage process in the Golgi apparatus [4,5]. Circulating PCSK9 binds to the extracellular epidermal growth factor-like repeat A (EGF-A) domain of LDLR, causing its internalization [2,6]. Followed, the PCSK9/LDLR complex translocates to the endosome-lysosomal compartment where LDLR is degraded [7,8]. Reduced cell-surface expression of LDLR leads to increased circulating levels of LDL-C. In human subjects, early studies have shown that gain-of-function mutations in the PCSK9 gene lead to familial hypercholesterolemia [9], while loss-of-function mutations are associated with hypocholesterolemia and protection against coronary artery disease [10], [11], [12]. Thus, decreasing circulating PCSK9 level or activity to up-regulate hepatic LDLR level and to lower circulating LDL-C level will be beneficial for reducing the risk of cardiovascular disease in humans.
The pivotal role of PCSK9 in the metabolism of LDL-C as well as the verified safety of PCSK9 inhibition led to the development of PCSK9 inhibitors. Recently, two PCSK9 monoclonal antibodies (mAbs) alirocumab and evolocumab which were reported to disrupt the PCSK9-LDLR interaction have been approved by the U.S. Food and Drug Administration (FDA) [13]. Alirocumab and evolocumab can be potentially used as monotherapy or add-on therapies to statins in patients with familial hypercholesterolemia or atherosclerotic cardiovascular disease, effectively reducing LDL-C up to 70% and with well-tolerated profile [14], [15], [16], [17]. Besides, these monoclonal antibodies significantly suppress circulating PCSK9 levels and consistently reduce LDL-C levels in patients [18]. However, it is estimated that the cost of PCSK9 mAb inhibitors was ~$14,000 per person per year in the US which limits its widespread use [19]. The small-molecule inhibitors can offer potentially more convenient routes of administration and lower costs. However, small-molecule inhibitors specific to inhibit PCSK9 activity remains challenging because it is difficult for small molecules to blockade the fairly large size of the flat interface between the catalytic subunit of PCSK9 and the EGF-A domain of LDLR [20]. Other promising approaches have been explored to discover small molecules with potential to become a therapeutic option by inhibiting PCSK9 protein synthesis. PF-06446846 was identified by Petersen et al. through phenotypic screening of a small-molecule compound collection, which directly and selectively inhibits translation of PCSK9 by stalling the 80S ribosome in the proximity of codon region [21,22]. Orally-administered PF-06446846 reduces plasma PCSK9 and LDL-C levels without liver toxicity symptoms in vivo [23]. However, its development was discontinued for competitive reasons [24].
The gene expression of PCSK9 is controlled by various transcriptional regulators such as sterol regulatory element binding protein (SREBP), hepatocyte nuclear factor 1α (HNF1α), and forkhead box O3 (FoxO3). A sterol regulatory element (SRE), which forms the target site for SREBPs, is present 330 bp upstream of the translation start site and functions as an important cis-element regulating PCSK9 transcription [25]. Cholesterol-lowering drugs, statins, which activate the SREBP2 pathway by inhibiting HMG-CoA reductase and subsequently activating the expression of LDLR, have also been proven to stimulate PCSK9 gene expression, thus diminish the beneficial effects of statin treatment [26]. Insulin and docosahexaenoic acid (DHA) could activate PCSK9 transcription via SREBP-1c [27], [28], [29]. In addition to SREBP regulation, HNF1α, a liver-enriched transcription factor, was found to bind HNF1 element within the PCSK9 promoter and activate PCSK9 expression in hepatic cells [30] and livers [31,32]. HINFP was reported as a positive regulator of PCSK9 transcription through facilitation of histone H4 acetylation at PCSK9 promoter [33]. FoxO3, a forkhead transcription factor, has been identified as a negative regulator of PCSK9 gene expression through epigenetic modulation. FoxO3 interacts with the insulin-response element (IRE) within PCSK9 promoter, recruiting the sirtuin-6 (Sirt6) protein to deacetylate histones and reducing the promoter binding capacity of HNF1α, thereby suppressing the PCSK9 gene expression in hepatic cells [34]. In this work, we developed a strategy to discover small-molecule inhibitors of PCSK9 expression at transcription level, and described the identification and mechanistic characterization of 7030B-C5, an active small-molecule inhibitor of PCSK9 transcription. Oral administration of the compound significantly reduced atherosclerosis progression, and showed dual benefits in both lipid and glucose metabolism, providing a promising lead compound for further development of novel drug candidate for small-molecule PCSK9 modulators.
2. Materials and methods
2.1. Cell cultures and cell viability assay
HepG2 cells were cultured in Eagle minimal essential medium (MEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), 1% sodium pyruvate and 1% nonessential amino acids (Invitrogen). HEK-293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) supplemented with 10% (vol/vol) FBS (Gibco). HepG2 cells were transfected with luciferase reporter constructs contained the PCSK9 gene promoter sequence spanning the region from −2112 to −1 bp (nucleotide +1 corresponds to the A of ATG start codon, the same as below). The stable pGL4-PCSK9-P transfected HepG2 cells were established by selection with G418 (700 μg/mL, Invitrogen). The cells were cultured at 37 °C in a humidified 5% CO2 incubator.
Cell viability was determined using MTT assay. HepG2 cells were treated with vehicle or positive compounds (0.39–100 μM) for 4 h followed by incubation with MTT reagent (1 mg/mL) at 37 °C for 4 h. The medium was removed, and the purple formazan crystals were dissolved in DMSO. The absorbance was measured at 550 nm using a Victor X5 multilabel plate reader (PerkinElmer, Waltham, MA, USA).
2.2. Construction of luciferase reporter system and high-throughput screening
Human PCSK9 gene promoter was amplified by PCR using HepG2 genomic DNA as the template and cloned into pGL4-Basic vector (Promega, Madison, WI, USA) at Xho I and Hind III sites to get pGL4-PCSK9-P. And pGL4-PCSK9-P plasmid was used as the template for the PCR amplification of PCSK9 gene promoter fragments (D1-D7). The HNF1α mutant (HNF1α-mu) and HINFP mutant (HINFP-mu) were generated from construct D5 using the Fast MultiSite Mutagenesis System (TransGen, Beijing, China). Primers used for plasmid construction are listed in Supplemental Table I.
The cells were transfected with the human PCSK9 promoter plasmid using Lipofectamine 2000 (Thermo Scientific, Wilmington, DE, USA) according to the manufacturer's instructions. After transfection, the cells were treated with compounds or vehicle control (0.1% DMSO) for 24 h. The luciferase activities were measured using the Luciferase Assay System (Promega) and detected by a Victor X5 multilabel plate reader (PerkinElmer).
A collection of 6328 compounds was screened in the pGL4-PCSK9-P stably transfected cells at a concentration of 25 μg/mL for the primary screening. The screening compound library and the positive compounds 7030B-C5, 7031B-H9, 7045B-E7 and 7045B-F7 are from the center for National New Drug Screening (Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences, Beijing, China).
2.3. Transfection with small interference RNA (siRNA)
Three pre-designed siRNAs targeted to human HNF1α mRNA, FoxO3 mRNA, HINFP mRNA and one silencer negative control siRNA were obtained from Ambion. Transfection of siRNAs was carried out using the Lipofectamine RNAiMax reagent (Thermo Scientific) according to the manufacturer's instructions. The cells were incubated with serum-free OPTI-MEM medium, and transfected with the negative control (siNC) or specific siRNA at a final concentration of 100 nM. After 48 h of transfection, the cells were treated with the vehicle or 7030B-C5 (12.5 μM) for 24 h and harvested for further analysis.
2.4. Quantitative real-time PCR
Total RNAs were isolated from cultured cells or tissues with the SV total RNA isolation system (Promega) and quantified by Nanodrop spectrophotometry (Thermo Scientific). RNA was reverse transcribed using TransScript First-Strand cDNA Synthesis SuperMix (TransGen). Quantitative real-time PCR (qPCR) was performed in the Bio-Rad CFX96 real-time system (Bio-Rad, Hercules, CA, USA) using the SYBR green Master Kit (Roche, Mannheim, Germany) as previously described [35]. qPCR primers are listed in Supplemental Table II. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize the mRNA levels of target genes.
2.5. Western blotting
Whole-cell lysates were prepared using RIPA lysis buffer (50 mM Tris HCl, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, pH 7.4) containing complete EDTA-free protease inhibitor cocktail (Roche). Protein samples were separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto a 0.45-μm polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA). The membranes were incubated with primary antibodies to LDLR (Abcam, Cambridge, UK), PCSK9 (R&D Systems, Minneapolis, MN, USA) HNF1α (Cell Signaling Technology, Danvers, MA, USA), FoxO1 (Cell Signaling Technology), FoxO3 (Cell Signaling Technology), HINFP (Novus, Littleton, CO, USA) or GAPDH (ZSJQ-Bio, Beijing, China), followed by horse radish peroxidase (HRP)-conjugated secondary antibodies (ZSJQ-Bio). An enhanced chemiluminescence detection system (Millipore) was used for blot detection. The quantification of detection protein amounts was determined by Image J software, normalized to GAPDH.
2.6. DiI-LDL uptake assays
After incubated with positive compounds, HepG2 cells were incubated with 2 mg/mL 1,1′-dioctadecyl-3,3,3′,3′- tetramethylindocarbocyanine perchlorate (DiI)-labeled LDL (Biomedical Technologies, Stoughton, MA, USA) for 4 h at 37 °C. Then cells were harvested, washed and resuspended in phosphate buffered saline (PBS). DiI fluorescence measurements were performed using an Epics XL flow cytometer (Beckman Coulter, Miami, FL, USA) as described previously [36].
2.7. Chromatin immunoprecipitation assay (ChIP assay)
Cells cultured in MEM medium were treated with vehicle or 7030B-C5 (12.5 μM) for 24 h. The ChIP assay was carried out using the ChIP Assay Kit (Cell Signaling Technology) according to the manufacturer's instructions. After 7030B-C5 treating, the cells were fixed in 1% formaldehyde for 10 min at room temperature. The chromatin was sheared to an average length of 150–900 bp by sonication. The chromatin extracts were immunoprecipitated at 4 °C overnight with anti-HNF1α, anti-FoxO3, or control IgG (Cell Signaling Technology). Immunocomplexes were isolated by binding to protein A-agarose beads. Precipitated DNA was isolated after ethanol precipitation. Quantitative real-time PCR was performed to analyze the promoter binding levels determined using the specific PCSK9 primers 5′-TCCAGCCCAGTTAGGATTTG-3′ and 5′-CGGAAACCTTCTAGGGTGTG-3′.
2.8. Animal experiments and quantification of atherosclerosis
All experimental procedures involving animals were approved by the Institutional Laboratory Animal Care and Use Committee of Institute of Medicinal Biotechnology. C57BL/6 J mice, ApoE KO mice were purchased from Vital River Laboratory Animal Technology (Beijing, China).
To investigate the oral acute toxicity of active compounds, male C57BL/6 J mice (~8 weeks old) fed normal chow were divided into four groups (5 mice/group), and intragastric injected with active compounds dissolved in H2O (100 mg/kg bodyweight/day, 300 mg/kg bodyweight/day, 500 mg/kg bodyweight/day, 1000 mg/kg bodyweight/day, respectively). Survival and behavioral characteristics were immediately observed as well as 10 min, 15 min, 30 min, 1 h, and 2 h for the first day. Then, it was observed for once a day lasting for a total of 7 days.
To determine the effect of active compounds on PCSK9 expression in vivo, male C57BL/6 J mice (~8 weeks old) fed normal chow were divided into three groups (5 mice/group), and intragastric injected with vehicle and 7030B-C5 (30 mg/kg bodyweight/day), 7031B-H9 (50 mg/kg bodyweight/day), 7045B-E7 (100 mg/kg bodyweight/day), 7045B-F7 dissolved in H2O (100 mg/kg bodyweight/day) for 4 weeks, respectively. Mice were then sacrificed with 1% sodium pentobarbital (40 mg/kg) followed by collection of liver samples individually to determine PCSK9 and LDLR expression by western blot and real-time PCR.
To study the effects of active compounds on atherosclerosis, male ApoE KO mice (~8 weeks old) were fed a high-fat diet (HFD) containing 0.15% cholesterol and 20% lard or a regular rodent chow as a control. And the male ApoE KO mice (~8 weeks old) fed a HFD were divided into three groups (10 mice/group), and intragastric injected with vehicle and 7030B-C5 (10 mg/kg bodyweight/day, 30 mg/kg bodyweight/day), 7031B-H9 (50 mg/kg bodyweight/day, 100 mg/kg bodyweight/day). After 8 weeks of feeding, blood samples were collected for determination of lipid profiles by 7100 automatic biochemical analyzer (HITACHI, Tokyo, Japan). After 12 weeks of feeding, the animals were sacrificed followed by collection of mouse aorta, liver and blood samples. Liver samples were used to determine hepatic protein expression by Western blot. Liver tissues were fixed in 4% neutral buffered formalin and embedded in OCT freezing medium. The tissues were sliced and stained with hematoxylin-eosin (H&E) staining for morphological analysis, Oil Red O to detect lipid accumulation. Blood samples were collected and detected as described above. And serum PCSK9 levels were determined by an ELISA assay kit (Sino Biological, Beijing, China). Aortas were collected and stained with Oil Red O solution. To determine lesions in aortic root, frozen sections of aortic root were prepared and then stained with Oil Red O solution. The lesions in en face aortas and aortic root were quantitatively determined by computer-assisted image analysis method (Image J) and showed as percent lesion area [37].
2.9. Fast protein liquid chromatography (FPLC)
Mice serum was fractionated using FPLC (Akta FPLC; GE Healthcare, Pittsburgh, PA, USA) using Superose 6 Increase 10/300 GL columns. Phosphate buffered saline (PBS; pH 7.4) eluent buffer was first passed through a 0.22 μm filter, and then the column was equilibrated at a rate of 0.25 mL/min. Serum sample aliquots of 500 μL were then injected into the column. Elutions were then carried out at a flow rate of 0.25 mL/min. 63 fractions, 0.3 mL each, were collected in separate eluant eppendorf tubes for subsequent cholesterol content analysis by 7100 automatic biochemical analyzer (HITACHI).
2.10. Hepatic transcriptome analysis
Total RNA from ApoE KO mice livers were extracted using TRIzol reagent (Invitrogen). Subsequently, mRNA was purified from total RNA using the Oligotex mRNA Midi Kit (Qiagen, Hilden, Germany). Poly(A)+ mRNA was obtained using magnetic poly(A)+ Dynabeads (Invitrogen). For RNA-seq, the cDNA libraries were prepared with fragmented mRNA according to a TruseqTM RNA sample prep kit (Illumina, California, USA). The cDNA was then end-repaired, adenylated, and ligated to the sequencing adapters provided by Illumina. Finally, the libraries were enriched by PCR amplification and sequenced using an Illumina HiSeq 2000 platform (Majorbio Bio-Pharm, Shanghai, China). Sequence data has been deposited with the NCBI BioProject database under accession PRJNA602107.
2.11. Statistical analyses
All experiments were repeated at least three times and representative results are shown in figures. The values are expressed as the mean ± SEM. Student's t-test was applied for comparing two groups and one-way ANOVA for multiple groups, followed by Bonferroni's correction as applicable (GraphPad Prism Software; Graph-Pad). P value of < 0.05 was considered significant. Error bars denote SEM.
3. Results
3.1. Discovery of novel PCSK9 inhibitors using cell-based high-throughput screening (HTS) assays
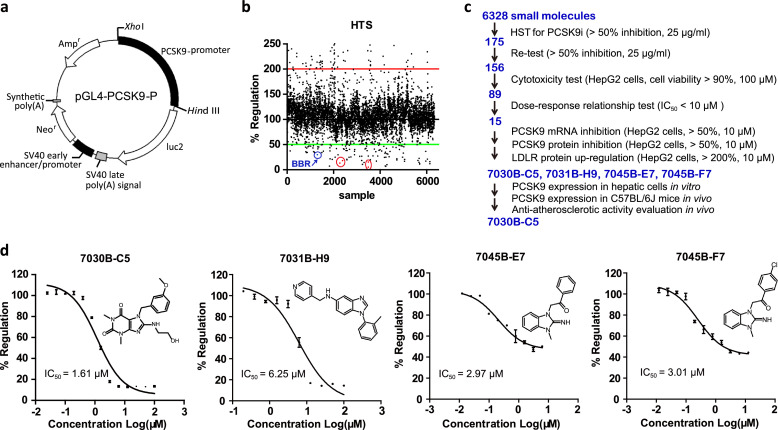
In order to establish a luciferase reporter-based HTS assay to find modulators targeting PCSK9 gene transcriptional expression, a 2112-bp fragment of PCSK9 gene promoter region was directionally inserted into the upstream of luciferase reporter gene of pGL4-Basic vector to construct the recombinant plasmid pGL4-PCSK9-P (Fig. 1a). Subsequently, the HTS assay was built by stably transfecting plasmid pGL4-PCSK9-P into HepG2 cells and quantitatively assessed by Z’ factor [38] using berberine (BBR) as a positive control. BBR is a known inhibitor of PCSK9 transcription [30] (Fig. 1b and Suppl Fig. 1), which regulated PCSK9 expression through the modulation of transcriptional factors SREBP2 and HNF1α in hepatic cells. In our assay, BBR significantly repressed PCSK9 transcriptional activity in a dose-dependent manner, with an IC50 of 10.26 μM (Suppl Fig. 1c). Besides, anacetrapib, a CETP inhibitor, which was reported to inhibit transcriptional activation of the PCSK9 gene by reducing the expression of mature form of SREBP2 [39], was used to evaluate the established in vitro HTS assay as well. The results showed that anacetrapib could also significantly reduce the PCSK9 transcriptional activity in a dose-dependent manner, with the IC50 of 33.16 μM (Suppl Fig. 1d). In addition, the HTS assay achieved a good signal-to-background ratio with a low percent coefficient of variation, indicating that the model is suitable for high-throughput screening (Suppl Table 3).
Fig. 1.
Identification of novel PCSK9 inhibitors using cell-based high-throughput screening (HTS) assays. (a) The construction of recombinant plasmid pGL4-PCSK9-P. Human PCSK9 promoter region spanning −2112 to −1 bp, relative to the ATG start codon, was amplified by PCR, verified by DNA sequencing and cloned into pGL4-Basic vector between the Xho I and Hind III sites to produce pGL4-PCSK9-P. (b) The samples in compound library were screened by the established cell-based HTS assay for their capability of inhibiting PCSK9 transcription. In total, 6328 compounds at 25 μg/mL were screened. The red dot represented PCSK9 inhibitors 7030B-C5, 7031B-H9, 7045B-E7 and 7045B-F7 we identified. The blue dot represented positive control berberine (BBR). (c) HTS hit triage workflow resulting in the identification of 7030B-C5, 7031B-H9, 7045B-E7 and 7045B-F7, with the number of compounds selected at each step. (d) The inhibitory activity and chemical structure of a novel PCSK9 inhibitor 7030B-C5, 7031B-H9, 7045B-E7 and 7045B-F7. The stable pGL4-PCSK9-P transfected HepG2 cells were treated with 7030B-C5, 7031B-H9, 7045B-E7 or 7045B-F7 for 24 h and the luciferase activities were measured. The data represent the mean ± SEM of at least three independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Then the screening was performed with the luciferase reporter-based HTS assay to identify small molecules that inhibited PCSK9 transcription. A collection of 6328 compounds at 25 μg/mL was screened and retested, and 156 hits were identified as PCSK9 transcriptional inhibitors by at least 50% suppression compared to vehicle (0.1% DMSO, Fig. 1b). Then these hits were evaluated by cell viability assays to early eliminate those with cytotoxicity. Among these compounds, cell viability of 89 compounds exceeded 90% when the treated concentration was up to 100 μM. Dose-response relationship analysis was performed on the established HTS assay and 15 positive compounds were obtained with IC50 < 10 μM. qPCR of mRNA level of PCSK9 and western blot detection of PCSK9 as well as LDLR protein level in HepG2 cells were also conducted and finally revealed 4 most effective positive compounds 7030B-C5, 7031B-H9, 7045B-E7 and 7045B-F7 (Fig. 1c). Accordingly, all the 4 active compounds significantly inhibited the PCSK9 promoter activity in a dose dependent manner, and their structures and IC50 were presented (Fig. 1d). Moreover, the 4 active compounds at a concentration range of 0.5 μM-100 μM caused no apparent effect on cell growth and survival in HepG2 cells (Suppl Fig. 2).
3.2. Effects of the active compounds on PCSK9 and LDLR expression in hepatic cells
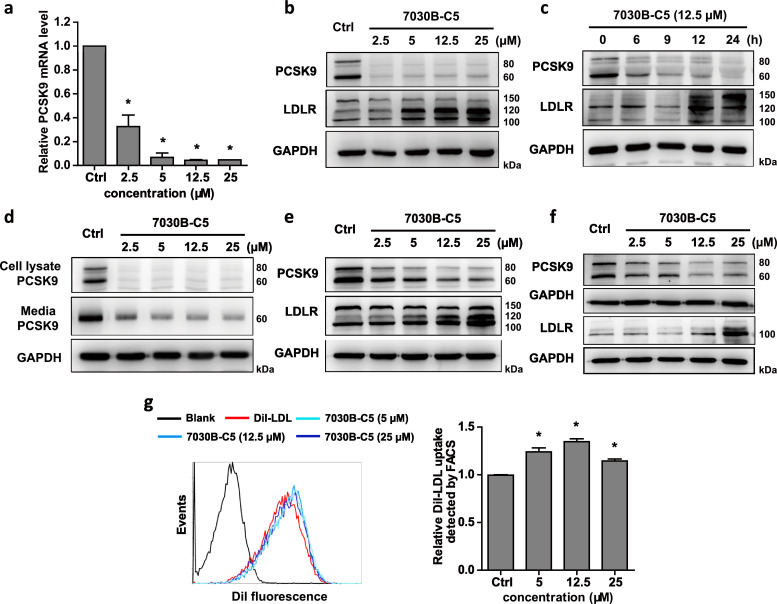
To confirm the inhibitory effect of the positive compounds on PCSK9 expression, biological activity was investigated in hepatic cells. HepG2 cells were firstly used for the evaluation, and the results showed that 7030B-C5 markedly suppressed the mRNA level of PCSK9 in a dose-dependent manner (Fig. 2a). Consistently, a concentration- and time-dependent decrease in PCSK9 protein expression upon 7030B-C5 treatment was also observed in cell lysates (Fig. 2b and c). In addition, the secreted form of PCSK9 protein was dramatically reduced with treatment of increasing concentrations of 7030B-C5 (Fig. 2d). As PCSK9 is a post-translational regulator of the degradation of LDLR in hepatic cells, we examined the impact of 7030B-C5 on LDLR protein level using western blot. As shown in Fig. 2b and c, 7030B-C5 significantly up-regulated LDLR protein levels compared to the vehicle group in a dose- and time-dependent manner. Similarly, 7030B-C5 decreased PCSK9 expression and elevated LDLR protein level in Huh7 cells and human primary hepatocytes (Fig. 2e and f), which indicates that the reduction of PCSK9 expression by 7030B-C5 is not cell or species specific. As a functional measure of the observed cellular LDLR increasement, DiI-labeled LDL (DiI-LDL) uptake by HepG2 cells was detected. Flow cytometry analysis showed that 7030B-C5 treatment led to a significant increase in DiI-LDL uptake compared to vehicle (Fig. 2g). 7031B-H9 showed similar activity as 7030B-C5 in down-regulating PCSK9 expression and elevating LDLR protein level and its mediated DiI-LDL uptake (Suppl Fig. 3). 7045B-E7 and 7045B-F7 decreased PCSK9 mRNA and protein levels in a concentration-dependent manner, however, the LDLR protein level was gradually increased only in the range of lower concentrations (Suppl Fig. 3). Consistently, 7030B-C5 and 7031B-H9 promoted DiI-LDL uptake into HepG2 cells by nearly 50%, while 7045B-E7 and 7045B-F7 reached no more than 20% at 12.5 μM. Taken together, these results suggested that 7030B-C5 and 7031B-H9 are superior to 7045B-E7 and 7045B-F7 with better effect on increasing the total cellular LDLR protein and its mediated LDL uptake.
Fig. 2.
Effects of 7030B-C5 on PCSK9 and LDLR expression in hepatic cells. (a) HepG2 cells were treated with 7030B-C5 in a series of concentration for 24 h. The mRNA level of PCSK9 was measured by RT-qPCR analysis. (b) HepG2 cells were treated with 7030B-C5 in a series of concentration for 24 h. Expression of PCSK9 and LDLR protein was measured by Western blot. (c) HepG2 cells were treated with 7030B-C5 in 12.5 μM with different times. After treatment, cellular proteins were extracted and used to determine PCSK9 protein by Western blot. (d) HepG2 cells were treated with 7030B-C5 in a series of concentrations for 24 h. Secreted form of PCSK9 protein and cellular PCSK9 proteins were determined. (e) Huh7 cells were treated with different concentrations of 7030B-C5 for 24 h. Expression of PCSK9 and LDLR protein was measured by western blot. (f) Human primary hepatocytes were treated with 7030B-C5 for 24 h. Expression of PCSK9 and LDLR protein was determined. (g) HepG2 cells were treated with vehicle or 7030B-C5 for 24 h. The cells were incubated with DiI-LDL (5 μg/mL) at 37 °C for 4 h, and then the LDL uptake activity was measured by flow cytometric analysis. Values are presented as means ± SEM (n = 3). *p < 0.05 vs. control in the corresponding group.
3.3. 7030B-C5 reduced atherosclerosis progression in ApoE KO mice
Given the effects of these 4 active compounds on PCSK9, LDLR expression and LDL uptake in HepG2 cells, we further explored their pharmacological activity in vivo. Firstly, we investigated their safety by oral acute toxicity assessment of 7030B-C5, 7031B-H9, 7045B-E7 and 7045B-F7 in C57BL/6 J mice, and LD50 of 7030B-C5 was approximately 300 mg/kg, while that of other 3 compounds (7031B-H9, 7045B-E7 and 7045B-F7) were more than 1000 mg/kg. Based on LD50 value, a maximum dose of 30 mg/kg 7030B-C5, 50 mg/kg 7031B-H9, 100 mg/kg 7045B-E7 and 7045B-E7 were applied in efficacy studies in vivo. C57BL/6 J mice on chow diet were intragastrically injected with 7030B-C5, 7031B-H9, 7045B-E7 or 7045B-F7 respectively for 4 weeks to confirm its regulatory effect on hepatic PCSK9 and LDLR. After the administration, 7030B-C5 and 7031B-H9 remarkably reduced PCSK9 protein level and led to an increase in the following LDLR expression in the liver as supposed (Suppl Fig. 4). However, 7045B-E7 and 7045B-F7 decreased both PCSK9 and LDLR protein level in the liver, implying more complex regulation mechanism of these 2 compounds (Suppl Fig. 4). Considering the potential capacity of LDLR in LDL-C clear-up and the further development of compounds for the treatment of cardiovascular diseases, 7030B-C5 and 7031B-H9 were investigated further in the following studies.
ApoE knockout (ApoE KO) mice fed with high-fat diet (HFD) for months can show obvious increasing serum cholesterol level and atherosclerosis symptoms resembling their human counterparts, which are extensively used in researches on cholesterol metabolism and atherosclerosis. In order to study the pharmacological effects of 7030B-C5 and 7031B-H9 on atheroprotection, male ApoE KO mice fed HFD were applied and intragastrically injected with 7030B-C5 or 7031B-H9. During the experiment, preliminary analysis of lipid profile assay was performed at the 8th week, and result showed that both plasma total cholesterol (TC) and LDL cholesterol (LDL-C) concentrations were significantly increased in HFD-fed mice compared with chow diet-fed mice (Suppl Fig. 5). 7031B-H9 (50 mg/kg) hardly reduced either serum TC or LDL-C level at the 8th week, and even significantly up-regulated LDL-C levels at dose of 100 mg/kg (Suppl Fig. 5b). Conversely, 7030B-C5 slightly decreased serum total cholesterol, triglyceride and glucose level (Suppl Fig. 5a), and was chosen in the subsequent anti-atherosclerotic activity evaluation.
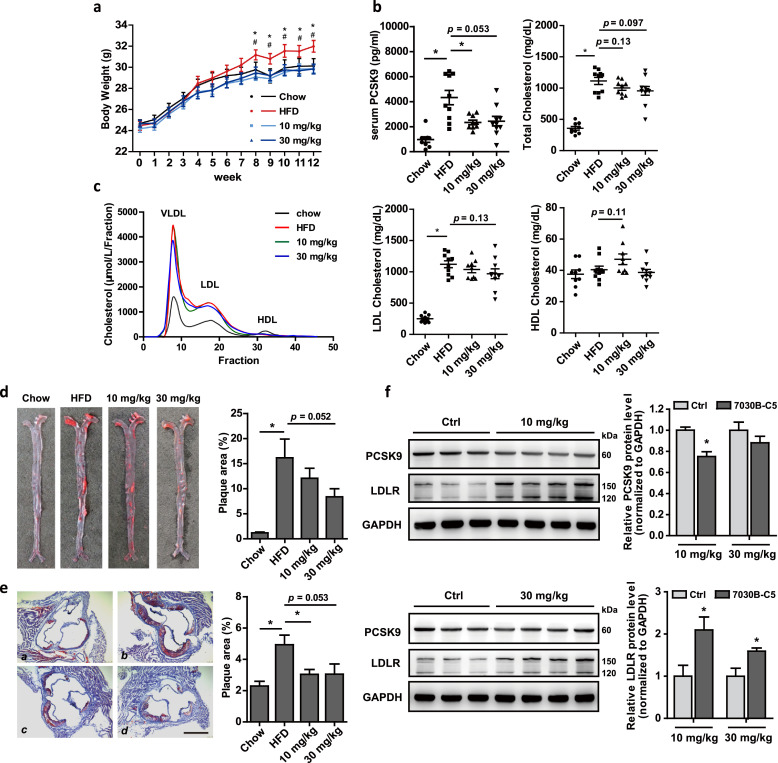
After 12 weeks' administration, both 10 and 30 mg/kg 7030B-C5 markedly abolished the increase of weight gain in the HFD fed mice compared with chow diet-fed mice since the 8th week till the end of the experiment (Fig. 3a). The serum PCSK9 level was significantly reduced by both 10 and 30 mg/kg 7030B-C5 treatment (Fig. 3b). Besides, 30 mg/kg 7030B-C5 administration decreased both TC and LDL-C by 15% at the 12th week compared with HFD group albeit not statistically significant (Fig. 3b). Further analysis by FPLC revealed ApoE KO mice had a distinct LDL and a VLDL peak, which were more pronounced in the HFD group, whereas 7030B-C5 treated mice displayed slightly decreased cholesterol levels in the LDL and VLDL fraction (Fig. 3c). Importantly, in assessing atherosclerotic lesion development, histomorphometry of aortae en face by Oil red O staining revealed a significant reduction in atherosclerotic plaque size in 7030B-C5 treated ApoE KO mice compared to HFD control group (Fig. 3d). And what's more, a similar reduction of atherosclerotic lesions in aortic root by 7030B-C5 treatment was observed as well (Fig. 3e). Meanwhile, we investigated the expression of PCSK9 and LDLR in the mice liver. As expected, the protein level of PCSK9 was diminished in 7030B-C5 treated mice compared with HFD group, while LDLR protein level was significantly increased (Fig. 3f), consistent with the dramatically decreased circulating PCSK9 level (Fig. 3b). These results were consistent with those observed in C57BL/6 J mice (Suppl Fig. 4). A group of mice treated with 15 mg/kg atorvastatin was applied as a positive control for the en face analysis, and atorvastatin effectively decreased atherosclerotic plaque accumulation in whole aorta with no differences in serum cholesterol levels as in our previous studies [40] (Suppl Fig. 6). Besides, the HFD induced increased serum alanine aminotransferase (AST)/aspartate aminotransferase (ALT) ratio was slightly diminished by 7030B-C5 administration, implying that 7030B-C5 displayed little liver toxicity, which was also proved by H&E staining of liver sections (Suppl Fig. 7). Pharmacokinetic analysis of 7030B-C5 in a single dose of 30 mg/kg was performed and the parameters were shown in Suppl Table 4 with an absorption rate of 23%. Taken together, these results suggested that 7030B-C5 is efficacious in regulating PCSK9 expression and the LDLR level in HFD-induced hyperlipidemic ApoE KO mice with low toxicity and acceptable bioavailability, which may ameliorate abnormal plasma cholesterol metabolism and provide atheroprotection.
Fig. 3.
Administration of 7030B-C5 to ApoE KO mice reduces atherosclerosis progression. Male ApoE KO mice were intragastrically injected with vehicle and 7030B-C5 (10 mg/kg per day, 30 mg/kg per day), respectively, for 12 weeks. At the end of experiment, aorta, serum and liver samples were individually collected and used for the following assays. (a) The body weight course of ApoE KO mice fed an HFD without (control) or with 7030B-C5. *p < 0.05 HFD vs. 10 mg/kg group, #p < 0.05 HFD vs. 30 mg/kg group, (b) Plasma total cholesterol, LDL-C, HDL-C collected from the ApoE KO mice at the 12th week after 7030B-C5 treatment. And serum PCSK9 levels were determined by an ELISA kit. (c) Plasma was pooled per group and the distribution of cholesterol over the individual lipoproteins was determined after separation by fast protein liquid chromatography (FPLC). (d and e) Atherosclerotic plaques in the en face aorta and aortic root were determined by Oil-Red-O staining, and the plaques area were quantified using ImageJ. a, chow; b, HFD; c, 7030B-C5 (10 mg/kg); d, 7030B-C5 (30 mg/kg). Scale bar = 500 μm. (f) Expression of PCSK9 and LDLR protein in the liver was determined by Western blot. Representative images are shown. Values are presented as means ± SEM (n = 10 per group). *p < 0.05 vs. control in the corresponding group.
3.4. 7030B-C5 transcriptionally suppressed PCSK9 expression through HNF1α and FoxO3 modulation
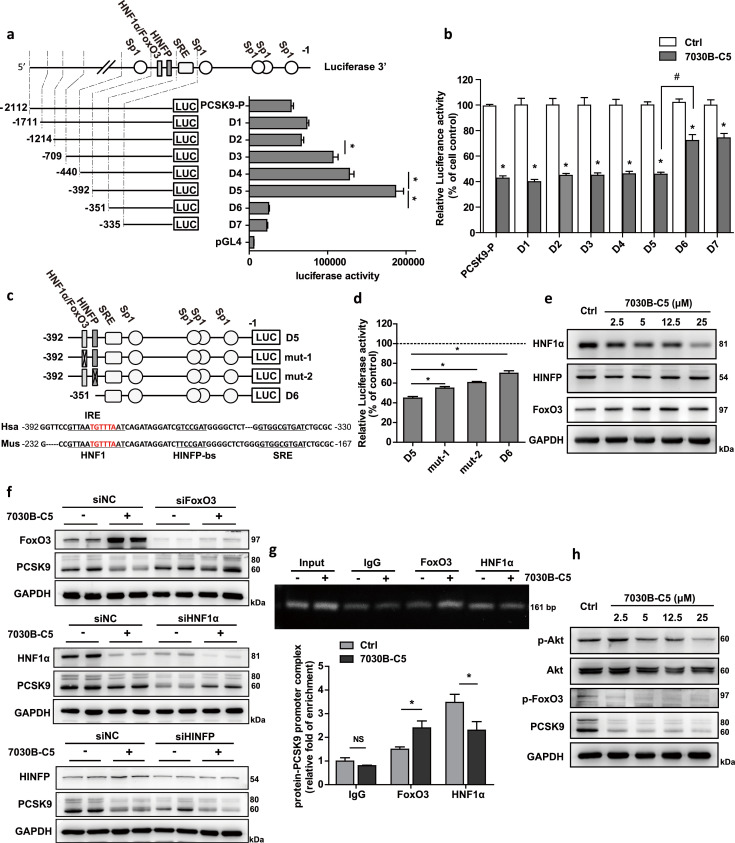
To explore the molecular mechanism underlying the 7030B-C5-mediated PCSK9 down-regulation, we firstly examined the effect of 7030B-C5 on the transcriptional activity of PCSK9 promoter in HepG2 cells. The luciferase-reporter plasmids containing different truncated promoter region (D1-D7, Fig. 4a) of PCSK9 gene were constructed as the 5' flanking full-length promoter region from −2112 to −1 bp (nucleotide +1 corresponds to the A of ATG start codon, the same as below). After transfected into HepG2 cells, the constructs D1-D5 showed gradual increases in the luciferase activities, which was drastically reduced upon removal of the bases between −392 bp and −351 bp (D5 vs. D6) (Fig. 4a). Previous reports have identified the binding sites for HNF1α, FoxO3 and HINFP on the PCSK9 promoter among the region from −392 bp to −351 bp [30,33,34]. The results indicated that the HNF1α recognition motif (HNF1), insulin-responsive element (IRE, recognized by FoxO family transcription factors) and HINFP binding site (HINFP-bs) within this region might be critical for the transcriptional activity of the whole PCSK9 promoter. Furthermore, in order to identify putative 7030B-C5 responsive site in PCSK9 promoter, we applied 7030B-C5 to these constructs transfected cells and detected the luciferase activity. The results showed that the luciferase activity of D1-D5 was markedly reduced by 7030B-C5, and the reduction was impaired when D6 was applied (Fig. 4b). This indicated that the region between −392 bp and −351 bp (D5 vs. D6) was important for 7030B-C5-suppressed PCSK9 promoter activity.
Fig. 4.
7030B-C5 reduces PCSK9 expression transcriptionally in a HNF1α and FoxO3-responsive element-dependent manner. (a) The constructs of human PCSK9 promoter-luciferase reporters. Position +1 was designated as the nucleotide preceding the ATG start codon. Position −1 is the 3′ end of PCSK9 promoter inserts in common to all promoter-reporter constructs. The 5′ ends of the promoters in each promoter-reporter construct are marked by numbers at left, and the name of each construct is shown at right. (b) HepG2 cells were transfected with the PCSK9 promoter-luciferase reporter constructs PCSK9-P D1-D7 for 24 h and then treated with vehicle (0.1% DMSO) or 7030B-C5 for 24 h. (c and d) Deletion analysis of PCSK9 promoter suggested that HNF1/IRE and HINFP-bs might be the key cis-regulatory elements responsible for the regulation of PCSK9 by 7030B-C5. Mut-1 represents core nucleotide sequence of the HNF1/IRE site mutation, mut-2 represents core nucleotide sequence of the HINFP-bs site mutation. (e) HepG2 cells were treated with vehicle or 7030B-C5 in a series of concentrations for 24 h. Expression of HNF1α HINFP and FoxO3 protein were measured by Western blot. (f) HepG2 cells was transfected with siRNA negative control (siNC) or siRNA for the knockdown of FoxO3 (siFoxO3), HNF1α (siHNF1α), HINFP (siHINFP) and the level of FoxO3, HNF1α and HINFP protein was determined by Western blot analysis. (g) HepG2 cells were treated with 7030B-C5 (12.5 μM) for 24 h. Chromatin was isolated followed by immunoprecipitation with normal IgG, anti-HNF1α or anti-FoxO3 antibody. The PCR was conducted with primers for HNF1 or IRE in the PCSK9 promoter. (h) Regulation of PI3K/Akt pathway by 7030B-C5 in HepG2. Representative images are shown. Values are presented as means ± SEM (n = 3). *p < 0.05, #p < 0.05 vs. control in the corresponding group.
To investigate which cis-element was responsible for the effect of 7030B-C5, we mutated the core nucleotide sequence of the HNF1/IRE site and HINFP-bs on the D5 construct respectively (Fig. 4c). The results from luciferase assays showed that 7030B-C5-induced suppression was significantly attenuated in both mutant constructs compared to D5, indicating that all of the three transcription factors HNF1α, FoxO3 and HINFP might be involved in 7030B-C5-mediated PCSK9 transcriptional suppression (Fig. 4d). To investigate the effect of these transcription factors on decreased PCSK9 expression by 7030B-C5, HNF1α, FoxO3 and HINFP protein levels were checked in HepG2 cells treated with or without 7030B-C5. As shown in Fig. 4e, 7030B-C5 treatment for 24 h decreased the level of HNF1α but increased FoxO3 significantly, while the HINFP was hardly altered. In addition, we analyzed PCSK9 expression with HNF1α, FoxO3 or HINFP knockdown respectively using their specific siRNAs. As shown in Fig. 4f, the 7030B-C5-induced inhibition of PCSK9 protein was abolished when FoxO3 or HNF1α siRNA was applied compared to siRNA negative control (siNC) treatment, while HINFP knockdown didn't affect the inhibition of PCSK9 by 7030B-C5. Together, these above data suggested that HNF1α and FoxO3 played a critical role in the 7030B-C5-mediated inhibition of PCSK9 gene expression. To further clarify the effect of 7030B-C5 on the interaction of transcription factors HNF1α and FoxO3 with PCSK9 promoter, the chromatin immunoprecipitation (ChIP) assay was performed. FoxO3 was known to repress PCSK9 transcription by suppressing HNF1α activity, which is important for PCSK9 transcription [34]. In our study, the HNF1α/PCSK9 promoter complex formation was markedly reduced in the 7030B-C5-treated cells (Fig. 4g). What's more, 7030B-C5 significantly increased the FoxO3/PCSK9 promoter complex compared to the vehicle control (Fig. 4g). These results suggested that 7030B-C5 enhanced the accumulation of FoxO3 as well as its interaction with PCSK9 promoter and attenuated both the expression and binding activity of HNF1α to PCSK9 promoter, leading to the reduction of PCSK9 transcription in hepatic cells.
Additionally, protein kinase B (Akt) plays an important role in the phosphorylation of FoxO3 [41]. Akt phosphorylates FoxO3 and causes its cytoplasmic retention leading to inhibition of its transcriptional activity [42]. To further elucidate the regulatory mechanism of 7030B-C5 on PCSK9, the relative level of p-FoxO3/FoxO3 and p-Akt/Akt in HepG2 cells under the treatment of 7030B-C5 were examined and result showed that 7030B-C5 significantly down-regulated p-FoxO3 (Fig. 4h) while increased FoxO3 protein level (Fig. 4e). Moreover, p-Akt level was suppressed by 7030B-C5 in a dose-dependent manner, while total Akt protein was hardly changed (Fig. 4h). These results indicated that 7030B-C5 possibly inhibit the phosphorylation of FoxO3 by inhibiting the Akt signaling pathway which prevented the regulatory function of FoxO3.
All together, these findings demonstrated that 7030B-C5 transcriptionally regulated PCSK9 through the transcription factors HNF1α and FoxO3. On one hand, 7030B-C5 reduced HNF1α protein levels and binding capacity towards PCSK9 promoter, leading to suppression of PCSK9 expression. On the other hand, 7030B-C5 promoted the interaction of FoxO3 with PCSK9 promoter by elevating total FoxO3 protein level and inhibiting the Akt induced FoxO3 phosphorylation, resulted in the reduction of PCSK9 gene transcription as well.
3.5. 7030B-C5 regulated glucose metabolism through FoxO1
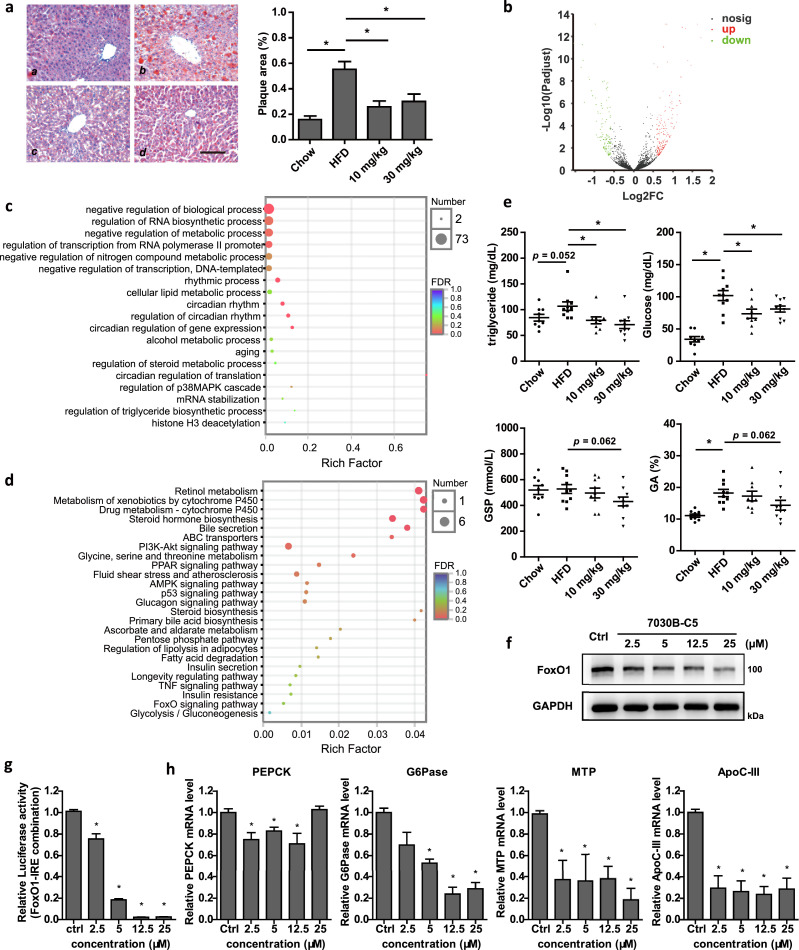
In order to further characterize the pharmacological effect of 7030B-C5 in the liver, we performed Oil Red O staining on histological liver sections to investigate hepatic lipid accumulation. As shown in Fig. 5a, there was a significant reduction of Oil Red O staining in the liver of 7030B-C5-treated mice, which is characteristic of a suppression of hepatic adipose infiltration. To characterize the mechanistic basis of 7030B-C5, transcriptome profiling in the liver of the ApoE KO mice administrated with 7030B-C5 (30 mg/kg) was performed. Results from RNA-seq analysis revealed that there were significant differences in the expression of 321 genes between two groups treated with or without 7030B-C5 (30 mg/kg) (fold change > ± 1.5, p < 0.05), with 204 genes up-regulated and 117 genes down-regulated (Fig. 5b), which were mainly involved in the modulation of metabolic process, transcriptional activator activity, circadian rhythm, etc. analyzed by KEGG and GO enrichment analysis (Fig. 5c and Suppl Fig. 8). Among these genes, we further identified 42 genes involved in glucose and lipid metabolism which were implicated in PI3K/Akt signaling pathway, glucagon signaling pathway, insulin secretion, FoxO signaling pathway, glycolysis/gluconeogenesis, etc. by KEGG and GO enrichment analysis (Fig. 5d and Suppl Fig. 8), indicating the possible involvement of 7030B-C5 in the regulation of glycometabolism.
Fig. 5.
7030B-C5 regulates glucose metabolism through FoxO1. (a) Male ApoE KO mice were intragastrically injected with vehicle and 7030B-C5 (10 mg/kg per day, 30 mg/kg per day), respectively, for 12 weeks. At the end of experiment, liver samples were collected. The liver lipid accumulation was determined by Oil-Red-O staining and quantified by using ImageJ. a, chow; b, HFD; c, 7030B-C5 (10 mg/kg); d, 7030B-C5 (30 mg/kg). Scale bar = 500 μm. Representative images are shown. (b) Volcano plots for all differentially expressed genes in comparison. Horizontal coordinate is the value of fold change between 7030B-C5 group/HFD difference genes and the vertical coordinates is the p value. The left side represents the differentially down-regulated gene, the right side represents the differentially up-regulated gene, and the green and red dots represent the differential gene p < 0.05, blank dots represent the non-significantly differential gene. (c) GO pathway enrichment analysis of 321 differentially expressed genes. (d) KEGG pathway enrichment analysis of 42 differentially expressed genes involved in glucose and lipid metabolism. (e) Male ApoE KO mice were intragastrically injected with vehicle and 7030B-C5 (10 mg/kg per day, 30 mg/kg per day), respectively, for 12 weeks. At the end of experiment, triglycerides, glucose, glycated serum protein (GSP) and glycated albumin (GA) in the serum were collected from the ApoE KO mice at the 12th week after 7030B-C5 treatment. (f) Effects of 7030B-C5 on FoxO1 expression in HepG2 cells. HepG2 cells were treated with 7030B-C5 in a series of concentrations for 24 h. Expression of FoxO1 protein was measured by Western blot. (g) The interaction of FoxO1 with IRE element was determined by luciferase reporter system. (h) HepG2 cells were treated with 7030B-C5 in a series of concentrations for 24 h. The mRNA level of PEPCK, G6Pase, MTP and ApoC-III was measured by RT-qPCR analysis. Values are presented as means ± SEM (n = 10 per group). *p < 0.05 vs. control in the corresponding group. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Interestingly, we found in our study that 7030B-C5 dramatically lowered triglycerides (TG) levels as well as glucose (Glu) levels, glycated serum protein (GSP) and glycated albumin (GA) in ApoE KO mice at the 12th week (Fig. 5e). The significantly lowered TG and Glu levels were already observed at the 8th week (Suppl Fig. 5a). FoxO family transcription factors play a major role in regulating the glucose metabolism and lipid metabolism, and the Insulin/PI3K/Akt/FoxO1 cascade is a classical regulatory pathway for glucose metabolism [43]. Subsequently, we further explored the effects of 7030B-C5 on FoxO1. As shown in Fig. 5f, 7030B-C5 treatment for 24 h significantly decreased FoxO1 protein level. Besides, a recombinant luciferase reporter plasmid named pc-IRE-LE [44] containing 5 × canonical FoxO1 binding site (IRE, 5′-TTGTTTTG-3′) was transfected into HEK293T cells. The luciferase activity was detected and the results indicated that 7030B-C5 markedly suppressed the interaction of FoxO1 with IRE element (Fig. 5g). Then we investigated the effect of 7030B-C5 on the FoxO1 target genes in HepG2 cells, including phosphoenolpyruvate carboxykinase (PEPCK) and glucose 6 phosphatase (G6Pase) which participated in gluconeogenesis process, microsomal triglyceride transfer protein (MTP) and Apolipoprotein C-III (ApoC-III) which were involved in lipid metabolism. As shown in Fig. 5h, G6Pase, MTP and ApoC-III mRNA exhibited a dose-dependent decrease with 7030B-C5 treatment, whereas PEPCK was markedly decreased at lower concentrations. Taken together, all the above results suggested a potential role of 7030B-C5 and a novel mechanism in the crosstalk of glucose and lipid metabolism via FoxO1, FoxO3 and HNF1α.
4. Discussion
Elevated LDL-C is a risk factor for cardiovascular disease [1]. Emerging approaches to inhibit PCSK9 function are currently under investigation for the modulation of lipid metabolism. PCSK9 monoclonal antibodies which prevent PCSK9 interaction with LDLR have been already approved on the market and been proven to reduce circulating LDL-C levels with good efficacy and tolerance, making PCSK9 emerge as a novel therapeutic target for lowering LDL-C levels recently. Several non-antibody treatments including antisense oligonucleotides, small interfering RNAs (siRNAs), mimetic peptides, adnectins and vaccination are also under development at different stages of clinical studies or preclinical researches. An injection of inclisiran (a novel siRNA-based silencer of PCSK9) once every six months was found to reduce PCSK9 level by more than 50%, and produce sustained suppression of LDL-C levels by ~40% among patients with high risk of cardiovascular disease [45]. The ORION-11 trial confirms the ability of inclisiran to reduced LDL-C effectively and safely on a background of statin therapy [46]. However, considering the high cost of mAbs or siRNAs, the small-molecule inhibitors of PCSK9 have attracted a great deal of attention for their cost-effectiveness and ease of administration [47]. Gustafsen et al. proposed that heparan sulfate (HS) mimetics directly interrupted the HS/PCSK9 interaction, which facilitated PCSK9 clustering to LDLR, but with low binding affinities [48]. A recent research on peptidomimetics was also performed to perturb the PCSK9-LDLR binding [49]. Last year, Hingez Therapeutics Inc. received a grant from the National Institutes of Health focusing on discovery of small molecules that block PCSK9-LDLR interactions using their novel high performance computing (HPC)-based platform. However, targeting PCSK9 with small-molecule approaches that can disrupt the interaction of PCSK9 with LDLR is still a great challenge due to the lack of druggable pockets on PCSK9, spurring interest in finding alternative accesses to antagonize PCSK9 function.
Recent years, some researches focused on small-molecule regulators targeting PCSK9 gene expression pathway which are controlled by diverse cellular processes. Pfizer has disclosed a new anti-PCSK9 small molecule which selectively binds to human 80S ribosomal complex to stall on mRNA and generate a dysfunctional PCSK9 peptide [21], [22], [23]. This strategy was innovative but the development of the active compound was stopped for some competitive reasons [24]. Some natural products, such as berberine (BBR) [50], curcumin [51] and tanshinone IIA [52], have been found to reduce PCSK9 transcription and expression, but none of them have been investigated as PCSK9 inhibitors further, probably due to their wide regulatory effects. Eli Lilly's Open Innovation Drug Discovery (OIDD) platform includes a screening module for PCSK9 synthesis inhibition. Its primary phenotypic assay is to evaluate the inhibition of PCSK9 secretion performed on HepG2 cells. The inhibition of non-specific secretion was tested through a luciferase reporter assay. The secondary assays were performed on primary rat hepatocyte [53]. In this study, we implement a PCSK9 transcription inhibitor discovery strategy which is different from that of Lilly. Primarily, we established the cell-based luciferase reporter HTS assay for PCSK9 inhibitor directly targeting PCSK9 transcriptional level and screened the institutional small-molecule compound library. Hits from the screen were then evaluated by the cytotoxicity and dose-effect relation tests. Secondarily, the compounds were verified by the effect on PCSK9 and LDLR protein level and LDL-C uptake in HepG2 cells. Four active compounds with good activities emerged from the screening funnel. For confirmation of the effectiveness of small molecules that inhibit PCSK9, hepatic cells besides HepG2 (Huh7 cells and human primary hepatocytes) in vitro and C57BL/6 J and ApoE KO mice in vivo were applied. 7030B-C5 (its structure and purity were confirmed as in Suppl Fig. 9) exhibited outstanding anti-atherosclerotic effect in ApoE KO mice. The active compounds discovered by this strategy might directly target the regulatory cascade of PCSK9 transcription and ultimately influence the PCSK9 expression. It is conceivable that they may target any key regulators in the PCSK9 transcriptional regulation cascade, e.g. HNF1α, FoxO3 and FoxO1. This is the case for compound 7030B-C5 that some hepatic genes related to lipid and glucose metabolism were affected through FoxO1, resulting multiple changes as shown in Fig. 6. However, to some extent the regulatory activity of 7030B-C5 is specific to the PCSK9 transcriptional regulation cascade, as it showed no inhibition of an ApoB (the structural protein component of LDL) gene promoter-reporter construct (Suppl Fig. 10). Although how to selectively target the transcription processes of PCSK9 without affecting off-target genes to improve safety is still intractable, this discovery strategy has been proved as a robust and pragmatic approach of step-by-step evaluation to facilitate the early elimination of unsuitable compounds and to accelerate the discovery of lead compounds for inhibition of PCSK9 transcription. Considering their druggability, the discovered transcriptional inhibitors of PCSK9 would be further structurally modified to be transformed into drug candidates with higher systemic bioavailability, higher effectiveness and lower toxicity.
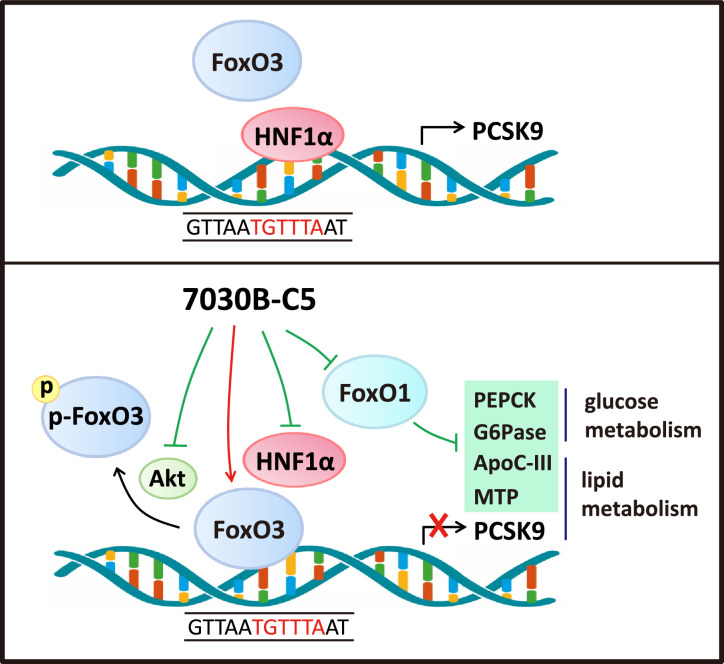
Fig. 6.
Schematic diagram of the regulatory mechanism of 7030B-C5 on lipid and glucose metabolism.
In this work, ApoE KO mice were applied to study the pharmacological effects of discovered transcriptional inhibitors of PCSK9. Although ApoE KO mice are widely used in the evaluation of serum cholesterol level, hepatic lipid accumulation and atherosclerosis progress in lots of researches [54], there are some paradoxical results on the impact of the inhibition of PCSK9 on atherosclerosis in ApoE KO mice. Ason et al. reported that PCSK9 KO in ApoE KO mice revealed little difference in either circulating cholesterol or hepatic LDLR protein levels [55]. Intriguingly, the researches by Denis et al. showed that the aortic CE accumulation was significantly reduced by 39% in PCSK9 KO/e mice, suggesting that “the absence of PCSK9 protects ApoE-deficient mice against atherosclerosis” [56]. Although the changes were not significant, a trend for increased LDLR expression, lower plasma cholesterol levels and plaque size was observed by the loss of PCSK9 [56]. Furthermore, Miranda et al. [57] reported that compound SRT3025 increased hepatic LDLR protein expression by reducing plasma PCSK9 in ApoE-deficient mice fed a high-cholesterol diet for 12 weeks. Their results showed that SRT3025 administration to ApoE KO mice significantly reduced plasma cholesterol level and the plaque accumulation in both thoraco-abdominal aortae en face and of aortic root cross sections, which confers atheroprotection. On the other hand, beyond cholesterol-lowering, PCSK9 have been reported to have pleiotropic effects to affect atherosclerosis lesion size and composition which are independent of ApoE [58,59]. In our study, it was found that daily oral administration of 7030B-C5 for 12 weeks significantly reduced hepatic and plasma PCSK9 level and increased hepatic LDLR expression. Most importantly, 7030B-C5 markedly inhibited lesions in en face aortas and aortic root in ApoE KO mice, suggesting the transcriptional inhibitor of PCSK9 identified here may exert their atheroprotective effect in ApoE KO mice. However, the inhibitory effects on serum cholesterol levels in our research are not significant either, which may be possibly due to a dominant effect of ApoE deficiency. To further investigate the cholesterol-lowering effect of active compound 7030B-C5, APOE*3Leiden.CETP mice with an intact ApoE-LDLR clearance pathway [55] will be a better choice in future studies.
Recently the mechanism of PCSK9 transcriptional regulation has been preliminarily elucidated. Within the region of PCSK9 promoter, several potential transcriptional factor binding sites have been identified, including the sterol regulatory element for SREBP1/SREBP2 interaction [25,27,29], the HNF1α binding element [30], FoxO3 binding site [34] and HINFP binding element [33]. In this study, 7030B-C5 was characterized to mediate PCSK9 transcriptional repression via suppressing HNF1α and promoting FoxO3 level simultaneously, which may compete the same binding site in PCSK9 promoter, thereby ameliorating atherosclerosis. Intriguingly, 7030B-C5 is quite distinctive that it down-regulates hepatic FoxO1, another FoxO family transcription factor while up-regulates FoxO3 level. FoxO family transcription factors are known to have both positive and negative effects on gene regulation. In mammals, there are four FoxOs (FoxO1, 3, 4 and 6) [60]. Although they may share the overlapped binding site in the promoter of the target genes, they show different biological functions in the body. Tao et al. have demonstrated that FoxO3 played a major role in the negative regulation of the PCSK9 gene [34]. In addition, FoxO3 is more prominently associated with tumor suppressor activity when overexpressed [61]. Moreover, FoxO3 is an important longevity factor in invertebrates, as well as mice and humans [62] which is beneficial to human health. FoxO1 is another important member of the FoxO family, which plays a major role in regulating hepatic glucose production response to insulin instead of LDL-C metabolism [63]. When FoxO1 is constitutively expressed in the liver, fasting blood glucose rises [64]. Conversely, liver specific FoxO1 knock-out mice develop fasting hypoglycemia [65]. Therefore, attenuating FoxO1 expression will help improve diabetes, which also inspires plenty of ongoing researches on anti-diabetic drug discovery targeting FoxO1. The dual modulation of lipid and glucose metabolism of 7030B-C5 through bidirectionally regulating FoxO3 and FoxO1 will make it an attractive agent for pharmacological intervention (Fig. 6), as this may contribute to the final effect in retarding the progress of atherosclerosis through mediating integration of hepatic glucose and lipid metabolism. However, the detailed mechanism required further exploration. The elucidation of the modulatory mechanism of the active compounds from the PCSK9 transcription inhibitor discovery strategy might dissect novel mechanisms and facilitate the discovery of new small-molecule PCSK9 inhibitors.
Interestingly, in the transcriptome profiling analysis of treatment with or without 7030B-C5, there are significant differences in the expression of clock, per1, per2, per3,etc. genes, which are mainly involved in the modulation of circadian rhythm (Fig. 5c and Suppl Fig. 8). Correspondingly, human circulating PCSK9 levels were found to display a diurnal rhythm closely synchronous with that of cholesterol synthesis [66]. Recent studies indicated that mutation of genes related to the circadian clock resulted in the disorder of rhythm of many nutrient metabolic-related genes, leading to metabolic disorders or metabolic-related diseases such as diabetes, atherosclerosis, obesity, inflammation and cancer [67,68]. Chaves et al. have confirmed that insulin/PI3K/FoxO3 signaling is required for circadian rhythmicity in the liver, implicating Clock as a transcriptional target of FoxO3 [69]. Given the regulation of 7030B-C5 on PCSK9 through FoxO3, whether 7030B-C5 would regulate circadian rhythm through FoxO3, thus improving the atherosclerosis, is worthy of further study. The results may provide a brand-new mechanism for investigating anti-atherosclerosis compounds.
In conclusion, the current study demonstrated that 7030B-C5, a small-molecule compound, with potential PCSK9 inhibitory activity could serve as an effective modulator to reduce plasma PCSK9 level and ameliorate atherosclerosis via a novel mechanism of action. The discovery and investigation of 7030B-C5 provide an intriguing new approach for small-molecule-mediated modulation of challenging targets such as PCSK9 and FoxOs involved in metabolic-related diseases. In addition, 7030B-C5 suppressed the hepatic lipid accumulation and ameliorated glycometabolism simultaneously, indicating that it has potential to be developed into drug candidate for the treatment of disorders of both lipid and glucose metabolism.
Declaration of Competing Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Acknowledgments
Funding Sources
This work was supported by grants from the National Natural Science Foundation of China (81473214, 81402929, and 81621064), the Drug Innovation Major Project of China (2018ZX09711001-003-006, 2018ZX09711001-007 and 2018ZX09735001-002), CAMS Innovation Fund for Medical Sciences (2016-I2M-2-002, 2016-I2M-1-011 and 2017-I2M-1-008), Beijing Natural Science Foundation (7162129). The funding sources had no role in the study design and execution, data analysis and interpretation, writing of the manuscript and decision to submit results, or any aspect pertinent to the study.
Acknowledgments
The authors are thankful to Xiuyong Fan and Xin Zhen from the Center of National New Drug Screening for their technical supports and stuffs from the animal laboratory for their help in animal feeding and preparation.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102650.
Contributor Information
Li Wang, Email: wangli@imb.pumc.edu.cn.
Bin Hong, Email: hongbin@imb.pumc.edu.cn.
Appendix. Supplementary materials
References
- 1.Bulbulia R., Armitage J. LDL cholesterol targets–how low to go? Curr Opin Lipidol. 2012;23(4):265–270. doi: 10.1097/MOL.0b013e3283556c1b. [DOI] [PubMed] [Google Scholar]
- 2.Zhang D.W., Lagace T.A., Garuti R., Zhao Z., McDonald M., Horton J.D. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat a of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem. 2007;282(25):18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 3.Peterson A.S., Fong L.G., Young S.G. PCSK9 function and physiology. J Lipid Res. 2008;49(7):1595–1599. doi: 10.1194/jlr.CX00001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem. 2004;279(47):48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham D., Danley D.E., Geoghegan K.F., Griffor M.C., Hawkins J.L., Subashi T.A. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat Struct Mol Biol. 2007;14(5):413–419. doi: 10.1038/nsmb1235. [DOI] [PubMed] [Google Scholar]
- 6.Lo Surdo P., Bottomley M.J., Calzetta A., Settembre E.C., Cirillo A., Pandit S. Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH. EMBO Rep. 2011;12(12):1300–1305. doi: 10.1038/embor.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nassoury N., Blasiole D.A., Tebon Oler A., Benjannet S., Hamelin J., Poupon V. The cellular trafficking of the secretory proprotein convertase PCSK9 and its dependence on the LDLR. Traffic. 2007;8(6):718–732. doi: 10.1111/j.1600-0854.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 8.Holla O.L., Cameron J., Berge K.E., Ranheim T., Leren T.P. Degradation of the LDL receptors by PCSK9 is not mediated by a secreted protein acted upon by PCSK9 extracellularly. BMC Cell Biol. 2007;8:9. doi: 10.1186/1471-2121-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abifadel M., Varret M., Rabes J.P., Allard D., Ouguerram K., Devillers M. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34(2):154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 10.Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 11.Cohen J.C., Boerwinkle E., Mosley T.H., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 12.Benn M., Nordestgaard B.G., Grande P., Schnohr P., Tybjaerg-Hansen A. PCSK9 R46L, low-density lipoprotein cholesterol levels, and risk of ischemic heart disease: 3 independent studies and meta-analyses. J Am Coll Cardiol. 2010;55(25):2833–2842. doi: 10.1016/j.jacc.2010.02.044. [DOI] [PubMed] [Google Scholar]
- 13.Chan J.C., Piper D.E., Cao Q., Liu D., King C., Wang W. A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proc Natl Acad Sci U S A. 2009;106(24):9820–9825. doi: 10.1073/pnas.0903849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raal FJ S.E., Dufour R., Turner T., Civeira F., Burgess L., Langslet G., Scott R., Olsson A.G., Sullivan D., Hovingh G.K., Cariou B., Gouni-Berthold I., Somaratne R., Bridges I., Scott R., Wasserman S.M., Gaudet D. PCSK9 inhibition with evolocumab (AMG 145) in heterozygous familial hypercholesterolaemia (RUTHERFORD-2): a randomised, double-blind, placebo-controlled trial. Lancet. 2015;385(9965):331–340. doi: 10.1016/S0140-6736(14)61399-4. [DOI] [PubMed] [Google Scholar]
- 15.Kastelein J.J., Ginsberg H.N., Langslet G., Hovingh G.K., Ceska R., Dufour R. Fast track: editor's choice: ODYSSEY FH I and FH II: 78 week results with alirocumab treatment in 735 patients with heterozygous familial hypercholesterolaemia. Eur Heart J. 2015;36(43):2996–3003. doi: 10.1093/eurheartj/ehv370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kereiakes D.J., Robinson J.G., Cannon C.P., Lorenzato C., Pordy R., Chaudhari U. Efficacy and safety of the proprotein convertase subtilisin/kexin type 9 inhibitor alirocumab among high cardiovascular risk patients on maximally tolerated statin therapy: the ODYSSEY COMBO I study. Am Heart J. 2015;169(6):906–915. doi: 10.1016/j.ahj.2015.03.004. e13. [DOI] [PubMed] [Google Scholar]
- 17.Blom D.J., Hala T., Bolognese M., Lillestol M.J., Toth P.D., Burgess L. A 52-week placebo-controlled trial of evolocumab in hyperlipidemia. N Engl J Med. 2014;370(19):1809–1819. doi: 10.1056/NEJMoa1316222. [DOI] [PubMed] [Google Scholar]
- 18.Desai N.R., Giugliano R.P., Wasserman S.M., Gibbs J.P., Liu T., Scott R. Association between circulating baseline proprotein convertase subtilisin kexin type 9 levels and efficacy of evolocumab. JAMA Cardiol. 2017;2(5):556–560. doi: 10.1001/jamacardio.2016.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazi D.S., Moran A.E., Coxson P.G., Penko J., Ollendorf D.A., Pearson S.D. Cost-effectiveness of PCSK9 inhibitor therapy in patients with heterozygous familial hypercholesterolemia or atherosclerotic cardiovascular disease. JAMA. 2016;316(7):743–753. doi: 10.1001/jama.2016.11004. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee Y., Santos R.D., Al-Rasadi K., Rizzo M. Targeting PCSK9 for therapeutic gains: have we addressed all the concerns? Atherosclerosis. 2016;248:62–75. doi: 10.1016/j.atherosclerosis.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 21.Petersen D.N., Hawkins J., Ruangsiriluk W., Stevens K.A., Maguire B.A., O'Connell T.N. A small-molecule anti-secretagogue of PCSK9 targets the 80S ribosome to inhibit PCSK9 protein translation. Cell Chem Biol. 2016;23(11):1362–1371. doi: 10.1016/j.chembiol.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 22.D.Disney M. Inhibiting translation one protein at a time. Trends Biochem Sci. 2017;42(6):412–413. doi: 10.1016/j.tibs.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Londregan A.T., Wei L., Xiao J., Lintner N.G., Petersen D., Dullea R.G. Small molecule proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors: hit to lead optimization of systemic agents. J Med Chem. 2018;61(13):5704–5718. doi: 10.1021/acs.jmedchem.8b00650. [DOI] [PubMed] [Google Scholar]
- 24.Clinicaltrials. Gov. https://clinicaltrials.gov/ct2/show/NCT02654899. Accessed 20171125.
- 25.Jeong H.J., Lee H.S., Kim K.S., Kim Y.K., Yoon D., Park S.W. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J Lipid Res. 2008;49(2):399–409. doi: 10.1194/jlr.M700443-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Dubuc G., Chamberland A., Wassef H., Davignon J., Seidah N.G., Bernier L. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2004;24(8):1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 27.Costet P., Cariou B., Lambert G., Lalanne F., Lardeux B., Jarnoux A.L. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. J Biol Chem. 2006;281(10):6211–6218. doi: 10.1074/jbc.M508582200. [DOI] [PubMed] [Google Scholar]
- 28.Jump D.B. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr Opin Lipidol. 2002;13(2):155–164. doi: 10.1097/00041433-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Ruscica M., Ricci C., Macchi C., Magni P., Cristofani R., Liu J. Suppressor of cytokine signaling-3 (SOCS-3) induces proprotein convertase subtilisin kexin type 9 (PCSK9) expression in hepatic HepG2 cell line. J Biol Chem. 2016;291(7):3508–3519. doi: 10.1074/jbc.M115.664706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Dong B., Park S.W., Lee H.S., Chen W., Liu J. Hepatocyte nuclear factor 1alpha plays a critical role in PCSK9 gene transcription and regulation by the natural hypocholesterolemic compound berberine. J Biol Chem. 2009;284(42):28885–28895. doi: 10.1074/jbc.M109.052407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong B., Wu M., Li H., Kraemer F.B., Adeli K., Seidah N.G. Strong induction of PCSK9 gene expression through HNF1alpha and SREBP2: mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J Lipid Res. 2010;51(6):1486–1495. doi: 10.1194/jlr.M003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shende V.R., Wu M., Singh A.B., Dong B., Kan C.F., Liu J. Reduction of circulating PCSK9 and LDL-C levels by liver-specific knockdown of HNF1alpha in normolipidemic mice. J Lipid Res. 2015;56(4):801–809. doi: 10.1194/jlr.M052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Liu J. The novel function of HINFP as a co-activator in sterol-regulated transcription of PCSK9 in HepG2 cells. Biochem J. 2012;443(3):757–768. doi: 10.1042/BJ20111645. [DOI] [PubMed] [Google Scholar]
- 34.Tao R., Xiong X., DePinho R.A., Deng C.X., Dong X.C. FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J Biol Chem. 2013;288(41):29252–29259. doi: 10.1074/jbc.M113.481473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao Y., Wang L., Xu Y., Yang Y., Wang L., Si S. Salvianolic acid B inhibits macrophage uptake of modified low density lipoprotein (mLDL) in a scavenger receptor CD36-dependent manner. Atherosclerosis. 2012;223(1):152–159. doi: 10.1016/j.atherosclerosis.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L., Jia X.J., Jiang H.J., Du Y., Yang F., Si S.Y. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol. 2013;33(10):1956–1964. doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H., Zhang J., Du Y., Jia X., Yang F., Si S. microRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis. 2015;243(2):523–532. doi: 10.1016/j.atherosclerosis.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J.H., Chung T.D.Y., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 39.Dong B., Singh A.B., Fung C., Kan K., Liu J. CETP inhibitors downregulate hepatic LDL receptor and PCSK9 expression in vitro and in vivo through a SREBP2 dependent mechanism. Atherosclerosis. 2014;235(2):449–462. doi: 10.1016/j.atherosclerosis.2014.05.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du Y., Li X., Su C., Xi M., Zhang X., Jiang Z. Butyrate protects against high-fat diet-induced atherosclerosis via up-regulating ABCA1 expression in apolipoprotein E-deficiency mice. Br J Pharmacol. 2019 doi: 10.1111/bph.14933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plas D.R., Thompson C.B. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem. 2003;278(14):12361–12366. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Chen W.R., Xing D. A pathway from JNK through decreased ERK and Akt activities for FOXO3a nuclear translocation in response to UV irradiation. J Cell Physiol. 2012;227(3):1168–1178. doi: 10.1002/jcp.22839. [DOI] [PubMed] [Google Scholar]
- 43.Dong X.C., Copps K.D., Guo S., Li Y., Kollipara R., DePinho R.A. Inactivation of hepatic FoxO1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 2008;8(1):65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang M., Zhang J., DU Y., He W., Wang X., Wang L. Construction and application of a cell-based high-throughput screening model for FoxO1 inhibitors. Chin Med Biotechnol. 2019;3(14):199–209. [Google Scholar]
- 45.Ray K.K., Landmesser U., Leiter L.A., Kallend D., Dufour R., Karakas M. Inclisiran in patients at high cardiovascular risk with elevated LDL cholesterol. N Engl J Med. 2017;376(15):1430–1440. doi: 10.1056/NEJMoa1615758. [DOI] [PubMed] [Google Scholar]
- 46.Jia X., Al Rifai M., Gluckman T.J., Birnbaum Y., Virani S.S. Highlights from selected cardiovascular disease prevention studies presented at the 2019 European society of cardiology congress. Curr Atheroscler Rep. 2019;21(12):46. doi: 10.1007/s11883-019-0813-7. [DOI] [PubMed] [Google Scholar]
- 47.Arnett D.K., Blumenthal R.S., Albert M.A., Buroker A.B., Goldberger Z.D., Hahn E.J. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: executive summary: a report of the American college of cardiology/American heart association task force on clinical practice guidelines. Circulation. 2019;140(11):e563–ee95. doi: 10.1161/CIR.0000000000000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gustafsen C., Olsen D., Vilstrup J., Lund S., Reinhardt A., Wellner N. Heparan sulfate proteoglycans present PCSK9 to the LDL receptor. Nat Commun. 2017;8(1):503. doi: 10.1038/s41467-017-00568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taechalertpaisarn J., Zhao B., Liang X., Burgess K. Small molecule inhibitors of the PCSK9 LDLR interaction. J Am Chem Soc. 2018;140(9):3242–3249. doi: 10.1021/jacs.7b09360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cameron J., Ranheim T., Kulseth M.A., Leren T.P., Berge K.E. Berberine decreases PCSK9 expression in HepG2 cells. Atherosclerosis. 2008;201(2):266–273. doi: 10.1016/j.atherosclerosis.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Tai M.H., Chen P.K., Chen P.Y., Wu M.J., Ho C.T., Yen J.H. Curcumin enhances cell-surface LDLR level and promotes LDL uptake through downregulation of PCSK9 gene expression in HepG2 cells. Mol Nutr Food Res. 2014;58(11):2133–2145. doi: 10.1002/mnfr.201400366. [DOI] [PubMed] [Google Scholar]
- 52.Chen H.C., Chen P.Y., Wu M.J., Tai M.H., Yen J.H. Tanshinone IIA modulates low density lipoprotein uptake via down-regulation of PCSK9 gene expression in HepG2 cells. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0162414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amico G., Basile L., Romeo G., Salerno L., Modica M., Siracusa M. Rescuing abandoned molecules as Nav1.7 and PCSK9 inhibitors. J Adv Med Pharm Sci. 2016;5(2):1–10. [Google Scholar]
- 54.Jawien J. The role of an experimental model of atherosclerosis: apoE-knockout mice in developing new drugs against atherogenesis. Curr Pharm Biotechnol. 2012;13(13):2435–2439. [PubMed] [Google Scholar]
- 55.Ason B., van der Hoorn J.W., Chan J., Lee E., Pieterman E.J., Nguyen K.K. PCSK9 inhibition fails to alter hepatic LDLR, circulating cholesterol, and atherosclerosis in the absence of ApoE. J Lipid Res. 2014;55(11):2370–2379. doi: 10.1194/jlr.M053207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Denis M., Marcinkiewicz J., Zaid A., Gauthier D., Poirier S., Lazure C. Gene inactivation of proprotein convertase subtilisin/kexin type 9 reduces atherosclerosis in mice. Circulation. 2012;125(7):894–901. doi: 10.1161/CIRCULATIONAHA.111.057406. [DOI] [PubMed] [Google Scholar]
- 57.Miranda M.X., van Tits L.J., Lohmann C., Arsiwala T., Winnik S., Tailleux A. The Sirt1 activator SRT3025 provides atheroprotection in Apoe-/- mice by reducing hepatic PCSK9 secretion and enhancing LDLR expression. Eur Heart J. 2015;36(1):51–59. doi: 10.1093/eurheartj/ehu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tavori H., Giunzioni I., Predazzi I.M., Plubell D., Shivinsky A., Miles J. Human PCSK9 promotes hepatic lipogenesis and atherosclerosis development via apoE- and LDLR-mediated mechanisms. Cardiovasc Res. 2016;110(2):268–278. doi: 10.1093/cvr/cvw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macchi C., Banach M., Corsini A., Sirtori C.R., Ferri N., Ruscica M. Changes in circulating pro-protein convertase subtilisin/kexin type 9 levels - experimental and clinical approaches with lipid-lowering agents. Eur J Prev Cardiol. 2019;26(9):930–949. doi: 10.1177/2047487319831500. [DOI] [PubMed] [Google Scholar]
- 60.Webb A.E., Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014;39(4):159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J.Y., Zong C.S., Xia W., Yamaguchi H., Ding Q., Xie X. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10(2):138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris B.J., Willcox D.C., Donlon T.A., Willcox B.J. FOXO3: a major gene for human longevity–a mini-review. Gerontology. 2015;61(6):515–525. doi: 10.1159/000375235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang W., Patil S., Chauhan B., Guo S., Powell D.R., Le J. FoxO1 regulates multiple metabolic pathways in the liver. J Biol Chem. 2006;281(15):10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 64.Matsumoto M., Han S., Kitamura T., Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest. 2006;116(9):2464–2472. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsumoto M., Pocai A., Rossetti L., Depinho R.A., Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor FoxO1 in liver. Cell Metab. 2007;6(3):208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 66.Persson L., Cao G., Stahle L., Sjoberg B.G., Troutt J.S., Konrad R.J. Circulating proprotein convertase subtilisin kexin type 9 has a diurnal rhythm synchronous with cholesterol synthesis and is reduced by fasting in humans. Arterioscler Thromb Vasc Biol. 2010;30(12):2666–2672. doi: 10.1161/ATVBAHA.110.214130. [DOI] [PubMed] [Google Scholar]
- 67.Turek F.W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oishi K., Ohkura N., Wakabayashi M., Shirai H., Sato K., Matsuda J. CLOCK is involved in obesity-induced disordered fibrinolysis in ob/ob mice by regulating PAI-1 gene expression. J Thromb Haemost. 2006;4(8):1774–1780. doi: 10.1111/j.1538-7836.2006.02032.x. [DOI] [PubMed] [Google Scholar]
- 69.Chaves I., van der Horst G.T., Schellevis R., Nijman R.M., Koerkamp M.G., Holstege F.C. Insulin-FOXO3 signaling modulates circadian rhythms via regulation of clock transcription. Curr Biol. 2014;24(11):1248–1255. doi: 10.1016/j.cub.2014.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.