Key Points
Roquin-1 in recruited mø alleviated liver IR injury.
Roquin-1 modulated the polarization of mø via AMPK–STAT3–mTOR.
Roquin-1 promoted the ubiquitination of AMPKα and inhibited its activation.
Abstract
With the development of liver surgery, ischemia–reperfusion (IR) injury has received increasing attention. Roquin-1 has been shown to play an important role in innate immune and immune balance. We demonstrate that Roquin-1 expression increased at 1 h after IR and then decreased in C57B/L mice. The immunofluorescence double-label showed that Roquin-1 was mainly expressed in macrophages (mø). Furthermore, we used clodronate liposomes to remove mø, and injected the bone marrow–derived mø (BMDM) through the tail vein in 1 h before IR. We found that liver IR injury was aggravated by Roquin-1 interference. The results of PCR and ELISA suggested that after interference with Roquin-1, mø increased toward M1 and decreased toward M2. Then, interference with Roquin-1 promoted the polarization of mø to M1 and inhibited the polarization of M2. By Western blot technology and AMPKα and mTOR inhibitors, we found that Roquin-1 promotes the phosphorylation of mTOR and STAT3 by inhibiting the phosphorylation of AMPKα. We used AICAR to activate AMPKα in mø and found that the level of ubiquitination of AMPKα was decreased after activation of AMPKα. Furthermore, by bioinformatics methods, we identified potential ubiquitination sites on AMPKα. By the point mutation experiments in vitro, we confirmed that the ubiquitination of these sites is regulated by Roquin-1. Meanwhile, Roquin-1 interference inhibited the activation and function of AMPKα. This topic describes the protection of liver IR injury by Roquin-1 and discusses its main mechanism for regulating AMPKα activity through ubiquitination and affecting the polarization of mø.
Introduction
Hepatocellular injury caused by ischemia–reperfusion (IR) is the result of a highly complex cascade of events that is triggered when the liver is exposed transiently to hypoxia then reperfused with oxygenated blood. It is an important clinical problem that affects hepatic surgeries and liver transplantation and frequently induces the injury of remote organs, including kidney, lung, and heart. In particular, acute kidney injury after liver IR is very common (40–85% incidence) and greatly increases patient mortality and morbidity during perioperative period (1–3). No specific treatment is available to prevent or reduce hepatic IR injury (IRI) and current management is based on supportive care. Thus, it is important to better understand the mechanisms that underlie liver IRI.
Macrophages (mø), the largest quantity parenchymal cells in liver, play an important role in hepatic pathophysiological process. Their proinflammatory immune response modulates diversity of hepatic sterile inflammatory disease, including IR, cirrhosis, and so on (4–6). Previous studies found that mø could be divided into two polarized phenotypes (M1 and M2) by its function. M1 cells secrete the proinflammatory facts, such as TNF-α, IL-6, IFN-γ reactive oxygen species, reactive nitrogen species, and chemotactic factor, to protect pathogen and promote inflammatory response. Meanwhile, M2 cells alleviate tissue inflammation and promote tissue repair by the secretion of anti-inflammatory facts (TGF-β, IL-10) (6, 7). In liver IRI, multiple-signal mechanisms play critical role in the polarization of mø. Recent research demonstrated that overexpression of HO-1 suppressed the polarization of M1 and promoted the polarization of M2, and then relieved liver IRI (8). Our previous study found that the endoplasmic reticulum stress activated the release of inflammatory facts and aggravated liver IRI (9, 10). As such, mø serve as a modulator in IRI and may be a therapeutic option for inhibition of liver IR injury.
Roquin-1, which was originally discovered in a mutational screen for genetic factors contributory to systemic lupus erythematosus-like symptoms in mice, is an E3 ubiquitin (11). Previous studies have uncovered a multifactorial process by which Roquin-1 contributes to the maintenance of immune homeostasis. Roquin-1 binds to several key immune-relevant mRNA targets through its ROQ domain modulating their stability and influencing mø function and the peripheral homeostasis of T cells and B cells (12). Roquin-1 in mø has been shown to regulate a set of target transcripts that is shared by Roquin-2 and the RNase REGNASE-1, including the proinflammatory cytokines IL-6 and TNF-α mRNA (13, 14). Moreover, Sanroque mice deficient in Roquin-1 RNA regulatory activity show increased susceptibility to LPS-induced systemic lethal inflammation, associated with abnormally high TNF-α production in CD11b high mø (15). To date, no data are available concerning the role of Roquin-1 in hepatic IRI. Understanding the mechanisms by which Roquin-1 regulate mø polarization and function might help in devising ways to modulate polarization and function in the therapeutic schedule of liver IRI.
In our study, we hypothesized that Roquin-1 in mø might be involved in the pathogenesis of liver IRI and that Roquin-1 might protect the liver from IRI by reducing the polarization of M1 and promoting the polarization of M2. Our data show that mø are activated and polarized to M1 after liver IR and that Roquin-1 modulate the balance between M1 and M2 in vitro. In vivo, the mø that knocks down the expression of Roquin-1 significantly promotes liver IR-induced damage, with associated in the up-polarization of M1 and down-polarization of M2. This represents a novel mechanism by which Roquin-1 modulates the immune homeostasis to alleviate the liver IRI.
Materials and Methods
Mice
Male wild-type, 6–8-wk-old mice (C57BL/6) were purchased from the model research center of Nanjing University. All animal use and procedures described were reviewed and approved by the Animal Care and Use Committee of Nanjing Medical University and following the National Institutes of Health guidelines. All animals and resulting samples were assigned a number that did not reveal the group allocation so that analyses were performed by blinded investigators. All the experiment protocols were approved by the Animal Use and Care Committee of Nanjing Medical University.
Liver IRI model and mø injection in mice
As previously described (16), we used the established nonlethal model of segmental (70%) hepatic warm IR. The 6–8 wk-old C57BL mice (n = 6 per group) were anesthetized by ether inhalation, and then interrupted the artery and portal venous blood supply to the left and middle liver lobes were interrupted for 60 min. A warming pad and heat lamp were used to maintain the body temperature at 37°C. At the end of the predetermined period following reperfusion, we anesthetized the mice via ether inhalation and euthanized by cervical dislocation for tissue and plasma collection. Then, we suspended 1 × 106 mø into 100 μl PBS and injected them through the tail vein at 1 h before liver IR. (n = 6 per group).
Depletion of resident peritoneal mø and Kupffer cells by cloronate liposomes
Mice were injected i.p. with clodronate liposomes (400 μl/mouse). To avoid infection of clodronate liposome-treated mice with commensal bacteria, mice were then fed with drinking water containing ampicillin (1 mg/ml) until sacrifice. Depletion of resident peritoneal mø and Kupffer cells were verified by using flow cytometry and immunohistochemistry, respectively, 48 h after injection (Supplemental Fig. 1A, 1B).
Histology and immunofluorescence staining in tissue
In histology, the mice liver was fixed in paraformaldehyde, embedded in paraffin, prepared in 5-μm-thick sections, and stained with H&E. To analyze the change in size and morphology of IR area, we used light microscopy and Suzuki score (17) to evaluate the tissue sections.
By immunofluorescence staining, primary rabbit anti-mouse RC3H1 mAb (Invitrogen), primary mouse anti-mouse F4/80 mAb/Ly6c mAb (Cell Signaling Technology), primary mouse anti-mouse Clec4F mAb (Abcam), and primary mouse anti-mouse CD11b mAb (Abcam) were used to identify the expression of Roquin-1 in mø. After incubation with secondary Ab (Cell Signaling Technology), the samples were premounted with VECTASHIELD medium with DAPI. Then, a Leica TCS SP8 Confocal Laser Scanning Microscope (Leica Microsystems) was used to analyze the samples.
Hepatocellular function assay
The level of serum alanine transaminase (ALT) and aspartate transaminase (AST) were measured by automatic biochemistry analyzer (Beckman Coulter).
Analysis of cytokines
According to the manufacturer’s protocols, ELISA kits from BD Bioscience (NJ) were used to examine the inflammatory cytokines in serum and supernatant.
mø immunomagnetic beads separation
mø separation was performed by the MACS Macrophage Isolation Kit. First, the mononuclear cell suspension was obtained by Ficoll density gradient centrifugation. Then, 36 μl of sorting buffer and 4 μl of FcR blocking were resuspended per 107 cells. An additional 10 μl of Macrophage (Peritoneum) Biotin-Ab Cocktail was added and incubated for 10 min at 4°C. We washed it twice by buffer and resuspended it to 80 μl. Furthermore, 20 μl of Anti-Biotin MicroBeads was added, incubated at 4°C for 15 min, washed twice by buffer, and resuspended to 108/500 μl. Last, we used the magnetic bead sorting column for sorting.
Bone marrow–derived mø isolation and in vitro transfection
Murine bone marrow–derived mø (BMDM) were used to isolate and culture primary hepatocytes by the protocol described by Yue et al. (18). Cells (1 × 106 per well) were cultured for 7 d and then transfected with 100 nM Rc3h1–small interfering RNA (siRNA) (GenePharma) using Lipofectamine 3000 regent (Invitrogen). The nonspecific siRNA was used as control.
Induction of the polarization in mø
We performed the polarization of mø in vitro. In brief, on day 7, we changed the medium to fresh stimulation medium: for M1 activation, we used DMEM containing 10% FBS and 100 ng/ml LPS (Sigma, St. Louis, MO), and for M2 activation, we used DMEM containing 10% FBS with 10 ng/ml IL-4 (eBioscience, CA). After 24 h, the mø were stained with anti-CD11b PE/cy7, anti-CD11c PerCP/cy5.5, anti-F4/80 PE, and anti-CD206 allophycocyanin (BioLegend) and then measured by the FACSCanto flow cytometer (BD Bioscience) and Critical Care Medicine Flow Cytometry Core.
Isolation and culture of primary hepatocytes
We used 6–8-wk-old male C57B/L mice (n = 6) to isolate and culture primary hepatocytes by the protocol described by Czaya et al. (19). In brief, we used ether to perform inhalation anesthesia. We then cannulated the inferior vena cava and transected the portal vein to allow for adequate drainage of blood and perfusion media. We perfused the liver with ∼30 ml of liver perfusion media (Life Technologies), then switched to liver digestion media (Life Technologies). Within a cell culture hood, we transferred the liver from the 50 ml polypropylene conical tube containing digestion media into a sterile tissue culture dish and removed undigested tissue particles and cell debris. The cells were then plated in 2 ml of Williams’ Media E 1× (Life Technologies) that contained primary hepatocyte thawing and plating supplements (Life Technologies) on a collagen-coated six-well plate. After 4 h, the medium was changed to Williams' Media E 1× that contained primary hepatocyte maintenance supplements (Life Technologies).
Simulation of the liver IR hepatocyte microenvironment
We used supernatants from primary hepatocytes exposed to hypoxia-reoxygenation (H/R) as described by Chen et al. (20) to mimic the liver IR microenvironment.
To generate the H/R conditions, we washed the primary hepatocytes three times with PBS and used the AnaeroPack-Anaero (Mitsubishi Gas Chemical, Tokyo, Japan) to absorb O2 and produce CO2. Following the manufacturer’s instructions, a pink oxygen indicator that turned blue when the O2 concentration was >5% was used to detect O2. We placed the culture plate in an incubator with 5% CO2/95% N2 under constant temperature and humidity for 3 h, then removed the plate from the hypoxia chamber and placed it in a normoxic environment for 6 h. Serum-free medium was applied to mimic the ischemic condition. At the conclusion of the reoxygenation period, we collected the supernatants to stimulate mø.
The use of AMPK/mTOR inhibitor
The AMPK inhibitor Compound C 10 mg (Selleck, Houston, TX) was dissolved in 0.2 ml of DMSO and thoroughly dissolved by shaking. After adding 9.8 ml of PBS, a working solution having a concentration of 1 mg/ml was placed. The mTORCl inhibitor Rapamycin 10 mg (Selleck) was dissolved in 0.2 ml of DMSO and diluted with PBS to prepare a 1 mg/ml working solution. BMDM were seeded in six-well plates (5 × 105 per well). The next step was to put them into the cell culture incubator and wait until the cell plating area was ∼70–80%, after which the six-well plates could be taken out to start the test. Next, the medium in the six wells was discarded and the prepared working solution was added (Compound C 2 μM, Rapamycin 10 nM) for 24 h.
Immunoprecipitation
The total protein in the cells is lysed. A corresponding amount of Agrose-protein A/G (Thermo Fisher Scientific) was taken into the tube, resuspended in 500 μl × PBS, and washed twice at 4°C, 5000 rpm × 2 min, and the supernatant was aspirated and used. The lysate was added to a new tube, and the appropriate amount of Ab and protein A/G beads were added according to the instructions and incubated overnight at 4°C. Next, the medium was centrifuged at 2500 rpm for 10 min at 4°C and the supernatant was removed. One milliliter of the lysate was washed five times. Loading buffer (2 μl) was added to a 20-μl, 100°C water bath for 10 min.
Plasmid construction
Vector sequence determination and primer synthesis were performed by Hanheng Biotechnology. The specific operation process is as follows (Table I):
Table I. Carrier clone sequence primers.
| Vector | Primer | Cleavage Site |
|---|---|---|
| pLenti-FLAG-AMPKα1 | 5′-CTAGCTAGCATGCGCAGACTCAGTTCC-3′ | NheI/AscI |
| 5′-TTGGCGCGCCTTATTGTGCAAGAATTTT-3′ | ||
| pLenti-FLAG-AMPKα2 | 5′-CTAGCTAGCATGGCTGAGAAGCAGAAG-3′ | NheI/AscI |
| 5′-TTGGCGCGCCTCAACGGGCTAAAGTAGT-3′ | ||
| pLenti-FLAG-Roquin-1 | 5′-GGCTCTAGAATTCCGCAATGCCTGTACAAGCTCC-3′ | NheI/AscI |
| pLenti-FLAG-Roquin-1133−1130 | 5′-ACTACTCAGAGTGTTCTGGAATTCCCAATGCAGAG GAAACTT-3′ | NheI/AscI |
The PCR target gene was designed by using cDNA library as template, and the target band was determined by agarose gel electrophoresis. The PCR product was recovered using a DNA recovery kit. The recovered PCR product and the plasmid were digested with restriction endonucleases at 37°C for 2–4 h. The enzyme-cut products were subjected to agarose gel electrophoresis, and the DNA gel recovery kit was used to recover the target gene fragment and the vector fragment. DNA recovery was detected by agarose gel electrophoresis. The fragment and the vector were added to the T4 ligase reaction system at a molar ratio of 3:1 and ligated overnight at 4°C. The ligation product was transformed into Escherichia coli competent cells and plated on resistant plates. Colony PCR detected the target fragment, amplified the positive clone, and extracted the plasmid for sequencing.
The full-length genes of AMPKα1, AMPKα2, Roquin-1, and Roquin-1133−1130 were cloned into plasmids such as pLenti-FLAG 6.3 for eukaryotic expression.
Point mutation plasmid construction
The construction of the point mutation plasmid was completed by Hanheng Biotechnology. The specific experimental steps are as seen in Table II:
Table II. PCR system and condition.
| PCR System | |
|---|---|
| Ingredient | Usage (μl) |
| DEPC water | Up to 50 |
| 10× buffer | 5 |
| 2 mM dNTPs | 4 |
| Primer | 2 |
| Template plasmid | 0.5 |
| KOD plus | 1 |
| Reaction Condition | ||
|---|---|---|
| Step | Condition | No. of Cycles |
| Denaturation | 95°C, 1 min | 1 |
| Extension | 95°C, 40 s | 35 |
| 60°C, 1 min | ||
| 68°C, 2 min | ||
| Extension | 68°C, 10 min | 1 |
One microliter of DpnI endonuclease was added to the PCR product and incubated for 2 h. Five microliters of transforming competent cells were plated on the resistant plates.
Lentivirus package
We used the Lentivirus (Hanheng Biotechnology) to package the plasmid. At first, we cultured 293T cells to a density of 90% and then passaged at a ratio of 1:3. Then, we replaced the fresh medium 1–2 h before transfection and transfected the cells until the cells grew to 90% density. According to l LipoFiter instruction (Hanheng Biotechnology), 10 μl of psPAX2, pMD2.G, and pHBLV (Hanheng Biotechnology) were added. We replaced fresh medium (DMEM + 10% FBS) after 6 h. Virus supernatants were collected twice, at 48 and 72 h after transfection (replacement of fresh medium after 48 h collection), collected, and filtered through a 0.45-μm filter in a 40-ml ultracentrifuge tube and centrifuged at 4°C, 72,000 × g for 120 min. The virus pellet was resuspended in 500 ml of fresh medium and stored at −80°C even in liquid nitrogen.
Lentiviral infection
Cells were plated at 37°C overnight as needed for experimentation, and cells were confluent to 70% within 24 h. The next step was to aspirate the original culture medium, add 1/2 vol of fresh medium, and add polybrene at a final concentration of 5–8 μg/ml to the fresh medium, and then convert the corresponding virus stock volume according to the selected multiplicity of infection (15), and Add the virus stock to the cells by volume and shake well. The virus solution was added to the cells and directly inoculated in a 37°C incubator for 4 h. The cells were removed, and an additional 1/2 vol of fresh medium was added directly to the virus-containing medium. After 12 h, the virus-containing medium was aspirated and replaced with fresh whole medium (without polybrene) and cultured at 37°C. After 72 h of infection, the fluorescence expression efficiency of GFP/RFP was detected by fluorescence microscopy.
Transfection of siRNA
One day prior to transfection, 4–5 × 104 cells were seeded in 24-well plates, and 0.5 ml of DMEM cell medium containing FBS and antibiotics was allowed to reach 70 confluence within 24 h. Next, 20 pmol of siRNA was added to 50 μl of DMEM serum-free medium and mixed gently. Then, we diluted 1 μl of Lipofectamine 2000 reagent (Thermo Fisher Scientific ) with 50 μl of serum-free DMEM, mixed gently, and let stand for 5 min at room temperature. The diluted siRNA was mixed with the RNAi-Mate reagent, gently mixed, and allowed to stand at room temperature for 20 min. One hundred microliters of the siRNA/Lipofectamine complex was added to the wells of the culture plate containing the cells and the medium, and the cell culture plate was gently shaken back and forth. The cells were incubated in a CO2 incubator at 37°C for 4–6 h, the complex was removed, and the medium was changed; after 24 h, other experimental steps after transfection were performed.
The small interfering Roquin-1 (si-Roquin-1) sequence (5′-3′) was as follows: 5′-GCACCAGAAUCCUCAGCAATTUUGCUGAGGAUUCUGGUGCTT-3′.
Qualitative RT-PCR analysis
According to the protocol recommended by the manufacturer, TRIzol reagent (Invitrogen) was used to extract total RNA from cells and tissues. Then RNA was reverse-transcribed into cDNA using HiScript Q RT SuperMix for qPCR (Vazyme). Real-time quantitative PCR analysis was performed using a SYBR RT-PCR kit (Vazyme) and the LightCycler 480II Real-Time PCR system (Roche). Primer sequences used for amplification were listed in Supplemental Table I.
Western blot analysis
RIPA lysis buffer (Beyotime, Shanghai, China) was used to isolated protein from cells and tissues. Total protein levels were quantified using a Bio-Rad (Hercules, CA) protein assay, and equal amounts of protein were loaded and separated using 10% (pPI3K, AMPK) or 8% (p-mTOR) SDS-PAGE (Beyotime), followed by immunoblotting with the appropriate Abs. After incubation with primary Ab (Cell Signaling Technology) overnight, we washed membrane and incubated it with HRP-conjugated secondary Abs (Cell Signaling Technology). Then, chemiluminescence kit (Thermo Fisher Scientific, CA) was used to detect the fluorescence. Western blots (WBs) were quantified using Image Lab (Bio-Rad).
Flow cytometer
For measuring differentiation of mø, the flow cytometer was used. We used anti-CD11b PE/cy7, anti-CD11c PerCP/cy5.5, anti-F4/80 PE, and anti-CD206 allophycocyanin (BioLegend) to stain BMDM. Staining was performed in PBS supplemented with 0.1% BSA and 0.02% sodium azide for 30 min at 4°C. Intracellular staining was performed by the protocol described by the manufacturer (BioLegend).
Cells were examined on the FACS Canto flow cytometer and analyzed by FlowJo v10.0.7 software. mø were defined as CD11b+F4/80+ cells, M1 mø were defined as CD11b+F4/80+CD11c+CD206− cells, and M2 mø were defined as CD11b+F4/80+CD11C−CD206+ cells.
Statistical analysis
Data were expressed as means ± SD. We used the Mann–Whitney U test, Student t test, and one-way ANOVA test to compare treatment groups. Statistical analysis was performed using SPSS (IBM, Armonk, NY) and GraphPad Prism 6 (GraphPad Software). A p value <0.05 was considered statistically significant.
Results
The function of Roquin-1 in liver IRI
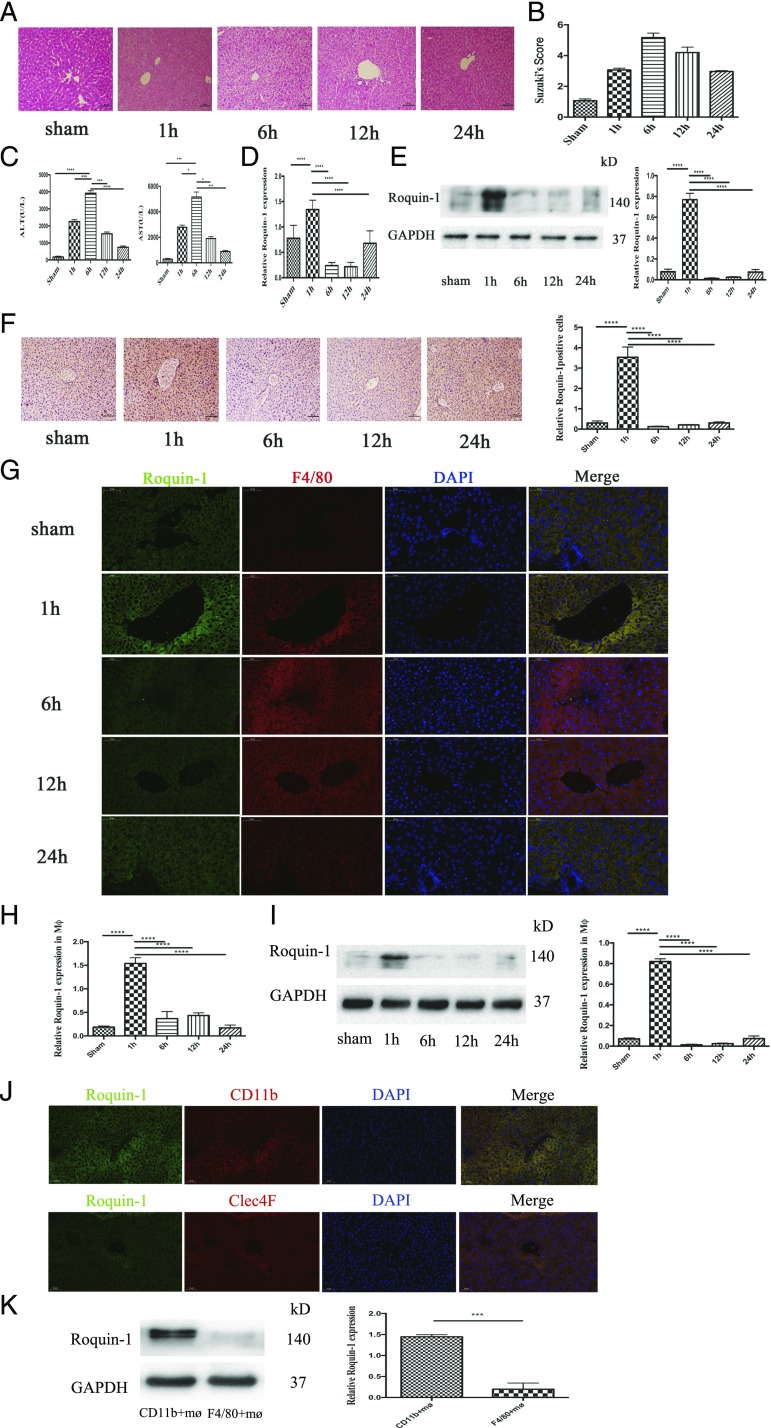
To investigate whether Roquin-1 is involved in the development of liver IR. We established a mouse liver IR model, using H&E staining and serum ALT and AST to test the success of the animal model (Fig. 1A–C). The change of Roquin-1 during liver IR was detected by PCR, WB, and immunohistochemistry (Fig. 1D–F). The study found that Rqouin-1 significantly decreased after 1 h of liver IR and recovered slightly at 24 h. Furthermore, to clarify which cells of the liver Roquin-1 are significantly expressed, we used immunofluorescence double labeling (Fig. 1G, Supplemental Fig. 1C). The results showed that Roquin-1 was expressed in liver mø. Then, we separated the mø from liver and used PCR and WB to detect the expression of Roquin-1. The results demonstrated that Roquin-1 expressions in mø from liver were equally to its expression in liver tissue (Fig. 1H, 1I). Furthermore, to distinguish whether the observations are from infiltrating mø or resident mø (Kupffer cells), we used CD11b (infiltrating mø marker) and Clec4F (resident mø marker) to isolate the infiltrating mø or intrahepatic mø. The immunofluorescence double labeling and WB results showed that Roquin-1 in 1 h after liver IR was mainly expressed in CD11b+ mø (Fig. 1J, 1K).
FIGURE 1.
The change of Roquin-1 in liver IRI. (A) H&E staining of liver in mice liver IR (n = 6). Scale bars, 100 μm. (B) Suzuki score in mice liver IR (n = 6). (C) Serum ALT and AST levels in mice liver IR (n = 6). (D–F) Quantitative RT-PCR, WB, and immunohistochemistry results suggest that Roquin-1 expression is increased 1 h after liver IR, and decreased at 6 and 12 h, 24 h restore (n = 6). Scale bars, 100 μm. (G) Immunofluorescence double-labeled Roquin-1 (green) and mø (red). Roquin-1 is primarily expressed in hepatic mø (n = 6). Scale bars, 50 μm. (H and I) Quantitative RT-PCR and WB in the mø suggest that Roquin-1 expression is increased 1 h after liver IR, and decreased at 6 and 12 h, 24 h after liver IR (n = 6). (J and K) The immunofluorescence double labeling and WB results showed that Roquin-1 in 1 h after liver IR was mainly expressed in CD11b+ mø. Scale bars, 50 μm. The data are expressed as the mean ± SD. *p < 0.01, **p < 0.01, ***p < 0.001, ****p < 0.0001.
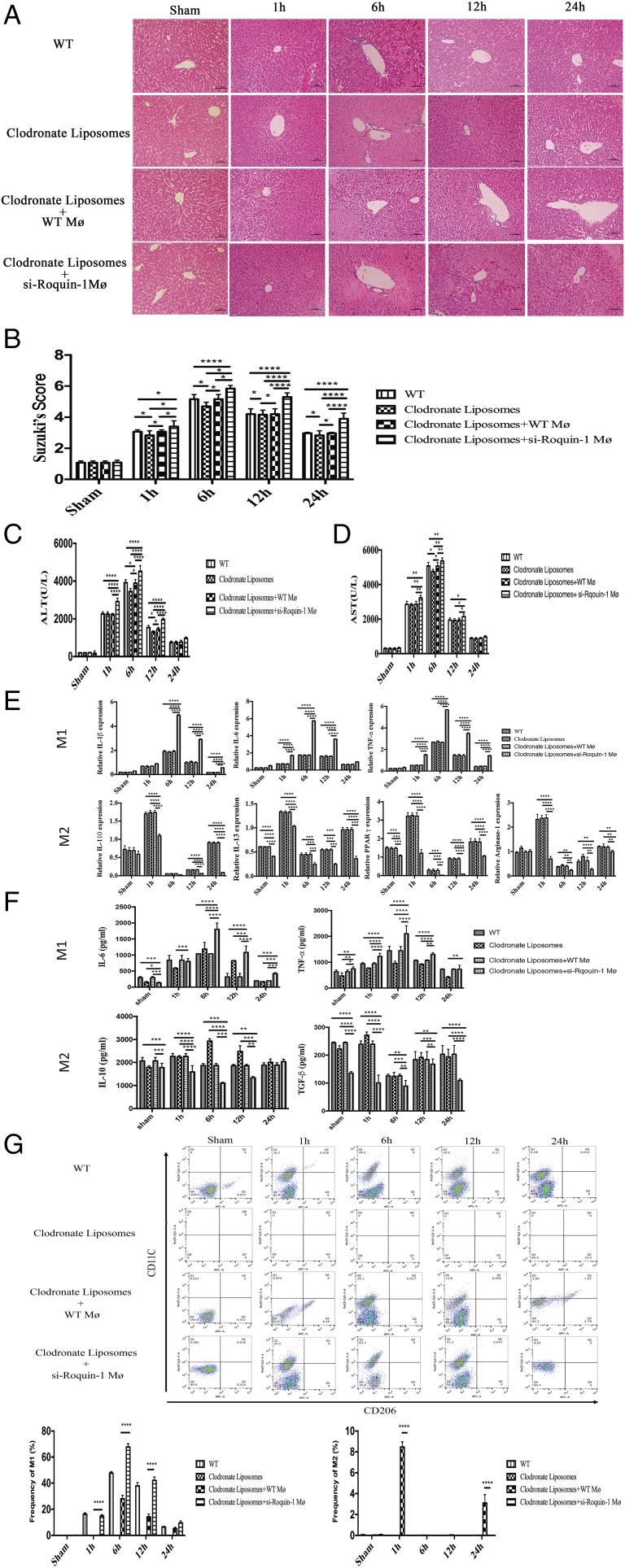
To confirm the role of mø in liver IR, we used liposomes to scavenge mø in mice and interfered with Roquin-1–expressing mø by tail vein injection 1 h before liver IR. H&E (Fig. 2A), Suzuki score (Fig. 2B), and serum ALT (Fig. 2C) and AST (Fig. 2D) results showed that treatment with liposomes alleviates liver IR injury, and injection of wild-type (WT) mø could remove the function of liposomes. Furthermore, we found that IRI in mice injected with si-Roquin-1 mø was significantly worse than that in other three groups (WT, liposome group, and liposome + WT mø group) (Tables I, II).
FIGURE 2.
The effect of Roquin-1 in liver IRI. (A and B) Representative images of liver IR damage and Suzuki score in the WT, clodronate liposomes group, clodronate liposomes + WT mø group, and clodronate liposomes + si-Roquin-1 mø group after liver IR (n = 6). Scale bars, 100 μm. (C and D) Quantification of serum ALT and AST levels in WT, clodronate liposomes group, clodronate liposomes + WT mø group, and clodronate liposomes + si-Roquin-1 mø group after liver IR (n = 6). (E) Quantification of the liver mRNA levels of M1 marker (IL-1β, IL-6, and TNF-α) and M2 marker (IL-10, IL-13, and PPARγ) in WT, clodronate liposomes group, clodronate liposomes + WT mø group, and clodronate liposomes + si-Roquin-1 mø group after liver IR (n = 6 mice per group). (F) ELISA in serum suggests that after injection of mø that interferes with Roquin-1 expression, M1 polarization and function are enhanced during liver IR, and M2 polarization and function are weakened (n = 6). (G) FACS analysis showed si-Roquin-1 promoted the polarization of M1 in the liver IR proceeding and inhibited the polarization of M2 in 1 and 24 h during liver IR (n = 6). The data are expressed as the mean ± SD. *p < 0.01, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To clarify the relationship between Roqiuin-1 and mø subtypes, we used PCR and ELISA to detect the changes of M1/M2-related markers and serum cytokines in the WT group, clodronate liposomes group, clodronate liposomes + WT mø, and clodronate liposomes + si-Roquin-1 mø group. Compared with the WT group, clodronate liposomes, and clodronate liposomes + WT mø groups at 1 h after IR, M1-related markers (IL-1β, IL-6, and TNF-α) and cytokines (IL-6 and TNF-α) in the clodronate liposomes + si-Roquin-1 mø group were significantly elevated; M2 markers (IL-10, IL-13, Arg-1, and PPARγ) and cytokines (IL-10, TGF-β) decreased significantly (Fig. 2E, 2F). Furthermore, we used FACS to analysis the population of M1 and M2 in liver IR. The results showed that si-Roquin-1 promoted the polarization of M1 in the liver IR proceeding and inhibited the polarization of M2 in 1 and 24 h during liver IR (Fig. 2G).
Roquin-1 regulate mø polarization and function via the AMPKα/mTOR/STAT3 pathway
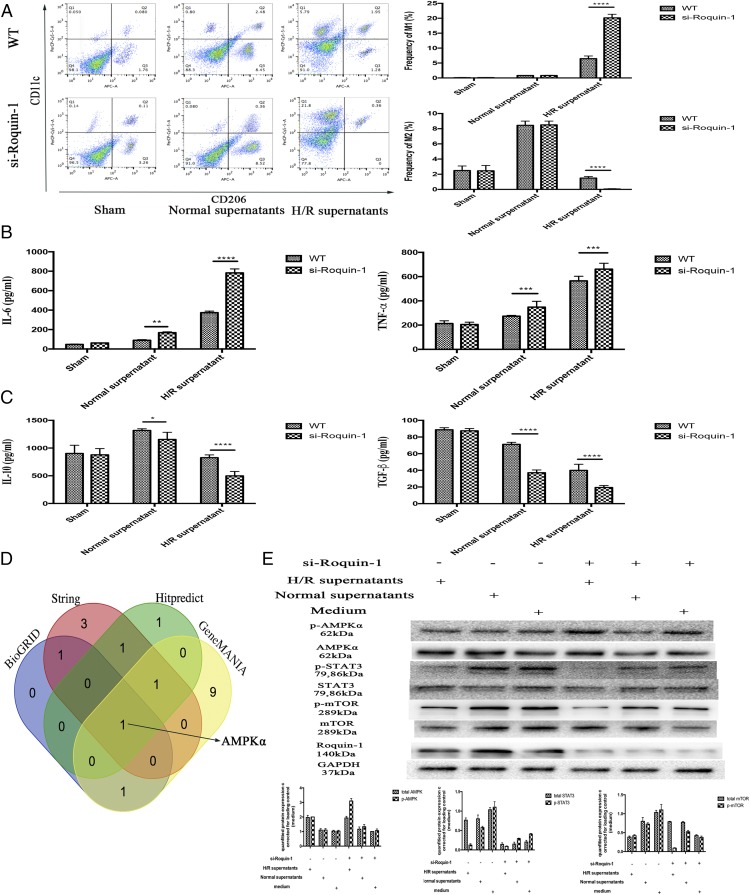
We established an H/R model of primary liver cells in vitro. The supernatant was used to stimulate BMDM for 24 h. Through flow cytometry and ELISA, mø polarization to M1 increased after H/R supernatant stimulation (Supplemental Fig. 2A–D). Furthermore, after interference with Roquin-1 expression, the H/R supernatant stimulated mø polarization to M1 significantly (Fig. 3A–C). At the same time, we predicted the target of Roquon-1 by String, BioGRID, GeneMANINA, and HitPredict (Fig. 3D, Supplemental Table II). Combined with previous research results (21, 22), we inferred that the target of Roquin-1 was AMPKα (Supplemental Fig. 2E), and Roquin-1 regulate AMPKα/mTOR singling pathway to modulate the polarization of mø. The results of WB detection of related signaling pathway proteins showed that H/R supernatant stimulation promoted the phosphorylation of AMPKα and inhibited the phosphorylation of mTOR and STAT3. After Roquin-1 interference, the phosphorylation of AMPKα increased and the phosphorylation of mTOR and STAT3 decreased (Fig. 3E).
FIGURE 3.
The effect of Roquin-1 deficiency on H/R-induced mø polarization. (A) Flow cytometry analysis of mø polarization (n = 6). (B and C) ELISA analysis of the cytokines of M1/M2 in supernatants (n = 6). (D) Venn diagram displaying Roquin-1 computationally predicted to target AMPKα by four different prediction algorithms: String, BioGRID, GeneMANINA, and Hitpredict. (E) WB analysis of the phosphorylation of AMPKα, mTOR, and STAT3 after H/R supernatants stimulation and/or Roquin-1 interference (n = 6). The data are expressed as the mean ± SD. *p < 0.01, **p < 0.01, ***p < 0.001, ****p < 0.0001.
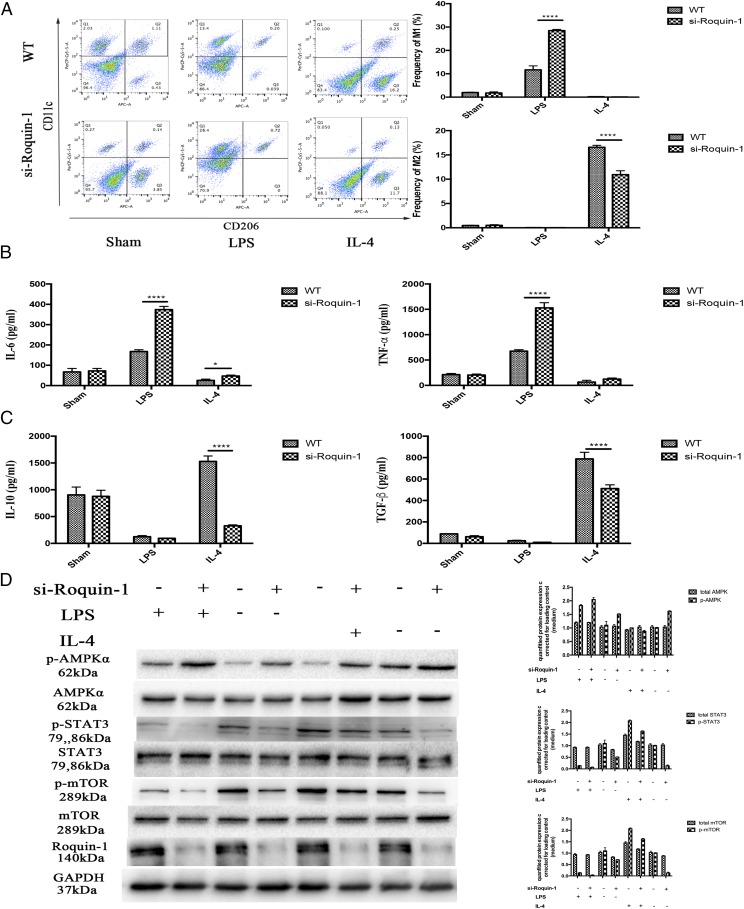
To investigate the mechanism by which Roquin-1 regulates mø polarization, we used LPS and IL-4 to stimulate mø to induce their polarization to M1/M2, respectively. First, we used si-Roquin-1 to transfect BMDM to interfere with the expression of Roquin-1. After 24 h, LPS and IL-4 were used to stimulate BMDM polarization to M1 and M2, respectively. Flow cytometer and ELISA results showed that Roquin-1 interference promoted the polarization and function of mø to M1, inhibiting polarization to M2, and function (Fig. 4A–C). At the same time, WB technology detected protein expression results that show that LPS can promote the phosphorylation of AMPKα and inhibit the phosphorylation of STAT3 and mTOR. Roquin-1 inhibits the phosphorylation of p-AMPKα and is enhanced by and promotes the phosphorylation of mTOR and STAT3 (Fig. 4D).
FIGURE 4.
The effect of Roquin-1 deficiency on M1/M2 polarization. (A) Flow cytometry analysis of M1/M2 polarization after LPS/IL-4 stimulation and/or Roquin-1 interference (n = 6). (B and C) ELISA analysis indicated that Roquin-1 interference promoted to M1 polarization and function, whereas it inhibited M2 polarization and function (6). (D) WB detects changes in signaling pathway proteins after LPS and IL-4 stimulation (n = 6). The data are expressed as the mean ± SD. *p < 0.01, **p < 0.01, ***p < 0.001, ****p < 0.0001.
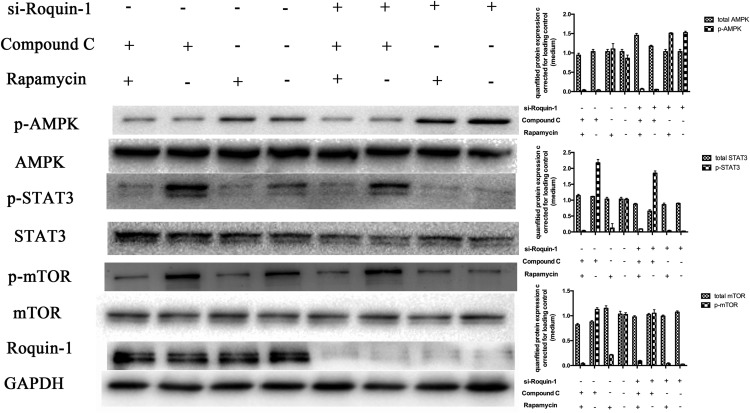
Furthermore, we used Compound C and rapamycin to inhibit the expression of AMPKα and mTOR (Fig. 5). WB showed that AMPKα phosphorylation was attenuated and mTOR and STAT3 phosphorylation were enhanced after interference with Roquin-1. Roquin-1 had no significant effect on mTOR and STAT3 phosphorylation after inhibiting AMPKα activity. If mTOR activity was inhibited, Roquin-1 could inhibit the phosphorylation of AMPKα and had no significant effect on phosphorylation of STAT3. The above results suggest that Roquin-1 further inhibits the phosphorylation of mTOR and STAT3 by promoting the phosphorylation of AMPKα.
FIGURE 5.
si-Roquin-1 was used to interfere with the expression of Roquin-1, and Compound C and rapamycin were used to inhibit the activity of AMPKα and mTOR, respectively. WB analysis indicated that inhibition of phosphorylation of mTOR and STAT3 was enhanced after APMK phosphorylation, and si-Roquin-1 had no effect on phosphorylation of mTOR and STAT3; phosphorylation of STAT3 was attenuated after inhibition of mTOR phosphorylation, and phosphorylation of AMPKα was absent. Effect, and si-Roquin-1 had no effect on phosphorylation of STAT3 (n = 6). The data are expressed as the mean ± SD. *p < 0.01, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Roquin-1 regulates ubiquitination of AMPKα and affects its activity
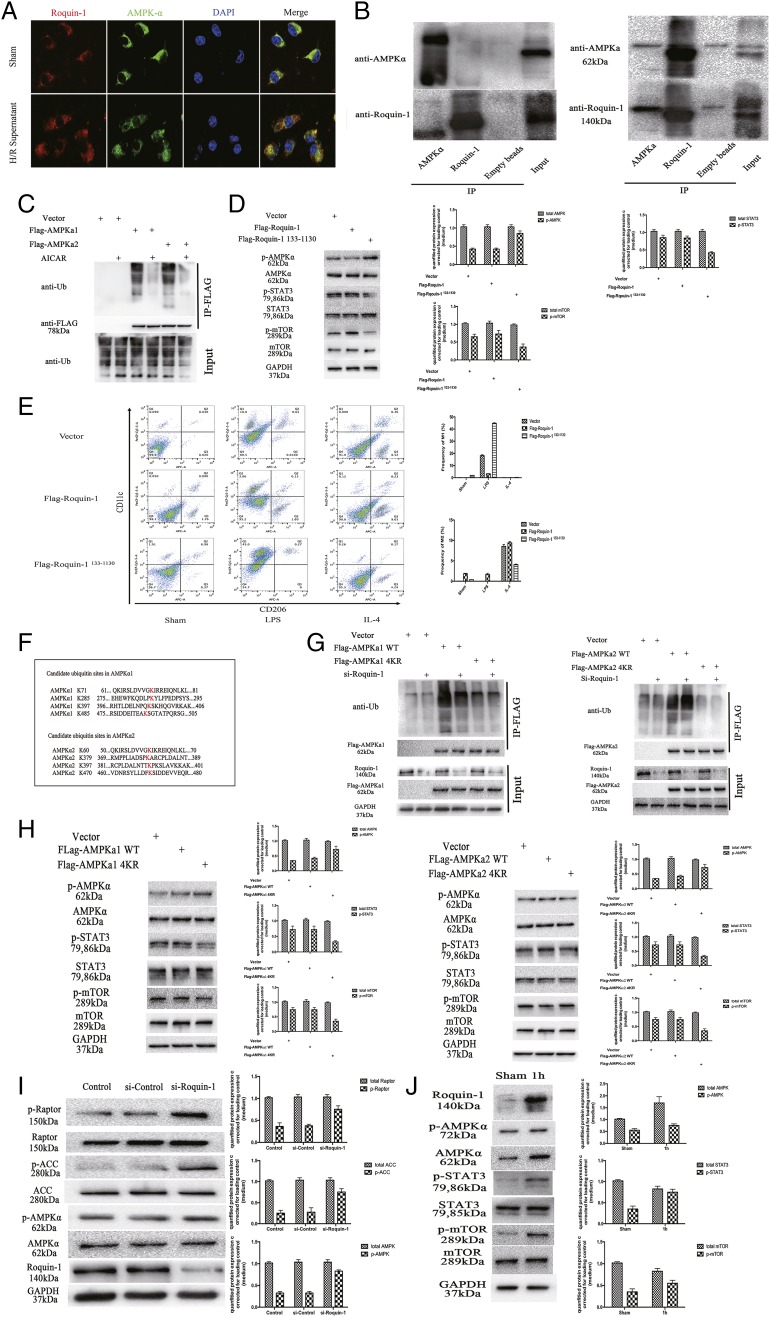
To demonstrate the direct role of Roquin-1 and AMPKα, we used immunofluorescence double labeling and coimmunoprecipitation. After treatment of mø with H/R hepatocyte supernatants, double immunofluorescent labels showed that Roquin-1 and AMPKα bound more after H/R supernatant treatment (Fig. 6A). Furthermore, after coimmunoprecipitation with Rqouin-1 and AMPKα Abs, respectively, WB results suggested that Roquin-1 and AMPKα were bound to each other under stimulation with H/R supernatants (Fig. 6B).
FIGURE 6.
The function of Roquin-1 in AMPKα activity. (A) Immunofluorescent labeling of Roquin-1 (red) and AMPKα (green) (n = 6). (B) The coimmunoprecipitation of Roquin-1 and AMPKα under stimulation with H/R supernatants (right) and without (left) (n = 6). (C) The cells were treated with 2 mM AICAR for 1 h, the cells were lysed, FLAG was coprecipitated, and WB was used to detect ubiquitination levels (n = 6). (D and E) Roquin-1133−1330, which was deleted the RING domain and lacked the ubiquitination activity, was used to investigate the relationship among Roquin-1’s ubiquitination activity, phosphorylation of STAT3 and mTOR, and M1/M2 polarization of mø. (F) Candidate ubiquitin sites on AMPKα. (G) The expression of AMPKα WT and 4KR mutant ubiquitination was compared by using si-Roquin-1 to interfere with its expression and coprecipitate FLAG (n = 6). (H) The relationship between the mutations in the ubiquitination sites of AMPKα (AMPKα-4KR mutants) and the phosphorylation of AMPKα, STAT3, and mTOR (n = 6). (I) si-Roquin-1 interferes with its expression and WB detects cell lysates (n = 6). (J) The relationship between Roquin-1 and AMPKa–STAT3–mTOR signal pathway in vivo (n = 6). The data are expressed as the mean ± SD. *p < 0.01, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To study the relationship between ubiquitination and AMPKα activity, we constructed the FLAG-tagged AMPKα1 and AMPKα2 lentiviruses using the lentiviral expression vector pLenti-FLAG 6.3 and infected 293T cells to express either FLAG-AMPKα1 or FLAG-AMPKα2. Further treatment with AICAR (2 mM) for 1 h activates AMPKα. After lysing the cells to obtain total protein, FLAG was immunoprecipitated and anti-Ub Ab was used to detect the level of ubiquitination of AMPKα. The results showed that the level of ubiquitination of AMPKα was significantly decreased after activation (Fig. 6C). To demonstrate that the role of Roquin-1 as E3 ubiquitin ligase is responsible for ubiquitination of AMPKα, we used Roquin-1133−1330, which was deleted the RING domain and lacked the ubiquitination activity, to investigate the relationship among Roquin-1’s ubiquitination activity, phosphorylation of STAT3 and mTOR, and M1/M2 polarization of mø. The result showed that the Roquin-1133−1330 promoted the phosphorylation of AMPKα and the polarization of M1 and inhibited the phosphorylation of STAT3 and mTOR and the polarization of M2 (Fig. 6D, 6E).
The RING domain of Roquin-1 is characteristic of E3 ubiquitin ligase, and E3 ubiquitin ligase plays an important role in ubiquitination. We speculate that Roquin-1 regulates the ubiquitination level of AMPKα. At the same time, we found that the potential ubiquitination sites of AMPKα (AMPKα1 are K71, K285, K396, and K485; AMPKα2 is K60, K379, K391, and K470) (Fig. 6F). We constructed the AMPKα mutant AMPKα4KR. Simultaneous use of si-Roquin-1 interfered with the expression of Roquin-1. The results showed that the ubiquitination of the AMPKα 4KR mutant was significantly lower than that of AMPKα WT. si-Roquin-1 inhibited ubiquitination of AMPKα WT but had no effect on the level of ubiquitination of the AMPKα 4KR mutant (Fig. 6G). The above results indicate that Roquin-1 promotes the ubiquitination of AMPKα and regulates the formation of ubiquitin chains through the above ubiquitination sites. Furthermore, we performed WB to verify the relationship between AMPKα ubiquitination and activity. The result showed that the mutations in the ubiquitination sites of AMPKα (AMPKα-4KR mutants) promoted phosphorylation of AMPKα and inhibited phosphorylation of STAT3 and mTOR (Fig. 6H).
To investigate the biological effects of Roquin-1 on promoting ubiquitination of AMPKα, we used siRNA to interfere with Roquin-1 expression and detect phosphorylated levels of AMPKα and its downstream substrates. The results showed that after interference with Roquin-1, there was no significant change in the level of AMPKα protein and the level of phosphorylation increased. Furthermore, there was no significant change in APC1 and Raptor protein levels downstream of AMPKα, and phosphorylation levels increased (Fig. 6I). Furthermore, to verify that Roquin-1 regulates the IRI severity by regulation of AMPKα–STAT3–mTOR signal pathway via inhibiting the AMPKα’s activity by ubiquitination of AMPKα, we performed WB to detect the ubiquitination level of AMPKα and phosphorylation level of signaling pathways in 1 h after liver IR. Compared with the sham group, the expression of Roquin-1, the ubiquitination level of AMPKα, and the phosphorylation of STAT3 and mTOR was increased. Meanwhile, the phosphorylation of AMPKα was decreased (Fig. 6J).
Discussion
Liver IRI is a result of inflammatory response, and mø activation plays a central role of the acute inflammatory response in liver IRI. In this respect, the use of mø modulator as a target to limit liver IRI has attracted attention. In this study, we demonstrated that the function of Roquin-1, which is known to a multifunctional regulator of immune homeostasis, would protect against liver IRI.
Previous studies indicate that Roquin-1 is involved in the inflammatory response in IR injury. First, the expression of Roquin-1 mRNA was found to increase in cardiac IR models established by inhibition of MCP-1; the expression of Roquin-1 was significantly increased in cardiac ischemic lesions (8, 12). Further mechanistic studies have shown that Roquin-1 induces cardiomyocyte apoptosis and ventricular dysfunction but can relieve toxin-induced cardiac dysfunction by inhibiting the activity of the NF-κB pathway (23, 24). In addition, Roquin-1 can be functionally dysregulated after myocardial infarction by alleviating NF-κB pathway activity and inhibiting the expression of inflammation-associated RNAs (e.g., miR-126, -146a, -155, -199a). At the same time, Roquin-1 can inhibit the expression of JNK pathway and thus suppress the expression of inflammatory factors (such as TNF-α, IL-1β, IL-6, MCP-1) and improve the tolerance of tissues to ischemia and hypoxia (25). Second, studies have shown that Roquin-1 negatively regulates NF-κB and JNK inflammatory signaling pathways by deubiquitylation (26). O’Neill et al. (27) found that Roquin-1 alleviates renal IR injury by inhibiting TLR4-mediated NF-κB pathway activity, and that Roquin-1 inhibits the activity of TLR4 downstream pathway. This study found that Roquin-1 liver is inhibited during IR and interferes with Roquin-1, and hepatic IR damage worsens. This shows that Roquin-1 plays a protective role in the process of liver IR.
H/R models were established in vitro, and mø were stimulated with H/R supernatant. We have found that H/R supernatants stimulate mø and induce their polarization to M1. Previous studies have shown that after liver IR, liver cells secrete HMGB1, histone/DNA, and ATP to stimulate mø and produce chemokines and cytokines (28). This is consistent with our experimental results. The M1 mø mainly promotes the inflammatory response, whereas the M2 mø inhibits the inflammatory response (29). Both of them can regulate the hepatic IRI (30). When Roquin-1 was interfered, M1 polarization increased and M2 polarization decreased. It demonstrated that Roquin-1 influenced hepatic IRI by regulating mø polarization. Furthermore, we used LPS and IL-4 to stimulate mø to induce their polarization to M1 and M2, respectively. When si-Roquin-1 interferes with the expression of Roquin-1, the mø increase toward M1 and decrease toward M2. This shows that Roquin-1 is crucial for mø polarization to M1/M2. Through an in vitro model, we again confirmed that Roquin-1 participates in hepatic IRI by regulating mø polarization and function.
Furthermore, we used vitro experiments to investigate the mechanism of Roquin-1 in mø polarization. We found that Roquin-1 has an effect on AMPKα activity in the database. Previous studies have also confirmed that in T cells, the RING domain of Roquin-1 can affect binding to AMPKα and affect its phosphorylation (23, 31, 32). This study found that phosphorylation of AMPKα was enhanced following interference with Roquin-1 expression. At the same time, mTOR plays an important role in mø polarization and is inhibited by AMPKα. The study found that IL-4 treatment of BMDMs significantly downregulated Cox-2 protein expression, whereas Cox-1 levels increased significantly and the polarization to M2 was promoted (33). AMPKα inhibits mTORC or CRISPR/Cas9-mediated tuberous sclerosis complex-2 (Tsc2) knockout by torin 1 and blocks Cox-1 expression (34). Our results showed that phosphorylation of mTOR in mø weakened after interference with Roquin-1. Further use of Compound C inhibited AMPKα activity, and the effect of Roquin-1 on mTOR phosphorylation disappeared. Additionally, STAT3 has been shown to play an important role in the polarization and function of mø (35). Decreased flagellin induced TNF-α and IL-6 expression and cell proliferation in mø with LY294002 and wortmannin. mTORC1 regulates the activation of NF-κB p65 and STAT3 by degradation of IκBα and phosphorylation of STAT3, respectively (34, 36). Our study found that the interference of Roquin-1 expression can inhibit the phosphorylation of STAT3; after inhibiting AMPKα and/or mTOR, phosphorylation of STAT3 is no longer affected by the expression of Roquin-1.
Roquin-1 includes three domains, called the ROQ domain; the C3H1 zinc finger domain, which bind to RNAs; and the RING domain, which have the function of E3 ubiquitin ligase (37). Studies have shown that Roquin-1 binds to AMPKα (23, 32). We have shown that Roquin-1 directly binds to AMPKα through coimmunoprecipitation and fluorescence confocal.
At present, most of the regulation of AMPKα is focused on phosphorylation (23, 38). Recently, studies of the impact of ubiquitination on AMPKα have been undertaken. Studies have shown that USP10 promotes AMPKα activity by inhibiting the ubiquitination of AMPKα and plays an important role in a variety of metabolic-deficient diseases (39). In addition, studies have found that Cidea and AMPKα are colocalized in the endoplasmic reticulum and ubiquitinated by AMPKα (40). Whether ubiquitination leads to decreased AMPKα activity or degradation and its mechanisms is not yet clear. In the current study, we used AICAR to activate AMPKα and detect levels of ubiquitination of AMPKα1 and AMPKα2. The activity of AMPKα1 and AMPKα2 was found to be contrary to its ubiquitination level. This result indicates that the ubiquitination of AMPKα1 and AMKα2 inhibited its activity.
Furthermore, Roquin-1 regulates the ubiquitination of AMPKα. Studies have shown that the RING domain of Roquin-1 is related to the ubiquitination of AMPKα1 (31). We used si-Roquin-1 to interfere with the expression of Roquin-1 and found that the ubiquitination of AMPKα1 and AMPKα2 was reduced. Therefore, it can be speculated that Roquin-1 can promote the ubiquitination of AMPKα, thereby reducing its activity.
Ubiquitination is an important posttranslational modification of proteins. Previous studies divided it into monoubiquitination and polyubiquitination (41). The polyubiquitin chains begin with the binding of the monoubiquitin to the substrate protein, whereas the subsequent ubiquitin molecules bind to the lysine residues of the previous ubiquitin molecule and form ubiquitin chains. Common lysine sites of common ubiquitin molecules include K6, K11, K26, K29, K33, K48, and K63. Studies have shown that K48 is mainly involved in protein degradation, whereas K63 is related to protein function and localization (42). We have found that Roquin-1 primarily regulates the ubiquitination of the ubiquitin chain of AMPKα1 K63.
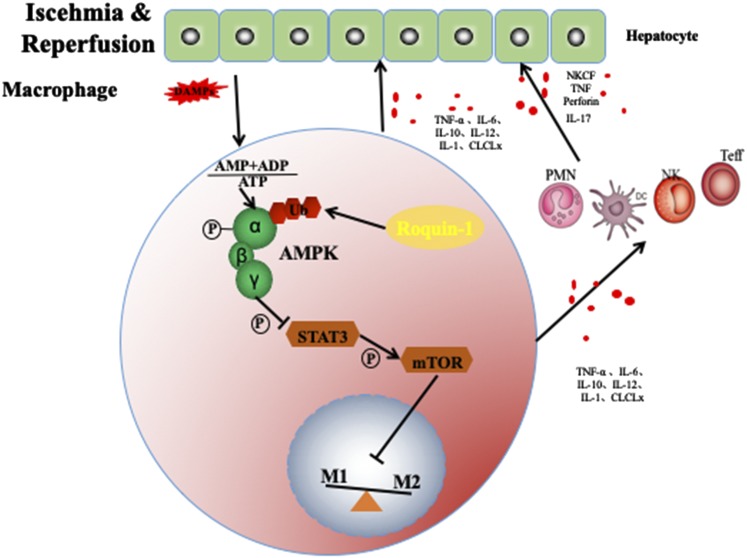
In summary, our results suggest that Roquin-1 plays an important role in liver IR. First, Roquin-1 is expressed in mø and inhibits the polarization of mø to Ml and promotes its polarization to M2. Furthermore, Roquin-1 inhibited the activity of AMPKα by promoting ubiquitination in mø, thereby further promoting the activity of mTOR and STAT3.This topic describes the protection of liver IRI by Roquin-1 and discusses its main mechanism for regulating AMPKα activity through ubiquitination and affecting the polarization of mø, providing a certain theoretical basis for the intervention of liver IRI (Fig. 7).
FIGURE 7.
Model of Roquin-1 in mø.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 81270483 and 81871260.
The online version of this article contains supplemental material.
- ALT
- alanine transaminase
- AST
- aspartate transaminase
- BMDM
- bone marrow–derived mø
- H/R
- hypoxia-reoxygenation
- IR
- ischemia–reperfusion
- IRI
- IR injury
- mø
- macrophage
- WB
- Western blot.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Honda M., Takeichi T., Hashimoto S., Yoshii D., Isono K., Hayashida S., Ohya Y., Yamamoto H., Sugawara Y., Inomata Y. 2017. Intravital imaging of neutrophil recruitment reveals the efficacy of FPR1 blockade in hepatic ischemia-reperfusion injury. J. Immunol. 198: 1718–1728. [DOI] [PubMed] [Google Scholar]
- 2.Ohana G., Cohen S., Rath-Wolfson L., Fishman P. 2016. A3 adenosine receptor agonist, CF102, protects against hepatic ischemia/reperfusion injury following partial hepatectomy. Mol. Med. Rep. 14: 4335–4341. [DOI] [PubMed] [Google Scholar]
- 3.Tsaroucha A. K., Tsiaousidou A., Ouzounidis N., Tsalkidou E., Lambropoulou M., Giakoustidis D., Chatzaki E., Simopoulos C. 2016. Intraperitoneal administration of apigenin in liver ischemia/reperfusion injury protective effects. Saudi J. Gastroenterol. 22: 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Possamai L. A., Thursz M. R., Wendon J. A., Antoniades C. G. 2014. Modulation of monocyte/macrophage function: a therapeutic strategy in the treatment of acute liver failure. J. Hepatol. 61: 439–445. [DOI] [PubMed] [Google Scholar]
- 5.Tacke F., Zimmermann H. W. 2014. Macrophage heterogeneity in liver injury and fibrosis. J. Hepatol. 60: 1090–1096. [DOI] [PubMed] [Google Scholar]
- 6.Abdullah Z., Knolle P. A. 2017. Liver macrophages in healthy and diseased liver. Pflugers Arch. 469: 553–560. [DOI] [PubMed] [Google Scholar]
- 7.Chistiakov D. A., Myasoedova V. A., Revin V. V., Orekhov A. N., Bobryshev Y. V. 2018. The impact of interferon-regulatory factors to macrophage differentiation and polarization into M1 and M2. Immunobiology 223: 101–111. [DOI] [PubMed] [Google Scholar]
- 8.Huang J., Shen X. D., Yue S., Zhu J., Gao F., Zhai Y., Busuttil R. W., Ke B., Kupiec-Weglinski J. W. 2014. Adoptive transfer of heme oxygenase-1 (HO-1)-modified macrophages rescues the nuclear factor erythroid 2-related factor (Nrf2) antiinflammatory phenotype in liver ischemia/reperfusion injury. Mol. Med. 20: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao J., Yue S., Fu Y., Zhu J., Wang X., Busuttil R. W., Kupiec-Weglinski J. W., Lu L., Zhai Y. 2014. ATF6 mediates a pro-inflammatory synergy between ER stress and TLR activation in the pathogenesis of liver ischemia-reperfusion injury. Am. J. Transplant. 14: 1552–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao J., Qian X., Li G., Pan X., Zhang C., Zhang F., Zhai Y., Wang X., Lu L. 2015. ATF3-mediated NRF2/HO-1 signaling regulates TLR4 innate immune responses in mouse liver ischemia/reperfusion injury. Am. J. Transplant. 15: 76–87. [DOI] [PubMed] [Google Scholar]
- 11.Athanasopoulos V., Barker A., Yu D., Tan A. H., Srivastava M., Contreras N., Wang J., Lam K. P., Brown S. H., Goodnow C. C., et al. 2010. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. FEBS J. 277: 2109–2127. [DOI] [PubMed] [Google Scholar]
- 12.Athanasopoulos V., Ramiscal R. R., Vinuesa C. G. 2016. ROQUIN signalling pathways in innate and adaptive immunity. Eur. J. Immunol. 46: 1082–1090. [DOI] [PubMed] [Google Scholar]
- 13.Leppek K., Schott J., Reitter S., Poetz F., Hammond M. C., Stoecklin G. 2013. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 153: 869–881. [DOI] [PubMed] [Google Scholar]
- 14.Mino T., Murakawa Y., Fukao A., Vandenbon A., Wessels H. H., Ori D., Uehata T., Tartey S., Akira S., Suzuki Y., et al. 2015. Regnase-1 and roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161: 1058–1073. [DOI] [PubMed] [Google Scholar]
- 15.Pratama A., Ramiscal R. R., Silva D. G., Das S. K., Athanasopoulos V., Fitch J., Botelho N. K., Chang P. P., Hu X., Hogan J. J., et al. 2013. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity 38: 669–680. [DOI] [PubMed] [Google Scholar]
- 16.Wang L., Li N., Lin D., Zang Y. 2017. Curcumin protects against hepatic ischemia/reperfusion induced injury through inhibiting TLR4/NF-κB pathway. Oncotarget 8: 65414–65420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., Ji H., Zhang Y., Shen X., Gao F., He X., Li G. A., Busuttil R. W., Kuchroo V. K., Kupiec-Weglinski J. W. 2015. Recipient T cell TIM-3 and hepatocyte galectin-9 signalling protects mouse liver transplants against ischemia-reperfusion injury. J. Hepatol. 62: 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue S., Zhu J., Zhang M., Li C., Zhou X., Zhou M., Ke M., Busuttil R. W., Ying Q. L., Kupiec-Weglinski J. W., et al. 2016. The myeloid heat shock transcription factor 1/β-catenin axis regulates NLR family, pyrin domain-containing 3 inflammasome activation in mouse liver ischemia/reperfusion injury. Hepatology 64: 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czaya B., Singh S., Yanucil C., Schramm K., Faul C., Grabner A. 2017. Induction of an inflammatory response in primary hepatocyte cultures from mice. J. Vis. Exp. DOI: 10.3791/55319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q., Kong L., Xu X., Geng Q., Tang W., Jiang W. 2013. Down-regulation of microRNA-146a in the early stage of liver ischemia-reperfusion injury. Transplant. Proc. 45: 492–496. [DOI] [PubMed] [Google Scholar]
- 21.Qing L., Fu J., Wu P., Zhou Z., Yu F., Tang J. 2019. Metformin induces the M2 macrophage polarization to accelerate the wound healing via regulating AMPK/mTOR/NLRP3 inflammasome singling pathway. Am. J. Transl. Res. 11: 655–668. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X., Tang X., Guo N., An Y., Chen X., Shi C., Wang C., Li Y., Li S., Xu H., et al. 2018. Biochanin a enhances the defense against Salmonella enterica infection through AMPK/ULK1/mTOR-mediated autophagy and extracellular traps and reversing SPI-1-dependent macrophage (MΦ) M2 polarization. Front. Cell. Infect. Microbiol. 8: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad N., Gardner C. R., Yurkow E. J., Laskin D. L. 1999. Inhibition of macrophages with gadolinium chloride alters intercellular adhesion molecule-1 expression in the liver during acute endotoxemia in rats. Hepatology 29: 728–736. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L., Azfer A., Niu J., Graham S., Choudhury M., Adamski F. M., Younce C., Binkley P. F., Kolattukudy P. E. 2006. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ. Res. 98: 1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang J., Wang J., Saad Y., Warble L., Becerra E., Kolattukudy P. E. 2011. Participation of MCP-induced protein 1 in lipopolysaccharide preconditioning-induced ischemic stroke tolerance by regulating the expression of proinflammatory cytokines. J. Neuroinflammation 8: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang J., Saad Y., Lei T., Wang J., Qi D., Yang Q., Kolattukudy P. E., Fu M. 2010. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 207: 2959–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Neill S., Humphries D., Tse G., Marson L. P., Dhaliwal K., Hughes J., Ross J. A., Wigmore S. J., Harrison E. M. 2015. Heat shock protein 90 inhibition abrogates TLR4-mediated NF-κB activity and reduces renal ischemia-reperfusion injury. Sci. Rep. 5: 12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsung A., Klune J. R., Zhang X., Jeyabalan G., Cao Z., Peng X., Stolz D. B., Geller D. A., Rosengart M. R., Billiar T. R. 2007. HMGB1 release induced by liver ischemia involves toll-like receptor 4 dependent reactive oxygen species production and calcium-mediated signaling. J. Exp. Med. 204: 2913–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santoni G., Morelli M. B., Amantini C., Santoni M., Nabissi M., Marinelli O., Santoni A. 2018. “Immuno-transient receptor potential ion channels”: the role in monocyte- and macrophage-mediated inflammatory responses. Front. Immunol. 9: 1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laban H., Weigert A., Zink J., Elgheznawy A., Schürmann C., Günther L., Abdel Malik R., Bothur S., Wingert S., Bremer R., et al. 2018. VASP regulates leukocyte infiltration, polarization, and vascular repair after ischemia. J. Cell Biol. 217: 1503–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramiscal R. R., Parish I. A., Lee-Young R. S., Babon J. J., Blagih J., Pratama A., Martin J., Hawley N., Cappello J. Y., Nieto P. F., et al. 2015. Attenuation of AMPK signaling by ROQUIN promotes T follicular helper cell formation. eLife 4: e08698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S. Y., Moon S. J., Kim E. K., Seo H. B., Yang E. J., Son H. J., Kim J. K., Min J. K., Park S. H., Cho M. L. 2017. Metformin suppresses systemic autoimmunity in Roquinsan/san mice through inhibiting B cell differentiation into plasma cells via regulation of AMPK/mTOR/STAT3. J. Immunol. 198: 2661–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shay A. E., Diwakar B. T., Guan B. J., Narayan V., Urban J. F., Jr., Prabhu K. S. 2017. IL-4 up-regulates cyclooxygenase-1 expression in macrophages. J. Biol. Chem. 292: 14544–14555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams D. H., Ju C., Ramaiah S. K., Uetrecht J., Jaeschke H. 2010. Mechanisms of immune-mediated liver injury. Toxicol. Sci. 115: 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shirakawa K., Endo J., Kataoka M., Katsumata Y., Yoshida N., Yamamoto T., Isobe S., Moriyama H., Goto S., Kitakata H., et al. 2018. IL (interleukin)-10-STAT3-galectin-3 axis is essential for osteopontin-producing reparative macrophage polarization after myocardial infarction. Circulation 138: 2021–2035. [DOI] [PubMed] [Google Scholar]
- 36.Bao W., Wang Y., Fu Y., Jia X., Li J., Vangan N., Bao L., Hao H., Wang Z. 2015. mTORC1 regulates flagellin-induced inflammatory response in macrophages. PLoS One 10: e0125910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaefer J. S., Klein J. R. 2016. Roquin--a multifunctional regulator of immune homeostasis. Genes Immun. 17: 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin S. C., Hardie D. G. 2018. AMPK: sensing glucose as well as cellular energy status. Cell Metab. 27: 299–313. [DOI] [PubMed] [Google Scholar]
- 39.Deng M., Yang X., Qin B., Liu T., Zhang H., Guo W., Lee S. B., Kim J. J., Yuan J., Pei H., et al. 2016. Deubiquitination and activation of AMPK by USP10. Mol. Cell 61: 614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akira S. 2013. Regnase-1, a ribonuclease involved in the regulation of immune responses. Cold Spring Harb. Symp. Quant. Biol. 78: 51–60. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda F. 2015. Linear ubiquitination signals in adaptive immune responses. Immunol. Rev. 266: 222–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dittmar G., Selbach M. 2017. Deciphering the ubiquitin code. Mol. Cell 65: 779–780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.